Abstract
Background
Human rhinoviruses (HRVs) are often concurrently detected with other viruses found in the respiratory tract because of the high total number of HRV infections occurring throughout the year. This feature has previously relegated HRVs to being considered passengers in acute respiratory infections. HRVs remain poorly characterized and are seldom included as a target in diagnostic panels despite their pathogenic potential, infection-associated healthcare expenditure and relatively unmoderated elicitation of an antiviral state.
Objectives
To test the hypothesis that respiratory viruses are proportionately more or less likely to co-occur, particularly the HRVs.
Study design
Retrospective PCR-based analyses of 1247 specimens for 17 viruses, including HRV strains, identified 131 specimens containing two or more targets. We investigated the proportions of co-detections and compared the proportion of upper vs. lower respiratory tract presentations in the HRV positive group. Both univariate contingency table and multivariate logistic regression analyses were conducted to identify trends of association among the viruses present in co-detections.
Results
Many of the co-detections occurred in patterns. In particular, HRV detection was associated with a reduced probability of detecting human adenoviruses, coronaviruses, bocavirus, metapneumovirus, respiratory syncytial virus, parainfluenza virus, influenza A virus, and the polyomaviruses KIPyV and WUPyV (p ≤ 0.05). No single HRV species nor cluster of particular strains predominated.
Conclusions
HRVs were proportionately under-represented among viral co-detections. For some period, HRVs may render the host less likely to be infected by other viruses.
Keywords: Rhinovirus, Interference, Acute respiratory tract infection, Innate immunity, Protective effect
1. Background
Acute respiratory tract infections (ARTIs) are the leading cause of morbidity in young children1 and may become one of the five most common causes of mortality in adults and children.2 Causality is often attributed to the first detection3 of one of a select group of viruses which include human respiratory syncytial virus (HRSV), metapneumovirus (HMPV), parainfluenza viruses (HPIVs) and influenza A viruses (IFAV). When assayed, human rhinoviruses (HRVs) are frequently detected in ARTIs4 often leading to confusion when the high total number of HRV co-detections is assumed to equate to an equally high proportion of HRVs in co-detections with other “respiratory viruses”. Symptomatic HRV infections are associated with the common cold syndrome and episodic expiratory wheezing early in life.5, 6, 7 Despite this, many relevant studies still do not include them during virus screening. Exposure to HRV strains begins early and continues throughout life. Unsurprisingly, the impact of HRV infections on healthcare expenditure is also significant8, 9 as is their broader role within the hygiene hypothesis. Frequent, mild infections by HRVs may be a major factor in the maturation of a robust innate and acquired antiviral immunity10 perhaps through the apparently unmoderated HRV activation of the antiviral state.11
Specific patterns have been ascribed to the seasonal and annual variations in respiratory virus epidemics with interference between viruses being a suggested reason for peak fluctuations.12, 13, 14 Interference may be a biological feature of RNA virus infections but for HRVs it was once exploited on a small scale as an in vitro diagnostic tool, whereby successful experimental HRV infection of organ cultures was indicated by blockading the replication of other, subsequently inoculated, respiratory viruses.15 However, because HRVs have been infrequently included in diagnostic testing panels, it is difficult to identify whether an interference effect may be acting on the epidemic patterns of other viruses. Recent identification of new diversity within the HRV super-group16, 17, 18, 19 has made study of these poorly characterized pathogens a particular priority.
We used statistical approaches to test the hypothesis that patterns of virus detection, whereby some viruses are more or less likely to concurrently occur, are commonplace, particularly when the HRVs are involved.
2. Methods
2.1. Clinical specimens and testing
Clinical specimens (n = 1247; 95% nasopharyngeal aspirates) were predominantly from children (59% male) with symptoms of ARTI who presented to hospitals or to general practitioners in Queensland, Australia, during all seasons of 2003.17 Nearly half the specimens (537, 43%) were from infants 0–1 year of age. Proportions of specimens from toddlers (1–2 years), children (2–14 years) and adults (over 14 years) were 19%, 22% and 16% respectively. Median and interquartile ranges for age were 1.32 and 0.5–4.3 years respectively.
To obtain a preliminary gauge of the impact of HRV detection as a cause of presentation to a hospital or clinic, HRV-positive individuals were classified according to whether they presented with upper or lower respiratory tract illness (URTI, n = 69 or LRTI n = 154). Those with chronic illness or with immunocompromise were excluded from classification.
As previously described,16, 20 specimens were screened using uniplex and multiplex conventional and real-time PCRs for 17 viruses or virus groups.
2.2. Statistical analyses
Where co-detections occurred, association between virus pairs was considered using 2 × 2 contingency tables with Fisher's exact test. Odds ratio (OR) and 95% confidence interval (CI) for the likelihood of co-detection were calculated (Table 1 ). Additionally, the observed number of co-detections of any particular pair was compared with the expected number of co-detections using the exact binomial test (Table 2 ). The expected number of co-detections was calculated as the product of prevalence of the two viruses in the study specimens multiplied by the number of specimens. Where a significant association was detected and the number of co-detections was >2, the effects of other selected viruses, adjusted for season (winter set as reference season), gender, age group (infant set as reference group) and their interactions, were examined using a separate logistic regression model with backward selection for each outcome (virus test) of interest. Results are presented as ORs with 95% CIs (Table 3 ). Differences in age according to number of co-detections and detection of HRV were compared using the Kruskal–Wallis test. Significance level was set at p = 0.05.
Table 1.
Measures of association between virus pairs where co-detections occurred as determined by 2 × 2 contingency tables with Fisher's exact test.
| HAdV | HCoV | IFAV | HBoV | HEV | HMPV | HRSV | HRV | KIPyV | HPIV | WUPyV | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total detections | 31 | 54 | 34 | 101 | 15 | 67 | 71 | 331 | 30 | 36 | 34 |
| Single detections number (% of total) | 16 (51.6%) | 39 (72.22%) | 25 (73.5%) | 39 (38.6%) | 7 (46.7%) | 51 (76.1%) | 51 (71.8%) | 250 (75.5%) | 12 (40%) | 32 (88.9%) | 9 (26.5%) |
| Co-detections number (% of total) | 15 (48.4%) | 15 (27%) | 9 (26.5%) | 62 (61.4%) | 8 (53.3%) | 16 (23.9%) | 20 (28.2%) | 78 (23.6%) | 18 (60%) | 4 (11.1%) | 25 (73.5%) |
| HAdV | 0.50 | 0.67 | 0.12 | 1.0000 | 0.06 | 0.07 | <0.0001 | 1.00 | 0.40 | 1.00 | |
| 0.36, 0.05–2.70 | 1.29, 0.29–5.63 | 2.00, 0.87–4.61 | 0.63, 0.04–10.76 | 0.13, 0.01–2.19 | 0.12, 0.01–2.05 | 0.18, 0.07–0.47 | 0.69, 0.09–5.23 | 0.26, 0.02–4.30 | 0.60, 0.08–4.550 | ||
| HCoV | 0.10 | 1.00 | 0.62 | 0.15 | 0.02 | <0.0001 | 0.16 | 0.06 | 0.10 | ||
| 0.15, 0.01–2.52 | 0.96, 0.44–2.10 | 0.35, 0.02–5.93 | 0.32, 0.08–1.34 | 0.14, 0.00–0.87 | 0.09, 0.03–0.22 | 0.17 0.01–2.87 | 0.14, 0.01–2.37 | 0.15, 0.01–2.52 | |||
| IFAV | 0.46 | 1.00 | 0.56 | 0.04 | <0.0001 | 1.00 | 0.25 | 1.00 | |||
| 0.52, 0.16–1.74 | 0.57, 0.03–9.76 | 0.54, 0.13–2.30 | 0.11, 0.00–0.92 | 0.06, 0.01–0.24 | 0.62, 0.08–4.72 | 0.23, 0.01–3.907 | 0.54, 0.07–4.10 | ||||
| HBoV | 0.49 | 0.006 | 0.004 | <0.0001 | 0.44 | 0.34 | 0.01 | ||||
| 0.38, 0.05–2.30 | 0.240.07–0.77 | 0.22, 0.07–0.72 | 0.40, 0.26–0.63 | 1.41, 0.56–3.54 | 0.49, 0.15–1.62 | 2.85, 1.34–6.04 | |||||
| HEV | 1.00 | 0.40 | 0.45 | 0.51 | 1.00 | 0.55 | |||||
| 0.63, 0.08-4.84 | 0.26, 0.01–4.38 | 0.65, 0.23–1.86 | 1.52, 0.19–11.949 | 0.54, 0.03–9.18 | 1.32, 0.17–10.38 | ||||||
| HMPV | 0.0006 | <0.0001 | 0.35 | 0.04 | 0.56 | ||||||
| 0.06, 0.003–0.88 | 0.10, 0.04–0.21 | 0.29, 0.04–2.20 | 0.11, 0.00–0.74 | 0.54, 0.13–2.30 | |||||||
| HRSV | <0.0001 | 0.55 | 0.02 | 0.16 | |||||||
| 0.17, 0.09–0.338 | 1.29, 0.44–3.81 | 0.11, 0.00–0.69 | 0.24, 0.032–1.78 | ||||||||
| HRV | 0.03 | <0.0001 | 0.30 | ||||||||
| 0.41, 0.18–0.91 | 0.03, 0.003–0.19 | 0.68, 0.34-–1.37 | |||||||||
| KIPyV | 0.40 | 0.06 | |||||||||
| 0.27, 0.01–4.45 | 3.07, 1.00–9.38 | ||||||||||
| HPIV | 1.00 | ||||||||||
| 0.51, 0.07–3.858 | |||||||||||
| WUPyV | |||||||||||
Probabilities of association by chance (p-values) are shown above the corresponding OR and 95% CI for each pair. Figures indicating significant associations (p < 0.05) are shown in bold. The number of each virus detected (n) is shown along the top. The total detections of each virus are divided into whether they occurred by themselves (single detections) or concurrently with one or more other viruses (co-detections; (> = 2 detections in one sample).
Table 2.
Observed frequency compared with expected frequency of virus co-detections using the exact binomial test.
| HAdV | HCoV | IFAV | HBoV | HEV | HMPV | HRSV | HRV | KIPyV | HPIV | WUPyV | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HAdV | 0.53 | 0.38 | 0.15 | 1.00 | 0.09 | 0.09 | 0.003 | 1.00 | 0.28 | 1.00 | |
| 1/3 | 2/1 | 8/5 | 0/1 | 0/3 | 0/3 | 5/16 | 1/1 | 0/2 | 1/1 | ||
| HCoV | 0.12 | 1.00 | 0.65 | 0.19 | 0.03 | <0.0001 | 0.12 | 0.12 | 0.12 | ||
| 0/3 | 8/9 | 0/1 | 2/5 | 1/6 | 5/27 | 0/3 | 0/3 | 0/3 | |||
| IFAV | 0.5 | 1.00 | 0.78 | 0.04 | <0.0001 | 1.00 | 0.28 | 1.00 | |||
| 3/5 | 0/1 | 2/3 | 0/4 | 2/17 | 1/1 | 0/2 | 1/2 | ||||
| HBoV | 0.73 | 0.01 | 0.01 | 0.004 | 0.48 | 0.5 | 0.02 | ||||
| 8/5 | 3/11 | 3/10 | 32/51 | 6/5 | 3/5 | 11/5 | |||||
| HEV | 1.00 | 0.65 | 0.85 | 0.48 | 1.00 | 0.14 | |||||
| 1/1 | 0/1 | 6/7 | 1/2 | 1/1 | 0/1 | ||||||
| HMPV | 0.001 | <0.0001 | 0.57 | 0.04 | 0.2 | ||||||
| 0/7 | 7/34 | 1/3 | 0/4 | 2/3 | |||||||
| HRSV | 0.001 | 0.56 | 0.02 | 0.2 | |||||||
| 12/36 | 4/3 | 0/4 | 1/4 | ||||||||
| HRV | 0.12 | <0.0001 | 0.54 | ||||||||
| 9/15 | 1/18 | 14/17 | |||||||||
| KIPyV | 0.65 | 0.04 | |||||||||
| 0/1 | 4/1 | ||||||||||
| HPIV | 1.00 | ||||||||||
| 1/2 | |||||||||||
| WUPyV |
Probability (p-values, significant associations, p < 0.05, shown in bold) of the observed number of co-detections compared with the expected number of co-detections, based on the expected frequency of co-detections were calculated as prevalence of first virus x prevalence of second virus. Observed/expected.
Table 3.
Eleven regression models examining factors that modify the probability of a positive test to other viruses.
| Outcome of interest | Factorsap < 0.05 | OR | CI | p-value |
|---|---|---|---|---|
| HBoV | HRV | 0.43 | 0.27–0.68 | 0.0003 |
| Autumn | 0.25 | 0.10–0.60 | 0.0003 | |
| Toddler | 1.78 | 1.11–2.85 | 0.02 | |
| HMPV | HRV | 0.10 | 0.04–0.22 | <0.0001 |
| ADV | HRV | 0.18 | 0.07–0.47 | 0.0005 |
| IFAV | HRV | 0.06 | 0.01–0.24 | 0.0001 |
| Child | 2.40b | 1.12–5.10 | 0.02 | |
| HPIV | HRV | 0.02 | 0.00–0.14 | 0.0001 |
| Spring | 7.25 | 2.34–22.11 | 0.0005 | |
| Summer | 8.78 | 2.50–30.94 | 0.0007 | |
| Autumn | 5.85 | 1.57–21.81 | 0.008 | |
| HRSV | HRV | 0.17 | 0.08–0.33 | <0.0001 |
| spring | 0.11 | 0.05–0.27 | <0.0001 | |
| Summer | 0.05 | 0.01–0.34 | 0.003 | |
| Toddler | 0.50 | 0.25–0.96 | 0.04 | |
| Child | 0.10 | 0.03–0.34 | 0.0002 | |
| Adult | 0.35 | 0.13–0.98 | 0.04 | |
| HCoV | HRV | 0.09 | 0.030.22 | <0.0001 |
| KIPyV | HRV | 0.43 | 0.19–0.95 | 0.04 |
| Child | 2.25 | 1.02–4.95 | 0.04 | |
| HBoV | HRSV | 0.23 | 0.07–0.76 | 0.02 |
| Autumn | 0.23 | 0.09–0.57 | 0.002 | |
| Toddler | 1.77 | 1.11–2.82 | 0.02 | |
| HBoV | HMPV | 0.23 | 0.07–0.77 | 0.02 |
| Autumn | 0.22 | 0.09–0.55 | 0.001 | |
| Toddler | 1.78 | 1.11–2.83 | 0.02 | |
| HBoV | WUPyV | 2.66 | 1.25–5.70 | 0.01 |
| Autumn | 0.23 | 0.09–0.57 | 0.001 | |
| Toddler | 1.80 | 1.12–2.86 | 0.01 |
Adjusted for gender (male is reference category), season (winter is reference category) and age group (infant 0–1 years (reference category), toddler 1–2 years, child 2–14 years).
Significant positive associations are shown in bold.
In accordance with the recommendation of Rothman21 no post-hoc adjustment for multiple comparisons was performed.
3. Results
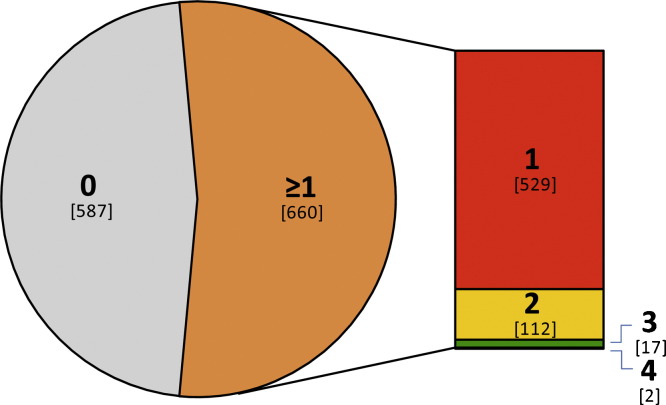
The 17 viruses sought16, 20 included some (human bocavirus, HBoV, and the polyomaviruses, KIPyV and WUPyV) with unclear aetiologies in the respiratory tract. In 660 of the specimens (52.9%), at least one virus was detected (Fig. 1 ).
Fig. 1.
Distribution of viral detections. Bold type indicates the number of viruses detected in each of the 1,247 specimen extracts. Co-detections of two or more viruses occurred in 131 of the 660 (20%) specimens in which a virus was detected.
HRVs were the most commonly detected virus group (Table 1; HRV primers from Arola et al.22, 23) and were involved in the highest total number of co-detections, being found with another virus in 78 specimens (23.6% of HRV detections). However, a higher proportion of HBoV, HRSV, human adenovirus (HAdV), human coronavirus (HCoV), human enterovirus (HEV) and PyV positives were involved in co-detections (29.6% to 73.5%; Table 1). There was no difference in age among the patients either according to number of co-detections (p = 0.27) or whether or not an HRV was present (p = 0.20). There was no difference in the proportion of co-detections involving an HRV according to presentation with URTI or LRTI (p = 0.34).
There was a consistent pattern of HRV detection associated with a reduced likelihood of co-detection with a number of other viruses. Both the univariate contingency table analyses and multivariate logistic regression analysis showed similar trends (Table 1, Table 2, Table 3). For example, from Table 1, an HRV detection resulted in a reduced likelihood of co-detecting eight viruses or virus groups (HAdV, HCoV, IFAV, HBoV, HMPV, HRSV, HPIV or KIPyV) as reflected by an odds ratio (OR) of <1. HRSV was involved with six negative associations, HMPV and HBoV with four and HPIV with only three. Of these, all are RNA viruses except HBoV. HRSV and HMPV exhibited different association patterns despite shared genetic features and clinical impact. HRSV, HMPV and HPIV did not occur together in our specimen set. The only positive association to reach significance was the pairing of HBoV and WUPyV.
Eleven virus pairs (models) were selected for logistic regression analysis (Table 3). All models were consistent with results from the univariate analyses, but some added information relating to season and age group. For example there was a reduced likelihood (OR <1) of an HBoV detection among toddlers, during autumn or if an HRV had been detected.
To investigate whether a particular HRV strain or group of strains drove the trends we observed, we randomly selected 10% of HRV PCR-positive specimens and sequenced a portion of the 5′ untranslated region.24 Nearest matches by comparative BLAST analysis indicated both classic and novel strains of HRV species A (HRV-78, 82, 49, 94, 44, 29, 65, 19, 56, NAT040, NAT049), only a few from species B (HRV-6, NAT066) and strains belonging to the putative new species, HRV C (similar to HRV-QPM,17 QPID03-0013,16 QPID03-0009, HRV-C024,18 HRV-C025, NAT08319 and Antwerp HRV strains25).
4. Discussion
Our hypothesis was supported by the finding that detection of HRV RNA in particular, but also that from HRSV and HMPV, are often associated with reduced co-detection of other respiratory virus RNA/DNA in statistically significant patterns. Our analyses showed similar patterns of virus association irrespective of which comparison method was used, notwithstanding minor variations in the p-values.
This is the first detailed analysis of respiratory virus co-detections among a comprehensively tested collection of specimens. Historically, a belief in the elevated involvement of HRVs in co-detections with other viruses in ARTI has been based solely on the high overall number of HRV detections and has provided a reason to diminish the causal role of HRV infections in clinical outcomes other than the common cold26 despite evidence to the contrary.27, 28, 29 Our findings, which dispute these superficial observations, indirectly indicate an aetiological role for HRVs and are supported by examples from the literature. In a study of infants, HRV strains were co-detected in only 65% of instances compared to 80% of HRSV detections.30 Among children, more than one third of HPIV, HMPV, HCoV-NL63 and HAdV positives were involved in co-detections whereas only 17% of picornaviruses were detected with another virus.31 Our observations are consistent with the findings of Brunstein et al. who identified significant associations between some viruses in the respiratory tract.3 However that study observed an unusually low frequency of HRV detection compared to ours, possibly due to the oligonucleotides they employed or their use of a multiplex PCR method which may have been adversely affected by the competitive amplification of multiple templates. Additionally, the study only collected specimens across a 4 month period compared to the 12 month period of our observations.
Some have ascribed the increased frequency of viral co-detection to simple coincidental overlap of epidemic seasons32 however our analyses indicate that these associations do not occur by chance, suggesting more complex mechanisms are responsible. HRVs are considered to mediate illness through immunological rather than cytopathic mechanisms. HRV replication, as for HRSV and IFV, produces double-stranded RNA (dsRNA) replicative intermediates33; molecules which mediate triggering of interferon (IFN)-stimulated genes inducing an antiviral state.11, 34, 35 It is possible that one mechanism behind the putative interference patterns we observed, strongest for HRVs but also in action among other viruses, may have been related to this innate IFN response which shields neighbouring cells from viral infection. Interestingly HCoV, IFAV, HEV and HPIV are also RNA viruses, but they had only 2, 2, 0 and 3 negative associations, respectively, compared to 3 for HBoV, a DNA virus (Table 1, Table 2), indicating that negative associations were not determined by nucleic acid type alone. Based on the literature available, there are no clearly defined mechanisms whereby HRVs antagonise the IFN cascade, indirectly apparent from the large body of literature describing their efficient induction of IFN and associated cytokines36, 37, 38, 39, 40; unusual among viruses and a possible reason for the scale of the statistical patterns we observed. Additionally, the IFN-antagonizing mechanisms employed by other viruses41, 42 may inadvertently permit higher frequencies of co-infection via attenuation of the innate response. A weak or absent IFN-antagonistic mechanism should generally result in the rapid suppression of HRV replication. This is not the case for asthmatic individuals because of diminished innate43, 43 and acquired44 antiviral defences resulting in more infections45 with higher HRV viral loads36 for longer periods associated with more severe symptoms.43, 46
In addition to the features we have examined, the local climate, reduced social cohorting (such as seen during holiday periods), immune status, co-morbidities and the urbanization of the population under study could bias the results. We did not seek the contribution of bacteria since it is difficult to distinguish a pathogenic versus colonizing role in the upper airways. Also, our study sample was drawn from individuals ill enough to require medical care and laboratory testing of nasopharyngeal specimens. This sample is not representative of the general population although it is drawn from it. The contribution of as yet unidentified respiratory viruses could not be gauged and furthermore, as with any PCR-based method, it was impossible to state that every possible strain of each target virus was detected. Nonetheless, in our hands the HRV assay22 successfully detected many strains from HRV A, B and the proposed C species.18, 47 Our observations require confirmation in other data sets, particularly across multiple years to account for the contribution of those viruses which were not epidemic in Queensland during 2003.
The patterns we report hint at a previously unrecognised aspect to HRV infections; the possibility of a leading role in vivo in interference with other virus detections. In light of this, we have formulated a new hypothesis, that HRV infection can protect its host from infection by other, often more cytopathic, viruses. We could not address the order of infection by the viruses detected here but future epidemiological studies should prospectively enrol and sample subjects regardless of symptomatic context, for at least 12 months, screening for all known respiratory viruses using PCR. If our new hypothesis is correct, serial sampling and testing will find that immediately after an HRV infection in the immunocompetent host, there is a period of time during which subjects are refractory to the acquisition of another respiratory virus. Such investigations should continue to find that proportionately, co-detections involving other virus pairings are relatively more prevalent.
While there are no HRV vaccines on the horizon, we suggest that any such prophylaxis could withdraw a valuable natural moderator of other viral infections, potentially leaving the individual at greater risk of clinically severe infections by more cytopathogenic viruses. Meanwhile, anti-picornaviral drug development is an active research area and while such compounds are not usually administered early enough48 to stop sufficient viral replication and interrupt immune engagement, they still reduce viral load and symptom severity,49 of particular benefit for wheezing exacerbations triggered by HRV infections. Our data are also interesting in the context of recent claims that HRVs may contribute to the spread of pandemic influenza through the mechanism of increased coughing and sneezing with subsequent increased aerosol spread of influenza virus.50 We suggest that acquisition of an HRV infection may paradoxically provide a natural protection against influenza infection.
Competing interests
All authors: no conflicts.
Acknowledgments
This work was supported by a National Health and Medical Research Committee grant (455905). We thank Pathology Services, Queensland Health for the provision of clinical specimens and Dr. Kirsten Spann for discussions on immunological matters.
References
- 1.Bryce J., Boschi-Pinto C., Shibuya K., Black R.E. WHO estimates of the causes of death in children. Lancet. 2005;365:1147–1152. doi: 10.1016/S0140-6736(05)71877-8. [DOI] [PubMed] [Google Scholar]
- 2.Murray C.J.L., Lopez A.D. Alternative projections of mortality and disability by cause 1990–2020: global burden of disease study. Lancet. 1997;349:1498–1504. doi: 10.1016/S0140-6736(96)07492-2. [DOI] [PubMed] [Google Scholar]
- 3.Brunstein J.D., Cline C.L., McKinney S., Thomas E. Evidence from multiplex molecular assays for complex multipathogen interactions in acute respiratory infections. J Clin Microbiol. 2008;46:97–102. doi: 10.1128/JCM.01117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jartti T., Lee W.-M., Pappas T., Evans M., Lemanske R.F., Gern J.E. Serial viral infections in infants with recurrent respiratory illnesses. Eur Respir J. 2008;32:314–320. doi: 10.1183/09031936.00161907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemanske R.F., Jackson D.J., Gangnon R.E., Evans M.D., Li Z., Shult P.A. Rhinovirus illnesses during infancy predict subsequent childhood wheezing. J Allergy Clin Immunol. 2005;116:571–577. doi: 10.1016/j.jaci.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 6.Sly P.D., Boner A.L., Bjorksten B., Bush A., Custovic A., Eigenmann P.A. Early identification of atopy in the prediction of persistent asthma in children. Lancet. 2008;372:1100–1106. doi: 10.1016/S0140-6736(08)61451-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson D.J., Gangnon R.E., Evans M.D., Roberg K.A., Anderson E.L., Pappas T.E. Wheezing rhinovirus illnesses in early life predict asthma development in high risk children. Am J Respir Crit Care Med. 2008;178:667–672. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertino J.S. Cost burden of viral respiratory infections: Issues for formulary decision makers. Am J Med. 2002;112:42S–49S. doi: 10.1016/s0002-9343(01)01063-4. [DOI] [PubMed] [Google Scholar]
- 9.Lambert S.B., Allen K.M., Carter R.C., Nolan T.M. The cost of community-managed viral respiratory illnesses in a cohort of healthy preschool-aged children. Respir Res. 2008;9:1–11. doi: 10.1186/1465-9921-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnston S.L., Openshaw P.J.M. The protective effect of childhood infections. Br Med J. 2001;322:376–377. doi: 10.1136/bmj.322.7283.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y., Hamati E., Lee P.K., Lee W.M., Wachi S., Schnurr D. Rhinovirus induces airway epithelial gene expression through double-stranded RNA and IFN-dependent pathways. Am J Respir Cell Mol Biol. 2006;34:192–203. doi: 10.1165/rcmb.2004-0417OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ånestad G. Interference between outbreaks of respiratory syncytial virus and influenza virus infection. Lancet. 1982;1:502. doi: 10.1016/s0140-6736(82)91466-0. [DOI] [PubMed] [Google Scholar]
- 13.Ånestad G., Vainio K., Hungnes O. Interference between outbreaks of epidemic viruses. Scand J Infect Dis. 2007;39:653–654. doi: 10.1080/00365540701253860. [DOI] [PubMed] [Google Scholar]
- 14.Glezen W.P., Paredes A., Taber L.H. Influenza in children relationship to other respiratory agents. JAMA. 1980;243:1345–1349. doi: 10.1001/jama.243.13.1345. [DOI] [PubMed] [Google Scholar]
- 15.Andrewes C.H. Rhinoviruses and common colds. Ann Rev Med. 1966;17:361–370. doi: 10.1146/annurev.me.17.020166.002045. [DOI] [PubMed] [Google Scholar]
- 16.Arden K.E., McErlean P., Nissen M.D., Sloots T.P., Mackay I.M. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol. 2006;78:1232–1240. doi: 10.1002/jmv.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McErlean P., Shackleton L.A., Lambert S.B., Nissen M.D., Sloots T.P., Mackay I.M. Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J Clin Virol. 2007;39:67–75. doi: 10.1016/j.jcv.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau S.K.P., Yip C.C.Y., Tsoi H.-W., Lee R.A., So L.-Y., Lau Y.-L. Clinical features and complete genome characterization of a distinct human rhinovirus genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J Clin Microbiol. 2007;45:3655–3664. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kistler A., Avila P.C., Rouskin S., Wang D., Ward T., Yagi S. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J Infect Dis. 2007;196:817–825. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bialasiewicz S., Whiley D.M., Lambert S.B., Jacob K.C., Bletchly C., Wang D. Presence of the newly discovered human polyomaviruses KI and WU in Australian patients with acute respiratory tract infection. J Clin Virol. 2008;41:63–68. doi: 10.1016/j.jcv.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothman K.I. No adjustments are needed for multiple comparisons. Epidemiology. 1990;1:43–46. [PubMed] [Google Scholar]
- 22.Arola A., Santti J., Ruuskanen O., Halonen P., Hyypiä T. Identification of enteroviruses in clinical specimens by competitive PCR followed by genetic typing using sequence analysis. J Clin Microbiol. 1996;34:313–318. doi: 10.1128/jcm.34.2.313-318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jartti T., Lehtinen P., Vuorinen P., Koskenvuo M., Ruuskanen O. Persistence of rhinovirus and enterovirus RNA after acute respiratory illness in children. J Med Virol. 2004;72:695–699. doi: 10.1002/jmv.20027. [DOI] [PubMed] [Google Scholar]
- 24.Gama R.E., Horsnell P.R., Hughes P.J., North C., Bruce C.B., Al-Nakib W. Amplification of rhinovirus specific nucleic acids from clinical samples using the polymerase chain reaction. J Med Virol. 1989;28:73–77. doi: 10.1002/jmv.1890280204. [DOI] [PubMed] [Google Scholar]
- 25.Loens K., Goossens H., de Laat C., Foolen H., Oudshoorn P., Pattyn S. Detection of rhinoviruses by tissue culture and two independent amplification techniques, nucleic acid sequence-based amplification and reverse transcription-PCR, in children with acute respiratory infections during a winter season. J Clin Microbiol. 2006;44:166–171. doi: 10.1128/JCM.44.1.166-171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stott E.J., Eadie M.B., Grist N.R. Rhinovirus infections of children in hospital; Isolation of three possibly new rhinovirus serotypes. Am J Epidemiol. 1969;90:45–52. doi: 10.1093/oxfordjournals.aje.a121048. [DOI] [PubMed] [Google Scholar]
- 27.Douglas R.G., Jr., Cate T.R., Gerone P.J., Couch R.B. Quantitative rhinovirus shedding patterns in volunteers. Am Rev Respir Dis. 1966;94:159–167. doi: 10.1164/arrd.1966.94.2.159. [DOI] [PubMed] [Google Scholar]
- 28.Glezen W.P., Loda F.A., Clyde W.A., Senior R.J., Sheaffer C.I., Conley W.G. Epidemiologic patterns of acute lower respiratory disease of children in a pediatric group practice. J Pediatr. 1971;78:397–406. doi: 10.1016/s0022-3476(71)80218-4. [DOI] [PubMed] [Google Scholar]
- 29.Papadopoulos N.G., Bates P.J., Bardin P.G., Papi A., Leir S.H., Fraenkel D.J. Rhinoviruses infect the lower airways. J Infect Dis. 2000;181:1875–1884. doi: 10.1086/315513. [DOI] [PubMed] [Google Scholar]
- 30.Richard N., Komurian-Pradel F., Javouhey E., Perret M., Rajoharison A., Bagnaud A. The impact of dual viral infection in infants admitted to a pediatric intensive care unit associated with severe bronchiolitis. Pediatr Infect Dis J. 2008;27:1–5. doi: 10.1097/INF.0b013e31815b4935. [DOI] [PubMed] [Google Scholar]
- 31.Lambert S.B., Allen K.M., Druce J.D., Birch C.J., Mackay I.M., Carlin J.B. Community epidemiology of human metapneumovirus, human coronavirus NL63, and other respiratory viruses in healthy preschool-aged children using parent-collected specimens. Pediatrics. 2007;120:e929–e937. doi: 10.1542/peds.2006-3703. [DOI] [PubMed] [Google Scholar]
- 32.Paranhos-Baccala G., Komurian-Pradel F., Richard N., Vernet G., Lina B., Floret D. Mixed respiratory virus infections. J Clin Virol. 2008 doi: 10.1016/j.jcv.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koliais S.I., Dimmock N.J. Replication of rhinovirus RNA. J Gen Virol. 1973;20:1–15. doi: 10.1099/0022-1317-20-1-1. [DOI] [PubMed] [Google Scholar]
- 34.Hewson C.A., Jardine D., Edwards M.R., Laza-Stanca V., Johnston S.L. Toll-like receptor 3 is induced by and mediates antiviral activity against rhinovirus infection of human bronchial epithelial cells. J Virol. 2005;79:12273–12279. doi: 10.1128/JVI.79.19.12273-12279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeda K., Akira S., Toll-like receptors. Curr Prot Immunol, 2007;Chapter 14:14.12.1-14.12.13. [DOI] [PubMed]
- 36.Wark P.A.B., Bucchieri F., Johnston S.L., Gibson P.G., Hamilton L., Mimica J. IFN-γ-induced protein 10 is a novel biomarker of rhinovirus-induced asthma exacerbations. J Allergy Clin Immunol. 2007;120:586–593. doi: 10.1016/j.jaci.2007.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Contoli M., Message S.D., Laza-Stanca V., Edwards M.R., Wark P.A.B., Bartlett N.W. Role of deficient type III interferon-λ production in asthma exacerbations. Nat Med. 2006;12:1023–1026. doi: 10.1038/nm1462. [DOI] [PubMed] [Google Scholar]
- 38.Brooks G.D., Buchta K.A., Swenson C.A., Gern J.E., Busse W.W. Rhinovirus-induced interferon-gamma and airway responsiveness in asthma. Am J Respir Crit Care Med. 2003;168:1091–1094. doi: 10.1164/rccm.200306-737OC. [DOI] [PubMed] [Google Scholar]
- 39.Yamaya M., Sekizawa K., Suzuki T., Yamada N., Furukawa M., Ishizuka S. Infection of human respiratory submucosal glands with rhinovirus: effects on cytokine and ICAM-1 production. Am J Physiol. 1999;277:L362–L371. doi: 10.1152/ajplung.1999.277.2.L362. [DOI] [PubMed] [Google Scholar]
- 40.Wark P.A., Grissell T., Davies B., See H., Gibson P.G. Diversity in the bronchial epithelial cell response to infection with different rhinovirus strains. Respirology. 2009 doi: 10.1111/j.1440-1843.2009.01480.x. [DOI] [PubMed] [Google Scholar]
- 41.Guerrero-Plata A., Baron S., Poast J.S., Adegboyega P.A., Casola A., Garofalo R.P. Activity and regulation of alpha interferon in respiratory syncytial virus and human metapneumovirus experimental infections. J Virol. 2005;79:10190–10199. doi: 10.1128/JVI.79.16.10190-10199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spann K.M., Tran K.C., Collins P.L. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-kappaB, and proinflammatory cytokines. J Virol. 2005;79:5353–5362. doi: 10.1128/JVI.79.9.5353-5362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wark P.A.B., Johnston S.L., Bucchieri F., Powell R., Puddicombe S., Laza-Stanca V. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med. 2005;201:937–947. doi: 10.1084/jem.20041901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papadopoulos N.G., Stanciu L.A., Papi A., Holgate S.T., Johnston S.L. A defective type 1 response to rhinovirus in atopic asthma. Thorax. 2002;57:328–332. doi: 10.1136/thorax.57.4.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minor T.E., Baker J.W., Dick E.C., DeMeo A.N., Ouellette J.J., Cohen M. Greater frequency of viral respiratory infections in asthmatic children as compared with their nonasthmatic siblings. J Pediatr. 1974;85:472–477. doi: 10.1016/S0022-3476(74)80447-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corne J.M., Marshall C., Smith S., Schreiber J., Sanderson G., Holgate S.T. Frequency, severity, and duration of rhinovirus infections in asthmatic and non-asthmatic individuals: a longitudinal cohort study. Lancet. 2002;359:831–834. doi: 10.1016/S0140-6736(02)07953-9. [DOI] [PubMed] [Google Scholar]
- 47.McErlean P., Shackleton L.A., Andrewes E., Webster D.R., Lambert S.B., Nissen M.D. Distinguishing Molecular Features and Clinical Characteristics of a Putative New Rhinovirus Species, Human Rhinovirus C (HRVC) PLoS ONE. 2008;3:e1847. doi: 10.1371/journal.pone.0001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Linder J.A. Improving care for acute respiratory infections: Better systems, not better microbiology. Clin Infect Dis. 2007;45:1189–1191. doi: 10.1086/522191. [DOI] [PubMed] [Google Scholar]
- 49.Hayden F.G., Herrington D.T., Coats T.L., Kim K., Cooper E.C., Villano S.A. Efficacy and safety of oral pleconaril for treatment of colds due to picornaviruses in adults: Results of 2 double-blind, randomized, placebo-controlled trials. Clin Infect Dis. 2003;36:1523–1532. doi: 10.1086/375069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merler S., Poletti P., Ajelli M., Caprile B., Manfredi P. Coinfection can trigger multiple pandemic waves. J Theor Biol. 2008 doi: 10.1016/j.jtbi.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]