Highlights
-
•
Stress granules (SGs) are induced upon infection with some DNA and RNA viruses.
-
•
Many viruses block the induction of SG, although some appear to benefit from SG formation.
-
•
Some innate immune sensors associate with SG, but how this affects function is unclear.
-
•
The evidence suggests that SG formation is an important aspect of the antiviral response.
Keywords: stress granules, virus infection, interferon, shut-off, innate immunity, dsRNA
Abstract
Viral infection triggers the activation of antiviral innate immune responses in mammalian cells. Viral RNA in the cytoplasm activates signaling pathways that result in the production of interferons (IFNs) and IFN-stimulated genes. Some viral infections have been shown to induce cytoplasmic granular aggregates similar to the dynamic ribonucleoprotein aggregates termed stress granules (SGs), suggesting that these viruses may utilize this stress response for their own benefit. By contrast, some viruses actively inhibit SG formation, suggesting an antiviral function for these structures. We review here the relationship between different viral infections and SG formation. We examine the evidence for antiviral functions for SGs and highlight important areas of inquiry towards understanding cellular stress responses to viral infection.
Viral infection and stress granules
Viral invasion and replication are detected by innate immune sensors in cells, triggering downstream signaling pathways that can ultimately result in the activation of systemic immune responses. Several innate immune sensors recognize cytoplasmic viral RNA [1], and lead to the production of IFNs which in turn trigger various antiviral pathways aimed at halting viral replication and spread. These antiviral effects include double-stranded (ds) RNA-dependent protein kinase (PKR)-dependent inhibition of mRNA translation, and 2′,5′-oligoadenylate synthetase (OAS)/RNase L-mediated RNA degradation [2]. Innate immune responses also trigger the activation of adaptive immunity in the form of T and B cell activation and proliferation, and modulate the phenotype and function of these adaptive responses 3, 4.
In some cases, viral infection also induces the formation of cytoplasmic granules similar to those induced by cellular stresses such as heat, oxidation, hypoxia, and osmotic pressure, which are referred to as stress granules (SGs). SGs are ribonucleoprotein (RNP) aggregates that contain translationally stalled mRNAs, 40S ribosomes, and various RNA-binding proteins 5, 6, 7 (Box 1 ).
Box 1. An overview of SG biology.
Cells respond to various insults including heat, oxidative stress, nutrient starvation, and proteotoxic stress by forming cytoplasmic nucleoprotein aggregates termed SGs 5, 68, 69, 70. Multiple RNA-binding proteins (RBPs) localize to SGs, and some have been used as markers for these cytoplasmic bodies (Table I ). Although the formation of SGs in live cells can be detected by monitoring fluorescence-tagged SG marker proteins, biochemical isolation of SGs is notoriously challenging because these are not membrane-sequestered compartments.
Table I.
Protein components of SGs and P-bodies.
| SG components | |||
|---|---|---|---|
| Factor | Full name | Functions | Refs |
| ADAR |
Adenosine deaminase, RNA-specific |
RNA editing, RNA stability |
[77] |
| Caprin-1 |
Cell cycle associated protein 1 |
Cell growth, SG assembly |
44, 78 |
| phospho-eIF2α |
Eukaryotic translation initiation factor 2A |
Initiation factor, SG assembly |
60, 79 |
| eIF3 |
Eukaryotic translation initiation factor 3 |
Multisubunit initiation factor |
[5] |
| eIF4G |
Eukaryotic translation initiation factor 4G |
Initiation factor |
[5] |
| G3BP1 |
Ras-GTPase-activating protein SH3-domain-binding protein 1 |
Endoribonuclease, ras signaling, SG assembly |
[75] |
| HDAC6 |
Histone deacetylase 6 |
Translation regulator, SG assembly |
[80] |
| HuR/ELAVL1 |
Hu antigen R/ELAV-like RNA-binding protein 1 |
mRNA stability, translation regulator |
5, 81 |
| OGFOD1 |
2-Oxoglutarate and iron-dependent oxygenase domain containing 1 |
Translation regulator, SG assembly |
[82] |
| PABP1 |
PolyA-binding protein 1 |
mRNA stability, translation regulator |
[74] |
| Pum1 |
Pumilio RNA-binding family member 1 |
Translation regulator, cell growth |
[83] |
| Pum2 |
Pumilio RNA-binding family member 2 |
Translation regulator, SG assembly |
[84] |
| RHAU/DHX36 |
RNA helicase associated with AU-rich element/DEAH box polypeptide 36 |
RNA helicase, SG assembly, antiviral activity |
49, 85 |
| SMN |
Survival of motor neuron |
RNA metabolism, SG assembly |
86, 87 |
| STAU1 |
Staufen dsRNA-binding protein 1 |
RNA transport, SG assembly |
[88] |
| TIA1 |
T cell restricted intracellular antigen-1 |
Translation regulator, SG assembly |
74, 76 |
| TIAR |
TIA-1-related protein |
Translation regulator |
[74] |
| ZBP1 |
Z-DNA-binding protein 1 |
DNA sensor, translational regulator |
11, 89 |
| 40S | Eukaryotic small ribosomal subunit | Ribosome | [5] |
| P-body components | |||
|---|---|---|---|
| CNOT6/CCR4 |
CCR4–NOT transcription complex, subunit 6 |
mRNA deadenylation, PB assembly |
90, 91 |
| DCP1a |
Decapping mRNA 1A |
mRNA decapping |
60, 92 |
| DCP2a |
Decapping mRNA 2A |
mRNA decapping |
60, 92 |
| EDC4/GE-1/Hedls |
Enhancer of mRNA decapping 4 |
Decapping coactivator, PB assembly |
93, 94 |
| TNRC6A/GW182 |
Trinucleotide repeat-containing 6A/GW bodies 182 |
RNA silencing, PB assembly |
95, 96 |
| Lsm1 | Lsm1, U6 small nuclear RNA associated | Decapping coactivator, PB assembly | 90, 92, 97 |
| SG and PB components | |||
|---|---|---|---|
| APOBEC3G |
Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G |
Antiviral activity |
98, 99 |
| Ago2 |
Argonaute RISC catalytic component 2 |
RNA silencing, PB assembly |
100, 101 |
| BRF1 |
Butyrate response factor 1 |
ARE-mediated mRNA decay |
[60] |
| CPEB |
Cytoplasmic polyadenylation element binding protein |
Polyadenylation, translation regulator |
[102] |
| DDX3 |
DEAD box helicase 3 |
RNA helicase, antiviral activity |
103, 104 |
| DDX6/RCK |
DEAD box helicase 6 |
RNA helicase, antiviral activity, PB assembly |
90, 102, 105 |
| FAST |
Fas-activated Ser/Thr kinase |
Splicing regulator |
[60] |
| RAP55/LSM14A |
RNA-associated protein 55 |
PB assembly, antiviral activity |
11, 106 |
| TTP |
Tristetraprolin |
ARE-mediated mRNA decay |
[60] |
| Xrn1 | 5′-3′ Exoribonuclease 1 | 5′–3′ exonuclease | 60, 92 |
SG formation has been interpreted as a response aimed at preventing the generation of abnormal proteins by transient stalling of translation in times of cellular stress. Stalled transcripts undergo translation upon recovery from stress, or alternatively they are degraded in another granular compartment termed the processing body (P-body, PB) 71, 72. Unlike SGs, PBs are present in the unstressed cell, and contain enzymes for mRNA degradation such as decapping enzymes and 5′–3′ exonucleases (Table I). It is thought that transcripts and proteins can move from SGs to PBs (and vice versa), and these aggregates share some of their components (Table I) [60]; however, the mechanisms underlying the proposed exchange of contents are unclear.
SG proteins, as defined by studies using proteins characteristic to SGs as markers [73], are either diffusely distributed in the cytoplasm or localized in the nucleus in normal conditions; stress triggers their aggregation in the cytoplasm 5, 74. A common event downstream from the aforementioned cellular stresses is phosphorylation of eIF2α at serine 51, which is considered to be an initial trigger for SG formation. Four eIF2α kinases, PKR, GCN2, PKR-like endoplasmic reticulum kinase (PERK), and heme-regulated eIF2α kinase (HRI), can phosphorylate eIF2α in mammals (Table II ). The mechanisms that connect eIF2α phosphorylation to SG formation remain to be elucidated.
Table II.
Kinases that target eIF2α.
| Kinase | Full name | Stress | Refs |
|---|---|---|---|
| PKR | dsRNA-dependent protein kinase | dsRNA, viral RNA, viral infection | 107, 108 |
| PERK | PKR-like ER kinase | ER stress, hypoxia | 109, 110, 111 |
| GCN2 | General control non-derepressible 2 | Nutrient deprivation, amino acid deprivation, viral infection | 28, 112 |
| HRI | Heme-regulated eIF2α kinase | Heat shock, oxidative stress, osmotic stress | 113, 114 |
Some proteins have been shown to be crucial for the formation and or stability of SGs. These include G3BP1, a phosphorylation-dependent endonuclease 30, 75, and T cell restricted intracellular antigen-1 (TIA1) and TIA-related protein (TIAR) that are collectively termed TIA1/TIAR 74, 76. Removal of these regulators by genomic deletion or RNAi blocks SG formation by sodium arsenite, and a mutant form of G3BP1 (S149E) acted as a dominant inhibitor of SG formation [75]. However, because of analytical constraints, the molecular machinery underlying the formation of SGs remains unclear.
Activation of the RLR signaling pathways by viral RNA
Innate immune responses are triggered upon recognition of pathogen-associated molecular patterns (PAMPs) which in the case of viruses are often nucleic acid-based, either RNA or DNA. Viral nucleic acid can be detected by sensors including Toll-like receptors (TLR)3, 7/8, and 9, retinoic acid inducible gene I (RIG-I)-like receptors (RLRs), and cytoplasmic DNA sensors such as DNA-dependent activator of IFN-regulator factors (DAI), stimulator of IFN genes (STING, also known as MITA/ERIS/MPYS), DEAD box polypeptide 41 (DDX41), and cGMP/cAMP synthase (cGAS) 8, 9, 10, 11, 12. RLRs, RIG-I, melanoma differentiation-associated protein 5 (MDA5), and DHX58 [DEXH (Asp-Glu-X-His) box polypeptide 58, also known as LGP2] are all RNA helicases and contain the signature DExD/H motif that characterized the DExD/H box family. These proteins are crucial for the detection of cytoplasmic RNA 13, 14. RLRs discriminate self from viral transcripts by recognizing specific biochemical signatures such as ds structure and the presence of a 5′-ppp moiety 15, 16, 17; self-transcipts lack these viral signatures.
The signaling pathways downstream of the founding member of the RLR family, RIG-I, are among the best understood. Upon viral RNA recognition by RIG-I, the signal is relayed to the adaptor protein IFN-β promoter stimulator 1 (IPS-1, also known as MAVS, VISA, or Cardif), which predominantly localizes to the mitochondrial outer membrane 18, 19. When viral RNA binds at the helicase and the C-terminal domain (CTD) of RIG-I, its N-terminal caspase recruitment domains (CARDs) are covalently modified with K63-linked polyubiquitin chains by the E3 ligase, tripartite motif-containing protein 25 (TRIM25) [20], and oligomerize [21]. The ubiquitinated and oligomerized CARDs bind to the CARD domain on IPS-1 on mitochondria, peroxisomes, and/or mitochondrion-associated membrane (MAM) regions in the endoplasmic reticulum (ER) [22]. The translocation of RIG-I to these locales is facilitated by the chaperone protein 14-3-3ɛ [23]. The requirement for K63-linked polyubiquitin chains is complex because several reports have demonstrated the importance of non-covalent interaction between RIG-I CARDs and the unanchored K63–ubiquitin chains 21, 24. Furthermore, it has been reported that RIG-I forms a signaling-competent filament on substrate dsRNA independently of ubiquitins [25]. Although a recent structural report suggests that the conformation of the active tetramer of RIG-I CARDs can be stabilized by covalent conjugation with ubiquitin chains, and that filament formation may partially compensate for ubiquitin-dependent RIG-I activation [26], further analysis will be necessary to clarify the molecular role of ubiquitin chains in RIG-I signaling. The RIG-I/IPS-1 interaction recruits signaling molecules including tumor necrosis factor (TNF) receptor-associated factors (TRAFs). Subsequent activation of TANK-binding kinase 1(TBK1)/inducible IκB kinase (IKKi), and IKKα/IKKβ induces downstream signaling via the IFN regulatory factor (IRF) and nuclear factor-κB (NF-κB) pathways, respectively. These pathways ultimately culminate in the activation of the transcription factors IRF-3, IRF-7, and NF-κB, which activate the transcription of IFN and proinflammatory cytokine genes [27]. These pathways are summarized in Figure 1 . Moreover, secreted IFN amplifies the expression of ISGs, such as RLRs, PKR, and OAS, as a host strategy to amplify antiviral signaling.
Figure 1.
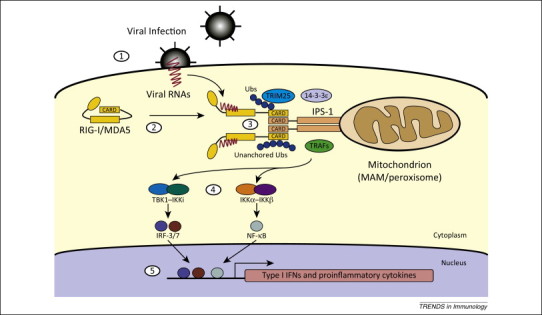
Detection of viral RNA by RLR [retinoic acid inducible gene I (RIG-I)-like receptor]. (1) Viral internalization and release of the RNA genome in the cytoplasm. (2) Autorepressed RIG-I or MDA5 bind to viral RNA and undergo a conformational change to expose CARD and associate with IPS-1 via CARD–CARD interactions. (3) Signaling proteins are recruited around the RLR/IPS-1 complex and (4) activate the TBK1/IKKi–IRF-3/7 and IKKα/IKKβ–NF-κB pathways, resulting in (5) the activation of type I IFNs and proinflammatory cytokines.
Induction of SGs by viral infection
Viruses, particularly RNA viruses, have been shown to induce the formation of SG-like cytoplasmic bodies (Table 1 and references therein). In some cases these bodies have been given different names in an attempt to distinguish them from SGs; in this review, however, we refer to virus-induced SG-like granules collectively as SGs. Many viruses induce SGs through the activation of the eukaryotic translation initiation factor (eIF)2α kinases PKR and, in some cases, general control non-depressible 2 (GCN2), which are both triggered by detection of RNA in the cytoplasm [28] (Figure 2 ). Depending on both the virus and the host cell, different patterns of SG formation have been observed upon infection: stable SG formation, no SG formation, transient SG formation, or alternating (oscillating) SG formation in which SGs form, disperse, and reform during the assays (Table 1).
Table 1.
Viral infection and SG formation.
| Family | Species | Genome | SG formation | Mechanism of inhibition or activation | Refs |
|---|---|---|---|---|---|
| Picornaviridae | Poliovirus | ssRNA (+ sense) | Yes (transient) | G3BP1 cleavage by 3C protease | 30, 67 |
| EMCV | ssRNA (+ sense) | Yes (transient) | G3BP1 cleavage by 3C protease | [29] | |
| Mengovirus | ssRNA (+ sense) | No | Inhibition by leader protein | [37] | |
| TMEV | ssRNA (+ sense) | No | Inhibition by leader protein | [37] | |
| Togaviridae | Sindbis virus | ssRNA (+ sense) | Yes (transient) | PKR-and GCN2-dependent. tRNA-like motifs in the genome are responsible for GCN2 activation | 28, 29 |
| SFV | ssRNA (+ sense) | Yes (transient) | [33] | ||
| Rubella virus | ssRNA (+ sense) | No | [115] | ||
| Flaviviridae | West Nile virus | ssRNA (+ sense) | No | Recruitment of TIA/TIAR to replication complexes | 48, 116 |
| Dengue virus | ssRNA (+ sense) | No | Recruitment of TIA/TIAR to replication complexes | [48] | |
| JEV | ssRNA (+ sense) | No | Core protein interacts with caprin 1 | [44] | |
| HCV | ssRNA (+ sense) | Yes | 5′-UTR of HCV genome activates PKR | 34, 35, 36, 117, 118 | |
| Nidoviridae | Coronavirus | ssRNA (+ sense) | Yes | Host polypyrimidine tract-binding protein is essential | [119] |
| Mouse hepatitis virus | ssRNA (+ sense) | Yes | [120] | ||
| Rhabdoviridae | VSV | ssRNA (− sense) | Yes or no? (strain-dependent?) | 29, 121 | |
| Paramyxoviridae | Sendai virus | ssRNA (− sense) | No | Inhibition of PKR activation by C and V proteins Viral trailer RNA also inhibits SG through interacting with TIAR |
45, 46 |
| Measles virus | ssRNA (− sense) | No | C protein activates ADAR, resulting in PKR inhibition | [39] | |
| RSV | ssRNA (− sense) | Yes | PKR-dependent. Trailer RNA inhibits SG formation | 122, 123, 124 | |
| Orthomyxoviridae | Influenza virus | ssRNA (− sense, segmented) | No | NS1 blocks PKR activation | 41, 42, 43 |
| Arenaviridae | Junin virus | ss Ambisense RNA | No | Nucleoprotein and glycoprotein precursor are implicated to inhibit eIF2α phosphorylation | [125] |
| Reoviridae | Reovirus | dsRNA (segmented) | Yes (Transient) | Involvement of ATF accumulation in SG disassembly is suggested | 32, 126, 127 |
| Rotavirus | dsRNA (segmented) | No | Inhibition by NSP3? | [47] | |
| Adenoviridae | Adenovirus | Linear dsDNA | No | Inhibition by E1A | [29] |
| Poxviridae | Vaccinia virus | dsDNA | No | Inhibition by E3L | 128, 129 |
| ? | Cricket paralysis virus | ssRNA (+ sense) | No | Viral infection inhibits SG by unknown mechanisms | [130] |
Figure 2.
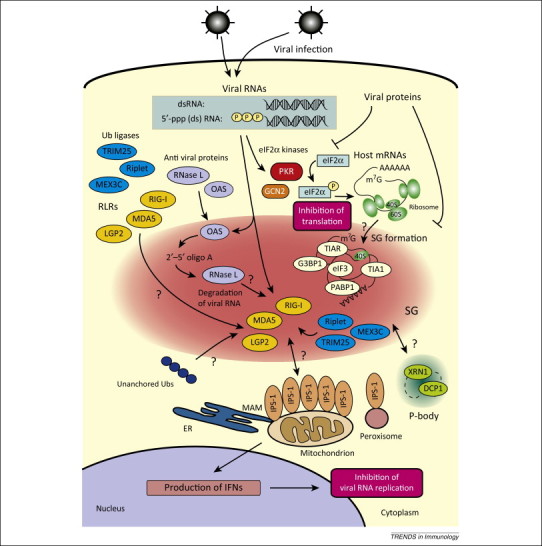
A model for antiviral function of stress granules (SG). In virus-infected cells, viral RNAs activate PKR (or GCN2, in the case of Sindbis virus) to initiate assembly of SG through eIF2α phosphorylation. eIF2α phosphorylation blocks translation of cellular mRNAs. Translation-stalled mRNAs may be transferred to a distinct cellular compartment, the P-body, to be degraded. Viral RNAs are also recognized by RLRs, which are recruited to SGs with several signaling molecules including antiviral proteins and ubiquitin ligases. The OAS–RNase L pathway cleaves viral RNAs, and the cleaved RNAs may act as ligands for RLRs. IPS-1, which is localized on mitochondria and/or MAM, forms prion-like aggregates, interacts with RLRs on SGs, and activates IFN-inducing signaling. Areas that require further investigation are highlighted with question marks.
Transient SG formation results from dissociation of key components in SGs by viral proteins 29, 30, 31, 32, 33. For instance, in the case of infection of several picornaviruses, such as poliovirus, coxsackievirus and encephalomyocarditis virus (EMCV), transient formation of SGs is associated with the cleavage of Ras-GAP SH3 domain binding protein-1 (G3BP1) by the viral 3C protease 29, 30, 31. This was confirmed by the observation that ectopic expression of cleavage-resistant G3BP1 leads to stable SG formation. On the other hand, recent studies have demonstrated that infection with hepatitis C virus (HCV) produces oscillating SGs [34]. HCV strongly activates PKR via the 5′-untranslated region (UTR) of its genome [35], thereby inducing SGs 34, 36, but stress-inducible expression of growth arrest DNA-damage-inducible 34 (GADD34), a regulatory component of host protein phosphatase 1 (PP1), leads to dephosphorylation of eIF2α and terminates SG formation. In the stress-recovered condition, GADD34 protein is rapidly downregulated by an unknown mechanism and the phosphorylated form of eIF2α reaccumulates in the cells, resulting in an oscillating pattern of SGs.
Inhibition of SG formation by viruses: an antiviral role for SGs?
In cases where viral infection appears to not induce SGs, accumulating evidence suggest that these viruses inhibit SG formation. Cells infected with mengovirus or Theiler's murine encephalomyelitis virus (TMEV), which belong to the Cardiovirus genus within the family of Picornaviridae, do not exhibit SGs, and this has been proposed to be due to complete inhibition of SG formation by the viral nonstructural protein, leader (L) protein [37]. Although L protein is known to block IFN production via inhibition of IRF-3 activation [38], it remains unknown how L limits SG formation.
Similarly, SGs do not form upon infection of cells with measles virus. In this case the mechanism proposed involves the viral C protein because C-deficient virus, but not the wild type, strikingly induces SGs [39]. Although measles C protein has diverse functions during infection, including modulation of viral polymerase activity and inhibition of IFN production [40], the molecular machinery for SG inhibition remains to be determined.
In the case of influenza A virus (IAV), the viral nonstructural protein 1(NS1) has been shown to inhibit eIF2α phosphorylation by blocking PKR activation via viral RNA sequestration and physical interaction 41, 42, 43. During IAV infection, the viral nucleocapsid initially accumulates in the nucleus and is then transported to the cytoplasm in the late phase of infection. However, in a mutant IAV lacking NS1, the viral nucleocapsid was reported to coincide with SGs in the cytoplasm, to which RIG-I is colocalized [42]. These findings suggest that SGs function as a platform for the detection of IAV genomic RNA by RIG-I.
As in the cases of mengovirus, TMEV, and measles virus, there appears to be a clear benefit, from the standpoint of the virus, to interfering with SG formation. An implication of these findings is that SGs have an antiviral role and that, accordingly, viruses have developed strategies to suppress their formation (Figure 2). Indeed, there are multiple examples of viruses actively inhibiting the formation of SGs through varied mechanisms 44, 45, 46, 47, 48. An inverse correlation between SG formation and viral propagation has been reported in multiple viral replication systems. In the case of Japanese encephalitis virus (JEV) infection, SG formation is inhibited by the viral core protein, which directly interacts with SG component, caprin 1 [44]. In cells infected with a mutant virus whose core protein fails to interact with caprin 1, inhibition of SG formation is abrogated and viral propagation is significantly impaired both in vitro and in vivo, suggesting that SGs impact negatively on viral replication. This notion is further supported by studies showing that RLRs localize to virus-induced SGs, suggesting that SGs may act as a platform for viral RNA sensing and the activation of downstream signaling pathways [42]. There is a strong correlation between PKR-dependent SG formation and IFN production in some viral infections 29, 42, 49 (Figure 2). However, a recent study has demonstrated that PKR-dependent accumulation of MDA5 in SGs is dispensable for triggering IFN responses [50]. Further investigations will be necessary to address this discrepancy.
Whether PKR is required for virus-induced IFN production is controversial. Some reports indicate that IFN production is significantly impaired in PKR-deficient cells 29, 42, 49, 51, 52, 53, 54, 55, whereas others show that deficiency of PKR has no effect 54, 56, 57, 58. One possible explanation for these observations is that there are differences in the viruses and cell types used in each study. In the early 2000s, several studies demonstrated that virus-induced activation of IFN is independent of PKR in plasmacytoid dendritic cells (pDCs), which are known to be ‘IFN-producing cells’. However, subsequent reports revealed that robust IFN induction by pDCs is exclusively induced by TLRs, suggesting a dispensable role for PKR in TLR-mediated signaling. By contrast, the PKR-dependency of RLR-mediated signaling is more complicated. Because viruses are extraordinary diverse in their genome structures and life cycles, different viruses are likely to produce different RNA species during viral replication at distinct locations in the infected cells. Thus, the ability of these viral RNAs to activate PKR and/or RLRs could be divergent. Moreover, as mentioned above, viruses employ a variety of strategies to terminate antiviral responses. For instance, it has been demonstrated that Sendai virus (SeV)-induced IFN production is independent of PKR 53, 54. Indeed, infection of SeV can activate IFN without SG formation [29]; however, infection by a mutant virus in which accessory protein C is deleted leads to significant activation of PKR and eIF2α phosphorylation, with concomitant upregulation of IFN 45, 59, suggesting that PKR is dispensable for SeV-induced IFN activation, but is responsible for the enhancement of IFN production. Thus, the PKR-dependency of IFN production might vary depending on the ability of each virus to modulate host responses. This notion is supported by reports in which IFN production following stimulation with a virus-mimetic synthetic dsRNA such as poly(I:C) showed significant PKR-dependency 29, 42, 49, 53, 55.
Understanding the relationship between mechanisms of viral detection and SGs
During viral infection, viral RNA, either incoming or produced as a replication intermediate, triggers a series of events. DsRNA activates PKR (or GCN2) to initiate assembly of SG via eIF2α phosphorylation, and this in turn blocks translation and leads to the recruitment of stalled transcripts into SGs (Box 1 and Figure 2). It has been proposed that, if the stress stimulus is not resolved, the stalled transcripts are transferred to processing bodies (P-bodies, PBs) for degradation [60]. This view requires reexamination because SG formation does not necessarily result in translational shut-off. Indeed, it is unclear whether SG formation results in total host cell translational shut-off or translational arrest at limited areas in the cytoplasm. Many viruses hijack host cellular compartments to form replication complexes for viral transcription and translation 61, 62. The fact that IFN is efficiently translated in virus-infected and SG-containing cells 29, 42, 49 suggests that SG formation does not necessarily correlate with total host translational shut-off.
Viral dsRNA contained in SG potentially activates OAS to catalyze the synthesis of 2′-5′ oligo A, which activates cytoplasmic endoribonuclease RNase L [63]. RNase L is also detected in SGs of virus-infected cells [42]. Activated RNase L may cleave viral RNA in SG to block viral transcription and translation, and some cleavage products may act as ligands for RLR [64]. Several ubiquitin ligases including TRIM25, RING finger protein leading to RIG-I activation (Riplet), and mex-3 RNA binding family member (MEX3C), that are known to regulate RIG-I activation, are also colocalized in virus-induced SG (Figure 2) 65, 66. Although several studies suggest that RLR might be activated in virus-induced SGs 29, 42, 49, direct evidence to this effect is lacking. RLR-mediated signaling is transmitted via homotypic interaction with IPS-1, which is localized on mitochondria, peroxisomes, and MAMs. It is unclear how RLR-containing SGs can communicate with these organelles and activate antiviral signaling (Figure 2).
A major challenge for SG research is that SGs are difficult entities to isolate for biochemical analyses. The composition of SGs induced by different viruses and in different host cells may vary 49, 67. The development of novel biochemical isolation approaches and molecular probes for cell biological analyses will be crucial in investigating the molecular events taking place in SGs during viral infection.
Concluding remarks and future directions
Stress responses are known to be crucial for maintaining the homeostasis of living organisms. In response to various stresses, eukaryotic cells initiate stress responses, including the formation of SGs, in which cytoplasmic mRNAs are compartmentalized to escape dysregulation. Accumulated lines of evidence show that viral infection can also induce stress responses including SG formation concurrently with the initiation of innate antiviral responses via pattern recognition receptors (PRRs). The observations that (i) there is a strong correlation between SG formation and IFN production, (ii) there is a reverse correlation between SG formation and viral propagation, and (iii) RLRs are localized in viral-induced SGs together with viral non-self RNAs, together strongly suggest that SGs have an antiviral role and possibly function as platform to initiate innate responses. Interestingly, this notion clearly indicates that the quality control machinery for ‘self RNA’ and the host defense mechanism against invasion of ‘non-self RNA’ are closely coordinated with each other. Moreover, it is interesting to note that these observations may help us to develop a novel therapeutic or preventive strategy for virus-induced infectious diseases.
Acknowledgments
We were supported by the following grants: the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan [Innovative Areas ‘Infection Competency’ (grants 24115004 and 25115503), Scientific Research ‘A’ (23249023) and ‘B’ (26293101)], the Ministry of Health, Labor and Welfare of Japan, the Uehara Memorial Foundation, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, the Takeda Science Foundation, the Naito Foundation, and Nippon Boehringer Ingelheim.
References
- 1.Gurtler C., Bowie A.G. Innate immune detection of microbial nucleic acids. Trends Microbiol. 2013;21:413–420. doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwasaki A., Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michallet M.C. Innate receptors for adaptive immunity. Curr. Opin. Microbiol. 2013;16:296–302. doi: 10.1016/j.mib.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Kedersha N. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kedersha N., Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 7.Anderson P., Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 8.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 9.Ramos H.J., Gale M., Jr RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr. Opin. Virol. 2011;1:167–176. doi: 10.1016/j.coviro.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pichlmair A., Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 11.Nie Y., Wang Y.Y. Innate immune responses to DNA viruses. Protein Cell. 2013;4:1–7. doi: 10.1007/s13238-012-2122-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ablasser A. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoneyama M. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 14.Yoneyama M. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 15.Schlee M. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt A. 5′-Triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornung V. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 18.Kawai T. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 19.Belgnaoui S.M. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr. Opin. Immunol. 2011;23:564–572. doi: 10.1016/j.coi.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Gack M.U. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 21.Jiang X. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horner S.M. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H.M. The mitochondrial targeting chaperone 14-3-3epsilon regulates a RIG-I translocon that mediates membrane association and innate antiviral immunity. Cell Host Microbe. 2012;11:528–537. doi: 10.1016/j.chom.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng W. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peisley A. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol. Cell. 2013;51:573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 26.Peisley A. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509:110–114. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honda K. Type I interferon gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Berlanga J.J. Antiviral effect of the mammalian translation initiation factor 2alpha kinase GCN2 against RNA viruses. EMBO J. 2006;25:1730–1740. doi: 10.1038/sj.emboj.7601073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng C.S. Encephalomyocarditis virus disrupts stress granules, the critical platform for triggering antiviral innate immune responses. J. Virol. 2013;87:9511–9522. doi: 10.1128/JVI.03248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White J.P. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe. 2007;2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Fung G. Production of a dominant-negative fragment due to G3BP1 cleavage contributes to the disruption of mitochondria-associated protective stress granules during CVB3 infection. PLoS ONE. 2013;8:e79546. doi: 10.1371/journal.pone.0079546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin Q. Mammalian orthoreovirus particles induce and are recruited into stress granules at early times postinfection. J. Virol. 2009;83:11090–11101. doi: 10.1128/JVI.01239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McInerney G.M. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell. 2005;16:3753–3763. doi: 10.1091/mbc.E05-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruggieri A. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe. 2012;12:71–85. doi: 10.1016/j.chom.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toroney R. Regulation of PKR by HCV IRES RNA: importance of domain II and NS5A. J. Mol. Biol. 2010;400:393–412. doi: 10.1016/j.jmb.2010.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garaigorta U. Hepatitis C virus (HCV) induces formation of stress granules whose proteins regulate HCV RNA replication and virus assembly and egress. J. Virol. 2012;86:11043–11056. doi: 10.1128/JVI.07101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borghese F., Michiels T. The leader protein of cardioviruses inhibits stress granule assembly. J. Virol. 2011;85:9614–9622. doi: 10.1128/JVI.00480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hato S.V. The mengovirus leader protein blocks interferon-alpha/beta gene transcription and inhibits activation of interferon regulatory factor 3. Cell. Microbiol. 2007;9:2921–2930. doi: 10.1111/j.1462-5822.2007.01006.x. [DOI] [PubMed] [Google Scholar]
- 39.Okonski K.M., Samuel C.E. Stress granule formation induced by measles virus is protein kinase PKR dependent and impaired by RNA adenosine deaminase ADAR1. J. Virol. 2013;87:756–766. doi: 10.1128/JVI.02270-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito M. Measles virus nonstructural C protein modulates viral RNA polymerase activity by interacting with host protein SHCBP1. J. Virol. 2013;87:9633–9642. doi: 10.1128/JVI.00714-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khaperskyy D.A. Influenza A virus inhibits cytoplasmic stress granule formation. FASEB J. 2012;26:1629–1639. doi: 10.1096/fj.11-196915. [DOI] [PubMed] [Google Scholar]
- 42.Onomoto K. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS ONE. 2012;7:e43031. doi: 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Y. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 44.Katoh H. Japanese encephalitis virus core protein inhibits stress granule formation through an interaction with Caprin-1 and facilitates viral propagation. J. Virol. 2013;87:489–502. doi: 10.1128/JVI.02186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeuchi K. Sendai virus C protein plays a role in restricting PKR activation by limiting the generation of intracellular double-stranded RNA. J. Virol. 2008;82:10102–10110. doi: 10.1128/JVI.00599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iseni F. Sendai virus trailer RNA binds TIAR, a cellular protein involved in virus-induced apoptosis. EMBO J. 2002;21:5141–5150. doi: 10.1093/emboj/cdf513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montero H. Rotavirus infection induces the phosphorylation of eIF2alpha but prevents the formation of stress granules. J. Virol. 2008;82:1496–1504. doi: 10.1128/JVI.01779-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emara M.M., Brinton M.A. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. U.S.A. 2007;104:9041–9046. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoo J.S. DHX36 enhances RIG-I signaling by facilitating PKR-mediated antiviral stress granule formation. PLoS Pathog. 2014;10:e1004012. doi: 10.1371/journal.ppat.1004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langereis M.A. MDA5 localizes to stress granules, but this localization is not required for the induction of type I interferon. J. Virol. 2013;87:6314–6325. doi: 10.1128/JVI.03213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diebold S.S. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature. 2003;424:324–328. doi: 10.1038/nature01783. [DOI] [PubMed] [Google Scholar]
- 52.Stewart M.J. PKR's protective role in viral myocarditis. Virology. 2003;314:92–100. doi: 10.1016/s0042-6822(03)00414-8. [DOI] [PubMed] [Google Scholar]
- 53.Gilfoy F.D., Mason P.W. West Nile virus-induced interferon production is mediated by the double-stranded RNA-dependent protein kinase PKR. J. Virol. 2007;81:11148–11158. doi: 10.1128/JVI.00446-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schulz O. Protein kinase R contributes to immunity against specific viruses by regulating interferon mRNA integrity. Cell Host Microbe. 2010;7:354–361. doi: 10.1016/j.chom.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McAllister C.S., Samuel C.E. The RNA-activated protein kinase enhances the induction of interferon-beta and apoptosis mediated by cytoplasmic RNA sensors. J. Biol. Chem. 2009;284:1644–1651. doi: 10.1074/jbc.M807888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Honda K. Selective contribution of IFN-alpha/beta signaling to the maturation of dendritic cells induced by double-stranded RNA or viral infection. Proc. Natl. Acad. Sci. U.S.A. 2003;100:10872–10877. doi: 10.1073/pnas.1934678100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith E.J. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or Ikappa B kinase but is blocked by Vaccinia virus E3L protein. J. Biol. Chem. 2001;276:8951–8957. doi: 10.1074/jbc.M008717200. [DOI] [PubMed] [Google Scholar]
- 58.Hornung V. Replication-dependent potent IFN-alpha induction in human plasmacytoid dendritic cells by a single-stranded RNA virus. J. Immunol. 2004;173:5935–5943. doi: 10.4049/jimmunol.173.10.5935. [DOI] [PubMed] [Google Scholar]
- 59.Komatsu T. C and V proteins of Sendai virus target signaling pathways leading to IRF-3 activation for the negative regulation of interferon-beta production. Virology. 2004;325:137–148. doi: 10.1016/j.virol.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 60.Kedersha N. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagy P.D., Pogany J. The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol. 2012;10:137–149. doi: 10.1038/nrmicro2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Friedel C.C., Haas J. Virus–host interactomes and global models of virus-infected cells. Trends Microbiol. 2011;19:501–508. doi: 10.1016/j.tim.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 63.Chakrabarti A. New insights into the role of RNase L in innate immunity. J. Interferon Cytokine Res. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malathi K. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oshiumi H. A distinct role of Riplet-mediated K63-Linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog. 2013;9:e1003533. doi: 10.1371/journal.ppat.1003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuniyoshi K. Pivotal role of RNA-binding E3 ubiquitin ligase MEX3C in RIG-I-mediated antiviral innate immunity. Proc. Natl. Acad. Sci. U.S.A. 2014;111:5646–5651. doi: 10.1073/pnas.1401674111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Piotrowska J. Stable formation of compositionally unique stress granules in virus-infected cells. J. Virol. 2010;84:3654–3665. doi: 10.1128/JVI.01320-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nover L. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol. Cell. Biol. 1989;9:1298–1308. doi: 10.1128/mcb.9.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Collier N.C., Schlesinger M.J. The dynamic state of heat shock proteins in chicken embryo fibroblasts. J. Cell Biol. 1986;103:1495–1507. doi: 10.1083/jcb.103.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anderson P., Kedersha N. Stressful initiations. J. Cell Sci. 2002;115:3227–3234. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- 71.Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 72.Eulalio A. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 73.Kedersha N., Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 74.Kedersha N.L. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tourriere H. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Gilks N. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weissbach R., Scadden A.D. Tudor-SN and ADAR1 are components of cytoplasmic stress granules. RNA. 2012;18:462–471. doi: 10.1261/rna.027656.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Solomon S. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell. Biol. 2007;27:2324–2342. doi: 10.1128/MCB.02300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kimball S.R. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 2003;284:C273–C284. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- 80.Kwon S. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 2007;21:3381–3394. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gallouzi I.E. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wehner K.A. OGFOD1, a novel modulator of eukaryotic translation initiation factor 2alpha phosphorylation and the cellular response to stress. Mol. Cell. Biol. 2010;30:2006–2016. doi: 10.1128/MCB.01350-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morris A.R. Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite evolution of diverse mRNA target sets. Mol. Cell. Biol. 2008;28:4093–4103. doi: 10.1128/MCB.00155-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vessey J.P. Dendritic localization of the translational repressor Pumilio 2 and its contribution to dendritic stress granules. J. Neurosci. 2006;26:6496–6508. doi: 10.1523/JNEUROSCI.0649-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chalupnikova K. Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J. Biol. Chem. 2008;283:35186–35198. doi: 10.1074/jbc.M804857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hua Y., Zhou J. Rpp20 interacts with SMN and is re-distributed into SMN granules in response to stress. Biochem. Biophys. Res. Commun. 2004;314:268–276. doi: 10.1016/j.bbrc.2003.12.084. [DOI] [PubMed] [Google Scholar]
- 87.Hua Y., Zhou J. Survival motor neuron protein facilitates assembly of stress granules. FEBS Lett. 2004;572:69–74. doi: 10.1016/j.febslet.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 88.Thomas M.G. Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes. Mol. Biol. Cell. 2005;16:405–420. doi: 10.1091/mbc.E04-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Deigendesch N. ZBP1 subcellular localization and association with stress granules is controlled by its Z-DNA binding domains. Nucleic Acids Res. 2006;34:5007–5020. doi: 10.1093/nar/gkl575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Andrei M.A. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cougot N. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ingelfinger D. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- 93.Yu J.H. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA. 2005;11:1795–1802. doi: 10.1261/rna.2142405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fenger-Gron M. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 95.Eystathioy T. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Eystathioy T. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sheth U., Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wichroski M.J. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2006;2:e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gallois-Montbrun S. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 2007;81:2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leung A.K. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. U.S.A. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sen G.L., Blau H.M. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 102.Wilczynska A. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- 103.Lai M.C. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol. Biol. Cell. 2008;19:3847–3858. doi: 10.1091/mbc.E07-12-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kasim V. Determination of the role of DDX3 a factor involved in mammalian RNAi pathway using an shRNA-expression library. PLoS ONE. 2013;8:e59445. doi: 10.1371/journal.pone.0059445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nathans R. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol. Cell. 2009;34:696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang W.H. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA. 2006;12:547–554. doi: 10.1261/rna.2302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Garcia M.A. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 108.Williams B.R. PKR; a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 109.Shi Y. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Harding H.P. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 111.Koumenis C. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol. Cell. Biol. 2002;22:7405–7416. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dong J. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell. 2000;6:269–279. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 113.McEwen E. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 2005;280:16925–16933. doi: 10.1074/jbc.M412882200. [DOI] [PubMed] [Google Scholar]
- 114.Chen J.J., London I.M. Regulation of protein synthesis by heme-regulated eIF-2 alpha kinase. Trends Biochem. Sci. 1995;20:105–108. doi: 10.1016/s0968-0004(00)88975-6. [DOI] [PubMed] [Google Scholar]
- 115.Matthews J.D., Frey T.K. Analysis of subcellular G3BP redistribution during rubella virus infection. J. Gen. Virol. 2012;93:267–274. doi: 10.1099/vir.0.036780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li W. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 2002;76:11989–12000. doi: 10.1128/JVI.76.23.11989-12000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pager C.T. Modulation of hepatitis C virus RNA abundance and virus release by dispersion of processing bodies and enrichment of stress granules. Virology. 2013;435:472–484. doi: 10.1016/j.virol.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ariumi Y. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J. Virol. 2011;85:6882–6892. doi: 10.1128/JVI.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sola I. The polypyrimidine tract-binding protein affects coronavirus RNA accumulation levels and relocalizes viral RNAs to novel cytoplasmic domains different from replication-transcription sites. J. Virol. 2011;85:5136–5149. doi: 10.1128/JVI.00195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Raaben M. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell. Microbiol. 2007;9:2218–2229. doi: 10.1111/j.1462-5822.2007.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dinh P.X. Induction of stress granule-like structures in vesicular stomatitis virus-infected cells. J. Virol. 2013;87:372–383. doi: 10.1128/JVI.02305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lindquist M.E. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J. Virol. 2010;84:12274–12284. doi: 10.1128/JVI.00260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lindquist M.E. Activation of protein kinase R is required for induction of stress granules by respiratory syncytial virus but dispensable for viral replication. Virology. 2011;413:103–110. doi: 10.1016/j.virol.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hanley L.L. Roles of the respiratory syncytial virus trailer region: effects of mutations on genome production and stress granule formation. Virology. 2010;406:241–252. doi: 10.1016/j.virol.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Baird N.L. Arenavirus infection induces discrete cytosolic structures for RNA replication. J. Virol. 2012;86:11301–11310. doi: 10.1128/JVI.01635-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Qin Q. Mammalian orthoreovirus escape from host translational shutoff correlates with stress granule disruption and is independent of eIF2alpha phosphorylation and PKR. J. Virol. 2011;85:8798–8810. doi: 10.1128/JVI.01831-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Smith J.A. Reovirus induces and benefits from an integrated cellular stress response. J. Virol. 2006;80:2019–2033. doi: 10.1128/JVI.80.4.2019-2033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Simpson-Holley M. Formation of antiviral cytoplasmic granules during orthopoxvirus infection. J. Virol. 2011;85:1581–1593. doi: 10.1128/JVI.02247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Katsafanas G.C., Moss B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe. 2007;2:221–228. doi: 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Khong A., Jan E. Modulation of stress granules and P bodies during dicistrovirus infection. J. Virol. 2011;85:1439–1451. doi: 10.1128/JVI.02220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


