Discovery of MHC Restriction
“Interaction between immune lymphocytes and cells expressing antigens expressed by LCM virus is, however, apparently confined to a histocompatible system perhaps because it is only in this situation that the necessary intimacy of contact is achieved…… An alternative possibility that must be considered in LCM is that the process of virus maturation through the cell membrane causes changes in self-components, which are recognized only within the syngeneic or semiallogeneic system….. The cytotoxic T cell may thus be recognizing altered self” (99)
So records the critical interpretation of experiments performed by Rolf M. Zinkernagel and Peter C. Doherty, which documented the ability of T cells from lymphocytic choriomeningitis virus (LCMV)-infected mice to kill LCMV-infected target cells in vitro, only if the T cells and the targets shared at least one H-2 antigen. Of course, we now understand the extraordinary insight of this interpretation. In 1996, the same year that Zinkernagel and Doherty were awarded the Nobel Prize for their discovery of major histocompatibility complex (MHC) restriction, the first structures of T cell receptor-peptide+MHC (TCR-pMHCI) complexes were reported, for mouse (26) and human (23), enabling the direct visualization of this interaction. Indeed, these initial structures and those following show simultaneous TCR binding of self-MHC and either self- or non-self-peptide for the express purpose of detection of altered self. In the joint recognition of self and non-self, this receptor-ligand interaction is unprecedented in biology, highlighting the perceptiveness of Doherty and Zinkernagel's interpretation. Of course, at the time Zinkernagel and Doherty's experiments were performed, both murine transplantation antigens and the human lymphocyte antigen (HLA) system had been discovered, and their influence on transplantation outcomes as well as differential susceptibility to infectious diseases had been noted [reviewed in Zinkernagel and Doherty (100)]. However, the purpose and mode of action of major histocompatibility antigens were unknown. It was speculated that the extreme polymorphism of the MHC served to protect the body from invasion by cells (e.g., tumor cells) from another individual (11), to facilitate cell-cell recognition for differentiation and morphogenesis of multicellular organisms (8), or to drive elimination of self-reactive antibody-producing cells (43), among other theories. In an attempt to address the question of whether the lytic activity of antiviral cytotoxic T lymphocytes (CTLs), which were responsible for fatal choriomeningitis, could be correlated with H-2 haplotype, Doherty and Zinkernagel tailored the 51Cr release assay to allow for analysis of cytolysis by relatively few LCMV-specific CTLs. In doing so, they made the striking finding that unveiled the mechanism of MHC restriction, namely that CTL effectors could kill virally infected targets only if the two cells were H-2 compatible.
Defining Characteristics of the TCR-pMHC Interaction
Over 40 years have passed since these original observations and our understanding of MHC restriction of TCR recognition has progressed significantly. Extreme diversity is inherent in all components of the interaction; generation of the TCR by a somatic recombination process allows a theoretical diversity of ∼1015–18 different TCRs, of which ∼2.5 × 107 can be found within any individual (17); the MHC genes are the most polymorphic genes of the human genome, and the peptide cargo that can be bound by MHC is virtually limitless. As a consequence, TCR recognition of pMHC is unparalleled in the diversity of the interacting surfaces. Our understanding of the nature of TCR-pMHC interactions can be almost completely attributed to structural studies—over 50 ternary structures of unique TCRs interacting with cognate pMHCI or pMHCII have been solved—which have provided direct, high-resolution visualization of these interactions, enabling conserved patterns of TCR-pMHC binding to be identified (reviewed in La Gruta et al., Rossjohn et al. (50,70).
Notably, the extreme diversity in the interacting ligands is reflected in considerable variation in the manner of the interaction; namely in the relative contribution to binding of the various molecules, in the angle at which TCR docks over pMHC (pMHCI: average ∼63°; pMHCII: average ∼76°), and in the extent of contact between the TCR and pMHC (buried surface area ranges from 1,240 to 2,400 A2) (70). Thus, the interaction is able to simultaneously maintain specificity, while accommodating the variable nature of the ligands. The diversity inherent in the TCR-pMHC interaction is perhaps best exemplified by the observation that within both MHCI and MHCII-restricted TCR recognition, the same TCR V gene elements can interact with multiple pMHC complexes, with no conservation in the interactions between TCR and MHC in the different contexts (70).
Despite the flexibility of the interaction, key structural characteristics have emerged, which are highly conserved across virtually all interactions. The TCR makes contact with both the peptide cargo and the MHC (co-recognition), the TCR binds over the top of the peptide, the peptide is contacted by at least one of the highly variable TCR CDR3 loops, and the TCR binds pMHC with a highly conserved polarity. That is, the TCR α chain sits over the top of the MHCI α2 helix or MHCII β chain, while the TCR β chain is positioned over the MHCI α1 helix or MHCII α chain (70,72). Recently, examples of deviations from these tenets of TCR recognition have been reported (discussed below) and have the potential to advance our understanding of the key requirements for TCR recognition of pMHC.
Models to Explain TCR Recognition of pMHC
Given the inherent random rearrangement process, and the consequent diversity of TCRs, one overarching question that is yet to be definitively answered is, what drives MHC restriction? Here we discuss our current understanding of the TCR's obsession with MHC by examining evidence from the perspective of the two current models: the germline encoded and selection theories of MHC restriction.
Germline-encoded hypothesis
The germline encoded model of TCR-MHC bias stems from an idea first proposed by Jerne (43), suggesting that the TCR and MHC genes have, over millions of years of co-evolution, developed an intrinsic bias for co-recognition. It suggests that the intrinsic bias of TCRs for MHC molecules is driven by evolutionarily conserved amino acid motifs encoded by both the TCR and MHC molecules. Recent reports of complete TCR-pMHC structures solved by X-ray crystallography have identified multiple conserved pairwise interactions between the TCR and the MHC molecule [termed “interaction codons” (25)] leading to the hypothesis that TCRs are genetically biased toward MHC recognition. The germline-encoded theory of MHC restriction relies on evolutionarily conserved pair-wise interactions between the TCR and pMHC, and suggests that any deviation from “interaction codons” would result in unfavorable or low- affinity interactions. The germline-encoded model is supported by data that show up to 30% of the preselection repertoire is reactive to MHC molecules (61,62,83,98). However, it does not support unconventional modes of antigen recognition, namely TCRs that recognize MHC-independent antigens (4,35,58,68,81,90) or reversed polarity recognition of TCRs on MHCI and MHCII (6,32).
Selection hypothesis
Some immunologists argue that germline-encoded bias of TCRs for MHC molecules is minimal and that MHC restriction is driven ultimately by constraints imposed on the TCR during positive selection and the nature of TCR signaling (67,92). TCRs themselves do not possess intrinsic signaling capacity, and instead, TCR signaling relies on the delivery of Lck to the CD3 complex when associated with the cytoplasmic tails of CD4 or CD8 co-receptors (87). Thus, the CD4 and CD8 co-receptors act to focus the TCR onto the MHC molecule. This requirement for co-receptor facilitation of signaling prevents development of T cells expressing TCRs that are not specific for MHC, ensuring MHC restriction. Early support for this model was found when mice lacking expression of co-receptors developed a diverse repertoire of αβ T cells that could react to conformational epitopes independent of MHC presentation (90), suggesting that co-receptors are critical for MHC restriction. This model is also supported by the fact that that at least 70% of the preselection repertoire has no intrinsic capacity to recognize MHC (61,62,83,98) and also supports the many reports of unconventional modes of antigen recognition, namely two distinct examples of reversed polarity docking over MHCII and MHCI (6,32), atypical binding modes such as recognition of super-bulged peptides (88) and TCR recognition of MHC-independent ligands (4,35,58,68,81,90).
Analysis of the Preselection Repertoire
Positive selection in the thymus, and therefore continued T cell development, depends on the TCR-pMHC interaction delivering a survival signal to T cells. As a consequence, the postselection TCR repertoire is altered to reflect “successful” TCRs. For this reason, immunologists have relied on analyses of αβ TCRs expressed by preselection thymocytes to understand intrinsic MHC reactivity without selection bias. Studies have used mice that lack the expression of MHCI and MHCII molecules such that thymocytes are blocked at the CD4+CD8+TCRαβ+ stage (62,86,98) or TCRα−/− mice (47), which are unable to express TCR on their cell surface. Both of these T cell populations have intact TCRαβ gene rearrangements that allow the sampling of “untested” preselection TCRs for the presence or absence of inherent MHC reactivity by germline-encoded recognition motifs. The ability to test the MHC reactivity of the preselection repertoire using MHC-deficient models has revealed that a substantial proportion (15–30%) of TCRs has the capacity to be activated by pMHC molecules (61,62,83,98), a relatively high frequency if we consider that there are 1015–20 theoretical TCR rearrangements possible, but arguably a low frequency if intrinsic affinity for MHC molecules is an evolutionarily encoded feature of TCRs. Similarly, the use of TCRα−/− mice to study MHC reactivity of unselected TCRα sequences was consistent with these findings and fascinatingly, pairing of unselected TCRα sequences with TCRβ sequences containing a modified germline region derived from frog, shark, or trout retained a high frequency of pMHC-reactive cells (47), suggestive of a highly evolutionarily conserved TCR bias for MHC reactivity.
Another approach used to resolve this biological question was the generation of mice lacking MHCI, MHCII, CD8, and CD4 (Quad-KO mice) or expressing a mutant variant of Lck that cannot associate with co-receptors (90,91), both of which circumvented the requirement for MHC recognition during T cell development. Surprisingly, in this context, when Lck was not constrained to a co-receptor, αβ T cells were positively selected, expressing TCRs that recognized non-MHC-associated epitopes in a manner akin to antibody recognition of foreign antigens. Specifically, Quad-KO mice were found to positively select αβ T cells specific for multiple conformational epitopes of CD155, an abundant molecule in the thymus, without any detectable reactivity toward MHC molecules. This study was the first evidence that MHC restriction was imposed at least, in part, by selection events and driven by co-receptor binding to MHC molecules. One thing to note is that CD155-reactive TCRs made up around 40% of the TCR repertoire in QuadKO mice. While this was taken as supporting evidence for the selection model, the fact that most of the repertoire was reactive to a single molecule was also interpreted as evidence for the importance of germline-encoded recognition, given the apparently limited capacity of TCRs to recognize non-MHC-restricted epitopes.
More recently, the MHC-independent TCR repertoires from Quad-KO mice were compared with MHC-restricted TCR repertoires from intact mice of different MHC haplotypes (B6 and B10.BR mice). Interestingly, while ∼33% of TCR sequences were shared between individual B6 and B10.BR mice, individual MHC-independent TCR repertoires derived from Quad-KO mice shared only 3% of TCR sequences, with the lack in sequence overlap between mice resembling the extent of overlap usually found in antibody repertoires (57). These data indicate that thymic selection samples from a very large repertoire of preselection TCRs and drives selection of a much smaller pool of MHC-restricted TCRs.
Ultimately, studies of the preselection repertoire have yielded results that provide evidence and support for both germline-encoded and selection models of MHC restriction. While it appears that germline-encoded amino acids are not strictly required for the development of αβ TCRs restricted to MHC molecules, evolutionary pressures appear to have predisposed a level of complementarity between the TCR and the MHC.
A Structural Perspective on TCR-pMHC Recognition
Conserved residues in the TCR-MHC interaction
The very first TCR-pMHC complexes reported showed that the germline-encoded CDR1 and CDR2 loops made contact with the MHC molecule, whereas the most variable CDR3 loop typically lay centrally over the peptide. These early structural studies revealed three key residues on MHCI (65, 69, and 155; termed the “restriction triad”) that were contacted by nearly all TCRs identified at that point in time, suggesting that these key positions predetermined, and were the minimum requirements for, MHC restriction of TCRs. However, mutation of any or all of these three residues did not abolish TCR recognition of MHC (12), suggesting that reproducible contacts to the “restriction triad” were dispensable and perhaps represented incidental recognition motifs due to the fairly reproducible nature of TCR-pMHC docking.
Other approaches to understanding the germline-encoded bias of TCRs have identified germline-encoded motifs in Vβ8.2+ TCRs (βY46, βY48, and βE54) make reproducible contacts with different pMHC molecules (60). Artificially modifying these conserved residues substantially diminished, but did not abolish, naive T cell development (75), indicative of an important but nonessential role in TCR-MHC recognition. Further evidence of the plasticity and flexibility of TCR-MHC recognition was demonstrated in an elegant study by Holland et al. (38), in which extensive modifications were made to the CDR1 and CDR2 loops of a Vα8.3 and Vβ11 TCR by redirecting Rag recombination to the germline-encoded residues. This approach generated substantially altered CDR1 and CDR2 loops in the TCR genes, yet allowed development of a high proportion of CD4+ and CD8+ T cells (38). Moreover, the importance of evolutionarily conserved residues in the MHCII molecule was tested by introducing amino acid substitutions into either the α (A64Q) or β chain (R70A and T77A) of MHCII in mice. These mutations resulted in minor shifts in the TRAV and TRBV usage of positively selected T cells, but surprisingly did not have any effect on the frequency or diversity of thymocytes that were positively selected (80).
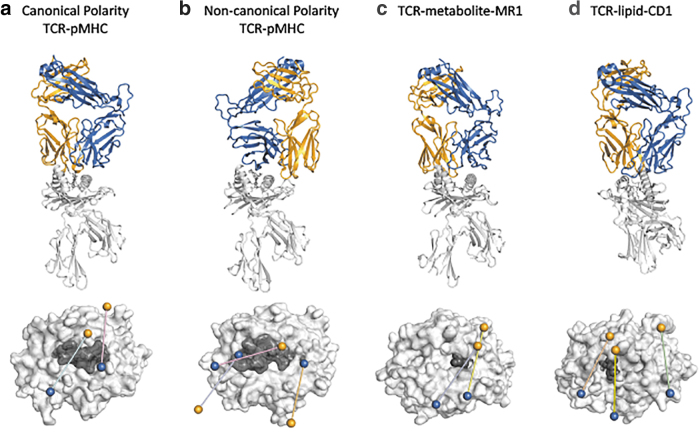
A key feature that has emerged from numerous TCR-pMHC structures is the highly reproducible docking orientation of the TCR over the pMHC, with the TCRα chain sitting over the MHCI α2 helix or MHCII β chain, and the TCRβ chain sitting over the MHCI α1 helix or MHCII β chain (Fig. 1a). The reproducible docking polarity has been interpreted as clear evidence of germline-encoded specificity between the TCR and MHC molecule. This was in agreement with the observation from multiple TCR-pMHC structures that have identified reproducible and conserved “interaction codons” in the CDR1 and CDR2 regions of αβTCRs. Indeed, studies have demonstrated conserved pairwise interaction between CDR1β and CDR2β of Vβ8.2 TCR in complex with pMHCII complexes (16,69,74,75,84,94). These conserved pairwise interactions between the TCR and pMHC were considered key drivers of MHC restriction and suggested that the docking polarity was “hardwired.”
FIG. 1.
Comparison of docking orientations for TCR-MHC and MHC-like interactions. Above: Ribbon structures of selected TCR-MHC and TCR-MHC-like interactions. Bottom: Top view of the TCR docking footprint on top of the MHC or MHC-like molecule. Orange and blue spheres represent the centers of mass for TCRα and TCRβ variable domains, respectively. The MHC molecule is colored light gray and the antigen is colored in dark gray. (a) Canonical docking topology of the LC13-HLA-B8EBV complex (above) and the footprints of the LC13 TCR (cyan; PDB: 1MI5) and the “superbulged” peptide-centric TCR SB27 over HLA-B*3508EBV (pink; PDB: 2AK4) (bottom). (b) Noncanonical topology of the reversed NP1-B17-H-2DbNP366 complex (above) and the footprints of the NP1-B17 TCR (blue; PDB: 5SWZ), the reversed FS18 TCR over HLA-DR4proinsulin (orange; PDB: 4Y19), and the nonsignaling 42F3 TCR over H-2:Ldp3A1 (pink; PDB: 3TJH) (bottom). (c) Unconventional MAIT M33.64-MR15-OP-RU complex (above) and the footprints of the MAIT M33.64 TCR (yellow; PDB: 5D5M) and MAV36 TCR over MR15-OP-RU (blue PDB: 5D7L) (bottom). (d) Unconventional 3C8-CD1cmonoacylglycerol complex (above), and the footprints of the 3C8 TCR (wheat; PDB: C09), the BK6 TCR over CD1aLPC (green, PDB: 4X6C), and the PG90 TCR over CD1bPG (blue; PDB: 5WKI) (bottom). EBV, Epstein-Barr Virus; HLA, human lymphocyte antigen; MAIT, mucosal associated invariant T; PDB, Protein Data Bank; TCR-MHC, T cell receptor-major histocompatibility complex.
These data were ostensibly supported by studies that used a library of pMHC molecules to determine the relative contribution of the TCR CDR3-peptide interface in driving TCR-pMHC docking polarity. In these studies, several alternative peptides were found to be recognized by a single TCR, all of which were recognized by the TCR in a conventional docking topology (9). Similar studies have been performed by modifying the CDR3α sequence of the TCR or by identifying TCRs cross-reactive to different pMHC. Modification of CDR3α sequence affected cross-reactivity to different ligands and its binding affinity, but did not affect TCR-pMHC docking orientation (45). Likewise, a single TCR has been shown to cross-react to two distinct pMHC, yet this TCR maintains the conventional docking orientation (13). These findings were taken as evidence that germline CDR1 and CDR2 loops, rather than the largely non-germline-encoded CDR3, played dominant roles in driving MHC restriction (24). These conclusions are muddied somewhat by the observation that the major conserved Y48, Y46, and E54 residues also made key contacts with MHC-independent conformational epitopes, including in the TCRs specific for CD155 found in Quad-KO mice (86), raising the argument that these residues have been evolutionarily conserved to drive enhanced reactivity of the TCR to ligands more globally.
Organization of the CD3-TCR-pMHC complex
The weight of evidence indicates that evolutionarily conserved complementarity between the TCR and the MHC predisposes, but does not mandate, MHC recognition by TCRs. The ultimate driver of MHC restriction by TCRs, as proposed by the selection model, is the requirement for the TCR-ligand interaction to result in a signal that is achieved by pMHC recognition by the simultaneous binding of MHC by the CD4 and CD8 coreceptors, which co-localizes Lck to the intracellular ITAMs of CD3. That is, the main driver for MHC restriction is TCR signaling, rather than TCR binding. The question that remains is how the requirement for this signal mandates the precise nature of the TCR-MHC interaction.
To understand how TCR-pMHC docking topology can influence TCR signaling, we must look at what is currently known about the structural assembly of the TCR-CD3 complex. Early mutational studies to understand the assembly of the TCR-CD3 complex have suggested that the CD3ɛδ and ɛγ heterodimers congregate on one side of the TCR (49). Later, the first ternary complex of the TCR-pMHC-CD4 was solved by the Mariuzza group (95); it was then suggested that the CD3 heterodimers are positioned inside the TCR-pMHC CD4 arch, thereby facilitating intracellular localization of Lck to the CD3 ITAMS (48,95). All of these early data are in agreement with the most recent structural assembly of the TCR-CD3 complex, first proposed from studies using solution NMR (36) and recently confirmed by the first cryo-electron microscopy structure of the complete TCR-CD3 complex (19), which similarly indicates a “sided” orientation of the CD3 complex in relationship to the TCR. Based on these observations, it has been proposed that the highly reproducible TCR-pMHC docking orientation may reflect the structural positioning that is most conducive to signaling either by facilitating Lck proximity to the CD3 ITAMS (48,67) or by affecting TCR clustering by dimer exclusion or partial occupancy (1).
Unorthodox TCR Recognition of pMHC
Much of what we know about TCR-pMHC complexes have come from TCRs that are well represented in immune or expanded repertoires, and therefore may actually represent a biased view of the fittest TCRs that have the capacity to induce strong TCR signaling. In this study, we look at unconventional TCR recognition of pMHC- or MHC-like ligands for further insight into how generalizable these observations are in similar, but distinct models of recognition. In 2005, the first unorthodox modes of TCR-pMHC recognition were identified for two TCRs specific for myelin basic protein-derived peptides presented by HLA-DR2, where, instead of sitting centrally over the peptide, the TCR lay over the N-terminal end of the peptide (34,52). In both of these cases, it was hypothesized that this unusual mode of docking negatively impacted on the affinity of the interaction, allowing these autoreactive T cells to escape negative selection (48,67).
More recently, there have been two discoveries of TCRs that recognize the pMHC complex in a completely reversed orientation (Fig. 1b). Before these structures, no reversed orientation TCRs had been observed, although their presence had been suggested (82). First, two reverse orientation human HLA-DR4-restricted TCRs were derived from Tregs induced by in vitro stimulation with tolerogenic proinsulin-pulsed DCs (6). More recently, two TCRs, bound in a reversed orientation to mouse H-2Db loaded with an immunodominant influenza A virus (IAV)-derived nucleoprotein peptide, were isolated from the preimmune repertoire (32). The identification of these reversed polarity TCRs is significant as they are the first examples of TCR-pMHC complexes to deviate from conventional docking polarity, and the mouse TCRs also represent the first TCR-pMHC complexes solved from the preimmune repertoire. The analysis of preimmune TCRs has allowed us to take an unbiased look at TCR-pMHC interactions, independent of their ability to support robust T cell activation. The two reversed MHCI-restricted TCRs interacted primarily through germline-encoded TCRβ framework regions and resulted in a TCR-pMHCI interaction of moderate affinity, suggesting that energetically favorable interactions can be achieved in the absence of conserved germline-encoded interactions (25). Reversed polarity TCR-pMHCI complexes from the preimmune repertoire was found to drive poor signaling and immune expansion after viral challenge, despite having moderate affinity for pMHC, providing further evidence for structural constraints imposed on the TCR-pMHC complex for effective signaling, which were independent of binding strength (32). It remains to be determined how a reversed TCR-pMHC docking topology might negatively impact signaling, and by extension, how canonical docking facilitates signaling. Another example of unusual TCR-pMHC recognition is the CD8 TCR recognition of an unusually long 13 amino acid Epstein-Barr Virus (EBV) peptide that bulges out of the MHC peptide binding groove. Structural analysis of this TCR-pMHC complex revealed a highly peptide centric mode of antigen recognition that made minimal contacts with MHCI. Interestingly, although the TCR made minimal contacts with the MHC, it retained the canonical docking orientation over the MHCI and was capable of transducing a TCR signal and killing target cells (Fig. 1a) (88). In addition, Adams et al. demonstrated that different peptides presented by the same MHCI molecule can alter the TCR-pMHC docking topology. Peptides that induced a canonical TCR-pMHC “fit” were capable of signaling, whereas one peptide, which resulted in a significantly altered TCR-pMHC docking orientation, reduced the capacity for the TCR to induce a signal, independent of binding affinity (Fig. 1b) (1). One explanation for the lack of signaling was that the unusual TCR-pMHC docking angle exceeds tolerances allowed for productive arrangement of the TCR-CD3-CD8 complex.
TCR Recognition of Unconventional MHCI-Like Ligands
Unlike classical MHCI and MHCII molecules, MHCI-like molecules such as CD1 and MR1 are monomorphic (63) and present lipid or metabolite antigens, rather than peptides, to TCRs expressed by “unconventional” T cells such as mucosal associated invariant T (MAIT) cells, natural killer T cells (NKT), and subsets of γδ T cells (reviewed in Godfrey et al. (30)). The three key tenets of conventional TCR-pMHC binding, namely the TCR binding over peptide, TCR co-recognition of both the MHC and the peptide cargo, and the conserved docking polarity of the TCR, have also been observed for TCR recognition of nonclassical MHC molecules. This includes recognition of HLA-E (37,85) and MR1, which presents metabolites to MAIT cells, and to atypical MR1-restricted T cells (14,21,27,28,65) (Fig. 1c).
However, analysis of TCR recognition of CD1 molecules provides an interesting exception. CD1 molecules are a family of MHCI-like antigen-presenting molecules (CD1a, CD1b, CD1c, and CD1d proteins) that are specialized in lipid antigen presentation. Typically, the hydrophobic chains are sequestered within the CD1 cleft, while the polar headgroups protrude from the cleft and are potentially available for TCR contact. There have been a number of TCR-CD1 ternary complexes solved (70,71), in which the TCR has been observed to co-recognize the CD1 protein and the surface-exposed polar headgroup, including recognition of mycobacterially derived lipids (33) and self-phospholipids (77,78). However, two distinct autoreactive TCRs in complex with CD1c (3C8) (93) and CD1a (BK6) (7) showed that the TCRs exclusively contacted the CD1 molecule and made no contact with the lipid ligand (Fig. 1d), contravening the co-recognition tenet. Human CD1 molecules have relatively enclosed ligand binding pockets, termed the A’ and F’ pockets, above which sit the closed A’ roof and the F’ portal, respectively. Small ligands can bind entirely within these pockets, while ligands with sufficiently large headgroups can protrude through the F’ portal. Although the BK6 TCR avoided lipid contact by assuming a left-shifted footprint over the CD1a A’ roof, the 3C8 TCR made critical contacts with the F’ portal, facilitated by the complete burial of the ligand within the pocket. This presents a scenario in which moieties with large headgroups may obstruct TCR-CD1 interactions, while small “headless” moieties, such as fatty acids, monoacylglycerols, and squalene, may facilitate the interaction and drive T cell activation (15). Thus, it appears that, unlike in conventional TCR-pMHC recognition, co-recognition may in some instances serve as an impediment to T cell activation.
In both modes of CD1 recognition, the TCR docks over the CD1 with the canonical topology defined by conventional TCR-pMHC interactions, that is, Vα-chain is positioned over the α2-helix and the Vβ-chain resides over the α1-helix. The docking angle is 66° for 3C8 and 110° for BK6 (Fig. 1d) and differs from the TCRs co-recognizing CD1d and polar headgroups (∼90°), but fits within the observations made for MHCI (37° to 90°), MHC II (44° to 115°), and MR1 [82° to 89°; (70)] (Fig. 1). Interestingly, even human γδ TCRs, which are known to recognize antigens independent of MHC (51), have been shown to recognize CD1d with a conserved polarity and docking angle (74° to 84°) (70,89). Such conservation in docking polarity in the absence of obvious utilization of conserved germline-encoded motifs provides further evidence that the docking polarity facilitates signaling in a manner unrelated to binding.
However, some extreme docking modalities that appear to support productive signaling have also been observed. For example, type I NKT cells, which use a largely invariant Vα14-Jα18 TCR in mice and Vα24-Jα18 TCR in humans, are known to dock over CD1d-αGalCer in an orientation that is almost parallel to the antigen binding groove (conserved angle of 5° to 17°) (10), although more “conventional” angles of TCR-MHC docking have also been shown for type II NKT, which are more diverse in their TCRα and TCRβ gene usage (3,29,66,71).
In addition, if we assume that the canonical docking orientation is mandated by the need for appropriate colocalization of coreceptor-associated Lck, the driver for the canonical docking polarity of TCRs expressed on coreceptor-negative NKT cell, MAIT cell, and γδ T cell raises becomes less clear. One likelihood is that, for NKT cells and MAIT cells, the recognition modes reflect those that are conducive to signaling during positive selection at the CD4+ CD8+ double-positive (DP) stage, when coreceptors are able to contribute to signaling (5,76). The situation is less clear with γδ T cells, which do not go through DP selection, but egress to the periphery after the DN3 stage (64). A greater understanding of unconventional T cell development and preselection TCR repertoires is needed to fully appreciate the drivers of TCR-MHC-like ligand recognition and their similarity to conventional TCR-pMHC recognition.
TCR-pMHC Mechanotransduction
Of course, TCR signaling is not binary and it is well established that the strength of the TCR-pMHCI interaction substantially impacts T cell activation and function (18,20,96,97). One of the key recent shifts in our appreciation of how TCR binding of pMHC drives T cell signaling has emerged from a change in the biophysical measurement of interaction “strength” and by accounting for the conditions of force under which physiological recognition of antigen occurs. The gold standard measure of TCR-pMHC interaction strength has long been Surface Plasmon Resonance, which utilizes isolated molecules (at least one of which is in the fluid phase) to determine the intrinsic or “three-dimensional” affinity of the TCR for pMHC. These three-dimensional (3D) measurements of off- and on-rates have also been used to yield the total “dwell time” of a TCR on a pMHC complex, which, along with affinity, have broadly, but not universally, shown correlations with the extent of T cell activation (2,31,46,59,73,97). More recently, two-dimensional (2D) measurements, which directly measure molecular interactions at cell-cell junctions, were thought to better characterize TCR-pMHC interaction strength in the context of the cellular membrane, and are proposed to better correlate with T cell signaling/activation (41,42,44,55). In addition, because physiological TCR-pMHC interactions occur under conditions of force (22,40,53,56), such 2D measurements are typically performed under conditions of applied mechanical force. Using such measurements, a number of recent studies have observed that productive (signal inducing) TCR-pMHC interactions correlate with the ability to form bonds that strengthen with increased force (catch-bonds), while unsuccessful TCR-pMHC interactions are thought to form slip-bonds, whose strength diminishes or is lost with applied force (41,55,79).
However, a couple of very recent studies have called into question whether catch- versus slip-bond formation is a cause or a consequence of a productive TCR-pMHC interaction (39,54). One study found that the formation of TCR-pMHC-CD8 catch bonds was dependent on the kinase activity of Lck and its ability to localize to CD8 and CD3, with inhibition of Lck kinase activity and mutation of several CD3ζ ITAMS resulting in a reduced ability of TCR and CD8 to form a catch bond with pMHC (39). A more recent study showed, in a cell-free system that precluded the contribution of the cellular response to catch bond formation, that intrinsic catch bonds were not formed by any of the five agonist TCR-pMHC ligand pairs studied, and that off-rates of binding were the best predictor of activation potency (54). Thus, our understanding of how the TCR-pMHC-coreceptor bond formation is initiated and changes over the duration of the encounter, and how that drives (or is driven) by downstream signaling events continues to evolve.
Summary
From the initial observations by Zinkernagel and Doherty over 40 years ago of the need for T cells to recognize “altered self,” our understanding of TCR recognition of peptide+MHC has made significant advances. However, alongside a more detailed understanding of the interaction comes additional questions around precisely why T cells must limit themselves to MHC, when a greater range of ligand binding is demonstrably possible, and mechanistically, how MHC restriction achieves the necessary T cell survival and activation signals. The answer may well lie in the study of noncanonical or poorly signaling TCRs to understand the absolute requirements for effective TCR-pMHC recognition, continued advances in structural biology providing resolution of multimolecular complexes, and cryo-EM providing information on the dynamics of molecular localization and organization before and after TCR ligation of pMHC.
Acknowledgments
The authors would like to thank J. Rossjohn for critical review of the article.
Author Disclosure Statement
No competing financial interests exist
Funding Information
Funded by the Australian National Health and Medical Research Council (NHMRC) and the Australian Research Council (ARC). N.L.L.G. would also like to extend her personal gratitude to Prof. Peter Doherty for many years of outstanding mentorship, support, and friendship.
References
- 1. Adams JJ, Narayanan S, Liu B, et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity 2011;35:681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allison KA, Sajti E, Collier JG, et al. Affinity and dose of TCR engagement yield proportional enhancer and gene activity in CD4+ T cells. Elife 2016;5:e10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Almeida CF, Sundararaj S, Le Nours J, et al. Distinct CD1d docking strategies exhibited by diverse Type II NKT cell receptors. Nat Commun 2019;10:5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barnd DL, Lan MS, Metzgar RS, et al. Specific, major histocompatibility complex-unrestricted recognition of tumor-associated mucins by human cytotoxic T cells. Proc Natl Acad Sci U S A 1989;86:7159–7163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med 1995;182:2091–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beringer DX, Kleijwegt FS, Wiede F, et al. T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex. Nat Immunol 2015;16:1153–1161 [DOI] [PubMed] [Google Scholar]
- 7. Birkinshaw RW, Pellicci DG, Cheng T-Y, et al. αβ T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nat Immunol 2015;16:258–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bodmer WF. Evolutionary significance of the HL-A system. Nature 1972;237:139–145 passim. [DOI] [PubMed] [Google Scholar]
- 9. Borbulevych OY, Piepenbrink KH, Gloor BE, et al. T cell receptor cross-reactivity directed by antigen-dependent tuning of peptide-MHC molecular flexibility. Immunity 2009;31:885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Borg NA, Wun KS, Kjer-Nielsen L, et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature 2007;448:44–49 [DOI] [PubMed] [Google Scholar]
- 11. Burnet FM. Multiple polymorphism in relation to histocompatibility antigens. Nature 1973;245:359–361 [DOI] [PubMed] [Google Scholar]
- 12. Burrows SR, Chen Z, Archbold JK, et al. Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability. Proc Natl Acad Sci U S A 2010;107:10608–10613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Colf LA, Bankovich AJ, Hanick NA, et al. How a single T cell receptor recognizes both self and foreign MHC. Cell 2007;129:135–146 [DOI] [PubMed] [Google Scholar]
- 14. Corbett AJ, Eckle SB, Birkinshaw RW, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 2014;509:361–365 [DOI] [PubMed] [Google Scholar]
- 15. Cotton RN, Shahine A, Rossjohn J, et al. Lipids hide or step aside for CD1-autoreactive T cell receptors. Curr Opin Immunol 2018;52:93–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dai S, Huseby ES, Rubtsova K, et al. Crossreactive T cells spotlight the germline rules for αβ T cell-receptor interactions with MHC molecules. Immunity 2008;28:324–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davis MM, and Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature 1988;334:395–402 [DOI] [PubMed] [Google Scholar]
- 18. Derby M, Alexander-Miller M, Tse R, et al. High-avidity CTL exploit two complementary mechanisms to provide better protection against viral infection than low-avidity CTL. J Immunol 2001;166:1690–1697 [DOI] [PubMed] [Google Scholar]
- 19. Dong D, Zheng L, Lin J, et al. Structural basis of assembly of the human T cell receptor-CD3 complex. Nature 2019;573:546–552 [DOI] [PubMed] [Google Scholar]
- 20. Dutoit V, Rubio-Godoy V, Dietrich PY, et al. Heterogeneous T-cell response to MAGE-A10(254–262): high avidity-specific cytolytic T lymphocytes show superior antitumor activity. Cancer Res 2001;61:5850–5856 [PubMed] [Google Scholar]
- 21. Eckle SB, Birkinshaw RW, Kostenko L, et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J Exp Med 2014;211:1585–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feng Y, Brazin KN, Kobayashi E, et al. Mechanosensing drives acuity of alphabeta T-cell recognition. Proc Natl Acad Sci U S A 2017;114:E8204–E8213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garboczi DN, Ghosh P, Utz U, et al. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 1996;384:134–141 [PubMed] [Google Scholar]
- 24. Garcia KC. Reconciling views on T cell receptor germline bias for MHC. Trends Immunol 2012;33:429–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garcia KC, Adams JJ, Feng D, et al. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol 2009;10:143–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Garcia KC, Degano M, Stanfield RL, et al. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science 1996;274:209–219 [DOI] [PubMed] [Google Scholar]
- 27. Gherardin NA, Keller AN, Woolley RE, et al. Diversity of T cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition. Immunity 2016;44:32–45 [DOI] [PubMed] [Google Scholar]
- 28. Gherardin NA, McCluskey J, Rossjohn J, et al. The diverse family of MR1-restricted T cells. J Immunol 2018;201:2862. [DOI] [PubMed] [Google Scholar]
- 29. Girardi E, Maricic I, Wang J, et al. Type II natural killer T cells use features of both innate-like and conventional T cells to recognize sulfatide self antigens. Nat Immunol 2012;13:851–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Godfrey DI, Uldrich AP, McCluskey J, et al. The burgeoning family of unconventional T cells. Nat Immunol 2015;16:1114–1123 [DOI] [PubMed] [Google Scholar]
- 31. Govern CC, Paczosa MK, Chakraborty AK, et al. Fast on-rates allow short dwell time ligands to activate T cells. Proc Natl Acad Sci U S A 2010;107:8724–8729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gras S, Chadderton J, Del Campo CM, et al. Reversed T cell receptor docking on a major histocompatibility class I complex limits involvement in the immune response. Immunity 2016;45:749–760 [DOI] [PubMed] [Google Scholar]
- 33. Gras S, Van Rhijn I, Shahine A, et al. T cell receptor recognition of CD1b presenting a mycobacterial glycolipid. Nat Commun 2016;7:13257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hahn M, Nicholson MJ, Pyrdol J, et al. Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat Immunol 2005;6:490–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hanada K, Wang QJ, Inozume T, et al. Molecular identification of an MHC-independent ligand recognized by a human {alpha}/{beta} T-cell receptor. Blood 2011;117:4816–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He Y, Rangarajan S, Kerzic M, et al. Identification of the docking site for CD3 on the T cell receptor beta chain by solution NMR. J Biol Chem 2015;290:19796–19805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hoare HL, Sullivan LC, Pietra G, et al. Structural basis for a major histocompatibility complex class Ib-restricted T cell response. Nat Immunol 2006;7:256–264 [DOI] [PubMed] [Google Scholar]
- 38. Holland SJ, Bartok I, Attaf M, et al. The T-cell receptor is not hardwired to engage MHC ligands. Proc Natl Acad Sci U S A 2012;109:E3111–E3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hong J, Ge C, Jothikumar P, et al. A TCR mechanotransduction signaling loop induces negative selection in the thymus. Nat Immunol 2018;19:1379–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hu KH, and Butte MJ. T cell activation requires force generation. J Cell Biol 2016;213:535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang J, Zarnitsyna VI, Liu B, et al. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature 2010;464:932–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huppa JB, Axmann M, Mortelmaier MA, et al. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature 2010;463:963–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jerne NK. The somatic generation of immune recognition. Eur J Immunol 1971;1:1–9 [DOI] [PubMed] [Google Scholar]
- 44. Jiang N, Huang J, Edwards LJ, et al. Two-stage cooperative T cell receptor-peptide major histocompatibility complex-CD8 trimolecular interactions amplify antigen discrimination. Immunity 2011;34:13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jones LL, Colf LA, Stone JD, et al. Distinct CDR3 conformations in TCRs determine the level of cross-reactivity for diverse antigens, but not the docking orientation. J Immunol 2008;181:6255–6264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kalergis AM, Boucheron N, Doucey MA, et al. Efficient T cell activation requires an optimal dwell-time of interaction between the TCR and the pMHC complex. Nat Immunol 2001;2:229–234 [DOI] [PubMed] [Google Scholar]
- 47. Krovi SH, Kappler JW, Marrack P, et al. Inherent reactivity of unselected TCR repertoires to peptide-MHC molecules. Proc Natl Acad Sci U S A 2019;116:22252–22261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kuhns MS, and Davis MM. TCR signaling emerges from the sum of many parts. Front Immunol 2012;3:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kuhns MS, Davis MM, and Garcia KC. Deconstructing the form and function of the TCR/CD3 complex. Immunity 2006;24:133–139 [DOI] [PubMed] [Google Scholar]
- 50. La Gruta NL, Gras S, Daley SR, et al. Understanding the drivers of MHC restriction of T cell receptors. Nat Rev Immunol 2018;18:467–478 [DOI] [PubMed] [Google Scholar]
- 51. Legut M, Cole DK, and Sewell AK. The promise of γδ T cells and the γδ T cell receptor for cancer immunotherapy. Cell Mol Immunol 2015;12:656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li Y, Huang Y, Lue J, et al. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J 2005;24:2968–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li YC, Chen BM, Wu PC, et al. Cutting edge: mechanical forces acting on T cells immobilized via the TCR complex can trigger TCR signaling. J Immunol 2010;184:5959–5963 [DOI] [PubMed] [Google Scholar]
- 54. Limozin L, Bridge M, Bongrand P, et al. TCR-pMHC kinetics under force in a cell-free system show no intrinsic catch bond, but a minimal encounter duration before binding. Proc Natl Acad Sci U S A 2019;116:16943–16948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu B, Zhong S, Malecek K, et al. 2D TCR-pMHC-CD8 kinetics determines T-cell responses in a self-antigen-specific TCR system. Eur J Immunol 2014;44:239–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu Y, Blanchfield L, Ma VP, et al. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc Natl Acad Sci U S A 2016;113:5610–5615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lu J, Van Laethem F, Bhattacharya A, et al. Molecular constraints on CDR3 for thymic selection of MHC-restricted TCRs from a random pre-selection repertoire. Nat Commun 2019;10:1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Magarian-Blander J, Ciborowski P, Hsia S, et al. Intercellular and intracellular events following the MHC-unrestricted TCR recognition of a tumor-specific peptide epitope on the epithelial antigen MUC1. J Immunol 1998;160:3111–3120 [PubMed] [Google Scholar]
- 59. Malherbe L, Hausl C, Teyton L, et al. Clonal selection of helper T cells is determined by an affinity threshold with no further skewing of TCR binding properties. Immunity 2004;21:669–679 [DOI] [PubMed] [Google Scholar]
- 60. Marrack P, Scott-Browne JP, Dai S, et al. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol 2008;26:171–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. McDonald BD, Bunker JJ, Erickson SA, et al. Crossreactive alphabeta T cell receptors are the predominant targets of thymocyte negative selection. Immunity 2015;43:859–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Merkenschlager M, Graf D, Lovatt M, et al. How many thymocytes audition for selection? J Exp Med 1997;186:1149–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mori L, Lepore M, and De Libero G. The immunology of CD1- and MR1-restricted T cells. Annu Rev Immunol 2016;34:479–510 [DOI] [PubMed] [Google Scholar]
- 64. Munoz-Ruiz M, Sumaria N, Pennington DJ, et al. Thymic determinants of gammadelta T cell differentiation. Trends Immunol 2017;38:336–344 [DOI] [PubMed] [Google Scholar]
- 65. Patel O, Kjer-Nielsen L, Le Nours J, et al. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat Commun 2013;4:2142. [DOI] [PubMed] [Google Scholar]
- 66. Patel O, Pellicci DG, Gras S, et al. Recognition of CD1d-sulfatide mediated by a type II natural killer T cell antigen receptor. Nat Immunol 2012;13:857–863 [DOI] [PubMed] [Google Scholar]
- 67. Rangarajan S, and Mariuzza RA. T cell receptor bias for MHC: co-evolution or co-receptors? Cell Mol Life Sci 2014;71:3059–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rao A, Ko WW, Faas SJ, et al. Binding of antigen in the absence of histocompatibility proteins by arsonate-reactive T-cell clones. Cell 1984;36:879–888 [DOI] [PubMed] [Google Scholar]
- 69. Reinherz EL, Tan K, Tang L, et al. The crystal structure of a T cell receptor in complex with peptide and MHC class II. Science 1999;286:1913–1921 [DOI] [PubMed] [Google Scholar]
- 70. Rossjohn J, Gras S, Miles JJ, et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol 2015;33:169–200 [DOI] [PubMed] [Google Scholar]
- 71. Rossjohn J, Pellicci DG, Patel O, et al. Recognition of CD1d-restricted antigens by natural killer T cells. Nat Rev Immunol 2012;12:845–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rudolph MG, Stanfield RL, and Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol 2006;24:419–466 [DOI] [PubMed] [Google Scholar]
- 73. Schmid DA, Irving MB, Posevitz V, et al. Evidence for a TCR affinity threshold delimiting maximal CD8 T cell function. J Immunol 2010;184:4936–4946 [DOI] [PubMed] [Google Scholar]
- 74. Scott-Browne JP, Crawford F, Young MH, et al. Evolutionarily conserved features contribute to ±≤ T cell receptor specificity. Immunity 2011;35:526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Scott-Browne JP, White J, Kappler JW, et al. Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature 2009;458:1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Seach N, Guerri L, Le Bourhis L, et al. Double positive thymocytes select mucosal-associated invariant T cells. J Immunol 2013;191:6002. [DOI] [PubMed] [Google Scholar]
- 77. Shahine A, Reinink P, Reijneveld JF, et al. A T-cell receptor escape channel allows broad T-cell response to CD1b and membrane phospholipids. Nat Commun 2019;10:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shahine A, Van Rhijn I, Cheng T-Y, et al. A molecular basis of human T cell receptor autoreactivity toward self-phospholipids. Sci Immunol 2017;2:eaao1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sibener LV, Fernandes RA, Kolawole EM, et al. Isolation of a structural mechanism for uncoupling T cell receptor signaling from peptide-MHC binding. Cell 2018;174:672–687. e627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Silberman D, Krovi SH, Tuttle KD, et al. Class II major histocompatibility complex mutant mice to study the germ-line bias of T-cell antigen receptors. Proc Natl Acad Sci U S A 2016;113:E5608–E5617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Siliciano RF, Hemesath TJ, Pratt JC, et al. Direct evidence for the existence of nominal antigen binding sites on T cell surface Ti alpha-beta heterodimers of MHC-restricted T cell clones. Cell 1986;47:161–171 [DOI] [PubMed] [Google Scholar]
- 82. Sim BC, Travers PJ, and Gascoigne NR. V alpha 3.2 selection in MHC class I mutant mice: evidence for an alternate orientation of TCR-MHC class I interaction. J Immunol 1997;159:3322–3329 [PubMed] [Google Scholar]
- 83. Sinclair C, Bains I, Yates AJ, et al. Asymmetric thymocyte death underlies the CD4:CD8 T-cell ratio in the adaptive immune system. Proc Natl Acad Sci U S A 2013;110:E2905–E2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Stadinski BD, Trenh P, Smith RL, et al. A role for differential variable gene pairing in creating T cell receptors specific for unique major histocompatibility ligands. Immunity 2011;35:694–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sullivan LC, Walpole NG, Farenc C, et al. A conserved energetic footprint underpins recognition of Human Leukocyte Antigen-E by two distinct alphabeta T cell receptors. J Biol Chem 2017;292:21149–21158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tikhonova AN, Van Laethem F, Hanada K, et al. αβ T cell receptors that do not undergo major histocompatibility complex-specific thymic selection possess antibody-like recognition specificities. Immunity 2012;36:79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Turner JM, Brodsky MH, Irving BA, et al. Interaction of the unique N-terminal region of tyrosine kinase p56lck with cytoplasmic domains of CD4 and CD8 is mediated by cysteine motifs. Cell 1990;60:755–765 [DOI] [PubMed] [Google Scholar]
- 88. Tynan FE, Burrows SR, Buckle AM, et al. T cell receptor recognition of a “super-bulged” major histocompatibility complex class I-bound peptide. Nat Immunol 2005;6:1114–1122 [DOI] [PubMed] [Google Scholar]
- 89. Uldrich AP, Le Nours J, Pellicci DG, et al. CD1d-lipid antigen recognition by the gammadelta TCR. Nat Immunol 2013;14:1137–1145 [DOI] [PubMed] [Google Scholar]
- 90. Van Laethem F, Sarafova SD, Park JH, et al. Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC. Immunity 2007;27:735–750 [DOI] [PubMed] [Google Scholar]
- 91. Van Laethem F, Tikhonova AN, Pobezinsky LA, et al. Lck availability during thymic selection determines the recognition specificity of the T cell repertoire. Cell 2013;154:1326–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Van Laethem F, Tikhonova AN, and Singer A. MHC restriction is imposed on a diverse T cell receptor repertoire by CD4 and CD8 co-receptors during thymic selection. Trends Immunol 2012;33:437–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wun KS, Reijneveld JF, Cheng TY, et al. T cell autoreactivity directed toward CD1c itself rather than toward carried self lipids. Nat Immunol 2018;19:397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yin L, Huseby E, Scott-Browne J, et al. A single T cell receptor bound to major histocompatibility complex class I and class II glycoproteins reveals switchable TCR conformers. Immunity 2011;35:23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yin Y, Wang XX, and Mariuzza RA. Crystal structure of a complete ternary complex of T-cell receptor, peptide-MHC, and CD4. Proc Natl Acad Sci U S A 2012;109:5405–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zeh HJ 3rd, Perry-Lalley D, Dudley ME, et al. High avidity CTLs for two self-antigens demonstrate superior in vitro and in vivo antitumor efficacy. J Immunol 1999;162:989–994 [PubMed] [Google Scholar]
- 97. Zehn D, Lee SY, and Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature 2009;458:211–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zerrahn J, Held W, and Raulet DH. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell 1997;88:627–636 [DOI] [PubMed] [Google Scholar]
- 99. Zinkernagel RM, and Doherty PC. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature 1974;248:701–702 [DOI] [PubMed] [Google Scholar]
- 100. Zinkernagel RM, and Doherty PC. The discovery of MHC restriction. Immunol Today 1997;18:14–17 [DOI] [PubMed] [Google Scholar]