Abstract
Neonatal calf diarrhea remains one of the most important problems faced by livestock, causing great economic losses. This study investigated the prevalence of Salmonella and Escherichia coli, especially enterotoxigenic E. coli (ETEC), in diarrheic calves. Fecal samples were collected from 127 diarrheic calves up to 3 months of age at 12 farms from different governorates in Egypt. 119 bacterial isolates (93.7%) were recovered and the prevalences of Salmonella and E. coli in diarrheic calves were 18.1% and 75.6%, respectively. Serotyping of Salmonella isolates revealed that S. Enteritidis and S. Typhimurium were the most prevalent serotypes, representing 60.9% and 30.4%, respectively, while S. Dublin was 8.7%. Serogrouping of E. coli isolates showed that 10 O-serogroups were obtained where O26 and O103 were the most prevalent (17.7% of each). Salmonella serotypes showed positive results with PCR test using oligonucleotide primer amplifying 521 bp fragment of invA gene of Salmonella while 70% of E. coli serogroups possessed ETEC virulent gene (K99). The in-vitro antibiotic sensitivity test indicated that Salmonella serotypes showed high sensitivity against enrofloxacin, spectinomycin and neomycin while E. coli isolates showed high sensitivities against marbofloxacin, spectinomycin and neomycin only.
Keywords: Neonatal calf diarrhea (NCD), Salmonella, enterotoxigenic E. coli (ETEC)
1. Introduction
Neonatal calf diarrhea (NCD) is one of the most common diseases in young animals, causing huge economic and productivity losses to bovine industry worldwide (Cho and Yoon, 2014). In 2007, The National Animal Health Monitoring System (NAHMS) for U.S. dairy reported that 57% of unweaned calf mortality was due to diarrhea especially in calves less than one month old (USDA Dairy, 2007). In Egypt, NCD continues to be the 1st cause of calf mortality, which ranges between 27.4% and 55% of the total deaths in young calves (Ahmed, 1980). The economic losses occur not only from mortality but also from other costs including treatment, diagnostics, labor, veterinary intervention and decreased number of herd replacements as well as subsequent chronic ill thrift and impaired growth performance (Bazeley, 2003).
NCD is a multifactorial syndrome including pathogen (infectious NCD) as well as non-infectious factors related to the animal (immunological and nutritional status), the environment or the management (Izzo et al., 2011). Because of the multifactorial nature of NCD, it is difficult to be controlled effectively (Cho and Yoon, 2014).
Infectious diarrhea is the most significant cause of morbidity and mortality in neonatal dairy calves throughout the world and it can be caused by many pathogens including viruses (coronavirus and rotavirus), protozoa (Cryptosporidium parvum) and bacteria (Izzo et al., 2011). Among bacteria, enterotoxigenic Escherichia coli (ETEC) and Salmonellae are the most economically important pathogens (Achá et al., 2004), although other bacteria have also been identified as causes of enteric disease and NCD, e.g. Campylobacter species (Myers et al., 1984) and Clostridium species (Cho et al., 2010). Although co-infection is considerably observed in diarrheic calves, sometimes single infection is recorded. Farm geographic location, management practices, as well as herd size affect considerably the prevalence of the pathogen. (Cho and Yoon, 2014). Identification of the possible causative agent in outbreaks of diarrhea is important to allow targeted preventative measures, such as vaccination, and identification of possible risk factors or sources of infection (Izzo et al., 2011).
Salmonella enterica colonizes the digestive of both adult cattle and calves, but the infection is often recorded in the first 3 months of age and often causing severe symptoms (Fossler et al., 2005). S. enterica serovar typhimurium (S. Typhimurium) and serovar dublin (S. Dublin) are the most common causative agents of salmonellosis in cattle causing acute and systemic diarrheal diseases, respectively (Cho and Yoon, 2014). Diarrhea due to Salmonella infection is watery and mucoid with the presence of blood and fibrin (Fossler et al., 2005). Calves can shed Salmonella for variable periods of time and intermittently depending on the degree of infection (Cho and Yoon, 2014). Furthermore, after the disappearance of the organism from the intestinal tract, up to 5% of recovered animals may become carriers shedding the organism in their feces (Jay, 2000). Infected cattle and carriers can serve as source of infection for other animals or even human through food-borne routes or direct contact and so the determination of Salmonella strains in fecal samples is not only important for the diagnosis of salmonellosis, but also essential to identify the carriers (Warnick et al., 2003).
Among E. coli pathogroups, the most common cause of NCD is ETEC stains that produce the K99 (F5) adhesion antigen (E. coli K99+) and heat-stable (STa or STb) and/or heat-labile (LT1 or LT2) enterotoxins (Kaper et al., 2004). E. coli causes a watery diarrhea and weakness in 1–4 day old newborn calves. Death usually occurred within 24 hours due to severe dehydration (Cho et al., 2010). The fimbrial adhesion F5 (K99) promotes the adhesion of bacterial cells to glycoproteins on the epithelial surface of the jejunum and/or ileum, and bacterial enterotoxin also causes damage to the epithelial cells, resulting in fluid secretion and diarrhea (Acres, 1985).
The present study aimed to investigate the prevalence of Salmonella and E. col, especially ETEC, in diarrheic calves in Egypt.
2. Material and methods
2.1. Samples
Fecal samples were collected every 4 weeks for a period of 5 months from the rectum of 127 cross-breed diarrheic calves up to 3 months of age at 12 farms from different governorates in Egypt along the period of April to November 2013. All samples were transferred in an ice box to the laboratory with minimal delay for bacteriological examination.
2.2. Bacteriological examination
One gram of each fecal sample was diluted in 3 mL of sterile saline. Samples were cultured and identified according to Quinn et al. (2002).
For isolation of Salmonella strains, a loopful from the diluted specimens was inoculated into selenite F broth with overnight incubation at 37 °C. Then, a loopful was streaked out onto MacConkey's agar, xylose lysine deoxycholate and Salmonella–Shigella agar media and incubated at 37 °C for 18–24 hours. Suspected colonies were subjected to biochemical testing according to Collee et al. (1996).
For isolation of E. coli strains, a loopful from the diluted specimens was inoculated into MacConkey's agar and incubated at 37 °C for 18–24 hours. Lactose fermenter (pink) colonies were streaked onto and eosin methylene blue agar and confirmed as E. coli using the standard biochemical tests according to Collee et al. (1996).
2.3. Serological identification
2.3.1. Serotyping of Salmonella
Salmonella serovars were identified serologically by slide agglutination test using diagnostic polyvalent and monovalent O and H Salmonella antisera according to Kauffman–White scheme (Kauffmann, 1974).
2.3.2. Serogrouping of E. coli isolates
E. coli serogroups were identified serologically by slide agglutination test using standard polyvalent and monovalent E. coli antisera according to Edwards and Ewing (1972).
2.4. Polymerase chain reaction
PCR was applied on 3 Salmonella isolates representing all the recovered serotypes as well as 10 E. coli isolates representing all the recovered serogroups. In case of Salmonella, oligonucleotide primers that amplify a 521 bp fragment for the invA (invasion A) gene in Salmonella serovars was used. Meanwhile, in case of E. coli, oligonucleotide primers that amplify a 314 bp fragment for the K99 (F5) gene in E. coli species was used (Table 1 ).
Table 1.
Primers used in PCR.
| Gene | Primer Sequence 5′-3′ | Size of product | Reference |
|---|---|---|---|
|
invA(F) invA(R) |
TTGTTACGGCTATTTTGACCA CTGACTGCTACCTTGCTGATG |
521 bp | Swamy et al. (1996) |
|
F5(F) F5(R) |
TATTATCTTAGGTGGTATGG GGTATCCTTTAGCAGCAGTATTTC |
314 bp | Frank et al. (1998) |
2.5. Antibiotic susceptibility testing
All Salmonellae (23 isolates) and E. coli (96 isolates) recovered from diarrheic calves were tested for their antimicrobial susceptibility to 12 different antimicrobial discs including enrofloxacin (10 µg), marbofloxacin (10 µg), gentamycin (10 µg), erythromycin (15 µg), cefotaxime sodium (30 µg), amoxicillin (10 µg), penicillin (10 µg), tetracycline (30 µg), streptomycin (10 µg), trimethoprim-sulphamethoxazol (1.25 + 23.75 µg), spectinomycin (20 µg) and neomycin (20 µg) (Oxoid, Basing Stoke, UK). Antimicrobial susceptibility testing was performed using disc diffusion method on Muller Hinton agar according to Clinical and Laboratory Standards Institute (CLSI) (2012). The antibiotic susceptibility was based on the induced inhibition zones according to the guidelines of the CLSI (2012).
3. Results
3.1. Bacterial isolation
Out of 127 collected fecal samples from diarrheic calves, 119 (93.7%) bacterial isolates were recovered, including 23 (18.1%) Salmonella serovars and 96 (75.6%) E. coli strains (Table 2 ).
Table 2.
Prevalence of Salmonella and E. coli in diarrheic calves.
| No. of samples | Salmonella | E. coli | Total | Negative isolation | ||||
|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | |
| 127 | 23 | 18.1 | 96 | 75.6 | 119 | 93.7 | 8 | 6.3 |
%: was calculated according to the number of the collected samples.
3.2. Serological identification
3.2.1. Serotyping of Salmonella isolates
The 23 Salmonella isolates were serotyped as 14 S. Enteritidis (60.9%), 7 S. Typhimurium (30.4%) and 2 S. Dublin (8.7%).
3.2.2. Serogrouping of E. coli isolates
Out of 96 E. coli isolates, 10 O-serogroups were identified. The serogroups O26 and O103 were the most prevalent representing 17 isolates (17.7%) for each followed by serogroups O127; 14 isolates (14.6%) and O119; 13 isolates (13.6%). While the serogroups O86, O111 and O157 represented 5 isolates (5.2%) for each, meanwhile O44 and O158 serogroups represented 4 isolates (4.2%) and finally serogroup O78; 3 isolates (3.1%). Moreover, there were 9 (9.4%) isolates were untyped with the available antisera.
3.3. PCR results
3.3.1. PCR of Salmonellaserovars
PCR results revealed that all the tested isolates confirmed morphologically, biochemically and serologically as being Salmonella showed positive results with PCR assay with oligonucleotide primer that amplified a 521 bp fragment of invA gene (Fig. 1 ).
Fig. 1.
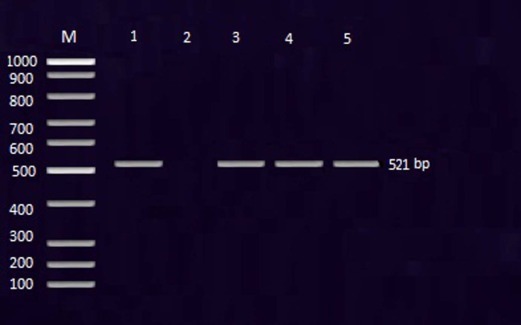
The PCR amplification of invA gene of Salmonella serovars showing positive amplicons at 521 bp. DNA size marker (M). Lane 1: positive control (S. Typhimurium strain). Lane 2: negative control. Lane 3: S. Enteritidis. Lane (4): S. Typhimurium. Lane (5): S. Dublin.
3.3.2. PCR of E. coli serogroups
PCR results revealed that out of the tested 10 E. coli serogroups, 7 serogroups (70%) possessed ETEC virulence gene (K99) including O26, O44, O78, O86, O111, O119 and O127 while O103, O157 and O158 not possessed K99 gene (Fig. 2 ).
Fig. 2.

The PCR amplification of K99 virulence gene of E. coli showing positive amplicons at 314 bp. M: DNA size marker (100–1000 bp). Lane 1: Negative control. Lane 2: Positive control (E. coli serogroup O86). Lane (3–12): The tested serogroups (O26, O44, O78, O86, O103, O111, O119, O127, O157 and O158, respectively).
3.4. Antibiotic susceptibility testing
Results of in-vitro sensitivity tests of Salmonella isolates against 12 antimicrobial agents revealed that S. Typhimurium and S. Enteritidis isolates showed high sensitivities against marbofloxacin, enrofloxacin, trimethoprim-sulphamethoxazol, spectinomycin and neomycin. On the other hand, high resistances were observed against amoxicillin, gentamycin, cefotaxime sodium, streptomycin, erythromycin, tetracycline and amoxicillin. Meanwhile, S. Dublin isolates were sensitive to enrofloxacin, cefotaxime sodium, penicillin, tetracycline, streptomycin, spectinomycin and neomycin. On the contrary, they were resistant to marbofloxacin, gentamycin, erythromycin, amoxicillin and trimethoprim-sulphamethoxazol. Concerning E. coli isolates, the different serogroups as well as the untyped group showed different degrees of sensitivity against the tested antibiotics. Mostly, E. coli isolates showed high resistances against the majority of antimicrobials, meanwhile, high sensitivity was observed against marbofloxacin, spectinomycin and neomycin only.
4. Discussion
Neonatal calf diarrhea remains as one of the most important problems faced by livestock, causing great economic losses. Calves are at greatest risk of diarrhea within the first month of life and the incidence of diarrhea decreases with age (Garcia et al., 2000).
The multifactorial nature of NCD makes this disease hard to control effectively. Therefore, prevention and control of such disease must be based on a good understanding of those disease complexities during the calving period before disease outbreaks (Cho and Yoon, 2014). Identification of the possible causative agent in outbreaks of diarrhea is important to allow targeted preventative measures, such as vaccination, and identification of possible risk factors or sources of infection (Izzo et al., 2011). E. coli and Salmonella are the most common identified pathogens in scouring calves less than 2 months of age (Achá et al., 2004). Their prevalences vary by geographical location of the farms, farm management practices, and herd size (Cho and Yoon, 2014).
In the present study, the prevalences of E. coli and Salmonella in neonatal diarrheic calf were 18.1% and 75.6%, respectively (Table 2). The negative bacterial isolation in some fecal samples (6.3%) did not mean absence of microbial infection but may attribute to infection with bacteria requiring specific or enriched culture media and can't grow on the used culture media such as Clostridium species (Cho et al., 2010) and Campylobacter species (Myers et al., 1984) or viral infection; requiring tissue culture, or even protozoal infection. Fricker (1987) elucidated that bacteria were not detectable due to the number of colony forming units are below the detection limit of the assay (especially in case of Salmonella in feces). Additionally, the presence of antibiotic residues may explain falsely negative bacteriological results because the withdrawal time is not regarded in our herds.
The prevalence of Salmonella in the present study was similar to those of other studies in Egypt (Riad et al., 1998: 18.2%; Seleim et al., 2004: 17.5%; and Youssef and El-Haig, 2012: 18.66%) while higher than that reported by Haggag and Khaliel (2002) (4%), and Younis et al. (2009) (4.09%). On contrary, this result was much lower than that reported by Moussa et al. (2010) (43.53%). Varying prevalences of salmonellosis in calves were recorded in diarrheic calves in African countries such as Mozambique (5%; Achá et al., 2004) and Algeria (66.6% in calves at the end of the first month; Akam et al., 2004), and other countries such as India (5%; Joon and Kaura, 1993) and Australia (23.8%; Izzo et al., 2011). Differences of the prevalence rates of Salmonella in diarrheic calves in comparison to the previous studies could be explained in the light of species and geographical locations and hygienic measures, and these factors significantly influence the prevalence of salmonellosis in calves (Younis et al., 2009).
The prevalence of E. coli in the current study had nearly coincided with the findings of Awad et al. (1979: 80%) and Haggag and Khaliel (2002: 82%), while higher than those of Azzam et al. (2006: 5.4%), El-Shehedi et al. (2013: 35.83%), Osman et al. (2013: 63.6%), and Hassan (2014: 50%), and lower than that obtained by Ibrahim (1995: 100%). In comparison to other countries, the present results were similar to that of Hemashenpagam et al. (2009) in India (75%), while it was higher than those obtained by Joon and Kaura (1993) in India (23%), Viring et al. (1993) in Sweden (11.5%), Khan and Khan (1997) in Pakistan (54%), Steiner et al. (1997) in Germany (42%), Bendali et al. (1999) in France (20.3%), Valdivia-Andy et al. (2000) in Mexico (63.7%), Oporto et al. (2008) in Northern Spain (35.9%), and Izzo et al. (2011) in Australia (17.4%). On contrary, this result was lower than that of Pourtaghi et al. (2013) in Iran (86.7%). The differences of the prevalence rates of E. coli in diarrheic calves may be attributed also to the geographical locations and management practice as well as hygienic measures where ETEC infection occurs mainly through ingestion of contaminated food or water (Cho and Yoon, 2014).
In the current study, serotyping of Salmonella isolates revealed that S. Enteritidis (SE) and S. Typhimurium (ST) were the most prevalent serotypes with 60.9% and 30.4%, respectively, while S. Dublin (SD) represented 8.7%. Salmonella serovars isolation frequencies vary from one location to another due to different management and hygienic measures as well as geographical, environmental and individual variations (Veling et al., 2002). The predominance of ST and SE serovars among diarrheic calves detected in the present study was supported by many previous reports in Egypt (Seleim et al, 2004, Younis et al, 2009, Moussa et al, 2010 and Youssef and El-Haig, 2012). In addition; this finding substantiated the reports from the other countries (Murray, 1994, in Australia, and Smith-Palmer et al., 2003, in Scotland).
S. Typhimurium and S. Enteritidis are non-host-adapted serovars and so they are less likely to establish a “carrier state” in the recovered animals. Infection with a non-host-adapted Salmonella results in transient shedding of organisms (3–16 weeks). The most common sources of infection are contaminated food and water. Approximately 40% of all animal-origin feed additives (bone or fish meals) are contaminated with Salmonella. Human sewage has also been tracked down as a source of infection in some herd outbreaks. Rodents, birds, and other animals spread infection through their feces and their carcasses (Donaldson et al., 2006). On contrary, S. Dublin is host-adapted in cattle, and commonly produces a carrier state in the recovered animals. It can survive intracellularly inside the mononuclear phagocytic cells in the face of high serum neutralizing antibody titers. Intermittent shedding occurs, which is maximized by stress factors. In outbreaks of S. Dublin infection, carriers are the most likely source. These asymptomatic carriers can shed organisms in their feces and milk for the remainder of their lives. Calves, which are fed raw, contaminated milk from the bulk tank, can be infected by this route (Lance et al., 1992).
Serogrouping of E. coli isolates showed that 10 O-serogroups were identified; of them O26 and O103 were the most prevalent groups (17.7% each) followed by O127 (14.6%) and O119 (13.6%). These results run hand to hand with those of Ibrahim (1972) and Abd-Elrahman (2013). The present findings were also nearly similar to that obtained by El-Shehedi et al. (2013) and Osman et al. (2013). Concerning the other countries, Tamaki et al. (2005) in Japan found that the most common E. coli serotypes isolated from diarrheic fecal samples of calves were O119, O111, O126, and O78. Similarly, Badouei et al. (2010) recovered O157:H7, O111, and O26 serotypes from diarrheic and non-diarrheic calves and the most common serogroups was O26 (18.4%).
Salmonella invA gene has become one of the most popular PCR target sequences (Rahn et al., 1992) and its amplification now has been recognized as an international standard for detection of Salmonella (Malorny et al., 2003). The invA gene encodes a protein in the inner membrane of bacteria, which is responsible for invasion to the epithelial cells of the host (Darwin and Miller, 1999).
In the present study, PCR was applied on 3 isolates of Salmonella representing the recovered serovars to determine the virulence invA gene. The results revealed that all the tested isolates of Salmonella showed positive results with PCR assay using oligonucleotide primer that amplified a 521 bp fragment of invA gene of Salmonella. These results were similar to those obtained by Soliman (2014), who determined the virulence invA gene in all recovered Salmonella serogroups from bovine and ovine; S. Typhimurium, S. Enteritidis, S. Virchow, S. Montividio and S. Rubislow, by PCR.
Concerning E. coli, PCR was applied on 10 isolates representing all the recovered serogroups. Results revealed that 7 serogroups (70%) possessed ETEC virulent gene (K99) including O26, O44, O78, O86, O111, O119 and O127 while O103, O157 and O158 not possessed K99 gene. Although Sherwood et al. (1983) confirmed the close correlation between enterotoxigenicity and the K99 antigen presence and Pourtaghi et al. (2013) reported that most bovine ETEC produce K99 fimbriae, although Moon et al. (1976) have reported K99 antigen in non-enterotoxigenic E. coli.
Despite the increased availability of vaccines against ETEC and other pathogens associated with NCD and continued emphasis on optimizing colostral transfer of passive immunity, improved treatment protocols for calf diarrhea are necessary. Although the administration of intravenous fluids and oral electrolyte solutions plays the main role in treatment, the efficacy of antimicrobial drugs in treating calf diarrhea is argumentative (Constable, 2004). Diarrheic calves are more likely to have failure or partial failure of passive transfer, and so they are more likely to be bacteraemic (Constable, 2004) and this is an additional cause that antimicrobial agents might be indicated in the treatment of calf diarrhea. The type of antibiotic drug should better be selected on the basis of its sensitivity, which could be detected by laboratory examination, and the antimicrobial treatment of diarrheic calves should therefore be focused against bacteria in the two sites of infection: the small intestine and the blood (Constable, 2004).
In the present study, the results of in-vitro antibiotic sensitivity test showed that the different serotypes of Salmonella showed different degrees of sensitivity against the tested antibiotics but in general they all showed high sensitivities against enrofloxacin, spectinomycin and neomycin. On contrary, they were resistant to gentamycin, erythromycin and amoxicillin. On the other hand, the different E. coli serogroups as well as the untyped group showed different degrees of sensitivity against the tested antibiotics. Mostly, E. coli isolates showed high resistances against the majority of antimicrobials, meanwhile, high sensitivities were observed against marbofloxacin, spectinomycin and neomycin only. The high occurrence of resistance can be predicted since a large proportion of the animals are probably treated with antimicrobial drugs (de Verdier et al., 2012). Call et al. (2008) discussed the epidemiology of resistant E. coli in calves as multifactorial, complex and e.g. influenced by co-selection due to linkage of resistance genes. Walk et al. (2007) supposed that regardless of use of antimicrobials, antibiotic resistance in E. coli is co-selected in calves by an unknown “beneficial mutation.”
5. Conclusion
Neonatal calf diarrhea remains one of the most important problems faced by livestock, causing great economic losses. The prevalences of Salmonella and E. coli in diarrheic calves were 18.1% and 75.6%, respectively. S. Enteritidis and S. Typhimurium were the most prevalent Salmonella serotypes while 70% of E. coli was ETEC.
References
- Abd-Elrahman A.H. Colibacillosis in newly born buffalo calves and role of lacteol fort in preventing recurrence of calf diarrhea. Life Sci J. 2013;8(4) [Google Scholar]
- Achá S.J., Kuhn I., Jonsson P., Mbazima G., Katouli M., Mollby R.S. Studies on calf diarrhoea in Mozambique: prevalence of bacterial pathogens. Acta Vet Scand. 2004;45:27–36. doi: 10.1186/1751-0147-45-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acres S.D. Enterotoxigenic Escherichia coli infections in newborn calves: a review. J Dairy Sci. 1985;68:229–256. doi: 10.3168/jds.S0022-0302(85)80814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A.A. 1980. Calf sources in Egyptian buffalo-cows; pp. 19–21. Egyptian German Seminar on the Mortality of Newly-born Calves. [Google Scholar]
- Akam A., Khelef D., Kaidi R., Othmani A., Lafri M., Tali-Maamar H. Frequency of Cryptosporidium parvum, Escherichia coli K99 and Salmonella spp. isolated from healthy and unhealthy calves in six breeding farms from Mitidja, Algeria (preliminary results) Rev Sci Parasitol. 2004;5:13–21. [Google Scholar]
- Awad F.I., Farrag I., Shawkat M.E., Abeid M.H. Studies on enterotoxaemia in young buffalo calves. Egypt J Vet Sci. 1979;14(1):24–29. [Google Scholar]
- Azzam R.A., Hassan W.H., Ibrahim M.A., Khaled M.S. Prevalence of verocytotoxigenic E. coli O157: H7 in cattle and man in Beni-Sueif Government. Alex J Vet. 2006;24(1):111–122. [Google Scholar]
- Badouei M.A., Salehi T.Z., Khorasgani M.R., Tadjbakhsh H., Brujeni G.N., Nadalian M.G. Virulence gene profiles and intimin subtypes of Shiga toxin-producing Escherichia coli isolated from healthy and diarrhoeic calves. Vet Rec. 2010;167(22):858–861. doi: 10.1136/vr.c4009. [DOI] [PubMed] [Google Scholar]
- Bazeley K. Investigation of diarrhoea in the neonatal calf. In Pract. 2003;25:152–159. [Google Scholar]
- Bendali F., Bichet H., Schelcher F., Sanaa M. Pattern of diarrhoea in new born calves in South west France. Vet Res. 1999;30:61–74. [PubMed] [Google Scholar]
- Call D.R., Davis M.A., Sawant A.A. Antimicrobial resistance in beef and dairy cattle production. Anim Health Res Rev. 2008;9:159–167. doi: 10.1017/S1466252308001515. [DOI] [PubMed] [Google Scholar]
- Cho Y., Yoon K.J. An overview of calf diarrhea-infectious etiology, diagnosis, and intervention. J Vet Sci. 2014;15(1):1–17. doi: 10.4142/jvs.2014.15.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y., Kim W., Liu S., Kinyon J.M., Yoon K.J. Development of a panel of multiplex real-time polymerase chain reaction assays for simultaneous detection of major agents causing calf diarrhea in feces. J Vet Diagn Invest. 2010;22:509–517. doi: 10.1177/104063871002200403. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) Approved Standard – 11th ed. vol. 32. 2012. (Performance standards for antimicrobial disk susceptibility tests). M02-A11. [Google Scholar]
- Collee J.G., Fraser A.G., Marmion B.P., Simmons A. 14th ed. 1996. Practical medical microbiology. [Google Scholar]
- Constable P.D. Review: antimicrobial use in the treatment of calf diarrhea. J Vet Intern Med. 2004;18:8–17. doi: 10.1111/j.1939-1676.2004.tb00129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Verdier K., Nyman A., Greko C., Bengtsson B. Antimicrobial resistance and virulence factors in Escherichia coli from Swedish dairy calves. Acta Vet Scand. 2012;54:2. doi: 10.1186/1751-0147-54-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin K.H., Miller V.L. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin Microbiol Rev. 1999;12:405–428. doi: 10.1128/cmr.12.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson S.C., Straley B.A., Hegde N.V., Sawant A.A., DebRoy C., Jayarao B.M. Molecular epidemiology of ceftiofur-resistant Escherichia coli: isolates from dairy calves. Appl Environ Microbiol. 2006;72(6):3940–3948. doi: 10.1128/AEM.02770-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards P.R., Ewing N.H. 3rd ed. Burgeon Publishing, Co.; Atlanta, GA: 1972. Identification of enterobacteriaceae; pp. 208–339. [Google Scholar]
- El-Shehedi M.A., Eraqi M.M., Ali A.R. Characterization of Escherichia coli from diarrheic calves with special reference to plasmid profile. J Am Sci. 2013;9(7) [Google Scholar]
- Fossler C.P., Wells S.J., Kaneene J.B., Ruegg P.L., Warnick L.D., Bender J.B. Herd-level factors associated with isolation of Salmonella in a multi-state study of conventional and organic dairy farms II. Salmonella shedding in calves. Prev Vet Med. 2005;70:279–291. doi: 10.1016/j.prevetmed.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Frank S.M., Bosworth B.T., Moon H.W. Multiplex PCR for enterotoxigenic, attaching and effacing, and Shiga toxin-producing Escherichia coli strains from calves. J ClinMicrobiol. 1998;36:1795–1797. doi: 10.1128/jcm.36.6.1795-1797.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker C.R. The isolation of salmonellas and campylobacters. J ApplBacteriol. 1987;63:99–116. doi: 10.1111/j.1365-2672.1987.tb02692.x. [DOI] [PubMed] [Google Scholar]
- Garcia A., Ruiz-Santa-Quiteria J.A., Orden J.A., Cid D., Sanz R., Gómez-Bautista M. Rotavirus and concurrent infections with other enteropathogens in neonatal diarrheic dairy calves in Spain. Comp ImmunolMicrobiol Infect Dis. 2000;23:175–183. doi: 10.1016/S0147-9571(99)00071-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggag Y.N., Khaliel S.A. Public health importance of certain bacteria isolated from calves and small ruminants. 2nd Vet Cong, Fac Vet Med, Minufiya Univ, Egypt. 2002;2(1):173–184. [Google Scholar]
- Hassan A.M. Some studies on bacteriological causes of enteritis in calves. J Vet Adv. 2014;4(5):503–507. [Google Scholar]
- Hemashenpagam N., Kiruthiga B., Selvaraj T., Panneerselvam A. Isolation, identification and characterization of bacterial pathogens causing calf diarrhea with special reference to Escherichia coli. Internet J Microbiol. 2009;7(2) [Google Scholar]
- Ibrahim A.M. 1995. Sanitary studies on newly born calves. Ph. D. thesis (Animal Hygiene); Faculty of Veterinary Medicine, Suez Canal University, Egypt. [Google Scholar]
- Ibrahim M.S. 1972. Bacteriological studies of colibacillosis in calves. M.V.Sc. thesis (Microbiology); Faculty of Veterinary Medicine, Cairo University, Egypt. [Google Scholar]
- Izzo M.M., Kirkland P.D., Mohler V.L., Perkins N.R., Gunna A.A., House J.K. Prevalence of major enteric pathogens in Australian dairy calves with diarrhea. Aust Vet J. 2011;89(5):167–173. doi: 10.1111/j.1751-0813.2011.00692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay J.M. Modern food microbiology. 2000. Foodborne gastroenteritis caused by Salmonella and Shigella; pp. 511–528. [Google Scholar]
- Joon D.S., Kaura Y.K. Isolation and characterization of same of the enterobacteria from diarrhoeic and non-diarrhoeic calves. Ind J Anim Sci. 1993;63:373–383. [Google Scholar]
- Kaper J.B., Nataro J.P., Mobley H.L. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Kauffmann F. Kauffmann White scheme WHO-BD/72, 1, Rev. 1. Acta Path Microbiol Scand. 1974;61:385–387. [Google Scholar]
- Khan A., Khan M. Bacteria isolated from natural cases of buffalo and bovine neonatal calf diarrhoea, pneumonia and pneumoenteritis. Vet Arch. 1997;67(4):161–167. [Google Scholar]
- Lance S.E., Miller G.Y., Hancock D.D., Bartlett P.C., Heider L.E. Salmonella infections in neonatal dairy calves. J Am Vet Assoc. 1992;201(6):864–868. [PubMed] [Google Scholar]
- Malorny B., Tassios P.T., Rådström P., Cook N., Wagner M., Hoorfar J. Standardization of diagnostic PCR for the detection of food borne pathogens. Int J Food Microbiol. 2003;83:39–48. doi: 10.1016/s0168-1605(02)00322-7. [DOI] [PubMed] [Google Scholar]
- Moon H.W., Whipp S.C., Skartvedt S.M. Etiologic diagnosis of diarrheal diseases of calves: frequency and methods for detecting enterotoxin and K99 production by Escherichia coli. Am J Vet Res. 1976;37:1025–1029. [PubMed] [Google Scholar]
- Moussa I.M., Ashgan M.H., Mahmoud M.H., Mohamed K.F.H., Al-Doss A.A. Rapid detection of Salmonella species in new-borne calves by polymerase chain reaction. Int J Gen Mol Biol. 2010:62–66. [Google Scholar]
- Murray C.J. Salmonella serovars and phage types in humans and animals in Australia 1987–1992. Aust Vet J. 1994;71:78–81. doi: 10.1111/j.1751-0813.1994.tb03332.x. [DOI] [PubMed] [Google Scholar]
- Myers L.L., Firehammer B.D., Border M.M., Shop D.S. Prevalence of enteric pathogens in the feces of healthy beef calves. Am J Vet Res. 1984;45:1544–1548. [PubMed] [Google Scholar]
- Oporto B., Esteban J.I., Aduriz G., Juste R.A., Hurtado A. Escherichia coli O157:H7 and non- O157 Shiga toxin-producing E. coli in healthy cattle, sheep and swine herds in Northern Spain. Zoonoses Pub Health. 2008;52:411–550. doi: 10.1111/j.1863-2378.2007.01080.x. [DOI] [PubMed] [Google Scholar]
- Osman K.M., Mustafa A.M., Elhariri M., Abdelhamed G.S. The distribution of Escherichia coli serovars, virulence genes, gene association and combinations and virulence genes encoding serotypes in pathogenic E. coli recovered from diarrhoeic calves, sheep and goat. Transbound Emerg Dis. 2013;60(1):69–78. doi: 10.1111/j.1865-1682.2012.01319.x. [DOI] [PubMed] [Google Scholar]
- Pourtaghi H., Dahpahlavan V., Momtaz H. Virulence genes in Escherichia coli isolated from calves with diarrhoea in Iran. Comp ClinPathol. 2013;22:513–515. [Google Scholar]
- Quinn P.J., Markey B.K., Carter M.E., Donnelly W.J.C., Leonard F.C., Maguire D. Blackwell; 2002. Veterinary microbiology and microbial disease; pp. 113–116. [Google Scholar]
- Rahn K., Grandis S.A., Clarke R.C., McEwen S.A., Galan J.E., Ginocchio C. Amplification of an invA gene sequence of Salmonella Typhimurium by polymerase chain reaction as specific method of detection of Salmonella. Mol Cell Probes. 1992;6:271–279. doi: 10.1016/0890-8508(92)90002-f. [DOI] [PubMed] [Google Scholar]
- Riad E.M., Tanios A.I., El-Moghny A.F.A. Antigen capture ELISA technique for rapid detection of Salmonella typhimurium in fecal samples of diarrheic cow calves. Assiut Vet Med J. 1998;39(78):324–333. [Google Scholar]
- Seleim R.S., Sahar R.M., Novert M.H., Gobran R.A. Salmonella infection in calves: virulence proteins and its immunogenic properties. J Vet. 2004 http://www.priory.com/vet/salmonella.htm online. [Google Scholar]
- Sherwood D., Snodgrass D.R., Lawson G.H.K. Prevalence of enterotoxigenic Escherichia coli in calves in Scotland and northern England. Vet Rec. 1983;113:208–212. doi: 10.1136/vr.113.10.208. [DOI] [PubMed] [Google Scholar]
- Smith-Palmer A., Stewart W.C., Mather H., Greig A., Cowden J.M., Reilly W.J. Epidemiology of Salmonella enterica serovars enteritidis and typhimurium in animals and people in Scotland between 1990 and 2001. Vet Rec. 2003;153:517–520. doi: 10.1136/vr.153.17.517. [DOI] [PubMed] [Google Scholar]
- Soliman H.A. 2014. Conventional and advanced techniques for identification of bovine and ovine salmonellae. Ph.D. thesis (Microbiology); Faculty of Veterinary Medicine, Beni-Suef University, Egypt. [Google Scholar]
- Steiner L., Busato A., Burnen S.A., Gaillard C. Frequency and aetiology of calf losses and calf diseases before weaning in cow calf farms. II. Microbiological and Parasitological diagnosis in diarrhoeic calves. Dtch Tieraztl Wochenschr. 1997;104(5):169–173. [PubMed] [Google Scholar]
- Swamy S.C., Barnhard H.M., Lee H.D., Dresen D.W. Virulence determinant invA and spvC in salmonella isolated from poultry product, waste water, and human sources. App Environ Microbiol. 1996;62:3768–3771. doi: 10.1128/aem.62.10.3768-3771.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki Y., Narimatsu H., Miyazato T., Nakasone N., Toma C., Iwanaga M. The relationship between O antigens and pathogenic genes of diarrhea- associated E. coli. Jpn J Infect Dis. 2005;58:65–69. [PubMed] [Google Scholar]
- USDA Dairy Part II: changes in the U.S. dairy cattle industry, 1991–2007. 2007. http://nahms.aphis.usda.gov/dairy/index.htm Pp. 57–61, USDA-APHIS-VS, CEAH, Fort Collins, February 2008.
- Valdivia-Andy G., Rosales C., Soriano-becerrili D.M., Alba-Hurtado F., Montaraz-Crespo J.A., Tortora-Prez J.L. Interaction of E. coli verocytotoxin strains and rotavirus in outbreak of calf's diarrhoea. Vet Mex. 2000;31:293–300. [Google Scholar]
- Veling J., Barkema H.W., van der Schans J., van Zijjderveld F., Verhoeff J. Herd level diagnosis for Salmonella enterica subsp. Enterica serovars Dublin infection in bovine dairy herds. Prev Vet Med. 2002;53:31–42. doi: 10.1016/s0167-5877(01)00276-8. [DOI] [PubMed] [Google Scholar]
- Viring S., Olsson S.O., Alenius S., Emanuelsson U., Jacobsson S.O., Larsson B. Studies of enteric pathogens and gamma-globulin levels of neonatal calves in Sweden. Acta Vet Scand. 1993;34(3):271–279. doi: 10.1186/BF03548191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walk S.T., Mladonicky J.M., Middleton J.A., Heidt A.J., Cunningham J.R., Bartlett P. Influence of antibiotic selection on genetic composition of Escherichia coli populations from conventional and organic dairy farms. Appl Environ Microbiol. 2007;73:5982–5989. doi: 10.1128/AEM.00709-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnick L.D., Kanistanon K., McDonough P.L., Power L. Effect of previous antimicrobial treatment on fecal shedding of Salmonella enterica subsp. Enterica serogroup b in New York dairy herds with recent clinical salmonellosis. Prev Vet Med (Praha) 2003;56:285–297. doi: 10.1016/s0167-5877(02)00210-6. [DOI] [PubMed] [Google Scholar]
- Younis E.E., Ahmed A.M., El-Khodery S.A., Osman S.A., El-Naker Y.F. Molecular screening and risk factors of enterotoxigenic Escherichia coli and Salmonella spp. in diarrheic neonatal calves in Egypt. Res Vet Sci. 2009;87:373–379. doi: 10.1016/j.rvsc.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef A.I., El-Haig M.M. Herd problems and occupational zoonoses of Salmonella enterica serovars Typhimurium and Enteritidis infection in diarrheic cattle and buffalo calves in Egypt. Int J Bioflux Soc. 2012;4(3):118–123. [Google Scholar]


