ABSTRACT
Exhaustion cripples T cell effector responses against metastatic cancers and chronic infections alike. There has been considerable interest in understanding the molecular and cellular mechanisms driving T cell exhaustion in human cancers fueled by the success of immunotherapy drugs especially the checkpoint receptor blockade (CRB) inhibitory antibodies that reverses T cell functional exhaustion. The current understanding of molecular mechanism of T cell exhaustion has been elucidated from the studies utilizing murine models of chronic viral infections. These studies have formed the basis for much of our understanding of the process of exhaustion and proven vital to developing anti-exhaustion therapies against human cancers. In this review, we discuss the T cell exhaustion differentiation pathway in cancers and chronic viral infections and explore how the transcription factors expression dynamics play role in T cell exhaustion fate choices and maturation. Finally, we summarize the role of some of the most important transcription factors involved in T cell functional exhaustion and construct exhaustion specific signaling pathway maps.
KEYWORDS: CD8 T cells, two axes of exhaustion, checkpoint receptors, cancer, chronic viral infections
Introduction
T cell exhaustion is at the heart of success of checkpoint receptor blockade (CRB) inhibitory drugs (i.e., Opdivo® [nivolumab], Keytruda® [pembrolizumab]). These drugs function by reversing functional exhaustion in antigen-specific T cells inducing remissions in stage III and IV melanoma, non-small lung cancer (NSLC), and urothelial cancers.1–3 Despite the success of CRB therapies, the clinical data have also highlighted limitations with these therapies as only 20% to 40% of patients respond to single agent CRB therapies, others have shown resistance with progression, indicating a huge scope for improvement.4–7 One of the reasons impeding improvements in these therapies otherwise possible may be due to lack of understanding of the molecular and cellular mechanisms underlying T cell exhaustion lineage growth and maturation in human cancers. One among many means to improve CRB therapies could be investing resources in next generation immunotherapies. For example, developing new immunotherapy drugs that does not rely entirely on antibodies to inhibit checkpoint receptor blockade but also target signaling pathways regulating T cell exhaustion growth, and epigenetic regulators of exhaustion. Therefore, in-depth understanding of molecular mechanisms of T cell exhaustion is of great interest both from a clinical and a basic science perspective. Several papers have been published recently that present new data on the mechanisms underlying T cell exhaustion developmental fates and maintenance in murine chronic viral infection and human cancer models. These papers elegantly describe the role of novel transcription factors, transcription regulators, epigenetic remodeling enzymes, and metabolic regulators involved in the development and stability of T cell exhaustion subsets {Reviewed in ref.8–11}. Table 1 summarizes the role of some of these transcription factors in T cell exhaustion development and maturation. As noted in the table, almost all transcription factors implicated in T cell exhaustion have overlapping functions in T effector and/or T cell memory differentiation. Therefore, it can be strongly argued that molecular mechanisms operational in T cell exhaustion can be expected to be operational at some level in T effector and T memory cells as well. In the following sections, we discuss how the transcription factors expression dynamics play instructive roles in T cell exhaustion, T effector, and T cell memory differentiation. We first begin with defining CD8 T cell exhaustion.
Table 1.
Transcription factor activities in various T cell subtypes.
| TEffector | TMemory precursors | TExhaustion | |
|---|---|---|---|
| NFAT1 | Moderately expressed | Expressed and has a role | Highly expressed |
| BLIMP1 | Moderately expressed | Not expressed | Highly expressed |
| Eomes | Moderately expressed | Highly expressed | Highly expressed |
| T-bet | Highly expressed | Expressed and has a role | Expressed and has a role |
| IRF4 | Essential for effector functions | Not expressed | Highly expressed |
| BATF | Moderately expressed and essential for effector functions | Expressed and has a role | Highly expressed |
| YY1 | Moderately expressed | Expressed and has a role | Highly expressed |
| FOXO1 | Opposes effector functions | Expressed and has a role | Highly expressed |
Defining CD8 T cell exhaustion in the context of chronic viral infections and cancer
Several studies have provided critical insights into the developmental fates of CD8 T cells.12–14 One established view in the context of murine model of acute infections that resolve successfully is that the naïve CD8 T cells become activated after interacting with professional antigen presenting cells that present viral antigens via canonical endogenous pathway or non-canonical cross presentation. This reaction occurs in draining lymph nodes.15 These activated T cells react with virally infected tissues and differentiate into short-lived effector T cells generating alongside a long-lived memory T cell pool.16 The short-lived effector T cells, according to the linear differentiation model, become terminally differentiated and subsequently undergo apoptosis with the elimination of acute infections (Figure 2). As Figure 2 demonstrates, 3-signals, the TCR, costimulatory, and cytokine signals are vital for CD8 T cell differentiation process.
Figure 2.
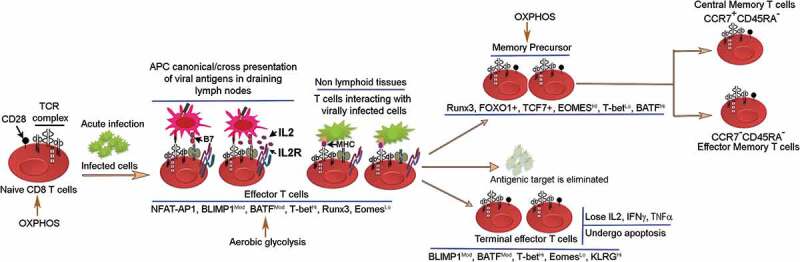
The CD8 T cell linear differentiation model. Naïve T cells in acute viral infections become activated in lymphoid tissues via canonical and cross presentation of viral antigens by antigen-presenting cells. The activation process ensues with the delivery of signal 1 + 2 and IL2 production. IL2 subsequently diffuses locally and binds IL2 receptor to generate high affinity IL2-R that promotes IL2-R mediating signaling pathway, which is important for proliferation and survival of antigen-specific CD8 T cells. Activated CD8 T cells react with virally infected cells and undergo proliferative expansion and differentiate into terminal T effector cells generating alongside memory precursors that differentiate further into central memory and T effector memory subsets. These memory subsets persist at various sites in vivo.17–20 The transcription factor expression pattern in CD8 T effector, T cell exhaustion and T cell memory subsets are shown.
The developmental fates of CD8 T cells in chronic viral infections and cancers have begun to be understood only recently. In murine models of chronic actively replicating viral infections and in immunogenic cancers, the naïve/resting CD8 T cells also follow the linear differentiation model and generate effector T cell pool. But, due to the presence of persistent antigens T effector cells fail to properly activate terminal differentiation program instead differentiate into a hypofunctional quasi-differentiated chronically stimulated state termed exhaustion. The memory precursors generated in chronic viral infections are unstable and not properly maintained in vivo.17,21,22 The antigen-presenting cells (APCs) engaged in antigen presentation of viral antigens via canonical endogenous pathway or cross presentation have been identified. For example, in chronic HIV macrophages harbor HIV viruses and also scavenge HIV proteins from plasma thereby presenting antigens to T cells.23,24 Likewise, in human cancers, several studies have established that APCs especially migratory dendritic cells that immigrate from tissues, and lymphoid resident conventional dendritic cells process and present tumor antigens to CD8 T cells via cross presentation in the draining lymph nodes.15,25-28 The activated viral antigen or tumor antigen-specific T cell subsequently react with virally infected tissues or tumor tissues, respectively (Figure 3a,B). Accordingly, exhausted T cells have been found to be present in lymphoid tissue T cell zones and at the sites of infection and cancer tissues with features ranging from partial to terminal exhaustion phenotypes.29–31 It is noteworthy, in chronic viral infections the antigen-specific effector T cells lose specificity for immunodominant epitopes instead develop specificity for subdominant epitopes .32,33 Moreover, using lineage tracing techniques, it has recently become clear that during early chronic viral infections, a precursor population exist within T terminal-effector population that become lineage committed to T cell exhaustion fate and maturation34,35 (Figure 3b). T cell exhaustion precursors with stem cell line properties have also been identified in cancers, but if they commit to T cell exhaustion fate is unclear.36 Taken together, these studies provide strong evidence that T cell exhaustion development follows a linear differentiation model.
Figure 3.
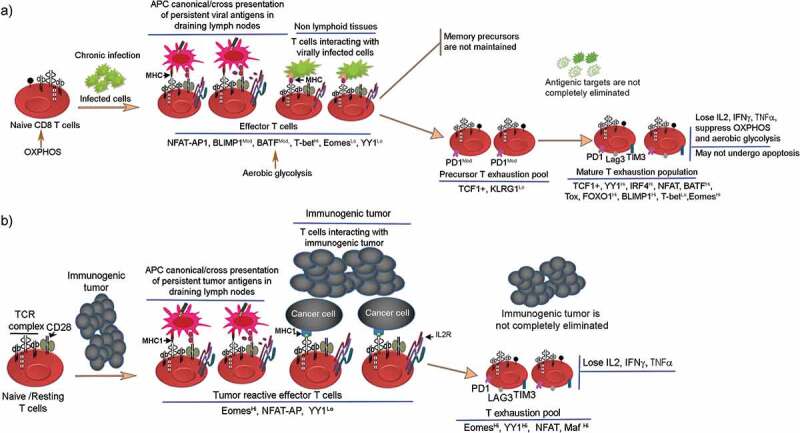
Transcription factors involved in regulating lineage differentiation of naïve CD8 T cells into effector and T cell exhaustion fates in chronic viral infections and immunogenic cancers. (a) Illustrates naïve CD8 T cells upon exposure to chronic viral infections initially differentiate into effector T cells, but with persistent antigen presence, effector T cells fail to activate terminal T effector program instead generate a PD1ModTCF1+ precursor T exhaustion population. This precursor population gives rise to mature pool of exhausted T cells that are hyporesponsive, undergo cytokine failure, and upregulate exhaustion specific markers. Exhausted T cells lose the capacity to eliminate chronic infections. (b) Illustrates the role of antigen-presenting cells especially tissue resident conventional dendritic cells in acquiring tumor antigens, processing and presenting to CD8 T cells. The reaction occurs in draining lymph nodes. The list of transcription factors involved in T cell exhaustion to tumors is provided.
The hallmarks of T cell exhaustion are presented in Figure 1. One of the hallmarks of exhausted T cells is the upregulation of Checkpoint Receptor (CR) Axis, i.e.,, PD1, Lag3, Tim3, TIGIT, and downregulation of Cytokine Axis, i.e.,, IL2, IFNγ, TNFα. These represent two vital axes of exhaustion. By addressing therapeutically the checkpoint receptor blockade (CRB), the quasi-differentiated T cell exhaustion pool in cancers can reverse some aspects of functional exhaustion such as cytokine secretion, proliferation, and killing activities generating alongside the memory precursors.29,37-39 The origin of CRB therapy-related T cell exhaustion memory precursors in vivo is poorly understood (Figure 4). The notion that T cell exhaustion subsets are quasi-differentiated and chronically stimulated is supported by the following: 1) Their partial restoration of effector functions after therapeutic interventions such as antibody-mediated blockade of PD1:PD-L1 interaction or through attenuation of regulators that sustain exhaustion ;40–42 2) The upregulation of CD69 activation marker and CR axis in vivo is an evidence that mature exhausted T cells are chronically activated. The upregulation of CR axis in exhausted T cells could be a negative feedback mechanism for preventing generation of deleterious autoimmune responses.37,43 This view is supported by the cases of toxic autoimmune responses reported in patients receiving CRB therapy, as much as the toxic responses in patients are correlated with the regression of tumors.44,45
Figure 1.
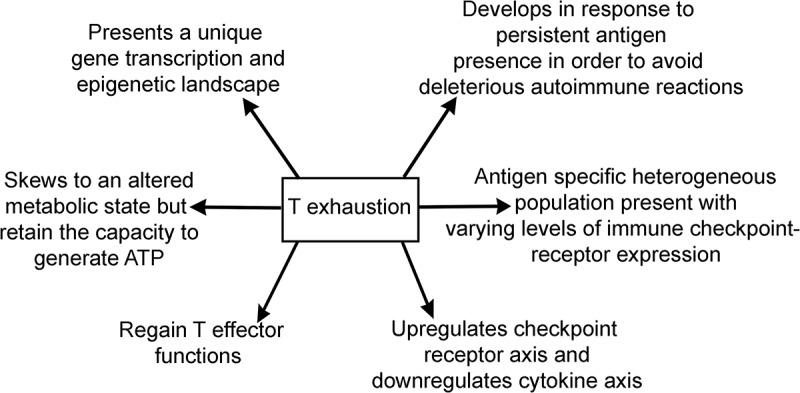
Summary of hallmarks of T cell exhaustion.
Figure 4.
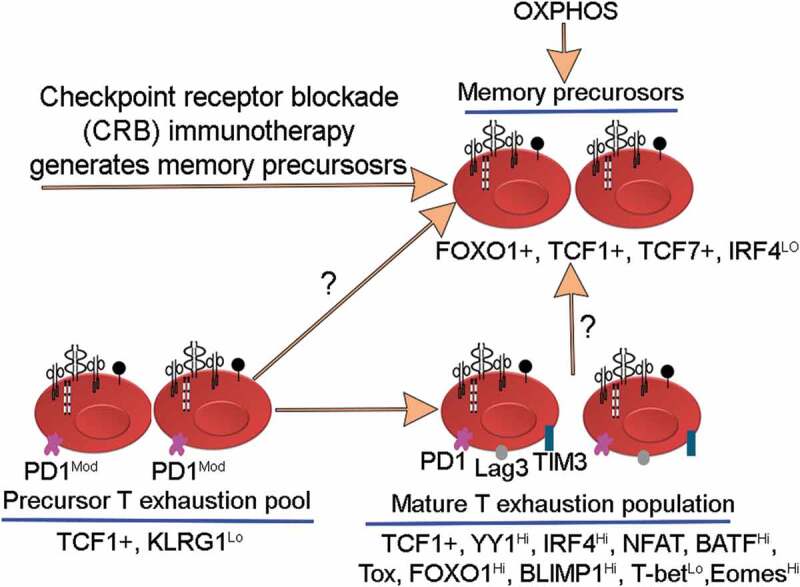
Illustrates differentiation of memory precursors in vivo after checkpoint receptor blockade (CRB) immunotherapy. The origin of these memory precursors is unknown. For example, it is not known if these memory precursors arise from T exhaustion precursors or from mature T-exhausted pool.
The heterogeneity of T cell exhaustion pool
Exhausted T cells present at the sites of immunogenic tumors and chronic viral infections have been recognized to be heterogeneous population of cells. A specific subset of T cells within the larger pool of heterogeneous subsets of T cell exhaustion have been shown to respond vigorously to the anti-exhaustion therapies, 41 and even without therapy, a subset of T cells have also been shown to retain effector functions. Therefore, examining the heterogeneity of T cell exhaustion pool is vital not only from a clinical perspective but also for our understanding of the mechanism of T cell exhaustion. Several subsets in T exhaustion pool have been identified expressing varying levels of exhaustion-specific markers alongside the markers of memory and progenitor T cell lineage. Among the heterogeneous CD8 T cell exhaustion pool, a short-lived proliferating and long-lived non-proliferating subsets were discovered to be present in chronic human HIV1 and LCMV infections.21,46,47 The short-lived proliferating CD8 T cells subsets were confirmed to be Eomeshi whereas long-lived non-proliferating subsets were confirmed to be T-bethi.47 Another subset of cells found to be present within the T cell exhaustion pool include TCF1+ memory T cell like precursors, and CXCR5+ follicular cytotoxic CD8 T cell like that expresses exhaustion-specific inhibitory receptors notably PD1 albeit at lower levels.30,42,48,49 Importantly, the Runx-family transcription factor Runx-3 has been shown to be important for repressing development of CXCR5+ CD8 T cells with follicular T helper cell-like phenotypes, and for regulating T-bet/Eomes functions as well as for promoting effector and tissue-resident memory CTL differentiation.50–52 The development of heterogeneous subsets of T cell exhaustion in the settings of cancer and chronic viral infections also seem to determine the response to therapy. The two subsets of T cells within the heterogeneous T cell exhaustion pool were identified to respond differently to the anti-PD1/PD-L1 therapy. For example, the PD-1Int CD44Hi population were shown to respond vigorously whereas PD-1Hi CD44Int respond poorly to the anti-PD1/PD-L1 therapy.41 In a recent study, Thommen et al., utilizing a gene expression profile reported a remarkable heterogeneity in the intratumoral CD8+ TILs population in human non-small lung cancer patients.53 Thommen et al. demonstrated the distinct functional and metabolic gene signature existed for intratumoral CD8+PD1Hi, CD8+PD1Int and CD8+PD1− T cell populations. Importantly, the differential gene signature could predict the response to anti-PD1 therapy in a small cohort of NSLC patients with PD1Hi being the strong predictor of response and survival.53 As to the origin of CD8 T cell exhaustion subsets, little is known so far. For example, it is unclear if it is the effector CD8 T cells that after experiencing persistent chronic viral antigens or immunogenic tumor antigens in vivo differentiate into different subsets of exhausted CD8 T cells or whether these subtypes arise following a linear differentiation program.
Reversal of T cell exhaustion
T cell exhaustion is partially reversible process, i.e.,, subset of cells within T cell exhaustion pool regain the antigenic target killing functions with therapeutic interventions. That makes T cell exhaustion subsets distinct from the terminal T effector cells which cannot regain effector functions and are programmed to undergo apoptosis. In a sharp contrast to T cell exhaustion, memory T cell differentiation and for generating strong central memory recall responses both require the downregulation of immune checkpoint receptors.54,55 Another marked distinction with the T cell memory is that the latent memory T cells become reactivated if reexposed to the same strain of infectious pathogens. In contrast, exhausted T cells can regain antigen-specific effector functions especially cancer-killing functions in vivo with the drug interventions such as checkpoint receptor blockade (CRB) inhibitory antibodies56,57 or with high doses of IL2.58–62 In chronic viral infections and cancer, IL2 has been shown to reduce the expression of PD1 and enhance the proliferation of resting memory T cell pool.38,63 The need for extrinsic interventions to regain killing functions is one of the hallmarks of exhausted T cells. The key molecular, phenotypic, and functional differences between various CD8 T cell subtypes are provided in the Box.
Metabolism in exhausted CD8 T cells
Altered metabolism is a hallmark of exhausted T cells. Choice of metabolic pathways plays an important role in the differentiation and activation of antigen-specific T cells. T cells adopt two choices for ATP generation: 1. Glycolysis and mitochondrial mediated oxidative phosphorylation, 2. Aerobic glycolysis. Naïve, memory, effector, and exhausted T cells have different bioenergy demands and substrate requirements. The quiescent T cells, i.e.,, naïve and memory T cells under normal oxygen levels metabolize glucose through glycolysis and via oxidative phosphorylation (OXPHOS) generates ATP utilizing electron transport chain complexes I–V in the mitochondria, which is an efficient mechanism for generating ATP.64,65 The antigen-activated T cells synthesize rapidly effector molecules and effector cytokines (e.g., IL2, IFNγ), and have increased proliferation rates. Accordingly, effector T cells under normal oxygen levels switch to aerobic glycolysis, a process also referred to as Warburg effect.66,67 In aerobic glycolysis, glucose is converted into lactic acid and even though only fewer molecules of ATP are generated, it represents a rapid mechanism of ATP and metabolic substrate intermediate synthesis including nucleotides, non-essential amino acids and fatty acids. Therefore, aerobic glycolysis helps to catch up with the demand for enhanced anabolism, proliferation, and effector T cell activities.65,66,68 It is presently unclear how the quiescent memory T cells switch to aerobic glycolysis after antigen rechallenge and maintain OXPHOS after antigen clearance. It appears that memory T cells unlike naïve or effector T cells maintain mitochondrial functions especially fatty acid oxidation, and IL15 seem to play a vital role in promoting mitochondrial biogenesis and respiratory capacity.69 The metabolic requirements of T cell exhaustion subsets may be distinct in the hyporesponsive state versus reactivation phases, i.e.,, during reactivation of T cell exhaustion subsets induced by CRB inhibitory antibodies. Exhausted T cells, as reported in both chronic viral infections and cancers present with significant suppression of glycolysis, OXPHOS, and glucose deprivation due to GLUT1 downregulation, and repression of mitochondrial biogenesis.70–72 There may be additional candidates for how the change in metabolism in this transition works, such as downregulation of mTOR or activation of the AMPK pathway, although it is unclear how the choice for such pathways occurs, some works suggest asymmetrical division of effector T cells may play a role. In the LCMV exhaustion model, the metabolic dysfunction of T cells can be detected immediately after the infection and appears to be PD1 regulated and driven by the enhanced mTOR activity. Among the metabolic changes that were noted in chronic LCMV, specific CD8 T cells include reduced glucose uptake and glycolysis, reduced OXPHOS and mitochondrial respiration, but enhanced mitochondrial mass, membrane depolarization, ROS production, and ultrastructural changes such as elongated mitochondria cristae.71 Part of the metabolic dysregulation driven by PD1 was found to be through repression of the key metabolic regulator, the peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α (PGC1α).73 PGC1α is a transcriptional activator that induces expression of genes important for mitochondrial biogenesis, and when reexpressed in exhausted T cells recovers glucose uptake, lowers mitochondrial biomass and depolarization, and improves effector functions.71
The metabolic dysfunctions such as the reduced glucose uptake and mitochondrial biogenesis as seen in chronic LCMV specific CD8 T cells were also noted in the CD8 TILs in B16 melanoma mouse model and metastatic melanoma genetic mouse model, BrafV600E/Pten−/- with the exception of mitochondrial mass that was found to be reduced.72,74,75 The intratumoral TILs remain exposed to glucose deprivation in the tumor microenvironment that restricts aerobic glycolysis and also negatively affect T cell effector functions. In glucose sufficient environment, the glucose gets transported via GLUT1 into cytoplasm where it is subjected to glycolysis generating several substrate intermediates such as phosphoenol pyruvate (PEP). The PEP inhibits endoplasmic reticulum membrane Sarco/ER Ca2+-ATPase (SERCA) activity that prevents ER uptake of cytosolic calcium allowing optimal TCR →NFAT signaling pathway to proceed normally thereby helping mediate optimal effector functions. Correspondingly, glucose deficiency in tumor microenvironment produces PEP insufficiency promoting SERCA mediated calcium uptake that attenuates TCR signaling and as a result the NFAT activity.76,77 Interestingly, in mouse sarcoma tumor model, the systemic analysis of the metabolic changes associated with reactivation of exhausted T cells with CRB inhibitory antibodies PD1, CTLA4, and PD-L1 have shown to restore glucose in tumor microenvironment by restricting glucose uptake in tumor cells and triggering glycolysis and enhanced IFNγ production in antigen-specific T cells.75 These data indicate that similar metabolic changes could be taking place in the reactivated antitumor T cells in humans responding to CRB therapy. Confirmatory studies using human samples will be needed to prove this point.
Gene expression signature in exhausted T cells
Several attempts have been made to elucidate the transcription profile of exhausted T cells.70,78-80 The T cell exhaustion transcription profile often contains genes that overlapped with the T effector and T cell memory gene expression modules. That essentially could be due to the various subsets of cells present within the T cell exhaustion pool. The composition of heterogeneous subpopulation within T cell exhaustion pool can vary significantly with infections from different strains of viruses or between different lineages of cancers. Moreover, isolating a homogenous population of tumor antigen-specific T cells for obtaining T cell exhaustion specific gene expression profile can be technically challenging and labor intensive. One of the challenges is designing unique MHC-tumor peptide tetramers that TCRs could recognize and bind in sufficient numbers.81 Until today, very few groups have been successful in isolating tumor antigen-specific T cells in vivo with the exception of melanoma antigens.79 Overall, the number of tumor antigen-specific CD8 T cells can be extremely low. They may represent as little as 0.002% of the peripheral T cell population in humans, and approximately 0.1% of TIL ex vivo culture.82 Accordingly, several recent studies have performed whole-exome sequencing and RNA-seq analysis on human tumors to identify novel tumor antigens that TCRs could bind in sufficient numbers.83–86 These new technologies can prove extremely valuable in identifying unique tumor antigens and in designing MHC-tumor peptide tetramers that eventually could help isolating and analyzing TCR-specific tumor-infiltrating T cells. Moreover, powerful new technologies such as single-cell RNA-seq analysis of tumor-infiltrating T cells has proven powerful resource in revealing a reliable transcriptional profile of homogenous T cell exhaustion pool.87–91
Exhaustion specific transcription factors
Several new transcription factors have recently been identified to be associated with exhaustion in murine chronic viral infections in vivo. Table 2 summarizes the activities of these transcription factors. Some of these transcription factors have opposing or overlapping functions in T effector, T cell memory, and T cell exhaustion subsets, which is due to the heterogeneity that exists in these cell types especially in T effector and T cell exhaustion pool. Both T effectors and exhausted T cells contain subsets with varying degrees of differentiation, some present with markers of terminal differentiation and other express markers of T cell memory. In this review, we focus mostly on the transcription factors that have been revealed to regulate the T cell exhaustion fate choices, maturation, metabolism, and memory precursor generation.
Table 2.
Transcription factors involved in regulating T cell exhaustion-specific genes.
| YY1 | NFAT-AP1 | NFAT1 | BLIMP1 | T-bet | Eomes | IRF4 | BATF | FOXO1 | |
|---|---|---|---|---|---|---|---|---|---|
| PD1 | hB↑ | ↓ | B↑ | B↓↑ | B↓ | - | B↑ | B↑ | B↑ |
| LAG3 | hB↑ | ↓ | B | - | - | - | B | B | - |
| TIM3 | h↑* | ↓ | B | - | - | - | B | B | - |
| IL2 | hB↓ | hB↑ | B↓ | B↓ | - | - | ↓ | hB↓ | − |
| IFNγ | hB↓ | B↑ | B↑ | B↓ | ↑ | B↑ | ↓ | h↓ | − |
B indicates binds on consensus transcription factor sites present in genes.
h denotes in humans.
↑ indicate enhances gene expression.
↓ indicate represses gene transcription.
* Indicate indirect binding.
IRF4
IRF4 transcription factor was identified in a gene expression analysis to be expressed in the chronic LCMV specific CD8 T exhaustion pool.70 The functional role of IRF4 in T cell exhaustion has recently been revealed. Using LCMV chronic infection model, Man et al., showed that IRF4 expression is elevated in exhausted T cells. Furthermore, through in silico ChIP-seq and ChIP assays, they showed that IRF4 binds to immune checkpoint receptor genes, PD1, Lag3, and Tim3.42 And, part of the mechanism through which IRF4 exerts its exhaustion specific role was through the disruption of metabolic regulation affecting mitochondrial size and production of reactive oxygen species (ROS). IRF4 disrupted glycolysis and oxidative phosphorylation in exhausted T cells.42 In addition, IRF4 forms a transcriptional circuit with NFAT1 and BATF transcription factors. IRF4 and NF-AT1 also form autoregulatory loop with IRF4 promoting expression of NF-AT1. The NF-AT1 in turn binds to IRF4 promoter.92,93 It appears from the insilico ChIP-seq data that IRF4, BATF, and NFAT1 bind together on the PD1 gene. However, it was not clear whether these composite consensus regulatory sites on the PD1 were conserved on the human PD1. Interestingly, reducing the IRF4 levels attenuates T cells exhaustion, corrected mitochondrial size and ROS production, and promoted differentiation of TCF1+ memory like cell types.42 From the data, it is not clear if the T cells in mice infected with chronic viral infections and having lost the IRF4 allele convert to memory like T cells from a reactivating pool of exhausted T cells or represent a separate lineage of cells differentiating from recent effector T cell emigrants. Several lines of evidence support that reactivated exhausted T cells after CRB therapy can generate heterogeneous subset of cells some resembling TCF1+ expressing memory precursors and others the follicular T cells.30,42,48,49 Overall, IRF4 is required for the maintenance of exhaustion phenotype along with the NFAT1 and BATF. These three molecules seem to accumulate with TCR activation. Whether CD28 stimulation (signal 2) was also necessary for their accumulation was not examined. Interestingly, the IRF4 binding site are present on human PD1, Lag3, and Tim3 promoters. In my search for transcription factors binding to human IRF4 promoter using publicly available promoter database http://epd.vital-it.ch/cgibin/, revealed several IRF4 binding site in ~2kb upstream of TSS on human PD1, Lag3, and Tim3 promoter. Whether these sites are functional and important for human T cell exhaustion to chronic infections and in cancer needs elaborate analysis. Furthermore, in CD4 T cells, IRF4 is known to coordinate with AP1, and IRF4-AP1 bind on composite elements on IL10 gene to promote transcription.94 IL10 is one cytokine that increases during exhaustion to chronic LCMV infections.95,96 Whether IRF4:AP1 play any role in IL10 gene transcription during exhaustion remains unknown. It is important to note that intratumoral IL10 released by Tregs into tumor microenvironment contribute to T cell exhaustion. Correspondingly, targeting IL10 or Tregs in combination with checkpoint receptor blockade (CRB) anti-PD1 therapy reverses some aspects of exhaustion to chronic LCMV infection.97
T-bet and Eomes
T-bet and Eomes are T-box transcription factors that play a crucial role in effector and memory functions of T cells.98,99 The physiologically significant role of T-bet in protective Immunity and effector functions was revealed in T-bet−/- deficient mice. These mice demonstrated the compromised protection against intracranial LCMV infection.100 T-bet and its paralogue Eomes appear to have redundant and cooperative functions in effector T cell differentiation. For example, T-bet−/- CD8 T cells secrete reduced levels of effector cytokine, IFNγ. Whereas Eomes overexpression rescues IFNγ production in T-bet−/- CD8 T cells. Correspondingly, haploinsufficient Eomes± mice do not produce any defect in IFNγ production that could be due to haploinsufficiency being compensation by the normal T-bet expression.101 The inverse kinetics of T-bet and Eomes expression appear to regulate lineage differentiation of T effector versus T cell memory and T cell exhaustion.16,102 The high expression of T-bet and Eomes appears to be important for the effector functions of CD8 T cells in acute infection model.99,103 The high T-bet expression in effector T cells during acute infections progressively declines with memory T cell differentiation; however, an inverse kinetics was observed with respect to Eomes104 (Figure 2). In chronic LCMV infection exhaustion model, a low T-bet expression is crucial for maintaining exhaustion phenotype because T-bet is revealed to be a repressor of PD1 and was shown to bind directly on PD1 promoter.102 Consistent with the murine data, the human chronic HIV antigen-specific exhausted T cells have lower T-bet expression but maintained higher Eomes expression, and these expression kinetics correlated with upregulation of inhibitory immune checkpoint receptor PD1.105 It remains unclear how reuse of T-bet and Eomes in exhausted T cells in the same kinetics as in memory T cells contribute to the exhaustion state. One explanation could be that the exhausted T cells like memory T cells remain quiescent with potential for regaining effector activities; therefore, T-bet and Eomes exist in the same kinetics in these two cell types to regulate the quiescence and reactivation programs. The role of Eomes and T-bet to T cell exhaustion in cancers remains unknown; however, similar to chronic infections, in autochthonous melanoma mouse model and in patients with metastatic melanoma expression of Eomes was detected to be upregulated in tumor antigen-specific exhausted T cells.43,79
Blimp1
Blimp1 has been extensively studied for its role in CD8 T effector cells and T cell memory differentiation during acute infections. It emerged from the studies involving acute viral infections that Blimp1 expression increased during T effector cell differentiation and was essential for the effector activities.106–108 Blimp1 expression though important for T effector differentiation appears to restrict memory T cell development as the Blimp1 deficient effector T cells showed unrestricted capacity to differentiate into memory precursors.107 Blimp1 has also emerged as an important player in T cell exhaustion. The gene expression dosage of Blimp1 appears to be crucial for instructing lineage choices to T effector versus T cell exhaustion. The moderate Blimp1 expression appears to favor CD8 T effector functions. However, in chronic LCMV infections where Blimp1 expression was detected to be elevated promote upregulation of immune checkpoint receptor molecules associated with exhaustion while repressing the genes involved in memory T cell differentiation.109 Intriguingly, Blimp1 was also reported to repress PD1 expression in acute infection model through a mechanism involving direct transcriptional repression of PD1 as well as, indirectly, through inhibition of NFAT1, which is a direct transcriptional activator of PD1.110 Why Blimp1 does not repress PD1 and NFAT1 in exhausted T cells where Blimp1 is detected to be elevated remains unclear. One explanation could be that other transcription factors that activate PD1 transcription override Blimp 1 repressive functions directed at PD1. Blimp1 has been proposed to have a role in IL2 shutdown.111 That assumption was based on, 1) A Blimp1 binding site on IL2 promoter 2kb upstream from the transcription start site (TSS), 2) Blimp1 knockout studies in mice.112 However, Blimp1 mediated IL2 repression in the above studies was not evaluated through promoter mutation analysis, and the site for Blimp1 binding is also not conserved in the human IL2 promoter. Whether Blimp1 can be a direct mediator of human IL2 failure during T cell exhaustion require further investigations. It is possible that Blimp1 has a role in mouse T cells which is distinct from its role in human CD8 T cell exhaustion (Figure 3a).
NFAT
NFAT is a regulator of T cell activation.113,114 NFAT1 (human ortholog NFATc2) is a founding member of the NFAT family which was elucidated to have a prominent role in T cell activation, and exhaustion to chronic infections (bacterial and viral) and cancer.93,115 NFAT1 (NFATc2) including its other members NFAT2 (NFATc1) and NFAT4 (NFATc3) get activated through TCR signaling and triggers biphasic calcium mobilization and binding of cytosolic calcium to calmodulin calcium binding protein. Ca2+/calmodulin activates serine/threonine phosphatase calcineurin, which dephosphorylate cytoplasmic NFAT1 allowing it to translocate to the nucleus to initiate gene transcription program.116 However, for the effector cytokine synthesis such as IL2, NFAT1 cooperates and interacts with AP1 transcription factor that binds to the NFAT-AP1 composite DNA binding sites on IL2 promoter.117–120 NFAT1 is extensively studied for its role in regulating the gene transcription program associated with anergy and exhaustion.80,93,121 In anergy, NFAT1 mediated gene expression is driven primarily by TCR activation in the absence of costimulation that prevents AP1 activation and interaction with NFAT inhibiting in the process the NFAT:AP1 regulated gene transcription program. That includes among others, IL2.122 Moreover, anergic T cells in vivo show reduced calcium flux, and as a result inhibited NFAT1 nuclear localization.123
Through gene knockout studies, Martinez et al. showed that NFAT1 and 2 were required for driving the gene transcription program associated with exhaustion. For example, mice deficient in NFAT1−/- or NFAT2−/- in CD4 T cells when exposed to chronic LCMV infection fail to upregulate PD1, Lag3, and Tim3 genes. Furthermore, NFAT1 was shown to directly bind on PD1, Tim3, Lag3, and IFNγ genes to upregulate their expression.93,124,125 It is clear from these studies that costimulation and NFAT interaction with AP1 could interfere with the exhaustion specific role of NFAT. In the in vivo settings, T cells encountering chronic viral or immunoreactive tumor antigens as also shown recently by the Mognol et al.80 may also receive costimulatory signals. It is unclear how the stoichiometry would shift to NFAT monomer activation over NFAT:AP1 dimerization to coordinate exhaustion program when the costimulatory signaling may be unhindered and the calcium flux sustained.126 One explanation could be the negative regulation of AP1 by transcription factor BATF, which has been reported to increase significantly in exhausted CD8 T cells, e.g.,, in human HIV infections.127,128 Future studies utilizing CD28 knockout mutant mice129 with intact TCR signaling could very well clear the role of CD28 costimulation and NFAT signaling to initiation and maintenance of T cell exhaustion to immunogenic cancers and chronic viral infections. Nevertheless, inducing NFAT:AP1 interaction might have therapeutic benefits for reversing exhaustion. Promoting CD28 costimulatory signaling pathway should be able to promote AP1: NFAT interactions that in turn could reinvigorate effector T cell program.
BATF
BATF belong to AP1 transcription factor family and is essential for antigen-specific CD8 effector T cell differentiation.130 In the acute LCMV infection, expression of BATF increases in effector T cells and its expression also remains high in memory T cells.127 The role of BATF in exhaustion has been reported both in humans and mouse models. The expression of BATF increased significantly in exhausted T cells during chronic HIV and LCMV infection model where it represses IL2 expression and together with IRF4 regulates PD1 expression.42,127 Yet, major role of BATF appears to be coordinating T effector differentiation and for that its target appeared to be other transcription factors rather than the effector cytokines. BATF directly binds on T-bet and Blimp1 genes to upregulate their expression. Moreover, BATF functionally cooperates with IRF4 and Jun in a cell type specific manner to activate gene transcription program.94 BATF forms heterodimer with Jun but unlike Jun/Fos heterodimers, BATF/Jun dimers acts as a negative regulator of AP1 gene transcription and represses gene transcription such as inflammatory cytokine IFNγ.128,130
TCF1, TCF7 and FOXO1
Several studies have established the role of transcription factor T cell factor (TCF1) in vivo in T cell exhaustion fate using gene deletion studies.30,49 Recently, Chen et al. defined TCF1+Ly108+PD1+ CD8 T cells as precursor population of exhausted T cells using mouse model of LMCV infection.35 TCF1 appears to orchestrate early T cell exhaustion fate differentiation by suppressing KLRG1Hi terminal effector T cells. Interestingly, TCF1+ also maintains T cell exhaustion program by promoting Eomes and cMyb expression. cMyb regulates Bcl2 expression promoting survival of early T exhaustion population.34,35 In murine melanoma model and human melanoma patients, the TCF1+ PD1+ population was identified in T cell exhaustion pool that generate proliferative burst after CRB therapy and demonstrate memory like properties.36
The forkhead O transcription factors 1 (FOXO1) and T cell factor 7 (TCF7) play important roles in maintenance of exhaustion and functional T cell memory phenotype. The FOXO1 has been reported to be important for viral antigen-specific CD8 T cell survival, expansion as well as memory T cell identity and differentiation.131 In murine chronic LCMV infections, FOXO1 transcription factor was shown to activate PD1 expression in CD8 T cells.132 The FOXO1 activity was shown to be controlled through AKT and mTOR signaling. Accordingly, suppression of AKT-mTOR circuit in antigen-specific CD8 T cells was demonstrated to be necessary for enhanced FOXO1 nuclear export and its activity, and for sustaining PD1 expression, therefore, linking the metabolic regulatory signaling pathways to FOXO1 and to T cells exhaustion.132 As opposed to continuous activity, FOXO1 loss in CD8 T cells lead to functional anergy in response to latent murine cytomegalovirus (MCMV) infection characterized by decline in the effector cytokine production and reduced proliferation rates.133 Here again, continuous activity of FOXO1 for maintenance of exhaustion and memory phenotype contrasts with its role in anergy pointing to the heterogeneous nature of T cell subsets generated in response to different latent viruses. The FOXO1 regulates expression of TCF7 in virally infected cells but not in naïve T cells .134 In acute infection models, both TCF7 and FOXO1 orchestrate memory differentiation. TCF7 has been shown to downregulate granzyme-B (GZMB) expression and reduce the KRG1hi effector T cell population. And, its expression is highly elevated in naïve T cells and then downregulate upon pathogen activation but gets reexpressed in the memory precursors.134 Likewise, in chronic viral infections, FOXO1 sustains terminal exhaustion phenotype besides maintaining TCF7+ memory like cells. Loss of FOXO1 in these infections led to the loss of TCF7+ memory like cells .135 The TCF7+ CD8 memory precursors in vivo in chronic infections and cancer were recently found to represent two distinct subpopulations of exhausted T cells, the TCF7+ PD1+ and TCF7-PD1 + . The TCF7+ PD1+ were found to be with stem cell like properties, and the cells responsible for generating proliferative burst with anti-PD1 immunotherapy mediating tumor control as well as control of chronic viral infections.30,36 Whether these two subsets arise from T effector, T exhaustion precursors or from mature pool of exhausted T cells is presently unclear (Figure 4).
Yin Yang 1
The stability of T cell exhaustion involves a unique gene transcription program in which expression of some genes upregulate, i.e., immune checkpoint receptors and others downregulate, i.e.,, class I cytokines. Perhaps, the transcription factor(s) poised to be an important regulator of exhaustion must have the capacity to regulate both gene activation and repression programs intrinsically, epigenetically, and cooperatively by interacting with other transcription factors or cofactors. Yin Yang 1 (YY1) along with Blimp1 embodies such dual functionalities. YY1 is a ubiquitous multifunctional zinc finger transcription factor that is known to play crucial roles in B cell differentiation, development, and cellular proliferation.136–138 Several studies have elucidated the role of YY1 in T cell functions in murine models. For example, CD4-specific knockout of YY1 has revealed its requirement for Th2 cytokine expression and cell-type specific binding to Th2 cytokine locus.139 YY1 is also vital for T effector differentiation to acute LCMV infections in mice. Yu et al., using ATAC-seq and Page Rank analysis revealed chromatin accessibility of YY1 binding motifs to be highly enriched in antigen-specific terminal-effector (TE) cells in mice.140 However, YY1 has not been recognized to play a significant role in antigen-specific immune responses or in the mechanism of T cell exhaustion and is little expressed in resting normal human T cells. YY1 was recently shown to serve as the master regulator of T cell functional exhaustion in vitro by participating in both activation and repression of a broad array of exhaustion’s presently defined elements (Table 2). YY1 was shown to be central for promoting the exhaustion response upon “chronic stimulation” of human T cells through repeat exposure to CD3/CD28 beads or to tumor antigens in vitro. In the repeat exposure system, YY1 was shown uniquely to participate in virtually all defined components of the exhaustion process mediating downregulation of class I and II cytokines, i.e.,, IL2 and IFNγ, respectively, and upregulation of immune checkpoint receptors (PD1, Tim3, Lag3), with a concomitant decline in functional killing of human antigenic targets. YY1 own transcription and the ensuing exhaustion process was shown to depend on two signals (Signal 1 + 2) that activates p38MAPK/JNK kinase pathway and transcription factor complex composed of phospho-ATF2/cJun that binds to YY1 promoter activating YY1 transcription, YY1 finally orchestrate functional exhaustion of human T cells.37 YY1 may meet the standard of “exhaustion-specific” when expressed at high levels in vivo and “representing homogenous cell-type.” The recently published literature on homogenous population of dysfunctional T cells in murine syngeneic tumor model seem to corroborate with the exhaustion specific nature of YY1. YY1 is shown to physically interact with GATA3 in T cells and is further shown to be required for recruiting GATA3 on regulatory elements of several Th2 genes.139 Singer et al. identified GATA3 through single-cell RNA-seq analysis of intratumoral CD8+ TILs to be involved in T cell dysfunction. GATA3 was found to be expressed in highly dysfunction CD8+ TILs in tumor bearing mice.91 Further in vivo studies would be needed to identify a definitive connection and collaboration between YY1 and GATA3 in the initiation, maintenance, and lineage differentiation to a pool of highly dysfunctional exhausted T cells.
Tox
The thymocyte selection-associated high mobility group-box protein (Tox) has been revealed to be vital for maintenance of CD8 T cell exhaustion phenotype in chronic LCMV and human HCV viral infections and cancer.141 Tox belongs to high mobility group (HMG)-box proteins that can bind to DNA in a manner which is dependent on the structure but independent of specific DNA sequence motif.142 Tox appears to be important for CD4 T cell lineage development program in thymus. CD4 T cell lineage development is disrupted in Tox−/- deficient mice without disrupting lineage growth of CD8 T cells .143 Moreover, Tox−/- CD8 T cells in vitro appear to retain proliferative capacity when exposed to two signal activation as well as cytolytic activity.143 Tox is revealed to be highly expressed in dysfunctional tumor specific T cells and exhausted T cells in chronic viral infections. Interestingly, utilizing two commonly employed LCMV strains, the Armstrong and Clone 13, Tox was shown to be induced during chronic infections but not in acute infections marking Tox to be the first exhaustion specific molecule with in vivo relevance to exhaustion. The in vivo experiments demonstrated that CD8 T cell specific deletion of Tox downregulates PD1 expression and upregulated cytokines, however, Tox deleted tumor specific dysfunctional “exhausted” CD8 T cells did not regain effector activities.144 The attenuation of PD1 with Tox deficiency or inversely PD1 enhanced expression with Tox overexpression does explain a role for Tox in maintenance of CD8 T cell exhaustion to chronic infections. However, Tox deficient CD8 T cells failure to regain antitumor effector activity also indicate a role which appears not analogous to exhaustion in cancers. Apart from CD8 T cells, CD4 T cells also become exhausted in cancer, chronic viral infections as well as chronic bacterial and parasitic infections. Correspondingly, the instability of Tox deficient CD4 T cells suggests that utility of targeting Tox for reversing T cell functional exhaustion may be limited.
The transcriptional regulators involved in migration of exhausted CD8 T cells
Apart from the above-mentioned transcription factors, there are several other transcription regulators in particular HIFs, Id2, Id3, that play an important role in T cell exhaustion. The transcriptional regulators Id2 and Id3 belong to inhibitor of DNA binding family and play important roles in differentiation of various lymphocyte subsets such as NK, B and, T cell subsets.145 Id2 and Id3 during chronic HIV and EBV infections affect migration and differentiation of a subset of antigen-specific exhausted T cells, the CXCR5+ follicular cytotoxic T cells (TFC).48 Id2 and Id3 were shown to suppress the expression of CXCR5 chemokine receptor gene through blocking E protein family transcription factor E2A. Accordingly, CXCR5+ TFC cells deficient in Id2 and Id3 genes upon exposure to chronic HIV and EBV infections in humans and mice retained the capacity to migrate to B cell follicles and eliminate the Follicular helper T cells (FTH) HIV and EBV B cell reservoir.48 Therefore, Id2 and Id3 by controlling CXCR5 expression regulate the lineage differentiation of TFC subset of exhausted T cells. Likewise, the VHL-HIF axis plays a crucial role in the control of chronic infections and B16 melanoma tumors in vivo. Under hypoxic conditions of chronic viral infections and malignancy, hypoxia inducible factors (HIFs) and its negative regulator the von Hippel–Lindau tumor suppressor VHL control CTL responses. Under normal oxygen tension (normoxic conditions), the HIFs are constantly degraded in a process that is dependent on VHL tumor suppressor protein complex.146 The CD8 + T cells deficient in VHL gene succumb to chronic LCMV infections due to increased immunopathology triggered by higher expression of effector molecules and cytokines such as granzymes, perforins, TNFα, and IFNγ. The expression of these genes in turn depend on enhanced HIF1 activity. As expected, CD8 T cells deficient in VHL gene with higher expression of effector molecules show remarkable control of implanted B16 tumors.147 Overall, these data demonstrate that enhanced HIF1 activity can maintain effector activity and resist exhaustion despite sustained immune checkpoint receptors expression.
Epigenetic landscape of exhausted CD8 T cells
The epigenetic landscape of exhausted CD8 T cells is distinct from T effector and T cell memory subtypes.40,148,149 Even within the exhausted T cell pool, the epigenetic landscape varies between partially versus terminally versus therapeutically reactivated exhausted T cell subtypes.150–152 The reason why a distinct epigenetic landscape exists is that a distinctive gene expression patterns exist in T cell exhaustion, T effector and T cell memory subtypes indicating that T cell exhaustion represents a distinct cell type. Even though the lineage growth of T cell exhaustion appears to be derived from reprogramming of T effector population in murine chronic infection model, 35 the transcription factors that mediate distinct epigenetic landscape in T cell exhaustion subtypes has begun to be revealed only recently. The epigenetic landscape of T cell exhaustion subsets in vivo has been defined utilizing chronic LCMV infection model and method of choice has been the chromatic accessibility assays. The high throughput assay known as assay for transposase-accessible chromatin (ATAC) is used to identify regulatory regions in the genome.149 By motif enrichment analysis of the regulatory elements, the transcription factors involved in the process can be revealed. Using these techniques, several transcription factors have been identified to be involved in epigenetic reprogramming of exhausted T cell subtypes. Several thousand chromatin-accessible regions were identified to be unique in T exhausted compared to T effector and T cell memory subtypes. These open chromatin-accessible regions were present in intronic or intergenic regions containing enhancer elements of target genes with binding activity for transcription factors NFAT2, Nr4a, Nur77, Eomes, Egr2, Sox3, T-bet, retinoic acid receptor (RAR).40,148,149 Interestingly, upon PD1 treatment, the epigenetic landscape is partially reprogrammed to support effector activity. The T effector population open chromatin-accessible regions contain target genes with binding activity of transcription factors NF-κB, IRF1, IRF2, Nur77, Blimp1.40 Recently, the high-mobility group protein TOX was revealed to be vital for epigenetic reprogramming of T cell exhaustion to chronic viral infections. Using the ATAC assay, Khan et al., revealed that TOX deletion significantly altered chromatin-accessible regions of genes associated with exhaustion.153
The chimeric antigen receptor (CAR) T cell exhaustion
CAR-T cells are chimeric antigen receptor-modified T cells that recognize tumor antigens through chimeric receptor antibody. CAR-T cells have demonstrated remarkable success in eradicating refractory B cell malignancies, 154–156 but have so far shown limited success against solid tumors. The major reasons for this limited impact among others derives from CAR-T cell exhaustion.157 Similar to unmodified effector T cells, chimeric antigen receptor (CAR)-modified T cells also undergo functional exhaustion in presence of persistent tumor antigens.58,158-161 The detailed molecular mechanisms of CAR-T cell exhaustion are yet unknown. However, the 2nd generation CAR modified T cells incorporating costimulatory molecules have provided some vital clues. The second generation (CAR)-T cells incorporating CD28 costimulatory molecules and receiving persistent antigenic stimulation from tumors grown in the in vitro cultures have been shown to differentiate into dysfunctional state with features consistent with perturbation of dual axes of exhaustion, i.e.,, failure to sustain cytokine production and upregulation of CR axis especially PD1.37,160,162-164 A paper recently published by Zolov et al. demonstrated through a direct comparison between CD123-CD28-ζ-CAR, CD123-4IBB-ζ-CAR or CD123-ζ-CAR that CD123 CAR with CD28 costimulatory domain was prone to PD1/PD-L1 mediated exhaustion and failure of antitumor activity. Interestingly, both CD28-CAR and 4IBB-CAR enhanced exhaustion specific markers PD1, Lag3, Tim3 (CR axis), however, it was only CD28-CAR that when exposed to antigenic targets showed IL2, IFNγ (Cytokine axis) failure as well as demonstrated reduced proliferative capacity in consistent with the terminal exhaustion phenotype.164 These observations are clinically significant as majority of the active clinical trials with CARs listed in NIH clinical research trials database have been utilizing second generation CARs incorporating CD28 costimulatory molecule. https://clinicaltrials.gov/ct2/results?cond=&term=CAR&cntry=&state=&city=&dist= . Furthermore, the second generation CARs incorporating CD28 costimulatory domain targeting neuroblastoma, ovarian cancer, and metastatic renal carcinoma have reported partial clinical responses in the clinical trials.157–159,165,166 It remains unclear if persistent signal 2 contributed to the reported partial clinical responses with second generation CARs versus other factors such as restricted tumor access and immunosuppressive tumor microenvironment. It is likely that the mechanisms driving CAR-T cell exhaustion are different from the mechanisms contributing to exhaustion of antigen-specific unmodified T cells.
T cell exhaustion specific signaling pathways in cancer and chronic viral infections
Not much is known about the signaling pathways driving T cell exhaustion in cancer or chronic infections. For example, there is limited understanding if signaling pathways driving exhaustion to chronic infections and cancers are alike or different. Or, if any synergy exists in the signaling pathways between murine models and humans. Therefore, knowledge is limited to construct a comprehensive signaling pathway map specific to exhausted T cells. Nevertheless, based on known activities of transcription factors in T cell exhaustion and the knowledge of their signaling pathways in T cell functions can help constructing exhaustion specific signaling pathway maps. The two important evidences exist that point to activities of two distinct pathways in exhaustion: 1) TCR mediated NFAT signaling pathway, which has emerged as a common link to exhaustion in cancer and chronic viral infections in vivo, 2) The in vitro two-signal model of human T cell exhaustion which is centered around YY1 and is relevant to CAR-T cell exhaustion.
TCR →NFAT1 signaling pathway
Significant amount of data provide strong evidences linking TCR signaling with NFAT1, IRF4, and BATF activation to T cell exhaustion. All three are induced in response to TCR signaling to murine chronic infections and are essential components of maintaining exhaustion specific transcriptional profile. The exact membrane proximal TCR events associated during persistent antigenic activation in vivo is unknown (Figure 5a). Data exist, however, that suggested the occurrence of TCR responsive calcium flux and calcineurin activation in T cells experiencing chronic infections, thus, indicating an involvement of NFAT activation.93 NFAT1 has emerged as a crucial player of TCR responsive exhaustion specific gene transcription program both in cancer and chronic infections. Therefore, a signaling model can be designed based on constitutive NFAT1 nuclear activity driven by constitutive TCR proximal signaling complex, calcium influx and calcineurin phosphatase activity (Figure 5a). The nuclear NFAT1 directly binds on PD1 promoter and activates its transcription. NFAT1 also contributes to PD1 transcription indirectly through IRF4. IRF4 bind on PD1 promoter enhancing its transcription. Another component of TCR exhaustion pathway is the BATF and Jun that form heterodimer and repress effector cytokine expression. In addition, BATF also potentially bind on PD1 promoter to promote its transcription. Taken together, NFAT1, and BATF in response to persistent TCR activation and calcium mobilization accumulate in the nucleus promoting a cumulative effect on PD1 transcription by binding on its promoter (Figure 5a). However, the scheme of signaling pathway presented in Figure 5a is not without a caveat. As data exist that shows in response to chronic LCMV and HIV infections, NFAT1 paralog NFAT2 translocation to the nucleus was inhibited in the setting of persistent TCR activation and calcium flux producing profound defect on IFNγ production.126 Additional studies will help clear the apparent dichotomy that exist between NFAT1 and NFAT2 in exhaustion.
Figure 5.
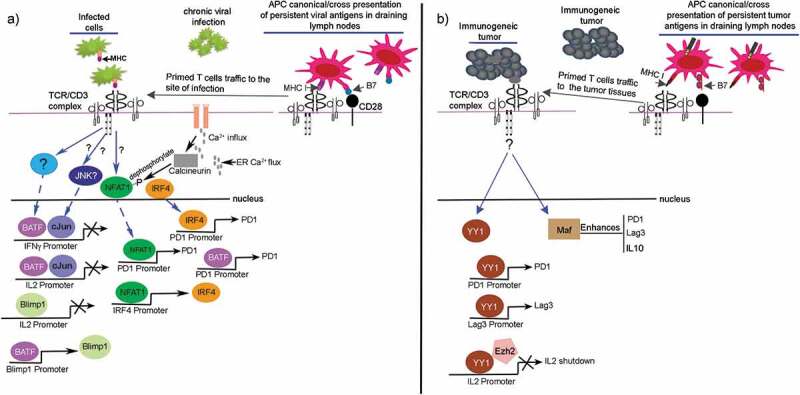
T cell exhaustion specific signaling pathways. (a) TCR →NFAT1→exhaustion, which is relevant to chronic viral infections in vivo. The antigen-specific T cells receive activation from APCs in draining lymph nodes followed by their reactivity against infected cells. (b) TCR→YY1 →exhaustion of human T cells, which is relevant to immunogenic cancers. The tumor antigen-specific T cells in vivo receive activation from APCs in draining lymph nodes followed by their reactivity with immunogenic tumors, e.g.,, melanoma. The YY1 along with c-Maf are highly expressed in T cells reacting with immunogenic melanoma, however, the upstream signaling pathways regulating their transcription during development of T cell exhaustion are unknown. The YY1 is shown to upregulate CR axis and suppresses IL2 production in cooperation with Ezh2.37 Maf enhances PD1, Lag3, and IL10.
TCR→ YY1 → exhaustion
The signaling pathways active in exhausted T cells in human cancers is unknown. YY1 was revealed to be highly elevated in melanoma-infiltrating tumors.37 The upstream signaling pathway regulating YY1 expression in vivo in immunogenic tumors is not known. However, the in vitro human T cell exhaustion model that uses CD3/CD28 beads to repeatedly stimulate T cells have revealed activation of MAPK/JNK signaling pathway leading to heterodimerization of phospho-cJun and phospho-ATF2 that bind to the YY1 promoter to initiate its transcription.37,167 YY1 binds directly to PD1 and Lag3 genes in vitro to upregulate their transcription. Moreover, YY1 binds directly on IL2 promoter and recruits Ezh2 that catalyzes H3K27me3 marks repressing IL2 transcription (Figure 5b). Extensive RNA-seq data exist that shows enrichment of the elements of cJun/ATF signaling pathway in T cell exhaustion in vivo cancer models and chronic viral infections providing relevance to MAPK/JNK signaling pathway in exhaustion.93,141,153 Apart from YY1, the c-MAF transcription factor was also revealed to be highly elevated in melanoma antigen-specific T cells in mouse model and melanoma draining lymph node in humans.31,43 Maf enhances expression of CR axis as well as immune-suppressive cytokine IL10.43 However, the signaling pathway regulating Maf expression in T cell exhaustion is also unknown.
Conclusion
As evident in this review, knowledge is still evolving about the transcriptional regulation and developmental fates of T cell exhaustion both in human cancers and chronic viral infections. Exhaustion is revealed to be a time sensitive process that occurs via perturbation of two axes of exhaustion, i.e.,, failure of cytokine axis and the upregulation of checkpoint receptor axis that include among others the PD1. The PD1 recruits Src homology 2 (SH2) domain-containing tyrosine phosphatase SHP2 to dephosphorylates CD3ζ chain168 as well as phospho-CD28169 leading to the inhibition of both TCR and CD28 costimulatory signaling. Finally, once the T cells are fully exhausted, the pressure to maintain exhaustion associated genetic and epigenetic program remains high as the process itself is reversible proportional to the pool of chronically infected cells or tumor volume. With the shifting dynamics of antigen pool, the exhaustion specific genes expression may be altered to reflect the change.
In chronic viral infections, the development of T cell exhaustion appears to be a linear differentiation process. If the rise of T cell exhaustion in immunogenic cancers also follows a linear differentiation model will require further investigation. The lineage tracing techniques may help define molecularly the stages of exhaustion. Taken together, T cell exhaustion is dynamic, transcriptionally, and metabolically active and energy-consuming state in which transcription factors may often alter their expression kinetics to reflect the state of differentiation of T cell exhaustion pool. The T cell exhaustion pool responding to the therapeutic interventions could provide vital clues regarding the pattern of genomic and epigenetic changes associated with the suppression of T effector functions. Moreover, immunotherapy induced reactivation of T cell exhaustion pool can also help to identify the origin of reinvigorated population whether these being pre-exhausted CD8 population or recent emigrants to antigen rich environment.
BOX
TNaïve: Naïve T cells are recent thymic emigrants, self-antigen tolerant and having undergone TCR rearrangements. Typically, after their development from thymus and subsequent positive and negative selection, T cells circulate through blood and secondary lymphoid organs as naïve T cells and have not yet responded to antigens.
TEffector: CD8 T Effector cells arise during acute viral infections. For T effector differentiation, naïve T cells must react with professional antigen presenting cells that present viral antigens to CD8 TCR through MHC Class I and coengages costimulatory, adhesion, and cytokine receptor molecules. T effector are antigen specific and have huge potential to proliferate and can synthesize and secrete effector molecules such as granules containing granzymes and perforins, cytokines and chemokines. Effector T cells effectively perform antigen target killings. After the pathogenic target is elimination, effector T cells contract leaving behind a small percentage of long-lived memory precursors.170,171
TMemory: T effector cells responding to pathogenic targets differentiate into terminal T effector population and generate memory precursor cells.172,173 The generation of T cell memory precursors are regulated through epigenetic mechanisms involving changes in DNA methylation patterns orchestrated by methyltransferase Dnmt3a173 and histone methylation notably histone H3 lysine 9 catalyzed by histone methyltransferase Suv39h1.174 Memory precursor differentiate into central memory (TCM) and effector memory T cells (TEM). TCM reside and travel between lymphoid organs whereas TEM reside and travel to tertiary tissues, e.g.,, mucosal surfaces, lungs, liver, skin, intestine, etc.175 Both TCM and TEM retain effector functions including lytic activity upon antigenic re-challenge.17–19,176
TExhaustion: T cell exhaustion is quasi-terminally differentiated state, presented with altered metabolic, and epigenetic state that develops in response to chronic infections and immune reactive cancers.8,40,148,149,177 Exhaustion is initiated with persistent delivery of TCR signaling and maintained by the checkpoint PD1/PDL1 signaling, as well as by immune suppressive cellular, molecular and metabolic intermediators, DNA and histone methylation. The activity of transcription factors such as YY1, IRF4, NFAT, Maf, Blimp1, Tox have been shown to be vital for the exhaustion phenotype. The inhibitory PD1/PDL1 signaling in exhausted T cells may be a response to control the proliferation of persistently activated antigen-specific T cells helping to avoid autoimmune responses and promoting peripheral tolerance.178,179 Therefore, exhausted T cells even though consistently expressing activation markers have lost the capacity to proliferate and have poor effector functions. One of the distinctive characteristics of exhausted T cells is that they can regain effector functions especially their killing functions against antigenic targets with both immune checkpoint receptor blockade inhibitors as well as with non-checkpoint therapeutic interventions such as cytokines.
Acknowledgments
I would like to acknowledge Dr. Richard P. Junghans at Immune Therapy Bio for his valuable inputs in the interpretation of novel concepts elaborated in this review.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Availability of data and material
I agree to share all the data and content of this review.
Author contributions
M.Y.B. conceived, planned, and wrote the review article.
Consent for publication
I consent to publication.
References
- 1.Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob -J-J, Cowey CL, Lao CD, Wagstaff J, Schadendorf D, Ferrucci PF.. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2017. October 5;377(14):1345–18. doi: 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pai‐Scherf L, Blumenthal GM, Li H, Subramaniam S, Mishra‐Kalyani PS, He K, Zhao H, Yu J, Paciga M, Goldberg KB. FDA approval summary: pembrolizumab for treatment of metastatic non-small cell lung cancer: first-line therapy and beyond. The Oncologist. 2017. November;22(11):1392–1399. doi: 10.1634/theoncologist.2017-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sul J, Blumenthal GM, Jiang X, He K, Keegan P, Pazdur R. FDA approval summary: pembrolizumab for the treatment of patients with metastatic non-small cell lung cancer whose tumors express programmed death-ligand 1. The Oncologist. 2016. May;21(5):643–650. doi: 10.1634/theoncologist.2015-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato S, Goodman A, Walavalkar V, Barkauskas DA, Sharabi A, Kurzrock R. Hyperprogressors after immunotherapy: analysis of genomic alterations associated with accelerated growth rate. Clin Cancer Res. 2017. August 1;23(15):4242–4250. doi: 10.1158/1078-0432.CCR-16-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E. Nivolumab versus docetaxel in advanced nonsquamous non–small-cell lung cancer. N Engl J Med. 2015. October 22;373(17):1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C, Kalinka-Warzocha E. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015. January 22;372(4):320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 7.Yan B, Noone A-M, Yee C, Banerjee M, Schwartz K, Simon MS. Racial differences in colorectal cancer survival in the detroit metropolitan area. Cancer. 2009. August 15;115(16):3791–3800. doi: 10.1002/cncr.24408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashimoto M, Kamphorst AO, Im SJ, Kissick HT, Pillai RN, Ramalingam SS, Araki K, Ahmed R. CD8 T cell exhaustion in chronic infection and cancer: opportunities for interventions. Annu Rev Med. 2018. January 29;69(1):301–318. doi: 10.1146/annurev-med-012017-043208. [DOI] [PubMed] [Google Scholar]
- 9.Pauken KE, Wherry EJ. Overcoming T cell exhaustion in infection and cancer. Trends Immunol. 2015. April;36(4):265–276. doi: 10.1016/j.it.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Philip M, Schietinger A. Heterogeneity and fate choice: T cell exhaustion in cancer and chronic infections. Curr Opin Immunol. 2019. June;58:98–103. doi: 10.1016/j.coi.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Picarda E, Ren X, Zang X. Tumor cholesterol up, T cells down. Cell Metab. 2019. July 2;30(1):12–13. doi: 10.1016/j.cmet.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Buchholz VR, Flossdorf M, Hensel I, Kretschmer L, Weissbrich B, Graf P, Verschoor A, Schiemann M, Hofer T, Busch DH. Disparate individual fates compose robust CD8+ T cell immunity. Science. 2013. May 3;340(6132):630–635. doi: 10.1126/science.1235454. [DOI] [PubMed] [Google Scholar]
- 13.Gerlach C, Rohr JC, Perie L, van Rooij N, van Heijst JWJ, Velds A, Urbanus J, Naik SH, Jacobs H, Beltman JB. Heterogeneous differentiation patterns of individual CD8+ T cells. Science. 2013. May 3;340(6132):635–639. doi: 10.1126/science.1235487. [DOI] [PubMed] [Google Scholar]
- 14.Kakaradov B, Arsenio J, Widjaja CE, He Z, Aigner S, Metz PJ, Yu B, Wehrens EJ, Lopez J, Kim SH. Early transcriptional and epigenetic regulation of CD8+ T cell differentiation revealed by single-cell RNA sequencing. Nat Immunol. 2017. April;18(4):422–432. doi: 10.1038/ni.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joffre OP, Segura E, Savina A, Amigorena S. Cross-presentation by dendritic cells. Nat Rev Immunol. 2012. July 13;12(8):557–569. doi: 10.1038/nri3254. [DOI] [PubMed] [Google Scholar]
- 16.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012. November;12(11):749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wherry EJ, Ahmed R. Memory CD8 T-cell differentiation during viral infection. J Virol. 2004. June;78(11):5535–5545. doi: 10.1128/JVI.78.11.5535-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farber DL, Yudanin NA, Restifo NP. Human memory T cells: generation, compartmentalization and homeostasis. Nat Rev Immunol. 2014. January;14(1):24–35. doi: 10.1038/nri3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Remakus S, Sigal LJ. Memory CD8(+) T cell protection. Adv Exp Med Biol. 2013;785:77–86. [DOI] [PubMed] [Google Scholar]
- 20.Romero P, Zippelius A, Kurth I, Pittet MJ, Touvrey C, Iancu EM, Corthesy P, Devevre E, Speiser DE, Rufer N. Four functionally distinct populations of human effector-memory CD8+ T lymphocytes. J Immunol. 2007. April 1;178(7):4112–4119. doi: 10.4049/jimmunol.178.7.4112. [DOI] [PubMed] [Google Scholar]
- 21.Shin H, Blackburn SD, Blattman JN, Wherry EJ. Viral antigen and extensive division maintain virus-specific CD8 T cells during chronic infection. J Exp Med. 2007. April 16;204(4):941–949. doi: 10.1084/jem.20061937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wherry EJ, Barber DL, Kaech SM, Blattman JN, Ahmed R. Antigen-independent memory CD8 T cells do not develop during chronic viral infection. Proc Natl Acad Sci U S A. 2004. November 9;101(45):16004–16009. doi: 10.1073/pnas.0407192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giguere J-F, Bounou S, Paquette J-S, Madrenas J, Tremblay MJ. Insertion of host-derived costimulatory molecules CD80 (B7.1) and CD86 (B7.2) into human immunodeficiency virus type 1 affects the virus life cycle. J Virol. 2004. June;78(12):6222–6232. doi: 10.1128/JVI.78.12.6222-6232.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kedzierska K, Crowe SM. The role of monocytes and macrophages in the pathogenesis of HIV-1 infection. Curr Med Chem. 2002. November;9(21):1893–1903. doi: 10.2174/0929867023368935. [DOI] [PubMed] [Google Scholar]
- 25.Huang AY, Golumbek P, Ahmadzadeh M, Jaffee E, Pardoll D, Levitsky H. Role of bone marrow-derived cells in presenting MHC class I-restricted tumor antigens. Science. 1994. May 13;264(5161):961–965. doi: 10.1126/science.7513904. [DOI] [PubMed] [Google Scholar]
- 26.Gardner A, Ruffell B. Dendritic cells and cancer immunity. Trends Immunol. 2016. December;37(12):855–865. doi: 10.1016/j.it.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bottcher JP, Reis ESC. The role of type 1 conventional dendritic cells in cancer immunity. Trends in Cancer. 2018. November;4(11):784–792. doi: 10.1016/j.trecan.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saxena M, Bhardwaj N. Re-emergence of dendritic cell vaccines for cancer treatment. Trends in Cancer. 2018. February;4(2):119–137. doi: 10.1016/j.trecan.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jadhav RR, Im SJ, Hu B, Hashimoto M, Li P, Lin J-X, Leonard WJ, Greenleaf WJ, Ahmed R, Goronzy JJ. Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc Natl Acad Sci U S A. 2019. July 9;116(28):14113–14118. doi: 10.1073/pnas.1903520116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016. September 15;537(7620):417–421. doi: 10.1038/nature19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verdeil G. MAF drives CD8 (+)T-cell exhaustion. Oncoimmunology. 2016. February;5(2):e1082707. doi: 10.1080/2162402X.2015.1082707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJD, Suresh M, Altman JD, Ahmed R. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med. 1998. December 21;188(12):2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalia V, Sarkar S, Ahmed R. CD8 T-cell memory differentiation during acute and chronic viral infections. Adv Exp Med Biol. 2010;684:79–95. [DOI] [PubMed] [Google Scholar]
- 34.Chen Z, Stelekati E, Kurachi M, Yu S, Cai Z, Manne S, Khan O, Yang X, Wherry EJ. miR-150 regulates memory CD8 T cell differentiation via c-Myb. Cell Rep. 2017. September 12;20(11):2584–2597. doi: 10.1016/j.celrep.2017.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Z, Ji Z, Ngiow SF. TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity. 2019;October 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, Carmona SJ, Scarpellino L, Gfeller D, Pradervand S. Intratumoral Tcf1+PD-1+CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity. 2019. January 15;50(1):195–211 e10. doi: 10.1016/j.immuni.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 37.Balkhi MY, Wittmann G, Xiong F, Junghans RP. YY1 upregulates checkpoint receptors and downregulates type I cytokines in exhausted, chronically stimulated human T cells. iScience. 2018. April 27;2:105–122. doi: 10.1016/j.isci.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.West EE, Jin H-T, Rasheed A-U, Penaloza-MacMaster P, Ha S-J, Tan WG, Youngblood B, Freeman GJ, Smith KA, Ahmed R. PD-L1 blockade synergizes with IL-2 therapy in reinvigorating exhausted T cells. J Clin Invest. 2013. June;123(6):2604–2615. doi: 10.1172/JCI67008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamphorst AO, Wieland A, Nasti T, Yang S, Zhang R, Barber DL, Konieczny BT, Daugherty CZ, Koenig L, Yu K. Rescue of exhausted CD8 T cells by PD-1–targeted therapies is CD28-dependent. Science. 2017. March 31;355(6332):1423–1427. doi: 10.1126/science.aaf0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pauken KE, Sammons MA, Odorizzi PM, Manne S, Godec J, Khan O, Drake AM, Chen Z, Sen DR, Kurachi M. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 2016. December 2;354(6316):1160–1165. doi: 10.1126/science.aaf2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blackburn SD, Shin H, Freeman GJ, Wherry EJ. Selective expansion of a subset of exhausted CD8 T cells by alphaPD-L1 blockade. Proc Natl Acad Sci U S A. 2008. September 30;105(39):15016–15021. doi: 10.1073/pnas.0801497105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Man K, Gabriel SS, Liao Y, Gloury R, Preston S, Henstridge DC, Pellegrini M, Zehn D, Berberich-Siebelt F, Febbraio MA. Transcription factor IRF4 promotes CD8+ T cell exhaustion and limits the development of memory-like T cells during chronic infection. Immunity. 2017. December 19;47(6):1129–1141 e5. doi: 10.1016/j.immuni.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 43.Giordano M, Henin C, Maurizio J, Imbratta C, Bourdely P, Buferne M, Baitsch L, Vanhille L, Sieweke MH, Speiser DE. Molecular profiling of CD8 T cells in autochthonous melanoma identifies Maf as driver of exhaustion. Embo J. 2015. August 4;34(15):2042–2058. doi: 10.15252/embj.201490786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thangavelu G, Gill RG, Boon L, Ellestad KK, Anderson CC. Control of in vivo collateral damage generated by T cell immunity. J Immunol. 2013. August 15;191(4):1686–1691. doi: 10.4049/jimmunol.1203240. [DOI] [PubMed] [Google Scholar]
- 45.Bakacs T, Moss RW, Kleef R. Exploiting autoimmunity unleashed by low-dose immune checkpoint blockade to treat advanced cancer. Scand J Immunol. 2019. October 7:e12821. [DOI] [PubMed] [Google Scholar]
- 46.Hellerstein MK, Hoh RA, Hanley MB, Cesar D, Lee D, Neese RA, McCune JM. Subpopulations of long-lived and short-lived T cells in advanced HIV-1 infection. J Clin Invest. 2003. September;112(6):956–966. doi: 10.1172/JCI200317533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, Bikoff EK, Robertson EJ, Lauer GM, Reiner SL. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science. 2012. November 30;338(6111):1220–1225. doi: 10.1126/science.1229620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leong YA, Chen Y, Ong HS, Wu D, Man K, Deleage C, Minnich M, Meckiff BJ, Wei Y, Hou Z. CXCR5+ follicular cytotoxic T cells control viral infection in B cell follicles. Nat Immunol. 2016. October;17(10):1187–1196. doi: 10.1038/ni.3543. [DOI] [PubMed] [Google Scholar]
- 49.Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, Danilo M, Alfei F, Hofmann M, Wieland D. T cell factor 1-expressing memory-like CD8+ T cells sustain the immune response to chronic viral infections. Immunity. 2016. August 16;45(2):415–427. doi: 10.1016/j.immuni.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 50.Shan Q, Zeng Z, Xing S, Li F, Hartwig SM, Gullicksrud JA, Kurup SP, Van Braeckel-Budimir N, Su Y, Martin MD. The transcription factor Runx3 guards cytotoxic CD8+ effector T cells against deviation towards follicular helper T cell lineage. Nat Immunol. 2017. August;18(8):931–939. doi: 10.1038/ni.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang D, Diao H, Getzler AJ, Rogal W, Frederick MA, Milner J, Yu B, Crotty S, Goldrath AW, Pipkin ME. The transcription factor Runx3 establishes chromatin accessibility of cis-regulatory landscapes that drive memory cytotoxic T lymphocyte formation. Immunity. 2018. April 17;48(4):659–674 e6. doi: 10.1016/j.immuni.2018.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, Groner Y, Rao A. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med. 2009. January 16;206(1):51–59. doi: 10.1084/jem.20081242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thommen DS, Koelzer VH, Herzig P, Roller A, Trefny M, Dimeloe S, Kiialainen A, Hanhart J, Schill C, Hess C. A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med. 2018. July;24(7):994–1004. doi: 10.1038/s41591-018-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Allie SR, Zhang W, Fuse S, Usherwood EJ. Programmed death 1 regulates development of central memory CD8 T cells after acute viral infection. J Immunol. 2011. June 1;186(11):6280–6286. doi: 10.4049/jimmunol.1003870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shin H, Wherry EJ. CD8 T cell dysfunction during chronic viral infection. Curr Opin Immunol. 2007. August;19(4):408–415. doi: 10.1016/j.coi.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 56.Nguyen LT, Ohashi PS. Clinical blockade of PD1 and LAG3–potential mechanisms of action. Nat Rev Immunol. 2015. January;15(1):45–56. doi: 10.1038/nri3790. [DOI] [PubMed] [Google Scholar]
- 57.Rosenberg SA, Yang JC, Sherry RM, Kammula US, Hughes MS, Phan GQ, Citrin DE, Restifo NP, Robbins PF, Wunderlich JR. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011. July 1;17(13):4550–4557. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emtage PC, Lo AS, Gomes EM, Liu DL, Gonzalo-Daganzo RM, Junghans RP. Second-generation anti-carcinoembryonic antigen designer T cells resist activation-induced cell death, proliferate on tumor contact, secrete cytokines, and exhibit superior antitumor activity in vivo: a preclinical evaluation. Clin Cancer Res. 2008. December 15;14(24):8112–8122. doi: 10.1158/1078-0432.CCR-07-4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosenberg SA, Mule JJ, Spiess PJ, Reichert CM, Schwarz SL. Regression of established pulmonary metastases and subcutaneous tumor mediated by the systemic administration of high-dose recombinant interleukin 2. J Exp Med. 1985. May 1;161(5):1169–1188. doi: 10.1084/jem.161.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol. 2003. April;77(8):4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lo AS, Ma Q, Liu DL, Junghans RP. Anti-GD3 chimeric sFv-CD28/T-cell receptor zeta designer T cells for treatment of metastatic melanoma and other neuroectodermal tumors. Clin Cancer Res. 2010. May 15;16(10):2769–2780. doi: 10.1158/1078-0432.CCR-10-0043. [DOI] [PubMed] [Google Scholar]
- 62.Liao W, Lin J-X, Leonard WJ. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. 2013. January 24;38(1):13–25. doi: 10.1016/j.immuni.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu CY, Zhang YH, Wang T, Chen L, Gong ZH, Wan YS, Li QJ, Li YS, Zhu B. Interleukin-2 reverses CD8(+)T cell exhaustion in clinical malignant pleural effusion of lung cancer. Clin Exp Immunol. 2016. October;186(1):106–114. doi: 10.1111/cei.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31(1):259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krauss S, Brand MD, Buttgereit F. Signaling takes a breath – new quantitative perspectives on bioenergetics and signal transduction. Immunity. 2001. October;15(4):497–502. doi: 10.1016/S1074-7613(01)00205-9. [DOI] [PubMed] [Google Scholar]
- 66.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009. May 22;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Warburg O. On the origin of cancer cells. Science. 1956. February 24;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 68.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013. April 18;38(4):633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van der Windt GW, Everts B, Chang C-H, Curtis J, Freitas T, Amiel E, Pearce E, Pearce E. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012. January 27;36(1):68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wherry EJ, Ha S-J, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007. October;27(4):670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 71.Bengsch B, Johnson AL, Kurachi M, Odorizzi PM, Pauken KE, Attanasio J, Stelekati E, McLane LM, Paley MA, Delgoffe GM. Bioenergetic insufficiencies due to metabolic alterations regulated by the inhibitory receptor PD-1 are an early driver of CD8(+) T cell exhaustion. Immunity. 2016. August 16;45(2):358–373. doi: 10.1016/j.immuni.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scharping NE, Menk AV, Moreci RS, Whetstone RD, Dadey RE, Watkins SC, Ferris RL, Delgoffe GM. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity. 2016. August 16;45(2):374–388. doi: 10.1016/j.immuni.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Austin S, St-Pierre J. PGC1alpha and mitochondrial metabolism - emerging concepts and relevance in ageing and neurodegenerative disorders. J Cell Sci. 2012. November 1;125(21):4963–4971. doi: 10.1242/jcs.113662. [DOI] [PubMed] [Google Scholar]
- 74.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky Jr WE, You MJ, DePinho RA, McMahon M, Bosenberg M. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009. May;41(5):544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chang C-H, Qiu J, O’Sullivan D, Buck M, Noguchi T, Curtis J, Chen Q, Gindin M, Gubin M, van der Windt GW. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015. September 10;162(6):1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McKinney EF, Smith KGC. Metabolic exhaustion in infection, cancer and autoimmunity. Nat Immunol. 2018. March;19(3):213–221. doi: 10.1038/s41590-018-0045-y. [DOI] [PubMed] [Google Scholar]
- 77.Ho P-C, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui Y-C, Cui G, Micevic G, Perales J. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell. 2015. September 10;162(6):1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crawford A, Angelosanto JM, Kao C, Doering T, Odorizzi P, Barnett B, Wherry E. Molecular and transcriptional basis of CD4+ T cell dysfunction during chronic infection. Immunity. 2014. February 20;40(2):289–302. doi: 10.1016/j.immuni.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baitsch L, Baumgaertner P, Devevre E, Raghav SK, Legat A, Barba L, Wieckowski S, Bouzourene H, Deplancke B, Romero P. Exhaustion of tumor-specific CD8(+) T cells in metastases from melanoma patients. J Clin Invest. 2011. June;121(6):2350–2360. doi: 10.1172/JCI46102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mognol GP, Spreafico R, Wong V, Scott-Browne JP, Togher S, Hoffmann A, Hogan PG, Rao A, Trifari S. Exhaustion-associated regulatory regions in CD8(+) tumor-infiltrating T cells. Proc Natl Acad Sci U S A. 2017. March 28;114(13):E2776–E2785. doi: 10.1073/pnas.1620498114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hadrup SR. The antigen specific composition of melanoma tumor infiltrating lymphocytes? Oncoimmunology. 2012. September 1;1(6):935–936. doi: 10.4161/onci.20303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cohen CJ, Gartner JJ, Horovitz-Fried M, Shamalov K, Trebska-McGowan K, Bliskovsky VV, Parkhurst MR, Ankri C, Prickett TD, Crystal JS. Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J Clin Invest. 2015. October 01;125(10):3981–3991. doi: 10.1172/JCI82416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robbins PF, Lu Y-C, El-Gamil M, Li YF, Gross C, Gartner J, Lin JC, Teer JK, Cliften P, Tycksen E. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013. June;19(6):747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lu Y-C, Yao X, Crystal JS, Li YF, El-Gamil M, Gross C, Davis L, Dudley ME, Yang JC, Samuels Y. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res. 2014. July 01;20(13):3401–3410. doi: 10.1158/1078-0432.CCR-14-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer. 2014. February;14(2):135–146. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 86.Stevanovic S, Pasetto A, Helman SR, Gartner JJ, Prickett TD, Howie B, Robins HS, Robbins PF, Klebanoff CA, Rosenberg SA. Landscape of immunogenic tumor antigens in successful immunotherapy of virally induced epithelial cancer. Science. 2017. April 14;356(6334):200–205. doi: 10.1126/science.aak9510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perrotti D, Jamieson C, Goldman J, Skorski T. Chronic myeloid leukemia: mechanisms of blastic transformation. J Clin Invest. 2010. July 1;120(7):2254–2264. doi: 10.1172/JCI41246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bjorklund ÅK, Forkel M, Picelli S, Konya V, Theorell J, Friberg D, Sandberg R, Mjösberg J. The heterogeneity of human CD127+ innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol. 2016. April;17(4):451–460. doi: 10.1038/ni.3368. [DOI] [PubMed] [Google Scholar]
- 89.Villani AC, Shekhar K. Single-cell RNA sequencing of human T cells. Methods Mol Biol. 2017;1514:203–239. [DOI] [PubMed] [Google Scholar]
- 90.Proserpio V, Piccolo A, Haim-Vilmovsky L, Kar G, Lönnberg T, Svensson V, Pramanik J, Natarajan KN, Zhai W, Zhang X. Single-cell analysis of CD4+ T-cell differentiation reveals three major cell states and progressive acceleration of proliferation. Genome Biol. 2016. May 12;17(1):103. doi: 10.1186/s13059-016-0957-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Singer M, Wang C, Cong L, Marjanovic ND, Kowalczyk MS, Zhang H, Nyman J, Sakuishi K, Kurtulus S, Gennert D. A distinct gene module for dysfunction uncoupled from activation in tumor-infiltrating T cells. Cell. 2017. November 16;171(5):1221–1223. doi: 10.1016/j.cell.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Concepcion AR, Vaeth M, Wagner LE 2nd, Eckstein M, Hecht L, Yang J, Crottes D, Seidl M, Shin HP, Weidinger C. Store-operated Ca2+ entry regulates Ca2+-activated chloride channels and eccrine sweat gland function. J Clin Invest. 2016. November 1;126(11):4303–4318. doi: 10.1172/JCI89056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Martinez GJ, Pereira RM, Aijo T, Kim EY, Marangoni F, Pipkin ME, Togher S, Heissmeyer V, Zhang YC, Crotty S. The transcription factor NFAT promotes exhaustion of activated CD8 + T cells. Immunity. 2015. February 17;42(2):265–278. doi: 10.1016/j.immuni.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li P, Spolski R, Liao W, Wang L, Murphy TL, Murphy KM, Leonard WJ. BATF–JUN is critical for IRF4-mediated transcription in T cells. Nature. 2012. October 25;490(7421):543–546. doi: 10.1038/nature11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brooks DG, Lee AM, Elsaesser H, McGavern DB, Oldstone MBA. IL-10 blockade facilitates DNA vaccine-induced T cell responses and enhances clearance of persistent virus infection. J Exp Med. 2008. March 17;205(3):533–541. doi: 10.1084/jem.20071948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ejrnaes M, Filippi CM, Martinic MM, Ling EM, Togher LM, Crotty S, von Herrath MG. Resolution of a chronic viral infection after interleukin-10 receptor blockade. J Exp Med. 2006. October 30;203(11):2461–2472. doi: 10.1084/jem.20061462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Penaloza-MacMaster P, Kamphorst AO, Wieland A, Araki K, Iyer SS, West EE, O’Mara L, Yang S, Konieczny BT, Sharpe AH. Interplay between regulatory T cells and PD-1 in modulating T cell exhaustion and viral control during chronic LCMV infection. J Exp Med. 2014. August 25;211(9):1905–1918. doi: 10.1084/jem.20132577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Intlekofer AM, Takemoto N, Kao C, Banerjee A, Schambach F, Northrop JK, Shen H, Wherry EJ, Reiner SL. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J Exp Med. 2007. September 3;204(9):2015–2021. doi: 10.1084/jem.20070841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pearce EL, Mullen AC, Martins GA. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003. November 7;302(5647):1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 100.Sullivan BM, Juedes A, Szabo SJ, von Herrath M, Glimcher LH. Antigen-driven effector CD8 T cell function regulated by T-bet. Proc Natl Acad Sci U S A. 2003. December 23;100(26):15818–15823. doi: 10.1073/pnas.2636938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Glimcher LH, Townsend MJ, Sullivan BM, Lord GM. Recent developments in the transcriptional regulation of cytolytic effector cells. Nat Rev Immunol. 2004. November;4(11):900–911. doi: 10.1038/nri1490. [DOI] [PubMed] [Google Scholar]
- 102.Kao C, Oestreich KJ, Paley MA, Crawford A, Angelosanto JM, Ali MAA, Intlekofer AM, Boss JM, Reiner SL, Weinmann AS. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nat Immunol. 2011. May 29;12(7):663–671. doi: 10.1038/ni.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007. August;27(2):281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Joshi NS, Cui W, Dominguez CX, Chen JH, Hand TW, Kaech SM. Increased numbers of preexisting memory CD8 T cells and decreased T-bet expression can restrain terminal differentiation of secondary effector and memory CD8 T cells. J Immunol. 2011. October 15;187(8):4068–4076. doi: 10.4049/jimmunol.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Buggert M, Tauriainen J, Yamamoto T, Frederiksen J, Ivarsson MA, Michaëlsson J, Lund O, Hejdeman B, Jansson M, Sönnerborg A. T-bet and Eomes are differentially linked to the exhausted phenotype of CD8+ T cells in HIV infection. PLoS Pathog. 2014. July;10(7):e1004251. doi: 10.1371/journal.ppat.1004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8+ T cells and memory responses. Immunity. 2009. August 21;31(2):283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 107.Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, Jacob J, Calame K, Kaech SM. Transcriptional repressor Blimp-1 promotes CD8+ T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009. August 21;31(2):296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xin A, Masson F, Liao Y, Preston S, Guan T, Gloury R, Olshansky M, Lin J-X, Li P, Speed TP. A molecular threshold for effector CD8+ T cell differentiation controlled by transcription factors Blimp-1 and T-bet. Nat Immunol. 2016. April;17(4):422–432. doi: 10.1038/ni.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shin H, Blackburn SD, Intlekofer AM, Kao C, Angelosanto JM, Reiner SL, Wherry EJ. A Role for the Transcriptional Repressor Blimp-1 in CD8+ T Cell Exhaustion during Chronic Viral Infection. Immunity. 2009. August 21;31(2):309–320. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lu P, Youngblood BA, Austin JW, Rasheed Mohammed AU, Butler R, Ahmed R, Boss JM. Blimp-1 represses CD8 T cell expression of PD-1 using a feed-forward transcriptional circuit during acute viral infection. J Exp Med. 2014. March 10;211(3):515–527. doi: 10.1084/jem.20130208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wherry EJ. T cell exhaustion. Nat Immunol. 2011. June;12(6):492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 112.Martins GA, Cimmino L, Liao J, Magnusdottir E, Calame K. Blimp-1 directly represses Il2 and the Il2 activator Fos, attenuating T cell proliferation and survival. J Exp Med. 2008. September 1;205(9):1959–1965. doi: 10.1084/jem.20080526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002. April;109(Suppl):S67–79. doi: 10.1016/S0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- 114.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 115.Klein-Hessling S, Muhammad K, Klein M, Pusch T, Rudolf R, Flöter J, Qureischi M, Beilhack A, Vaeth M, Kummerow C. NFATc1 controls the cytotoxicity of CD8+ T cells. Nat Commun. 2017. September 11;8(1):511. doi: 10.1038/s41467-017-00612-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005. June;5(6):472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 117.Bengsch B, Wherry EJ. The importance of cooperation: partnerless NFAT induces T cell exhaustion. Immunity. 2015. February 17;42(2):203–205. doi: 10.1016/j.immuni.2015.01.023. [DOI] [PubMed] [Google Scholar]
- 118.Shaw JP, Utz PJ, Durand DB, Toole J, Emmel E, Crabtree G. Identification of a putative regulator of early T cell activation genes. Science. 1988. July 8;241(4862):202–205. doi: 10.1126/science.3260404. [DOI] [PubMed] [Google Scholar]
- 119.Jain J, McCafffrey PG, Miner Z, Kerppola TK, Lambert JN, Verdine GL, Curran T, Rao A. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature. 1993. September 23;365(6444):352–355. doi: 10.1038/365352a0. [DOI] [PubMed] [Google Scholar]
- 120.Macian F, Garcia-Rodriguez C, Rao A. Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun. Embo J. 2000. September 1;19(17):4783–4795. doi: 10.1093/emboj/19.17.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Abe BT, Shin DS, Mocholi E, Macian F. NFAT1 supports tumor-induced anergy of CD4+ T cells. Cancer Res. 2012. September 15;72(18):4642–4651. doi: 10.1158/0008-5472.CAN-11-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Muller MR, Rao A. NFAT, immunity and cancer: a transcription factor comes of age. Nat Rev Immunol. 2010. September;10(9):645–656. doi: 10.1038/nri2818. [DOI] [PubMed] [Google Scholar]
- 123.Srinivasan M, Frauwirth KA. Reciprocal NFAT1 and NFAT2 nuclear localization in CD8+ anergic T cells is regulated by suboptimal calcium signaling. J Immunol. 2007. September 15;179(6):3734–3741. doi: 10.4049/jimmunol.179.6.3734. [DOI] [PubMed] [Google Scholar]
- 124.Oestreich KJ, Yoon H, Ahmed R, Boss JM. NFATc1 regulates PD-1 expression upon T cell activation. J Immunol. 2008. October 1;181(7):4832–4839. doi: 10.4049/jimmunol.181.7.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Teixeira LK, Fonseca BP, Vieira-de-Abreu A, Barboza BA, Robbs BK, Bozza PT, Viola JPB. IFN-γ production by CD8+ T cells depends on NFAT1 transcription factor and regulates Th differentiation. J Immunol. 2005. November 1;175(9):5931–5939. doi: 10.4049/jimmunol.175.9.5931. [DOI] [PubMed] [Google Scholar]
- 126.Agnellini P, Wolint P, Rehr M, Cahenzli J, Karrer U, Oxenius A. Impaired NFAT nuclear translocation results in split exhaustion of virus-specific CD8+ T cell functions during chronic viral infection. Proc Natl Acad Sci U S A. 2007. March 13;104(11):4565–4570. doi: 10.1073/pnas.0610335104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, Julg B, Jesneck JL, Brosnahan K, Imam S. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat Med. 2010. October;16(10):1147–1151. doi: 10.1038/nm.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Williams KL, Nanda I, Lyons GE, Kuo C, Schmid M, Leiden J, Kaplan M, Taparowsky E. Characterization of murine BATF: a negative regulator of activator protein-1 activity in the thymus. Eur J Immunol. 2001. May;31(5):1620–1627. doi:. [DOI] [PubMed] [Google Scholar]
- 129.Shahinian A, Pfeffer K, Lee KP, Kundig T, Kishihara K, Wakeham A, Kawai K, Ohashi P, Thompson C, Mak T. Differential T cell costimulatory requirements in CD28-deficient mice. Science. 1993. July 30;261(5121):609–612. doi: 10.1126/science.7688139. [DOI] [PubMed] [Google Scholar]
- 130.Kurachi M, Barnitz RA, Yosef N, Odorizzi PM, DiIorio MA, Lemieux ME, Yates K, Godec J, Klatt MG, Regev A. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat Immunol. 2014. April;15(4):373–383. doi: 10.1038/ni.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hess Michelini R, Doedens AL, Goldrath AW, Hedrick SM. Differentiation of CD8 memory T cells depends on Foxo1. J Exp Med. 2013. June 3;210(6):1189–1200. doi: 10.1084/jem.20130392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Staron MM, Gray SM, Marshall HD, Parish I, Chen J, Perry C, Cui G, Li M, Kaech S. The transcription factor FoxO1 sustains expression of the inhibitory receptor PD-1 and survival of antiviral CD8+ T cells during chronic infection. Immunity. 2014. November 20;41(5):802–814. doi: 10.1016/j.immuni.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Delpoux A, Michelini RH, Verma S, Lai C-Y, Omilusik KD, Utzschneider DT, Redwood AJ, Goldrath AW, Benedict CA, Hedrick SM. Continuous activity of Foxo1 is required to prevent anergy and maintain the memory state of CD8+ T cells. J Exp Med. 2018. February 5;215(2):575–594. doi: 10.1084/jem.20170697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Delpoux A, Lai C-Y, Hedrick SM, Doedens AL. FOXO1 opposition of CD8(+) T cell effector programming confers early memory properties and phenotypic diversity. Proc Natl Acad Sci U S A. 2017. October 17;114(42):E8865–E8874. doi: 10.1073/pnas.1618916114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Utzschneider DT, Delpoux A, Wieland D, Huang X, Lai C-Y, Hofmann M, Thimme R, Hedrick SM. Active maintenance of T cell memory in acute and chronic viral infection depends on continuous expression of FOXO1. Cell Rep. 2018. March 27;22(13):3454–3467. doi: 10.1016/j.celrep.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene. 1999. August 20;236(2):197–208. doi: 10.1016/S0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 137.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006. February 23;25(8):1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 138.Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta. 1997. April 18;1332(2):F49–66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 139.Hwang SS, Kim YU, Lee S, Jang SW, Kim MK, Koh BH, Lee W, Kim J, Souabni A, Busslinger M. Transcription factor YY1 is essential for regulation of the Th2 cytokine locus and for Th2 cell differentiation. Proc Natl Acad Sci U S A. 2013. January 2;110(1):276–281. doi: 10.1073/pnas.1214682110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yu B, Zhang K, Milner JJ, Toma C, Chen R, Scott-Browne JP, Pereira RM, Crotty S, Chang JT, Pipkin ME. Erratum: epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat Immunol. 2017. May 18;18(6):705. doi: 10.1038/ni0617-705b. [DOI] [PubMed] [Google Scholar]
- 141.Alfei F, Kanev K, Hofmann M, Wu M, Ghoneim HE, Roelli P, Utzschneider DT, von Hoesslin M, Cullen JG, Fan Y. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature. 2019. June 17;571(7764):265–269. doi: 10.1038/s41586-019-1326-9. [DOI] [PubMed] [Google Scholar]
- 142.O’Flaherty E, Kaye J. TOX defines a conserved subfamily of HMG-box proteins. BMC Genomics. 2003. April 2;4(1):13. doi: 10.1186/1471-2164-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Aliahmad P, Kaye J. Development of all CD4 T lineages requires nuclear factor TOX. J Exp Med. 2008. January 21;205(1):245–256. doi: 10.1084/jem.20071944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Scott AC, Dundar F, Zumbo P, Chandran SS, Klebanoff CA, Shakiba M, Trivedi P, Menocal L, Appleby H, Camara S. TOX is a critical regulator of tumour-specific T cell differentiation. Nature. 2019. June 17;571(7764):270–274. doi: 10.1038/s41586-019-1324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shaw LA, Belanger S, Omilusik KD, Cho S, Scott-Browne JP, Nance JP, Goulding J, Lasorella A, Lu L-F, Crotty S. Id2 reinforces TH1 differentiation and inhibits E2A to repress TFH differentiation. Nat Immunol. 2016. July;17(7):834–843. doi: 10.1038/ni.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Latif F, Tory K, Gnarra J, Yao M, Duh F, Orcutt M, Stackhouse T, Kuzmin I, Modi W, Geil L. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993. May 28;260(5112):1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 147.Doedens AL, Phan AT, Stradner MH, Fujimoto JK, Nguyen JV, Yang E, Johnson RS, Goldrath AW. Hypoxia-inducible factors enhance the effector responses of CD8+ T cells to persistent antigen. Nat Immunol. 2013. November;14(11):1173–1182. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, Tsao H-W, Godec J, LaFleur MW, Brown FD. The epigenetic landscape of T cell exhaustion. Science. 2016. December 2;354(6316):1165–1169. doi: 10.1126/science.aae0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Scott-Browne JP, Lopez-Moyado IF, Trifari S, Wong V, Chavez L, Rao A, Pereira RM. Dynamic changes in chromatin accessibility occur in CD8 + T cells responding to viral infection. Immunity. 2016. December 20;45(6):1327–1340. doi: 10.1016/j.immuni.2016.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Alfei F, Zehn D. T cell exhaustion: an epigenetically imprinted phenotypic and functional makeover. Trends Mol Med. 2017. September;23(9):769–771. doi: 10.1016/j.molmed.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 151.Philip M, Fairchild L, Sun L, Horste EL, Camara S, Shakiba M, Scott AC, Viale A, Lauer P, Merghoub T. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 2017. May 25;545(7655):452–456. doi: 10.1038/nature22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Doering TA, Crawford A, Angelosanto JM, Paley M, Ziegler C, Wherry E. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity. 2012. December 14;37(6):1130–1144. doi: 10.1016/j.immuni.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Khan O, Giles JR, McDonald S, Manne S, Ngiow SF, Patel KP, Werner MT, Huang AC, Alexander KA, Wu JE. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature. 2019. June 17;571(7764):211–218. doi: 10.1038/s41586-019-1325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Brentjens RJ, Davila ML, Riviere I, Park J, Wang X, Cowell LG, Bartido S, Stefanski J, Taylor C, Olszewska M. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med. 2013. March 20;5(177):177ra38. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, Teachey DT, Chew A, Hauck B, Wright JF. Chimeric Antigen Receptor–Modified T Cells for Acute Lymphoid Leukemia. N Engl J Med. 2013. April 18;368(16):1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor–modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011. August 25;365(8):725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Junghans RP. The challenges of solid tumor for designer CAR-T therapies: a 25-year perspective. Cancer Gene Ther. 2017. March;24(3):89–99. doi: 10.1038/cgt.2016.82. [DOI] [PubMed] [Google Scholar]
- 158.Kershaw MH, Westwood JA, Parker LL, Wang G, Eshhar Z, Mavroukakis SA, White DE, Wunderlich JR, Canevari S, Rogers-Freezer L. A phase I study on adoptive immunotherapy using gene-modified T cells for ovarian cancer. Clin Cancer Res. 2006. October 15;12(20):6106–6115. doi: 10.1158/1078-0432.CCR-06-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lamers CH, Sleijfer S, van Steenbergen S, van Elzakker P, van Krimpen B, Groot C, Vulto A, den Bakker M, Oosterwijk E, Debets R. Treatment of metastatic renal cell carcinoma with CAIX CAR-engineered T cells: clinical evaluation and management of on-target toxicity. Mol Ther. 2013. April;21(4):904–912. doi: 10.1038/mt.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Long AH, Haso WM, Shern JF, Wanhainen KM, Murgai M, Ingaramo M, Smith JP, Walker AJ, Kohler ME, Venkateshwara VR. 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat Med. 2015. May 4;21(6):581–590. doi: 10.1038/nm.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fraietta JA, Lacey SF, Orlando EJ, Pruteanu-Malinici I, Gohil M, Lundh S, Boesteanu AC, Wang Y, O’Connor RS, Hwang W-T. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat Med. 2018. May;24(5):563–571. doi: 10.1038/s41591-018-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cherkassky L, Morello A, Villena-Vargas J, Feng Y, Dimitrov DS, Jones DR, Sadelain M, Adusumilli PS. Human CAR T cells with cell-intrinsic PD-1 checkpoint blockade resist tumor-mediated inhibition. J Clin Invest. 2016. August 1;126(8):3130–3144. doi: 10.1172/JCI83092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.John LB, Devaud C, Duong CP, Yong CS, Beavis PA, Haynes NM, Chow MT, Smyth MJ, Kershaw MH, Darcy PK. Anti-PD-1 antibody therapy potently enhances the eradication of established tumors by gene-modified T cells. Clin Cancer Res. 2013. October 15;19(20):5636–5646. doi: 10.1158/1078-0432.CCR-13-0458. [DOI] [PubMed] [Google Scholar]
- 164.Zolov SN, Rietberg SP, Bonifant CL. Programmed cell death protein 1 activation preferentially inhibits CD28.CAR–T cells. Cytotherapy. 2018. October 8;20(10):1259–1266. doi: 10.1016/j.jcyt.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 165.Park JR, Digiusto DL, Slovak M, Wright C, Naranjo A, Wagner J, Meechoovet HB, Bautista C, Chang W-C, Ostberg JR. Adoptive transfer of chimeric antigen receptor re-directed cytolytic T lymphocyte clones in patients with neuroblastoma. Mol Ther. 2007. April;15(4):825–833. doi: 10.1038/sj.mt.6300104. [DOI] [PubMed] [Google Scholar]
- 166.Till BG, Jensen MC, Wang J, Qian X, Gopal AK, Maloney DG, Lindgren CG, Lin Y, Pagel JM, Budde LE. CD20-specific adoptive immunotherapy for lymphoma using a chimeric antigen receptor with both CD28 and 4-1BB domains: pilot clinical trial results. Blood. 2012. April 26;119(17):3940–3950. doi: 10.1182/blood-2011-10-387969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Emran AA, Chatterjee A, Rodger EJ, Tiffen JC, Gallagher SJ, Eccles MR, Hersey P. Targeting DNA methylation and EZH2 activity to overcome melanoma resistance to immunotherapy. Trends Immunol. 2019. April;40(4):328–344. doi: 10.1016/j.it.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 168.Salmond RJ, Alexander DR. SHP2 forecast for the immune system: fog gradually clearing. Trends Immunol. 2006. March;27(3):154–160. doi: 10.1016/j.it.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 169.Hui E, Cheung J, Zhu J, Su X, Taylor MJ, Wallweber HA, Sasmal DK, Huang J, Kim JM, Mellman I. T cell costimulatory receptor CD28 is a primary target for PD-1–mediated inhibition. Science. 2017. March 31;355(6332):1428–1433. doi: 10.1126/science.aaf1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Oehen S, Brduscha-Riem K. Differentiation of naive CTL to effector and memory CTL: correlation of effector function with phenotype and cell division. J Immunol. 1998. November 15;161(10):5338–5346. [PubMed] [Google Scholar]
- 171.Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu Rev Immunol. 2007;25(1):171–192. doi: 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- 172.Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, Youngblood BA, Abdelsamed HA, McGuire DJ, Cohen KW. Origin and differentiation of human memory CD8 T cells after vaccination. Nature. 2017. December 21;552(7685):362–367. doi: 10.1038/nature24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Youngblood B, Hale JS, Kissick HT, Ahn E, Xu X, Wieland A, Araki K, West EE, Ghoneim HE, Fan Y. Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature. 2017. December 21;552(7685):404–409. doi: 10.1038/nature25144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pace L, Goudot C, Zueva E, Gueguen P, Burgdorf N, Waterfall JJ, Quivy J-P, Almouzni G, Amigorena S. The epigenetic control of stemness in CD8(+) T cell fate commitment. Science. 2018. January 12;359(6372):177–186. doi: 10.1126/science.aah6499. [DOI] [PubMed] [Google Scholar]
- 175.Reinhardt RL, Khoruts A, Merica R, Zell T, Jenkins MK. Visualizing the generation of memory CD4 T cells in the whole body. Nature. 2001. March 1;410(6824):101–105. doi: 10.1038/35065111. [DOI] [PubMed] [Google Scholar]
- 176.Masopust D. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001. March 23;291(5512):2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- 177.Balkhi MY, Ma Q, Ahmad S, Junghans RP. T cell exhaustion and Interleukin 2 downregulation. Cytokine. 2015. February;71(2):339–347. doi: 10.1016/j.cyto.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 178.Okazaki T, Honjo T. The PD-1-PD-L pathway in immunological tolerance. Trends Immunol. 2006. April;27(4):195–201. doi: 10.1016/j.it.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 179.Keir ME, Liang SC, Guleria I, Latchman YE, Qipo A, Albacker LA, Koulmanda M, Freeman GJ, Sayegh MH, Sharpe AH. Tissue expression of PD-L1 mediates peripheral T cell tolerance. J Exp Med. 2006. April 17;203(4):883–895. doi: 10.1084/jem.20051776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
I agree to share all the data and content of this review.


