Abstract
In the present study, the role of microRNA-663b (miR-663b) in cardiomyocyte injury was examined. Reverse transcription-quantitative PCR (RT-qPCR) was performed to detect miR-663b expression in hypoxia-induced H9c2 cells. The results revealed that miR-663b expression was significantly upregulated in hypoxia-induced H9c2 cells compared with control cells. TargetScan analysis and dual-luciferase reporter assays demonstrated that miR-663b directly targeted the B-cell lymphoma 2 like 1 (BCL2L1) gene. RT-qPCR and western blotting data indicated that BCL2L1 expression was significantly downregulated in hypoxia-induced H9c2 cells compared with control cells. Under hypoxic conditions, H9c2 cells were transfected with miR-663b inhibitor, inhibitor control, miR-663b inhibitor + control small interfering (si)RNA or miR-663b inhibitor + BCL2L1-siRNA for 48 h. ELISA against creatine kinase-muscle/brain (CK-MB) and cardiac troponin 1 (cTnI) demonstrated that the miR-663b inhibitor reduced CK-MD and cTnI release and increased mitochondrial viability when compared with hypoxia-treated cells. Additionally, the miR-663b inhibitor significantly increased H9c2 cell viability and decreased cell apoptosis under hypoxic conditions. The results of ELISA further revealed that the miR-663b inhibitor decreased the release of various inflammatory factors, including tumour necrosis factor α, interleukin (IL) 1β and IL-6 in H9c2 cells under hypoxic conditions. These changes were reversed following BCL2L1 knockdown. In conclusion, miR-663b inhibition protected cardiomyocytes against hypoxia-induced injury by targeting BCL2L1 and may potentially be a novel target for the treatment of patients with myocardial infarction.
Keywords: myocardial infarction, microRNA-663b, B-cell lymphoma 2 like 1, hypoxia
Introduction
Coronary heart disease is a major cause of death and disability that causes serious debilitation in patients, with an incidence rate that is increasing on a yearly basis (1,2). Acute myocardial infarction (MI) is a coronary heart disease with high worldwide incidence and mortality rates that leads to myocardial tissue injury and ultimately disability or death (3). Hypoxia is one of the primary causes of MI. Under hypoxic conditions, changes in energy metabolism occur during mitochondrial respiration in which anaerobic glycolysis results in numerous physiological and pathological reactions leading to acidosis and necrosis (4). Therefore, understanding the pathogenesis of MI may highlight potentially novel targets to limit damage.
MicroRNAs (miRNAs or miRs) are small single-stranded non-coding RNAs that are 22-25 nucleotides in length (5,6). miRNAs post-transcriptionally regulate gene expression (7-9) and mediate the degradation of or inhibit the translation of mRNAs by directly binding to their 3' untranslated region (3'-UTR) (10,11). Numerous studies have demonstrated that miRNAs serve an important role in the pathogenesis of many types of cancer (12-14), including miR-663b, indicating that they may be involved in the occurrence and development of tumors (15).
Pan et al (16) demonstrated that miR-663b is downregulated in gastric cancer and Jiao et al (17) revealed that expression was increased in castration-resistant prostate cancer. Additionally, miR-663b is upregulated in nasopharyngeal carcinoma (18). Liang et al (19) demonstrated that miR-663b promoted proliferation by targeting tumor suppressor candidate 2 and that the resulting upregulation was associated with advanced clinical stage cancer and lymph node metastasis. miR-663b expression is also upregulated in patients with MI (20). However, its role and function in MI remain unknown.
The aim of the present study was to determine whether miR-663b served a role in hypoxia-induced cardiomyocyte injury in vitro and to study the specific underlying molecular mechanisms. The present study may provide a theoretical basis for the development of novel strategies for the treatment of MI.
Materials and methods
Cell culture and transfection
Rat cardiomyocyte H9c2 cells were purchased from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences. H9c2 cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and incubated at 37˚C with 5% CO2. To simulate hypoxic damage, cells were incubated for 48 h at 37˚C with 94% N2, 5% CO2 and 1% O2 (21). Cells in the control group were incubated at 37˚C with 5% CO2.
Under hypoxic conditions, H9c2 cells were transfected with 50 nM miR-663b inhibitor (5'-GCGGUCCCGCGGCGCCCCGCCU-3'; Shanghai GenePharma Co., Ltd.), 50 nM inhibitor control (5'-UUGUACUACACAAAAGUACUG-3'; Shanghai GenePharma Co., Ltd.), 50 nM miR-663b inhibitor + 1 µM control-small interfering (si)RNA (cat. no. sc-36869; Santa Cruz Biotechnology, Inc.) or 50 nM miR-663b inhibitor + 1 µM B-cell lymphoma 2 like 1 (BCL2L1)-siRNA (cat. no. sc-29216; Santa Cruz Biotechnology, Inc.) for 48 h using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Transfection efficiency was assessed using reverse transcription quantitative PCR (RT-qPCR) 48 h after experimentation.
RT-qPCR
Total RNA was extracted from H9c2 cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). RNA was reverse transcribed into cDNA using a cDNA Reverse Transcription kit (Vazyme, Biotech Co., Ltd.) at a temperature of 42˚C for 60 min and 75˚C for 5 min. qPCR was performed using a SYBR™ Green PCR Master kit (Vazyme Biotech Co., Ltd.). The thermocycling conditions were as follows: Initial denaturation at 95˚C for 3 min; followed by 40 cycles of 95˚C for 30 sec, 56˚C for 30 sec and 72˚C for 30 sec. Primer sequences used for PCR were listed as follows: miR-663b forward, 5'-CGCTAACAGTCTCCAGTC-3' and reverse 5'-GTGCAGGGTCCGAGGT-3'; BCL2L1 forward, 5'-GACTGAATCGGAGATGGAGACC-3' and reverse 5'-GCAGTTCAAACTCGTCGCCT-3'; U6, 5'-GCTTCGGCAGCACATATACTAAAAT-3'; reverse, 5'-CGCTTCACGAATTTGCGTGTCAT-3'; GAPDH forward, 5'-CTTTGGTATCGTGGAAGGACTC-3' and reverse, 5'-GTAGAGGCAGGGATGATGTTCT-3'. The fold change of gene expression was calculated using the 2-ΔΔCq method (22). U6 and GAPDH were used as the internal controls for normalizing expression of miRNA and mRNA, respectively. All experiments were performed in triplicate.
Dual-luciferase reporter assay
TargetScan analysis (version 7.2; http://www.targetscan.org/vert_72/) was performed to examine the association between miRNA-663b and BCL2L1. The results indicated that miR-663b binding sites were present in BCL2L1. To confirm binding between miR-663 and BCL2L1, a dual luciferase reporter assay was performed.
Luciferase reported plasmids (psi-CHECK2) containing BCL2L1 wild-type (WT-BCL2L1) or mutant (MUT-BCL2L1) 3'-UTRs were synthesized by TsingKe Biotech Co. H9c2 cells were co-transferred with WT-BCL2L1 or MUT-BCL2L1 luciferase reporter plasmids and the miR-663b mimic or mimic control, respectively, using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were harvested 48 h after transfection and luciferase activity was measured using a Dual-Luciferase® Reporter Assay system (Promega Corporation) according to the manufacturer's protocol. Firefly luciferase was used to normalize data.
Western blotting
Total protein was extracted from H9c2 cells using RIPA lysis buffer (Beijing Solarbio Science & Technology Co., Ltd.) at room temperature for 30 min, after which samples were stored at -20˚C. A bicinchoninic acid assay kit (Pierce; Thermo Fisher Scientific, Inc.) was used to quantify protein concentration. Equal quantities of protein (30 µg/lane) were loaded onto a 15% gel, resolved using SDS-PAGE and transferred onto PVDF membranes (EMD Millipore). Membranes were blocked with 5% skimmed milk in TBS containing 0.1% Tween at room temperature for 2 h. Subsequently, the membranes were incubated with the following primary antibodies: Anti-BCL2L1 (cat. no. sc-70418; 1:1,000; Santa Cruz Biotechnology, Inc.), anti-cleaved-Caspase-3 (cat. no. ab2302; 1:1,000; Abcam) or anti-GAPDH (cat. no. 5174; 1:1,000; Cell Signaling Technology, Inc.) at 4˚C overnight. The membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G secondary antibodies (cat. no. 7074; 1:2,000; Cell Signaling Technology, Inc.) at room temperature for 2 h. Signals were visualized using enhanced chemiluminescence reagent (EMD Millipore) and GAPDH was used as the loading control.
Flow cytometry analysis
Transfected cells (1x106 cells) were harvested, centrifuged at a low temperature at a high speed (1,000 x g; 4˚C; 5 min) and re-suspended in 200 µl binding buffer containing 10 µl fluorescein isothiocyanate-annexin V. After centrifugation, cells were re-suspended in 5 µl propidium iodide (PI) and 300 µl PBS, and incubated for 30 min at room temperature in the dark. Cells were then stained using annexin-V/PI Apoptosis Detection kit (BD Biosciences) according to the manufacturer's protocol. Fluorescence was assessed using a BD FACSCalibur™ flow cytometer (Becton, Dickinson and Company) and flow data was analyzed using FlowJo (version 7.6.1; FlowJo LLC).
MTT assay
H9c2 cells were transfected with miR-663b inhibitor, inhibitor control, miR-663b inhibitor + control-siRNA or miR-663b inhibitor + BCL2L1-siRNA for 48 h under hypoxic conditions. Transfected cells were then plated in a 96-well plate at a density of 5x103 cells/well. After 24 h of incubation at 37˚C, 20 µl MTT reagent (Sigma-Aldrich; Merck KGaA) was added to each well. Following a further 4 h of incubation at 37˚C, 150 µl DMSO was added to each well and agitated for 15 min in the dark. Optical density values were measured at 490 nm using a microplate reader.
ELISA
Levels of certain inflammatory cytokines, including tumour necrosis factor α (TNF-α; cat. no. PT516), interleukin (IL) 1β (cat. no. PI303) and IL-6 (cat. no. PI328) in cell supernatants were detected using specific ELISA kits (Beyotime Institute of Biotechnology) according to the manufacturer's protocol.
Assessment of cellular injury
Following treatment, the release of various cardiomyocyte injury biomarkers, including creatine kinase-muscle/brain (CK-MB; cat. no. CSB-E14403r; Cusabio Technology LLC) and cardiac troponin I (cTnI; cat. no. CSB-E08594r; Cusabio Technology LLC) was determined using specific ELISA kits according to the manufacturer's protocol. Mitochondrial viability was measured using a mitochondrial viability staining kit (cat. no. ab129732; Abcam) according to the manufacturer's protocol.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software (version 5; GraphPad Software, Inc.). Statistical differences between groups were determined using a Student's t-test or one-way ANOVA followed by Tukey's post hoc test. Data are expressed as the mean ± standard deviation of at least three independent experiments. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-663b expression is significantly increased in hypoxia-induced H9c2 cells
To examine the role of miR-663b in hypoxia-induced H9c2 cells, miR-663b expression was assessed using RT-qPCR. The results revealed that miR-663b expression was significantly upregulated in H9c2 cells compared with the control group (Fig. 1), indicating that miR-663b may be associated with the development of MI.
Figure 1.
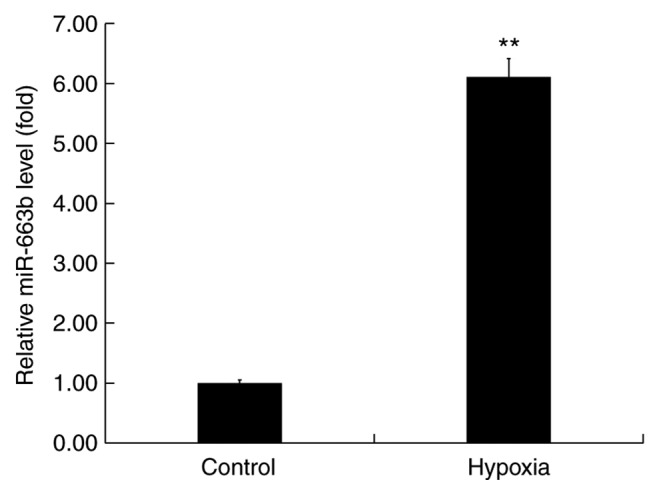
Expression of miR-663b in hypoxia-induced H9c2 cells. H9c2 cells were cultured in hypoxic conditions for 48 h. Reverse transcription-quantitative PCR was performed to detect the expression of miR-663b. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01 vs. control group. miR, microRNA.
BCL2L1 is a direct target gene of miR-663b
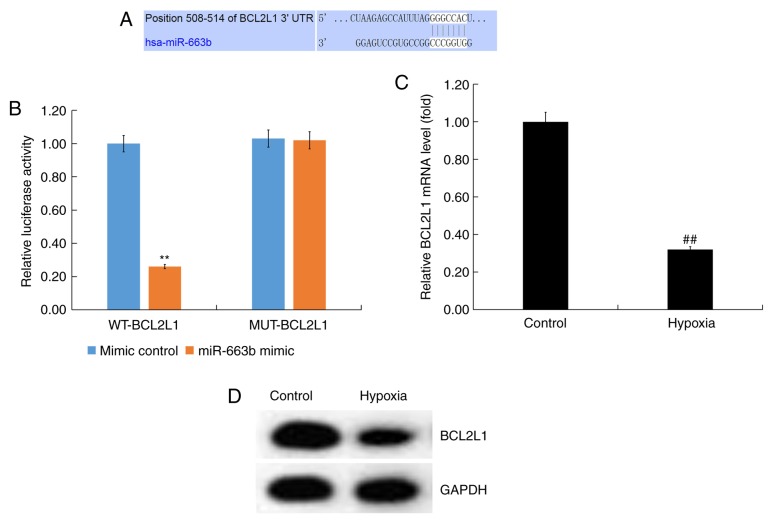
To determine the role and mechanism by which miR-663 exerted its effects, bioinformatics analysis was performed using TargetScan to predict miR-663b targets. BCL2L1 was predicted to be a target gene of miR-663b (Fig. 2A).
Figure 2.
BCL2L1 is a target gene of miR-663b. (A) Prediction of miR-663b binding to BCL2L1 using TargetScan. (B) A dual luciferase reporter assay was performed to confirm binding between miR-663b and BCL2L1. (C) Reverse transcription-quantitative PCR and (D) western blotting were performed to determine the expression of BCL2L1. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01 vs. mimic control group; ##P<0.01 vs. control group. BCL2L1, B-cell lymphoma 2 like 1; WT, wild type; MUT, mutant; miR, microRNA.
The 3'-UTR (WT or MUT) of BCL2L1 was inserted into the pmiR luciferase reporter and H9c2 cells were co-transfected with the miR-663b mimic and luciferase reporter plasmid. The results revealed that miR-663b mimic co-transfection with the WT-BCL2L1 3'-UTR reporter inhibited luciferase activity while miR-663b mimic had no effect on the reporter containing the MUT-BCL2L1 3'-UTR (Fig. 2B). These data suggested that BCL2L1 was a direct target gene of miR-663b.
Subsequently, the expression of BCL2L1 in hypoxia-induced H9c2 cells was determined. RT-qPCR and western blotting demonstrated that BCL2L1 expression was downregulated in hypoxia-induced H9c2 cells (Fig. 2C and D).
Effect of miR-663b inhibitor on H9c2 cell injury induced by hypoxia
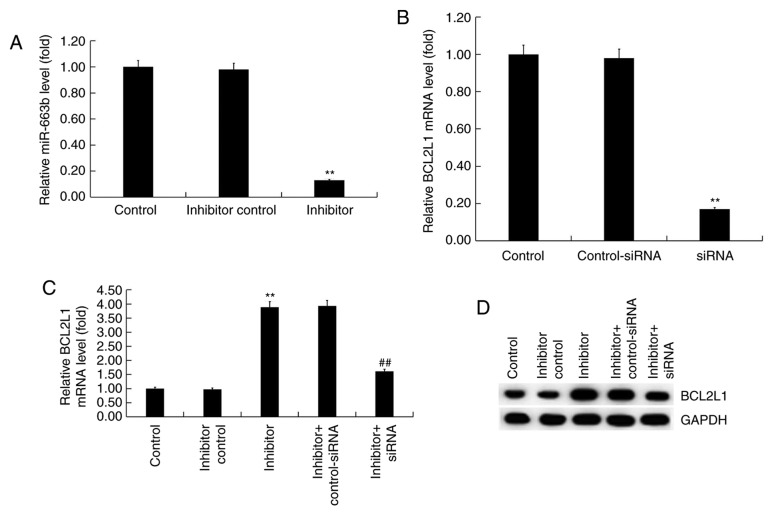
H9c2 cells were transfected with inhibitor control or miR-663b inhibitor for 48 h. RT-qPCR revealed that the miR-663b inhibitor significantly reduced miR-663 expression in H9c2 cells compared with controls (Fig. 3A). Additionally, H9c2 cells were transfected with BCL2L1-siRNA or control-siRNA. RT-qPCR results revealed that BCL2L1-siRNA significantly decreased the expression of BCL2L1 at the mRNA level in H9c2 cells compared with controls (Fig. 3B). Subsequently, H9c2 cells were co-transfected with miR-663b inhibitor + control-siRNA, or miR-663b inhibitor + BCL2L1-siRNA. RT-qPCR and western blotting results revealed that miR-663b inhibitor significantly increased the expression of BCL2L1 at the mRNA level and that this increase was reversed by BCL2L1-siRNA (Fig. 3C). Similar results were observed at the protein level as determined via western blotting (Fig. 3D).
Figure 3.
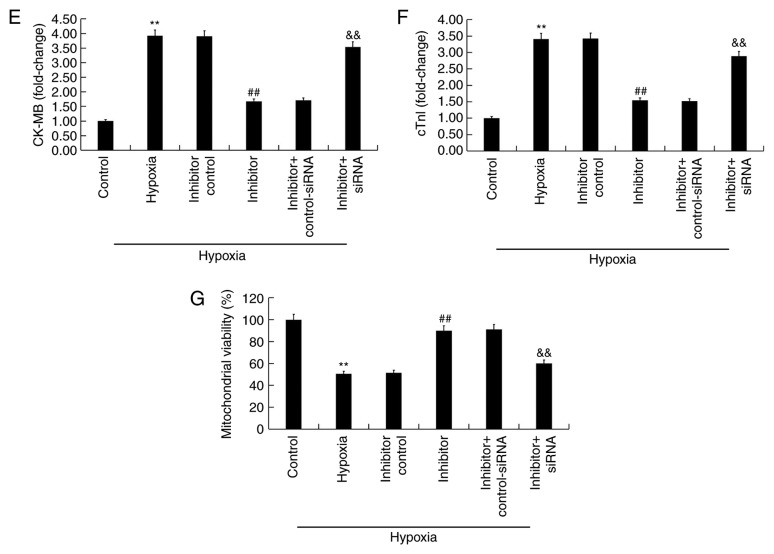
Effect of the miR-663b inhibitor on H9c2 hypoxia-induced cell injury. (A) miR-663b expression in transfected H9c2 cells. (B) mRNA expression of BCL2L1 in transfected H9c2. (C) BCL2L1 mRNA and (D) protein levels in transfected H9c2 cells. Release of (E) CK-MB and (F) cTnI. (G) Mitochondrial viability. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01 vs. control group; ##P<0.01 vs. hypoxia group; &&P<0.01 vs. hypoxia + inhibitor group. miR, microRNA; BCL2L1, B-cell lymphoma 2 like 1; siRNA, small interfering RNA; CK-MB, creatine kinase-muscle/brain; CTn1, Cardiac troponin 1.
The effect of miR-663b on hypoxia-induced cell damage was determined by detecting the release of two biomarkers associated with cardiomyocyte injury, CK-MB and cTnI. Compared with the controls, hypoxia significantly increased the release of CK-MB and cTnI whereas the miR-663b inhibitor significantly decreased CK-MB (Fig. 3E) and cTnI release (Fig. 3F). Transfection of BCL2L1-siRNA abolished the aforementioned changes. Furthermore, hypoxia also significantly reduced mitochondrial viability while miR-663b inhibitor improved mitochondrial viability in H9c2 cells (Fig. 3G). These changes were reversed by transfection of BCL2L1-siRNA.
Effect of miR-663b inhibitor on cell growth of hypoxia-induced H9c2 cells
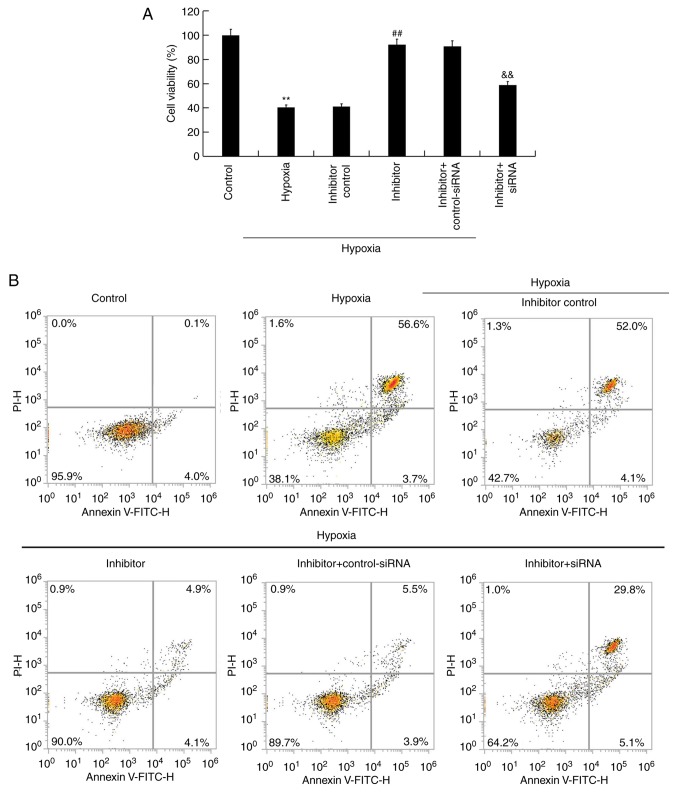
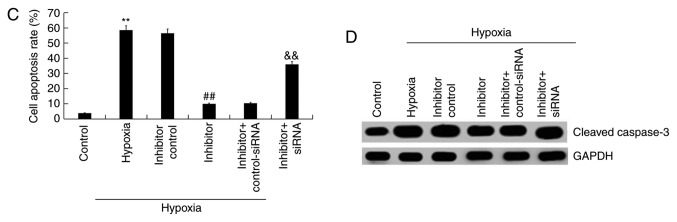
Under hypoxic conditions, H9c2 cells were transfected with inhibitor control, miR-663b inhibitor, miR-663b inhibitor + control-siRNA or miR-663b inhibitor + BCL2L1-siRNA for 48 h, after which an MTT assay was performed to measure cell viability. The results revealed that hypoxia significantly decreased the viability of H9c2 cells, whereas miR-663b inhibitor increased cell viability. This change was reversed by BCL2L1-siRNA when compared with the controls (Fig. 4A). Flow cytometry was subsequently performed to detect the proportion of apoptotic cells. The results revealed that hypoxia induced cell apoptosis and that the miR-663b inhibitor decreased cell apoptosis in the hypoxic cells compared with controls. These changes were reversed following BCL2L1-siRNA transfection (Fig. 4B and C).
Figure 4.
miR-663b inhibitor increases hypoxia-induced H9c2 cell viability and reduces apoptosis. (A) An MTT assay was used to detect cell viability. (B) Flow cytometry combined with (C) annexin V/PI staining was performed to measure apoptosis. (D) Western blotting was used to demonstrate cleaved-Caspase-3 protein expression levels in H9c2 cells. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01 vs. control group; ##P<0.01 vs. hypoxia group; &&P<0.01 vs. hypoxia + inhibitor group. PI, propidium iodide; miR, microRNA; si, small interfering.
The protein expression of cleaved-Caspase-3 was determined via western blotting. The results indicated that the hypoxia-induced upregulation of cleaved-Caspase-3 protein expression in H9c2 cells was significantly reduced in cells transfected with miR-663b inhibitor. This reduction was reversed following transfection with BCL2L1-siRNA (Fig. 4D). Together, the results indicated that the miR-663b inhibitor increased the viability of hypoxia-induced H9c2 cells and decreased cell apoptosis.
Effect of miR-663b inhibitor on the secretion of inflammatory factors in hypoxia-induced H9c2 cells
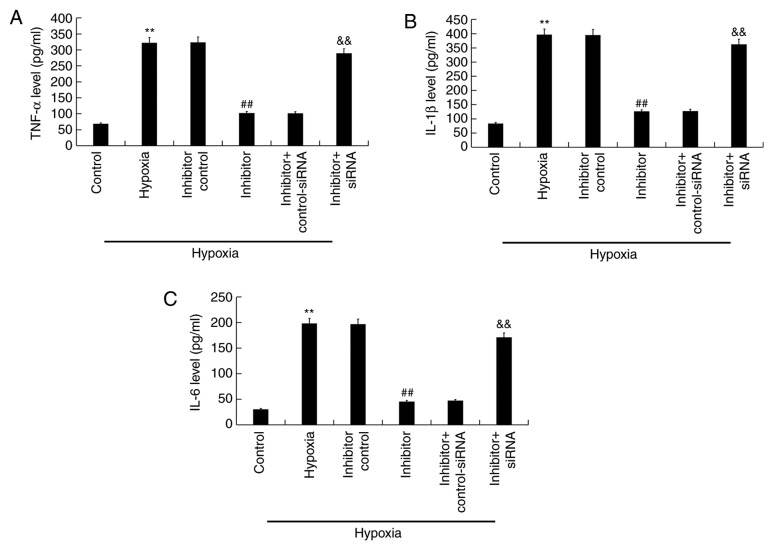
ELISA was performed to detect the effects of miR-663b inhibitor on the secretion of certain inflammatory factors in hypoxia-induced H9c2 cells. Compared with the control group, hypoxia increased the release of TNF-α, IL-1β and IL-6 while miR-663b inhibitor transfection significantly decreased their release (Fig. 5A-C). Knocking down BCL2L1 expression using siRNA reversed these effects.
Figure 5.
miR-663b inhibitor decreases TNF-α, IL-1β and IL-6 expression in H9c2 cells under hypoxic conditions. ELISA was performed to measure the levels of (A) TNF-α, (B) IL-1β (C) and IL-6. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01 vs. control group; ##P<0.01 vs. hypoxia group; &&P<0.01 vs. hypoxia + inhibitor group. miR, microRNA; TNF-α, tumor necrosis factor α; IL, interleukin; siRNA, small interfering RNA.
Discussion
Acute MI is characterized by ischemic injury and cardiomyocyte cell death, which leads to cardiac dysfunction and eventual heart failure (3). Cardiomyocyte apoptosis is one of the primary types of cardiac cell death during MI (23). Protecting cardiomyocytes against hypoxia-induced damage may therefore be a promising therapeutic target for the treatment of MI (24-26).
miRNAs serve an important role in the development of MI. Xiao et al (27) demonstrated that miR-24-3p decreased apoptosis in mouse cardiomyocytes in response to ischemia/reperfusion (I/R) injury. Zhang et al (2) also revealed that miR-103a-3p serves a protective role in myocardial I/R injury. Liu et al (28) demonstrated that miR-199a-5p expression affects hypoxia/reoxygenation-induced cytotoxicity in cardiomyocytes. Additionally, Peng et al (20) demonstrated that miR-133, miR-1291 and miR-663b expression was significantly increased in AMI compared with controls. However, the role and function of miR-663b in MI is yet to be fully elucidated. A previous study indicated that miR-663b is a tumor-associated miRNA (29). Cai et al (30) also demonstrated that miR-663b exerts a tumour suppressive effect by targeting insulin-like growth factor in pancreatic cancer. It has been reported that miR-663b expression was upregulated in osteosarcoma (31). In the present study, it was demonstrated that the expression of miR-663b was upregulated in hypoxia-induced H9c2 cells.
BCL2L1 was determined to be a direct target gene of miR-663b. Bcl2 genes are a mammalian family of genes that encode several proteins that directly regulate the activation of caspase-mediated apoptosis (32). As a member of this family, BCL2L1 acts as an antiapoptotic protein, with decreased expression activating a caspase cascade that ultimately promotes apoptosis. However, increased expression of BCL2L1 inhibits cell apoptosis (33). Sun and Pei (34) revealed that BCL2L1 is a target of miR-66 and increases vulnerability to propofol treatment in developing astrocytes. Lin et al (35) indicated that BCL2L1 was a target gene of miR-184 and that the miR-184 antagomir promoted apoptosis of osteosarcoma cells.
In the present study, BCL2L1 expression was downregulated at the mRNA and protein level in hypoxia-induced H9c2 cells. The effects of miR-663b on cell injury, viability, apoptosis and the release of inflammatory factors in H9c2 cells were also determined. Under hypoxic conditions, H9c2 cells were transfected with miR-663b inhibitor, inhibitor control, miR-663b inhibitor + control-siRNA or miR-663b inhibitor + BCL2L1-siRNA for 48 h. The miR-663b inhibitor decreased the release of cardiomyocyte injury biomarkers CK-MB and cTnI and increased mitochondrial viability and cell viability while decreasing cell apoptosis and the secretion of the inflammatory factors IL-1β, TNF-α and IL-6. These effects were reversed by BCL2L1 knockdown.
In conclusion, the miR-663b inhibitor protected cardiomyocytes against hypoxia-induced damage by targeting BCL2L1. These data suggested that miR-663b may be a novel target for the treatment of MI, exerting its effects by directly targeting BCL2L1.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
FY and XZ designed the study, collected the data, performed statistical analysis and data interpretation, and wrote the manuscript. CS (qRT-PCR, MTT data collection), WX (flow cytometry analysis and ELISA data collection) and JX (ELISA and MTT data collection) collected the data and prepared the manuscript. All authors read and approved the final version of this manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Henriksson R, Björklund F, Mooe T. The introduction of ticagrelor is associated with lower rates of recurrent ischemic stroke after myocardial infarction. PLoS One. 2019;14(e0216404) doi: 10.1371/journal.pone.0216404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang C, Zhang C, Wang H, Qi Y, Kan Y, Ge Z. Effects of miR-103a-3p on the autophagy and apoptosis of cardiomyocytes by regulating Atg5. Int J Mol Med. 2019;43:1951–1960. doi: 10.3892/ijmm.2019.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeymer U. Diagnosis and initial management of acute myocardial infarction. MMW Fortschr Med. 2019;161:34–36. doi: 10.1007/s15006-019-0223-3. [DOI] [PubMed] [Google Scholar]
- 4.Michiels C. Physiological and pathological responses to hypoxia. Am J Pathol. 2004;164:1875–1882. doi: 10.1016/S0002-9440(10)63747-9. (In German) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specifc microRNA regulates insulin secretion. Nature. 2004;432:226–30. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q, Liu N, Yang X, Tu L, Zhang X. Small RNA-mediated responses to low- and high-temperature stresses in cotton. Sci Rep. 2016;6(35558) doi: 10.1038/srep35558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Chen F, Zhao M, Yang Z, Zhang S, Ye L, Gao H, Zhang X. MiR-107 suppresses proliferation of hepatoma cells through targeting HMGA2 mRNA 3'UTR. Biochem Biophys Res Commun. 2016;480:455–460. doi: 10.1016/j.bbrc.2016.10.070. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Q, Gong L, Wang J, Tu Q, Yao L, Zhang JR, Han XJ, Zhu SJ, Wang SM, Li YH, Zhang W. miR-10b exerts oncogenic activity in human hepatocellular carcinoma cells by targeting expression of CUB and sushi multiple domains 1 (CSMD1) BMC Cancer. 2016;16(806) doi: 10.1186/s12885-016-2801-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 14.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 15.Liang S, Zhang N, Deng Y, Chen L, Zhang Y, Zheng Z, Luo W, Lv Z, Li S, Xun T. Increased serum level of MicroRNA-663 is correlated with poor prognosis of patients with nasopharyngeal carcinoma. Dis Markers. 2016;2016(7648215) doi: 10.1155/2016/7648215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan J, Hu H, Zhou Z, Sun L, Peng L, Yu L, Sun L, Liu J, Yang Z, Ran Y. Tumor-suppressive mir-663 gene induces mitotic catastrophe growth arrest in human gastric cancer cells. Oncol Rep. 2010;24:105–112. doi: 10.3892/or_00000834. [DOI] [PubMed] [Google Scholar]
- 17.Jiao L, Deng Z, Xu C, Yu Y, Li Y, Yang C, Chen J, Liu Z, Huang G, Li LC, Sun Y. miR-663 induces castration-resistant prostate cancer transformation and predicts clinical recurrence. J Cell Physiol. 2014;229:834–844. doi: 10.1002/jcp.24510. [DOI] [PubMed] [Google Scholar]
- 18.Yi C, Wang Q, Wang L, Huang Y, Li L, Liu L, Zhou X, Xie G, Kang T, Wang H, et al. MiR-663, a microRNA targeting p21 (WAF1/CIP1), promotes the proliferation and tumorigenesis of nasopharyngeal carcinoma. Oncogene. 2012;31:4421–4433. doi: 10.1038/onc.2011.629. [DOI] [PubMed] [Google Scholar]
- 19.Liang S, Zhang N, Deng Y, Chen L, Zhang Y, Zheng Z, Luo W, Lv Z, Li S, Xu T. miR-663b promotes tumor cell proliferation, migration and invasion in nasopharyngeal carcinoma through targeting TUSC2. Exp Ther Med. 2017;14:1095–1103. doi: 10.3892/etm.2017.4608. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Peng L, Chun-guang Q, Bei-fang L, Xue-zhi D, Zi-hao W, Yun-fu L, Yan-ping D, Yang-gui L, Wei-guo L, Tian-yong H, Zhen-wen H. Clinical impact of circulating miR-133, miR-1291 and miR-663b in plasma of patients with acute myocardial infarction. Diagn Pathol. 2014;9(89) doi: 10.1186/1746-1596-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong L, Chang H, Zhang J, Guo G, Shi J, Xu H. Astragaloside IV protects rat cardiomyocytes from hypoxia-Induced injury by down-regulation of miR-23a and miR-92a. Cell Physiol Biochem. 2018;49:2240–2253. doi: 10.1159/000493827. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta c(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Thygesen K, Alpert JS, Jaffe AS, Simoons ML, Chaitman BR, White HD. Thygesen K, Alpert JS, White HD, et al. Third universal definition of myocardial infarction. Eur Heart J. 2012;33:2551–2567. doi: 10.1093/eurheartj/ehs184. Writing Group on the Joint ESC/ACCF/AHA/WHF Task Force for the Universal Definition of Myocardial Infarction. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Zhao Y, Chen W, Xie L, Zhao ZA, Yang J, Chen Y, Lei W, Shen Z. MicroRNA-133 overexpression promotes the therapeutic efficacy of mesenchymalstem cells on acute myocardial infarction. Stem Cell Res Ther. 2017;8(268) doi: 10.1186/s13287-017-0722-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu G, Ma L, Dong F, Hu X, Liu S, Sun H. Inhibition of microRNA-124-3p protects against acute myocardial infarction by suppressing the apoptosis of cardiomyocytes. Mol Med Rep. 2019;20:3379–3387. doi: 10.3892/mmr.2019.10565. [DOI] [PubMed] [Google Scholar]
- 26.Luo Q, Guo D, Liu G, Chen G, Hang M, Jin M. Exosomes from MiR-126-overexpressing adscs are therapeutic in relieving acute myocardial ischaemic injury. Cell Physiol Biochem. 2017;44:2105–2116. doi: 10.1159/000485949. [DOI] [PubMed] [Google Scholar]
- 27.Xiao X, Lu Z, Lin V, May A, Shaw DH, Wang Z, Che B, Tran K, Du H, Shaw PX. MicroRNA miR-24-3p reduces apoptosis and regulates Keap1-Nrf2 pathway in mouse cardiomyocytes responding to ischemia/reperfusion injury. Oxid Med Cell Longev. 2018;2018(7042105) doi: 10.1155/2018/7042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu DW, Zhang YN, Hu HJ, Zhang PQ, Cui W. Downregulation of microRNA-199a-5p attenuates hypoxia/reoxygenation-induced cytotoxicity in cardiomyocytes by targeting the HIF-1α-GSK3β-mPTP axis. Mol Med Rep. 2019;19:5335–5344. doi: 10.3892/mmr.2019.10197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang M, Jia M, Yuan K. MicroRNA-663b promotes cell proliferation and epithelial mesenchymal transition by directly targeting SMAD7 in nasopharyngeal carcinoma. Exp Ther Med. 2018;16:3129–3134. doi: 10.3892/etm.2018.6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai H, An Y, Chen X, Sun D, Chen T, Peng Y, Zhu F, Jiang Y, He X. Epigenetic inhibition of miR-663b by long non-coding RNA HOTAIR promotes pancreatic cancer cell proliferation via up-regulation of insulin-like growth factor 2. Oncotarget. 2016;7:86857–86870. doi: 10.18632/oncotarget.13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao H, Li M, Li L, Yang X, Lan G, Zhang Y. MiR-133b is down-regulated inhuman osteosarcomaand inhibits osteosarcoma cells proliferation, migrationand invasion, and promotes apoptosis. PLoS One. 2013;8(e83571) doi: 10.1371/journal.pone.0083571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fagundes DJ, Carrara FL, Teixeira WA, Simões RS, Taha MO. The role of the exogenous supply of adenosine triphosphate in the expression of Bax and Bcl2L1 genes in intestinal ischemia and reperfusion in rats1. Acta Cir Bras. 2018;33:889–895. doi: 10.1590/s0102-865020180100000003. [DOI] [PubMed] [Google Scholar]
- 33.Ureshino RP, Bertoncini CR, Fernandes MJ, Abdalla FM, Porto CS, Hsu YT, Lopes GS, Smaili SS. Alteratons in calcium signaling and a decrease in Bcl-2 expression: Possible correlaton with apoptosis in aged striatum. J Neurosci Res. 2010;88:438–447. doi: 10.1002/jnr.22214. [DOI] [PubMed] [Google Scholar]
- 34.Sun WC, Pei L. rno-miR-665 targets BCL2L1 (Bcl-xl) and increases vulnerability to propofol in developing astrocytes. J Neurochem. 2016;138:233–242. doi: 10.1111/jnc.13647. [DOI] [PubMed] [Google Scholar]
- 35.Lin BC, Huang D, Yu CQ, Mou Y, Liu YH, Zhang DW, Shi FJ. MicroRNA-184 modulates doxorubicin resistance in osteosarcoma cells by targeting BCL2L1. Med Sci Monit. 2016;22:1761–1765. doi: 10.12659/msm.896451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.