Abstract
Ultrasonic microbubbles in combination with microRNA (miRNAs/miRs) exhibited promising effects on cancer treatments. The aim was to investigate the role of miR-378 in hepatoma cells and the efficiency of it in combination with ultrasonic irradiation and SonoVue® microbubbles method for cell transfection. HuH-7, Hep3B and SK-Hep1 cells were transfected with an miR-378 mimic using only Lipofectamine® 3000 or combined with SonoVue microbubbles and ultrasonic irradiation at 0.5 W/cm2 for 30 sec. mRNAs and protein levels of Cyclin D1, Bcl-2, Bax, Akt, p53 and Survivin were detected by reverse transcription-quantitative PCR and western blotting, respectively. Cell survival rate, proliferation, cell cycle and apoptosis were determined by Cell Counting Kit-8, cell double cytochemical staining and flow cytometry, respectively. It was found that using a combination of ultrasonic irradiation and the SonoVue microbubbles method increased the effectiveness of miR-378 transfection into hepatocellular carcinoma (HCC) cells, and increased the inhibition of cell survival and proliferation. Moreover, miR-378 increased the rate of apoptosis and upregulated the expression of Bax and p53, and suppressed the cell cycle and downregulated the expression of Cyclin D1, Bcl-2, Akt, β-catenin and Survivin much more effectively in the HCC cell line by applying the combined method. Thus, miR-378 was shown to be a suppressive factor to reduce proliferation and increase apoptosis in HCC cells. Additionally, the combination of ultrasonic irradiation and SonoVue microbubbles method was more efficient in the transfection of miRNA.
Keywords: miRNA, hepatoma, ultrasonic irradiation, microbubbles
Introduction
The pathogenesis of hepatocellular carcinoma (HCC) has not yet been fully clarified and there is still a lack of targeted therapies for HCC (1,2). For patients with the disease, radical resection of HCC is the main treatment in its early stages, but >50% of post-operative patients show metastasis and recurrence within 5 years (1,2). Targeted drugs for HCC are emerging, providing possible new therapies to prevent and cure malignant cancer (3). However, its therapeutic effects are still not satisfactory, due to the side effects it has on normal cells (4). Thus, the current treatments for HCC still face several challenges.
Studies conducted to investigate HCC genes at molecular level indicate that a number of genes and proteins are closely related to the malignant characteristics of HCC staging, recurrence and metastasis, and these molecules may play important roles in the occurrence and development of HCC (5,6). Thousands of microRNAs (miRNAs/miRs) exist in the human genome and each miRNA can directly regulate ~200 target genes, and nearly 33% of the protein-coding genes in humans are controlled by miRNAs (5–7). Therefore, miRNAs occupy an indispensable position in the spectrum of human gene, thus, studying gene therapy for the treatment of hepatoma from the aspect of miRNA regulation has great potential.
Continuous research and development of ultrasound molecular imaging and biomedical engineering, especially development of targeted contrast microbubbles carrying various drugs and genes, have laid a solid foundation for building up ultrasonic microbubble technology for therapeutic use (8–11). Ultrasonic microbubbles are microbubbles composed of a core gas surrounded by a shell membrane with the size of 2–8 µm or even smaller (12). As lipids (represented by SonoVue®) have higher efficiency, fewer side effects and high stability, it has become the most widely used microbubbles in most studies (12–15). The mechanism of ultrasound-mediated targeted delivery mainly relies on cavitation and sonication effects of ultrasonic microbubbles generated under the irradiation of ultrasonic field strength (16). Ultrasound-targeted microbubbles show various modifications in their shell membranes and they can carry a variety of drugs or genes and can bind to specific antigens or genes expressed by specific cells in the body, thus providing the possibility of ultrasound-mediated targeted treatment (17).
Currently, the delivery of nucleic acid via microbubbles and ultrasound method has attracted much attention due to the discovery of the potentials of miRNAs in modulating target genes (18). In view of these valuable prospects, the present study aimed to investigate the role of miR-378 in hepatoma cells and the efficiency of combining ultrasonic irradiation and SonoVue microbubbles for cell transfection.
Materials and methods
Cell incubation
HuH-7 cell line was purchased from the Japanese Collection of Research Bioresources Cell Bank, while Hep3B cell line was purchased from American Type Culture Collection and SK-Hep1 cell line was purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. All the cell lines were cultured in DMEM (D0819; Sigma-Aldrich; Merck KGaA) containing 10% FBS (F8192; Sigma-Aldrich; Merck KGaA) and penicillin-streptomycin reagent (V900929; Sigma-Aldrich; Merck KGaA) in 5% CO2 at 37°C.
Grouping and transfection
In order to explore the role of miR-378 and the efficiency of the combined method for cell transfection, the cells were grouped as Blank, miR-378 control, L group (HCC cells transfected with miR-378) and LUS groups (HCC cells treated with miR-378 mimic combined with ultrasonic irradiation and SonoVue microbubbles). Briefly, HuH-7, Hep3B and SK-Hep1 cells at 1×106 cells/ml in the LUS or miR-378 control groups were plated in a 96-well plate and then respectively transfected with 100 nmol/l miR-378 mimic (Shanghai GenePharma Co., Ltd.), or miR-378 mimic control vector (Shanghai GenePharma Co., Ltd.) in a mixture with Lipofectamine® 3000 (L3000015; Thermo Fisher Scientific, Inc.) and 2.5 µg/µl SonoVue microbubbles (Bracco Suisse SA) under the irradiation of ultrasonic transfer apparatus via ultrasound couplant (Anhui Deepblue Medical Technology Co., Ltd.) at the parameters of 0.5 W/cm2 for 30 sec. The cells in the L group were transfected with miR-378 mimic using Lipofectamine 3000 only, while those in the Blank group were treated with medium only. All the cells were cultured for another 72 h after the transfection (19). The sequence of miR-378 mimic was 5′-AGGCUCUGACUCCAGGUCC-3′; The sequence of miR-378 mimic control was 5′-UUCUCCGAACGUGUCACGUTT-3′.
Reverse transcription-quantitative (RT-q)PCR
Total RNAs from HuH-7, Hep3B and SK-Hep1 cells at 1×106 cells/ml were obtained using TRIzol reagent (15596018; Thermo Fisher Scientific, Inc.) and further reverse-transcribed into cDNAs following the instructions of PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.), with the conditions for reverse transcription being: 30°C for 60 min; 30°C for 60 min; and 95°C for 60 min. The cells were then cultured for 72 h following the treatments for HuH-7 cells. A total of 0.5 µl forward primer, 0.5 µl reverse primer, 3 µl cDNA template, 5 µl 2X SYBR Green master mix (4913850001; Roche Diagnostics) and 1 µl ddH2O were mixed together and reacted for 40 cycles in the following conditions: Initial denaturation at 95°C for 60 sec, at 95°C for 20 sec, at 65°C for 30 sec, and at 72°C for 40 sec in Bio-Rad IQ5 thermocycler (Bio-Rad Laboratories, Inc.). The sequences of primers used are listed in Table I. Relative expression of miR-378, Cyclin D1, Bcl-2, Bax, Akt, p53 and Survivin were normalized to that of U6 by 2−ΔΔCq method (20).
Table I.
Primers used in the study.
| Primer name | Sequence (5′-3′) |
|---|---|
| miR-378 | F: CCTGACTCCAGGTCCT |
| R: GAACATGTCTGCGTATCTC | |
| Cyclin D1 | F: GTCTTCCCGCTGGCCATGAACTAC |
| R: GGAAGCGTGTGAGGCGGTAGTAGG | |
| Bcl-2 | F: GCCTTCTTTGAGTTCGGTG |
| R: CAGAGACAGCCAGGAGAAATC | |
| Bax | F: GCAAACTGGTGCTCAAGG |
| R: CGCCACAAAGATGGTCAC | |
| Akt | F: TGGACTACCTGCACTCGGAGAA |
| R: GTGCCGCAAAAGGTCTTCATGG | |
| p53 | F: TAAAAGATGTTTTGAATG |
| R: ATGTGTGTGATGTTGTAGATG | |
| Survivin | F: CCACTGAGAACGAGCCAGACTT |
| R: GTATTACAGGCGTAAGCCACCG | |
| U6 | F: GCTTCGGCAGCACATATACTAAAAT |
| R: GAAGATGGTGATGGGATTTC |
miR, microRNA.
Cell Counting Kit (CCK)-8
After 72 h, relative cell survival rates of HuH-7, Hep3B and SK-Hep1 cells were detected by CCK-8 following the manufacturer's protocols (96992-100TESTS-F; Sigma-Aldrich; Merck KGaA). Briefly, 1×106 cells were collected and tested after the transfection of miR-378 mimic. Optic density (OD) values were read on a microplate reader (Multiskan; Thermo Fisher Scientific, Inc.) at 450 nm and relative cell survival rates of cells were calculated according to the standard curve of OD. The experiment was conducted in triplicate.
Double cytochemical staining
Cells (~4×103−1×106) were cultured in 96-well plates. EdU solution (A10044; Thermo Fisher Scientific, Inc.) was diluted by culture medium at 1,000:1 and 100 µl of 50 µm EdU medium was added to each well and maintained for 2 h. The cells were washed once or twice using PBS for 5 min. Then, 100 µl cell fixative (PBS containing 4% paraformaldehyde) was added to each well and maintained for 30 min at room temperature. A total of 2 mg/ml glycine was added to each well for decolorization and then further incubated for 5 min on a shaker. PBS (100 µl) was added to each well for, held for 5 min and then 100 µl of penetrant (0.5% Triton X-100 in PBS) was added to each well for 10 min. After rinsing in PBS, 1X Hoechst 33342 reaction solution (H3570; Thermo Fisher Scientific, Inc.) was diluted by deionized water at 100:1 and added to each well for a 30 min incubation at room temperature in the dark. The staining result was immediately observed by using a fluorescence microscope (BX53T, Olympus Corporation; magnification, ×100) after washing the cells with PBS one to three times.
Evaluation of apoptosis
Briefly, 5×105 cells were resuspended in 1 ml cold PBS and collected into 100 ml binding buffer using an Annexin V-FITC kit (Sigma-Aldrich; Merck KGaA). A total of 10 µl FITC-labeled Annexin V and 5 µl propidium iodide were added to the cells for 20 min at room temperature. Cell apoptosis was detected using flow cytometry (BD FACSVerse Z200; FCAP Array software v3.0, Becton, Dickinson and Company) after the cells were mixed with 200 µl binding buffer and washed.
Determination of cell cycle
Followed by the detection of apoptosis rate, 1×106 cells were gathered and digested by 0.25% trypsin to determine the cell cycle in each phase. After cell transfection, Vybrant™ DyeCycle™ Violet Stain (Thermo Fisher Scientific, Inc.) was used for flow cytometry (BD FACSVerse Z200; FCAP Array software v3.0, Becton, Dickinson and Company) to determine the phases in the cell cycle after incubation at 4°C for 72 h.
Western blotting
Protein expression of Cyclin D1, Bcl-2, Bax, Akt, p53 and Survivin were detected 72 h later. Cells (1×106) were obtained and lysed in order to extract total proteins using a mixture of RIPA lysate (R0278; Sigma-Aldrich; Merck KGaA) with protease inhibitor (S8830; Sigma-Aldrich; Merck KGaA). The total proteins (50 µg per lane) from each sample was separated on 10% SDS-PAGE at 120 V for ~1.5 h. ReBlot Plus kit (2500; Sigma-Aldrich; Merck KGaA) was used to strip antibodies. In brief, 1X Antibody Solution was added to the protein suspension and incubated at room temperature for 15 min, and then transferred to a PVDF membrane, which was then blocked by 5% milk (non-fat) at room temperature for 1 h. Next, primary antibodies against Cyclin D1 (cat. no. ab16663), Bcl-2 (cat. no. ab59348), Bax (cat. no. ab32503), Akt (cat. no. ab8805), p53 (cat. no. ab26), Survivin (cat. no. ab469), β-catenin (cat. no. ab32572) and GAPDH (cat. no. ab8245; all purchased from Abcam) were diluted to 1:1,000 by 5% milk (non-fat) and used to incubate with the PVDF membrane at 4°C overnight. The membrane was then probed with a goat anti-rabbit horseradish peroxidase conjugated-secondary antibody (1:2,000; cat. no. ab6721; Abcam) at room temperature for 1 and washed by PBST (PBS with 0.1% Tween). A SignalFire™ ECL reagent (cat. no. 6883; Cell Signaling Technology, Inc.) was used for the detection of proteins. Image Lab™ Software (version 3.0) was used for densitometric analysis and quantification of the western blot data (Bio-Rad Laboratories Inc.).
Statistics
Results from the present study were analyzed by GraphPad Prism v8.0 (GraphPad Software, Inc.). The mean value in each group was compared by one-way ANOVA. Tukey's test was used as a post-hoc test following ANOVA. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-378 inhibits the proliferation of HuH-7 cells more efficiently using a combination of ultrasonic irradiation and SonoVue microbubbles method
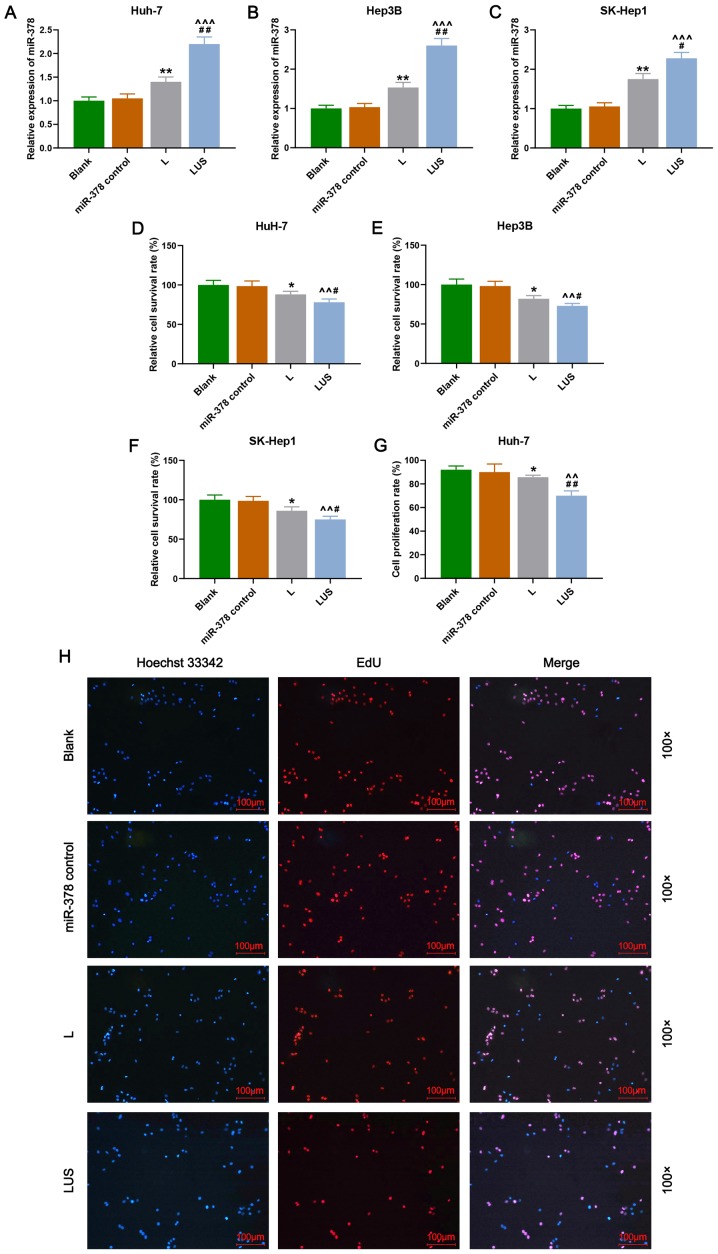
To investigate the role of miR-378 on the proliferation of HuH-7, Hep3B and SK-Hep1 cells, and the efficiency of transfection using ultrasonic irradiation in combination with SonoVue microbubbles, cell survival rate was detected by performing CCK-8 and cell proliferation rate was measured using double cytochemical staining. In the present study, the results revealed that the relative expression of miR-378 in the L group was significantly increased compared with that in the Blank group (P<0.01), and in the LUS group this expression was significantly increased compared with that in the miR-378 control and L groups (P<0.001; Fig. 1A-C). Moreover, the relative cell survival rate (%) of HuH-7, Hep3B and SK-Hep1 cells in the L group was reduced compared with that in the Blank group, and was also reduced in the LUS group compared with that in the miR-378 control and L groups (P<0.05 and P<0.01, respectively; Fig. 1D-F). Furthermore, the cell proliferation rate (%) of Huh-7 cells in the L and LUS groups demonstrated a similar trend to that of the cell survival rate (P<0.05 and P<0.01, respectively; Fig. 1G and H). Thus, these data suggested that miR-378 expression contributed to the suppression of the proliferation of HuH-7 cells and that the combination of ultrasonic irradiation and SonoVue microbubbles method was more effective in the transfection of miRNA.
Figure 1.
miR-378 inhibits the proliferation of HuH-7 cells much more efficiently using a combination of ultrasonic irradiation and the SonoVue microbubbles method. Relative expression of miR-378 in (A) Huh-7, (B) Hep3B and (C) SK-Hep-1 cells. Cell survival rate (%) was detected by Cell Counting Kit-8 in (D) HuH-7, (E) Hep3B and (F) SK-Hep1 cells. (G) Cell proliferation rate (%) of Huh-7 cells in Blank, miR-378 control, L and LUS groups. (H) Double cytochemical staining of Hoechst 33342 (blue) and EdU (red) in each group. Bars indicate mean ± SD. *P<0.05 and **P<0.01, vs. Blank; ^^P<.0.1 and ^^^P<0.001 vs. miR-378 control; #P<0.05 and ##P<0.01 vs. L. L group, the hepatocellular carcinoma cells were transfected with miR-378; LUS group, the hepatocellular carcinoma cells were treated with miR-378 mimic combined with Ultrasonic irradiation and SonoVue microbubbles. miR, microRNA.
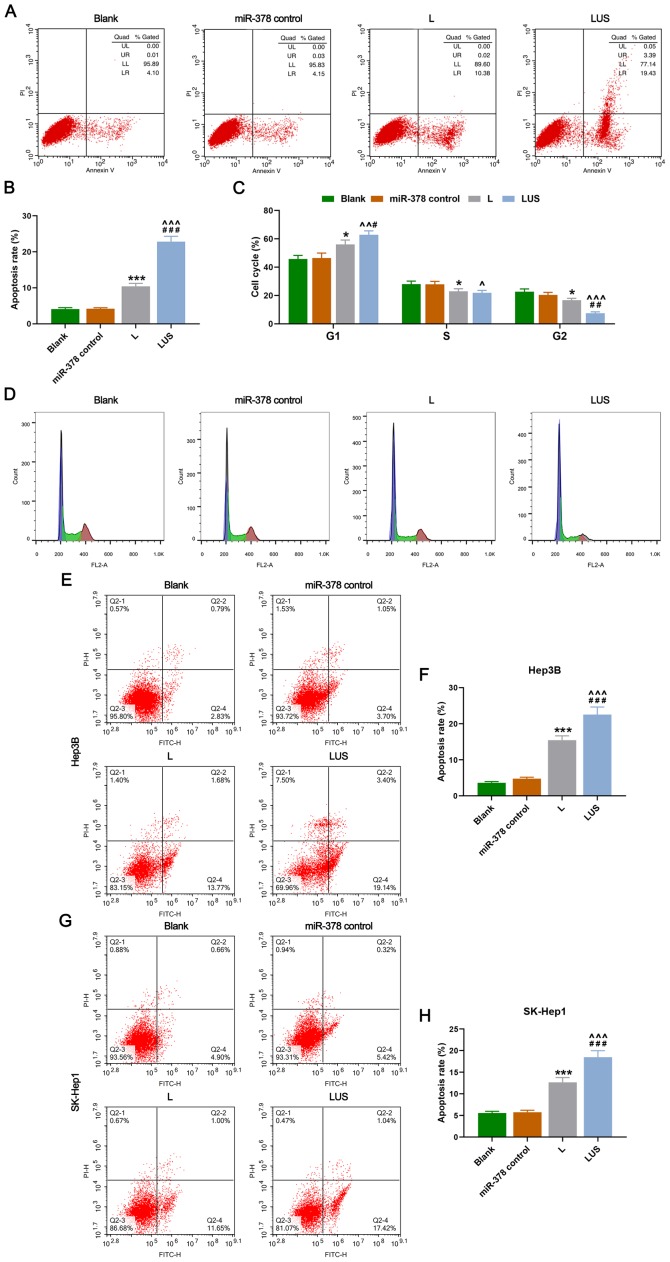
miR-378 increases the apoptosis rate and arrests the cell cycle of HuH-7 cells more efficiently using a combined method of ultrasonic irradiation and SonoVue microbubbles
Cell apoptosis and cell cycle of each phase were then evaluated. As expected, the apoptosis rate (%) of HuH-7 cells in the L group was increased compared with that in the Blank group, moreover, it was significantly elevated in the LUS group compared with that in the miR-378 control and L groups (P<0.001; Fig. 2A and B). Furthermore, the percentage of HuH-7 cells in the G1 phase in the L group was increased compared with that in the Blank group, but it was much higher in the LUS group than that in the miR-378 control and L groups (P<0.05 and P<0.01, respectively; Fig. 2C and D). However, the S and G2 phase in the L group were significantly lower than that in the miR-378 control group, while the two phases in the LUS group were reduced compared with those in the miR-378 control and L groups, and no significant difference between the L and LUS groups on S phase was observed (P<0.05, P<0.01 and P<0.001; Fig. 2C and D). Thus, miR-378 expression could increase the rate of apoptosis and arrest the cell cycle of HuH-7 cells in G1 phase more efficiently using a combination of ultrasonic irradiation and SonoVue microbubbles method. Similarly, the apoptosis rate (%) of Hep3B cells in the L group was increased compared with that in the Blank group and it was significantly higher in the LUS group compared with that in the miR-378 control and L groups (P<0.001; Fig. 2E and F). Moreover, the apoptosis rate (%) of SK-Hep1 cells in the L group was higher than that in the Blank group and it was significantly increased in the LUS group compared with that in the miR-378 control and L groups (P<0.001; Fig. 2G and H).
Figure 2.
miR-378 increases the rate of apoptosis and arrests the cell cycle of HuH-7 cells more efficiently using the combined method. (A) Rate of apoptosis of HuH-7 cells in Blank, miR-378 control, L and LUS groups detected by detection flow cytometry. (B) Apoptosis rate (%) in each group. (C) Cell cycle (%) of each phase in each group. (D) Cell cycle assessment using flow cytometry in each group. Flow cytometry was used to detect cell apoptosis in (E and F) Hep3B and (G and H) SK-Hep1 cells. Bars indicate mean ± SD. *P<0.05 and ***P<0.001 vs. Blank; ^P<0.05, ^^P<0.01 and ^^^P<0.001 vs. miR-378 control; #P<0.05, ##P<0.01 and ###P<0.001 vs. L. L group, the hepatocellular carcinoma cells were transfected with miR-378; LUS group, the hepatocellular carcinoma cells were treated with miR-378 mimic combined with Ultrasonic irradiation and SonoVue microbubbles. miR, microRNA.
miR-378 regulates genes related to apoptosis and proliferation of HuH-7 cells
The present study also explored whether genes related to apoptosis and the proliferation of HuH-7 cells could be affected by miR-378 expression, and the efficiency of the combined transfection method. In the current study, the data revealed that the protein and mRNA expression levels of Cyclin D1, Bcl-2, Akt and Survivin in the L group were lower than those in the miR-378 control group, and the levels were significantly decreased in the LUS group compared with those in the L and miR-378 control groups (P<0.01 and P<0.001; Fig. 3A-C). On the other hand, the expression of Bax and p53 exhibited a different trend, expression increased in the L and LUS groups compared with the miR-378 control and Blank groups, with the highest expression found in the LUS group (P<0.05 and P<0.001; Fig. 3A-C). In addition, β-catenin expression was significantly reduced in the LUS group compared with that in the miR-378 control and L groups (P<0.05 and P<0.001; Fig. 3D-E). Taken together, these results indicated that miR-378 expression led to a decrease in the expression of proliferation-related genes and increased the expression of apoptosis-related genes. Additionally, it was demonstrated that the use of combined ultrasonic irradiation and SonoVue microbubbles method for the transfection of miR-378 was more effective.
Figure 3.
miR-378 regulates genes related to apoptosis and proliferation of HuH-7 cells more efficiently. (A) Western blot images showing protein expression of Cyclin D1, Bcl-2, Bax, Akt, p53 and Survivin in Blank, miR-378 control, L and LUS groups. Histograms showing (B) relative protein and (C) mRNA expression levels in each group. (D) Western blotting was used to measure the expression of β-catenin. (E) Relative expression of β-catenin was quantified via western blotting. Bars indicate mean ± SD. *P<0.05, **P<0.01 and ***P<0.001 vs. Blank; ^^P<0.01 and ^^^P<0.001 vs. miR-378 control; #P<0.05, ##P<0.01 and ###P<0.001 vs. L. L group, the hepatocellular carcinoma cells were transfected with miR-378; LUS group, the hepatocellular carcinoma cells were treated with miR-378 mimic combined with Ultrasonic irradiation and SonoVue microbubbles. miR, microRNA.
Discussion
At present, miRNAs have been increasingly found to have important roles in the development of cancer. It has been shown that miR-146a not only has a tumor-suppressive effect, but also plays a critical role in the growth of HCC cells and that genetic variation of miR-146a may be a risk factor for developing HCC (21). The expression of miR143HG was significantly downregulated in HCC cells and tissues, and was associated with the staging and prognosis of patients with HCC (22). Furthermore, previous research has also suggested that the upregulation of miR-21-5p may be a functional regulator of HCC apoptosis and could be a new tumor marker for early diagnosis of HCC (23). The current study examined the effects of miR-378 overexpression on HuH-7 cells and explored the efficiency of the combined transfection method of ultrasonic irradiation and SonoVue microbubbles. The results demonstrated that miR-378 functioned as a suppressor in the proliferation of HuH-7, Hep3B and SK-Hep1 cells, and this effect of miR-378 could be enhanced by using the combined method. Thus, these results indicated that a combined transfection approach is useful and may have potential in gene therapy.
Overexpressed miR-378 could be transfected into HuH-7, Hep3B and SK-Hep1 cells, which are representative of hepatoma cells. Furthermore, it was found that miR-378 overexpression reduced the proliferation ability and increased the apoptosis rate of HuH-7, Hep3B and SK-Hep1 cells. The cell cycle is a process during which cells undergo a series of controls and regulations to maintain an orderly progression of cell growth and proliferation (24–26). Cyclin/CDK is a promoter of the cell cycle (24–26). Cyclin D1, a member of the Cyclin family and also a proto-oncogene, promotes cell G1/S phase via CDK, thereby promoting cancer development (24–26). p53 is a tumor suppressor gene located at 17p in human chromosomes and is normally expressed at low levels in the nucleus (27–30). Mutated p53 genes have been found in various human tumors, including in HCC (27–30). p53 proteins are involved in several cellular processes, including gene transcription, DNA repair, cell cycle, genome stabilization, chromosome segregation, senescence, apoptosis and angiogenesis (27–30). The p53 signal transduction pathway is an important signaling pathway regulating G1/S phase (27–30). The role of the Bcl-2 family in apoptosis has received much attention (31), Bcl-2 is an inhibitor of apoptosis protein, while Bax is an apoptosis-promoting protein, these two factors are closely related to the regulation of apoptosis (31–34). Specifically, Bax forms a heterodimer with Bcl-2, thereby inhibiting the function of Bcl-2 (35–37), moreover, the susceptibility of cell apoptosis depends on the ratio of Bax/Bcl-2, which also determines cell survival after receiving apoptosis signals, thus, the ratio plays an important role in tumor occurrence (35–37). Akt is a serine/threonine protein kinase (38–40) and when cells are stimulated by growth factors, PI3K activates and produces PIP3 that binds to the PH domain of Akt to trigger the recruitment of Akt to the cell membrane (38–40). This process is involved in resistance to apoptosis, glucose metabolism, protein synthesis and could promote cell growth and proliferation (38–40). Survivin is one of the members of the IAP family with the lightest molecular weight (41,42) and it specifically binds to mitotic spindles during G2/M phase and can inhibit caspase-3, caspase-7, caspase-8 and cytochrome C, which are the key factors in apoptosis signaling pathways, therefore it can promote cancer development (41,42). In the present study, expression of Cyclin D1, Bcl-2, Akt and Survivin were reduced in HuH-7 cells transfected with miR-378, while Bax and p53 expression was elevated, suggesting that the proliferation of HuH-7 cells was downregulated and apoptosis was increased by the overexpression of miR-378.
Several researchers have investigated the role of miR-378 in hepatoma. An et al (43) reported that in a Chinese population, mutant miR-378 was related to the prognosis of HCC. Similarly, Hyun et al (44) revealed that the activation of hepatic stellate cells and the liver fibrosis process could be suppressed by miR-378. In addition, Zhou et al (45) found that miR-378 could be overexpressed by metformin to reduce proliferation of HCC. Thus, miR-378 may be a suppressor in the process of hepatoma.
Interestingly, in the present study it was found that when miR-378 was transfected into HuH-7, Hep3B and SK-Hep1 cells by the combined method of ultrasonic irradiation and SonoVue microbubbles, the suppressive effect of miR-378 was more significant. Ran et al (19) indicated that interference of the target gene by short interfering RNA using the combined method of ultrasonic irradiation and SonoVue microbubbles was more efficient than using the lipidosome method for cell transfection. Li et al (46) also showed that SonoVue and ultrasound irradiation may have promising effects in breast cancer therapy by delivering the target genes effectively.
Generally, traditional viral transfection and lipofection methods limit the functionality of target genes in target cells, due to their low safety and poor transfection efficiency (47,48). However, SonoVue differs from traditional gene vector microbubbles and can overcome such limitations (49). When SonoVue microbubbles are irradiated by ultrasound, they can produce continuous compression or expansion, leading to the rupture of microbubbles with accompanied cavitation effect (50). Thus, the permeability of the target tissue or cell membrane is increased, which promotes the entry of the gene into the target cells (50).
In conclusion, miR-378 is shown to be a suppressive factor in HCC, as it suppresses the proliferation and increases the apoptosis of HCC cells. Moreover, the combination of ultrasonic irradiation and SonoVue microbubbles method is more efficient in the transfection of miRNA. Therefore, the present findings contribute to the current gene therapy for hepatoma, however, the feasibility of such a method should be further explored.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JW and YL made substantial contributions to study conception and design, drafted the article and critically revised it for important intellectual content. QM and JH were involved in data acquisition, analysis and interpretation. All authors gave final approval of the version to be published and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of the work are appropriately investigated and resolved. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, Luo R, Feng J, Ye S, Yang TS, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: A phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl L Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 3.Bruix J, Sherman M, American Association for the Study of Liver Diseases Management of hepatocellular carcinoma: An update. Hepatology. 2011;53:1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llovet JM, Bruix J. Systematic review of randomized trials for unresectable hepatocellular carcinoma: Chemoembolization improves survival. Hepatology. 2003;37:429–442. doi: 10.1053/jhep.2003.50047. [DOI] [PubMed] [Google Scholar]
- 5.Nam SW, Park JY, Ramasamy A, Shevade S, Islam A, Long PM, Park CK, Park SE, Kim SY, Lee SH, et al. Molecular changes from dysplastic nodule to hepatocellular carcinoma through gene expression profiling. Hepatology. 2005;42:809–818. doi: 10.1002/hep.20878. [DOI] [PubMed] [Google Scholar]
- 6.Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, Gupta S, Moore J, Wrobel MJ, Lerner J, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Roosbroeck K, Calin GA. Cancer hallmarks and MicroRNAs: The therapeutic connection. Adv Cancer Res. 2017;135:119–149. doi: 10.1016/bs.acr.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Wang G, Song L, Hou X, Kala S, Wong KF, Tang L, Dai Y, Sun L. Surface-modified GVs as nanosized contrast agents for molecular ultrasound imaging of tumor. Biomaterials. 2020;236:119803. doi: 10.1016/j.biomaterials.2020.119803. [DOI] [PubMed] [Google Scholar]
- 9.Qiu L, Leng QY, Luo Y. Progress of ultrasound microbubble contrast technology in the diagnosis and treatment of clinical diseases. Sichuan Da Xue Xue Bao Yi Xue Ban. 2014;45:974–978. (In Chinese) [PubMed] [Google Scholar]
- 10.Liu Y, Miyoshi H, Nakamura M. Encapsulated ultrasound microbubbles: Therapeutic application in drug/gene delivery. J Control Release. 2006;114:89–99. doi: 10.1016/j.jconrel.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 11.Price RJ, Kaul S. Contrast ultrasound targeted drug and gene delivery: An update on a new therapeutic modality. J Cardiovasc Pharmacol Ther. 2002;7:171–180. doi: 10.1177/107424840200700307. [DOI] [PubMed] [Google Scholar]
- 12.Wallace N, Wrenn SP. Ultrasound triggered drug delivery with liposomal nested microbubbles. Ultrasonics. 2015;63:31–38. doi: 10.1016/j.ultras.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Mountford PA, Sirsi SR, Borden MA. Condensation phase diagrams for lipid-coated perfluorobutane microbubbles. Langmuir. 2014;30:6209–6218. doi: 10.1021/la501004u. [DOI] [PubMed] [Google Scholar]
- 14.Kooiman K, van Rooij T, Qin B, Mastik F, Vos HJ, Versluis M, Klibanov AL, de Jong N, Villanueva FS, Chen X. Focal areas of increased lipid concentration on the coating of microbubbles during short tone-burst ultrasound insonification. PLoS One. 2017;12:e0180747. doi: 10.1371/journal.pone.0180747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myrset AH, Fjerdingstad HB, Bendiksen R, Arbo BE, Bjerke RM, Johansen JH, Kulseth MA, Skurtveit R. Design and characterization of targeted ultrasound microbubbles for diagnostic use. Ultrasound Med Biol. 2011;37:136–150. doi: 10.1016/j.ultrasmedbio.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 16.Lammertink B, Deckers R, Storm G, Moonen C, Bos C. Duration of ultrasound-mediated enhanced plasma membrane permeability. Int J Pharm. 2015;482:92–98. doi: 10.1016/j.ijpharm.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 17.Yang PS, Tung FI, Chen HP, Liu TY, Lin YY. A novel bubble-forming material for preparing hydrophobic-agent-loaded bubbles with theranostic functionality. Acta Biomater. 2014;10:3762–3774. doi: 10.1016/j.actbio.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Rychak JJ, Klibanov AL. Nucleic acid delivery with microbubbles and ultrasound. Adv Drug Deliv Rev. 2014;72:82–93. doi: 10.1016/j.addr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ran LW, Wang H, Lan D, Jia HX, Yu SS. Effect of RNA interference targeting STAT3 gene combined with ultrasonic irradiation and SonoVue microbubbles on proliferation and apoptosis in keratinocytes of psoriatic lesions. Chin Med J (Engl) 2018;131:2097–2104. doi: 10.4103/0366-6999.239297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Li X, Li T, Wang L, Wu X, Liu J, Xu Y, Wei W. Multiple roles of microRNA-146a in immune responses and hepatocellular carcinoma. Oncol Lett. 2019;18:5033–5042. doi: 10.3892/ol.2019.10862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin X, Xiaoqin H, Jiayu C, Li F, Yue L, Ximing X. Long non-coding RNA miR143HG predicts good prognosis and inhibits tumor multiplication and metastasis by suppressing mitogen-activated protein kinase and Wnt signaling pathways in hepatocellular carcinoma. Hepatol Res. 2019;49:902–918. doi: 10.1111/hepr.13344. [DOI] [PubMed] [Google Scholar]
- 23.Zhong XZ, Deng Y, Chen G, Yang H. Investigation of the clinical significance and molecular mechanism of miR-21-5p in hepatocellular carcinoma: A systematic review based on 24 studies and bioinformatics investigation. Oncol Lett. 2019;17:230–246. doi: 10.3892/ol.2018.9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JK, Diehl JA. Nuclear cyclin D1: An oncogenic driver in human cancer. J Cell Physiol. 2009;220:292–296. doi: 10.1002/jcp.21791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qie S, Diehl JA. Cyclin D1, cancer progression, and opportunities in cancer treatment. J Mol Med (Berl) 2016;94:1313–1326. doi: 10.1007/s00109-016-1475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witzel II, Koh LF, Perkins ND. Regulation of cyclin D1 gene expression. Biochem Soc Trans. 2010;38:217–222. doi: 10.1042/BST0380217. [DOI] [PubMed] [Google Scholar]
- 27.Hayman L, Chaudhry WR, Revin VV, Zhelev N, Bourdon JC. What is the potential of p53 isoforms as a predictive biomarker in the treatment of cancer? Expert Rev Mol Diagn. 2019;19:149–159. doi: 10.1080/14737159.2019.1563484. [DOI] [PubMed] [Google Scholar]
- 28.Tiwari B, Jones AE, Abrams JM. Transposons, p53 and genome security. Trends Genet. 2018;34:846–855. doi: 10.1016/j.tig.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue Y, San Luis B, Lane DP. Intratumour heterogeneity of p53 expression; causes and consequences. J Pathol. 2019;249:274–285. doi: 10.1002/path.5328. [DOI] [PubMed] [Google Scholar]
- 30.Chabeda A, Yanez RJR, Lamprecht R, Meyers AE, Rybicki EP, Hitzeroth II. Therapeutic vaccines for high-risk HPV-associated diseases. Papillomavirus Res. 2018;5:46–58. doi: 10.1016/j.pvr.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Neill JW, Hockenbery DM. Bcl-2-related proteins as drug targets. Curr Med Chem. 2003;10:1553–1562. doi: 10.2174/0929867033457241. [DOI] [PubMed] [Google Scholar]
- 32.Brown LM, Hanna DT, Khaw SL, Ekert PG. Dysregulation of BCL-2 family proteins by leukemia fusion genes. J Biol Chem. 2017;292:14325–14333. doi: 10.1074/jbc.R117.799056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peña-Blanco A, García-Sáez AJ. Bax, Bak and beyond-mitochondrial performance in apoptosis. FEBS J. 2018;285:416–431. doi: 10.1111/febs.14186. [DOI] [PubMed] [Google Scholar]
- 34.Renault TT, Dejean LM, Manon S. A brewing understanding of the regulation of Bax function by Bcl-xL and Bcl-2. Mech Ageing Dev. 2017;161:201–210. doi: 10.1016/j.mad.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Stefanaki C, Antoniou C, Stefanaki K, Petrikos G, Argyrakos T, Constantinidou CV, Karentzou O, Stratigos A, Katsambas A. Bcl-2 and Bax in congenital naevi. Br J Dermatol. 2006;154:1175–1179. doi: 10.1111/j.1365-2133.2006.07177.x. [DOI] [PubMed] [Google Scholar]
- 36.Abu Zeid EH, Hussein MMA, Ali H. Ascorbic acid protects male rat brain from oral potassium dichromate-induced oxdative DNA damage and apoptotic changes: The expression patterns of caspase-3, P 53, Bax, and Bcl-2 genes. Environ Sci Pollut Res Int. 2018;25:13056–13066. doi: 10.1007/s11356-018-1546-9. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Yang X, Ge X, Zhang F. Puerarin attenuates neurological deficits via Bcl-2/Bax/cleaved caspase-3 and Sirt3/SOD2 apoptotic pathways in subarachnoid hemorrhage mice. Biomed Pharmacother. 2019;109:726–733. doi: 10.1016/j.biopha.2018.10.161. [DOI] [PubMed] [Google Scholar]
- 38.Lien EC, Dibble CC, Toker A. PI3K signaling in cancer: Beyond AKT. Curr Opin Cell Biol. 2017;45:62–71. doi: 10.1016/j.ceb.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Driessen GJ, IJspeert H, Wentink M, Yntema HG, van Hagen PM, van Strien A, Bucciol G, Cogulu O, Trip M, Nillesen W, et al. Increased PI3K/Akt activity and deregulated humoral immune response in human PTEN deficiency. J Allergy Clin Immunol. 2016;138:1744–1747.e1745. doi: 10.1016/j.jaci.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Fu LX, Zhu WL, Shi H, Chen LJ, Ye B. Blockade of CXCR6 reduces invasive potential of gastric cancer cells through inhibition of AKT signaling. Int J Immunopathol Pharmacol. 2015;28:194–200. doi: 10.1177/0394632015584502. [DOI] [PubMed] [Google Scholar]
- 41.Han G, Gong H, Wang Y, Guo S, Liu K. AMPK/mTOR-mediated inhibition of survivin partly contributes to metformin-induced apoptosis in human gastric cancer cell. Cancer Biol Ther. 2015;16:77–87. doi: 10.4161/15384047.2014.987021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu JL, Gao W, Kang QM, Zhang XJ, Yang SG. Prognostic value of survivin in patients with gastric cancer: A systematic review with meta-analysis. PLoS One. 2013;8:e71930. doi: 10.1371/journal.pone.0083350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.An J, Liu J, Liu L, Liu Y, Pan Y, Huang M, Qi F, Wen J, Xie K, Ma H, et al. A genetic variant in primary miR-378 is associated with risk and prognosis of hepatocellular carcinoma in a Chinese population. PLoS One. 2014;9:e93707. doi: 10.1371/journal.pone.0093707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hyun J, Wang S, Kim J, Rao KM, Park SY, Chung I, Ha CS, Kim SW, Yun YH, Jung Y. MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat Commun. 2016;7:10993. doi: 10.1038/ncomms10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou J, Han S, Qian W, Gu Y, Li X, Yang K. Metformin induces miR-378 to downregulate the CDK1, leading to suppression of cell proliferation in hepatocellular carcinoma. OncoTargets Ther. 2018;11:4451–4459. doi: 10.2147/OTT.S167614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li XH, Zhou P, Wang LH, Tian SM, Qian Y, Chen LR, Zhang P. The targeted gene (KDRP-CD/TK) therapy of breast cancer mediated by SonoVue and ultrasound irradiation in vitro. Ultrasonics. 2012;52:186–191. doi: 10.1016/j.ultras.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 48.Ding B, Li T, Zhang J, Zhao L, Zhai G. Advances in liver-directed gene therapy for hepatocellular carcinoma by non-viral delivery systems. Curr Gene Ther. 2012;12:92–102. doi: 10.2174/156652312800099625. [DOI] [PubMed] [Google Scholar]
- 49.Delalande A, Bastié C, Pigeon L, Manta S, Lebertre M, Mignet N, Midoux P, Pichon C. Cationic gas-filled microbubbles for ultrasound-based nucleic acids delivery. Biosci Rep. 2017;37:BSR20160619. doi: 10.1042/BSR20160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin P, Han T, Yu ACH, Xu L. Mechanistic understanding the bioeffects of ultrasound-driven microbubbles to enhance macromolecule delivery. J Control Release. 2018;272:169–181. doi: 10.1016/j.jconrel.2018.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.