Abstract
Accurate segmentation of organs-at-risk is important inprostate cancer radiation therapy planning. However, poor soft tissue contrast in CT makes the segmentation task very challenging. We propose a deep convolutional neural network approach to automatically segment the prostate, bladder, and rectum from pelvic CT. A hierarchical coarse-to-fine segmentation strategy is used where the first step generates a coarse segmentation from which an organ-specific region of interest (ROI) localization map is produced. The second step produces detailed and accurate segmentation of the organs. The ROI localization map is generated using a 3D U-net. The localization map helps adjusting the ROI of each organ that needs to be segmented and hence improves computational efficiency by eliminating irrelevant background information. For the fine segmentation step, we designed a fully convolutional network (FCN) by combining a generative adversarial network (GAN) with a U-net. Specifically, the generator is a 3D U-net that is trained to predict individual pelvic structures, and the discriminator is an FCN which fine-tunes the generator predicted segmentation map by comparing it with the ground truth. The network was trained using 100 CT datasets and tested on 15 datasets to segment the prostate, bladder and rectum. The average Dice similarity (mean±SD) of the prostate, bladder and rectum are 0.90±0.05, 0.96±0.06 and 0.91±0.09, respectively, and Hausdorff distances of these three structures are 5.21±1.17, 4.37±0.56 and 6.11±1.47(mm), respectively. The proposed method produces accurate and reproducible segmentation of pelvic structures, which can be potentially valuable for prostate cancer radiotherapy treatment planning.
Keywords: Prostate Cancer, Male Pelvic Organ, CT image, Hierarchical Segmentation, Radiotherapy
1. INTRODUCTION
Prostate cancer is the most prevalent malignancy in men, and the second leading cause of cancer death in the US with approximately 174,650 new cases and 31,620 deaths estimated in 2019.1 Standard treatment options for prostate cancer include radiotherapy such as external beam radiation therapy (EBRT) and high/low-dose-rate brachytherapy. The efficacy of radiotherapy depends on accurate delivery of therapeutic radiation dose to the target while sparing adjacent healthy tissues. In image-guided radiotherapy planning, accurate segmentation of the prostate and other organs at risk (OAR) is a necessary step. Manual segmentation is still considered as the gold standard in current clinical practice, but it is very time consuming and the quality of the segmentation varies depending on the expert’s knowledge and experience. In addition, poor soft tissue contrast in CT images makes the contouring process challenging and thus yields to large inter-and intra-observer contouring variability.2–4
Automatic organ segmentation is an active research area for the last few decades. Existing automatic segmentation methods can be broadly categorized into three classes: atlas-based, model-based, and learning-based methods. In atlas-based methods, an atlas dataset is created from a set of CT images and manually segmented labels, which are registered to the image to be segmented followed by atlas contour propagation to the target image to get final segmentation.5–8 The segmentation accuracy of atlas-based method largely depends on the accuracy of image registration used for atlas matching and the selection of optimal atlases. Since a single atlas cannot perfectly fit to every patients, using multiple atlases has become a standard baseline for atlas-based segmentation, where multiple sets of contours transferred from different atlases will be fused into a single set of consensus segmentations by using a label fusion technique.9–14 While multi-atlas-based segmentation has been widely adopted with state-of-the-art segmentation quality, it requires a significant amount of time as it involves multiple registrations between the atlas and the target volumes. Model-based segmentation utilizes a priori knowledge of the target such as shape, intensity, and texture to constrain the registration process.15–21 It is often used in combination with other method, e.g., multi-atlas auto-segmentation, to further improve the segmentation. Such hybrid approaches that combine the atlas-and model-based segmentations have been applied for the delineation of head and neck structures on CT images, showing promising results.16, 22–24 Other model-based segmentation approaches include deformable model-based segmentations that initialize a deformable model into the image to be segmented and then the segmentation proceeds by deforming the initial model using image-specific knowledge.25, 26 These model-based methods require fine-tuned parameters for every structure to be segmented, and are sensitive to structures and image quality variations. Learning-based models train a classifier or regressor from a pool of training image. Then the segmentation is generated by predicting the likelihood map.27, 28 Even though this class of segmentation is more flexible than the other two classes as it does not require any priori knowledge, this class is limited to the hand-crafted feature extraction.
In the last few years, deep learning-based automatic segmentation demonstrated its potential in accurate and consistent organ delineation. Particularly, convolutional neural network (CNN) became a state-of-the-art in solving challenging problems in image classification and segmentation due to its automatic deep feature extraction.29 Ronnebergeret al. proposed the 2D U-net which is a fully convolutional neural network (FCN) with skip connection and capable of extracting contextual features from contacting layers and structural information from expansion layers30.A 3D volumetric segmentation using FCN, called V-net, is proposed to segment prostate in MR data.31 Zhu et al. incorporated deep supervision in hidden layers of U-net to segment prostate in MR images.32 Wang et al. developed a prostate segmentation in CT where a dilated convolution, along with deep supervision is combined with FCN.33 Segmentation of male pelvic structures (bladder, prostate and rectum) is performed using 2D U-net34 and using FCN with boundary sensitive information.35 General adversarial network (GAN) is a learning based network that estimates a generative model through an adversarial process. GANs have attracted much interest for their success in unsupervised learning36, 37 and have been used in detecting prostate cancer.38
In this paper, we propose a hierarchical volumetric segmentation of male pelvic CT where a coarse segmentation is produced using a multiclass 3DU-net to determine organ-specific regions of interest (ROIs)and fine segmentation is performed utilizing GAN with a 3D U-net as generator and FCN as discriminator. GAN contributes by providing learned parameters of accurate segmentation by distinguishing between real and fake segmentations and thus globally improving segmentation accuracy.
2. METHODS
We propose a fully automatic segmentation method to segment pelvic structures such as the prostate, bladder, and rectum from male pelvic CT images. The pelvic CT contains larger background compared to the small organs. Such irrelevant background information burdens on computation efficiency. To improve performance, we design a hierarchical coarse-to-fine segmentation approach as shown in Figure 1.
Figure 1.
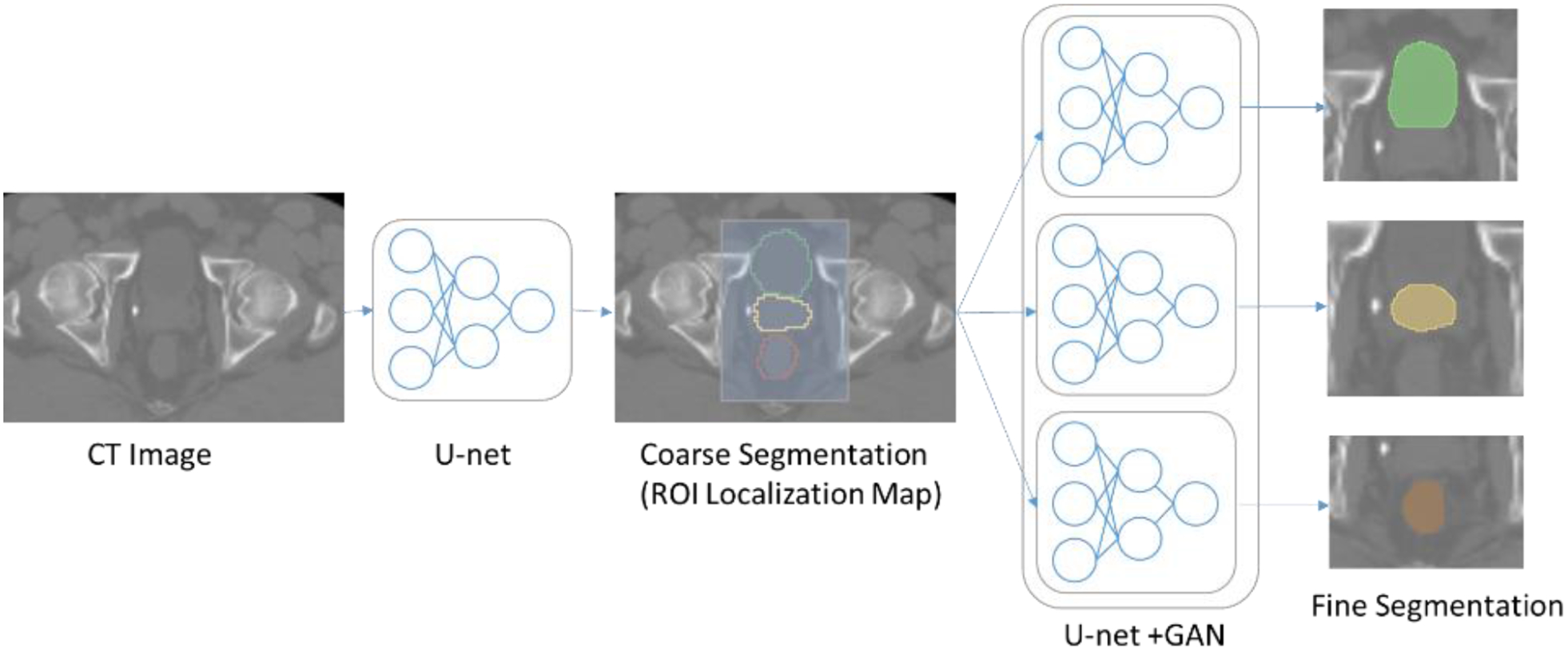
Workflow of the proposed hierarchical segmentation method.
2.1. Coarse Segmentation
In the coarse segmentation step, a ROI localization map is generated to extract only the ROIs that contain the organs of interest. The original CT image and manual contours are used to train a multiclass.3D U-net.30 Similar to the standard U-net, it has contraction and expansion paths each with four layers. Each layer of contraction path is composed of two 3×3×3 39convolutions each followed by a leaky rectifier linear activation function39, after that a 2×2×2 max pooling is performed. In each layer, we add batch normalization to speed up learning by reducing the internal covariate shift40 and dropout to prevent overfitting41. After each layer, the number of feature map is doubled. The expansion path has the similar architecture to the contraction path except that in each layer it has an upsampling followed by a 2×2×2 deconvolution that halves the number of feature map. In the last layer, a 1×1×1 convolutionis used to map the output feature to the desired number of labels. Then the copy layers are used to transfer features extracted from the early contraction path to the expansion path. To save training cost, the ROI localization network is trained to segment organs all together instead of segmenting each organ separately.
We have down-sampled the original CT image to a lower resolution to train the coarse segmentation network. Once this first network is trained, it produces a coarse segmentation of pelvic structures of interest, which can be used to automatically extract ROIs of the prostate, bladder and rectum. Based on the coarse segmentation, the centroid of each organ is calculated and then the ROI of each organ is cropped from the original image where the centroid is the center of the cropped ROI.The cropped ROI are large enough to sufficiently cover each organ and enough background context.
2.2. Fine Segmentation
For the fine segmentation, three CNNs are trained based on the cropped (using the ROIs from the coarse segmentation) CT and manual segmentations separately for the prostate, bladder, and rectum. The CNNs in this step are designed using GAN and 3D U-net as shown in Figure 2. The GAN network consists of a generator and discriminator networks where the two networks compete with each other to produce an accurate and optimal segmentation map. A 3D U-net is used as a generator, and trained using cropped CT and manual segmentations. After training, the generator produces predicted label of the OAR. On the other hand, an FCN is used as the discriminator and trained using manually contoured labels to determine if the generator predicted labels are real or fake.
Figure 2.
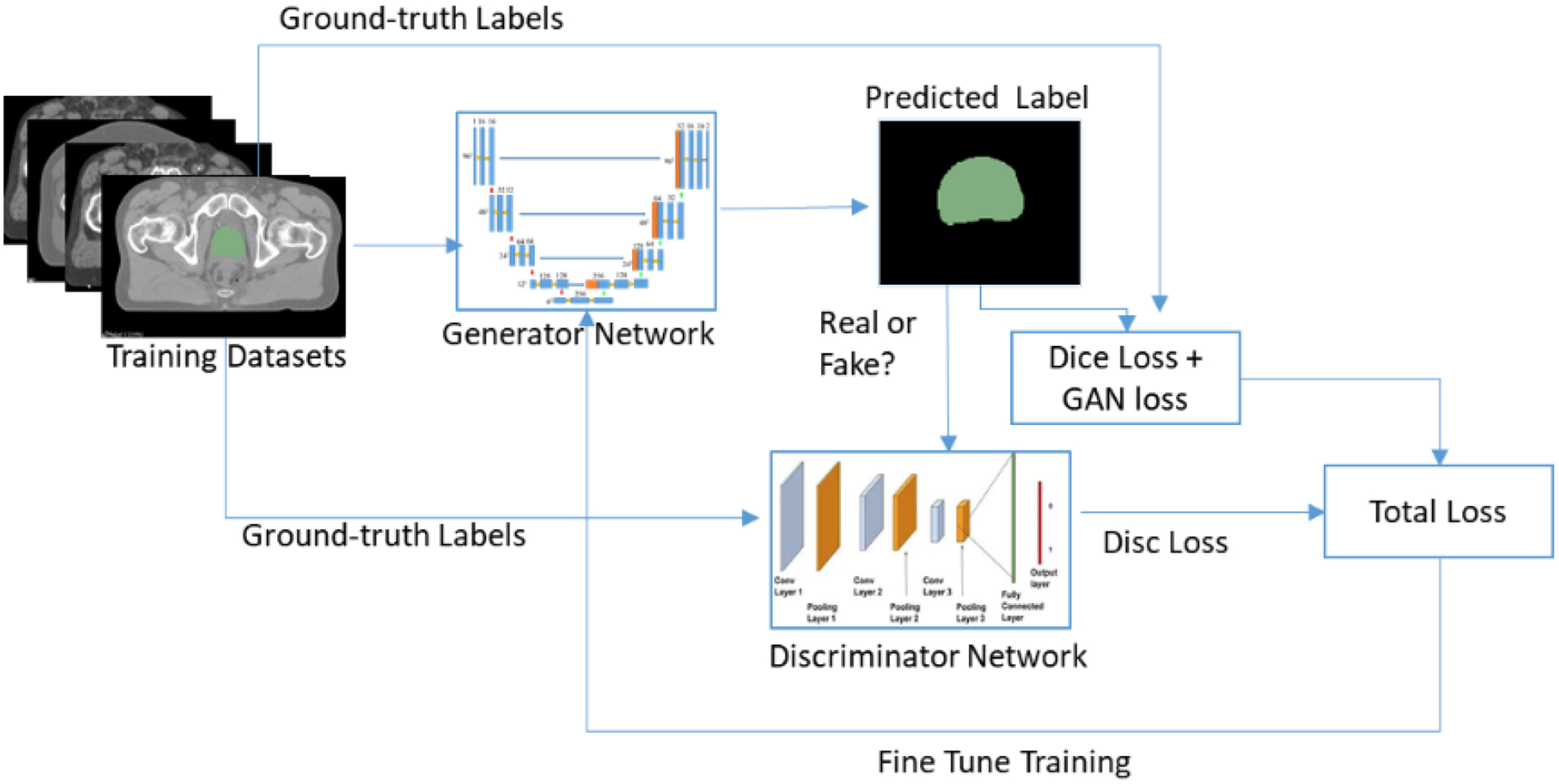
The U-net + GAN architecture for pelvic structure segmentation.
In GANs both the generator network G and discriminator network D are trained simultaneously. The objective of G is to learn the distribution px from the dataset x and then samples a variable z from the uniform or Gaussian distribution pz.(z). The purpose of D is to classify whether an image comes from training datasets or from G. To define the cost functionz of the GAN, let lfake and lreal denote labels for fake and real data, respectively. Then the cost function for D and G are defined using least square loss function42 as follows:
| (1) |
| (2) |
where c is constant value that generator wants discriminator to believe for fake labels.
In the proposed method, for fine segmentation we defined the objective function of the generator as the sum of weighted dice loss function and least squared generator loss function defined in equation (2).The final objective function to optimize the network is defined as:
| (3) |
We have used a weighted dice loss define as:
| (4) |
where for each class i, wi is the class weight and bi(x)and is the binary map at each pixel x and p(x,i) is the predicted probability.
3. RESULTS
We evaluated the proposed segmentation method using 115 CT datasets obtained from prostate cancer patients who were treated by EBRT. We used 100 datasets for training and remaining 15 datasets for testing. Each patient had CT images of pelvic region and manual contouring of the prostate, bladder, and rectum drawn by a radiation oncologist. The size of the original CT image was 512×512 ×[80–120] voxels with a voxel size of 1.17×1.17×3 mm3. We downsampled the original CT image to5×5×5 mm3 voxel resolution with a dimension of 118×118×[48–72] which were then used to train the coarse segmentation network. The coarse segmentation network produced a rough segmentation of the organs from which we extracted the ROIs of the prostate, bladder, and rectum. The ROIs were 96×96×32, 112×112×64, and 96×96×64 voxels for the prostate, bladder, and rectum, respectively, which were cropped from the original CT for the successive fine segmentations. The same 100 patient data were used to train the fine segmentation networks after ROI cropping from the original CT (not downsampled). In order to quantitatively assess the performance of the proposed method we used Dice similarity coefficient (DSC), Mean Surface Distance (MSD) and Hausdorff Distance (HD). An example segmentation from one of the test cases is shown in Figure 3 and the 3D rendering of the segmentations is shown in Figure 4.
Figure 3.
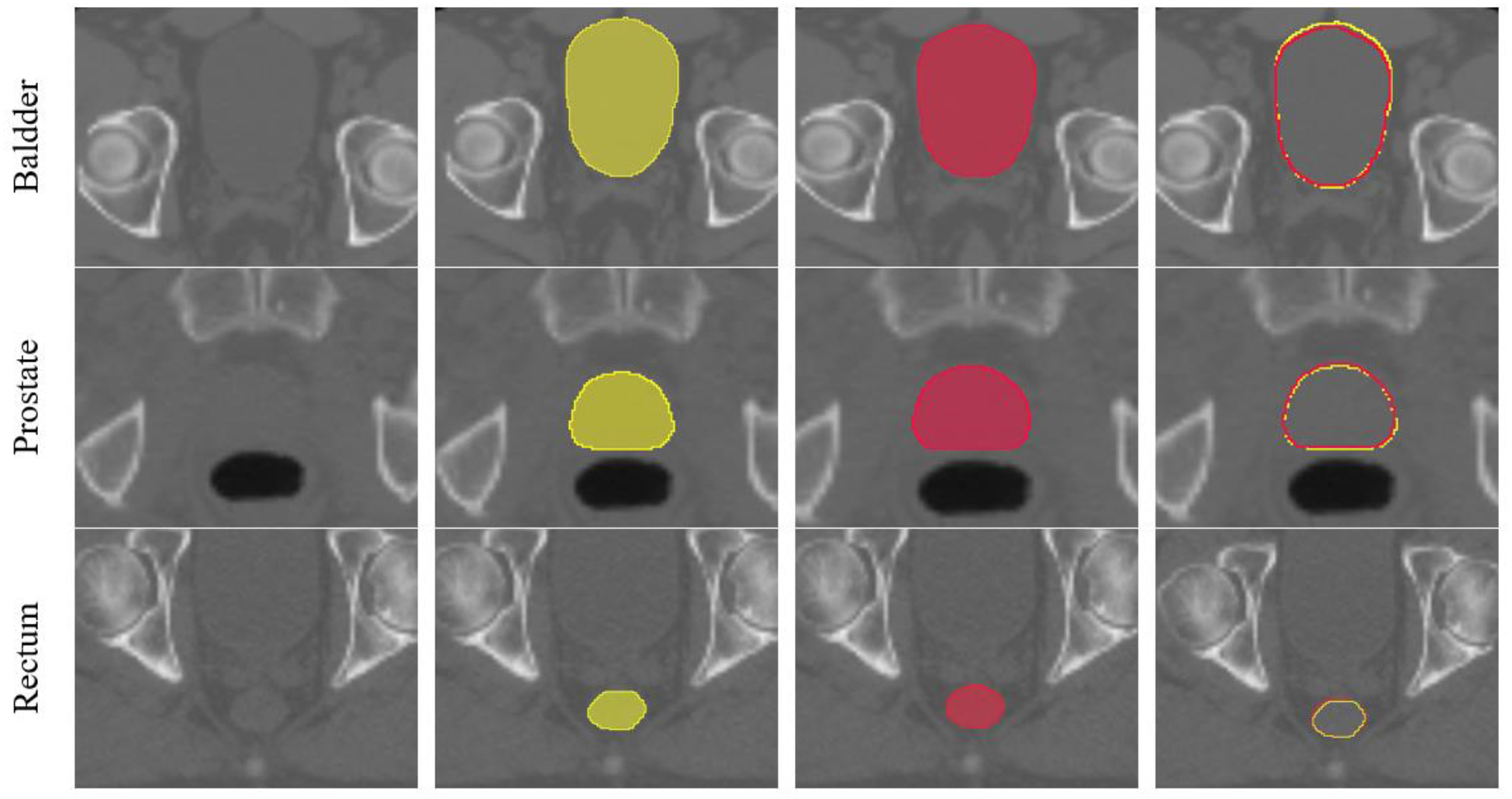
An example segmentation result. The first column shows the ROI of each organ. The second column shows the ground truth segmentations. The third column is the proposed automatic segmentations and the fourth column shows overlaid contours of the ground truth (yellow) and automatic segmentation (red).
Figure 4.
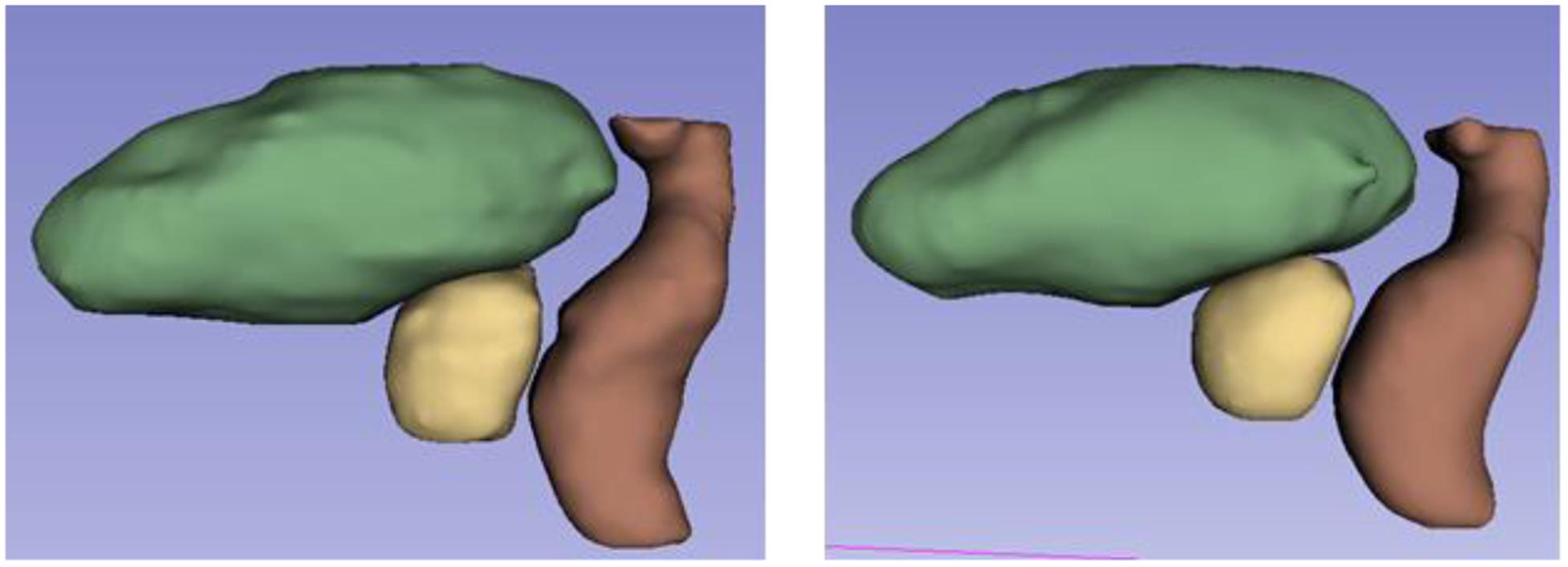
3D representation of the segmentations (yellow: prostate, green: bladder, brown: rectum). (Left) Ground truth. (Right) Proposed automatic segmentation.
Class-imbalance is a common problem in multiclass segmentation where small structures are prone to being underrepresented. We compared our proposed hierarchical segmentation with segmentation where the network was trained with all the multiclass labels simultaneously to produce segmentation label maps of the prostate, bladder, and rectum altogether. In terms of segmentation accuracy, the proposed method with single-class training always performed better than the multiclass segmentation. In addition, we also compared to the 3DU-net based segmentation which was trained for multiclass segmentation. Table 1 shows comparison results. The proposed method obtains significant improvement over the multiclass U-net+GAN and U-net-based segmentations with a respective Dice (mean ±SD)of 0.90 ± 0.05, 0.96 ± 0.06and 0.91 ± 0.09for the prostate, bladder and rectum, respectively. The MSD for all three structures are less than 1.8 mm, which is within the expected random error given the image resolution of 1.17×1.17×3 mm3. The proposed method also achieves lower HD than the other two methods.
Table 1.
Quantitative comparison of segmentation results from different methods.
| Metrics | Method | OAR | ||
|---|---|---|---|---|
| Prostate (mean ± SD) | Bladder (mean ± SD) | Rectum (mean ± SD) | ||
| DSC | Hierarchical U-net+GAN | 0.90 ± 0.05 | 0.96 ± 0.06 | 0.91 ± 0.09 |
| U-net+GAN (Multiclass) | 0.86 ± 0.07 | 0.92 ± 0.05 | 0.87 ± 0.13 | |
| U-net | 0.84 ± 0.05 | 0.88 ± 0.06 | 0.83 ± 0.16 | |
| MSD (mm) | Hierarchical U-net+GAN | 1.56 ± 0.37 | 0.95 ± 0.15 | 1.78 ± 1.13 |
| U-net+GAN (Multiclass) | 2.28 ± 0.78 | 2.11 ± 0.45 | 3.45 ± 0.95 | |
| U-net | 2.89 ± 1.15 | 2.34 ± 0.91 | 3.91 ± 0.47 | |
| HD (mm) | Hierarchical U-net+GAN | 5.21 ± 1.17 | 4.37 ± 0.56 | 6.11 ± 1.47 |
| U-net+GAN (Multiclass) | 6.55 ± 1.53 | 6.55 ± 1.53 | 7.55 ± 1.53 | |
| U-net | 8.20 ± 1.90 | 7.20 ± 1.90 | 9.20 ± 1.90 | |
Table 2 shows quantitative comparison between our method with existing state-of-the-art methods that used different datasets. Our method outperforms both multi-atlas-based methods and machine learning-based method using regression forest-based deformable segmentation.27, 28, 43 Kazemifar et al. proposed a deep learning based segmentation of CT male pelvic organs using 2D U-Net with DSC of 0.88± 0.10, 0.95± 0.04 and 0.92± 0.10 for the prostate, bladder and rectum, respectively. Another CNN-based automatic segmentation of the same pelvic organs by incorporating boundary information in network training is recently proposed where the segmentation accuracy in terms of DSC are 0.89 ± 0.03, 350.94 ± 0.03 and 0.89 ± 0.04 for the prostate, bladder and rectum, respectively.35 Although these methods are capable of producing satisfactory segmentation results, our method achieves better performance in segmenting pelvic organs from CT images.
Table 2.
Comparison with previous studies based on DSC
| Method | Prostate | Bladder | Rectum | |
|---|---|---|---|---|
| DSC (mean ± SD) | DSC (mean ± SD) | DSC (mean ± SD) | ||
| Shao et al.28 | Learning-based using regression Forest | 0.88 ± 0.02 | – | 0.84 ± 0.05 |
| Gao et al.27 | 0.86 ± 0.41 | 0.92 ± 0.47 | 0.88 ± 0.48 | |
| Acosta et al.43 | Multi-atlas | 0.85 ± 0.40 | 0.92 ± 0.20 | 0.80 ± 0.70 |
| Kazemifar et al.34 | Deep learning using CNN | 0.88 ± 0.10 | 0.95 ± 0.04 | 0.92 ± 0.10 |
| Wang et al.35 | 0.89 ± 0.03 | 0.94 ± 0.03 | 0.89 ± 0.04 |
4. CONCLUSION
We present an end-to-end, deep-learning-based automatic segmentation of male pelvic OARs using U-net+GAN with an automatic ROI localization of the organs to be segmented. The automatic ROI extraction improves computational efficiency and the segmentation accuracy by allowing the fine segmentation networks to focus only on the region of each organ. The fine segmentation of each organ is performed using U-net+GAN where the generator and discriminator competes with each other to improve segmentation accuracy. To avoid class imbalance problem of multiclass segmentation, we performed single-class training, which significantly improved segmentation accuracy over the multiclass training-based segmentation. We performed experimental validation using clinical data and showed that the proposed method outperformed other state-of-the-art methods. The proposed method can potentially improve the efficiency of radiation therapy planning of prostate cancer treatment.
ACKNOWLEDGEMENTS
This work was supported by the NIH/NCI under the grant R01CA151395.We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan Xp GPU.
REFERENCES
- [1].Siegel RL, Miller KD, and Jemal A, “Cancer statistics, 2019,” CA: a cancer journal for clinicians, 69(1), 7–34 (2019). [DOI] [PubMed] [Google Scholar]
- [2].Steenbergen P, Haustermans K, Lerut E et al. , “Prostate tumor delineation using multiparametric magnetic resonance imaging: Inter-observer variability and pathology validation,” Radiotherapy and Oncology, 115(2), 186–190 (2015). [DOI] [PubMed] [Google Scholar]
- [3].Fiorino C, Reni M, Bolognesi A et al. , “Intra-and inter-observer variability in contouring prostate and seminal vesicles: implications for conformal treatment planning,” Radiotherapy and oncology, 47(3), 285–292 (1998). [DOI] [PubMed] [Google Scholar]
- [4].Lee WR, Roach III M, Michalski J et al. , “Interobserver variability leads to significant differences in quantifiers of prostate implant adequacy,” International Journal of Radiation OncologyBiologyPhysics, 54(2), 457–461 (2002). [DOI] [PubMed] [Google Scholar]
- [5].Martin S, Daanen V, and Troccaz J, “Atlas-based prostate segmentation using an hybrid registration,” International Journal of Computer Assisted Radiology and Surgery,3(6), 485–492 (2008). [Google Scholar]
- [6].Klein S, Van Der Heide U, Raaymakers BW et al. , “Segmentation of the prostate in MR images by atlas matching,” 2007 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 1300–1303 (2007). [Google Scholar]
- [7].Sjöberg C, Lundmark M, Granberg C et al. , “Clinical evaluation of multi-atlas based segmentation of lymph node regions in head and neck and prostate cancer patients,” Radiation Oncology,8(1), 229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Acosta O, Simon A, Monge F et al. , “Evaluation of multi-atlas-based segmentation of CT scans in prostate cancer radiotherapy,” 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 1966–1969 (2011). [Google Scholar]
- [9].Han X, Hibbard L, O’Connell N et al. , “Automatic segmentation of head and neck CT images by GPU-accelerated multi-atlas fusion,” Proceedings of Head And Neck Auto-Segmentation Challenge Workshop, 219 (2009). [Google Scholar]
- [10].Heckemann RA, Hajnal JV, Aljabar P et al. , “Automatic anatomical brain MRI segmentation combining label propagation and decision fusion,” NeuroImage, 33(1), 115–126 (2006). [DOI] [PubMed] [Google Scholar]
- [11].Rohlfing T, Russakoff DB, and Maurer CR, “Performance-based classifier combination in atlas-based image segmentation using expectation-maximization parameter estimation,” IEEE transactions on medical imaging, 23(8), 983–994 (2004). [DOI] [PubMed] [Google Scholar]
- [12].Han X, Hibbard LS, O’Connell NP et al. , “Automatic segmentation of parotids in head and neck CT images using multi-atlas fusion,” Medical Image Analysis for the Clinic: A Grand Challenge, 297–304 (2010). [Google Scholar]
- [13].Wang H, Suh JW, Das SR et al. , “Multi-atlas segmentation with joint label fusion,” IEEE transactions on pattern analysis and machine intelligence, 35(3), 611–623 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Asman AJ, and Landman BA, “Non-local statistical label fusion for multi-atlas segmentation,” Medical image analysis, 17(2), 194–208 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sultana S, Agrawal P, Elhabian S et al. , “Medial axis segmentation of cranial nerves using shape statistics-aware discrete deformable models,” International Journal of Computer Assisted Radiology and Surgery, 1–13 (2019). [DOI] [PubMed] [Google Scholar]
- [16].Qazi AA, Pekar V, Kim J et al. , “Auto-segmentation of normal and target structures in head and neck CT images: A feature-driven model-based approach,” Medical physics, 38(11), 6160–6170 (2011). [DOI] [PubMed] [Google Scholar]
- [17].Huang J, Jian F, Wu H et al. , “An improved level set method for vertebra CT image segmentation,” Biomedical engineering online,12(1), 48 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Qian X, Wang J, Guo S et al. , “An active contour model for medical image segmentation with application to brain CT image,” Medical physics,40(2), 021911 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cootes TF, Taylor CJ, Cooperet al DH., “Active shape models-their training and application,” Computer vision and image understanding, 61(1), 38–59 (1995). [Google Scholar]
- [20].Pekar V, McNutt TR, and Kaus MR, “Automated model-based organ delineation for radiotherapy planning in prostatic region,” International Journal of Radiation Oncology* Biology* Physics, 60(3), 973–980 (2004). [DOI] [PubMed] [Google Scholar]
- [21].Ragan D, Starkschall G, McNutt T et al. , “Semiautomated four-dimensional computed tomography segmentation using deformablemodels,” Medical physics, 32(7Part1), 2254–2261 (2005). [DOI] [PubMed] [Google Scholar]
- [22].Chen A, Deeley MA, Niermann KJ et al. , “Combining registration and active shape models for the automatic segmentation of the lymph node regions in head and neck CT images,” Medical physics, 37(12), 6338–6346 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gorthi S, Duay V, Houhou N et al. , “Segmentation of head and neck lymph node regions for radiotherapy planning using active contour-based atlas registration,” IEEE Journal of selected topics in signal processing, 3(1), 135–147 (2009). [Google Scholar]
- [24].Pekar V, Allaire S, Qazi A et al. , “Head and neck auto-segmentation challenge: segmentation of the parotid glands,” Medical Image Computing and Computer Assisted Intervention (MICCAI), 273–280 (2010). [Google Scholar]
- [25].Sultana S, Blatt JE, Gilles B et al. , “MRI-based medial axis extraction and boundary segmentation of cranial nerves through discrete deformable 3D contour and surface models,” IEEE transactions on medical imaging, 36(8), 1711–1721 (2017). [DOI] [PubMed] [Google Scholar]
- [26].Costa MJ, Delingette H, Novellas S et al. , “Automatic segmentation of bladder and prostate using coupled 3D deformable models,” International conference on medical image computing and computer-assisted intervention, 252–260 (2007). [DOI] [PubMed] [Google Scholar]
- [27].Gao Y, Shao Y, Lian J et al. , “Accurate segmentation of CT male pelvic organs via regression-based deformable models and multi-task random forests,” IEEE transactions on medical imaging, 35(6), 1532–1543 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Shao Y, Gao Y, Wang Q et al. , “Locally-constrained boundary regression for segmentation of prostate and rectum in the planning CT images,” Medical image analysis, 26(1), 345–356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rawat W, and Wang Z, “Deep convolutional neural networks for image classification: A comprehensive review,” Neural computation, 29(9), 2352–2449 (2017). [DOI] [PubMed] [Google Scholar]
- [30].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” International Conference on Medical image computing and computer-assisted intervention, 234–241 (2015). [Google Scholar]
- [31].Milletari F, Navab N, and Ahmadi S-A, “V-net: Fully convolutional neural networks for volumetric medical image segmentation,” 2016 Fourth International Conference on 3D Vision (3DV), 565–571 (2016). [Google Scholar]
- [32].Zhu Q, Du B, Turkbey B et al. , “Deeply-supervised CNN for prostate segmentation,” 2017 International Joint Conference on Neural Networks (Ijcnn), 178–184 (2017). [Google Scholar]
- [33].Wang B, Lei Y, Wang T et al. , “Automated prostate segmentation of volumetric CT images using 3D deeply supervised dilated FCN,” Medical Imaging 2019: Image Processing,10949, 109492S (2019). [Google Scholar]
- [34].Kazemifar S, Balagopal A, Nguyen D et al. , “Segmentation of the prostate and organs at risk in male pelvic CT images using deep learning,” Biomedical Physics & Engineering Express,4(5), 055003 (2018). [Google Scholar]
- [35].Wang S, He K, Nie D et al. , “CT male pelvic organ segmentation using fully convolutional networks with boundary sensitive representation,” Medical image analysis, 54, 168–178 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Radford A, Metz L, and Chintala S, “Unsupervised representation learning with deep convolutional generative adversarial networks,” arXiv preprint arXiv:1511.06434, (2015). [Google Scholar]
- [37].Goodfellow I, “NIPS 2016 tutorial: Generative adversarial networks,” arXiv preprint arXiv:1701.00160, (2016). [Google Scholar]
- [38].Kohl S, Bonekamp D, Schlemmer H-P et al. , “Adversarial networks for the detection of aggressive prostate cancer,” arXiv preprint arXiv:1702.08014, (2017). [Google Scholar]
- [39].Xu B, Wang N, Chen T et al. , “Empirical evaluation of rectified activations in convolutional network,” arXiv preprint arXiv:1505.00853, (2015). [Google Scholar]
- [40].Ioffe S, and Szegedy C, “Batch normalization: Accelerating deep network training by reducing internal covariate shift,” arXiv preprint arXiv:1502.03167, (2015). [Google Scholar]
- [41].Srivastava N, Hinton G, Krizhevsky A et al. , “Dropout: a simple way to prevent neural networks from overfitting,” The journal of machine learning research, 15(1), 1929–1958 (2014). [Google Scholar]
- [42].Mao X, Li Q, Xie H et al. , “Least squares generative adversarial networks,” Proceedings of the IEEE International Conference on Computer Vision, 2794–2802 (2017). [Google Scholar]
- [43].Acosta O, Dowling J, Drean G et al. , [Multi-atlas-based segmentation of pelvic structures from CT scans for planning in prostate cancer radiotherapy] Springer, (2014). [Google Scholar]


