Summary
The GTPase Ran has a key role in nuclear import and export, mitotic spindle assembly and nuclear envelope formation. The cycling of Ran between its GTP- and GDP-bound forms is catalyzed by the chromatin-bound guanine nucleotide exchange factor RCC1 and the cytoplasmic Ran GTPase-activating protein RanGAP. The result is an intracellular concentration gradient of RanGTP that equips eukaryotic cells with a ‘genome-positioning system’ (GPS). The binding of RanGTP to nuclear transport receptors (NTRs) of the importin β superfamily mediates the effects of the gradient and generates further downstream gradients, which have been elucidated by fluorescence resonance energy transfer (FRET) imaging and computational modeling. The Ran-dependent GPS spatially directs many functions required for genome segregation by the mitotic spindle during mitosis. Through exportin 1, RanGTP recruits essential centrosome and kinetochore components, whereas the RanGTP-induced release of spindle assembly factors (SAFs) from importins activates SAFs to nucleate, bind and organize nascent spindle microtubules. Although a considerable fraction of cytoplasmic SAFs is active and RanGTP induces only partial further activation near chromatin, bipolar spindle assembly is robustly induced by cooperativity and positive-feedback mechanisms within the network of Ran-activated SAFs. The RanGTP gradient is conserved, although its roles vary among different cell types and species, and much remains to be learned regarding its functions.
Keywords: Ran, Importin, Exportin, Mitotic spindle, Cancer
Introduction
The Ras-related GTPase Ran controls many aspects of genome compartmentalization in the nucleus in eukaryotes, including the regulation of nucleo-cytoplasmic transport and formation of the nuclear envelope, as well as genome segregation to daughter cells by the mitotic spindle (Clarke and Zhang, 2004; Goodman and Zheng, 2006; Pemberton and Paschal, 2005; Terry et al., 2007). The cycling of Ran between its GTP- and GDP-bound forms is controlled by a pair of exquisitely Ran-specific regulators with distinct localizations: Ran’s guanine nucleotide exchange factor (GEF) regulator of chromatin condensation 1 (RCC1) is imported to the nucleus and binds to chromatin, whereas its GTPase-activating protein RanGAP is cytoplasmic. RanGAP accelerates GTP hydrolysis by purified Ran 105_fold, and RCC1 increases the rate of GTP-for-GDP exchange on Ran by about the same factor in vitro (Klebe et al., 1995). The localization of its regulators, therefore, dynamically regulates Ran’s guanine-nucleotide charge – RanGTP is formed at a higher rate around chromosomes than at the cell periphery, where more GTP hydrolysis on Ran occurs. In consequence, a diffusion-limited gradient of RanGTP surrounds chromosomes in metazoan cells that are undergoing open mitosis following nuclear envelope breakdown, and a steep gradient of RanGTP exists across the nuclear envelope during interphase (Hetzer et al., 2002). Because the concentration of RanGTP increases with proximity to chromatin, such a gradient can be said to act as a ‘genome-positioning system’ (GPS).
The functions of Ran are mediated by the binding of RanGTP to nuclear transport receptors (NTRs) of the importin β superfamily (Mans et al., 2004; Pemberton and Paschal, 2005). NTRs are structurally diverse proteins that are responsible for the Ran-regulated transport of proteins and of several classes of RNAs through the nuclear pore channel (Conti et al., 2006; Mans et al., 2004; Rodriguez et al., 2004; Stewart, 2007). NTRs are classified as either importins or exportins, depending on whether their cargos are imported into or exported from the nucleus. For simplicity, the term ‘cargo’ will be used here for any molecule or complex having an interaction with NTRs that is regulated by RanGTP binding (Table 1). The binding of RanGTP to importins dissociates nuclear import complexes that contain nuclear localization signal (NLS)-bearing cargos, resulting in the nuclear accumulation of the cargos. RanGTP binding is required to stabilize the interaction of exportins with their nuclear export signal (NES)-containing cargo. Upon entry into the cytoplasm, the complex is disassembled owing to GTP hydrolysis on Ran that is stimulated by RanGAP (Weis, 2003).
Table 1.
Functions and interactions of RanGTP-regulated NTR cargos
| Cargo | Class of protein | Nuclear transport receptors | Interacting spindle components or regulators* | RanGTP- and NTR-regulated function | Reference |
|---|---|---|---|---|---|
| RCCI | RanGEF | Importin α3, importin β | Histones H2A and H2B, DNA, Ran | Binding to histones H2A/H2B | (Nemergut et al., 2001; Chen et al.,2007) |
| TPX2 | Spindle MAP | Importin αl, importin β | Microtubules, Xklp2, Aurora A, BRCAl/BARDl, RHAMM,Eg5, XMAP215, HURP | MT nucleation, Aurora A activation | (Gruss et al., 2001; Groen et al., 2004; Joukov et al., 2006; Koffa et al., 2006) |
| NuMA | Spindle pole MAP | Importin αl, importin β | Dynein, BRCAl/BARDl, Rael | Unmown | (Nachury et al., 2001; Wiese et al., 2001; Joukov et al., 2006; Wong et al., 2006) |
| XXnf7 | MAP | Importin αl, importin β | Microtubules, anaphase- promoting complex | Unmown | (Maresca et al., 2005) |
| XCTK2 | Spindle kinesin | Importin αl, importin β | Microtubules | Microtubule binding | (Ems-McClung et al., 2004) |
| Kid | Chromosomal kinesin | Importin αl, importin β | Chromosomes, microtubules | Microtubule binding, loading on chromosomes | (Trieselman et al., 2003; Tahara et al., 2008) |
| Lamin B | Nuclear envelope scaffold | Importin αl, importin β | Spindle membranes, NuMA, TPX2, Eg5, PAR | Spindle matrix | (Tsai et al., 2006) |
| Cdkll | Cyclin-L- dependent kinase | Importin α, importin β | Unknown | Microtubule stabilization microtubule- kinetochore interaction | (Yokoyama et al., 2008) |
| HURP | MAP | Importin β | TPX2, AuroraA, XMAP215, Eg5 | Microtubule binding, stabilization of k-fibers (kinetochore microtubules), spindle bipolarization | (Koffa et al., 2006; Sillje et al., 2006; Wong et al., 2006; Wong et al., 2008) |
| Maskin (TACC, Alp7) | Spindle pole MAP | Importin β | XMAP2l5 (TOG), Aurora A, Rael, TPX2, Eg5 | Phosphorylation by Aurora A; mitotic nuclear import of Alp7 in S.pombe | (Blower et al., 2005; Albee et al., 2006; Koffa et al., 2006; Sato et al., 2007) |
| Rael | RNP adaptor, nucleoporin | Importin β, Nup98 | RNP, Maskin, NuMA | Indirect: microtubule polymerization | (Blower et al., 2005; Wong et al., 2006) |
| NuSAP | Spindle MAP | Importin αl, importin β and importin 7 | Chromosomes, microtubules | Microtubule stabilization and crosslinking; chromatin binding | (Ribbeck et al., 2006; Ribbeck et al., 2007) |
| CRB3-CLPI | Ciliar membrane protein | Importin β (possibly through an adaptor) | Unknown | Importin-P-dependent targeting to spindle pole | (Fan et al., 2007) |
| RanBP2-RanGAP-SUMO | Multifunctional | Exportin 1 | Kinetochore-bound Nupl07-Nupl60 complex | Formation of k-fibers | (Amaoutov et al., 2005) |
| NPMI | Multifunctional | Exportin 1 | Centrosome | Centrosome integrity | (Wang et al., 2005) |
| Survivin | CPC component, Antiapoptotic factor | Exportin l | Aurora B, INCENP | CPC recruitment; indirect: microtubule polymerization | (Knauer et al., 2006) |
Direct interactions are shown in red.
RanGTP-gradient-regulated NTR-cargo-complex assembly and disassembly reactions continue after nuclear envelope breakdown in Metazoa and perform essential mitotic roles. This Commentary focuses on how the RanGTP gradient is generated in mitotic cells and how it functions in spindle assembly by spatially directing the activity and/or localization of spindle assembly factors (SAFs) and mitotic regulators (Table 1). We discuss both conserved and cell type-specific mechanisms of mitotic Ran function and highlight the emerging connection between the Ran pathway and cancer cell proliferation. Unless otherwise specified, the discussion refers to open mitosis in animal cells. For the functions of Ran in the mitotic checkpoint we refer readers to recent reviews (Dasso, 2006; Goodman and Zheng, 2006).
Binding of RCC1 to chromatin drives RanGTP-gradient formation
RCC1 binds directly to dsDNA through its unusually processed N-terminal tail (in which the N-terminal serine or proline residue is methylated) (Chen et al., 2007) and to histones H2A or H2B on the nucleosome (Nemergut et al., 2001) by an adjacent domain (Hutchins et al., 2004; Li and Zheng, 2004a). Both modes of RCC1-chromatin interaction are required for correct spindle assembly in tissue culture cells (Chen et al., 2007; Hutchins et al., 2004; Li and Zheng, 2004a) and probably function simultaneously to strengthen the interaction of RCC1 with chromatin during the nucleotide-exchange reaction (Chen et al., 2007). Ran binds directly to histones H3 and H4 on chromatin (Bilbao-Cortes et al., 2002), promoting its interaction with RCC1. Fluorescence recovery after photobleaching experiments in live cells indicate that cooperative binding of RanGDP and RCC1 to chromatin is followed by the release of RanGTP after the completion of GDP/GTP exchange (Li and Zheng, 2004b), thereby heightening the peak of the RanGTP gradient and focusing it to a thin volume at the interface of chromatin and cytoplasm.
Interestingly, the histone-binding domain of RCC1 overlaps with its NLS, which is recognized by a complex between importin α3 and importin β that imports RCC1 into the nucleus during interphase. In mitosis, bound importins can block the interaction between RCC1 and chromatin in the absence of mitotic cyclin-dependent kinase 1 (Cdkl)-dependent phosphorylation of serine residues that are adjacent to the NLS (Hutchins et al., 2004; Li and Zheng, 2004a). However, human cells express three isoforms of RCC1 with variations in their N-termini that impose strikingly different regulation by phosphorylation and importins. Only RCC1γ is a substrate of mitotic kinases, but it does not bind well to importins; conversely, RCC1α is not mitotically phosphorylated and binds to importins well, indicating that these two isoforms may be specialized for mitotic and interphase functions, respectively (Hood and Clarke, 2007). In addition, all the isoforms are expressed in a complex tissue-specific manner (Hood and Clarke, 2007). Clearly, much remains to be learned about RCC1 function and regulation.
Cytoplasmic RanGAP catalyzes the conversion of RanGTP to RanGDP
After RCC1-catalyzed nucleotide exchange, RanGTP that diffuses from the chromatin can either be rapidly converted to RanGDP by RanGAP, or bind to abundant NTRs in the cytoplasm. RanGTP that is bound to an NTR is protected from RanGAP until it is extricated by proteins that contain a Ran-binding domain (RBD) (Bischoff and Gorlich, 1997) that presents it for RanGAP-catalyzed GTP hydrolysis. A large fraction of the cytoplasmic RanGTP that is detected with monoclonal antibodies in fixed mitotic cells (Ciciarello et al., 2004; Tedeschi et al., 2007) probably corresponds to the NTR-bound form. Human cells have two RBD proteins: the small cytoplasmic protein RanBP1 and the large nucleoporin RanBP2 (Nup358), which contains four RBDs. In mammalian cells, a fraction of RanBP1 associates tightly with centrosomes (Guarguaglini et al., 2000), whereas a fraction of RanBP2 is found on spindle microtubules and kinetochores in a complex with SUMOylated RanGAP (RanGAP-SUMO) (Joseph et al., 2004; Salina et al., 2003). Mitotic defects that result from RNA interference (RNAi)-mediated suppression of RanBP1 (Tedeschi et al., 2007) and RanBP2 (Salina et al., 2003) are dissimilar, suggesting that they possess specialized mitotic functions. The dissociation of RanGTP from importin β by RanBP1 is accelerated by importin α (Bischoff and Gorlich, 1997), which probably enhances the Ran-recycling function of the non-abundant RBD proteins. Together, these RanGAP cofactors are essential to generate dynamic Ran-NTR interactions. A simplified network of mitotic Ran-regulated reactions is summarized in Fig. 1A.
Fig. 1.
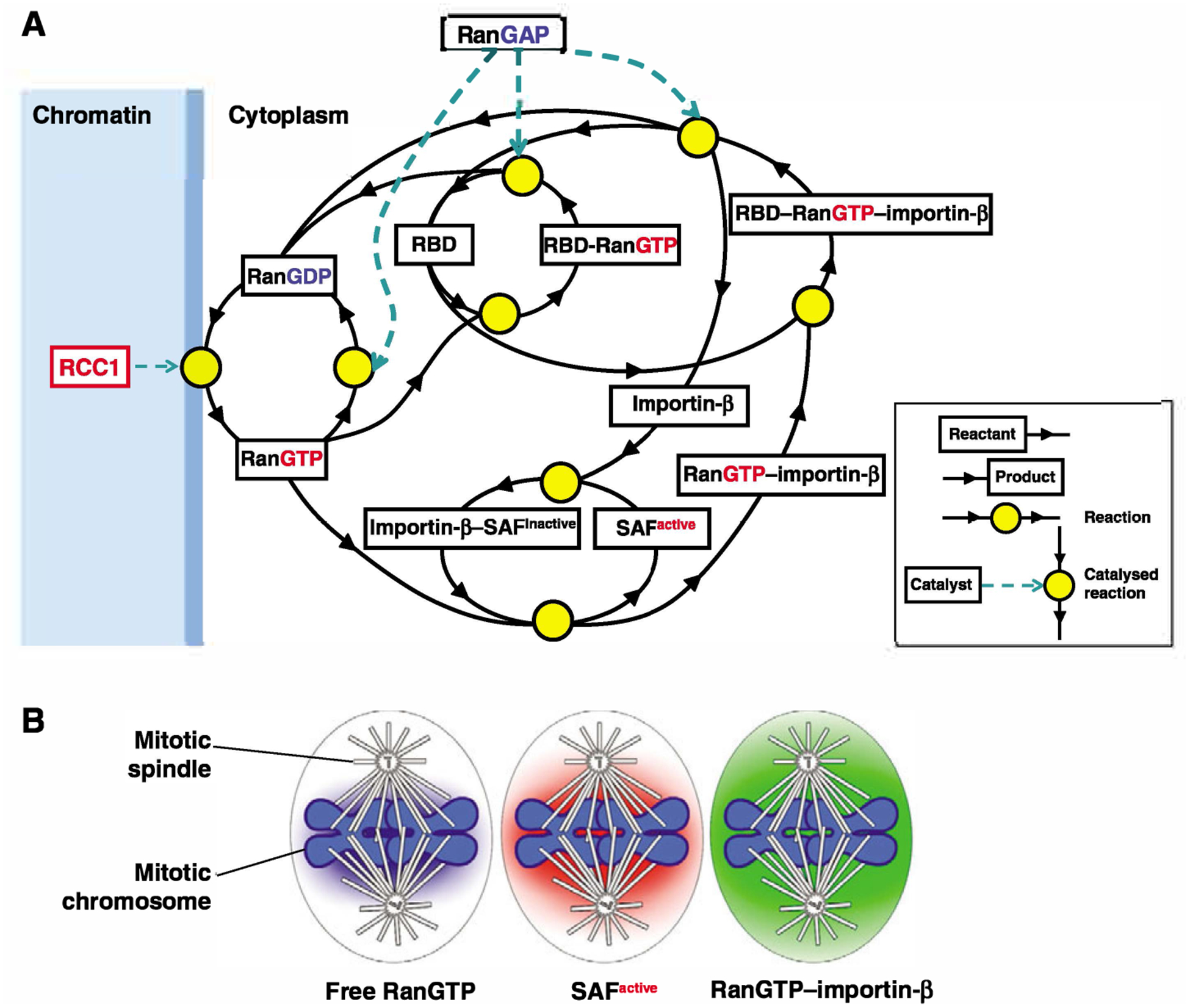
Network of interactions and reactions in the Ran pathway and the downstream cascade of gradients. (A) Coupling of the RanGTP signal to the dynamics of NTR-cargo interactions in the mitotic cell. The driving force is the steep gradient of free RanGTP, which is produced by RCC1 on the chromatin and dissipated by diffusion-limited reactions that include RanGAP-catalyzed GTP hydrolysis on Ran and the interactions of free RanGTP with NTRs. For simplicity, multi-step reactions are condensed into one step and only one NTR, importin β, is included. Ran is charged with GTP in a RCC1-catalyzed reaction on the surface of the chromatin and diffuses to the cytoplasm where it is either immediately converted to RanGDP in a reaction catalyzed by RanGAP, or interacts with abundant competing cytoplasmic NTRs. Binding ofRanGTP to the importin β-SAF complex produces a RanGTP-importin β complex and liberates an active SAF. RBDs dissociate RanGTP from its complex with importin β and present it for RanGAP-catalyzed GTP hydrolysis. The diagram is based on the Virtual Cell (http://vcell.org) model of the RanGTP gradient (Kalab et al., 2006). (B) The components of the Ran-NTR system form chromatin-centered concentration gradients that exist in parallel in the mitotic cytoplasm and are related to each other in a distinct regulatory order. As the spatial extent of the gradients is defined by the reactions that create and dissipate the individual molecular species and the diffusion rate of those species (Bastiaens et al., 2006; Caudron et al., 2005), the concentration gradient of the more stable RanGTP–importin-β complex is broader than that of active SAFs that have been liberated by RanGTP from importin β-SAF complexes. Note that the extent of the RanGTP–importin-β gradient is expected to be similar to that of the RanGTP–exportin-1–NES cargo complex.
Modeling and visualization of mitotic RanGTP-regulated gradients
The driving force of Ran-regulated events in mitosis is the concentration gradient of RanGTP in its free (unbound) form, which is determined by three parameters: the loading of Ran with GTP by chromatin-bound RCC1, the diffusion of RanGTP into the cytoplasm, and the conversion of RanGTP to RanGDP by RanGAP or the binding of RanGTP to ligands that include abundant NTRs. Advances in imaging and the design of fluorescent resonance energy transfer (FRET) reporters for the Ran nucleotide state have provided direct evidence that a RanGTP concentration gradient exists and functions in mitotic spindle assembly (Caudron et al., 2005; Kalab et al., 2006; Kalab et al., 2002; Li and Zheng, 2004a). Computational models of the Ran-NTR system have aided this progress (Caudron et al., 2005; Gorlich et al., 2003; Kalab et al., 2006; Smith et al., 2002). A mathematical model, which assumes the polarized spatial distribution of RanGAP and RCC1, and the free diffusion of a minimal set of Ran-NTR reactions in space, predicted that a steeply declining gradient of RanGTP forms around mitotic chromatin and induces a shallower gradient of cargos that are liberated from importin β through the binding of RanGTP (Bastiaens et al., 2006; Caudron et al., 2005). A gradient ofRanGTP–importin-β complexes was predicted to diffuse even further into the cytoplasm than the liberated cargos in this model. Finally, RanGTP–importin-β complexes either dissociate or interact with RanBP1, giving rise to a gradient of importin-β–RanGTP–RanBP1 (Bastiaens et al., 2006; Caudron et al., 2005) that is dissipated by RanGAP-catalyzed hydrolysis of RanGTP. The extent of the gradients (that is, the distance from chromatin at which the concentration of a given molecular species decreases to constant levels) is defined by the reactions that create and dissipate the individual molecular species and by their diffusion rate (Fig. 1B).
A variety of experiments using FRET reporters support the above model of ordered gradients. Although no direct detection of free RanGTP has been achieved, a FRET sensor that monitors the interaction between RanGTP and an RBD was created by flanking an RBD with YFP (FRET acceptor) and CFP (FRET donor), and was termed YRC (YFP-RBD-CFP) (Kalab et al., 2002) In the presence of RanGDP, YRC remains unbound and emits a FRET signal. If YRC binds to RanGTP, the N- and C-termini of the RBD are pushed apart (Vetter et al., 1999) and the FRET signal decreases. Imaging of YRC added to Xenopus laevis egg extracts revealed a gradient of low-FRET YRC surrounding mitotic chromosomes, which visualized the RanGTP-RBD gradient (Kalab et al., 2002). A second FRET sensor, which reports on RanGTP-induced liberation of importin-α–importin-β cargos, was inspired by the structure of the N-terminal importin-β-binding (IBB) domain of importin α bound to importin β(Cingolani et al., 1999), in which the IBB domain behaves as an importin β cargo. As the structure predicted, an importin β cargo probe consisting of IBB flanked with YFP and CFP showed a low FRET signal when bound to importin β and a high FRET signal upon the binding of RanGTP to importin β, releasing the probe. The chromatin in mitotic spindles assembled in Xenopus laevis egg extracts was surrounded by a cargo-probe gradient displaying a high FRET signal, which visualized the gradient of importin-α-importin-β cargos that were released by the mitotic RanGTP gradient (Kalab et al., 2002). When the cargo gradient was abolished by adding an importin β mutant – deficient in RanGTP binding – to extracts, the RanGTP-RBD gradient around chromatin persisted, although the spindles rapidly disassembled. When RCC1 was inhibited by a dominant-negative Ran mutant, both gradients as well as the spindle were destroyed (Kalab et al., 2002). The RanGTP gradient therefore induces ‘downstream’ gradients, including that of liberated importin-α–importin-β cargos, which is essential for spindle formation.
The gradient of RanGTP-RBD that was detected using YRC reached ~17 μm away from the chromatin (P.K., unpublished), compared with the gradient of ~25 μm of liberated importin-α-importin-β cargos (Kalab et al., 2006; Kalab et al., 2002). FRET between fluorescent Ran and importin β added to mitotic X. laevis egg extracts revealed a gradient of RanGTP–RanBP1–importin-β that reached up to 30–35 μm from the chromatin (Caudron et al., 2005) The observed size of the gradients, therefore, was consistent with the order described by the mathematical model (Caudron et al., 2005) (Fig. 1B). These examples illustrate how the combination of quantitative imaging and modeling has become essential to our understanding of the functions of the RanGTP gradient.
Many remaining questions could benefit from combining these approaches. For example, in mitotic mammalian cells, SUMO-modified RanGAP in a complex with RanBP2 binds to spindle microtubules and concentrates at kinetochores (Joseph et al., 2002; Salina et al., 2003), where it participates in microtubule attachment (Arnaoutov et al., 2005; Joseph et al., 2004). However, it is not known whether the role of the RanBP2-RanGAP-SUMO complex in kinetochore-microtubule interactions involves any local increase of the GTP hydrolysis on Ran owing to the local concentration of RanGAP or some other activity associated with the complex. Determining the functional and biochemical activity of this complex, and applying spatial modeling as well as high-resolution imaging of FRET reporters, will address this question.
RanGTP regulates recruitment of NTR cargos to the mitotic spindle
More than 20 NTRs are present in humans (Pemberton and Paschal, 2005) but only four are known to regulate the function of cargo proteins in the mitotic spindle (importin α1, importin β, importin 7 and exportin 1; see Table 1). RanGTP regulates the activity of NTRs to promote the recruitment of soluble cytoplasmic cargos to mitotic spindle structures by two different mechanisms that are not mutually exclusive. In the first mechanism, RanGTP-induced release of cargos from importins allows the cargos to associate with the spindle and function as SAFs, either directly or through other SAFs. In the second mechanism, the binding of cargos to NTRs is required to recruit the cargos to specific sites within the mitotic spindle apparatus. For example, RanGTP-dependent exportin 1 complexes are delivered to centrosomes, kinetochores and centromeres, whereas binding to the importin-α–importin-β complex promotes the loading of the chromokinesin Kid onto mitotic chromosomes (Tahara et al., 2008). At the same time, the binding of Kid to spindle microtubules is promoted by its RanGTP-induced release from importins (Tahara et al., 2008; Trieselmann et al., 2003), suggesting that dynamic spatial regulation of both mechanisms by the RanGTP gradient is essential for Kid function in the microtubule-driven movement of chromosome arms.
The function of importins in mitotic spindle assembly
All known RanGTP- and importin-regulated mitotic functions involve importinβ (Table 1). This S-shaped molecule consists of HEAT repeats and can undergo large conformational changes to fit its diverse cargos and rapidly release them upon the high-affinity binding of RanGTP (Stewart, 2006; Stewart, 2007). Importin β associates with the mitotic spindle in mammalian tissue culture cells (Ciciarello et al., 2004; Fan et al., 2007) and centrosomal defects are induced by the overexpression of importin β(Ciciarello et al., 2004), which might reflect the inactivation of several different cargos that localize to spindle poles, including Crumbs3 (CRB3) (Fan et al., 2007), a protein that functions in the primary cilium and in centrosome cohesion. In addition to carrying cargos directly, importin β also associates with cargos through adaptor proteins, including the NTRs importin α and importin 7 (Fried and Kutay, 2003) and the nucleoporin Nup98, which stabilizes the association of importin β with Rael, a SAF cargo that functions in a ribonucleoprotein complex (Blower et al., 2005).
Many mitotic cargos interact with importin β through importin α. The C-terminus of importin α contains two NLS cargo-binding sites that are sequestered by its N-terminal IBB domain unless the IBB domain binds to importin β (Cingolani et al., 1999). Several structurally and functionally specialized importin α isoforms exist in animals (Quensel et al., 2004). Only the homologues of the most conserved isoform importin α1 (KNPA2 in huans) (Mans et al., 2004) have been identified as regulators of mitotic spindle assembly (Askjaer et al., 2002; Mason et al., 2002; Nachury et al., 2001), and these might represent an isoform that is specialized to promote proliferation of embryonic and undifferentiated cells by both its interphase (Yasuhara et al., 2007) and mitotic functions (Askjaer et al., 2002). In HeLa cells the levels of importin α1 mRNA increase dramatically during mitosis, similar to those of cyclin B and distinct from those of other importin α isoforms (Whitfield et al., 2002), which suggests a key role for this cargo adapter in mitosis.
A diverse set of importin cargos is therefore recruited to the mitotic spindle through the Ran GPS (Table 1). In addition to the SAF proteins, these cargos include non-protein components, such as RNA in the Rael-ribonucleoprotein complex discussed above (Blower et al., 2005), as well as membranes associated with nuclear lamin B (Travis, 2007; Tsai et al., 2006). Thus, the Ran-NTR system integrates many different cellular factors within the spindle.
The mitotic importin-β-cargo gradient – not an on-off switch
The simplest model of mitotic-spindle-assembly regulation by the RanGTP gradient predicts that SAF cargos are inhibited by binding to importins in the cytoplasm and activated around chromatin by their RanGTP-induced release (Dasso, 2001; Weis, 2003). A more refined model has been proposed in which the individual spindle-assembly processes are locally activated by specific threshold concentrations of RanGTP. In this model, microtubule nucleation that requires high RanGTP concentration occurs close to chromatin, whereas the centrosomal microtubules that are located further away would be stabilized at lower RanGTP levels (Bastiaens et al., 2006; Caudron et al., 2005).
However, quantitative FRET imaging of the importin-β-cargo sensor in live HeLa cells indicates a more complex scenario (Kalab et al., 2006). First, most importin β cargos (~70%) were found to exist freely in the mitotic cytoplasm, suggesting that cytoplasmic SAFs are not completely inhibited. Second, only ~15% more cargos were released around chromosomes, indicating that the RanGTP gradient locally activates only a fraction of SAFs. However, the titration of RanGTP into X. laevis mitotic egg extract revealed that an increase in cargo release by less than 10% induces microtubule polymerization (Kalab et al., 2006), suggesting that low-level local SAF activation is physiologically relevant in mitotic cells. These observations raise a number of questions – namely: what prevents microtubule polymerization throughout the mitotic cytoplasm (which contains active SAFs), and how does partial activation of SAFs around chromatin induce spindle assembly?
Cooperativity and positive feedback mechanism among SAF drive switch-like activation of spindle assembly by the Ran GPS
RanGTP-regulated mechanisms function in the context of other mitotic events and activities to promote spindle formation and function. Microtubule destabilization at the onset of mitosis is essential to disassemble the interphase array and allow chromosome-associated activities to promote spindle morphogenesis (Zhai et al., 1996). Overall, these microtubule-destabilizing activities raise the threshold of SAF activation that is required to generate microtubules in the mitotic cytoplasm.
The spatial cue for spindle assembly that is provided by the RanGTP gradient is amplified by three major factors. First, upon nuclear envelope breakdown, major microtubule-nucleating sites at centrosomes, kinetochores and chromatin are located within the peak of the RanGTP gradient. Second, synergy and positive-feedback mechanisms within the mitotic Ran network stimulate microtubule assembly in a switch-like manner. Third, the RanGTP gradient acts cooperatively with other chromatin-centered spindle-assembly pathways, including the establishment of phosphorylation gradients of the microtubule destabilizers stathmin (Op18) and kinesin 13 (MCAK/XKCM1) by the kinase Aurora B (Kelly et al., 2007; Niethammer et al., 2004; Zhang et al., 2007) (Fig. 2).
Fig. 2.
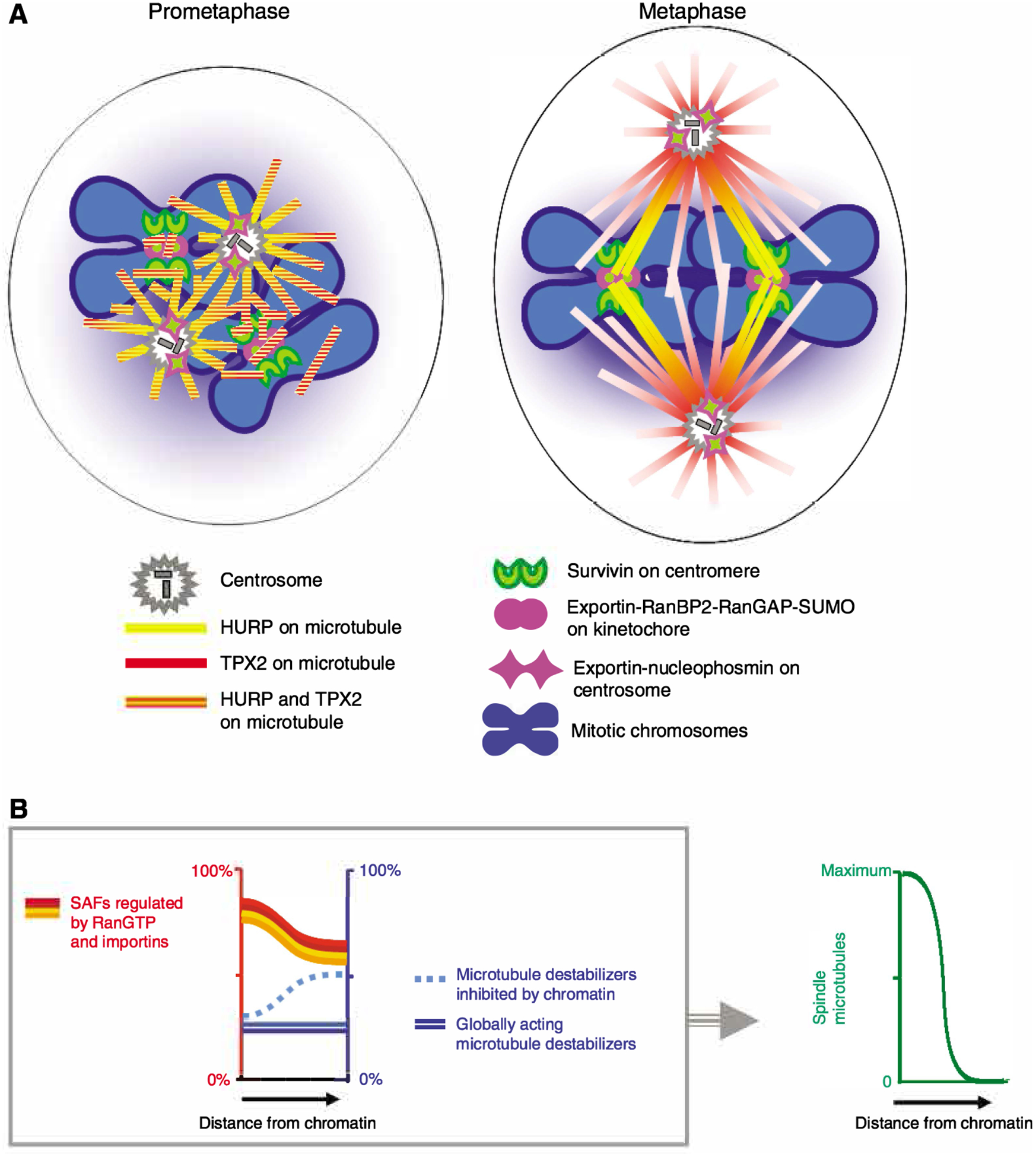
Function of the mitotic Ran GPS in spindle assembly. (A) In prometaphase, major centrosomal and non-centrosomal microtubule-organizing centers are located close to mitotic chromosomes and are exposed to high RanGTP concentrations, strongly promoting the local nucleation and stabilization of microtubules that is induced by RanGTP- and importin-regulated SAFs. The SAFs bind to and become concentrated on the nascent microtubules and promote their reorganization into bipolar spindles. During spindle bipolarization, distribution of some SAFs is biased towards specific sites on spindle microtubules. The two examples shown are TPX2, which concentrates at the spindle poles, and HURP, which relocates to kinetochore fibers close to the mitotic chromosomes in bipolar spindles. RanGTP- and exportin-1-regulated mitotic cargos are recruited by cargo-specific mechanisms to centromeres (survivin), kinetochores (RanBP2-RanGAP-SUMO) and centrosomes (NPM1) – either alone (survivin) or colocalizing with exportin 1 (RanBP2-RanGAP-SUMO and NPM1). (B) Switch-like induction of mitotic spindle assembly by multiple SAFs that are incrementally activated by the RanGTP gradient. (Left) SAFs that are inhibited by binding to importins in the cytoplasm are partially activated around the chromatin by the RanGTP-gradient-induced disassembly of SAF-importin complexes (red, orange and yellow lines). As a result, multiple Ran-GPS-directed overlapping gradients of active SAFs surround the mitotic chromosomes and cooperatively promote spindle assembly as described in A. There is only partial inhibition of SAFs in the cytoplasm, but this is balanced by the activity of microtubule depolymerizers (blue lines). Some microtubule depolymerizers, such as stathamin/Op18, are inhibited by chromatin signals, further promoting the localized polymerization of microtubules. (Right) RanGTP-induced local incremental activation of SAFs, in combination with microtubule destabilizers, produces a robust switch-like activation of the mitotic spindle assembly around chromatin.
All RanGTP-regulated SAFs act on microtubules, yet their functions in spindle assembly are distinct and often depend on one other. By nucleating microtubules, a SAF such as TPX2 produces substrates for other cargos and associated factors that stabilize microtubules, including TACC/maskin, NuSAP, HURP and XMAP215, as well as for microtubule-organizing kinesin 5, kinesin 10 and kinesin 14 (Eg5, Kid and XCTK2, respectively; see Table 1). Several of these interactions promote the localization and activation of the kinase Aurora A at spindle poles, which in phosphorylates several SAFs (Table 1).
Incremental activation of multiple SAFs could, therefore, induce the assembly of spindle subsystems of increasing complexity: microtubules, followed by an aster and then a spindle pole, the formation of which is not achievable by individual SAFs acting in isolation (Fig. 3). These highly cooperative interactions are probably enhanced by the association of SAFs in complexes that can simultaneously deliver multiple activities to the polymerizing microtubules. For example, TPX2 forms a complex with XRHAMM, BRCA1/BARD1 and NuMA (Joukov et al., 2006), NuMA interacts with Rael (Wong et al., 2006) and TACC binds to XMAP215 (Kinoshita et al., 2005; O’Brien et al., 2005; Peset et al., 2005). Remarkably, a complex containing HURP, XMAP215, Eg5, TPX2 and Aurora A has been isolated from X. laevis egg extracts (Koffa et al., 2006). Thus, once microtubules are generated in this system, they provide a substrate for more SAFs, many of which have been incrementally activated by the RanGTP gradient. The resulting cascade of RanGTP-regulated reactions leads to spindle assembly.
Fig. 3.
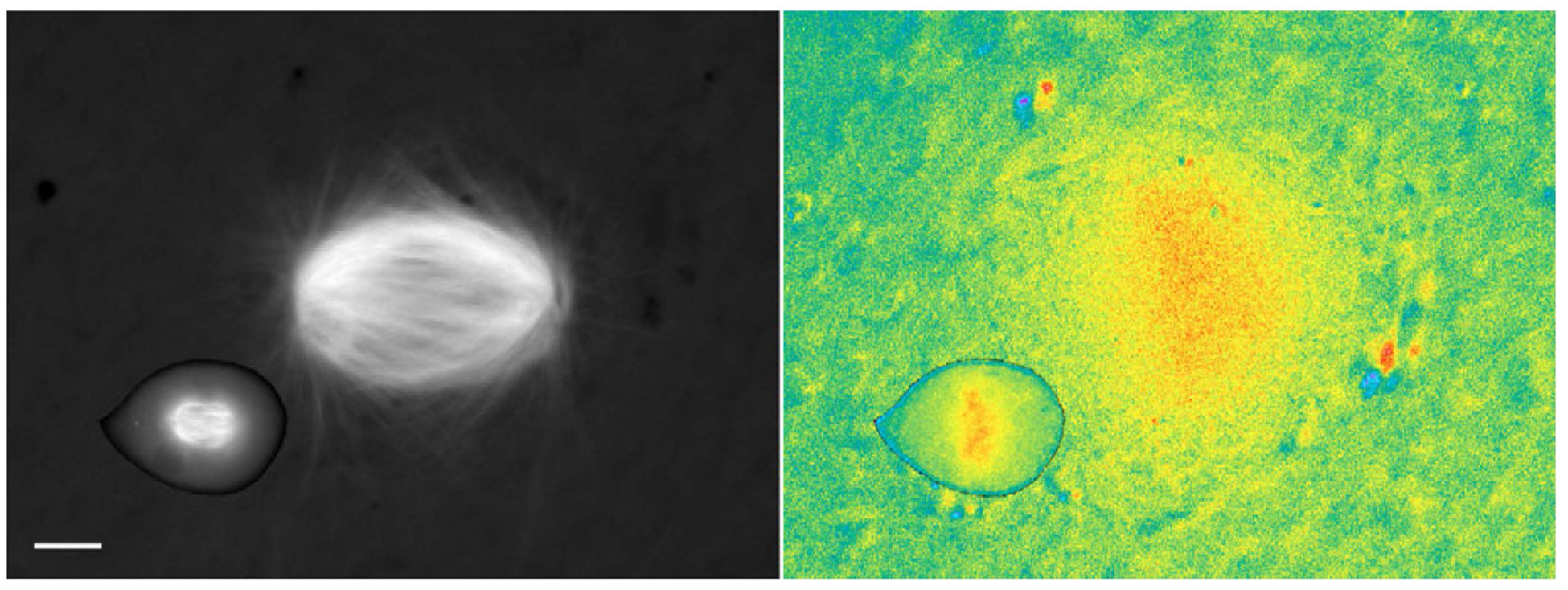
Mitotic importin β cargo gradients scale with the size of the mitotic spindle. (Left) Mitotic spindles visualized in a live HeLa cell and in X. laevis egg extract using incorporated Rhodamine-labeled tubulin. (Right) Pseudocolor image showing the signal of the Rango FRET sensor, which reports on the RanGTP-induced release of importin α and importin β cargos. The images are displayed at the same scale (scale bar, 10 μm). Adapted from Kalab et al. (Kalab et al., 2006) with permission.
The sufficiency of RanGTP, together with Aurora A activity, for spindle-pole formation has been demonstrated in X. laevis egg extracts, in which Aurora-A-coated beads induce bipolar spindle formation with a bead at each pole if RanGTP is added (Tsai and Zheng, 2005). Aurora A phosphorylation promotes spindle-pole formation by activating HURP binding to microtubules (Wong et al., 2008), by inhibiting the ubiquitylation activity of BRCA1 (Sankaran et al., 2007) and by enhancing the promotion of microtubule growth at spindle poles by TACC/maskin; (which occurs through its interaction with XMAP215) (reviewed in Barr and Gergely, 2007). The availability of TACC/maskin as a substrate also depends on its release from importins (Albee et al., 2006), augmenting the local effect of RanGTP. RanGTP-induced release from importin β is also required for the function of Cdk11 in spindle-pole stabilization (Yokoyama et al., 2008), which might be required for the efficient recruitment of Plkl and Aurora A to centrosomes (Petretti et al., 2006). Finally, Aurora A is an essential component of the above-mentioned HURP complex, which is required for the formation of mitotic spindle poles in X. laevis egg extracts (Koffa et al., 2006). It is unclear whether this complex also exists in somatic cells, where HURP is thought to function primarily in microtubule-kinetochore interactions (Sillje et al., 2006; Wong and Fang, 2006). However, HURP localizes to the centers of prometaphase microtubule asters, and relocates to microtubules adjacent to chromatin in metaphase cells only after bipolarity is achieved (Sillje et al., 2006; Wong and Fang, 2006). Altogether, this network of RanGTP-regulated synergistic interactions and positive-feedback mechanisms promotes the transition of spindle microtubules from prometaphase to metaphase (Fig. 2).
Mitotic functions of exportin 1
In contrast to the release of cargo in a RanGTP-gradient-dependent manner from importins, mitotic functions that are mediated by RanGTP-exportin result from binding interactions that target key components to the mitotic spindle apparatus. Once formed, RanGTP-exportin cargo complexes require an RBD protein and RanGAP for disassembly (Bischoff and Gorlich, 1997) and could therefore diffuse far from chromatin before RanGTP hydrolysis, similar to the RanGTP–importin-β complex (Caudron et al., 2005) (Fig. 1B). Therefore, it is possible that no significant concentration gradients of these complexes are formed in somatic cells, and although RanGTP is required for the formation of the complexes, its gradient may not be of importance. Instead, exportin 1 uses cargo-specific mechanisms to deliver its mitotic cargos to kinetochores, centromeres and centrosomes (Fig. 3).
At the kinetochore, exportin 1 functions as a ‘trap’, ready to bind the cargo that consists of the RanBP2-RanGAP-SUMO complex. A fraction of exportin 1 binds to kinetochores through its association with the Nup107-Nup160 nucleoporin complex (Zuccolo et al., 2007) in a RanGTP- and cargo-independent manner (Arnaoutov et al., 2005), and then recruits RanBP2-RanGAP-SUMO, generating a complex that is required for correct kinetochore-microtubule interactions. The formation of this large kinetochore complex requires exportin function, RanGTP, microtubules and SUMOylation of RanGAP (Arnaoutov et al., 2005). Disruption of RanBP2-RanGAP-SUMO complex formation as well as of its recruitment to kinetochores leads to defective kinetochore fibers, chromosome misalignment and mis-segregation (Arnaoutov et al., 2005; Joseph et al., 2004; Salina et al., 2003).
Exportin also functions at the centromeric chromatin that underlies the kinetochore, where it has been found to promote the localization of survivin (Knauer et al., 2006), an anti-apoptotic factor (Altieri, 2006), and a component of the chromosome passenger complex (CPC) that is required for spindle biorientation and contains Aurora B, INCENP, and borealin (Dasra in X. laevis) in addition to survivin (Ruchaud et al., 2007). Survivin apparently binds to centromeres after it is unloaded from exportin l (Knauer et al., 2006). Whether localization of the entire CPC is controlled by Ran through the exportin-1–survivin connection is unknown, but in mitotic X. laevis egg extracts CPC binds to chromatin through Dasra independently of Ran (Kelly et al., 2007).
Exportin l also associates with mitotic centrosomes (Wang et al., 2005), where it is thought to bind to nucleophosmin 1 (NPM1) (Budhu and Wang, 2005; Wang et al., 2005), a conserved nucleolar protein. How NPM1 functions at centrosomes is not clear, although experiments in tissue culture cells indicate that its NES-dependent centrosomal localization is required for centrosomal integrity (Wang et al., 2005). Also unclear is whether there is any connection between NPM1, exportin and a pool of Ran that has been found to be associated with centrosomal AKAP 450 (Keryer et al., 2003).
Species- and cell-type-specific functions of the Ran GPS in mitosis
The contribution of the Ran GPS to mitosis is organism- and cell-type-specific, which is not surprising given the evolutionary diversity of NTRs and the vast differences in cellular size that impose physical constraints on spindle assembly (Fig. 3). The large mitotic spindles assembled in X. laevis egg extract were disrupted by the addition of dominant-negative importin β mutants (Kalab et al., 2002; Nachury et al., 2001), but the same proteins microinjected into HeLa cells did not perturb spindles once they had formed (Kalab et al., 2006). Exportin functions also appear to vary, because the exportin-1–RanBP2–RanGAP–SUMO kinetochore complex is found in mammalian cells, but is absent from kinetochores in both X. laevis egg extracts and tissue culture cells (Arnaoutov and Dasso, 2005). Differences even exist in the requirement for the Ran gradient between consecutive meiotic cell divisions in mouse oocytes. Meiosis I spindles can recover from RCC1 inhibition, but meiosis II spindles are obliterated by the same treatment (Dumont et al., 2007; Schuh and Ellenberg, 2007). Therefore, generalizations about the functions of the mitotic Ran GPS must be made with caution.
However, some of the key mechanisms by which the RanGTP gradient functions to promote mitotic spindle assembly are conserved, even among organisms that undergo closed mitosis, such as yeast. For example, in Schizosaccharomyces pombe the RanGTP gradient is required for the import of A1p7 (TACC/maskin homologue) into the nucleus at the onset of mitosis. A1p7 is thought to be co-imported with Alp14 (an XMAP215 homologue) and the complex concentrates on microtubules to perform an essential function in bipolar spindle assembly (Sato and Toda, 2007). The emerging conserved paradigm therefore is that cells undergoing either open or closed mitosis depend on Ran-regulated SAFs to concentrate on microtubules within the peak of the RanGTP gradient in order to build a bipolar spindle.
An intermediate between the diffusion-limited RanGTP gradient in mitotic cells and the discrete gradient of RanGTP across the nuclear envelope may be found in syncytial Drosophila melanogaster embryos. Unlike labeled Ran in X. laevis eggs, starfish embryos and mammalian tissue culture cells (Hinkle et al., 2002), GFP-Ran concentrates within the mitotic spindles in the common syncytial cytoplasm (Trieselmann and Wilde, 2002), revealing the presence of some form of diffusion barrier despite the absence of a nuclear envelope.
New functions for the RanGTP gradient are being discovered that further extend its roles in cell division. A post-metaphase task for Ran in midbody formation has been described in D. melanogaster embryos (Silverman-Gavrila and Wilde, 2006). In mouse meiotic oocytes the RanGTP gradient marks the site at the plasma membrane where the first polar body will be extruded (Deng et al., 2007), suggesting that, in addition to directing spindle assembly around chromosomes, a longer-range Ran GPS signals the position of the spindle to the meiotic cell. RanGTP also determines the size of the products of meiosis I (Dumont et al., 2007). Whether Ran functions in spindle positioning and asymmetric cell divisions in somatic cells is unknown.
Are intracellular and embryonic morphogen gradients similar?
In the syncytial blastoderm of D. melanogaster embryos, a concentration gradient of bicoid directs the formation of the anterior and the position of posterior gene expression (Driever et al., 1989; Driever and Nusslein-Volhard, 1988; Gregor et al., 2007a; Gregor et al., 2007b; Lander, 2007). How does the RanGTP gradient, which has been called an ‘intracellular morphogen’ (Macara, 2002), compare with the embryonic morphogen gradient? The peak cytoplasmic level of the mitotic importin–β-cargo gradient in HeLa cells varies from cell to cell, although the amplitude of the gradient is more consistent (Kalab et al., 2006). By contrast, the bicoid gradient can deliver precise local morphogen concentrations to individual syncytial nuclei (Gregor et al., 2007a; Gregor et al., 2007b). To achieve a consistent local threshold concentration of liberated importin-β-regulated SAFs, cells would have to tightly control the expression of all NLS-containing proteins – a challenging requirement given the stochastic nature of gene expression in individual cells (Kaufmann and van Oudenaarden, 2007). However, in embryos in which the bicoid-gene dosage was experimentally upregulated up to sixfold, the cell-fate map shifted backwards but the embryos developed normally (Driever et al., 1989; Driever and Nusslein-Volhard, 1988). The formation of the embryo head was therefore robustly insensitive to the concentration of the morphogen, which is possibly analogous to the ability of the RanGTP gradient to direct spindle assembly despite fluctuations in protein expression.
Ran and cancer
The list of mitotic targets of Ran resembles a ‘Who’s Who’ of genes that are subject to frequent mutation, rearrangement or deletion in cancer (BRCA1, NPM1, survivin) or that are highly expressed in some cancers (NPM1, HURP, TPX2, Aurora A, TACC3, survivin, RHAMM; see Table 1). Although diverse mechanisms are probably at work, common themes are emerging, such as the misregulation in cancer cells of a Ran-regulated network that promotes spindle pole assembly. All five human homologues of the X. laevis HURP complex (Eg5, hTOG/XMAP215, TPX2, HURP, Aurora A) (Koffa et al., 2006; Sillje et al., 2006) are expressed coordinately with each other and with mitotic markers in cells derived from 22 different human tumors (Tsou et al., 2003; Wong and Fang, 2006), showing that, in some cancer cells, key Ran-regulated SAFs are upregulated at the same time. Rather than causing cell transformation directly, Ran-regulated SAFs are thought to contribute to tumor formation by overriding mitotic checkpoints, thereby inducing genomic instability (Castillo et al., 2007; Maxwell et al., 2008; Raff, 2002; Wong and Fang, 2006) It remains possible that aneuploidy, in turn; induces high levels of Ran-regulated SAFs such as HURP (Wong and Fang, 2006) promoting cancer-cell proliferation.
Expression levels of Ran-pathway components appear to be important in cancer. Ran and TPX2 were identified as two of the three most significant hits among 3700 genes tested in an RNAi screen for factors, the loss of which induced human tumor-cell death (Morgan-Lappe et al., 2007). Substantially elevated levels of Ran have been observed in a number of human tumors, and six tumor-derived tissue culture cell lines (including HeLa) contained Ran at concentrations many times higher than those found in three non-transformed human fibroblast lines (Xia et al., 2008). Importantly, the suppression of Ran by RNAi was tolerated by non-transformed cells but induced cell death in cancer cell lines. However, this could be reversed by the overexpression of survivin (Xia et al., 2008), itself a Ran- and exportin-1-regulated anti-apoptotic factor and mitotic regulator (Knauer et al., 2006; Knauer et al., 2007).
Interestingly, many Ran-related cancer pathways seem to involve importin al (KPNA2 in humans), which was identified as one of three most commonly overexpressed genes in undifferentiated cancer cells among six cancer types (Rhodes et al., 2004). Rather than acting directly on SAFs, importin α1 promotes the proliferation of undifferentiated cells by driving the nuclear import of the transcription factor Oct4 (also known as Oct3 or POU5F1) (Yasuhara et al., 2007), raising the question of whether its transcription targets include Ran-regulated SAFs. It is noteworthy that, although high overall expression of importin α1 correlates with a high risk of developing breast cancer, it is its nuclear accumulation that correlates with particularly poor prognosis (Dahl et al., 2006).
Conclusions
Since its discovery less than a decade ago, studies of the mitotic Ran GPS have provided many important insights into the amazingly complex interactions between mitotic chromosomes and cytoplasmic components during spindle assembly. The well-characterized and tractable components of the Ran-NTR system have served as a powerful discovery tool, enabling the biochemical discovery of mitotic regulators and facilitating the visualization of mitotic chromosome signals. The enzymes that control the Ran nucleotide state are dynamically regulated in space, and the mitotic function of Ran is subject to species- and cell-type-specific variations. Nevertheless, in mitosis, the localized production and diffusion-limited dissipation of the RanGTP signal appears to be conserved, and has similarities to embryonic morphogen gradients.
Until now, much of the research on Ran has relied on meiotic, embryonic and cancer-cell-based systems. It is becoming apparent that the powerful mitotic Ran GPS is characteristic of mitosis in non-differentiated and embryonic cells, where it may underlie their rapid proliferation. Increasing evidence suggests that aberrant or amplified Ran pathways can overwhelm mitotic controls in differentiated cells, thereby contributing to cancer. Important future directions are to investigate the function of the Ran GPS in differentiated somatic cells, and to exploit its role in cancer for the development of novel therapeutics.
We thank Karsten Weis, Sarah Munchel, Rose Loughlin and Jon Soderholm for helpful discussions and comments on the manuscript.
References
- Albee AJ, Tao W and Wiese C (2006). Phosphorylation of maskin by Aurora-A is regulated by RanGTP and importin beta. J. Biol. Chem 281, 38293–38301. [DOI] [PubMed] [Google Scholar]
- Altieri DC (2006). The case for survivin as a regulator of microtubule dynamics and cell-death decisions. Curr. Opin. Cell Biol 18, 609–615. [DOI] [PubMed] [Google Scholar]
- Arnaoutov A and Dasso M (2005). Ran-GTP regulates kinetochore attachment in somatic cells. Cell Cycle 4, 1161–1165. [DOI] [PubMed] [Google Scholar]
- Arnaoutov A, Azuma Y, Ribbeck K, Joseph J, Boyarchuk Y, Karpova T, McNally J and Dasso M (2005). Crml is a mitotic effector of Ran-GTP in somatic cells. Nat. Cell Biol 7, 626–632. [DOI] [PubMed] [Google Scholar]
- Askjaer P, Galy V, Hannak E and Mattaj IW (2002). Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos. Mol. Biol Cell 13, 4355–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr AR and Gergely F (2007). Aurora-A: the maker and breaker of spindle poles. J Cell Sci 120, 2987–2996. [DOI] [PubMed] [Google Scholar]
- Bastiaens P, Caudron M, Niethammer P and Karsenti E (2006). Gradients in the self-organization of the mitotic spindle. Trends Cell Biol 16, 125–134. [DOI] [PubMed] [Google Scholar]
- Bilbao-Cortes D, Hetzer M, Langst G, Becker PB and Mattaj IW (2002). Ran binds to chromatin by two distinct mechanisms. Curr. Biol 12, 1151–1156. [DOI] [PubMed] [Google Scholar]
- Bischoff FR and Gorlich D (1997). RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 419, 249–254. [DOI] [PubMed] [Google Scholar]
- Blower MD, Nachury M, Heald R and Weis K (2005). A Rael-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell 121, 223–234. [DOI] [PubMed] [Google Scholar]
- Budhu AS and Wang XW (2005). Loading and unloading: orchestrating centrosome duplication and spindle assembly by Ran/Crml. Cell Cycle 4, 1510–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo A, Morse HC 3rd, Godfrey VL, Naeem R and Justice MJ (2007). Overexpression of Eg5 causes genomic instability and tumor formation in mice. Cancer Res. 67, 10138–10147. [DOI] [PubMed] [Google Scholar]
- Caudron M, Bunt G, Bastiaens P and Karsenti E (2005). Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science 309, 1373–1376. [DOI] [PubMed] [Google Scholar]
- Chen T, Muratore TL, Schaner-Tooley CE, Shabanowitz J, Hunt DF and Macara IG (2007). N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat. Cell Biol 9, 596–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciciarello M, Mangiacasale R, Thibier C, Guarguaglini G, Marchetti E, Di Fiore B and Lavia P (2004). lmportin beta is transported to spindle poles during mitosis and regulates Ran-dependent spindle assembly factors in mammalian cells. J. Cell Sci 117, 6511–6522. [DOI] [PubMed] [Google Scholar]
- Cingolani G, Petosa C, Weis K and Muller CW (1999). Structure of importin-beta bound to the IBB domain of importin-alpha. Nature 399, 221–229. [DOI] [PubMed] [Google Scholar]
- Clarke PR and Zhang C (2004). Spatial and temporal control of nuclear envelope assembly by Ran GTPase. Symp. Soc. Exp. Biol 2004, 193–204. [PubMed] [Google Scholar]
- Conti E, Muller CW and Stewart M (2006). Karyopherin flexibility in nucleocytoplasmic transport. Curr. Opin. Struct. Biol 16, 237–244. [DOI] [PubMed] [Google Scholar]
- Dahl E, Kristiansen G, Gottlob K, Klaman I, Ebner E, Hinzmann B, Hermann K, Pilarsky C, Durst M, Klinkhammer-Schalke M et al. (2006). Molecular profiling of laser-microdissected matched tumor and normal breast tissue identifies karyopherin alpha2 as a potential novel prognostic marker in breast cancer. Clin. Cancer Res 12, 3950–3960. [DOI] [PubMed] [Google Scholar]
- Dasso M (2001). Running on Ran: nuclear transport and the mitotic spindle. Cell 104, 321–324. [DOI] [PubMed] [Google Scholar]
- Dasso M (2006). Ran at kinetochores. Biochem. Soc. Trans 34, 711–715. [DOI] [PubMed] [Google Scholar]
- Deng M, Suraneni P, Schultz RM and Li R (2007). The Ran GTPase mediates chromatin signaling to control cortical polarity during polar body extrusion in mouse oocytes. Dev. Cell 12, 301–308. [DOI] [PubMed] [Google Scholar]
- Driever W and Nusslein-Volhard C (1988). The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell 54, 95–104. [DOI] [PubMed] [Google Scholar]
- Driever W, Ma J, Nusslein-Volhard C and Ptashne M (1989). Rescue of bicoid mutant Drosophilaembryos by bicoid fusion proteins containing heterologous activating sequences. Nature 342, 149–154. [DOI] [PubMed] [Google Scholar]
- Dumont J, Petri S, Pellegrin F, Terret ME, Bohnsack MT, Rassinier P, Georget V, Kalab P, Gruss OJ and Verlhac MH (2007). A centriole- and RtranGTP- independent spindle assembly pathway in meiosis I of vertebrate oocytes. J. Cell Biol 176, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ems-McClung SC, Zheng Y and Walczak CE (2004). Importin alpha/beta and Ran-GTP regulate XCTK2 microtubule binding through a bipartite nuclear localization signal. Mol. Biol. Cell 15, 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Fogg V, Wang Q, Chen XW, Liu CJ and Margolis B (2007). A novel Crumbs3 isoform regulates cell division and ciliogenesis via importin {beta} interactions. J. Cell Biol 178, 387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H and Kutay U (2003). Nucleocytoplasmic transport: taking an inventory. Cell. Mol. Life Sci 60, 1659–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman B and Zheng Y (2006). Mitotic spindle morphogenesis: ran on the microtubule cytoskeleton and beyond. Biochem. Soc. Wans 34, 716–721. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Seewald MJ and Ribbeck K (2003). Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J. 22, 1088–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, Tank DW, Wieschaus EF and Bialek W (2007a). Probing the limits to positional information. Cell 130, 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, Wieschaus EF, McGregor AP, Bialek W and Tank DW (2007b). Stability and nuclear dynamics of the bicoid morphogen gradient. Cell 130, 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen AC, Cameron LA, Coughlin M, Miyamoto DT, Mitchison TJ and Ohi R (2004) XRHAMM functions in ran-dependent microtubule nucleation and pole formation during anastral spindle assembly. Curr. Biol 14, 1801–1811. [DOI] [PubMed] [Google Scholar]
- Guarguaglini G, Renzi L, D’Ottavio F, Di Fiore B, Casenghi M, Cundari E and Lavia P (2000). Regulated Ran-binding protein 1 activity is required for organization and function of the mitotic spindle in mammalian cells in vivo. Cell Gmwth Differ. 11, 455–465. [PubMed] [Google Scholar]
- Hetzer M, Gruss OJ and Mattaj IW (2002). The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat. Cell Biol 4, E177–E184. [DOI] [PubMed] [Google Scholar]
- Hinkle B, Slepchenko B, Rolls MM, Walther TC, Stein PA, Mehlmann LM, Ellenberg J and Terasaki M (2002). Chromosomal association of Ran during meiotic and mitotic divisions. J. Cell Sci 115, 4685–4693. [DOI] [PubMed] [Google Scholar]
- Hood FE and Clarke PR (2007). RCC1 isoforms differ in their affinity for chromatin, molecular interactions and regulation by phosphorylation. J. Cell Sci 120, 3436–3445. [DOI] [PubMed] [Google Scholar]
- Hutchins JR, Moore WJ, Hood FE, Wilson JS, Andrews PD, Swedlow JR and Clarke PR (2004). Phosphorylation regulates the dynamic interaction of RCC1 with chromosomes dmng mitosis. Curr. Biol 14, 1099–1104. [DOI] [PubMed] [Google Scholar]
- Joseph J, Tan SH, Karpova TS, McNally JG and Dasso M (2002). SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol 156, 595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J, Liu ST, Jablonski SA, Yen TJ and Dasso M (2004). The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol 14, 611–617. [DOI] [PubMed] [Google Scholar]
- Joukov V, Groen AC, Prokhorova T, Gerson R, White E, Rodriguez A, Walter JC and Livingston DM (2006). The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell 127, 539–552. [DOI] [PubMed] [Google Scholar]
- Kalab P, Weis K and Heald R (2002). Visualization of a Ran-GTP gradient in inteiphase and mitotic Xenopus egg extracts. Science 295, 2452–2456. [DOI] [PubMed] [Google Scholar]
- Kalab P, Pralle A, Isacoff EY, Heald R and Weis K (2006). Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature 440, 697–701. [DOI] [PubMed] [Google Scholar]
- Kaufmann BB and van Oudenaarden A (2007). Stochastic gene expression: from single molecules to the proteome. Curr. Opin. Genet. Dev 17, 107–112. [DOI] [PubMed] [Google Scholar]
- Kelly AE, Sampath SC, Maniar TA, Woo EM, Chait BT and Funabiki H (2007). Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev. Cell 12, 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keryer G, Di Fiore B, Celati C, Lechtreck KF, Mogensen M, Delouvee A, Lavia P, Bornens M and Tassin AM (2003). Part of Ran is associated with AKAP450 at the centrosome: involvement in microtubule-organizing activity. Mol Biol. Cell 14, 4260–4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Noetzel TL, Pelletier L, Mechtler K, Drechsel DN, Schwager A, Lee M, Raff JW and Hyman AA (2005). Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J. Cell Biol 170, 1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe C, Prinz H, Wittinghofer A and Goody RS (1995). The kinetic mechanism of Ran-nucleotide exchange catalyzed by RCC1. Biochemistry 34, 12543–12552. [DOI] [PubMed] [Google Scholar]
- Knauer SK, Bier C, Habtemichael N and Stauber RH (2006). The Survivin-Crml interaction is essential for chromosomal passenger complex localization and function. EMBO Rep. 7, 1259–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer SK, Kramer OH, Knosel T, Engels K, Rodel F, Kovacs AF, Dietmaier W, Klein-Hitpass L, Habtemichael N, Schweitzer A et al. (2007). Nuclear export is essential for the tumor-promoting activity of survivin. FASEB J. 21, 207–216. [DOI] [PubMed] [Google Scholar]
- Koffa MD, Casanova CM, Santarella R, Kocher T, Wilm M and Mattaj IW (2006). HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol 16, 743–754. [DOI] [PubMed] [Google Scholar]
- Lander AD (2007). Morpheus unbound: reimagining the morphogen gradient. Cell 128, 245–256. [DOI] [PubMed] [Google Scholar]
- Li HY and Zheng Y (2004a). Phosphorylation of RCC1 in mitosis is essential for producing a high RanGTP concentration on chromosomes and for spindle assembly in mammalian cells. Genes Dev. 18, 512–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HY and Zheng Y (2004b). The production and localization of GTP-bound ran in mitotic mammalian tissue culture cells. Cell Cycle 3, 993–995. [PubMed] [Google Scholar]
- Macara IG (2002). Why FRET about Ran? Dev. Cell 2, 379–380. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Anantharaman V, Aravind L and Koonin EV (2004). Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 3, 1612–1637. [DOI] [PubMed] [Google Scholar]
- Maresca TJ, Niederstrasser H, Weis K and Heald R (2005). Xnf7 contributes to spindle integrity through its microtubule-bundling activity. Curr. Biol 15, 1755–1761. [DOI] [PubMed] [Google Scholar]
- Mason DA, Fleming RJ and Goldfarb DS (2002). Drosophila melanogaster importin alpha1 and alpha3 can replace importin alpha2 during spermatogenesis but not oogenesis. Genetics 161, 157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell CA, McCarthy J and Turley E (2008). Cell-surface and mitotic-spindle moonlighting or dual oncogenic functions? J. Cell Sci 121, 925–932. [DOI] [PubMed] [Google Scholar]
- Morgan-Lappe SE, Tucker LA, Huang X, Zhang Q, Sarthy AV, Zakula D, Vernetti L, Schurdak M, Wang J and Fesik SW (2007). Identification of Ras-related nuclear protein, targeting protein for xenopus kinesin-like protein 2, and stearoyl-CoA desaturase l as promising cancer targets from an RNAi-based screen. Cancer Res. 67, 4390–4398. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R and Weis K (2001). Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell 104, 95–106. [DOI] [PubMed] [Google Scholar]
- Nemergut ME, Mizzen CA, Stukenberg T, Allis CD and Macara IG (2001). Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science 292, 1540–1543. [DOI] [PubMed] [Google Scholar]
- Niethammer P, Bastiaens P and Karsenti E (2004). Stathmin-tubulin interaction gradients in motile and mitotic cells. Science 303, 1862–1866. [DOI] [PubMed] [Google Scholar]
- O’Brien LL, Albee AJ, Liu L, Tao W, Dobrzyn P, Lizarraga SB and Wiese C (2005). The Xenopus TACC homologue, maskin, functions in mitotic spindle assembly. Mo/. Biol. Cell 16, 2836–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton LF and Paschal BM (2005). Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6, 187–198. [DOI] [PubMed] [Google Scholar]
- Peset I, Seiler J, Sardon T, Bejarano LA, Rybina S and Vernos I (2005). Function and regulation of Maskin, a TACC family protein, in microtubule growth during mitosis. J. Cell Biol 170, 1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petretti C, Savoian M, Montembault E, Glover DM, Prigent C and Giet R (2006). The PITSLRE/CDK11p58 protein kinase promotes centrosome maturation and bipolar spindle formation. EMBO Rep. 7, 418–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quensel C, Friedrich B, Sommer T, Hartmann E and Kohler M (2004). In vivo analysis of importin alpha proteins reveals cellular proliferation inhibition and substrate specificity. Mol. Cell. Biol 24, 10246–10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff JW (2002). Centrosomes and cancer: lessons from a TACC. Trends Cell Biol. 12, 222–225. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM (2004). Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc. Natl. Acad. Sci. USA 101, 9309–9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Groen AC, Santarella R, Bohnsack MT, Raemaekers T, Kocher T, Gentzel M, Gorlich D, Wilm M, Carmeliet G et al. (2006). NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol. Biol. Cell 17, 2646–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Raemaekers T, Carmeliet G and Mattaj IW (2007). A role for NuSAP in linking microtubules to mitotic chromosomes. Curr. Biol 17, 230–236. [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C and Stutz F (2004). Nuclear export of RNA. Biol. Cell 96, 639–655. [DOI] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M and Earnshaw WC (2007). Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol 8, 798–812. [DOI] [PubMed] [Google Scholar]
- Salina D, Enarson P, Rattner JB and Burke B (2003). Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol 162, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran S, Crone DE, Palazzo RE and Parvin JD (2007). Aurora-A kinase regulates breast cancer associated gene I inhibition of centrosome-dependent microtubule nucleation. Cancer Res. 67, 11186–11194. [DOI] [PubMed] [Google Scholar]
- Sato M and Toda T (2007). A1p7/TACC is a crucial target in Ran-GTPase-dependent spindle formation in fission yeast. Nature 447, 334–337. [DOI] [PubMed] [Google Scholar]
- Schuh M and Ellenberg J (2007). Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell 130, 484–498. [DOI] [PubMed] [Google Scholar]
- Sillje HH, Nagel S, Korner R and Nigg EA (2006). HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol 16, 731–742. [DOI] [PubMed] [Google Scholar]
- Silverman-Gavrila RV and Wilde A (2006). Ran is required before metaphase for spindle assembly and chromosome alignment and after metaphase for chromosome segregation and spindle midbody organization. Mol. Biol. Cell 17, 2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AE, Slepchenko BM, Schaff JC, Loew LM and Macara IG (2002). Systems analysis of Ran transport. Science 295, 488–491. [DOI] [PubMed] [Google Scholar]
- Stewart M (2006). Structural basis for the nuclear protein import cycle. Biochem. Soc. Trans 34, 701–704. [DOI] [PubMed] [Google Scholar]
- Stewart M (2007). Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol 8, 195–208. [DOI] [PubMed] [Google Scholar]
- Tahara K, Takagi M, Ohsugi M, Sone T, Nishiumi F, Maeshima K, Horiuchi Y, Tokai-Nishizumi N, Imamoto F, Yamamoto T et al. (2008). Importin-beta and the small guanosine triphosphatase Ran mediate chromosome loading of the human chromokinesin Kid. J. Cell Biol 180, 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi A, Ciciarello M, Mangiacasale R, Roscioli E, Rensen WM and Lavia P (2007). RANBP1 localizes a subset of mitotic regulatory factors on spindle microtubules and regulates chromosome segregation in human cells. J. Cell Sci 120, 3748–3761. [DOI] [PubMed] [Google Scholar]
- Terry LJ, Shows EB and Wente SR (2007). Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science 318, 1412–1416. [DOI] [PubMed] [Google Scholar]
- Travis J (2007). Return of the matrix. Science 318, 1400–1401. [DOI] [PubMed] [Google Scholar]
- Trieselmann N and Wilde A (2002). Ran localizes around the microtubule spindle in vivo during mitosis in Drosophila embryos. Curr. Biol 12, 1124–1129. [DOI] [PubMed] [Google Scholar]
- Teieselmann N, Armstrong S, Rauw J and Wilde A (2003). Ran modulates spindle assembly by regulating a subset of TPX2 and Kid activities including Aurora A activation. J. Cell Sci 116, 4791–4798. [DOI] [PubMed] [Google Scholar]
- Tsai MY and Zheng Y (2005). Aurora A kinase-coated beads function as microtubule-organizing centers and enhance RanGTP-induced spindle assembly. Curr. Biol 15, 2156–2163. [DOI] [PubMed] [Google Scholar]
- Tsai MY, Wang S, Heidinger JM, Shumaker DK, Adam SA, Goldman RD and Zheng Y (2006). A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science 311, 1887–1893. [DOI] [PubMed] [Google Scholar]
- Tsou AP, Yang CW, Huang CY, Yu RC, Lee YC, Chang CW, Chen BR, Chung YF, Fann MJ, Chi CW et al. (2003). Identification of a novel cell cycle regulated gene, HURP, overexpressed in human hepatocellular carcinoma. Oncogene 22, 298–307. [DOI] [PubMed] [Google Scholar]
- Vetter IR, Nowak C, Nishimoto T, Kuhlmann J and Wittinghofer A (1999). Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature 398, 39–46. [DOI] [PubMed] [Google Scholar]
- Wang W, Budhu A, Forgues M and Wang XW (2005). Temporal and spatial control of nucleophosmin by the Ran-Crml complex in centrosome duplication. Nat. Cell Biol 7, 823–830. [DOI] [PubMed] [Google Scholar]
- Weis K (2003). Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112, 441–451. [DOI] [PubMed] [Google Scholar]
- Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO et al. (2002). Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13, 1977–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Wilde A, Moore MS, Adam SA, Merdes A and Zheng Y (2001). Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science 291, 653–656. [DOI] [PubMed] [Google Scholar]
- Wong J and Fang G (2006). HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J. Cell Biol 173, 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Lerrigo R, Jang CY and Fang G (2008). Aurora A regulates the activity of HURP by controlling the accessibility of its microtubule-binding domain. Mol Biol. Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RW, Blobel G and Coutavas E (2006). Rael interaction with NuMA is required for bipolar spindle formation. Proc. Natl. Acad. Sci. USA 103, 19783–19787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia F, Lee CW and Altieri DC (2008). Tumor cell dependence on Ran-GTP-directed mitosis. Cancer Res. 68, 1826–1833. [DOI] [PubMed] [Google Scholar]
- Yasuhara N, Shibazaki N, Tanaka S, Nagai M, Kamikawa Y, Oe S, Asally M, Kamachi Y, Kondoh H and Yoneda Y (2007). Triggering neural differentiation of ES cells by subtype switching of importin-alpha. Nat Cell Biol. 9, 72–79. [DOI] [PubMed] [Google Scholar]
- Yokoyama H, Gruss OJ, Rybina S, Caudron M, Scheider M, Wilm M, Mattaj IW and Karsenti E (2008). Cdk11 is a RanGTP-dependent microtubule stabilization factor that regulates spindle assembly rate. J. Cell Biol 180, 867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Simon PM and Borisy GG (1996). Microtubule dynamics at the G2/M transition: abrupt breakdown of cytoplasmic microtubules at nuclear envelope breakdown and implications for spindle morphogenesis. J. Cell Biol 135, 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lan W, Ems-McClung SC, Stukenberg PT and Walczak CE (2007). Aurora B Phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Mol. Biol. Cell 18, 3264–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, Sibarita JB, Fukagawa T, Shiekhattar R, Yen T et al. (2007). The human Nup107–160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 26, 1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]


