Abstract
A seminal discovery of major histocompatibility complex (MHC) restriction in T cell recognition by Peter Doherty and Rolf Zinkernagel has led to 45 years of exciting research on the mechanisms governing peptide MHC (pMHC) recognition by T cell receptors (TCRs) and their importance in health and disease. T cells provide a significant level of protection against viral, bacterial, and parasitic infections, as well as tumors, hence, the generation of protective T cell responses is a primary goal for cell-mediated vaccines and immunotherapies. Understanding the mechanisms underlying generation of optimal high-avidity effector T cell responses, memory development, maintenance, and recall is of major importance for the rational design of preventative and therapeutic vaccines/immunotherapies. In this review, we summarize the lessons learned over the last four decades and outline our current understanding of the basis and consequences of pMHC/TCR interactions on T cell development and function, and TCR diversity and composition, driving better clinical outcomes and prevention of viral escape. We also discuss the current models of T cell memory formation and determinants of immunodominant T cell responses in animal models and humans. As TCR composition and diversity can affect both the protective capacity of T cells and protection against viral escape, defining the spectrum of TCR selection has implications for improving the functional efficacy of effector T cell responsiveness and memory formation.
Keywords: MHC, TCR, disease
Introduction
Adaptive immunity forms the basis of successful long-lasting protective immune responses, with humoral immunity being mediated by antibodies produced by B cells, while T cells drive cellular immunity (7). Cytotoxic CD8+ T cells can recognize virus-infected or cancerous cells, induce cell lysis, and produce antiviral cytokines. Helper CD4+ T cells predominantly provide critical help to B cells and CD8+ T cells (16). What distinguishes T cells and B cells from innate immunity is the antigen specificity and generation of long-lasting immunological memory. While antibodies bind to three-dimensional epitopes on proteins, T cell receptors (TCRs) recognize epitopes formed by small foreign peptide fragments presented by a self-encoded glycoprotein called major histocompatibility complexes (MHCs) (16). The requirement of the TCR to engage with both peptide and MHC concurrently is called MHC restriction and was first described by Zinkernagel and Doherty in 1974 (147). Their demonstration that T cells require interactions with both self-encoded antigen-presenting molecules and foreign peptides has been instrumental for our understanding of effective T cell immunity against pathogens and cancers, as well as the mechanisms underlying successful transplantation and development of autoimmune diseases. This key discovery was recognized by the Nobel Prize of Medicine and Physiology in 1996.
In this review, we summarize the lessons learned over the last 45 years since the seminal discovery of MHC-restriction by Doherty and Zinkernagel, and outline our current understanding of the basis and consequences of peptide MHC (pMHC)/TCR interactions on T cell development and function.
T Cell Development and Priming
T cells originate from bone marrow-derived hematopoietic progenitors, then traffic to the thymus, where double-negative immature CD4−CD8− T cells undergo TCR gene rearrangement, and after successfully doing so, become double-positive (DP) CD4+CD8+ immature T cells. As DP thymocytes undergo positive and negative selection, they commit to the CD4+ or CD8+ T cell lineage (67). The processes of thymic selection generate a diverse TCR repertoire of naive CD4+ or CD8+ restricted by the individual's MHC allotypes, but do not react to self-peptides. These mature naive T cells then circulate between secondary lymphoid organs (SLOs: spleen and lymph nodes), surveying for foreign antigens.
Encounter with a cognate antigen on professional antigen-presenting cells (APCs) in lymph nodes leads to T cell priming, a process requiring three key signals. The first is provided by the TCR-pMHC interaction and activation of the signaling pathways downstream of the TCR complex (40). The second signal is mediated by co-stimulatory molecules like B7 (CD80/CD86) on APCs and their interaction with their respective ligands, like CD28, on T cells (142). The third signal relies on cytokines like interferons (IFNs) or interleukin (IL)-12 released predominantly by APCs (30,34). The events of T cell priming result in epigenetic and transcriptional changes that underlie the well-orchestrated processes of T cell proliferation and differentiation (51,60), subsequently leading to generation of large pools of antigen-specific effector and memory T cell subsets.
T Cell Memory Differentiation
Following antigen clearance, the effector T cell pool contracts by 90–95%, with the remaining 5–10% of antigen-specific T cells forming a population of long-lasting memory T cells capable of responding more rapidly and with greater numbers to antigenic re-encounter. A hallmark of memory T cells is their longevity, with human smallpox-specific CD8+ T cells being detectable even at 75 years postvaccination (48). Elegant human studies by Ahmed's group using in vivo deuterium labeling following vaccination showed that Yellow Fever Virus-specific CD8+ T cells, generated within the first 2 weeks following vaccination, were detectable for as long as 750 days later, estimated to divide once every 485 days (1). Similarly, influenza-specific CD8+ T cells can be detected up to 13 years after an individual's last recorded natural influenza A virus (IAV) infection in humans (133) or for a life-span of a laboratory mouse (65,130). The maintenance of memory CD8+ T cells depends on survival signals provided by cytokines like IL-15 and IL-7, but not by antigen [reviewed by Raeber et al. (99)].
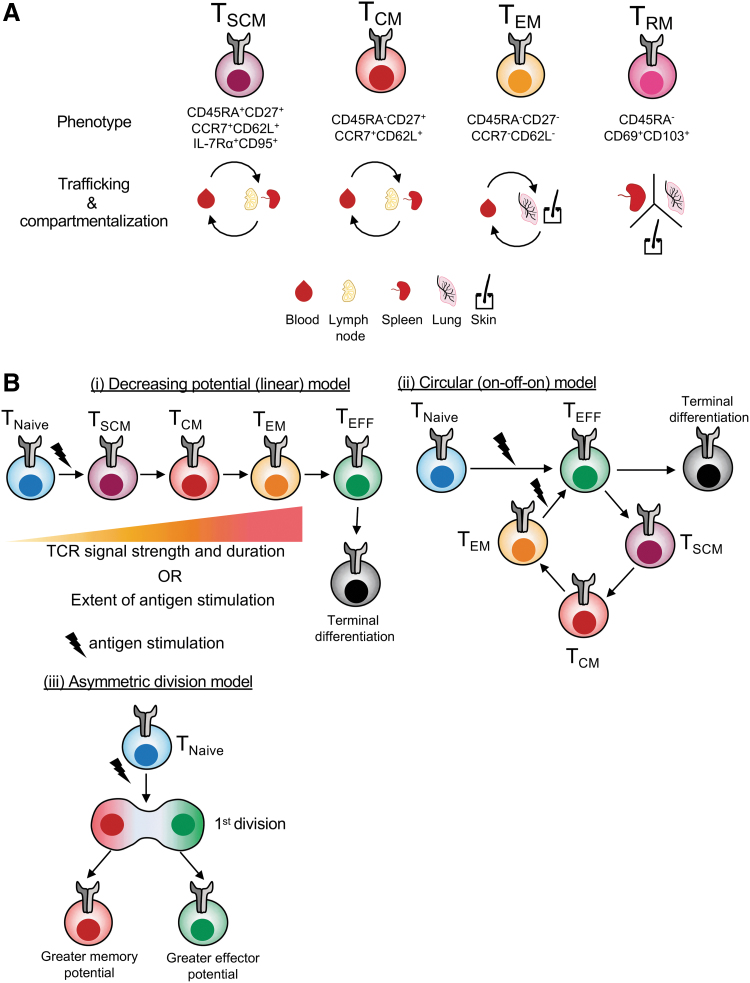
Antigen-specific memory CD8+ T cells are greatly heterogenous, with four main memory subsets being identified, namely T cell stem cell memory (TSCM, conventionally defined as CD45RA+CD27+CCR7+CD62LhiIL-7Rα+CD95+), T cell central memory (TCM, conventionally defined as CD45RA−CCR7+CD62LhiCD95+), T cell effector memory (TEM, conventionally defined as CD45RA−CCR7−CD62LloCD95+), and tissue-resident memory (TRM, conventionally defined as CD45RA−CD69+CD103−/+ present in tissues) (Fig. 1A). These subsets differ with regard to their proliferation, cytokine production, and tissue circulation potential [reviewed in refs. (37,55)]. For instance, TSCM and TCM CD8+ T cells have higher proliferative potential, produce high levels of IL-2, and recirculate between the blood and SLOs. Conversely, effector memory (TEM) CD8+ T cells have a limited proliferation potential, produce mainly IFNγ, and recirculate between blood and peripheral tissue. The fourth subset of memory, TRM, is retained within peripheral tissues where they can provide rapid and potent protection after antigen re-encounter [reviewed by Gebhardt et al. (41)].
FIG. 1.
CD8+ T cell memory subsets and differentiation models. (A) At least four different memory CD8+ T cell subsets have been proposed: stem cell memory (TSCM), central memory (TCM), effector memory (TEM), and tissue-resident memory (TRM) cells. Memory subsets display distinct circulations and tissue compartmentalization patterns. (B) Three proposed models of memory differentiation: (i) the linear model proposes the progressive loss of memory potential as the CD8+ T cells acquire effector functions according to the strength/duration of TCR signaling or the extent of antigenic stimulation. (ii) The circular model proposes memory CD8+ T cells undergo an obligatory effector stage before de-differentiating in memory CD8+ T cells. (iii) The asymmetric division model proposes an unequal distribution of regulatory molecules, with one daughter cell displaying a greater memory potential, while the other daughter cells have a greater effector potential. TCR, T cell receptor.
Different models exist on how CD8+ T cell memory is formed in relationship to the effector subset (Fig. 1B). The linear model (also called “decreasing potential” model) proposes that T cells progressively go through the memory and effector phases (SCM → CM → EM → EFF) in a process that decreases memory potential and increases effector differentiation (Fig. 1B[i]) (23,51). According to this model, the progression through the different stages is influenced by TCR signal strength/duration and/or the extent of antigenic stimulation on the T cell. The opposing model is called the circular model (or the “on–off–on” model) (Fig. 1B[ii]) (60). This model proposed that following antigen encounter, CD8+ T cells differentiate into effector cells and, upon contraction of the response, the same effector cells de-differentiate into memory T cells of different subsets, which then can be recalled and re-differentiated into effector cells after re-encountering the same antigen. Some studies propose an alternative model, whereby the fate of a naive CD8+ T cell is determined as early as the first cell division, with the asymmetric distribution of key transcriptional and/or epigenetic regulators between two daughter cells, whereby one displays increased memory potential, while the other has a greater effector capacity (Fig. 1B[iii]) (23,60,61).
These different models are supported, and refuted, by different lines of evidence in various models of infection (23,51,60). Thus, the formation of immunological T cell memory is incredibly complex, with knowledge gaps remaining to be addressed. For instance, although molecular cues and signaling pathways that drive TRM formation have been characterized, the exact origins of how TRM CD8+ T cells are developed are far from clear, that is, whether they are derived from effector cells or memory cells, or both. With the advent of new high-throughput single-cell technologies, novel insights are to be gained into the developmental pathways of memory CD8+ T cells. Given that life-long immune memory is the foundation of effective vaccination, such understanding of memory establishment and recall at different anatomical sites is of outmost importance.
CD8+ T Cell Effector Function
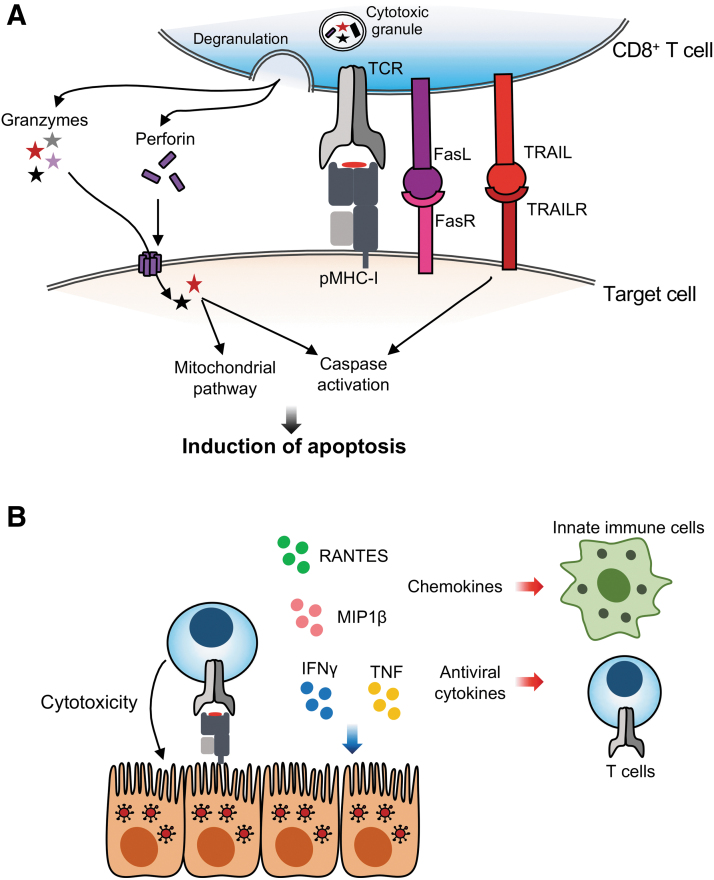
Differentiated effector CD8+ T cells traffic to the site of infection, where they recognize their cognate pMHC on target cells by their TCR and subsequently engage in their effector function. Preloaded cytotoxic granules are released following TCR engagement and deliver their contents in the immunological synapse between the T cells and the target cell (Fig. 2A). The granules predominantly release granzymes (A, B, H, K, and M in humans), perforin and granulysin. Perforin forms a pore in the membranes of the target cells for granzyme delivery to the cytosol of infected or cancerous cells (17,138). Granzymes, which are serine proteases, then induce cell-mediated apoptosis of the virus-infected cells. The most studied granzyme B causes a rapid induction of caspase-dependent apoptosis through a number of mechanisms, including the cleavage of caspases or antiapoptotic and proapoptotic factors (17). Granzymes A and K induce caspase-independent apoptosis through mitochondrial pathways, although the molecular details are yet to be fully elucidated (17,138). Granzyme M-induced cell death although ill defined, is caspase and mitochondria independent (17,138). Additional mechanisms of apoptosis induction occur through the engagement of the death receptor Fas or TRAILR on target cells with the Fas ligand or TRAIL, respectively, from/on cytotoxic CD8+ T cells (Fig. 2A). Signaling through these death receptors results in the activation of caspases and subsequent apoptosis. Regardless of the exact molecular mechanism, the effects of different granzymes and death receptor signaling converge to the aim at hand: death of the target cell. The importance of these pathways can be demonstrated in the context of IAV infection in mice. Following IAV infection, CD8+ T cells acquire a cytotoxic profile that increases with time and cell division (57), and is maintained up to at least 1 year after infection (56). Importantly, following IAV infection, mice lacking TRAIL (21), Fas, or perforin (123) in CD8+ T cells show increased morbidity and delayed viral clearance. Interestingly, mice lacking granzymes A and B show normal viral clearance, which may imply a redundancy in the mechanisms of cytotoxicity (58).
FIG. 2.
CD8+ T cell effector function. (A) Cytotoxicity can be induced through a delivery of granzymes and perforin. Perforin forms pores in target cell membranes, allowing delivery of granzymes to the cytosol, where granzymes induce apoptosis through activation of caspases and the mitochondrial pathway. Binding of Fas and TRAIL on the CD8+ T cell to their receptors FasL and TRAILR on the target cells also induced caspase activation. (B) Recognition of pMHC-I by the TCR results in cytotoxicity of the target cell as well as the release of antiviral cytokine and chemokines, which act on target cells and other immune cells. pMHC, peptide major histocompatibility complex.
TCR engagement also results in the release of antiviral cytokines like IFNγ and TNF (Fig. 2B). IFNγ is a pleiotropic cytokine that can enhance the recruitment and activation of immune cells like macrophages and NK cells (110) and the cytolytic capacity of T cells (11), and induce an antiviral state through the expression of antiviral IFN-stimulated genes (12,107). These include genes important for T cell-mediated immunity, like MHC-I and the genes to form the immunoproteasome (107). Proinflammatory cytokines like RANTES and MIP1β are also released by effector CD8+ T cells, which attract innate immune cells as well as T cells, and overall may aid to viral clearance (Fig. 2B) (116).
Overall, while the defining feature of CD8+ T cells is the lysis of the target cells, the response of the effector T cell is multifaceted and involves both the delivery of cytotoxic molecules as well as the release of antiviral/inflammatory cytokines and chemokines. Importantly, evidence suggests that polyfunctionality (producing multiple cytokines and cytotoxic molecules) as well as the antigen sensitivity of CD8+ T cells are important correlates of protection in infectious diseases (112). In HIV-1 elite controllers, an association was found between highly polyfunctional CD8+ T cells (5+ functions) and lower viral load (10). Polyfunctional CD8+ T cells produced more IFNγ and were superior effectors than monofunctional CD8+ T cells (94,112). In addition, HIV-specific human leukocyte antigen (HLA)-B27-restricted CD8+ T cells, which correlate with disease protection, are of higher polyfunctionality and functional avidity (antigen sensitivity) (4). An association between antigen sensitivity, polyfunctionality, and the ability of CD8+ T cell to suppress HIV-1 has also been demonstrated (5). Overall, the quality of the response is an important factor in CD8+ T cell-mediated protection and needs to be considered in the design of effective CD8+ T cell-based vaccines.
CD8+ T Cell Immunodominance
While the quality of the CD8+ T cell response can be a strong correlate of protection (4,5,82,112), the relative magnitude of different T cell specificities within a response can still be important in determining immune protection. From the thousands of potential peptides, which can be generated from the processing of viral proteins during an infection, the CD8+ T cell response is generally focused to a relatively small subset of epitopes. This can be exemplified by influenza B-derived peptides binding to HLA-A*02:01 (as detected by mass spectrometry), while only three of them (BHA543–551, BNS1266–274, and BNS1264–274) being consistently immunogenic across donors (69). Within this immunogenic subset, CD8+ T cells against different specificities vary in magnitude and accordingly fall in a reproducible immunodominance hierarchy (124).
A large number of factors dictate these hierarchies, including, but not limited to, those relating to antigen presentation (e.g., antigen abundance and processing, the affinity of peptide for MHC-I, the stability of the pMHC complex, and the presentation pathway) and those relating to the responding T cells [e.g., naive precursor frequency, extent of their recruitment and expansion, TCR affinity, and competition between T cells of different specificities, termed immunodomination (124)]. These factors contribute to immunodominance in a complex and often context-specific manner. It is important to note that subdominant epitopes of lower magnitude can still be highly functional and provide protection against viral disease in both humans (38,49) and mouse models (39,53,54,100,103) of viral infection. Thus, understanding the drivers of immunodominance may be important in ensuring sufficient priming of subdominant specificities to increase epitope coverage and response breadth, which can correlate with superior protection (104) and/or re-focusing the immune response to superior (e.g., highly conserved) epitopes (54).
CD4+ T Cell Subsets
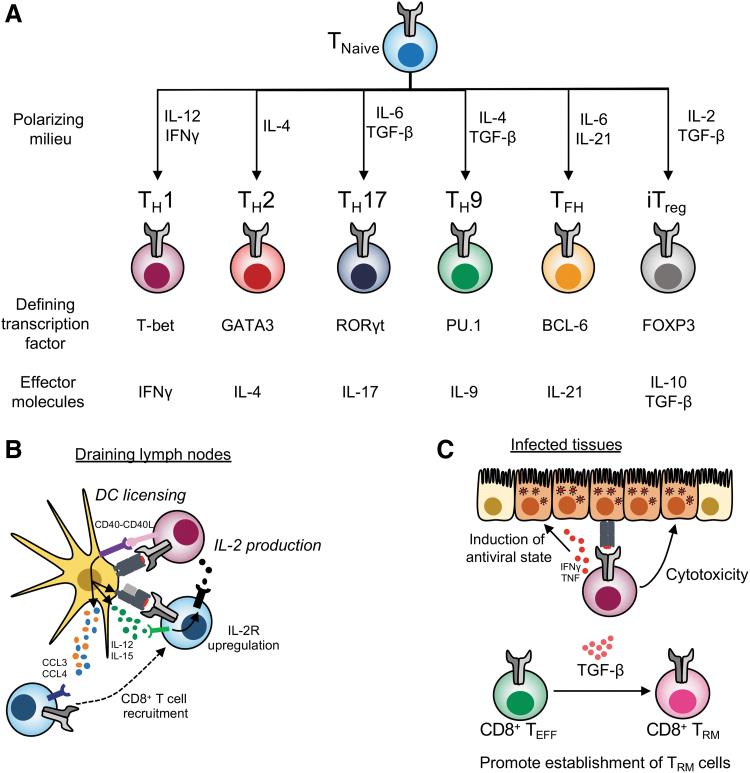
Helper CD4+ T cells develop in a manner similar to CD8+ T cells and many of the basic principles of T cell priming, memory and immunodominance, described for CD8+ T cells, largely apply to CD4+ T cell pools. However, a hallmark feature of CD4+ T cells is their great heterogeneity of effector subsets. Indeed, depending on the environment in which CD4+ T cell priming occurs, a naive CD4+ T cell can take multiple differentiation trajectories, defined by the expression of specific transcription factors and effector cytokines (105,120) (Fig. 3A). For instance, in the presence of IL-12, type I IFN, and IFNγ, induced by viral infections, naive CD4+ T cells develop toward the TH1 lineage, defined by the expression of T-bet and IFNγ (105). Conversely, exposure to IL-4, for example, induced by helminth infection, polarizes CD4+ T cells toward the TH2 lineage, characterized by expression of the transcriptional GATA-3 and IL-4 production (105). Importantly, different lineages are specialized in exerting “optimal” functions for the invading pathogen. For instance, a TH1 response promotes cellular immunity and the activation of macrophages needed for the killing and clearance of cells infected with intracellular pathogens (105).
FIG. 3.
CD4+ T cell heterogeneity and functions during viral infections. (A) CD4+ T cells may differentiate toward one of many lineages depending on the polarizing milieu they are exposed to. Different TH lineages are characterized by the expression of defining transcription factors and effector molecules. (B) Helper and non-helper functions of CD4+ T cells. In draining lymph nodes, CD4+ T cells provide help to CD8+ T cells indirectly by dendritic cell licensing and directly by the production of IL-2. (C) In infected tissues, CD4+ T cells can release antiviral cytokines like IFNγ that induce an antiviral state in epithelial cells. They can also directly induce cytotoxicity. Through the release of TGFβ, they can also promote the establishment of CD8+ TRM. IFN, interferon; TGF, transforming growth factor; IL, interleukin.
CD4+ T Cell Functions
CD4+ T cells can exert different helper and effector functions during an immune response [reviewed in refs. (75,108,120)]. This can be broadly divided into the following: (i) recruitment of immune cells in SLOs or the site of infection, (ii) provision of help for expansion or effector function, or (iii) direct antiviral functions (Fig. 3B, C). For example, in the draining lymph nodes, interactions between CD4+ T cells and APCs through CD40-CD40L binding induce signaling in the dendritic cell (DC) that results in DC “licensing” or maturation. This results in the release of chemokines like CCL3 and CCL4 by the licensed DC to attract CD8+ T cells at the site of priming (26), and upregulation of molecules involved in CD8+ T cell priming, like IL-12, by the licensed DCs (143). IL-12 can, in turn, increase expression of the IL-2Rα on CD8+ T cells (143). CD4+ T cells may also provide a source of IL-2 important for the expansion of CD8+ T cells (Fig. 3B) (143,145). CD4+ T cells are also found in the infected tissues where they can display numerous effector roles (Fig. 3C). For instance, they can act on epithelial cells through the release of antiviral cytokines like IFNγ to induce an antiviral state (75,120). In addition, CD4+ T cells can promote conversion of CD8+ T cells to TRM by the release of transforming growth factor β (75,76). It has also become evident that CD4+ T cells can exert direct cytotoxic function against virally infected cells mediated by perforin and FAS (20,22). It is important to note that many of these mechanisms may not be universally applicable, as studies with different viruses may provide different results [discussed in refs. (75,120)]. Finally, CD4+ T cell help is of course not limited to that provided to CD8+ T cells. In fact, the most studied function of CD4+ T cells is provision of help to B cells [recently review by Koutsakos et al. (70)]. Overall, CD4+ T cells are a critical component of both cellular and humoral immunity through both helper and non-helper functions.
Human Leukocyte Antigens: Genetics, Structure, and Diversity
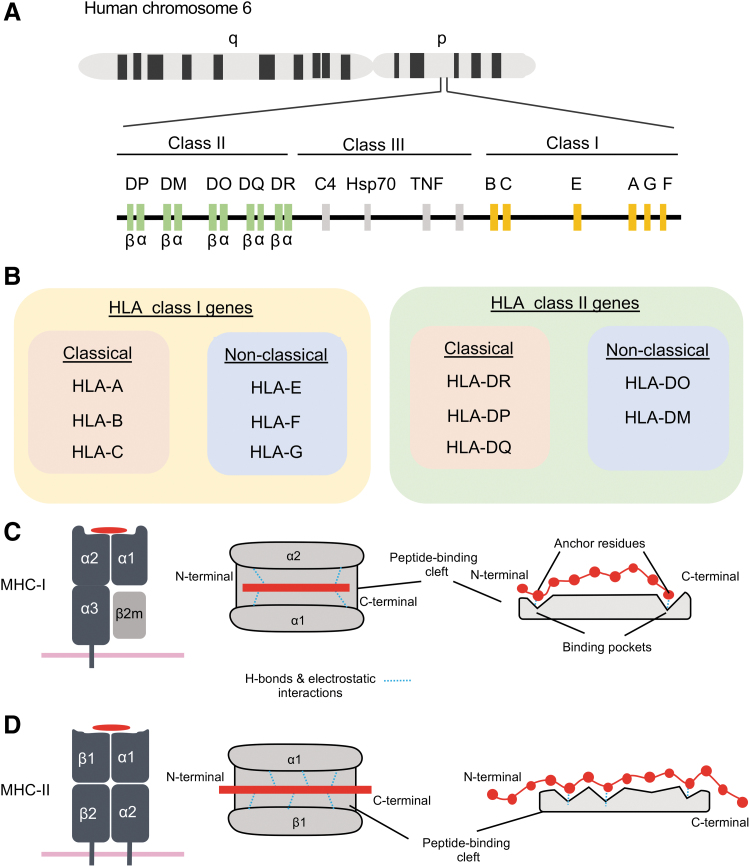
The basis of both CD8+ and CD4+ T cell priming, differentiation, and function is the cognate interaction between the TCR and pMHC. As demonstrated by Zinkernagel and Doherty in 1974 (147), this activation of T cells depends on simultaneous recognition of both foreign peptide fragments and self-MHC molecules, a phenomenon called MHC restriction. CD8+ T cells are restricted by MHC-I and CD4+ T cells are restricted by MHC-II. MHC glycoproteins are divided into three classes: MHC-I, MHC-II, and MHC-III. The MHC-III class comprises of many proteins of the complement system, cytokines, and heat shock proteins. In humans, the MHC molecules are termed HLA and are encoded on chromosome 6 of the human genome (Fig. 4A). HLA Class I (HLA-I) and HLA Class II (HLA-II), which correspond to MHC-I and MHC-II, respectively, are further divided into classical and nonclassical genes (102). Classical HLA-I genes include HLA-A, HLA-B, and HLA-C, while nonclassical alleles include HLA-E, HLA-F, and HLA-G. Classical HLA-II genes include HLA-DR, HLA-DP, and HLA-DQ, while nonclassical alleles include HLA-DO and HLA-DM (Fig. 4B) (102). Human CD8+ T cells recognize peptides presented by classical HLA-A and HLA-B, and to a lesser extend HLA-C (13), although rare populations of HLA-E-restricted CD8+ T cells have also been described (59). Human CD4+ T cells recognize peptides presented by HLA-DR, HLA-DQ, and HLA-DP.
FIG. 4.
MHC genes and structure. (A) The HLA locus encoded on Chr 6 of the human genome and is divided into Class I, Class II, and Class III genes. (B) Classification of HLA-I and HLA-II genes in classical and nonclassical groups. (C) Structure of MHC-I protein. A heavy chain made of three extracellular domains (α1, α2, and α3) is noncovalently attached to a β2-microglobulin molecule. The α1 and α2 domains form the peptide-binding cleft, where small peptides can bind. The peptides interact through two anchor residues with the binding pockets of the MHC-I molecule. (D) Structure of MHC-II protein. MHC-II is a heterodimer made of an α and a β chain, each of which has two subdomains. The α1 and β1 subdomains form the peptide binding cleft, where peptides can bind. Peptides that bind MHC II are longer than MHC-I peptides and also have anchor residues that interact with binding pockets in the peptide-binding cleft of MHC-II. HLA, human leukocyte antigen.
HLA molecules are surface membrane-bound glycoproteins. HLA-I comprises of a heavy chain made of three extracellular domains (α1, α2, and α3) noncovalently attached to a β2 microglobulin molecule (Fig. 4C) (102). HLA-II is a heterodimer consisting of an α and a β chain (Fig. 4D). The extracellular domains of HLA form an antigen-binding cleft, consisting of two α-helices surrounding an antiparallel β-sheet floor. This generates a platform that can cradle a short amino acid (aa) fragment, termed peptide. These peptides are bound to the floor of the binding groove through the interactions of specific amino acids (known as anchor residues) with specific peptide-binding pockets. Due to the closed structure of the HLA-I binding cleft, small peptides of 8–10 aa are typically presented, although longer peptides of up to 14 aa can be accommodated (Fig. 4C). HLA-II presents longer peptides of >11 aa, with N- and C-terminal “overhang” regions (Fig. 4D) (102).
The pockets and the peptide binding groove of the MHC molecules are highly polymorphic. Variation in those peptide-anchoring residues of the HLA means that different HLA allotypes (the HLA protein encoded by an HLA allele) will present a different spectrum of peptides. Despite this diversity, HLA alleles can be divided into superfamilies (or supertypes) that share peptide binding specificity, based on shared binding motifs within those peptides. For instance, members of the HLA-A2 superfamily bind to peptides with aliphatic hydrophobic residues (e.g., leucine and isoleucine) in position 2 and at the C-terminus (113). Furthermore, each individual may express up to two different HLA-A, HLA-B, and HLA-C alleles, for a total of six HLA-I alleles. The frequency of alleles and HLA superfamilies varies across ethnic groups. For example, the HLA-A*02:01 allele can be found between 1% and 54.5% of different populations around the world (121). It is worth noting that HLA allotypes can differ by >30 aa or by a handful of amino acids (44). However, even small amino acid changes can have a substantial impact on the peptide repertoire and its recognition by TCRs. This is best exemplified by the HLA-B*35:01 and HLA-B*35:08 allotypes, which only differ by one amino acid (position 156, leucine or arginine, respectively) in the α2 MHC-I helix. This single amino acid alters the conformation of the α2 MHC-I helix, in a manner that allows the prototypical SB27 TCR to recognize a 13-mer peptide from the EBV BZLF1 protein, in the context of HLA-B*35:08, but not HLA-B*35:01 (126). Thus, an appreciation of how MHC polymorphism may affect antigen presentation and recognition is crucial for our understanding of the basic principles that may underlie interindividual variation and aberrant T cell responses.
Antigen Presentation Pathways
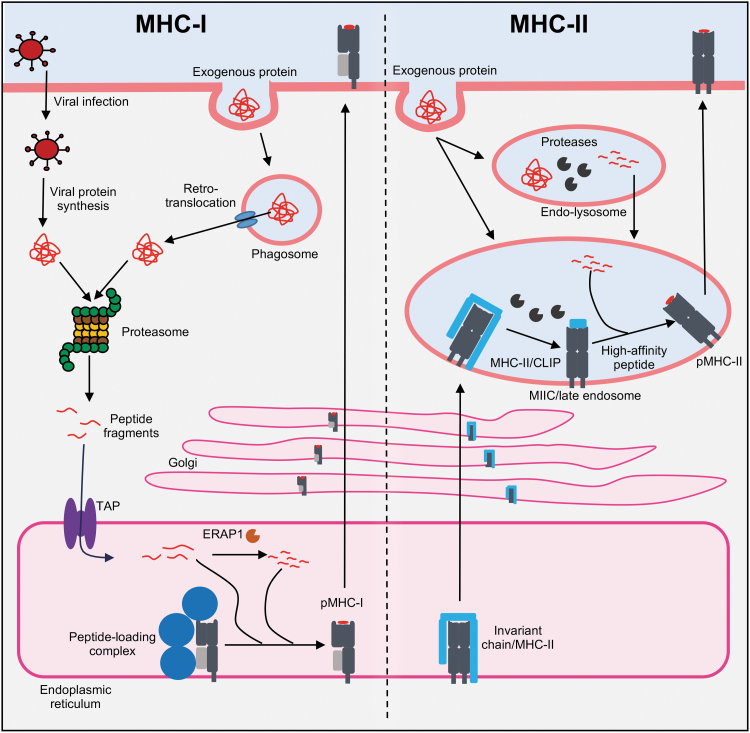
MHC-I proteins present peptides that predominantly originate from within the cell. During homeostasis, cellular proteins are degraded by the proteasome and small peptides are generated. The proteasome is a multimeric protein complex that degrades proteins into small peptide fragments (15,101). During a viral infection, the effects of IFNs induce the formation of an alternative proteasome complex, termed the immunoproteasome, which enhances the generation of peptides for MHC-I presentation. Thus, as viral proteins are synthesized, they are targeted for degradation by the immunoproteasome, either as fully folded proteins or as defective ribosomal products (15,101). This results in the generation of small peptide fragments that originate from the infecting virus, which can be further trimmed by cytoplasmic aminopeptidases. These peptide fragments are then transported into the endoplasmic reticulum (ER) through a protein called transporter associated with antigen processing (TAP), which spans the ER membranes (15,101). After translocation by TAP, any peptide that may be too long for MHC-I binding can be further trimmed by an ER-resident aminopeptidase (ERAP1). In the ER, empty MHC-I molecules are associated with the peptide loading complex (PLC), which includes chaperone proteins like Tapasin and Calnexin (15,101). The PLC holds the empty MHC-I in a peptide-receptive conformation that promotes the binding of peptides as they are translocated by TAP, which is also part of the PLC. Peptide binding stabilizes the MHC-I protein so that it is released from the ER quality control chaperones and is transported through the Golgi apparatus to the cell surface (Fig. 5) (15,101). This allows CD8+ T cells to survey the intracellular proteome of a cell for signs of infection or malignancy. This pathway also operates in professional APCs, enabling the priming of CD8+ T cells, and requires infection of the APC. In specialized subsets of dendritic cells, however, alternative pathways exist (15,101). This process, called cross-presentation, involves the uptake of extracellular antigens, its retrotranslocation from phagosomes into the cytoplasm, and subsequent degradation by the proteasome and MHC-I loading in the ER, although other pathways of delivery to MHC exist (15,101).
FIG. 5.
Antigen presentation pathways. Viral proteins produced during viral replication or antigens taken up by phagocytosis and retrotranslocated into the cytosol are degraded by the proteasome, releasing small peptide fragments. These are transported into the ER by TAP. Peptide length can be further optimized by an ER-resident aminopeptidase (ERAP1), before the peptide loading complex that promotes peptide binding to MHC-I. After peptide binding, MHC-I traffics to the cell surface through the Golgi. MHC-II in the ER is associated with an invariant chain that promotes its trafficking to a late endosomal compartment called MIIC. Proteins in endolysosomes can be broken into peptide fragments by resident proteases and delivered to the MIIC. There proteases cleave the invariant chain to a small peptide fragment (CLIP), which is exchanged for high-affinity peptides. pMHC-II complexes then traffic to the cell surface. ER, endoplasmic reticulum; TAP, transporter associated with antigen processing.
A different pathway operates for the generation of MHC-II-presented peptides. These peptides originate from extracellular sources or from proteins degraded by the endocytic pathway (15,101). Proteins in the endolysosomal compartments can be cleaved by resident proteases into peptide fragments, which can then be delivered to a late endosomal compartment called the MIIC (MHC-II compartment). As MHC-II is synthesized in the ER, it is associated with an invariant chain that promotes its trafficking to the MIIC. The proteases of the MIIC cleave the invariant chain to a small peptide fragment called the class II-associated invariant chain peptide, which can then be exchanged for high-affinity peptides with the assistance of a nonclassical MHC-II protein called HLA-DM in humans. The p/MHC-II complex will then traffic to the cell surface for CD4+ T cell recognition (Fig. 5) (15,101).
TCRs: Structure, Recombination, and Selection
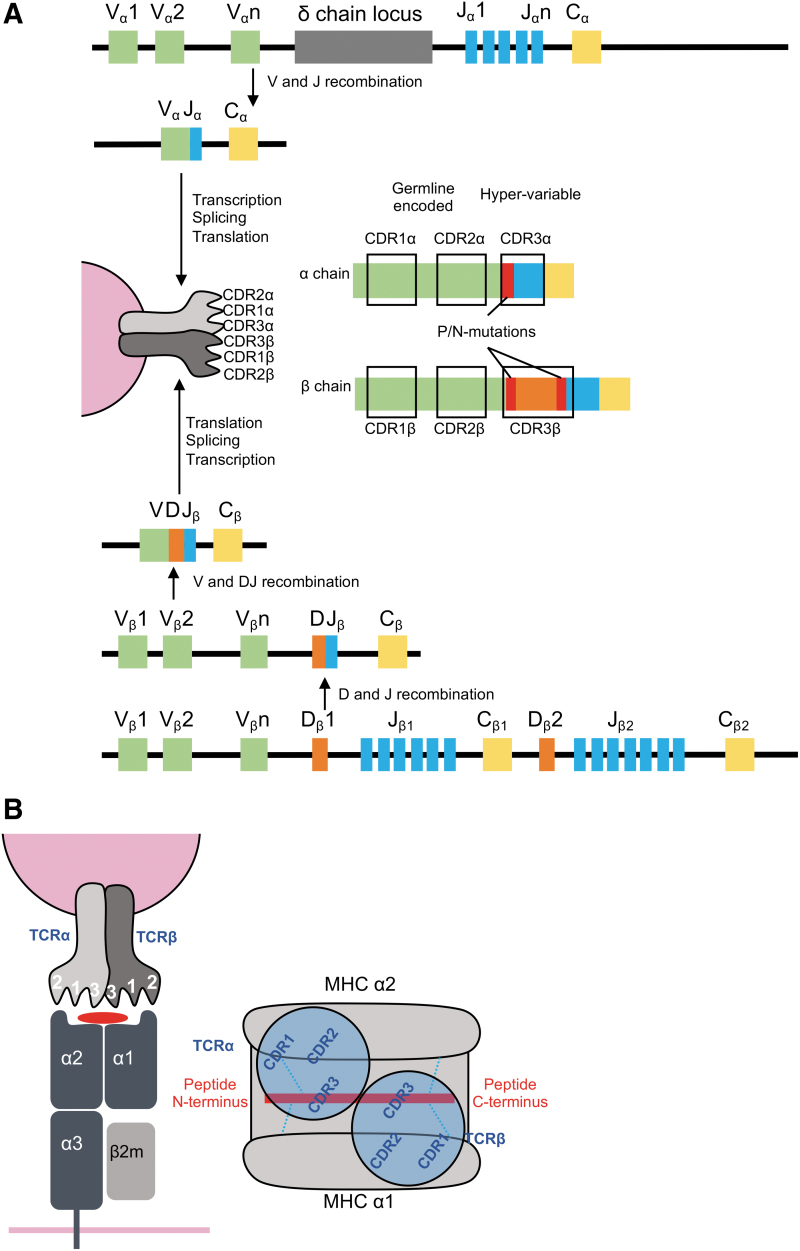
Recognition of pMHC complexes by αβ T cells is mediated by the TCR. The TCR consists of an α and a β chain (TRA and TRB). Each chain consists of several gene segments: TRA variable (TRAV), TRA joining (TRAJ), and TRA constant (TRAC) for the α chain (human chromosome 14), and TRBV, TRB diverse (TRBD), TRBJ, and TRBC for the β chain (human chromosome 7), which join during somatic recombination in the thymus. Each chain contains three variable loops called complementarity-determining regions (CDRs). While CDR1 and CDR2 are germline encoded in the V gene segments, the CDR3 loops are generated from the junction of V(D)J segments and thus display a greater level of diversity (83,102) (Fig. 6A).
FIG. 6.
TCR genetics, structure, and MHC-I recognition. (A) TCR comprises an α and a β chain, generated through the recombination of V(D)J genes. Each chain contains three CDRs. CDR1 and 2 are germline encoded, but V-segment-specific, while CDR3 is hypervariable as a result of combinatorial and junctional diversity. (B) The TCR docks over the pMHC in a conserved mode that positions the TCRα chain over the MHC α2 domain and the N-terminus of the peptide, while the TCRβ sits over the MHC α1 domain and the C-terminus of the peptide. CDR, complementarity-determining region.
Diversity in TCR sequences can arise from the following: (i) the permutations of different V(D)J segments for the α (47 TRAV and 61 TRAJ genes) and β (54 TRBV and 14 TRBJ and 2 TRBD genes) chains, (ii) the imprecise joining of gene segments during recombination by recombination activation genes, (iii) the random addition of nontemplate nucleotides at the junction sites by terminal deoxynucleotidyl, and (iv) the combination of α and β chains. This collectively results in a theoretical repertoire of 1015–1020 potential αβ TCRs that can be generated (35,83). However, after thymic selection, ∼2 × 107 distinct TCRs are maintained per individual in humans (6). An additional source of diversity can be provided by polymorphisms in TCR genes, which can have significant effects in the TCR repertoire. For instance, TCRs with the TRBV9*01 allele, but not the TRBV9*02 allele, recognize the HLA-B*35:01-restricted EBV peptide (HPVG), due to a single amino acid change between the two alleles (Gln55His). While the Gln55 in TRBV9*01 easily binds the P7-Asp of the peptide, the electropositive His of the TRBV9*02 allele cannot accommodate the P7-Asp (46).
The process of thymic selection considerably reduces the available TCRαβ repertoire by many orders of magnitude. The positive and negative selection of immature thymocytes are based on the strength of their interaction with self-pMHC expressed in the thymic medulla (67). A weak interaction rescues developing thymocytes from death by neglect and promotes their development into either CD4+ or CD8+ T cells. Conversely, a strong interaction for self-pMHC results in clonal deletion and thus removal of autoreactive T cells (67).
Overall, through the vast diversity enabled by TCR genetics, the immune system can recognize the extremely large number of potential peptide antigens, which can be derived from the plethora of host and viral proteins. Concomitantly, the intricate processes of thymic selection ensure that self-derived antigens are not targeted by T cells during an immune response.
TCR Recognition of pMHC: Geometry and Restriction
The six CDR loops of the TCR are critical for recognition of the pMHC complex. Overall, the TCR docks over the pMHC complex with a conserved geometry. The TCR binds in a diagonal manner on top of the peptide binding groove with an angle of ∼45°, although this can range from 20° to 70° (102). Two notable exceptions described in the last few years represent two TCRs, which dock over their cognate pMHC with a reverse geometry (9,45). The conventional docking mode positions the TCRα chain over the α2 MHC-I helix and the TCRβ over the α1 helix. In addition, the CDR3α is positioned over the N-terminus of the peptide and the CDR3β over the C-terminus (Fig. 6B). Consequently, the germline-encoded CDR1 and CDR2 loops interact with the MHC and the hypervariable CDR3 engages the peptide. However, it is pertinent to note that generally, the CDR1/2 loops can account for ∼40% of the buried surface area (a measure of the size of the interface between two interacting molecules) between the TCR and the peptide, and the CDR3 loops account for ∼35% of the contacts with the MHC-I (24).
The requirement of T cells to simultaneously bind the self-MHC as well as peptides is intriguing, and its functional and structural bases have been the focus of intense research [reviewed by La Gruta et al. (71)]. Briefly, two proposed theories explain what drives MHC restriction of TCRs. First, the germline-encoded theory proposes that TCRs are hardwired, through evolutionary selection of germline-encoded motifs, to recognize MHC molecules. This model is mostly supported by studies identifying conserved TCR-MHC interactions, like the conserved Tyr48 in CDR2β (111) and the intrinsic MHC reactivity of many (∼30%) TCRs before thymic selection (79,81,146). The second model, called selection theory, proposes that the ability of TCRs to recognize MHC is imposed by selection during T cell development in the thymus, through the requirement of CD4 or CD8 co-receptor binding for efficient signaling through the TCR. A third, unifying model proposes that (i) a predisposition of TCRs to bind MHC ensures that enough premature T cells in the thymus will develop into mature T cells in the periphery, despite the enormous diversity of potential MHC allotypes within a species, and (ii) the process of positive selection then endows the mature T cell pool with reactivity toward the specific MHC allotypes expressed within the individual. As more structural information becomes available and with the advent of system immunology that enables high-throughput analysis of TCR repertoires, a deeper understanding of MHC restriction is to be gained.
TCR Repertoire Diversity and Sharing
Although the theoretical/potential diversity of the human peripheral TCR repertoire is estimated to be around 1012, the actual TCR diversity of a naive individual is ∼2.5 × 107. This striking difference in numbers means that the likelihood of the same TCRαβ being found in two individuals would be extremely low (83,90) and indeed inbred mice only share 20–25% of their naive TCRβ repertoire (19). Some TCR clonotypes may be shared across unrelated individuals. Yet, many T cell responses are characterized by one or a few TCRαβ clonotypes that are shared across large numbers of genetically unrelated individuals, which is extraordinarily specific, given the vast diversity and number of potential TCRs generated between individuals (64,83,128). This raises the question of how the same TCRs can be found across individuals, given the numerical paradox described above.
Two key factors that may contribute to this phenomenon are TCR genetics and the structure of the pMHC-I/TCR interface (83). With regard to genetics, many public TCRs, that is, TCRs shared across different individuals, can be generated by convergent recombination (135–137), a process by which sequences converge to produce the same TCR amino acid sequence. Convergence may result from recombining TCR segments converging to the same nucleotide sequence or different nucleotide sequences converging to the same amino acid sequence due to codon redundancy. This, in combination with germline likeliness (low number of N-insertions), is thought to contribute to the generation of public TCRs by increasing the number of naive precursors of a specific TCR clonotype. This is supported by studies by Venturi et al. in the murine IAV model (137), Simian immunodeficiency virus (SIV) infection in macaques (136), and human Cytomegalovirus (CMV) and Epstein Barr virus (EBV) responses (135), revealing that the extent of TCR sharing correlated with a low number of N-insertions and a large number of nucleotide sequences encoding for the same amino acid sequence.
The structure underlying the TCR-pMHC-I interaction is another critical factor that can account for the presence of public TCRs. This is supported by structural studies of multiple public TCRs (44,102,119), which highlight that biases in V/J gene usage and CDR3 motifs (which define public TCRs) have structural foundations, whereby residues encoded within the biases are critical for pMHC engagement. This can be exemplified by the public HLA-A2-restricted M158–66 (A2/M158)-specific TCRs, which are strongly biased for TRBV19 and a non-germline-encoded arginine in the CDR3β loop. Based on the prototypical A2/M158-specific JM22 TCR structure (119), the TRBV19 gene segment dominates contacts with the MHC through its CDR1β and CDR2β loops, enabling the TCR to adopt the correct orientation over the peptide, so that the conserved arginine from CDR3β can form the “peg-in-notch” created by the peptide and the MHC α2-helix (119).
Overall, the public TCR clonotypes can be generated more efficiently, which result in a numeric advantage in the naive repertoire. These TCRs then need to optimally interact with their cognate epitope and outcompete other TCRs. Indeed, both criteria need to be met, as more efficiently generated TCRs still need to expand and clonally dominate the response. Conversely, TCRs capable of optimal pMHC engagement and clonal dominance need to be efficiently generated by recombination and preselected for in the thymus.
TCR Repertoire Diversity and Selection During Immune Responses
The majority of evidence on generation and persistence of antigen-specific TCRs comes from easily manipulated mouse models of viral infection or peptide immunization. The development of a tetramer-magnetic enrichment approach (86) has facilitated a dissection of naive antigen-specific TCR repertoires and the understanding of how particular clonotypes are recruited into an immune response. Using this method, the naive precursor frequencies of CD4+ T cells directed against peptides restricted by I-Ab were estimated between ∼20 and 200 per mouse (∼190 for 2W1S:I-Ab, ∼20 for FliC:I-Ab, and ∼16 for OVA:I-Ab specificities), when spleen and lymph node were analyzed (86). The naive population size correlated with the magnitude of the primary immunodominant CD4+ T cell responses. The TCRβ diversity of the naive antigen-specific CD4+ T cells, as measured by Vβ antibody staining and fluorescence-activated cell sorting, was highly reflective of the TCR repertoire on d8 after peptide + lipopolysaccharide immunization.
Adapting the enrichment method to CD8+ T cells, the numbers of naive tetramer-specific CD8+ T cells per mouse range from 10s [∼30 naive precursors for immunodominant influenza-specific DbNP366 (72)] to 100s [∼600 for murine CMV DbM45 (91)] and 1,000s in one extreme example (∼3,000 for malaria DbGAP5040) (131). Although the size of the naive CD8+ T cell pools can predict the size of the immunodominant effector CD8+ T cell populations (91), it is CD8+ T cell quality, rather than merely quantity, which plays a key role in determining immune response magnitude (33,72). Using tetramer enrichment, single-cell sorting, and reverse transcritpion-polymerase chain reaction (RT-PCR), the composition and diversity of naive and effector TCR repertoires were compared across four different CD8+ specificities (DbNP366+CD8+, DbPA224+CD8+, DbPB1-F262+CD8+, and KbNS2114+CD8+) in B6 mice (72,122). While the main TCR characteristics (Jβ region and CDR3β length) were highly comparable between the naive and effector TCR repertoires, selective expansion of TCR clonotypes and differential clonotype distribution was evident. This most likely results from both the intrinsic and extrinsic (antigen driven) characteristics of the naive TCRs (122). Notably, the subdominant T cell specificities (DbPB1-F262+CD8+ and KbNS2114+CD8+) demonstrated a failure to fully recruit and expand the naive precursor pool, which correlated with a lower prevalence of high-avidity T cells compared to immunodominant T cell specificities (DbNP366+CD8+ and DbPA224+CD8+) (33,72).
The notion of biased TCR recruitment is further supported by the use of a sophisticated transgenic TCRαβ barcoding approach to track T cells during an immune response. Although the majority of the naive TCR repertoire was recruited into the immune response (134), the different T cell families expanded at different rates (42). While the dominant transgenic OT-I TCR clones expanded 400-fold more than an average naive clone, subdominant clones, corresponding to 50% of OT-I families, had fewer than 200 daughter cells. An inverse correlation between CD62L expression and the clonal size suggests that despite a single TCR specificity, naive clones differ markedly in their clonal expansion and differentiation.
Using a human-adapted tetramer enrichment, the naive T cell precursor frequencies for HLA-A*0201-restricted NY-ESO-11157–165, WT1126–134, HIV Gag p1777–85, HCV Core132–140, and CMV pp65 antigens in cancer-free and seronegative individuals have been estimated between 0.6 × 10−6 and 5.3 × 10−6 CD8+ T cells (2,88). In comparison, frequencies of epitope-specific memory CD8+ T cells are in the range of 1 antigen-specific cells in 104 CD8+ T cells (1 × 10−4) (2,69,88). TCRαβ signatures within naive human antigen-specific T cells are still to be elucidated. Overall, these studies shed light on how the naive dominant and subdominant TCR clonotypes are mobilized into an immune response, important for designing improved vaccines and immunotherapies that aim to promote the magnitude and breadth of the T cell response.
The exact mechanism underlying preferential selection of specific TCR clones over the others is unclear; however, published evidence suggests that the avidity for pMHC is a key determinant of TCR selection and clonotype representation in the immune repertoire. As high-avidity CD8+ T responses can kill virus-infected cells ∼1,000 more efficiently than low-avidity T cell pools (3), high-avidity TCR clonotypes are clearly important for more favorable clinical outcomes. An approach combining single-cell RT-PCR and affinity measurements directly linked pMHC-II binding affinities and clonal diversity of responding CD4+ T cells (77). Although naive CD4+ T cells specific for the pigeon cytochrome c (PCC) antigen displayed a wide array of TCR affinities, TCR clonotypes with low-affinity TCRs were lost early after antigen exposure. Thus, it appears that the affinity threshold for pMHC class II is required for the transition of PCC-specific CD4+ T cells from naive to fully expanded immune effectors. This strategy enables the best-fit TCRs to have a selective advantage over less-fit clonotypes in the naive T cell repertoire (77).
Although only TCRs with the “adequate” threshold pMHC affinity get recruited into the immune response, TCR clonotypes within any given response display a wide range of pMHC avidities, all undoubtedly above the recruitment-threshold level. Clonotypic dissection of high pMHC avidity T cell pools performed by single-cell RT-PCR analysis combined with tetramer dilution (63) and tetramer dissociation (85) probed the link between pMHCI avidity and specific effector TCR clonotypes. In comparison to the total pool of antigen-specific CD8+ T cells (stained with tetramers at saturating levels and no anti-H2Db antibody), a subset of immunodominant DbPA224+Vβ7+CD8+ TCRs was preferentially selected under conditions of limited tetramer availability. This resulted in particular DbPA224+TCRβ clonotypes being overrepresented in the “high-avidity” population. Conversely, prominent public DbNP366+Vβ8.3+CD8+ T cell clonotypes were present at equivalent prevalence in immune T cells when sorted from the total antigen-specific CD8+ T cells. As DbPA224+Vβ7+CD8+ T cells are of much higher pMHCI avidity than DbNP366+Vβ8.3+CD8+ pools (73), there appears to be a pattern of differential TCRβ clonotype distribution within high and low pMHC avidity populations. Thus, a higher degree of clonal TCRβ diversity within a particular epitope leads to more scope for pMHCI avidity segregation between the clonotypes. In contrast, the finding of the same public DbNP366+Vβ8.3+CD8+ clonotypes being present in both high- and low-avidity subsets suggests that other intrinsic factors, apart from pMHC–TCR interactions, promote the overall effectiveness for T cell specificities with lower overall pMHC-I avidity.
Active repertoire selection and clonal dominance in immune repertoires are based on TCR avidity for pMHC. This was supported by a subsequent human study showing that the emergence of dominant TCR clonotypes during infection correlates with higher TCR avidity for pMHCI tetramers in individuals with chronic CMV or EBV infection (96). High pMHCI TCRs are more preferably driven toward extensive expansion and terminal differentiation (95).
While high pMHCI avidity allows rapid recruitment of the “best-fit” clonotypes into an immune response, lower pMHC avidity clonotypes can use a number of intrinsic mechanisms to enhance overall avidity and functional sensitivity, enabling their competitive recruitment and maintenance within an immune response. These mechanisms can be divided into (i) co-receptors that contribute to the TCR-pMHC interaction (through binding/stabilization or synapse formation, e.g., CD8β) and (ii) co-receptors that provide the necessary signals for entering the effector or memory pathways (CD27 and CD28 co-receptors or cytokine receptors, including IL-2R and IL-7R).
Differential recruitment into the immune response of barcoded transgenic OT-I cells with a single TCR specificity, as discussed above, provides clear evidence that preferential selection of TCR clonotypes does not solely depend on a TCRαβ signature (42). Similarly, the distribution of public TCRβ clonotypes among both low and high pMHCI avidity subsets within the influenza-specific DbNP366+CD8+ T cell response (63,85) suggests that intrinsic factors other than CDR3β sequence contribute to T cell avidity. The main intrinsic contributor to pMHC avidity is the dependence of specific T cells on a CD8β co-receptor for stable pMHC binding. Experimentally, this is assessed with either CD8β-null 227 tetramers, which prevent CD8β co-receptor binding and thereby detect only high avidity CD8+ T cells (96), or by using a blocking anti-CD8β mAb in combination with peptide stimulation in an ex vivo ICS (127). It is well established that TCR clonotypes with lower pMHCI avidities have higher dependence on CD8β for recruitment into the immune response (85,95,96). Thus, the CD8β co-receptor contributes greatly to the overall avidity of TCRs and serves to promote clonotypic diversity within an immune response.
Second, TCRαβ heterodimer expression levels and adjacent membrane organization also play an important role in determining T cell fitness during the immune response. The efficiency of TCRαβ surface expression depends highly on the intrinsic qualities of the clonally expressed TCR-CD3 complex and strongly correlates with functional activity of T cells (50). Early in the response, the affinity of CD8+ T cells can also be increased by optimization of the TCR-mediated signaling machinery, specifically Lck expression (114). Higher Lck protein levels lead to markedly increased IFN-γ production (functional avidity), thus allowing “affinity maturation” without any genetic modification in actual TCRαβ signatures. The cholesterol organization of a T cell membrane also contributes to pMHC avidity and the capacity to respond to a low antigen dose. Membrane TCR reorganization upon activation allows antigen-specific TCRs to bind multimeric pMHC complexes with increased avidity. This enhances responsiveness mediated through increased cross-linking of TCRs (36). Furthermore, the avidity of CD8+ T cells can be generated by co-localization of TCRs together with CD8αα and CD8αβ in the lipid rafts (27). Such optimal TCR-CD8 organization requires relatively fewer TCR engagements when compared to low-avidity TCRs. Finally, the expression (and/or levels) of co-stimulatory molecules like CD27 and CD28 as well as cytokine receptors like IL-2Rα and IL-7Rα can promote more efficient recruitment of T cells into the effector differentiation pathway (62,92).
Thus, while TCRαβ sequence appears to be the major contributor to pMHC avidity in diverse virus-specific T cell populations, other intrinsic mechanisms promoting CD8β and TCR–pMHC interactions play an important role in avidity determination, especially in the absence of TCR diversity.
TCR Repertoire Diversity Across Distinct Stages of CD8+ T Cell Differentiation
The frequency of memory T cells is <1% at birth and greatly increases with age. In humans, memory CD4+ and CD8+ T cells constitute ∼50% of all the circulating peripheral blood T cells in a healthy adult (89). Established memory T cell populations provide protection against recurrent infections, leading to milder clinical outcomes and rapid recovery of the host. For example, even in the absence of neutralizing antibodies to the newly emerged influenza viruses, preexisting memory CD8+ and CD4+ T cell pools promote recovery from experimental H3N2, natural H1N1-2009, or avian H7N9 influenza (118,140,144).
The frequency of epitope-specific memory T cells is up to 200–1,000 times (31) greater than the naive precursor pool (72), allowing rapid responses and enhanced pathogen clearance upon recall. In general, the TCR clonotypic composition of memory T cell pools reflects that of the naive and early or late effector T cell sets. In the B6 mouse model of influenza infection, longitudinal analyses of the same animals sampled repeatedly (using blood) have allowed dissection of individual clonotypes from acute primary effectors through to early and long-term memory using single-cell sorting, RT-PCR, and CDR3β sequencing (64,125). The prominent TCRβ clonotypes found at the acute effector phase for two immunodominant responses, DbPA224+Vβ7+CD8+ and DbNP336+Vβ8.3+CD8+ populations, persisted into both short- and long-term memory, and could be subsequently expanded after the secondary challenge (32,64,125). Furthermore, both TCRβ diversity and clonotypic composition within the antigen-specific memory pools are equivalent to those found at other time points after infection, including early antigen exposure. Such clonotypic stability of the long-term memory T cell populations is supported by earlier data from LCMV infection or CW3 immunization (14,78,117). In contrast, however, TCR narrowing in the memory pool has been reported for Listeria monocytogenes-specific CD8+ T cells and PCC-specific CD4+ T cells (25,80). In this study, the preferential selection of high-avidity TCR clones into the memory pool has been linked to TCR affinity/avidity maturation. Such differential levels of TCR clonotype persistence into memory might be related to a certain pMHCI-TCR avidity threshold required for survival into the long-term memory.
Memory T cells are highly heterogeneous with distinct lymph node homing properties (CD62L and CCR7), anatomical localizations and functions (106). An obvious question emerges as to how different TCR clonotypes contribute to the specific memory subsets and their functions. Experiments utilizing clonotypic analysis of TCR signatures in humans (8) and mice (18,66) showed that common TCRs are distributed across the main memory T cell subsets, central T cell memory (TCM) and effector T cell memory (TEM) populations. However, strikingly, the TCM sets contain additional TCRβ clonotypes (diversity increased by ∼30%) not found within the TEM populations. TCR diversity within both the TCM and TEM subsets was consistently stable, starting from the early acute phase of infection (d8) through to d28 early and >d500 long-term memory (66). The main conclusion here is that different T cell memory subsets contain stable TCRβ signatures from the early days after antigen exposure, with the memory TCM pool preserving clonal diversity and preventing “overdominance” by a few greatly expanded TCR clones. Together, these findings support the idea that any antigen-specific T cell (with the sufficient pMHCI avidity) that has not achieved a terminally differentiated stage can become a memory T cell. As the establishment of immunological memory is critical for a rational design of any cell-mediated vaccine, understanding the capacity of the optimal and suboptimal TCR clonotypes to transition between functionally different naive/effector/memory subsets is important.
The Importance of TCR Repertoire Diversity on Disease Outcome
TCR clonotype selection during an antigen-driven T cell response can influence biological outcome, with diversity and composition of the peripheral TCR repertoire having major consequences for subsequent immune responses in both animal models and human disease (83). Nikolich-Zugich and colleagues presented the first elegant evidence for the link between TCR repertoire diversity and immune protection (82). Although the herpes virus (HSV)-derived SL8495–501 peptide is presented equally well by both the wt H2Kb and the mutant H2-Kbm8 MHC class I glycoproteins, mice expressing the wt H2Kb allele are much more susceptible to HSV infection. Dissection of the SL8-specific TCR repertoire established that SL8-H2Kbm8 selects more diverse and higher-avidity TCRs (82). Thus, TCR repertoire diversity, pMHCI avidity, and protection are interrelated and influence resistance profiles.
A subsequent link between TCR repertoire diversity, avidity, and protection from viral escape was established in the rhesus macaque SIV model (96,97). SIV-specific CD8+ T cell responses are directed against two prominent epitopes characterized by different TCR repertoires. The Tat28–35 (TL8)-Mamu-A*01-specific CD8+ T cells are characterized by expression of TCRs with conserved CDR3β sequences, while the Gag181–189 (CM9)-Mamu-A*01-specific clonotypes display diverse CDR3β sequences. CD8+ T cell responses to the SIV-TL8 peptide with a restricted TCR repertoire were associated with the rapid emergence viral escape mutants at p5 of the TL8 peptide, while the diverse TCR repertoire of SIV CM9-specific CD8+ T cells led to a high level of conservation within the wt CM9 peptide sequence. Although mutations may occur within the CM9 peptide, the CM9-Mamu-A*01-specific TCRs, especially the public clonotypes (95), have the breadth to recognize escape variants, thereby preventing the outgrowth of mutant viruses. Thus, the diverse CM9-specific TCR repertoire allows the preferential selection of high-avidity TCR clonotypes, recognizing the newly emerging viral variants (96).
As with persistent infections, TCR diversity protects from viral escape in acute, readily resolved infections (129). In B6 mice, the diverse and high pMHCI avidity DbPA224-specific TCR repertoire protects against viral escape during influenza infection. Conversely, sequential emergence of escape variants occurs preferentially within the DbNP366+ epitope, which is recognized by CD8+ T cells expressing a restricted TCRβ repertoire (64). The selection of CD8+ T cell escape mutants is driven by selective pressure from DbNP366+CD8+ T cells as the variants revert to the wt sequence in the absence of immune pressure in MHC-mismatched mice (129). Preemptive priming against the influenza viral mutants, however, can expand T cells expressing additional distinct TCRs that are capable of mounting robust recall responses against the viral variants (129).
Overall, the data from animal models strongly suggest that the extent of TCR diversity, and the resultant selection of TCR clonotypes with high avidity for pMHCI, generates broadly protective optimal responses capable of recognizing both the wt virus and the newly emerging mutants.
Studies point to the importance of TCR selection for viral control and disease-associated morbidity in HIV, hepatitis C virus (HCV), CMV, and influenza. The most solid evidence comes from HIV infection, where HLA association with the disease outcome is well documented. Given the key role of MHC molecules in TCR selection during thymocyte ontogeny, differences in MHC haplotype are likely to shape the available TCR repertoire. While some HLA alleles, HLA-B*27, B*57, and B*58, have been identified as “protective”’ against HIV-1, others, including HLA-B*35 and HLA-B*53, are considered susceptibility factors for more rapid disease progression. This implies the importance of CD8+ T cells (and specific TCRs involved) in the control of HIV infection. However, as not all HLA-B*2705 individuals control HIV infection, the composition of the specific TCR clonotypes in a selected HLA-B*2705 population drives HIV control. Indeed, a comparative study of five elite controllers and five progressors found that differential control of HIV-1 is mediated through specific TCR clonotypes directed toward the immunodominant Gag epitope, KK10 (28). Superior control of HIV replication by “protective” TCR clonotypes was linked to the cross-reactive capacity to recognize the wt and the “escape” L6M variant, combined with their optimal capacity to rapidly deliver lytic granules (granzymes/perforin) to the pMHC-I-TCR immunological synapse and kill virally infected targets. In controllers, such effective TCRs are consistently of high prevalence (immunodominant), while the progressors possess only functionally ineffective clonotypes (immunodominant and subdominant). Interestingly, the CDR3 sequences of KK10-specific CD8+ T cells in the HIV controllers are close to germline and have been shown previously to efficiently recognize emerging viral variants, and thus control HIV infection (68).
An independent study also showed that cross-reactive KK10-specific TCR clonotypes, as shown by double-tetramer staining and similar docking sites within ternary pMHCI-TCR complexes, can be rapidly generated and recruited to control both and the wt virus and the newly emerged L6M mutant (74). Similarly, the antiviral efficacy and cross-reactivity of the TW10 epitope (Gag 240–246) were superior in HLA-B57-expressing controllers compared to progressors (28). Clearly, such associations between MHC haplotype, TCR repertoire, and disease control are of key interest as we seek to understand and manipulate T cell immunity through vaccination.
Similar to HIV, protective HLA alleles (A*0201, A*0301, B*5701, B*1801, and B*0801) are associated with universal cross-strain CD8+ T cell immunity against acute influenza A virus infections, including the newly emerged H7N9 subtype (98). In contrast, individuals expressing A*2402, A*6801, and B*1501 show poor influenza-specific CD8+ T cell response profiles (52,98,132). The molecular mechanism underlying the universal HLA-mediated protection is mainly unknown. To date, TCR repertoire for influenza-specific CD8+ T cells has been reported across different HLAs (29,43,89,93,109,128,141). The CD8+ T cells specific for A2-M158 are exclusively restricted to TRBV19 usage and express similar CDR3β sequences. A dominant public TCRαβ clone (TRAV27 GGSQGNL; TRBV19 SIRSSYEQ) can be found at high frequency within M158+CD8+ T cell responses in all HLA-A*0201+ individuals tested. This public A2+M158 TCR can also recognize the naturally occurring variants of the M158 peptide (128), which contributes, at least in part, to the high conservation rate of the M158 epitope within the influenza viruses in human circulation. This further suggests that the HLA-A2+ population (up to 50% population coverage) has universal T cell immunity to any M158 variant. In contrast, the immunodominant influenza-specific NP418–426 peptide presented by the HLA-B7 superfamily (including HLA-B*3501, B*3503, B*0702) has sequentially mutated at four different TCR contact positions, generating over 20 different peptide sequences over the past 90 years (47). However, depending on mutations occurring at the most solvent-exposed residues at p4 and p5, B7-NP418-specific CD8+ T cell responses show two distinct profiles of cross-reactivity toward either seasonal (1947–2005) or pandemic (1918, 2009) influenza-derived NP418 peptides. TCR clonotypes within the B7+NP418+CD8+ T cell populations differentially recognize distinct peptides from either the pandemic or seasonal influenza viruses (128). Thus, a preemptive influenza vaccine that elicits TCR pools, which recognize distinct antigenic variants, can provide universal immunity against any future influenza strain or subtype.
In CMV infection, the diversity of TCRαβ repertoire (rather than the magnitude of the CD8+ T cell pool) against the HLA-A*0201-restricted pp65495–503 (NLV) epitope is associated with viral control (139). TCRαβ diversity, analyzed by unbiased single-cell multiplex RT-PCR, inversely correlated with the circulating antibody levels, a hallmark of viral load in patients. Further modification of this technique to utilize IFN-γ capture in clinical settings found a novel TCR signature (TRAV3TRAJ31-TRBV12-4TRBJ1-1) within the HLA-A*0201-restricted CMV-NLV+CD8+ response capable of recognizing alloreactive peptide/s in the context of HLA-B27 alleles (B*2705, B*2707, and B*2709) (87). This clearly indicates the importance of any cross-reactive viral-specific TCRs in HLA-mismatched solid organ transplants. Cross-reactive CMV-specific CD8+ TCR clonotypes capable of recognizing distinct viral variants evolve over time, as shown for the immunodominant HLA-B8-restricted CMV IE-1 epitope (115). This is in contrast to CD8+ T cells directed toward a single genetic variant with oligoclonal private TCR repertoires.
Although there is much less understanding of the involvement of TCRs in controlling the HCV disease, comparative analysis of two immunodominant A*0101- and B*0801-restricted epitopes showed that TCR repertoire diversity might be associated with HCV persistence or clearance in humans (84). Such diversity generated by switching of TRBV and TRBJ genes around a conserved CDR3β loop in HCV-specific TCRs suggests a potential mechanism to control antigenic variations in HCV. Recent studies have also focused on TCR clonotype usage in cancer and autoimmunity and found common features between virus- and cancer-recognizing TCRs, including TCR bias and CDR3β length (83).
Concluding Remarks
Studies in the last four decades have made significant progress in our understanding of T cell-mediated immunity and dissecting the complexities of T cell populations, and have generated critical questions that need to be addressed if we are to harness T cells by vaccination and immunotherapies. Published evidence from both animals and humans shows the importance of TCR repertoire in the disease outcome, with a diverse TCR repertoire providing a greater range of clonotypes with scope for the preferential selection of high-avidity TCRs into the immune response. Of a particular advantage for disease control are diverse TCR repertoires with high pMHC avidity TCR clonotypes capable of promoting superior CD8+ T cell function and recognizing viral escape mutants, as well as the public prominent TCR clonotypes. However, the exact mechanisms underlying recruitment, persistence, and recall of dominant TCR clonotypes across the human life span, from early childhood to the aged years, are far from being clear and need to be taken into consideration when developing T cell-based vaccination strategies to provide effective and optimal protection across the life span of an individual. Similarly, the immunodominance of T cells directed at numerous epitopes across different HLAs within the same individual should be addressed, and so should identification of novel T cell epitopes across different ethnicities so the T cell-mediated vaccines can reach a substantial global coverage. Finally, while we now understand how TCRs and pMHCs interact at the molecular level, the very basis of MHC restriction remains enigmatic and highly debatable (71). All this knowledge gained and all the arising questions, which span every aspect of T cell biology and have been pioneered by work by Peter Doherty and colleagues, are only possible because of the 1974 groundbreaking discovery of MHC restriction by Doherty and Zinkernagel.
Acknowledgments
The authors would like to thank Bridie Clemens for a critical review of the article. K.K. would like to express her immense gratitude and appreciation to Peter Doherty for his invaluable mentorship and support over the last 17 years.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
K.K. is supported by an NHMRC Senior Research Fellow (#1102792).
References
- 1. Akondy RS, Fitch M, Edupuganti S, et al. Origin and differentiation of human memory CD8 T cells after vaccination. Nature 2017;552:362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alanio C, Lemaitre F, Law HK, et al. Enumeration of human antigen-specific naive CD8+ T cells reveals conserved precursor frequencies. Blood 2010;115:3718–3725 [DOI] [PubMed] [Google Scholar]
- 3. Alexander-Miller MA, Leggatt GR, and Berzofsky JA. Selective expansion of high- or low-avidity cytotoxic T lymphocytes and efficacy for adoptive immunotherapy. Proc Natl Acad Sci U S A 1996;93:4102–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Almeida JR, Price DA, Papagno L, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med 2007;204:2473–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Almeida JR, Sauce D, Price DA, et al. Antigen sensitivity is a major determinant of CD8+ T-cell polyfunctionality and HIV-suppressive activity. Blood 2009;113:6351–6360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arstila TP, Casrouge A, Baron V, et al. A direct estimate of the human alphabeta T cell receptor diversity. Science 1999;286:958–961 [DOI] [PubMed] [Google Scholar]
- 7. Auladell M, Jia X, Hensen L, et al. Recalling the future: immunological memory toward unpredictable influenza viruses. Front Immunol 2019;10:1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baron V, Bouneaud C, Cumano A, et al. The repertoires of circulating human CD8(+) central and effector memory T cell subsets are largely distinct. Immunity 2003;18:193–204 [DOI] [PubMed] [Google Scholar]
- 9. Beringer DX, Kleijwegt FS, Wiede F, et al. T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex. Nat Immunol 2015;16:1153–1161 [DOI] [PubMed] [Google Scholar]
- 10. Betts MR, Nason MC, West SM, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 2006;107:4781–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bhat P, Leggatt G, Waterhouse N, et al. Interferon-gamma derived from cytotoxic lymphocytes directly enhances their motility and cytotoxicity. Cell Death Dis 2017;8:e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Billiau A, and Matthys P. Interferon-gamma: a historical perspective. Cytokine Growth Factor Rev 2009;20:97–113 [DOI] [PubMed] [Google Scholar]
- 13. Blais ME, Dong T, and Rowland-Jones S. HLA-C as a mediator of natural killer and T-cell activation: spectator or key player? Immunology 2011;133:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blattman JN, Sourdive DJ, Murali-Krishna K, et al. Evolution of the T cell repertoire during primary, memory, and recall responses to viral infection. J Immunol 2000;165:6081–6090 [DOI] [PubMed] [Google Scholar]
- 15. Blum JS, Wearsch PA, and Cresswell P. Pathways of antigen processing. Annu Rev Immunol 2013;31:443–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bonilla FA, and Oettgen HC. Adaptive immunity. J Allergy Clin Immunol 2010;125:S33–S40 [DOI] [PubMed] [Google Scholar]
- 17. Bots M, and Medema JP. Granzymes at a glance. J Cell Sci 2006;119:5011–5014 [DOI] [PubMed] [Google Scholar]
- 18. Bouneaud C, Garcia Z, Kourilsky P, et al. Lineage relationships, homeostasis, and recall capacities of central- and effector-memory CD8 T cells in vivo. J Exp Med 2005;201:579–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bousso P, Casrouge A, Altman JD, et al. Individual variations in the murine T cell response to a specific peptide reflect variability in naive repertoires. Immunity 1998;9:169–178 [DOI] [PubMed] [Google Scholar]
- 20. Brien JD, Uhrlaub JL, and Nikolich-Zugich J. West Nile virus-specific CD4 T cells exhibit direct antiviral cytokine secretion and cytotoxicity and are sufficient for antiviral protection. J Immunol 2008;181:8568–8575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brincks EL, Katewa A, Kucaba TA, et al. CD8 T cells utilize TRAIL to control influenza virus infection. J Immunol 2008;181:4918–4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brown DM, Dilzer AM, Meents DL, et al. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J Immunol 2006;177:2888–2898 [DOI] [PubMed] [Google Scholar]
- 23. Buchholz VR, Schumacher TN, and Busch DH. T cell fate at the single-cell level. Annu Rev Immunol 2016;34:65–92 [DOI] [PubMed] [Google Scholar]
- 24. Burrows SR, Chen Z, Archbold JK, et al. Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability. Proc Natl Acad Sci U S A 2010;107:10608–10613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Busch DH, Pilip I, and Pamer EG. Evolution of a complex T cell receptor repertoire during primary and recall bacterial infection. J Exp Med 1998;188:61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Castellino F, Huang AY, Altan-Bonnet G, et al. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature 2006;440:890–895 [DOI] [PubMed] [Google Scholar]
- 27. Cawthon AG, Lu H, and Alexander-Miller MA. Peptide requirement for CTL activation reflects the sensitivity to CD3 engagement: correlation with CD8alphabeta versus CD8alphaalpha expression. J Immunol 2001;167:2577–2584 [DOI] [PubMed] [Google Scholar]
- 28. Chen H, Ndhlovu ZM, Liu D, et al. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nat Immunol 2012;13:691–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Clemens EB, Grant EJ, Wang Z, et al. Towards identification of immune and genetic correlates of severe influenza disease in indigenous Australians. Immunol Cell Biol 2017;95:648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cox MA, Harrington LE, and Zajac AJ. Cytokines and the inception of CD8 T cell responses. Trends Immunol 2011;32:180–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Croom HA, Denton AE, Valkenburg SA, et al. Memory precursor phenotype of CD8+ T cells reflects early antigenic experience rather than memory numbers in a model of localized acute influenza infection. Eur J Immunol 2011;41:682–693 [DOI] [PubMed] [Google Scholar]
- 32. Cukalac T, Chadderton J, Handel A, et al. Reproducible selection of high avidity CD8+ T-cell clones following secondary acute virus infection. Proc Natl Acad Sci U S A 2014;111:1485–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cukalac T, Chadderton J, Zeng W, et al. The influenza virus-specific CTL immunodominance hierarchy in mice is determined by the relative frequency of high-avidity T cells. J Immunol 2014;192:4061–4068 [DOI] [PubMed] [Google Scholar]
- 34. Curtsinger JM, and Mescher MF. Inflammatory cytokines as a third signal for T cell activation. Curr Opin Immunol 2010;22:333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Davis MM, and Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature 1988;334:395–402 [DOI] [PubMed] [Google Scholar]
- 36. Fahmy TM, Bieler JG, Edidin M, et al. Increased TCR avidity after T cell activation: a mechanism for sensing low-density antigen. Immunity 2001;14:135–143 [PubMed] [Google Scholar]
- 37. Farber DL, Yudanin NA, and Restifo NP. Human memory T cells: generation, compartmentalization and homeostasis. Nat Rev Immunol 2014;14:24–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Frahm N, Kiepiela P, Adams S, et al. Control of human immunodeficiency virus replication by cytotoxic T lymphocytes targeting subdominant epitopes. Nat Immunol 2006;7:173–178 [DOI] [PubMed] [Google Scholar]
- 39. Gallimore A, Dumrese T, Hengartner H, et al. Protective immunity does not correlate with the hierarchy of virus-specific cytotoxic T cell responses to naturally processed peptides. J Exp Med 1998;187:1647–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gaud G, Lesourne R, and Love PE. Regulatory mechanisms in T cell receptor signalling. Nat Rev Immunol 2018;18:485–497 [DOI] [PubMed] [Google Scholar]
- 41. Gebhardt T, Palendira U, Tscharke DC, et al. Tissue-resident memory T cells in tissue homeostasis, persistent infection, and cancer surveillance. Immunol Rev 2018;283:54–76 [DOI] [PubMed] [Google Scholar]
- 42. Gerlach C, Rohr JC, Perie L, et al. Heterogeneous differentiation patterns of individual CD8+ T cells. Science 2013;340:635–639 [DOI] [PubMed] [Google Scholar]
- 43. Grant EJ, Josephs TM, Loh L, et al. Broad CD8(+) T cell cross-recognition of distinct influenza A strains in humans. Nat Commun 2018;9:5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gras S, Burrows SR, Turner SJ, et al. A structural voyage toward an understanding of the MHC-I-restricted immune response: lessons learned and much to be learned. Immunol Rev 2012;250:61–81 [DOI] [PubMed] [Google Scholar]
- 45. Gras S, Chadderton J, Del Campo CM, et al. Reversed T cell receptor docking on a major histocompatibility class I complex limits involvement in the immune response. Immunity 2016;45:749–760 [DOI] [PubMed] [Google Scholar]
- 46. Gras S, Chen Z, Miles JJ, et al. Allelic polymorphism in the T cell receptor and its impact on immune responses. J Exp Med 2010;207:1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gras S, Kedzierski L, Valkenburg SA, et al. Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses. Proc Natl Acad Sci U S A 2010;107:12599–12604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hammarlund E, Lewis MW, Hansen SG, et al. Duration of antiviral immunity after smallpox vaccination. Nat Med 2003;9:1131–1137 [DOI] [PubMed] [Google Scholar]
- 49. Hancock G, Yang H, Yorke E, et al. Identification of effective subdominant anti-HIV-1 CD8+ T cells within entire post-infection and post-vaccination immune responses. PLoS Pathog 2015;11:e1004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Heemskerk MH, Hagedoorn RS, van der Hoorn MA, et al. Efficiency of T-cell receptor expression in dual-specific T cells is controlled by the intrinsic qualities of the TCR chains within the TCR-CD3 complex. Blood 2007;109:235–243 [DOI] [PubMed] [Google Scholar]
- 51. Henning AN, Roychoudhuri R, and Restifo NP. Epigenetic control of CD8(+) T cell differentiation. Nat Rev Immunol 2018;18:340–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hertz T, Oshansky CM, Roddam PL, et al. HLA targeting efficiency correlates with human T-cell response magnitude and with mortality from influenza A infection. Proc Natl Acad Sci U S A 2013;110:13492–13497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Holtappels R, Simon CO, Munks MW, et al. Subdominant CD8 T-cell epitopes account for protection against cytomegalovirus independent of immunodomination. J Virol 2008;82:5781–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Im EJ, Hong JP, Roshorm Y, et al. Protective efficacy of serially up-ranked subdominant CD8+ T cell epitopes against virus challenges. PLoS Pathog 2011;7:e1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jameson SC, and Masopust D. Understanding subset diversity in T cell memory. Immunity 2018;48:214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jenkins MR, Kedzierska K, Doherty PC, et al. Heterogeneity of effector phenotype for acute phase and memory influenza A virus-specific CTL. J Immunol 2007;179:64–70 [DOI] [PubMed] [Google Scholar]
- 57. Jenkins MR, Mintern J, La Gruta NL, et al. Cell cycle-related acquisition of cytotoxic mediators defines the progressive differentiation to effector status for virus-specific CD8+ T cells. J Immunol 2008;181:3818–3822 [DOI] [PubMed] [Google Scholar]
- 58. Jenkins MR, Trapani JA, Doherty PC, et al. Granzyme K expressing cytotoxic T lymphocytes protects against influenza virus in granzyme AB−/− mice. Viral Immunol 2008;21:341–346 [DOI] [PubMed] [Google Scholar]
- 59. Joosten SA, Sullivan LC, and Ottenhoff TH. Characteristics of HLA-E restricted T-cell responses and their role in infectious diseases. J Immunol Res 2016;2016:2695396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kaech SM, and Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol 2012;12:749–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kakaradov B, Arsenio J, Widjaja CE, et al. Early transcriptional and epigenetic regulation of CD8(+) T cell differentiation revealed by single-cell RNA sequencing. Nat Immunol 2017;18:422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kalia V, Sarkar S, Subramaniam S, et al. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity 2010;32:91–103 [DOI] [PubMed] [Google Scholar]
- 63. Kedzierska K, La Gruta NL, Davenport MP, et al. Contribution of T cell receptor affinity to overall avidity for virus-specific CD8+ T cell responses. Proc Natl Acad Sci U S A 2005;102:11432–11437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kedzierska K, Turner SJ, and Doherty PC. Conserved T cell receptor usage in primary and recall responses to an immunodominant influenza virus nucleoprotein epitope. Proc Natl Acad Sci U S A 2004;101:4942–4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kedzierska K, Valkenburg SA, Doherty PC, et al. Use it or lose it: establishment and persistence of T cell memory. Front Immunol 2012;3:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kedzierska K, Venturi V, Field K, et al. Early establishment of diverse T cell receptor profiles for influenza-specific CD8(+)CD62L(hi) memory T cells. Proc Natl Acad Sci U S A 2006;103:9184–9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Klein L, Kyewski B, Allen PM, et al. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see). Nat Rev Immunol 2014;14:377–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kosmrlj A, Read EL, Qi Y, et al. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature 2010;465:350–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Koutsakos M, Illing PT, Nguyen THO, et al. Human CD8(+) T cell cross-reactivity across influenza A, B and C viruses. Nat Immunol 2019;20:613–625 [DOI] [PubMed] [Google Scholar]
- 70. Koutsakos M, Nguyen THO, and Kedzierska K. With a little help from T Follicular helper friends: humoral immunity to influenza vaccination. J Immunol 2019;202:360–367 [DOI] [PubMed] [Google Scholar]
- 71. La Gruta NL, Gras S, Daley SR, et al. Understanding the drivers of MHC restriction of T cell receptors. Nat Rev Immunol 2018;18:467–478 [DOI] [PubMed] [Google Scholar]
- 72. La Gruta NL, Rothwell WT, Cukalac T, et al. Primary CTL response magnitude in mice is determined by the extent of naive T cell recruitment and subsequent clonal expansion. J Clin Invest 2010;120:1885–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. La Gruta NL, Turner SJ, and Doherty PC. Hierarchies in cytokine expression profiles for acute and resolving influenza virus-specific CD8+ T cell responses: correlation of cytokine profile and TCR avidity. J Immunol 2004;172:5553–5560 [DOI] [PubMed] [Google Scholar]
- 74. Ladell K, Hashimoto M, Iglesias MC, et al. A molecular basis for the control of preimmune escape variants by HIV-specific CD8+ T cells. Immunity 2013;38:425–436 [DOI] [PubMed] [Google Scholar]
- 75. Laidlaw BJ, Craft JE, and Kaech SM. The multifaceted role of CD4(+) T cells in CD8(+) T cell memory. Nat Rev Immunol 2016;16:102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Laidlaw BJ, Zhang N, Marshall HD, et al. CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity 2014;41:633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Malherbe L, Hausl C, Teyton L, et al. Clonal selection of helper T cells is determined by an affinity threshold with no further skewing of TCR binding properties. Immunity 2004;21:669–679 [DOI] [PubMed] [Google Scholar]
- 78. Maryanski JL, Jongeneel CV, Bucher P, et al. Single-cell PCR analysis of TCR repertoires selected by antigen in vivo: a high magnitude CD8 response is comprised of very few clones. Immunity 1996;4:47–55 [DOI] [PubMed] [Google Scholar]
- 79. McDonald BD, Bunker JJ, Erickson SA, et al. Crossreactive alphabeta T cell receptors are the predominant targets of thymocyte negative selection. Immunity 2015;43:859–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. McHeyzer-Williams LJ, Panus JF, Mikszta JA, et al. Evolution of antigen-specific T cell receptors in vivo: preimmune and antigen-driven selection of preferred complementarity-determining region 3 (CDR3) motifs. J Exp Med 1999;189:1823–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Merkenschlager M, Graf D, Lovatt M, et al. How many thymocytes audition for selection? J Exp Med 1997;186:1149–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Messaoudi I, Guevara Patino JA, Dyall R, et al. Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science 2002;298:1797–1800 [DOI] [PubMed] [Google Scholar]
- 83. Miles JJ, Douek DC, and Price DA. Bias in the alphabeta T-cell repertoire: implications for disease pathogenesis and vaccination. Immunol Cell Biol 2011;89:375–387 [DOI] [PubMed] [Google Scholar]
- 84. Miles JJ, Thammanichanond D, Moneer S, et al. Antigen-driven patterns of TCR bias are shared across diverse outcomes of human hepatitis C virus infection. J Immunol 2011;186:901–912 [DOI] [PubMed] [Google Scholar]
- 85. Moffat JM, Handel A, Doherty PC, et al. Influenza epitope-specific CD8+ T cell avidity, but not cytokine polyfunctionality, can be determined by TCRbeta clonotype. J Immunol 2010;185:6850–6856 [DOI] [PubMed] [Google Scholar]
- 86. Moon JJ, Chu HH, Pepper M, et al. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 2007;27:203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Nguyen TH, Rowntree LC, Pellicci DG, et al. Recognition of distinct cross-reactive virus-specific CD8+ T cells reveals a unique TCR signature in a clinical setting. J Immunol 2014;192:5039–5049 [DOI] [PubMed] [Google Scholar]
- 88. Nguyen TH, Tan AC, Xiang SD, et al. Understanding CD8(+) T-cell responses toward the native and alternate HLA-A*02:01-restricted WT1 epitope. Clin Transl Immunol 2017;6:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Nguyen THO, Sant S, Bird NL, et al. Perturbed CD8(+) T cell immunity across universal influenza epitopes in the elderly. J Leukoc Biol 2018;103:321–339 [DOI] [PubMed] [Google Scholar]
- 90. Nikolich-Zugich J, Slifka MK, and Messaoudi I. The many important facets of T-cell repertoire diversity. Nat Rev Immunol 2004;4:123–132 [DOI] [PubMed] [Google Scholar]
- 91. Obar JJ, and Lefrancois L. Early signals during CD8(+) T cell priming regulate the generation of central memory cells. J Immunol 2010;185:263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Pipkin ME, Sacks JA, Cruz-Guilloty F, et al. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity 2010;32:79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Pizzolla A, Nguyen TH, Sant S, et al. Influenza-specific lung-resident memory T cells are proliferative and polyfunctional and maintain diverse TCR profiles. J Clin Invest 2018;128:721–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Precopio ML, Betts MR, Parrino J, et al. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J Exp Med 2007;204:1405–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Price DA, Asher TE, Wilson NA, et al. Public clonotype usage identifies protective Gag-specific CD8+ T cell responses in SIV infection. J Exp Med 2009;206:923–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Price DA, Brenchley JM, Ruff LE, et al. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med 2005;202:1349–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Price DA, West SM, Betts MR, et al. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity 2004;21:793–803 [DOI] [PubMed] [Google Scholar]
- 98. Quinones-Parra S, Grant E, Loh L, et al. Preexisting CD8+ T-cell immunity to the H7N9 influenza A virus varies across ethnicities. Proc Natl Acad Sci U S A 2014;111:1049–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Raeber ME, Zurbuchen Y, Impellizzieri D, et al. The role of cytokines in T-cell memory in health and disease. Immunol Rev 2018;283:176–193 [DOI] [PubMed] [Google Scholar]
- 100. Remakus S, Rubio D, Ma X, et al. Memory CD8+ T cells specific for a single immunodominant or subdominant determinant induced by peptide-dendritic cell immunization protect from an acute lethal viral disease. J Virol 2012;86:9748–9759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rock KL, Reits E, and Neefjes J. Present yourself! By MHC class I and MHC class II molecules. Trends Immunol 2016;37:724–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Rossjohn J, Gras S, Miles JJ, et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol 2015;33:169–200 [DOI] [PubMed] [Google Scholar]
- 103. Ruckwardt TJ, Luongo C, Malloy AM, et al. Responses against a subdominant CD8+ T cell epitope protect against immunopathology caused by a dominant epitope. J Immunol 2010;185:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sacre K, Carcelain G, Cassoux N, et al. Repertoire, diversity, and differentiation of specific CD8 T cells are associated with immune protection against human cytomegalovirus disease. J Exp Med 2005;201:1999–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sallusto F. Heterogeneity of human CD4(+) T cells against microbes. Annu Rev Immunol 2016;34:317–334 [DOI] [PubMed] [Google Scholar]
- 106. Sallusto F, Lenig D, Forster R, et al. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999;401:708–712 [DOI] [PubMed] [Google Scholar]
- 107. Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev 2001;14:778–809, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sant AJ, and McMichael A. Revealing the role of CD4(+) T cells in viral immunity. J Exp Med 2012;209:1391–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sant S, Grzelak L, Wang Z, et al. Single-cell approach to influenza-specific CD8(+) T cell receptor repertoires across different age groups, tissues, and following influenza virus infection. Front Immunol 2018;9:1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Schroder K, Hertzog PJ, Ravasi T, et al. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol 2004;75:163–189 [DOI] [PubMed] [Google Scholar]
- 111. Scott-Browne JP, White J, Kappler JW, et al. Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature 2009;458:1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Seder RA, Darrah PA, and Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol 2008;8:247–258 [DOI] [PubMed] [Google Scholar]
- 113. Sidney J, Peters B, Frahm N, et al. HLA class I supertypes: a revised and updated classification. BMC Immunol 2008;9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Slifka MK, and Whitton JL. Functional avidity maturation of CD8(+) T cells without selection of higher affinity TCR. Nat Immunol 2001;2:711–717 [DOI] [PubMed] [Google Scholar]
- 115. Smith C, Gras S, Brennan RM, et al. Molecular imprint of exposure to naturally occurring genetic variants of human cytomegalovirus on the T cell repertoire. Sci Rep 2014;4:3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Sokol CL, and Luster AD. The chemokine system in innate immunity. Cold Spring Harb Perspect Biol 2015;7:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Sourdive DJ, Murali-Krishna K, Altman JD, et al. Conserved T cell receptor repertoire in primary and memory CD8 T cell responses to an acute viral infection. J Exp Med 1998;188:71–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Sridhar S, Begom S, Bermingham A, et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med 2013;19:1305–1312 [DOI] [PubMed] [Google Scholar]
- 119. Stewart-Jones GB, McMichael AJ, Bell JI, et al. A structural basis for immunodominant human T cell receptor recognition. Nat Immunol 2003;4:657–663 [DOI] [PubMed] [Google Scholar]
- 120. Swain SL, McKinstry KK, and Strutt TM. Expanding roles for CD4(+) T cells in immunity to viruses. Nat Rev Immunol 2012;12:136–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Takeshita LY, Jones AR, Gonzalez-Galarza FF, et al. Allele frequencies database. Transfus Med Hemother 2014;41:352–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Thomas PG, Handel A, Doherty PC, et al. Ecological analysis of antigen-specific CTL repertoires defines the relationship between naive and immune T-cell populations. Proc Natl Acad Sci U S A 2013;110:1839–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Topham DJ, Tripp RA, and Doherty PC. CD8+ T cells clear influenza virus by perforin or Fas-dependent processes. J Immunol 1997;159:5197–5200 [PubMed] [Google Scholar]
- 124. Tscharke DC, Croft NP, Doherty PC, et al. Sizing up the key determinants of the CD8(+) T cell response. Nat Rev Immunol 2015;15:705–716 [DOI] [PubMed] [Google Scholar]
- 125. Turner SJ, Diaz G, Cross R, et al. Analysis of clonotype distribution and persistence for an influenza virus-specific CD8+ T cell response. Immunity 2003;18:549–559 [DOI] [PubMed] [Google Scholar]
- 126. Tynan FE, Elhassen D, Purcell AW, et al. The immunogenicity of a viral cytotoxic T cell epitope is controlled by its MHC-bound conformation. J Exp Med 2005;202:1249–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Valkenburg SA, Gras S, Guillonneau C, et al. Protective efficacy of cross-reactive CD8+ T cells recognising mutant viral epitopes depends on peptide-MHC-I structural interactions and T cell activation threshold. PLoS Pathog 2010;6:e1001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Valkenburg SA, Josephs TM, Clemens EB, et al. Molecular basis for universal HLA-A*0201-restricted CD8+ T-cell immunity against influenza viruses. Proc Natl Acad Sci U S A 2016;113:4440–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Valkenburg SA, Quiñones-Parra S, Gras S, et al. Acute emergence and reversion of influenza A virus quasispecies within CD8+ T cell antigenic peptides. Nat Commun 2013;4:2663. [DOI] [PubMed] [Google Scholar]
- 130. Valkenburg SA, Venturi V, Dang TH, et al. Early priming minimizes the age-related immune compromise of CD8(+) T cell diversity and function. PLoS Pathog 2012;8:e1002544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Van Braeckel-Budimir N, Gras S, Ladell K, et al. A T cell receptor locus harbors a malaria-specific immune response gene. Immunity 2017;47:835–847 e834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. van de Sandt CE, Grant EJ, Rowntree LC, et al. Challenging immunodominance of influenza-specific CD8+ T cell responses directed against the risk-associated HLA*68:01 allomorph. Nat Commun 2019;10:5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. van de Sandt CE, Hillaire ML, Geelhoed-Mieras MM, et al. Human influenza A virus-specific CD8+ T-cell response is long-lived. J Infect Dis 2015;212:81–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. van Heijst JW, Gerlach C, Swart E, et al. Recruitment of antigen-specific CD8+ T cells in response to infection is markedly efficient. Science 2009;325:1265–1269 [DOI] [PubMed] [Google Scholar]
- 135. Venturi V, Chin HY, Asher TE, et al. TCR beta-chain sharing in human CD8+ T cell responses to cytomegalovirus and EBV. J Immunol 2008;181:7853–7862 [DOI] [PubMed] [Google Scholar]
- 136. Venturi V, Chin HY, Price DA, et al. The role of production frequency in the sharing of simian immunodeficiency virus-specific CD8+ TCRs between macaques. J Immunol 2008;181:2597–2609 [DOI] [PubMed] [Google Scholar]
- 137. Venturi V, Kedzierska K, Price DA, et al. Sharing of T cell receptors in antigen-specific responses is driven by convergent recombination. Proc Natl Acad Sci U S A 2006;103:18691–18696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Voskoboinik I, Whisstock JC, and Trapani JA. Perforin and granzymes: function, dysfunction and human pathology. Nat Rev Immunol 2015;15:388–400 [DOI] [PubMed] [Google Scholar]
- 139. Wang GC, Dash P, McCullers JA, et al. T cell receptor alphabeta diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection. Sci Transl Med 2012;4:128ra142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Wang Z, Wan Y, Qiu C, et al. Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8(+) T cells. Nat Commun 2015;6:6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Wang Z, Zhu L, Nguyen THO, et al. Clonally diverse CD38(+)HLA-DR(+)CD8(+) T cells persist during fatal H7N9 disease. Nat Commun 2018;9:824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Ward-Kavanagh LK, Lin WW, Sedy JR, et al. The TNF receptor superfamily in co-stimulating and co-inhibitory responses. Immunity 2016;44:1005–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Wiesel M, Joller N, Ehlert AK, et al. Th cells act via two synergistic pathways to promote antiviral CD8+ T cell responses. J Immunol 2010;185:5188–5197 [DOI] [PubMed] [Google Scholar]
- 144. Wilkinson TM, Li CK, Chui CS, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med 2012;18:274–280 [DOI] [PubMed] [Google Scholar]
- 145. Wilson EB, and Livingstone AM. Cutting edge: CD4+ T cell-derived IL-2 is essential for help-dependent primary CD8+ T cell responses. J Immunol 2008;181:7445–7448 [DOI] [PubMed] [Google Scholar]
- 146. Zerrahn J, Held W, and Raulet DH. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell 1997;88:627–636 [DOI] [PubMed] [Google Scholar]
- 147. Zinkernagel RM, and Doherty PC. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature 1974;248:701–702 [DOI] [PubMed] [Google Scholar]