Highlights
► The mutation pressure and natural selection exists in the processes of evolution of EV71. ► The synonymous codon usage pattern of EV71 is a mixture of coincidence and antagonism to that of host. ► The genetic diversity of EV71 strains was observed. ► The geographic factors of selection of some synonymous codons in EV71 was also observed.
Keywords: Enterovirus 71, The relative synonymous codon usage value, Codon usage pattern, Effective number of codons, Mutation pressure, Natural selection
Abstract
To give a new perspective on the evolutionary characteristics shaping the genetic diversity of enterovirus 71 (EV71) and the effects of natural selection from its host on the codon usage pattern of the virus, the relative synonymous codon usage (RSCU) values, codon usage bias (CUB) values, effective number of codons (ENCs) values and nucleotide contents were calculated to implement a comparative analysis to evaluate the dynamics of the virus evolution. The characteristics of the synonymous codon usage patterns and nucleotide contents of EV71 and the comparison between ENC values for the whole coding sequence of EV71 and that of coding sequences for viral proteins of EV71 all indicate that the interaction between mutation pressure from virus and natural selection from host exists in the processes of evolution of EV71. The synonymous codon usage pattern of EV71 is a mixture of coincidence and antagonism to that of host cell. In addition, the genetic diversity of EV71 strains and the preferential selection of some synonymous codons in EV71 strains based on the different epidemic areas were observed, suggesting that geographic and social factors may play roles in influencing the evolution of this virus.
1. Introduction
Hand-foot-and-mouth disease (HFMD) is a general illness in children which usually is caused by some human enteroviruses (Cherry, 1992). There were many reports which indicated some pandemics of HFMD were associated with EV71 infection in the Asia-Pacific area (AbuBakar et al., 1999, Chumakov et al., 1979, Fujimoto et al., 2002, Ho et al., 1999, Lin et al., 2005, Liu et al., 2000, Zheng et al., 1995). EV71 belongs to members of the Enterovirus genus of the Picornaviridae family and is a positive-strand RNA virus with a genome size of about 7500 bp. The two non-translated regions (5′-NTR and 3′-NTR) flank the single open reading frame (ORF) of EV71 virus genome. The coding sequence encodes one polyprotein that is cleaved by viral proteases to generate 11 proteins, namely VP4, VP2, VP3, VP1, 2A, 2B, 2C, 3A, 3B, 3C and 3D. The structural proteins VP1-3 are exposed on EV71 surface. The VP1 gene contained the major antigenic sites and genetic diversity associated with serotypes (Oberste et al., 1999a, Oberste et al., 1999b). Non-structural proteins were involved in polyprotein processing, RNA replication and the shut-down of host cell protein synthesis. In addition, recombinations are well known to result in a genetic diversity and evolution of enteroviruses (Chan and AbuBakar, 2004, Chen et al., 2010, Yoke-Fun and AbuBakar, 2006). Due to various genetic diversities of EV71, the effect of the vaccine is limited to prevent children from EV71. This situation has made researchers aware of the importance of analysis of EV71 genetic diversity (Bible et al., 2008, Cardosa et al., 2003, Herrero et al., 2003, Huang et al., 2008, Lewis-Rogers et al., 2009, McMinn, 2002, Sanders et al., 2006). It is noticed that nucleotide composition comprising of EV71 coding sequence with various genetic diversities is selective rather than random, because the natural selection from host is responsible to select various strains shaped by mutation. In previous reports, translation selection and compositional constraints under the mutational pressure are thought to be the major factors accounting for codon usage variation among genomes in microorganisms (Gu et al., 2004, Karlin and Mrázek, 1996, Lesnik et al., 2000, Liu et al., 2010, Zhou et al., 2005, Zhou et al., 2006, Zhou et al., 2010). In some RNA viruses, compared with natural selection, mutation pressure plays a more important role in synonymous codon usage pattern (Jenkins and Holmes, 2003, Levin and Whittome, 2000). Although it is known that compositional constraints and translation selection are the more generally accepted mechanisms accounting for codon usage bias (Coleman et al., 2008, Karlin et al., 1990, Zhi et al., 2010, Zhou et al., 1999), other selection forces have also been proposed such as fine-tuning translation kinetics selection as well as escape of cellular antiviral responses (Aragones et al., 2008, Aragones et al., 2010, Karlin et al., 1994, Sugiyama et al., 2005). Thus, the codon usage pattern may be important to disclose the molecular mechanism and evolutionary process of EV71 avoiding host cell response. To our knowledge, it is the first study that the synonymous codon usage pattern and evolutional dynamics of EV71 were systemically analyzed and the relationship between codon usage pattern of EV71 and that of its host was also analyzed.
2. Materials and methods
2.1. Sequence data
The 74 complete RNA sequences of EV71 were downloaded from the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/Genbank/) and detailed information about the viruses were listed in Table S1. Each general nucleotide composition (U%, A%, C% and G%) and each nucleotide composition in the third site of codon (U3%, A3%, C3% and G3%) in EV71 coding sequence were calculated by biosoftware DNAStar 7.0 for windows.
2.2. Calculation of the relative synonymous codon usage (RSCU)
To investigate the characteristics of synonymous codon usage without the confounding influence of amino acid composition among different sequences, the relative synonymous codon usage (RSCU) values among different codons in the EV71 ORF was calculated according to the published equation (Sharp et al., 1986).
2.3. Analysis of codon usage bias
The ‘effective number of codons’ (ENCs), the useful estimator of absolute codon usage bias, was a measure quantifying the codon usage bias of the whole coding sequence of EV71. The ENC value ranges from 20 (when only one synonymous codon is chosen by the corresponding amino acid) to 61 (when all synonymous codons are used equally) (Wright, 1990). In this study, this measure was used to evaluate the degree of codon usage bias of coding sequences for proteins of EV71 and to calculate the degree of the codon bias for the whole coding sequence of EV71 and other picornaviruses.
Additionally, there is a simple method which is supposed that statistically equal and random usage of all available synonymous codons was the “neutral point” (RSCU0 = 1.00) for the development of group-specific codon usage (Zhou et al., 2010). This method was introduced in the study to investigate the discrepancy of the synonymous codon usage pattern among of EV71 strains based on the different isolated areas.
2.4. Principal component analysis
Principal component analysis (PCA), which was a commonly used multivariate statistical method (Jolliffe, 2002, Mardia et al., 1979), was carried out to analyze the major trend in codon usage pattern among different strains of EV71. PCA involves a mathematical procedure that transforms some correlated variable (RSCU values) into a smaller number of uncorrelated variables called principal components. Each strain was represented as a 59 dimensional vector, and each dimension corresponded to the RSCU value of each sense codon, which only included several synonymous codons for a particular amino acid, excluding the codon of AUG, UGG and three stop codons. In addition, PCA was also performed for analyzing the discrepancy between codon usage pattern of EV71 and that of host cell.
2.5. Correlation analysis
The relationship between each general nucleotide composition (U%, A%, C% and G%) and each nucleotide composition in the third site of codon (U3%, A3%, C3% and G3%) in EV71 coding sequence and the relationship between U3%, A3%, C3%, G3% and the codon usage pattern of EV71 were evaluated by the Pearson's rank.
All statistical processes were carried out by statistical software SPSS11.5 for windows.
3. Results
3.1. Synonymous codon usage in EV71
The A% and U% were higher than C% and G%, but A3% and U3% were lower than C3% and G3% in EV71 (Table S2). The overall nucleotide composition never affects the nucleotide contents in the third site of codon in EV71 coding sequence, suggesting that composition constraints may be one of the factors in affecting the codon usage pattern of EV71.
The optimal codons of Ala, Arg, Asp, Cys, Glu, Gly, Ile, Phe, Pro, Ser were A-ended or U-ended, while those of Asn, Glu, His, Leu, Lys, Thr, Tyr, Val were C-ended or G-ended (Table 1 ). EV71 does not depends on all optimal codons with either A/U-end or C/G-ended like influenza A virus subtype H5N1, sever acute respiratory syndrome Coronavirus or foot-and-mouth disease virus (FMDV) (Gu et al., 2004, Zhao et al., 2008, Zhou et al., 2010), but shapes the optimal codons with any types of nucleotide-ended. It is noted that although Asn, Leu, Tyr, Glu, Lys and Thr possessed optimal codon with C- or G-ended, they also contained some favored codon with U- or A-ended. Similarly, Asp, Phe and Cys also had favored codons with C- or G-ended (Table 1). These amino acids which choose optimal codons with any nucleotide-ended are affected under both mutation pressure by itself and natural selection from host, since natural selection from host ultimately allows those strains with good-fitness to possess a special codon usage patterns.
Table 1.
The synonymous codon usage patterns of EV71 virus and human cell.
| AAa | Codon | RSCUd | AAa | Codon | RSCUd |
|---|---|---|---|---|---|
| Alab | GCA | 1.223 | Leuc | CUA | 0.807 |
| GCCf | 0.996 | CUC | 1.131 | ||
| GCGg | 0.536 | CUGf | 0.976 | ||
| GCU | 1.245 | CUU | 1.119 | ||
| Argb | AGAf | 1.853 | UUAg | 0.805 | |
| AGG | 1.702 | UUG | 1.159 | ||
| CGAg | 0.568 | Lysc | AAAg | 0.907 | |
| CGC | 1.020 | AAGf | 1.093 | ||
| CGG | 0.388 | Pheb | UUCf | 0.979 | |
| CGU | 0.469 | UUUg | 1.020 | ||
| Asnc | AACf | 1.091 | Prob | CCA | 1.589 |
| AAUg | 0.909 | CCCf | 0.881 | ||
| Aspb | GACf | 0.900 | CCGg | 0.405 | |
| GAUg | 1.100 | CCU | 1.124 | ||
| Cysb | UGCf | 0.972 | Serb | AGCf | 0.994 |
| UGUg | 1.028 | AGU | 1.172 | ||
| Glnb | CAAg | 1.103 | UCA | 1.315 | |
| CAGf | 0.897 | UCC | 1.133 | ||
| Gluc | GAAg | 0.933 | UCGg | 0.367 | |
| GAGf | 1.067 | UCU | 1.019 | ||
| Glyb | GGA | 0.997 | Thrc | ACA | 1.201 |
| GGCf | 0.498 | ACCf | 1.221 | ||
| GGG | 0.994 | ACGg | 0.376 | ||
| GGUg | 1.499 | ACU | 1.202 | ||
| Hisc | CACf | 1.253 | Tyrc | UACf | 1.092 |
| CAUg | 0.747 | UAUg | 0.908 | ||
| Ileb | AUAg | 0.691 | Valc | GUAg | 0.471 |
| AUCf | 1.048 | GUC | 0.893 | ||
| AUU | 1.261 | GUGf | 1.678 | ||
| GUU | 0.958 |
Stands for amino acid.
Stands for amino acids whose optimal codons are A- or U-end.
Stands for amino acids whose optimal codons are C- or G-end.
Stands for the relative synonymous codon usage value.
Stands for the optimal codons used in human cells (Pintó et al., 2007, Sánchez et al., 2003).
Stands for the rare codons used in human cells (Pintó et al., 2007, Sánchez et al., 2003).
3.2. Genetic relationship based on synonymous codon usage in EV71
The PCA detected the first principal component (f 1′) which can account for 13.73% of the total synonymous codon usage variation, and the second principal component (f 2′) for 11.81% of the total variation. It appeared to be a little complex with some overlapping plots representing different epidemic areas (Fig. S1). The plots for strains isolated from China-Mainland, compared with that of strains isolated from other areas, could aggregate highly, while the plots for strains isolated from Malaysia and China-Taiwan scattered largely, the plots for strains from Singapore, USA, Japan and Switzerland did not indicate the genetic diversity obviously, due to the limited samples. For strains circulating in China-Mainland, social factors (public health, interpersonal communication, etc.) may play a role in influencing genetic diversity of these strains. However, for strains in China-Taiwan and Malaysia, geographic factors likely influence genetic diversity of those strains except for social factors.
3.3. Compositional properties of the whole coding sequence of EV71
The nucleotide contents of the whole coding sequence of EV71 were analyzed. In Table 2 , the significant positive correlations between A% and A3%, U% and U3%, C% and C3% and significant negative correlations among most of heterogeneous nucleotide contents indicated that composition constraints play a role in codon usage pattern of EV71; however, significant positive correlation between G% and A3%, C% and G3% and no correlation between G% and G3% might suggest that natural selection from host plays a role in codon usage pattern of EV71 as well. In addition, there were significant correlations between each nucleotide content in the third site of codon and codon usage indices (f 1 and f 2) (Table 3 ). Although the positive and negative correlations existed between C3% and f 1, and between C3% and f 2, respectively, the positive correlation play an important role in affecting the codon usage pattern due to f 1 being the first principal component.
Table 2.
Summary of correlation analysis between the A%, U%, C%, G% and A3%, U3%, C3%, G3% in the whole coding sequences of 74 EV71 strainsa.
| A3% | U3% | C3% | G3% | (C3 + G3)% | |
|---|---|---|---|---|---|
| A% | r = 0.869** | r = −0.346** | r = −0.316** | r = −0.316** | r = −0.102NS |
| U% | r = −0.307** | r = 0.918** | r = −0.703** | r = −0.703** | r = −0.882** |
| C% | r = −0.467** | r = −0.084NS | r = 0.875** | r = 0.875** | r = 0.341** |
| G% | r = 0.316** | r = −0.751** | r = −0.027NS | r = 0.027NS | r = 0.671** |
| (C + G)% | r = −0.198NS | r = −0.665** | r = 0.832** | r = 0.832** | r = 0.884** |
r value in this table is calculated in each correlation analysis.
NS means non-significant (p > 0.05).
Means p < 0.01.
Table 3.
Summary of correlation analysis between the first two axes in principle and nucleotide contents in EV71.
| Base compositions | f1′ | f2′ |
|---|---|---|
| A3% | r = 0.359** | r = 0.542** |
| U3% | r = −0.513** | r = −0.516** |
| C3% | r = 0.238* | r = −0.355** |
| G3% | r = 0.238* | r = −0.355** |
| (C3 + G3)% | r = 0.439** | r = 0.250* |
Means 0.01 < p < 0.05.
Means p < 0.01.
3.4. Qualitative evaluation of codon usage bias of the whole coding sequence of EV71
The strong discrepancy of the synonymous codon usage in strains based on the different isolated areas was observed. In details, in strains from China-Mainland, CGC for Arg, GAC for Asp, CUU for Leu, UUC for Phe were chosen by EV71 strains preferentially, while CGG for Arg, GAC for Asp, UUU for Phe were chosen poorly; in Singapore, AUU for Ile, CUG for Leu, UCA for Ser were chosen preferentially, while UUG for Phe and UCG for Ser were chosen poorly; in USA, AGA for Arg was used preferentially, while GGU for Gly was used poorly; in Japan, GCU for Ala, AAU for Asn, CAA for Gln, GAA for Glu were chosen preferentially, while GCG for Ala and CAG for Gln were poorly used; in Switzerland, AGG for Arg, UGC for Cys, CUC for Leu, UUG for Leu, AGU for Ser were chosen preferentially; while CGA for Arg, UGU for Cys, CUG for Leu, CUU for Leu, AGC for Ser were poorly used (Fig. S2). These results may suggest that with the development of evolution of EV71 strains, the discrepancy of some synonymous codon usage probably is formed in different epidemic regions.
3.5. Relationship between amino acids and codon usage pattern in EV71
In order to analyze whether the evolution of CUB was controlled by mutation effect or natural selection from host, the CUB values had been calculated based on data listed in Table 1. The transition from maximum-negative to maximum-positive values was smooth and there was no obvious or unambiguous border between the so-called dominant and prohibited codons (Fig. 1 ), namely, all synonymous codons were used. This result implied that the interaction between mutation pressure from EV71 and natural selection from host exists in the evolution of EV71.
Fig. 1.
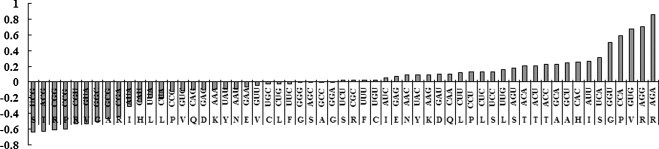
Distribution of the CUB of a codon for each amino acid. CUB was taken from Table 1 and sorted in ascending order.
3.6. Relationships between codon usage pattern of EV71 and that of the host cell
By comparing between the patterns of synonymous codon usage of human cell and that of EV71 virus, we found that the pattern of synonymous codon usage of EV71 strains is partially antagonistic to that of human cells. In detail, optimal codons of nine amino acids in EV71, including Ala, Asp, Cys, Gln, Gly, Leu, Phe, Pro, Ser, are the disfavored codons of the corresponding amino acids in its host. Among these non-coincidence patterns of synonymous codon usage of amino acids, the synonymous codon usage of Asp, Cys, Gln, Gly, Phe has evolved to be complementary to that of host cells (Table 1). In addition, the optimal and rare synonymous codon usage patterns of Arg, Asn, Glu, His, Lys, Thr, Tyr and Val of EV71 virus were in agreement with those of human cells (Table 1). Additionally, PCA was performed to examine the whole coding sequence of EV71 in this study. The method detected one major trend in the first axis (f 1′) which can account for 11.79% of the total synonymous codon usage variation, and another major trend in the second axis (f 2′) for 10.69% of the total variation. The plots for codon usage pattern of human are far from the plots for that of EV71 (Fig. S3).
The ENC values were calculated for FMDV, Cardiovirus, Hapatitis A virus (HAV), Poliovirus (PV) and compared with that of EV71 (Table S3). Among these virus examined, the ENC value for EV71 is highest, suggesting that EV71 has a most weak codon usage bias. In addition, we set up a plot which showed the relationship between GC3% and ENC values of all viral proteins (excluding the very small 3B protein) of EV71 virus, and found that the plots of coding sequences for VP1, VP2, VP3, 2A, 2C, 3A, 3C and 3D aggregated around the expected curve, but the plots of coding sequences for VP4 and 2B scattered highly under the expected curve (Fig. S4a–4c). It may be explained that the codon usage bias of VP4 and 2B genes is influenced by their small size. In addition, there is no obvious geographic factor in influencing codon usage bias of the coding sequences of EV71, implying that the natural selection from the geographic factor does not affect the codon usage patterns of specific coding sequences of EV71, but shape the pattern of the whole sequence of this virus. Furthermore, we found that some specific non-optimal codons are preferentially chosen in some coding sequences of EV71. In details, three non-optimal codons (UUA, CUA and GUU) are chosen in the VP4 gene, UUG in the VP3 gene, CUU in the VP1 and 3A genes, UUU, CUU and GUU in the 3C gene. It is also found that all coding sequences of EV71 contain some preferential codons. In details, GUG, CCC, ACA, GCC and GAC are preferentially chosen in the VP4 gene, GUG, CCA and AGG in the VP2 gene, CUG, UCA, ACC and AGA in the VP3 gene, UCA in the VP1 gene, CUC, CCA and AGA in the 2A gene, CCU, AGA and AGG in the 2B gene, GUG, UCU, CCA, ACA and AGA in the 2C gene, CCA, ACU, AGU, AGC, AGA and AGG in the 3A gene, AUU, CCU, ACA, GCA, AGU and AGG in the 3C gene, GUG and AGA in the 3D gene. Taken together, there is no obvious relationship between the distribution of non-optimal codons and the deviation of ENC value from the theoretical value.
4. Discussion
The pattern of codon usage is a genetic characteristic of various organisms. Previous reports have been focused on viruses in Picornavirdae family, such as FMDV, HAV, Poliovirus (Aragones et al., 2008, Aragones et al., 2010, Coleman et al., 2008, Zhong et al., 2007). Because A%, U%, G3% and C3% play roles in the formation of the different optimal codons with any nucleotide-ended, the codon usage pattern of EV71 is likely influenced by composition constraints. The codon usage pattern of PV is mostly coincident with that of its host, while the codon usage pattern of HAV is antagonistic to that of its host (Mueller et al., 2006, Sánchez et al., 2003). The codon usage pattern of EV71 is a mixture of the two types of codon usage. The coincident portion of codon usage pattern of EV71 enable the corresponding amino acids to be translated efficiently, the other antagonistic portion of codon usage pattern of EV71 may enable viral proteins to be folded properly, although the translation efficiency of the corresponding amino acids decreased. In Epstein-Barr virus latent genes deoptimize codon usage in order to evade competition for host protein translation (Karlin et al., 1990) and attenuation of PV activity was performed by rare codon pairs inducing poor translation for sequences of viral proteins (Coleman et al., 2008). These results suggest that disfavored codons coding for amino acids may not be deleterious factor for viruses to adapt to host cells. For codon usage patterns of the coding sequences of EV71, the VP2, 2A, 2B, 2C and 3D genes possess only some preferential codons and none of non-optimal codons is preferentially used, implying that translation of the whole coding sequence of EV71 is possibly regulated under the translation selection. Furthermore, the alternative translation is the possibility of fine-tuning the kinetics of protein translation by a combination of rare and optimal codons (Aragones et al., 2010, Komar, 2009). For codon usage patterns of VP4, VP3, VP1, 3A and 3C genes of EV71, these genes possess combination of some non-optimal codons and optimal ones which are preferentially used, implying that translation of the coding sequences of EV71 is possibly regulated under fine-tuning translation kinetics selection.
The sequences 5′NTR and VP1 are often used to analyze the genetic diversity of EV71 (Hagiwara et al., 1984, Hsu et al., 2007, Li et al., 2005). By analyzing the codon usage pattern of the whole coding sequence of strains from different areas, genetic diversity resulting from geographic and social factors is likely observed. The genetic diversity of the most strains from China-Mainland could indicate that a relatively independent area with geographic, public health and personal communication that enables the genetic diversity of EV71 to be sustained with little outside influence. Compared with the genetic diversity of strains from China-Mainland, that of strains from China-Taiwan and Malaysia also indicated that social factors play an important role in shaping the codon usage patterns of these strains. Based on the genetic diversities of China-Taiwan and Malaysia, social factors may play important roles in shaping codon usage patterns of EV71 from the two areas. These genetic diversities of EV71 strains from different areas give a sign that geographic and social factors should be noticed at genetic diversity of virus from different areas.
The ENC values calculated for some picornaviruses indicate that a significantly lower bias of codon usage exists in EV71 than in the other viruses. As for RNA viruses, previous study reported that the major factor in shaping codon usage patterns appears to be mutation pressure rather than natural selection (Liu et al., 2010, Zhao et al., 2008, Zhong et al., 2007, Zhou et al., 2005, Zhou et al., 2010). However, the genetic characteristics of EV71 suggest the interaction between mutation pressure and natural selection, although ENC values for the whole coding sequence of EV71 suggest mutation pressure is a factor in influencing codon usage pattern. Furthermore, in Fig. S4a–4c, the relationship between ENC data for EV71 proteins and CG3% indicated that natural selection probably play roles in genetic diversity of EV71 strains except for mutation pressure in order to adapt to host. A general mutational pressure, which affects the whole genome would certainly account for the majority of the codon usage among some RNA viruses (Jenkins and Holmes, 2003).
The genetic diversity and codon usage patterns results we proposed here are useful to understand the processes influencing the evolution of EV71, especially the roles played by natural selection from host and mutation pressure from virus. Additionally, such information might be helpful to understand the roles of geographic and social factors in influencing genetic diversity of EV71.
Acknowledgements
This work was supported in parts by grants from National Science & Technology Key Project (2009ZX08007-006B) and International Science & Technology Cooperation Program of China (No. 2010DFA32640) and Science and Technology Key Project of Gansu Province (No. 0801NKDA034). This study was also supported by National Natural Science foundation of China (No. 30700597 and No. 31072143).
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.meegid.2011.02.018.
Appendix A. Supplementary data
Fig. S1.
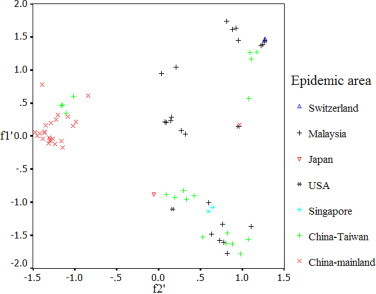
A plot of values of the first axis and the second axis of each EV71 strain for the principle component analysis.
Fig. S2.
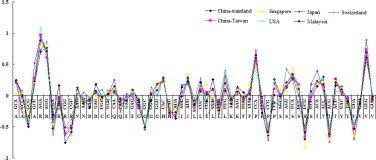
The CUB character corresponding to per codon for amino acid. Codon usage bias (CUB) values were calculated in strains based on the different isolated areas.
Fig. S3.
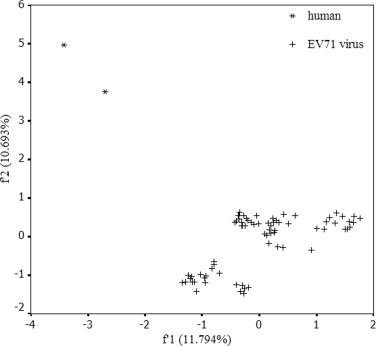
The discrepancy between codon usage pattern of human cells and that of EV71.
Fig. S4a.
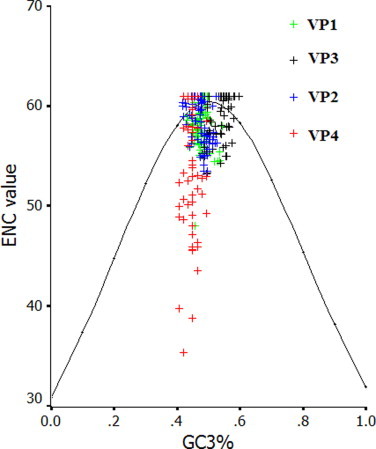
ENC vs. GC3% plot of the coding sequences for VP1-4 of EV71. ENC value denotes the effective number of codon of each coding sequence. The solid line represents the relationship between ENC and GC3% under random codon usage assumption.
Fig. S4b.
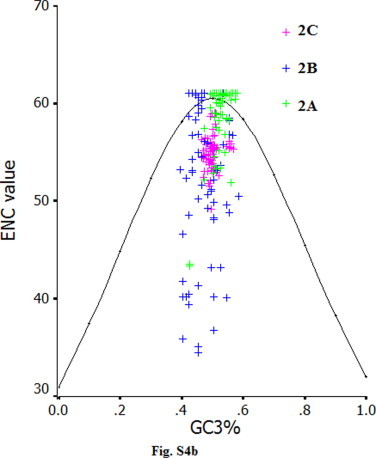
ENC vs. GC3% plot of the coding sequence for 2A, 2B and 2C of EV71.
Fig. S4c.
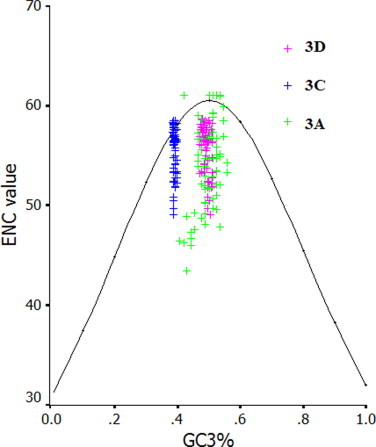
ENC vs. GC3% plot of the coding sequence for 3A, 3C and 3D of EV71.
References
- AbuBakar S., Chee H.Y., Al-kobaisi M.F., Xiaoshan J., Chua K.B., Lam S.K. Identification of enterovirus 71 isolates from an outbreak of hand, foot, and mouth disease (HFMD) with fatal cases of encephalomyelitis in Malaysia. Virus Res. 1999;61:1–9. doi: 10.1016/s0168-1702(99)00019-2. [DOI] [PubMed] [Google Scholar]
- Aragones L., Bosch A., Pinto R. Hepatitis A virus mutant spectra under the selective pressure of monoclonal antibodies: codon usage constrains limit capsid variability. J. Virol. 2008;82:1688–1700. doi: 10.1128/JVI.01842-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragones L., Guix S., Ribes E., Bosch A., Pinto R.M. Fine-tuning translation kinetics selection as the driving force of codon usage bias in the Hepatitis A virus capsid. PLoS Pathog. 2010;6:e1000797. doi: 10.1371/journal.ppat.1000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bible J.M., Iturriza-Gomara M., Megson B., Brown D., Pantelidis P., Earl P., Bendig J., Tong C.Y. Molecular epidemiology of human enterovirus 71 in the United Kingdom from 1998 to 2006. J. Clin. Microbiol. 2008;46:3192–3200. doi: 10.1128/JCM.00628-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardosa M.J., Perera D., Brown B.A., Cheon D., Chan H.M., Chan K.P., Cho H., McMinn P. Molecular epidemiology of human enterovirus 71 strains and recent outbreaks in the Asia-Pacific region: comparative analysis of the VP1 and VP4 genes. Emerg. Infect. Dis. 2003;9:461–468. doi: 10.3201/eid0904.020395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.F., AbuBakar S. Human enterovirus 71 in hand, foot and mouth disease patients. Emerg. Infect. Dis. 2004;10:1468–1470. doi: 10.3201/eid1008.040059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhang Q., Li J., Cao W., Zhang J.X., Zhang L., Zhang W., Shao Z.J., Yan Y. Analysis of recombination and natural selection in human enterovirus 71. Virology. 2010;398:251–261. doi: 10.1016/j.virol.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Cherry J.D. Enteroviruses: polioviruses (poliomylitis), coxsackieviruses, echoviruses and enteroviruses. In: Feign R.D., Cherry J.D., editors. Textbook of Pediatric Infectious Diseases. 3rd ed. The W.B. Saunders Co.; Philadelphia: 1992. pp. 1705–1752. [Google Scholar]
- Chumakov M., Voroshilova M., Shindarov L., Lavrova I., Gracheva L., Koroleva G.S. Enterovirus 71 isolated from cases of epidemic poliomyelitis-like disease in Bulgaria. Arch. Virol. 1979;60:329–340. doi: 10.1007/BF01317504. [DOI] [PubMed] [Google Scholar]
- Coleman J.R., Papamichail D., Skiena S., Futcher B., Wimmer E., Mueller S. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;27:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto T., Chikahira M., Yoshida S., Ebira H., Hasegawa A., Totsuka A., Nishio O. Outbreak of central nervous system disease associated with hand, foot, and mouth disease in Japan during the summer of 2000: detection and molecular epidemiology of enterovirus 71. Microbiol. Immunol. 2002;46:621–627. doi: 10.1111/j.1348-0421.2002.tb02743.x. [DOI] [PubMed] [Google Scholar]
- Gu W.J., Zhou T., Ma J.M., Sun X., Lu Z.H. Analysis of synonymous codon usage in SARS coronavirus and other viruses in the Nidovirales. Virus Res. 2004;101:155–161. doi: 10.1016/j.virusres.2004.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara A., Yoneyama T., Takami S., Hashimoto I. Genetic and phenotypic characteristics of enterovirus 71 isolates from patients with encephalitis and with hand, foot and mouth disease. Arch. Virol. 1984;79:273–283. doi: 10.1007/BF01310816. [DOI] [PubMed] [Google Scholar]
- Herrero L.J., Lee C.S., Hurrelbrink R.J., Chua B.H., Chua K.B., McMinn P.C. Molecular epidemiology of enterovirus 71 in peninsular Malaysia, 1997–2000. Arch. Virol. 2003;148:1369–1385. doi: 10.1007/s00705-003-0100-2. [DOI] [PubMed] [Google Scholar]
- Ho M., Chen E.R., Hsu K.H., Twu S.J., Chen K.T., Tsai S.F., Wang J.R., Shih S.R. An epidemic of enterovirus 71 infection in Taiwan. N. Engl. J. Med. 1999;341:929–935. doi: 10.1056/NEJM199909233411301. [DOI] [PubMed] [Google Scholar]
- Hsu B.M., Chen C.H., Wan M.T. Genetic diversity of epidemic enterovirus 71 strains recovered from clinical and environmental samples in Taiwan. Virus Res. 2007;126:69–75. doi: 10.1016/j.virusres.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Huang S.C., Hsu Y.W., Wang H.C., Huang S.W., Kiang D., Tsai H.P., Wang S.M., Liu C.C., Lin K.H., Su I.J., Wang J.R. Appearance of intratypic recombination of enterovirus 71 in Taiwan from 2002 to 2005. Virus Res. 2008;131:250–259. doi: 10.1016/j.virusres.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Jenkins G.M., Holmes E.C. The extent of codon usage bias in human RNA virus and its evolutionary origin. Virus Res. 2003;92:1–7. doi: 10.1016/s0168-1702(02)00309-x. [DOI] [PubMed] [Google Scholar]
- Jolliffe I.T. 2nded. Springer-Verlag, Inc.; New York: 2002. Principal Component Analysis. [Google Scholar]
- Karlin S., Mrázek J. What drives codon choices in human genes? J. Mol. Biol. 1996;262:459–472. doi: 10.1006/jmbi.1996.0528. [DOI] [PubMed] [Google Scholar]
- Karlin S., Blaisdell B.E., Schachtd G.A. Constrasts in codon usage of latent versus productive genes of Epstein-Barr virus: data and hypotheses. J. Virol. 1990;64:4264–4273. doi: 10.1128/jvi.64.9.4264-4273.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Doerfler W., Cardon L.R. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J. Virol. 1994;68:2889–2897. doi: 10.1128/jvi.68.5.2889-2897.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar A.A. A pause for thought along the co-translational folding pathway. Trends Biochem. Sci. 2009;34:16–24. doi: 10.1016/j.tibs.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Lesnik T., Solomovici J., Deana A., Ehrlich R., Reiss C. Ribosome traffic in E. Coli and regulation of gene expression. J. Theor. Biol. 2000;202:175–185. doi: 10.1006/jtbi.1999.1047. [DOI] [PubMed] [Google Scholar]
- Levin D.B., Whittome B. Codon usage in nucleopolyhedroviruses. J. Gen. Virol. 2000;81:2313–2325. doi: 10.1099/0022-1317-81-9-2313. [DOI] [PubMed] [Google Scholar]
- Lewis-Rogers N., Bendall M.L., Crandall K.A. Phylogenetic relationships and molecular adaptation dynamics of human rhinoviruses. Mol. Biol. Evol. 2009;26:969–981. doi: 10.1093/molbev/msp009. [DOI] [PubMed] [Google Scholar]
- Li L., He Y., Yang H., Zhu J., Xu X., Dong J., Zhu Y., Jin Q. Genetic characteristics of human enterovirus 71 and Coxsackievirus A16 circulating from 1999 to 2004 in Shenzhen, People's Republic of China. J. Clin. Microbiol. 2005;43:3835–3839. doi: 10.1128/JCM.43.8.3835-3839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., He Y., Hong Y., Zhu J. Genetic characteristics of human enterovirus 71 and cosackievirus A16 circulating from 1999 to 2004 in Shenzhen, People's Republic of China. J. Clin. Microbiol. 2005;43:3835–3839. doi: 10.1128/JCM.43.8.3835-3839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.C., Tsen H.W., Wang S.M., Wang J.R., Su I.J. An outbreak of enterovirus 71 infection in Taiwan, 1998: epidemiologic and clinical manifestations. J. Clin. Virol. 2000;17:23–30. doi: 10.1016/s1386-6532(00)00068-8. [DOI] [PubMed] [Google Scholar]
- Liu Y.S., Zhou J.H., Chen H.T., Ma L.N., Ding Y.Z., Wang M., Zhang J. Analysis of synonymous codon usage in porcine reproductive and respiratory syndrome virus. Infect. Genet. Evol. 2010;10:797–803. doi: 10.1016/j.meegid.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardia K.V., Kent J.T., Bibby J.M. Academic Press; New York: 1979. Multivariate Analysis. [Google Scholar]
- McMinn P.C. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol. Rev. 2002;26:91–107. doi: 10.1111/j.1574-6976.2002.tb00601.x. [DOI] [PubMed] [Google Scholar]
- Mueller S., Papamichail D., Coleman J.R., Skiena S., Wimmer E. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J. Virol. 2006;80:9687–9696. doi: 10.1128/JVI.00738-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M.S., Maher K., Kilpatrik D.R., Pallansch M.A. Molecular evolution of the human enterovirus: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 1999;73:1941–1948. doi: 10.1128/jvi.73.3.1941-1948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M.S., Maher K., Kilpatrick D.R., Flemister M.R., Brown B.A., Pallansch M.A. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 1999;37:1288–1293. doi: 10.1128/jcm.37.5.1288-1293.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintó R.M., Aragonè L., Costafreda M.I., Ribes E., Bosch A. Codon usage and replicative strategies of hepatits A virus. Virus Res. 2007;127:158–163. doi: 10.1016/j.virusres.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Sánchez G., Bosch A., Pintó R.M. Genome variability and capsid structural constraints of Hepatitis A virus. J. Virol. 2003;77:452–459. doi: 10.1128/JVI.77.1.452-459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S.A., Herrero L.J., McPhie K., Chow S.S., Craig M.E., Dwyer D.E., Rawlison W., McMinn P.C. Molecular epidemiology of enterovirus 71 over two decades in an Australian urban community. Arch. Virol. 2006;151:1003–1013. doi: 10.1007/s00705-005-0684-9. [DOI] [PubMed] [Google Scholar]
- Sharp P.M., Tuohy T.M.F., Mosurski K.R. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986;14(13):5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T., Gursel M., Takeshita F., Coban C., Jacqueline C., Kaisho T., Akira S., Klinman D.M., Ishii K.J. CpG RNA: identification of novel single-stranded RNA that stimulates human CD14+ CD11c+ monocytes. J. Immunol. 2005;174:2273–2279. doi: 10.4049/jimmunol.174.4.2273. [DOI] [PubMed] [Google Scholar]
- Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Yoke-Fun C., AbuBakar S. Phylogenetic evidence for inter-typic recombination in the emergence of human enterovirus 71 subgenotypes. BMC Microbiol. 2006;6:74. doi: 10.1186/1471-2180-6-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Zhang Q., Liu X.L., Wang X.M., Zhang H.L., Wu Y., Jiang F. Analysis of synonymous codon usage in 11 Human Bocavirus isolates. Biosystems. 2008;92:207–214. doi: 10.1016/j.biosystems.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z.H., He P.J., Caueffield D., Neumann M., Specter S., Baker C.C., Bankowski M.J. Enterovirus 71 isolated from China is serologically similar to the prototype EV71 BrCr strain but differs in the 5′-noncoding region. J. Med. Virol. 1995;47:161–167. doi: 10.1002/jmv.1890470209. [DOI] [PubMed] [Google Scholar]
- Zhi N., Wan Z., Liu X., Wong S., Kim D.J., Young N.S., Kajigaya S. Codon optimization of human parvovirus B19 capsid genes greatly increases their expression in nonpermissive cells. J. Virol. 2010;84:13059–13062. doi: 10.1128/JVI.00912-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J.C., Li Y.M., Zhao S., Liu S., Zhang Z. Mutation pressure shapes codon usage in the GC-Rich genome of foot-and-mouth disease virus. Virus Genes. 2007;35:767–776. doi: 10.1007/s11262-007-0159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Liu W.J., Peng S.H., Sun X.Y., Frazer I. Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. J. Virol. 1999;73:4972–4982. doi: 10.1128/jvi.73.6.4972-4982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.H., Zhang J., Chen H.T., Ma L.N., Liu Y.S. Analysis of synonymous codon usage in foot-and-mouth disease. Vet. Res. Commun. 2010;34:393–404. doi: 10.1007/s11259-010-9359-4. [DOI] [PubMed] [Google Scholar]
- Zhou T., Gu W.J., Ma J.M., Sun X., Lu Z.H. Analysis of synonymous codon usage in H5N1 virus and other influenza A viruses. Biosystems. 2005;81:77–86. doi: 10.1016/j.biosystems.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Zhou T., Sun X., Lu Z.H. Synonymous codon usage in environmental Chlamydia UWE25 reflects an evolution divergence from pathogenic chlamydiae. Gene. 2006;368:117–125. doi: 10.1016/j.gene.2005.10.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


