Abstract
Among the many medical applications of systems biology, we contend that infectious disease is one of the most important and tractable targets. We take the view that the complexity of the immune system is an inevitable consequence of its evolution, and this complexity has frustrated reductionist efforts to develop host-directed therapies for infection. However, since hosts vary widely in susceptibility and tolerance to infection, host-directed therapies are likely to be effective, by altering the biology of a susceptible host to induce a response more similar to a host who survives. Such therapies should exert minimal selection pressure on organisms, thus greatly decreasing the probability of pathogen resistance developing.
A systems medicine approach to infection has the potential to provide new solutions to old problems: to identify host traits that are potentially amenable to therapeutic intervention, and the host immune factors that could be targeted by host-directed therapies. Furthermore, undiscovered sub-groups with different responses to treatment are almost certain to exist among patients presenting with life-threatening infection, since this population is markedly clinically heterogeneous. A major driving force behind high-throughput clinical phenotyping studies is the aspiration that these subgroups, hitherto opaque to observation, may be observed in the data generated by new technologies. Subgroups of patients are unlikely to be static – serial clinical and biological phenotyping may reveal different trajectories through the pathophysiology of disease, in which different therapeutic approaches are required.
We suggest there are two major goals for systems biology in infection medicine: (1) to identify subgroups of patients that share treatable features; and, (2) to integrate high-throughput data from clinical and in vitro sources in order to predict tractable therapeutic targets with the potential to alter disease trajectories for individual patients.
Keywords: Transcriptomics, Genomics, Endotypes, Treatable traits, Infectious disease
Highlights
-
•
High throughput technologies can reveal clinical patterns in infection that were previously opaque.
-
•
Host-targeted therapies have conceptual advantages but are difficult to develop.
-
•
Key clinically-relevant objectives are identification of disease endotypes and treatable traits.
-
•
Mechanistic understanding will reveal opportunities for drug design, repurposing and better targeting.
Introduction
Box 1: Terminology.
| Term | Definition |
|---|---|
| Host-directed therapy | Therapeutic intervention to modulate an aspect of the host response to infection to alter the biology of a susceptible host to induce a response more similar to a host who survives. |
| Clinical syndrome | A collection of clinical symptoms and signs that tend to occur together. Depth of characterisation is limited by the range of observations available. |
| Disease | A clinical syndrome for which at least some of the underlying pathophysiological processes are thought to be known. |
| Subgroup | A smaller set within any population of patients, who are linked by some clinical feature or group of features. |
| Endotype | A subgroup within a population of patients who are distinguished by a shared disease process. |
| Treatable trait | The pathophysiological feature (or, in a looser sense, a biomarker or group of biomarkers for that feature) that determines whether a given therapy will improve a given patient's outcome. The same trait may be present in many different clinical syndromes or disease processes. |
Infection is the largest single cause of death in humans worldwide and many infectious agents provide relevant in vitro model systems that are both amenable to study with high-throughput techniques, and recapitulate key events in disease pathogenesis. In this review, we consider how systems biology approaches may be leveraged to address the major unmet needs in infection medicine in the 21st century, with the aim of improving outcomes for patients with infection. In clinical practice we are unable to therapeutically modulate the host immune response to infection, largely due to its inevitable complexity. Despite this, we contend that host-directed therapies have a high probability of success, since there is already considerable innate variation in host responses to infectious disease, ranging from extreme susceptibility, to complete resistance, and tolerance. Infectious diseases are survivable if you have the right genetics. The challenge is to make the same diseases survivable for patients who would otherwise succumb.
A systems medicine approach to infection has the potential to combine and integrate relevant signals from clinical, genomic, transcriptomic, proteomic and pathogen biology data to draw inferences about disease pathogenesis. Below we discuss examples of aspects of this approach applied to various infectious diseases, and suggest future goals for the application of systems biology to infection medicine.
Unmet needs for treating patients with infection
More than 70 years after the discovery of penicillin [1], this same drug is still a prominent weapon in our antibacterial armamentarium. More broadly, the concept underlying this therapeutic approach - attempting to eradicate the pathogen from a patient's body using antimicrobial drugs - remains the only effective treatment. Although spectacularly successful, the focus on the pathogen has two limitations.
Firstly, death frequently occurs in infectious disease despite effective antimicrobial therapy. Alongside the direct effects of microbial virulence factors, tissue damage is also caused by the host immune response. Immune-mediated damage leading to respiratory, cardiovascular and renal failure (sepsis) continues even after eradication of the pathogen [2]. At present, no treatments exist to modify these deleterious aspects of the host immune response.
Secondly, antimicrobial resistance threatens to liberate pathogens from the range of our solitary weapon against them. Unless something changes, deaths from infection are predicted to soar, overtaking malignant disease even in developed countries by 2050 [3].
Therapies to modulate the host response to infection would have the theoretical advantage that, in addition to promoting survival in the presence of effective antimicrobials, a host-targeted therapy may exert a less powerful selection pressure on pathogens, and may be more difficult for a pathogen to evolve to overcome. In our view, the development of such therapies is well-suited to the application of systems approaches.
Inevitable complexity of the immune system
The human immune system is arguably the most complicated organ system in the body, encompassing numerous effectors, inter-related feedback loops and extensive redundancy. This complexity is unsurprising when considering that our immune system has evolved in the face of microbial virulence factors that directly interfere with regulatory and effector mechanisms. Examples of microbial interference with host immune mechanisms are numerous and diverse. For example, one of the first innate immune mechanisms encountered by many pathogens is phagocytosis, which serves to both prime the adaptive response and eliminate invading pathogens by intracellular killing. Pathogenic Yersinia species, a group of facultative intracellular pathogens, encode a type three secretion system to directly inject effector proteins into the host cell cytoplasm, modulating the cytoskeleton to prevent phagocytosis, and inducing apoptosis of immune cells and blocking the MAPK and NF-κB pathways to reduce cytokine production [4]. Another bacterium, Pseudomonas aeruginosa, secretes a protease that cleaves a host protein (corticosteroid-binding globulin) to release the corticosteroid hormone cortisol at the site of initial infection, incapacitating the local innate immune response [5]. Even the relatively tiny genome of the influenza A virus encodes a protein (NS1) which is non-essential for replication and seems to be dedicated to interfering with both the induction and action of the host antiviral interferon response by sequestering viral dsRNA, preventing activation of RIG-1 signalling and inhibiting protein kinase R and OAS/RNase L [6].
The adaptive response, mediated by T and B lymphocytes, is also a target. The human immunodeficiency virus encodes three proteins that each down-regulate cell surface MHC-1 expression by distinct mechanisms, preventing MHC-I signalling to activate the cytotoxic T-cell response to virus-infected cells [7]. To prevent B-cells mounting an antibody response to infection, the Staphylococcus aureus surface protein A binds to the Fc-γ portion of antibodies [8].
These examples cover a few of the mechanisms pathogens have evolved to extensively interfere with the host immune response. The animal innate immune system is thought to have evolved over 1000 million years, starting with amebae able to phagocytose external material for nutrition [9]. The adaptive immune system in mammals is thought to have arisen 500 million years ago in fish [10]. Since these initial events, the immune system in each host species has participated in a genetic arms race, evolving alongside relentless exposure to these microbial immune interference strategies from innumerable pathogens. Furthermore, the immune system must successfully distinguish self from non-self antigens, with deleterious consequences arising (i.e. autoimmune disease) when this fails. The requirement to overcome these microbial immune interference strategies whilst preserving the recognition of self-antigens has provided the necessary pressure to drive the human immune system to evolve into a hugely complex organ system. In the context of this inevitable complexity, it is no surprise that reductionist approaches to development of host-targeted therapies in infectious disease have largely failed. A systems medicine approach may offer significant advantages.
It is reasonable to expect that as-yet undiscovered therapeutic interventions could alter the host immune response to promote survival. We infer this from the simple fact that some hosts do better than others when confronted with the same pathogen: the host response to infection is, in all cases that we know to have been studied, heterogeneous. Furthermore, much of this variation is heritable [11]. We conclude that host factors exist that promote survival from specific infections, and that these must vary between individuals, and hence that it should be possible to identify and utilise these factors therapeutically to alter the biology of a susceptible host to induce a response more similar to a host who survives. The scale of the challenge of finding these targets is such that it is hard to imagine a solution being found without the power of systems approaches [12].
Conceptually, this could involve promoting resistance to (suppressing pathogen replication), or tolerance of (preventing damage associated with immune response to pathogen), an infecting pathogen [13]. Although theoretically attractive as a tool to limit damage to the host [14], inducing tolerance is not without potential dangers: a sustained high pathogen load could facilitate transmission (iatrogenic super-shedders) and, perhaps more worryingly in emerging zoonotic infections, provide the time required for the selection of mutants with better host adaptation.
Current failings in targeting the host
Due to their reliable and broad acting anti-inflammatory effects, corticosteroids represent an intuitively attractive strategy to reduce inflammation during infection. Indeed, a survival benefit has been demonstrated in a small number of uncommon infections (bacterial meningitis, tuberculous meningitis and pericarditis, hypoxaemic Pneumocystis jiroveci pneumonia) [15]. In contrast, there is uncertainty over safety and benefit in other infections (e.g. RSV bronchiolitis) and clear evidence of harm in others (viral hepatitis, cerebral malaria, influenza virus, SARS coronavirus, HIV-associated Cryptococcal meningitis) 15, 16, 17, 18, 19.
A more specific host-directed therapy, recombinant human activated protein C (rhAPC), was licensed for treatment of severe sepsis based on the results of a single clinical trial [20]. rhAPC has anti-inflammatory and anti-thrombotic properties, and circulating levels are low in patients with sepsis [21]. Sadly, a subsequent trial in an overlapping patient group showed a trend towards increased mortality [22]. A third trial, mandated by the regulatory authorities, did not detect any mortality benefit, and the drug was quickly withdrawn from the market [23].
These negative trial results in sepsis are not necessarily conclusive. The heterogeneity of the host response, and the diverse range of microbes involved, means that amongst all patients with sepsis (itself an unrealistically broad syndrome) there are likely to be numerous biologically-different sub-groups, with distinct immune responses and more importantly, different risk: benefit balance for a given therapy. Corticosteroids, for example, may save some patients but harm others. Our current understanding of infection does not allow clinical differentiation of these sub-groups, so potentially beneficial therapies may be falsely rejected in clinical trials. We believe that a major output of systems approaches to infection will be the elucidation of previously hidden therapeutically-important sub-groups of patients who share a ‘treatable trait’ (i.e. response to therapy).
High-dimensionality data from patients
We have discussed two central problems in infection medicine. Firstly, the development of therapies to modulate the host immune response is impeded by the inevitable complexity of the human immune system. Secondly, the range and degree of characterisation of clinical syndromes in medicine is constrained by the range of observations that are available; we likely fail to identify sub-groups of patients with treatable traits due to a paucity of relevant observations. The promise of systems approaches to infection medicine is that new technologies may provide solutions to these old problems. Large-scale biological data-sets from an increasing range of high-throughput technologies are becoming available, including (but not limited to) high-resolution transcriptional profiling [24], mass spectrometry-based proteomics [25] and metabolomics, genome-scale CRISPR-Cas9 knockout screening [26], and whole genome sequencing/genome-wide association studies. These modalities could provide new, biologically important observations that have not previously been observed in patients. It is very likely that some of these observations will be directly relevant to clinical management – the challenge will be to identify the ones that matter.
There are three broad categories of clinical utility for these new data sources in infection medicine:
-
•
identifying treatable traits and therapeutically-relevant subgroups;
-
•
identifying new therapeutic targets in the immune response; and,
-
•
improving prognostication.
The value of prognostication in current clinical practice is, in our view, limited by the range of therapeutic options. Put simply, it is only useful to predict the future if you have some capacity to change it. Particularly in the case of acute and immediately life-threatening infection, our range of therapeutic options is very limited, regardless of the degree of certainty attached to the prognosis, therefore we suggest the first two applications should be prioritised. In contrast, the identification of syndromes (collections of clinical observations that tend to occur in patients suffering the same disease) has been the primary mechanism for progress in the understanding of human disease since long before the time of Hippocrates [27]. Finding a syndrome is the first step towards identifying the common biological processes that define a disease, and ultimately to identifying effective treatments. We hope that the application of systems technologies will help us define treatable traits in infection, thus providing a starting point for new therapeutic interventions (Figure 1 ).
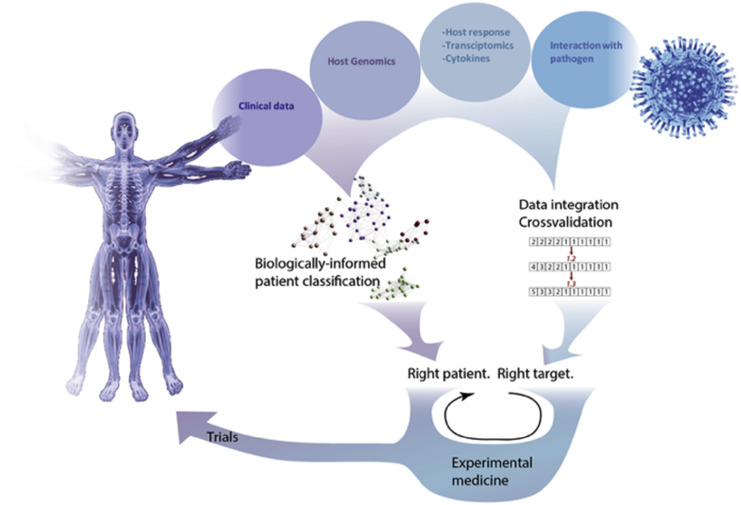
Figure 1.
Summary of a systems medicine approach to infection. A wide range of data sources can be combined using various methods (see text) to achieve two fundamental goals – clinically-informative phenotyping of patients, and identification of therapeutic targets.
Treatable traits and therapeutically-relevant subgroups
Importantly, a systems model of the disease process need not predict every transcript and metabolite in the massively multi-dimensional datasets generated by new technologies. The primary challenge is instead to identify those components of inter-host variation that are amenable to intervention; the evidence of disease processes that we can change. This is Agusti's concept of “treatable traits” [28], a term coined in the field of chronic obstructive lung disease but no less relevant here.
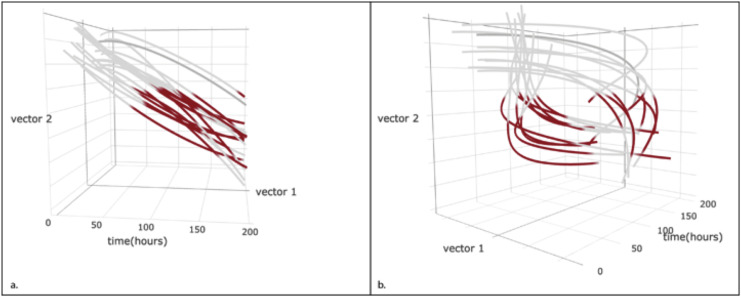
A computational model of infection could therefore include not only traditional measures of pathogen burden and evidence of systemic injury [29], but also independent components of the immune response or the metabolic consequences, detected using high-throughput technologies. The behaviour of the whole system may be impenetrably complex, but the components required to predict the effect of an intervention may be far simpler. The trajectory followed by each patient along an informative set of vectors may reveal different groups of patients that appear clinically similar, and may even have similar outcomes, but different disease processes (Figure 2 ). By mapping the “flight path” of each patient through disease, we anticipate that important similarities and differences in immune response will become apparent [29].
Figure 2.
Hypothetical trajectories of two groups of patients through multidimensional space. Each line indicates the path taken by a single patient, with periods of organ failure highlighted in red. A superficially similar group of patients may appear clinically indistinguishable (a), but different trajectories through illness are revealed by informative vectors derived from high-throughput data (b). It is reasonable to expect that such biological differences in disease process will underlie different responses to host-directed therapies.
Our ability to identify groups of patients sharing therapeutically-relevant similarities is dependent on measuring the relevant biological signal that determines classification. This is the fundamental attraction of high-throughput technologies – the probability of measuring important signals is greater if more signals are measured. That such groupings of patients, or disease endotypes, exist is already clear: therapeutically-important sub-classifications have recently been discovered that redefine the clinical syndromes of asthma 30, 31, ARDS [32], and acute mountain sickness [33]. In two related autoimmune conditions, ANCA-associated vasculitis (AAV) and systemic lupus erythematosus (SLE), a T-cell gene expression signature clearly delineates two distinct endotypes [34]. Subsequent work elucidated the immunological process underlying this sub-classification, CD8 T-cell exhaustion [35]. This process is associated with better outcomes in autoimmune disease, but poor clearance of viral infection. In the future it may be possible to manipulate this pathway therapeutically in patients with AAV or SLE, to prevent relapse, or in the opposite direction in patients with chronic viral infection, to promote clearance.
Therapeutic targets
Even in the best-case scenario, the distance between the identification of a tractable therapeutic target and successful exploitation in clinical practice is substantial, so it is no surprise that potential host-directed therapies discovered through high-dimensionality analytics have not yet been proven to be effective in clinical trials. Nonetheless there are some promising leads, a few of which are described here. These exemplify different approaches: integrating cell culture, animal and human data sequentially to identify a host anti-viral factor (influenza virus); and using computational predictions from transcriptional data with proteomic and genetically modified animal studies to identify host pathways involved in pathogenesis (SARS Coronavirus).
Influenza virus
Viruses are obligate parasites and undergo exclusively intracellular replication; properties that make them ideal for study in cell culture where host factors that affect viral replication can be expected to do the same in vivo. siRNA screening has been used as a genome-wide approach to identify such host factors for influenza virus infection. IFITM3 was identified as a novel host anti-viral factor by siRNA screening with confirmatory in vitro work (including exogenous interferon administration and stable IFITM3 expression) demonstrating it is required for an effective interferon response to inhibit influenza virus replication [36]. Work in Ifitm3–/– mice was then undertaken, confirming that in vivo, influenza virus-infected mice suffer fatal viral pneumonia, even when infected with a low-pathogenicity virus [37]. The GenISIS/MOSAIC groups identified a single nucleotide polymorphism (rs12252-C) within the IFITM3 coding region in humans that was strongly over-represented in patients hospitalised with influenza virus infection [37], the majority of whom required invasive ventilation. Meta-analysis of genomic studies of IFITM3 and influenza virus infection has confirmed this association between SNP rs12252-C and increased susceptibility to infection in humans [38]. Work is now underway in many groups to investigate the impact of IFITM proteins in antiviral defence, with a view to generating host-targeted antiviral therapies. This example highlights the potential of a systems-wide approach, integrating and cross-validating results from various experimental modalities (high-throughput screening in cell culture, followed by targeted studies in mice and humans) to converge from large-scale data onto a single critical host immune factor.
SARS coronavirus
Acute lung injury (ALI) and progression to acute respiratory distress syndrome are major features of the pathophysiology of SARS Coronavirus (and indeed other respiratory virus) infection. Pathological changes in mice are very similar to humans and there is a dose-response relationship between viral inoculum and the severity of ALI. Host responses to the virus that result in ALI are poorly understood. To investigate host responses associated with more severe acute lung injury in a murine model of SARS Coronavirus (SARS-CoV) infection, transcriptomic profiles of mice infected with lethal and sub-lethal doses of virus were compared and correlated with pathological data at multiple time points, then subject to bioinformatic network analysis [39]. A module of genes involved in cell adhesion, extracellular matrix remodelling and wound healing was significantly up-regulated in the lethal infection model, and components of the urokinase pathway were found to be the most enriched and differentially regulated. Mass-spectrometry proteomic analysis was then used to explore this transcriptional data further, demonstrating that SARS-CoV infection resulted in increased expression of fibrin β and γ chains, factor VIII and cytokeratins (all components of hyaline membranes), and reduced expression of surfactant proteins in the lung. These data are consistent with histological post-mortem findings in SARS-CoV infected humans, where extensive fibrin exudate, extensive hyaline membrane formation and alveolar collapse have been observed [40]. Serpine1 is part of the urokinase pathway and contributes to ECM remodelling. To confirm this finding from transcriptional and proteomic studies, Serpine1–/– knockout mice were infected with SARS-CoV and found to have a worse outcome compared to wild type mice. This systems-based approach to SARS-CoV, sequentially applying transcriptional, proteomic then genetic modification techniques, demonstrates that the urokinase pathway contributes to ALI, thus identifying a target for future experimental medicine work to determine if it can be therapeutically altered to benefit the host.
Conclusion
These examples give us confidence that we can expect more progress along similar lines as the potential of systems medicine approaches becomes more widely appreciated, as expertise in computational methodologies grows, and as the cost of generating relevant data falls.
Physicians have long made progress by recognising patterns of similar observations in groups of patients, and by determining which biological features of disease are amenable to therapy. What has changed is the unprecedented rate of advance in new resources and tools with which to tackle the ancient problems of diagnosis and therapy in infectious disease. Our responsibility is to ensure a similar acceleration in clinical progress.
This review comes from a themed issue on Cancer and systemic diseases (2017)
Edited by Dan Landau and Luonan Chen
References
- 1.Lawrie R. First clinical use of penicillin. Br Med J (Clin Res Ed) 1985;290:397. doi: 10.1136/bmj.290.6465.397-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angus D.C., van der Poll T. Severe sepsis and septic shock. N Engl J Med. 2013;369:840–851. doi: 10.1056/NEJMra1208623. [DOI] [PubMed] [Google Scholar]
- 3.O'Neill J. vol. 2016. 2016. (Tackling drug-resistant infections globally: final report and recommendations). Edited by: Review on antimicrobial resistance. [Google Scholar]
- 4.Cornelis G.R. The Yersinia Ysc-Yop 'type III' weaponry. Nat Rev Mol Cell Biol. 2002;3:742–752. doi: 10.1038/nrm932. [DOI] [PubMed] [Google Scholar]
- 5.Simard M., Hill L.A., Underhill C.M., Keller B.O., Villanueva I., Hancock R.E., Hammond G.L. Pseudomonas aeruginosa elastase disrupts the cortisol-binding activity of corticosteroid-binding globulin. Endocrinology. 2014;155:2900–2908. doi: 10.1210/en.2014-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hale B.G., Randall R.E., Ortin J., Jackson D. The multifunctional NS1 protein of influenza A viruses. J Gen Virol. 2008;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 7.Kamp W., Berk M.B., Visser C.J., Nottet H.S. Mechanisms of HIV-1 to escape from the host immune surveillance. Eur J Clin Invest. 2000;30:740–746. doi: 10.1046/j.1365-2362.2000.00697.x. [DOI] [PubMed] [Google Scholar]
- 8.Falugi F., Kim H.K., Missiakas D.M., Schneewind O. Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. MBio. 2013;4 doi: 10.1128/mBio.00575-13. e00575-00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchmann K. Evolution of innate immunity: clues from invertebrates via fish to mammals. Front Immunol. 2014;5:459. doi: 10.3389/fimmu.2014.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flajnik M., Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •11.Sorensen T.I., Nielsen G.G., Andersen P.K., Teasdale T.W. Genetic and environmental influences on premature death in adult adoptees. N Engl J Med. 1988;318:727–732. doi: 10.1056/NEJM198803243181202. [DOI] [PubMed] [Google Scholar]; This prospective cohort study of adoptees demonstrated that genetic background contributes more to the relative risk of death due to infection (i.e. biologic parent also died of infection) than for death due to cancer, vascular disease or natural causes, thus affirming that susceptibility to infection is a strongly heritable trait.
- 12.Baillie J.K. Translational genomics. Targeting the host immune response to fight infection. Science. 2014;344:807–808. doi: 10.1126/science.1255074. [DOI] [PubMed] [Google Scholar]
- •13.Raberg L., Sim D., Read A.F. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science. 2007;318:812–814. doi: 10.1126/science.1148526. [DOI] [PubMed] [Google Scholar]; This study utilises a murine malaria model to demonstrate that resistance to and tolerance of infection represent genetically distinct host defence mechanisms.
- 14.Medzhitov R., Schneider D.S., Soares M.P. Disease tolerance as a defense strategy. Science. 2012;335:936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGee S., Hirschmann J. Use of corticosteroids in treating infectious diseases. Arch Intern Med. 2008;168:1034–1046. doi: 10.1001/archinte.168.10.1034. [DOI] [PubMed] [Google Scholar]
- 16.Blum C.A., Nigro N., Briel M., Schuetz P., Ullmer E., Suter-Widmer I., Winzeler B., Bingisser R., Elsaesser H., Drozdov D. Adjunct prednisone therapy for patients with community-acquired pneumonia: a multicentre, double-blind, randomised, placebo-controlled trial. Lancet. 2015;385:1511–1518. doi: 10.1016/S0140-6736(14)62447-8. [DOI] [PubMed] [Google Scholar]
- 17.Beardsley J., Wolbers M., Kibengo F.M., Ggayi A.B., Kamali A., Cuc N.T., Binh T.Q., Chau N.V., Farrar J., Merson L. Adjunctive dexamethasone in HIV-associated cryptococcal meningitis. N Engl J Med. 2016;374:542–554. doi: 10.1056/NEJMoa1509024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodrigo C., Leonardi-Bee J., Nguyen-Van-Tam J., Lim W.S. Corticosteroids as adjunctive therapy in the treatment of influenza. Cochrane Database Syst Rev. 2016;3:Cd010406. doi: 10.1002/14651858.CD010406.pub2. [DOI] [PubMed] [Google Scholar]
- 19.Stockman L.J., Bellamy R., Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3:e343. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernard G.R., Vincent J.L., Laterre P.F., LaRosa S.P., Dhainaut J.F., Lopez-Rodriguez A., Steingrub J.S., Garber G.E., Helterbrand J.D., Ely E.W. Efficacy and safety of recombinant human activated protein C for severe sepsis. N Engl J Med. 2001;344:699–709. doi: 10.1056/NEJM200103083441001. [DOI] [PubMed] [Google Scholar]
- 21.Dettenmeier P., Swindell B., Stroud M., Arkins N., Howard A. Role of activated protein C in the pathophysiology of severe sepsis. Am J Crit Care. 2003;12:518–524. quiz 525–516. [PubMed] [Google Scholar]
- 22.Baillie J.K., Murray G. Drotrecogin alfa (activated) in severe sepsis. N Engl J Med. 2006;354:94–96. author reply 94–96. [PubMed] [Google Scholar]
- 23.Ranieri V.M., Thompson B.T., Barie P.S., Dhainaut J.F., Douglas I.S., Finfer S., Gardlund B., Marshall J.C., Rhodes A., Artigas A. Drotrecogin alfa (activated) in adults with septic shock. N Engl J Med. 2012;366:2055–2064. doi: 10.1056/NEJMoa1202290. [DOI] [PubMed] [Google Scholar]
- 24.Lizio M., Harshbarger J., Shimoji H., Severin J., Kasukawa T., Sahin S., Abugessaisa I., Fukuda S., Hori F., Ishikawa-Kato S. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015;16:22. doi: 10.1186/s13059-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aebersold R., Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- •26.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Genome editing using the CRISPR-Cas9 system can be applied to genome-wide knockout screening to identify genes with an important phenotype in an in vitro model, as exemplified in this study of genes required for cell viability in cancer and genes implicated in susceptibility to a therapeutic RAF inhibitor used in melanoma.
- 27.Schiefsky M.J. Brill; Leiden, Boston: 2005. Hippocrates on ancient medicine. [Google Scholar]
- 28.Agusti A. Phenotypes and disease characterization in chronic obstructive pulmonary disease. Toward the extinction of phenotypes? Ann Am Thorac Soc. 2013;10:S125–S130. doi: 10.1513/AnnalsATS.201303-055AW. [DOI] [PubMed] [Google Scholar]
- 29.Lough G., Kyriazakis I., Bergmann S., Lengeling A., Doeschl-Wilson A.B. Health trajectories reveal the dynamic contributions of host genetic resistance and tolerance to infection outcome. Proc Biol Sci. 2015:282. doi: 10.1098/rspb.2015.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinks T., Zhou X., Staples K., Dimitrov B., Manta A., Petrossian T., Lum P., Smith C., Ward J., Howarth P. Multidimensional endotypes of asthma: topological data analysis of cross-sectional clinical, pathological, and immunological data. Lancet. 2015;385(Suppl 1):S42. doi: 10.1016/S0140-6736(15)60357-9. [DOI] [PubMed] [Google Scholar]
- 31.Lotvall J., Akdis C.A., Bacharier L.B., Bjermer L., Casale T.B., Custovic A., Lemanske R.F., Jr., Wardlaw A.J., Wenzel S.E., Greenberger P.A. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol. 2011;127:355–360. doi: 10.1016/j.jaci.2010.11.037. [DOI] [PubMed] [Google Scholar]
- 32.Calfee C.S., Delucchi K., Parsons P.E., Thompson B.T., Ware L.B., Matthay M.A. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med. 2014;2:611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •33.Hall D.P., MacCormick I.J., Phythian-Adams A.T., Rzechorzek N.M., Hope-Jones D., Cosens S., Jackson S., Bates M.G., Collier D.J., Hume D.A. Network analysis reveals distinct clinical syndromes underlying acute mountain sickness. PLoS One. 2014;9:e81229. doi: 10.1371/journal.pone.0081229. [DOI] [PMC free article] [PubMed] [Google Scholar]; A systematic network analysis (using software originally designed for analysing microarray expression data) of symptoms associated with acute mountain sickness revealed three distinct sub-groups within the syndrome, resulting in a revision of international diagnostic criteria for what was previously considered as a single disease.
- ••34.McKinney E.F., Lyons P.A., Carr E.J., Hollis J.L., Jayne D.R., Willcocks L.C., Koukoulaki M., Brazma A., Jovanovic V., Kemeny D.M. A CD8+ T cell transcription signature predicts prognosis in autoimmune disease. Nat Med. 2010;16:586–591. doi: 10.1038/nm.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]; CD8+ T-cells were isolated from patients with ANCA-associated vasculitis and systemic lupus erythematosus. Transcriptional profiling identified two distinct sub-groups (related to T-cell survival and memory) which could then be differentiated on the basis of only three genes, providing a useful biomarker for clinical trials of immunosuppressant therapies and focussing further studies on these pathways.
- 35.McKinney E.F., Lee J.C., Jayne D.R., Lyons P.A., Smith K.G. T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature. 2015;523:612–616. doi: 10.1038/nature14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brass A.L., Huang I.C., Benita Y., John S.P., Krishnan M.N., Feeley E.M., Ryan B.J., Weyer J.L., van der Weyden L., Fikrig E. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••37.Everitt A.R., Clare S., Pertel T., John S.P., Wash R.S., Smith S.E., Chin C.R., Feeley E.M., Sims J.S., Adams D.J. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]; Data was integrated from cell culture work, a murine infection model, human genotyping and patient clinical variables to validate IFITM3 as a critical host defence against influenza virus infection and thus a tractable therapeutic target.
- 38.Yang X., Tan B., Zhou X., Xue J., Zhang X., Wang P., Shao C., Li Y., Li C., Xia H. Interferon-inducible transmembrane protein 3 genetic variant rs12252 and influenza susceptibility and severity: a meta-analysis. PLoS One. 2015;10:e0124985. doi: 10.1371/journal.pone.0124985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gralinski L.E., Bankhead A., 3rd, Jeng S., Menachery V.D., Proll S., Belisle S.E., Matzke M., Webb-Robertson B.J., Luna M.L., Shukla A.K. Mechanisms of severe acute respiratory syndrome coronavirus-induced acute lung injury. MBio. 2013:4. doi: 10.1128/mBio.00271-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu J., Gong E., Zhang B., Zheng J., Gao Z., Zhong Y., Zou W., Zhan J., Wang S., Xie Z. Multiple organ infection and the pathogenesis of SARS. J Exp Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]