Abstract
Colorectal cancer (CRC) is one of the most common malignant diseases in the world. Although mechanistic studies have been conducted on the pathogenesis of CRC, the molecular mechanism of CRC tumorigenesis remains unclear. In the present study, the weighted gene co-expression network analysis was performed for the Gene Expression Omnibus (GEO) dataset GSE87211, in order to analyze the key modules involved in the pathogenesis of CRC. Next, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses were performed on the key module genes to analyze the functional pathways involved. The hub genes were screened using the Cytoscape platform and verified by a second GEO dataset, GSE21510. Finally, 10 hub genes were identified in 2 key modules (the green and brown modules) as the genes most significantly associated with the tumorigenesis of CRC. The 5 hub genes from the green module included collagen type I α1 chain, collagen type XII α1 chain, collagen triple helix repeat containing 1, inhibin subunit βa (INHBA) and chromobox 2 (CBX2), while the 5 hub genes from the brown module included bestrophin 2 (BEST2), carbonic anhydrase 2, glucagon, solute carrier family 4 member 4 and gliomedin. The 2 key modules with the 10 hub genes identified may regulate the occurrence and development of CRC through the extracellular matrix pathway, PI3K-Akt and chemokine signaling pathways, thus providing a reference for understanding the complex mechanism of tumorigenesis in CRC. Of note, few studies have reported the pathogenesis of CRC with the 3 identified hub genes, INHBA, CBX2 and BEST2. Further investigation of the molecular mechanism of these genes in CRC is recommended.
Keywords: colorectal cancer, weighted gene co-expression network analysis, hub genes, bioinformatics
Introduction
Tumors that form in the colon or rectum are often referred to together as colorectal cancer (CRC). CRC is one of the most common types of digestive system tumors, with its mortality rate ranking 4th among all malignant tumors (1). A total of >90% of CRC cases occur after the age of 50 years, and the average age at diagnosis is 68 (2). The 5-year survival rate of CRC can reach 90% at early diagnosis, but is <10% when distant metastasis has developed (3). Unfortunately, CRC usually reveals no symptoms at early stages, so it is important to identify biomarkers for its earlier diagnosis to improve the outcome of this disease.
Various molecular pathways have been shown to be involved in CRC, such as the chromosomal instability (CIN), microsatellite instability and CpG island methylator phenotype pathways (4). These 3 CRC pathways overlap in complex ways (5). CIN is the most widespread in CRC, accounting for 65–70% of sporadic cases (6). In CIN, the Wnt/β-catenin signaling pathway, which includes adenomatous polyposis (APC), (pro) renin receptor [(P)RR] and axis inhibition protein 1 (Axin) (7), is the pathway most clinically associated with CRC. APC serves an anti-carcinogenic role by regulating canonical Wnt signal transduction mediated by cytoplasmic and nuclear mechanisms; mutations in the APC gene have been identified in ~80% patients with CRC (8). By contrast, the (P)RR, which is a component of the Wnt receptor complex, is usually overexpressed in CRC (9). The Axin gene can downregulate β-catenin and inhibit cell growth via its co-expression with APC5, when compared with cells transfected with Axin alone (10). However, the clinical approach to the CRC treatment of the CIN pathway is limited, suggesting that focusing on a single pathway is not sufficient to explain CRC pathobiology; a comprehensive consideration of multiple biological pathways is being suggested by an increasing number of studies (6,11). While studies into the molecular mechanism of CRC have focused on individual molecules rather than functional networks involving multiple pathways, weighted gene co-expression network analysis (WGCNA) may be used to analyze potential gene modules critically involved in gene expression. In the present study, WGCNA was performed on the Gene Expression Omnibus (GEO) dataset GSE87211 to further determine the molecular mechanisms of CRC. Key gene modules associated with CRC tumorigenesis were identified, and a series of biological functions and pathways were analyzed. A second GEO dataset, GSE21510, was used to validate the results, and The Cancer Genome Atlas (TCGA) database was used to further reveal the genetic information and clinical characteristics of CRC. The genes identified by WGCNA provided a more detailed insight into the molecular mechanism of CRC tumorigenesis, and could provide new targets for the diagnosis and treatment of the disease.
Materials and methods
Data extraction
A total of 1,014 series of human CRC were retrieved from the GEO database. Following careful screening of the content, discarding the datasets with incomplete information and those lacking control patients, the two datasets with the largest sample size (GSE87211 and GSE21510) were obtained. The GSE87211 dataset contained 230 CRC and 133 normal samples, and the platform used was GPL13497, Agilent-026652 Whole Human Genome Microarray 4×44K v2. Clinical information obtained from the dataset included sex, age and disease status (12). The GSE21510 dataset consisted of 123 CRC and 23 normal samples, and its platform was GPL570 Affymetrix Human Genome U133 Plus 2.0 Array (13). R packages were used to annotate the raw data, generate the expression matrix and match the probes targeted gene symbols.
Construction of WGCNA
Affy package (version 3.5.2 in R environment) (14) was used to pre-process and normalize (Robust Multiarray Averaging normalization) the original data of GEO database (.CEL file). Standard deviations (SDs) were arranged from large to small, and the expression of the top 5,000 genes with the greatest differences in case and control samples were selected for WGCNA. Using the pickSoftThreshold function in R language, the scale-free topology fitting index for several power was calculated, and the parameters that provided appropriate soft-threshold power for the construction of the network were obtained.
To measure the network connectivity of a gene defined as the sum of its adjacency with all other genes for the network generation, adjacency of the gene network was transformed into topological overlap. Hierarchical clustering was used to classify genes with similar expression profiles into the same modules, based on topical overlap matrix dissimilarity. As default, the minimum number of genes per gene module was set to 30 (14). The dynamicTreeCut algorithm of WGCNA was used to distinguish the gene co-expression modules by calculating the dissimilarity of the eigengenes. The MEDissThres, which is a parameter in the dynamicTreeCut algorithm of the WGCNA package, was used to select cutting lines merging some of the modules. Subsequently, visualization of eigengene network was performed.
Identification of modules association with clinical features
Module eigengenes (ME) is the first principal component of a given module and can be considered as a representative of the gene expression profile in a module (14). The association between ME and clinical features was calculated using linear regression, and modules significantly associated with clinical features were obtained. In addition, the logarithmic transformation of the P-value [gene significance (GS)=lgP] in the linear regression between genes and clinical features was calculated. GS was used to measure the correlation between gene expression and clinical features of CRC. The average value of GS in each module was defined as module significance (MS). The module with the highest absolute MS value was considered to be the module most significantly associated with the clinical information.
Functional enrichment analysis of key module genes
In order to investigate the function of genes in the selected module, genes of the most meaningful modules, the key modules, were uploaded to the online database DAVID (https://david.ncifcrf.gov/) for annotation. Gene Ontology (GO) and biological process analyses were performed using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis (15–17). False discovery ratio (FDR) <0.05 was considered to be statistically significant.
Identification of hub genes
The key module networks obtained from WGCNA analysis were imported into Cytoscape version 3.7.1 platform (18). Based on the degree of association, the top 30 hub genes of the 2 key module networks were selected as candidate genes for further analysis, verification and visualization.
Validation of hub genes
The limma algorithm of the R package (version 3.5.2) was used to screen the differentially expressed genes (DEGs) between CRC and normal samples in the dataset GSE21510, and the heatmap of DEGs was generated using the ggplot2 function of the R package (19,20). The significant DEGs were identified with the critical value of logFC≥|1.0|, and the adjusted P<0.05. A Venn diagram was drawn (http://bioinformatics.psb.ugent.be/webtools/Venn/) to obtain the overlapping genes in key modules from GSE87211 and DEGs from GSE21510. TCGA data of patients with CRC existing in the Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn) database were used to validate the expression of the hub genes (21). P<0.05 was considered to indicate a statistically significant difference.
Results
Gene selection and hierarchical clustering analysis
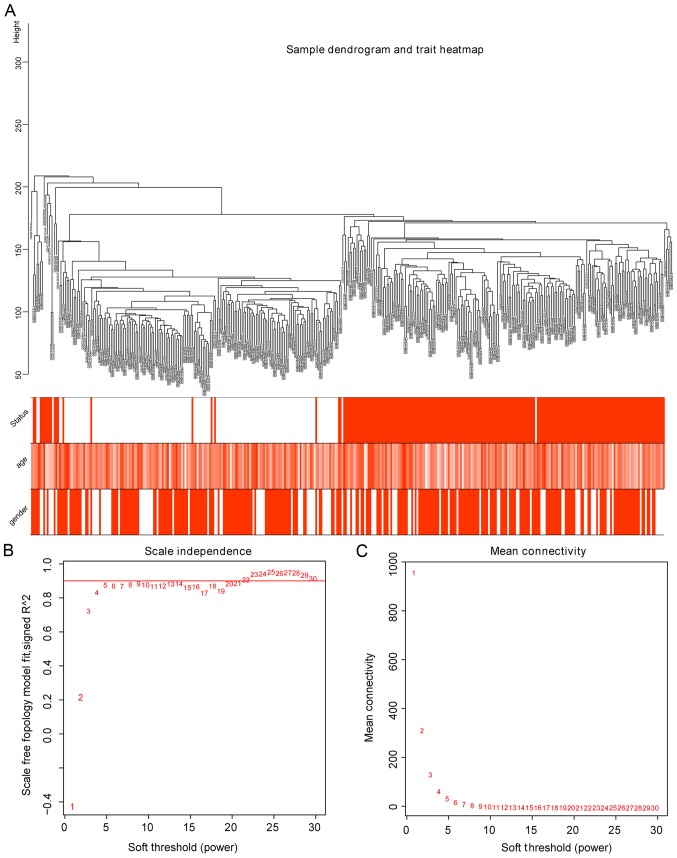
The R software was used to pre-process the original data for background correction and normalization. Probes used in the GEO datasets without corresponding annotation information, or probes matching multiple genes were removed; for genes matched by several probes, the median of that gene expression was selected. The expression profiles of 34,127 genes in 363 samples were obtained from the GSE87211 dataset. WGCNA was constructed by arranging the SDs from large to small, and the top 5,000 genes were selected. In order to further determine whether all samples obtained were suitable for WGCNA network analysis, samples with the clinical characteristics were analyzed. As a result, the 363 samples were divided into 2 clusters in the dendrogram (Fig. 1A). The threshold power (β), a key parameter for WGCNA, affects the independence and average connectivity of the co-expression module. The network topology of β from 1 to 30 was analyzed, and for further analysis, all samples with β=5 were selected, as the lowest power of the scale-free topological fitted an index of R2=0.9 at this point (Fig. 1B).
Figure 1.
Sample Clustering tree and soft threshold power analysis. (A) Sample dendrogram and trait heatmap were based on the GSE87211 dataset, which contained 163 CRC and 133 normal samples. The top 5,000 genes with the highest standard deviation values were used for WGCNA. Color intensity was proportional to disease status, sex and age. (B) Scale independence analysis of soft-thresholding power for the scale-free fit index of network topology. (C) Mean connectivity analysis for various soft-threshold powers. Determination of soft-thresholding power by analyzing network topology for thresholding powers from 1 to 30. WGCNA, weighted gene co-expression network analysis.
Construction and analysis of WGCNA with selected genes in CRC
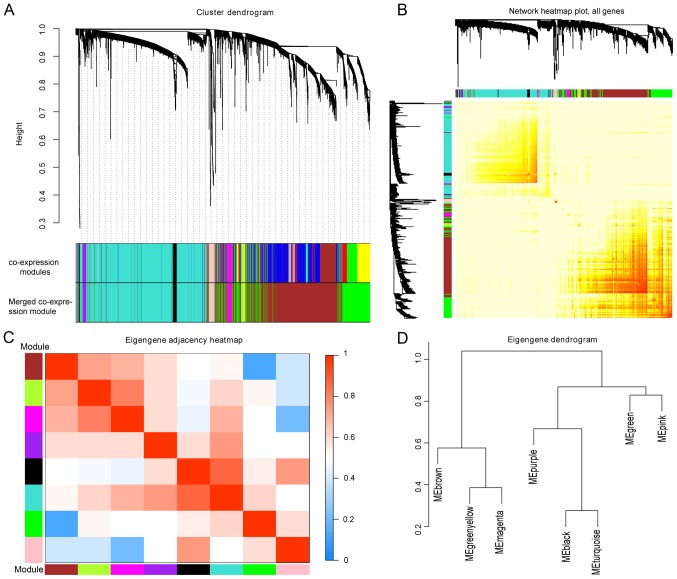
WGCNA was constructed based on computational heterogeneous hierarchical clustering. A total of 15 co-expression modules were generated in the GSE87211 dataset, and 9 merged co-expression modules were obtained by merging similar modules when the MEDissThres was set at 0.25 (Fig. 2A). As the gray module here indicated an unclassifiable eigengene cluster, the remaining 8 modules were selected for further analysis. The network heatmap of the 8 modules was plotted, and the results showed that each module was independent of one another. The modules and gene expression in each module showed a high relative independence level (Fig. 2B); similar results were observed by a heatmap plotted according to adjacencies (Fig. 2C). In addition, eigengenes in the 8 modules were calculated and clustered according to their correlations with each other, and the modules were divided into two groups (Fig. 2D).
Figure 2.
Analysis of co-expressing genes modules. (A) Hierarchical clustering analysis. Each branch was assigned a different color, signifying that different genes belonged to different gene co-expression modules. A total of 9 merged co-expression modules were obtained by merging similar modules when the MEDissThres was set as 0.25. (B) Heatmap of interaction analysis of co-expressing genes. Different colors represent different degrees of overlap. (C) Heatmap plot showing the adjacency in the eigengene. (D) Dendrogram showing the association among 8 co-expression modules.
Identification of key modules
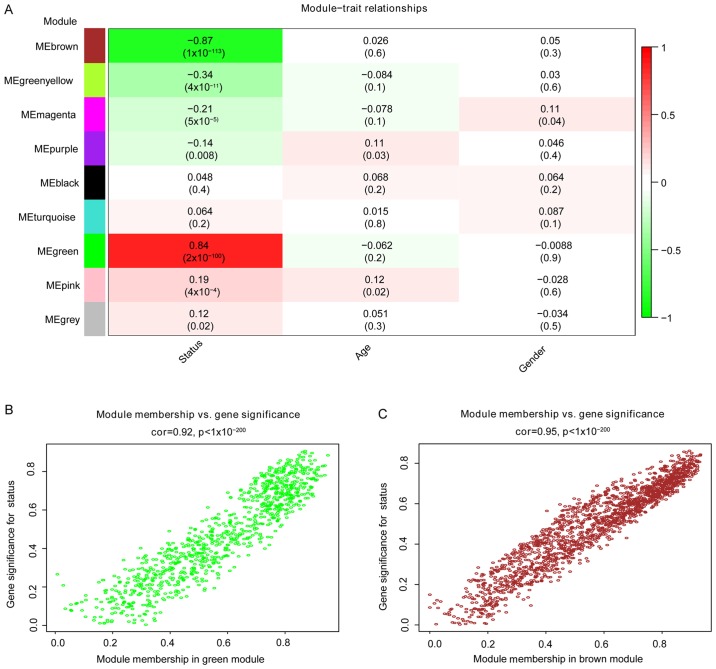
The correlation between modules and disease characteristics was examined, and the eigengene tree and thermogram demonstrated that the green and brown modules were highly correlated with disease status. The green module was identified to be significantly positively correlated with disease status (r=0.84, P=2e-100), while the brown module was markedly negatively correlated with disease status (r=0.87, P=1e-113; Fig. 3A). Therefore, the two key modules were identified as the modules most associated with CRC disease status. The correlations between module members and GS in the green and brown modules are demonstrated by scatter plots in Fig. 3B and C, respectively.
Figure 3.
Correlation of key modules with clinical stage. (A) Heatmap of the correlation between module eigengenes and clinical features of colorectal cancer. Numbers denote correlation (numbers in brackets are P-values). (B) Scatter plot of the correlation between MEgreen membership and gene significance. (C) Scatter plot of the correlation between MEbrown membership and gene significance.
GO enrichment and KEGG pathway analyses of key modules
Using the DAVID functional annotations tool, the GO function and KEGG pathway enrichment of genes in 2 key modules were analyzed, with detailed information listed in Tables I and II. The green module was mainly enriched in collagen catabolic process, cell adhesion, extracellular matrix (ECM) organization, chemotaxis and cell-cell signaling of biological processes, primarily regulation of the ECM, and the PI3K-Akt and Chemokine signaling pathways. These processes and pathways serve a key role in cancer progression, suggesting that genes in the green module participated in the progression of CRC. The enrichment analysis results of the brown module indicated that it was mainly enriched in the chemokine-mediated signaling pathway and negative regulation of growth and steroid metabolism, which were negatively correlated with tumorigenesis.
Table I.
GO enrichment analysis of green module and brown modules (only biological processes).
| Term | Description | Count | % | FDR |
|---|---|---|---|---|
| Green module | ||||
| GO:0030574 | Collagen catabolic process | 25 | 3.205128 | 1.50×10−14 |
| GO:0006954 | Inflammatory response | 55 | 7.051282 | 2.00×10−13 |
| GO:0007155 | Cell adhesion | 58 | 7.435897 | 1.81×10−11 |
| GO:0030198 | Extracellular matrix organization | 35 | 4.487179 | 4.26×10−10 |
| GO:0030593 | Neutrophil chemotaxis | 20 | 2.564103 | 8.23×10−9 |
| GO:0007267 | Cell-cell signaling | 38 | 4.871795 | 9.10×10−9 |
| GO:0030199 | Collagen fibril organization | 16 | 2.051282 | 1.31×10−9 |
| GO:0042060 | Wound healing | 21 | 2.692308 | 3.96×10−8 |
| GO:0070098 | Chemokine-mediated signaling pathway | 19 | 2.435897 | 3.11×10−7 |
| GO:0006935 | Chemotaxis | 24 | 3.076923 | 5.95×10−7 |
| GO:0060326 | Cell chemotaxis | 17 | 2.179487 | 5.16×10−6 |
| GO:0002548 | Monocyte chemotaxis | 13 | 1.666667 | 8.20×10−5 |
| GO:0008284 | Positive regulation of cell proliferation | 45 | 5.769231 | 1.24×10−4 |
| GO:0006955 | Immune response | 42 | 5.384615 | 1.51×10−4 |
| GO:0001501 | Skeletal system development | 22 | 2.820513 | 1.56×10−4 |
| GO:0071346 | Cellular response to interferon-γ | 14 | 1.794872 | 4.19×10−4 |
| GO:0071347 | Cellular response to interleukin-1 | 15 | 1.923077 | 9.83×10−4 |
| GO:0050900 | Leukocyte migration | 19 | 2.435897 | 2.31×10−3 |
| GO:0022617 | Extracellular matrix disassembly | 15 | 1.923077 | 2.34×10−3 |
| GO:0010628 | Positive regulation of gene expression | 29 | 3.717949 | 2.87×10−3 |
| Brown module | ||||
| GO:0007586 | Digestion | 20 | 1.380262 | 3.25×10−5 |
| GO:0034765 | Regulation of ion transmembrane transport | 25 | 1.725328 | 5.43×10−4 |
| GO:0071294 | Cellular response to zinc ion | 10 | 0.690131 | 2.60×10-3 |
| GO:0001764 | Neuron migration | 23 | 1.587302 | 2.94×10−3 |
| GO:0006730 | One-carbon metabolic process | 12 | 0.828157 | 3.87×10−3 |
| GO:1902476 | Chloride transmembrane transport | 21 | 1.449275 | 5.68×10−3 |
| GO:0015701 | Bicarbonate transport | 14 | 0.966184 | 7.57×10−3 |
| GO:0007218 | Neuropeptide signaling pathway | 21 | 1.449275 | 2.13×10−2 |
| GO:0070098 | Chemokine-mediated signaling pathway | 17 | 1.173223 | 2.90×10−2 |
| GO:0045926 | Negative regulation of growth | 9 | 0.621118 | 3.00×10−2 |
| GO:0008202 | Steroid metabolic process | 13 | 0.89717 | 3.38×10−2 |
| GO:0007267 | Cell-cell signaling | 37 | 2.553485 | 3.52×10−2 |
GO, Gene Ontology; FDR, false discovery ratio.
Table II.
KEGG pathway enrichment analysis of green module and brown modules.
| Category | Term | Description | Count | % | FDR |
|---|---|---|---|---|---|
| Green module | |||||
| KEGG | hsa04060 | Cytokine-cytokine receptor interaction | 40 | 5.128205 | 3.32×10−10 |
| KEGG | hsa04974 | Protein digestion and absorption | 22 | 2.820513 | 7.29×10−8 |
| KEGG | hsa04512 | ECM-receptor interaction | 18 | 2.307692 | 1.27×10−4 |
| KEGG | hsa05146 | Amoebiasis | 19 | 2.435897 | 4.96×10−4 |
| KEGG | hsa05323 | Rheumatoid arthritis | 16 | 2.051282 | 4.50×10−3 |
| KEGG | hsa04151 | PI3K-Akt signaling pathway | 33 | 4.230769 | 2.73×10−2 |
| KEGG | hsa04062 | Chemokine signaling pathway | 22 | 2.820513 | 4.61×10−2 |
| KEGG | hsa04668 | TNF signaling pathway | 14 | 1.794872 | 7.10×10−1 |
| KEGG | hsa04310 | Wnt signaling pathway | 16 | 2.05128 | 9.10×10−1 |
| Brown module | |||||
| KEGG | hsa00830 | Retinol metabolism | 24 | 1.656315 | 1.39×10−8 |
| KEGG | hsa05204 | Chemical carcinogenesis | 23 | 1.587302 | 1.30×10−5 |
| KEGG | hsa00982 | Drug metabolism-cytochrome P450 | 20 | 1.380262 | 1.02×10−4 |
| KEGG | hsa00910 | Nitrogen metabolism | 9 | 0.621118 | 8.46×10−3 |
| KEGG | hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 18 | 1.242236 | 9.57×10−3 |
| KEGG | hsa00140 | Steroid hormone biosynthesis | 15 | 1.035197 | 3.38×10−2 |
| KEGG | hsa04978 | Mineral absorption | 13 | 0.89717 | 3.44×10−2 |
FDR, false discovery ratio; KEGG, Kyoto Encyclopedia of Genes and Genomes; ECM, extracellular matrix; TNF, tumor necrosis factor.
Validation of hub genes
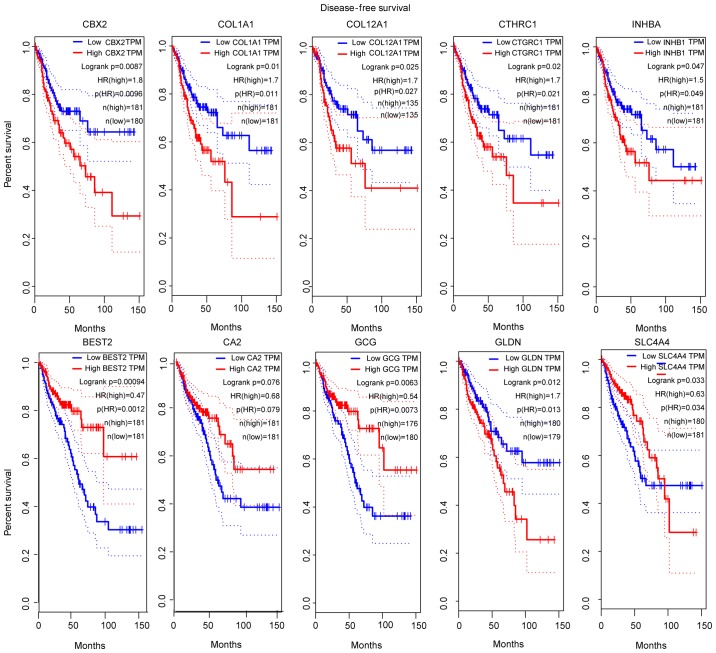
The gene network of 2 key modules was imported into Cytoscape and the scores of all genes were calculated by 11 different methods. Finally, the first 30 genes from each module were screened according to the degree of association among genes for further analysis. Survival analysis of patients with CRC in GEPIA, depending on the expression level of the selected genes, showed statistical significance; and the top 5 significant genes for survival analysis in each module were considered to be hub genes. Survival analysis of the hub genes was performed in GEPIA patients with CRC, the collagen type I α1 chain (COL1A1), collagen type XII α1 chain (COL12A1), collagen triple helix repeat containing 1 (CTHRC1), inhibin subunit βa (INHBA) and chromobox 2 (CBX2) genes were obtained from the green module; and the bestrophin 2 (BEST2), carbonic anhydrase 2 (CA2), glucagon (GCG), solute carrier family 4 member 4 (SLC4A4) and gliomedin (GLDN) genes were obtained from the brown module (Fig. 4).
Figure 4.
Survival analysis of 10 hub genes according to data from the TCGA database. P<0.05 was considered to indicate a statistically significant difference. COL1A1, collagen type I α1 chain; COL12A1, collagen type XII α1 chain; CTHRC1, collagen triple helix repeat containing 1; INHBA, inhibin subunit βa; CBX2, chromobox 2; BEST2, bestrophin 2; CA2, carbonic anhydrase 2; GCG, glucagon; SLC4A4, solute carrier family 4 member 4; GLDN, gliomedin.
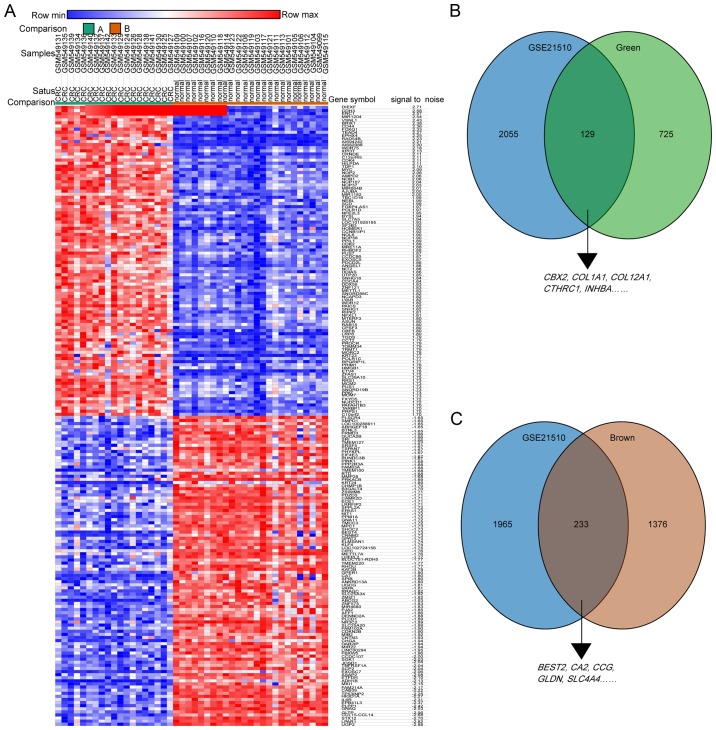
Next, the limma R package with a logFC≥|1.0| as the cutoff was used to screen for DEGs in another GEO dataset, GSE21510; the heatmap of the DEGs for this dataset is presented in Fig. 5A. The DEGs from GSE21510 and genes of the green modules or brown modules from GSE87211 were overlapped by Venn diagram. The results demonstrated that the 10 hub genes were validated in the Venn diagrams (Fig. 5B and C).
Figure 5.
Hub genes of GSE87211 were validated in the GSE21510 dataset. (A) Heatmap hierarchical clustering exhibiting DEGs between colorectal cancer and control groups in the GSE21510 dataset. (B) Overlapping genes between DEGs from the GSE21510 dataset and the green module from the GSE87211 dataset. (C) Overlapping genes between the DEGs from the GSE21510 dataset and the brown module from the GSE87211 dataset. DEGs, differentially expressed genes. COL1A1, collagen type I α1 chain; COL12A1, collagen type XII α1 chain; CTHRC1, collagen triple helix repeat containing 1; INHBA, inhibin subunit βa; CBX2, chromobox 2; BEST2, bestrophin 2; CA2, carbonic anhydrase 2; GCG, glucagon; SLC4A4, solute carrier family 4 member 4; GLDN, gliomedin.
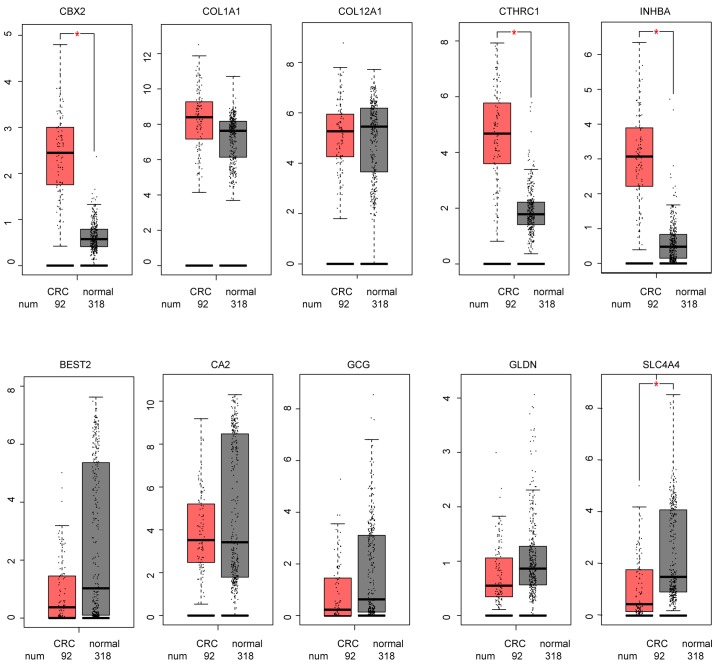
To further validate the hub genes, GEPIA analysis was conducted in TCGA data from patients with CRC. The results demonstrated that the expression levels of the 5 hub genes in the green module were all increased in CRC tissues, while those in the brown module were all decreased (Fig. 6).
Figure 6.
Hub genes expression of The Cancer Genome Atlas CRC data in GEPIA. *P<0.005. GEPIA, Gene Expression Profiling Interactive Analysis; CRC, colorectal cancer; COL1A1, collagen type I α1 chain; COL12A1, collagen type XII α1 chain; CTHRC1, collagen triple helix repeat containing 1; INHBA, inhibin subunit βa; CBX2, chromobox 2; BEST2, bestrophin 2; CA2, carbonic anhydrase 2; GCG, glucagon; SLC4A4, solute carrier family 4 member 4; GLDN, gliomedin.
Discussion
With the exception of patients with a family history, the majority of cases of CRC are sporadic (22). Although the mortality of this malignancy has been markedly decreased following the introduction of routine examinations, its incidence remains high (23). The development of innovative methods for the early diagnosis and treatment of CRC is essential (24). Multiple molecular studies have indicated that complex mechanisms are involved in CRC pathology; however, studies into the molecular mechanism commonly focus on individual signaling pathways in their attempt to determine the mechanism of CRC (25–27). WGCNA can be used to determine the expression of multiple genes in large sample datasets; disease analysis using WGCNA can ensure the investigation examines multiple signaling pathways and decreases the likelihood of excluding factors within the complex pathological mechanism (28).
In the present study, WGCNA was conducted in the GEO dataset GSE87211, and 8 independent modules with classifiable eigengenes were revealed. Following analyses of the correlations between modules and disease status, the green module was identified as the gene cluster most positively correlated with CRC status, and the brown module as the most negatively correlated.
The green module genes were more enriched in collagen catabolic process, cell adhesion, ECM organization, chemotaxis and cell-cell signaling, and participated primarily in the regulation of the ECM, and the PI3K-Akt and Chemokine signaling pathways. By contrast, the brown module was negatively correlated with CRC. Pathway analysis demonstrated that the brown module contained genes that were involved in the regulation of the chemokine-mediated signaling pathway, and negative regulation of growth and steroid metabolic process, which oppose cancer tissue expansion. CRC is an inflammation-associated type of cancer, and the chemokine expression pattern in CRC is similar to the cellular immune response involved in lymphocyte recirculation and the directed migration of leukocytes into mucosal tissues, which was consistent with the present results (29). A previous study has revealed that collective cell invasion of CRC tissue depends on cell-intrinsic mechanisms, but recently more evidence has indicated that it also depends on extracellular mechanisms involving bidirectional interplay between the tumor cell and the tumor environment, such as the ECM (30). The results of the present study emphasized the importance of the ECM in cancer. In addition, the Wnt signaling pathway has been widely studied as an important CRC pathway; however, its clinical application has a limited effect, as the pathogenesis of CRC involves other signaling pathways as well (9). The FDR value of tumor necrosis factor and Wnt signaling pathways in KEGG analysis of green module was not statistically significant. This result indicated that the popular pathway in CRC, Wnt signaling pathway, was unable to explain the pathological mechanism on its own. According to screening genes of Cytoscape platform and survival analysis in GEPIA, a total of 5 hub genes were identified in the green module, including COL1A1, COL12A1, CTHRC1, INHBA and CBX2, and 5 hub genes in the brown module, including BEST2, CA2, GCG, SLC4A4 and GLDN. Survival analysis indicated that the expression of all 10 hub genes was significantly associated with the survival of patients with CRC. The increased expression level of hub genes from the green module and decreased expression level of hub genes from the brown module in CRC were both verified by TCGA CRC data.
In light of previous studies, the correlation among these 7/10 hub genes and CRC have been explored. The ECM, the PI3K-Akt pathway and nitric oxide pathway served important roles in tumor initiation, invasion and progression (31,32). COL1A1 and CA2 genes contribute to ECM and the PI3K-Akt pathways, and have been reported to be associated with CRC metastasis (33). The CA2 gene encodes a member of the carbonic anhydrase family, which is significantly downregulated in the majority of colorectal tumors and associated with patient survival (34). The CA2 gene has been demonstrated to be involved in the nitrogen metabolism pathway and associated with lymph node metastasis in endometrial adenocarcinoma, and lymph node metastasis in gastric cancer and regulation of the pH regulatory system (35). Similarly, it has been reported that CA2 is associated with metastasis in CRC (36). COL1A1 and its homologous gene COL12A1 were revealed to be involved in ECM organization, and were identified be significantly upregulated in patients with CRC, which was consistent with previous studies (37,38). The CTHRC1 gene encodes an ECM-associated protein involved in extracellular space and proteinaceous ECM, which may contribute to tissue repair by limiting collagen matrix deposition and promoting cell migration (39). The SLC4A4 gene affects intracellular pH, which can regulate tumor progression in the hypoxic and acidic tumor environment (40). As for GLDN, also known as CRG-L2, it may serve an important role in extracellular structure or intercellular signaling, and it has been associated with CRC prognosis (41). The GCG gene is hypothesized to be involve in the regulation of incretin synthesis, secretion, inactivation and RET signaling, while some studies have identified that GCG is downregulated in both adenomas and CRC tissues (42,43).
Of note, a total of 3 hub genes (INHBA and CBX2 from the green module, BEST2 from the brown module) were identified in the present study, which had not been previously reported to be involved in CRC pathology, to the best of our knowledge. The INHBA protein belongs to the transforming growth factor β superfamily, which is associated with several types of human cancer (44). It has also been reported that high expression of INHBA gene in CRC may lead to poor survival (45). With regard to the CBX2 gene, to the best of our knowledge, there have been no studies on its association with CRC at present, although a growing body of evidence has suggested that CBX2 is overexpressed in breast cancer and advanced prostate cancer (46). The BEST2 gene, is a member of the bestrophin gene family of anion channels, which is mainly expressed in the retinal pigment epithelium and colon (47). To the best of our knowledge, there is no published research on the BEST2 gene in CRC at present.
The present study explored the potential pathogenic genes of CRC using data mining and data analysis, rather than focusing on a single signaling pathway. Multiple biological processes were identified to be involved with CRC progression, and the obtained hub genes provide a reference point for future studies. The present study reaffirmed the role of ECM, and the PI3K-Akt and chemokine signaling pathways in the development of CRC. To the best of our knowledge, few studies have reported the role of the INHBA, CBX2 and BEST2 genes in CRC; the present study highlighted these 3 genes as candidates for research of the molecular mechanism of CRC. To compensate for the limitations of a single dataset, another dataset was used for cross-validation, and the results were also validated in another public database. To validate the present results, analysis based on enlarged sample size and molecular research is under preparation. Further investigation of the molecular mechanism of the identified hub genes in CRC is recommended.
Acknowledgements
Not applicable.
Funding
This work was supported by the Natural Science Foundation of Zhejiang Province, China (grant no. LQ18H110001) and the National Natural Science Foundation of China (grant no. 81802771).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors contributions
LC and LW designed the project. LQ contributed to data analysis and prepared the main manuscript. JZ and NS were involved in revising the manuscript critically and analyzing the data. All authors reviewed the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 2.Janz T, Lu K, Povlow MR, Urso B. A review of colorectal cancer detection modalities, stool DNA, and fecal immunochemistry testing in adults over the age of 50. Cureus. 2016;8:e931. doi: 10.7759/cureus.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Advani S, Kopetz S. Ongoing and future directions in the management of metastatic colorectal cancer: Update on clinical trials. J Surg Oncol. 2019;119:642–652. doi: 10.1002/jso.25441. [DOI] [PubMed] [Google Scholar]
- 4.Muller MF, Ibrahim AE, Arends MJ. Molecular pathological classification of colorectal cancer. Virchows Arch. 2016;469:125–134. doi: 10.1007/s00428-016-1956-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marmol I, Sanchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: A general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18:E197. doi: 10.3390/ijms18010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138:2059–2072. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng X, Xu X, Chen D, Zhao F, Wang W. Therapeutic potential of targeting the Wnt/β-catenin signaling pathway in colorectal cancer. Biomed Pharmacother. 2019;110:473–481. doi: 10.1016/j.biopha.2018.11.082. [DOI] [PubMed] [Google Scholar]
- 8.Aghabozorgi AS, Bahreyni A, Soleimani A, Bahrami A, Khazaei M, Ferns GA, Avan A, Hassanian SM. Role of adenomatous polyposis coli (APC) gene mutations in the pathogenesis of colorectal cancer; current status and perspectives. Biochimie. 2019;157:64–71. doi: 10.1016/j.biochi.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Shibayama Y, Zhang A, Ohsaki H, Asano E, Suzuki Y, Kushida Y, Kobara H, Masaki T, Wang Z, Nishiyama A. (Pro)renin receptor promotes colorectal cancer through the Wnt/beta-catenin signalling pathway despite constitutive pathway component mutations. Br J Cancer. 2019;120:229–237. doi: 10.1038/s41416-018-0350-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu M, Liu X, Xu Y, Zhu S, Gao Y. Co-expression of Axin and APC gene fragments inhibits colorectal cancer cell growth via regulation of the Wnt signaling pathway. Mol Med Rep. 2017;16:3783–3790. doi: 10.3892/mmr.2017.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su YH, Tang WC, Cheng YW, Sia P, Huang CC, Lee YC, Jiang HY, Wu MH, Lai IL, Lee JW, Lee KH. Targeting of multiple oncogenic signaling pathways by Hsp90 inhibitor alone or in combination with berberine for treatment of colorectal cancer. Biochim Biophys Acta. 2015;1853:2261–2272. doi: 10.1016/j.bbamcr.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 12.Hu Y, Gaedcke J, Emons G, Beissbarth T, Grade M, Jo P, Yeager M, Chanock SJ, Wolff H, Camps J, et al. Colorectal cancer susceptibility loci as predictive markers of rectal cancer prognosis after surgery. Genes Chromosomes Cancer. 2018;57:140–149. doi: 10.1002/gcc.22512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsukamoto S, Ishikawa T, Iida S, Ishiguro M, Mogushi K, Mizushima H, Uetake H, Tanaka H, Sugihara K. Clinical significance of osteoprotegerin expression in human colorectal cancer. Clin Cancer Res. 2011;17:2444–2450. doi: 10.1158/1078-0432.CCR-10-2884. [DOI] [PubMed] [Google Scholar]
- 14.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Gene Ontology Consortium, corp-author. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Lim ma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wickham H. Springer International Publishing; New York, NY: 2016. ggplot2: Elegant Graphics for Data Analysis. ISBN 978-3-319-24277-4. [Google Scholar]
- 21.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer J, Walker LC, Robinson BA, Frizelle FA, Church JM, Eglinton TW. Clinical implications of the genetics of sporadic colorectal cancer. ANZ J Surg. 2019;89:1224–1229. doi: 10.1111/ans.15074. [DOI] [PubMed] [Google Scholar]
- 23.Maida M, Macaluso FS, Ianiro G, Mangiola F, Sinagra E, Hold G, Maida C, Cammarota G, Gasbarrini A, Scarpulla G. Screening of colorectal cancer: Present and future. Expert Rev Anticancer Ther. 2017;17:1131–1146. doi: 10.1080/14737140.2017.1392243. [DOI] [PubMed] [Google Scholar]
- 24.Willauer AN, Liu Y, Pereira AAL, Lam M, Morris JS, Raghav KPS, Morris VK, Menter D, Broaddus R, Meric-Bernstam F, et al. Clinical and molecular characterization of early-onset colorectal cancer. Cancer. 2019;125:2002–2010. doi: 10.1002/cncr.31994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pancione M, Remo A, Colantuoni V. Genetic and epigenetic events generate multiple pathways in colorectal cancer progression. Patholog Res Int. 2012;2012:509348. doi: 10.1155/2012/509348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagland HR, Berg M, Jolma IW, Carlsen A, Soreide K. Molecular pathways and cellular metabolism in colorectal cancer. Dig Surg. 2013;30:12–25. doi: 10.1159/000347166. [DOI] [PubMed] [Google Scholar]
- 27.Wang W, Kandimalla R, Huang H, Zhu L, Li Y, Gao F, Goel A, Wang X. Molecular subtyping of colorectal cancer: Recent progress, new challenges and emerging opportunities. Semin Cancer Biol. 2019;55:37–52. doi: 10.1016/j.semcancer.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W, Li L, Ye H, Tu W. Weighted gene co-expression network analysis in biomedicine research. Sheng Wu Gong Cheng Xue Bao. 2017;33:1791–1801. doi: 10.13345/j.cjb.170006. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 29.Roy I, Veldkamp CT, Volkman BF, Dwinell MB. Chemokines in colitis: MicroRNA control. Gut. 2014;63:1202–1204. doi: 10.1136/gutjnl-2013-305886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu JS, Sheng SR, Liang XH, Tang YL. The role of tumor microenvironment in collective tumor cell invasion. Future Oncol. 2017;13:991–1002. doi: 10.2217/fon-2016-0501. [DOI] [PubMed] [Google Scholar]
- 31.Mandal P. Insight of nitric oxide signaling: A potential biomarker with multifaceted complex mechanism in colorectal carcinogenesis. Biochem Biophys Res Commun. 2018;495:1766–1768. doi: 10.1016/j.bbrc.2017.12.051. [DOI] [PubMed] [Google Scholar]
- 32.Wu X, Cai J, Zuo Z, Li J. Collagen facilitates the colorectal cancer stemness and metastasis through an integrin/PI3K/AKT/Snail signaling pathway. Biomed Pharmacother. 2019;114:108708. doi: 10.1016/j.biopha.2019.108708. [DOI] [PubMed] [Google Scholar]
- 33.Qi L, Ding Y. Construction of key signal regulatory network in metastatic colorectal cancer. Oncotarget. 2018;9:6086–6094. doi: 10.18632/oncotarget.23710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viikila P, Kivela AJ, Mustonen H, Koskensalo S, Waheed A, Sly WS, Pastorek J, Pastorekova S, Parkkila S, Haglund C. Carbonic anhydrase enzymes II, VII, IX and XII in colorectal carcinomas. World J Gastroenterol. 2016;22:8168–8177. doi: 10.3748/wjg.v22.i36.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDonald PC, Winum JY, Supuran CT, Dedhar S. Recent developments in targeting carbonic anhydrase IX for cancer therapeutics. Oncotarget. 2012;3:84–97. doi: 10.18632/oncotarget.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi C, Hong L, Cheng Z, Yin Q. Identification of metastasis-associated genes in colorectal cancer using metaDE and survival analysis. Oncol Lett. 2016;11:568–574. doi: 10.3892/ol.2015.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou X, Feng B, Dong T, Yan G, Tan B, Shen H, Huang A, Zhang X, Zhang M, Yang P, et al. Up-regulation of type I collagen during tumorigenesis of colorectal cancer revealed by quantitative proteomic analysis. J Proteomics. 2013;94:473–485. doi: 10.1016/j.jprot.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 38.Mikula M, Rubel T, Karczmarski J, Goryca K, Dadlez M, Ostrowski J. Integrating proteomic and transcriptomic high-throughput surveys for search of new biomarkers of colon tumors. Funct Integr Genomics. 2011;11:215–224. doi: 10.1007/s10142-010-0200-5. [DOI] [PubMed] [Google Scholar]
- 39.Yang XM, You HY, Li Q, Ma H, Wang YH, Zhang YL, Zhu L, Nie HZ, Qin WX, Zhang ZG, Li J. CTHRC1 promotes human colorectal cancer cell proliferation and invasiveness by activating Wnt/PCP signaling. Int J Clin Exp Pathol. 2015;8:12793–12801. [PMC free article] [PubMed] [Google Scholar]
- 40.Parks SK, Pouyssegur J. The Na(+)/HCO3(−) co-transporter SLC4A4 plays a role in growth and migration of colon and breast cancer cells. J Cell Physiol. 2015;230:1954–1963. doi: 10.1002/jcp.24930. [DOI] [PubMed] [Google Scholar]
- 41.Chen L, Lu D, Sun K, Xu Y, Hu P, Li X, Xu F. Identification of biomarkers associated with diagnosis and prognosis of colorectal cancer patients based on integrated bioinformatics analysis. Gene. 2019;692:119–125. doi: 10.1016/j.gene.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 42.Spisak S, Kalmar A, Galamb O, Wichmann B, Sipos F, Peterfia B, Csabai I, Kovalszky I, Semsey S, Tulassay Z, Molnár B. Genome-wide screening of genes regulated by DNA methylation in colon cancer development. PLoS One. 2012;7:e46215. doi: 10.1371/journal.pone.0046215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Z, Liu Z, Ge W, Shou J, You L, Pan H, Han W. Analysis of potential genes and pathways associated with the colorectal normal mucosa-adenoma-carcinoma sequence. Cancer Med. 2018;7:2555–2566. doi: 10.1002/cam4.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okano M, Yamamoto H, Ohkuma H, Kano Y, Kim H, Nishikawa S, Konno M, Kawamoto K, Haraguchi N, Takemasa I, et al. Significance of INHBA expression in human colorectal cancer. Oncol Rep. 2013;30:2903–2908. doi: 10.3892/or.2013.2761. [DOI] [PubMed] [Google Scholar]
- 45.Yokota M, Kojima M, Higuchi Y, Nishizawa Y, Kobayashi A, Ito M, Saito N, Ochiai A. Gene expression profile in the activation of subperitoneal fibroblasts reflects prognosis of patients with colon cancer. Int J Cancer. 2016;138:1422–1431. doi: 10.1002/ijc.29851. [DOI] [PubMed] [Google Scholar]
- 46.Clermont PL, Crea F, Chiang YT, Lin D, Zhang A, Wang JZ, Parolia A, Wu R, Xue H, Wang Y, et al. Identification of the epigenetic reader CBX2 as a potential drug target in advanced prostate cancer. Clin Epigenetics. 2016;8:16. doi: 10.1186/s13148-016-0182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu K, Lujan R, Marmorstein A, Gabriel S, Hartzell HC. Bestrophin-2 mediates bicarbonate transport by goblet cells in mouse colon. J Clin Invest. 2010;120:1722–1735. doi: 10.1172/JCI41129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.