Abstract
Life on Earth emerged in a hydrogen sulfide-rich environment eons ago and with it protein persulfidation mediated by H2S evolved as a signaling mechanism. Protein persulfidation (S-sulfhydration) is a posttranslational modification of reactive cysteine residues, which modulate protein structure and/or function. Persulfides are difficult to label and study due to their reactivity and similarity with cysteine. Here, we report a facile strategy for chemoselective persulfide bioconjugation using dimedone-based probes, to achieve highly selective, rapid, and robust persulfide labeling in biological samples with broad utility. Using this method, we show persulfidation is an evolutionarily conserved modification and waves of persulfidation are employed by cells to resolve sulfenylation and prevent irreversible cysteine overoxidation preserving protein function. We report an age-associated decline in persulfidation which is conserved across evolutionary boundaries. Accordingly, dietary or pharmacological interventions to increase persulfidation associate with increased longevity and improved capacity to cope with stress stimuli.
Keywords: hydrogen sulfide, protein persulfidation, hydrogen peroxide, sulfenylation, sulfinylation, sulfonylation, redox signalling, aging, calorie restriction
INTRODUCTION
Although considered a toxic gas for more than a century, hydrogen sulfide (H2S) was one of the essential ingredients required for life to emerge on Earth (Patel et al., 2015). Early anaerobic bacteria flourished in H2S-rich environments and even used H2S instead of water for the first photosynthetic process (Filipovic et al., 2018; Wang, 2012). Two decades ago H2S re-emerged as an important signaling molecule produced by cells (Filipovic et al., 2018; Paul and Snyder, 2012; Szabó, 2007; Wang, 2012). Genetic deletion of the H2S producing enzyme cystathionine γ-lyase (CSE) can lead to hypertension (Yang et al., 2008). Animals exposed to H2S enter a suspended animation-like state (Blackstone, 2005), while dietary restriction-induced stress resistance and lifespan extension depends on intracellular H2S production (Hine et al., 2015). Despite beneficial effects of H2S, observed in a plethora of pathological states (Filipovic et al., 2018; Paul and Snyder, 2012; Szabó, 2007), the mechanism or mechanisms underlying these effects remain poorly characterized. However, its role in the oxidative posttranslational modification (oxPTM) of cysteine residues, known either as protein S-sulfhydration or persulfidation, is thought to be one of its main beneficial mechanisms of action (Paul and Snyder, 2015).
Cysteine is a rare amino acid residue that often occurs in functional sites of proteins and represents a site for redox control of protein function (Marino and Gladyshev, 2010; Paulsen and Carroll, 2013). For example, hydrogen peroxide (H2O2) signals via the oxidation of cysteine residues to sulfenic acids (PSOH), while part of nitric oxide signaling could be explained by cysteine S-nitrosation (Foster et al., 2009; Paulsen and Carroll, 2013). A variety of chemical approaches have been used to label and study cysteine modifications, leading to a broad range of fundamental and applied advances (Furdui and Poole, 2014; Paulsen and Carroll, 2013). However, protein persulfides (PSSH) and their role in cell signaling managed to remain understudied, due to the fact that PSSH are very reactive and their reactivity is similar to that of cysteine residues (Cuevasanta et al., 2015; Filipovic et al., 2018; Pan and Carroll, 2013). Consequently, this has made it difficult to design tools for selective labeling, hampering a better understanding of the function of this specific oxPTM. Current methods rely on blocking both thiols and persulfides with electrophiles and then releasing the latter with a reducing agent, but this approach is linked to many caveats (Dóka et al., 2016; Filipovic et al., 2018; Reisz et al., 2013).
Nonetheless, due to their enhanced nucleophilicity persulfides react readily with reactive oxygen species (ROS), while H2S itself is a poor ROS scavenger (Cuevasanta et al., 2015; Filipovic et al., 2018; Ono et al., 2014). Given the fact that ROS play an important role in signaling (D’Autréaux and Toledano, 2007; Finkel, 2011) and aging (Finkel and Holbrook, 2000), it is tempting to speculate that the general beneficial effects of H2S are evolutionary conserved and conveyed through protein persulfidation.
To better understand persulfidation in vivo, we explored the phenomenon that, if first transformed to mixed aromatic disulfides, protein persulfides could be selectively labeled by certain nucleophiles. Here we report the development of a new, dimedone-based method that enables chemoselective persulfide bioconjugation of proteins from a wide range of source materials in vitro and in vivo. Using this new method, we report that persulfidation plays an integral role in hydrogen peroxide-based signal transduction. We also found that protein persulfidation decreases with age and is maintained by interventions that increase lifespan across evolutionary boundaries, and may thus play a previously unrecognized protective role against aging.
RESULTS
Development of dimedone switch method
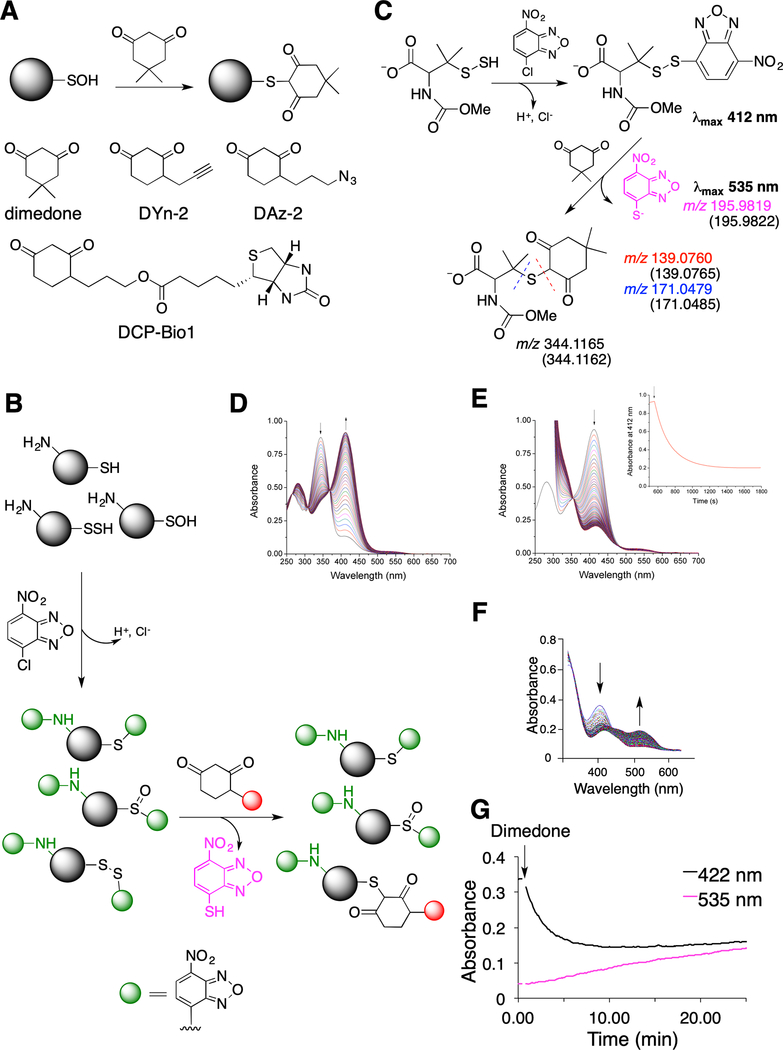
To be able to use nucleophilic substitution to tag persulfides, PSSH need to first be transformed into a mixed disulfide (Wedmann et al., 2016; Zhang et al., 2014) in such a way that one of the sulfurs in the S-S bond possesses a much more enhanced electrophilicity (Figure S1A). Despite their selectivity in sulfenic acid labeling (Klomsiri et al., 2010; Paulsen and Carroll, 2013; Yang et al., 2014), dimedone-based probes (Figure 1A) could be an excellent candidate in the second step, serving as the nucleophile. They are additionally attractive candidates since a plethora of those probes are commercially available with different reporting moieties and have been thoroughly tested (Furdui and Poole, 2014; Paulsen and Carroll, 2013). However, for dimedone-based probes to be used in PSSH labeling, the initial step needs to involve a reagent which not only reacts with PSSH and thiols but also blocks sulfenic acids. 4-chloro-7-nitrobenzofurazan (NBF-Cl) fulfils these criteria. It is used as a tool for the blocking and detection of thiols, amines and sulfenic acids (Bernal-Perez et al., 2012; Ellis and Poole, 1997) and it should form a mixed disulfide upon its reaction with PSSH (Figure 1B). We initiated our study by monitoring the labeling of the low molecular weight persulfide, N-methoxycarbonyl penicillamine persulfide (nmc-PSSH, (Artaud and Galardon, 2014)) (Figure 1C). Nmc-PSSH reacted readily with NBF-Cl resulting in a characteristic absorbance maximum at 412 nm (Figure 1D). Next, the addition of an equimolar amount of dimedone led to a fast disappearance of the 412 nm peak, suggesting that switching did occur (Figure 1E). ESI-TOF MS/MS analysis of the reaction mixture confirmed that the two main products are the NBF-SH and dimedone labeled nmc-PSSH (Figure 1C, Figure S1B–F).
Fig. 1. Probing dimedone switch strategy for persulfide labeling.
(A) (Upper) Labeling of sulfenic acids with dimedone. (Lower) Structures of dimedone-based probes.
(B) Proposed dimedone switch strategy for persulfide labeling. In the first step proteins react with 4-chloro-7-nitrobenzofurazan (NBF-Cl) to label persulfides, thiols, sulfenic acids, and amino groups. Reaction with amino groups gives characteristic green fluorescence. In the second step, NBF tag is switched by a dimedone-based probe, selectively labeling persulfides.
(C) Model switch reaction with 100 μM N-methoxycarbonyl penicillamine persulfide (nmc-PSSH) and 100 μM NBF-Cl, followed by 500 μM dimedone. MS analysis reveals formation of 4-thio-7-nitrobenzofurazan (535 nm) and dimedone labeled nmc-penicillamine, which under MS/MS conditions decomposes along the blue or red dash line. Numbers given in the brackets represent calculated m/z for the observed ions.
(D) Time-resolved spectra for the reaction of 100 μM nmc-PSSH with 100 μM NBF-Cl (pH 7.4, 23 °C). Arrows indicated disappearance of NBF-Cl and appearance of nmc-PSS-NBF adduct at 412 nm.
(E) Time-resolved spectral changes upon addition of 200 μM dimedone to a reaction mixture shown in (D) (pH 7.4, 23 °C). Inset: Kinetics of decay of 412 nm absorbance maximum after addition of dimedone.
(F-G) 23 μM HSA-SSH was left to react with 100 μM NBF-Cl over 30 min in phosphate buffer (50 mM, pH 7.4) with 1% SDS, at 37 °C and then 200 μM dimedone was added. UV-Vis spectral changes (F) and kinetic traces (G) show the decay of the 422 nm absorbance and the appearance of a 535 nm peak.
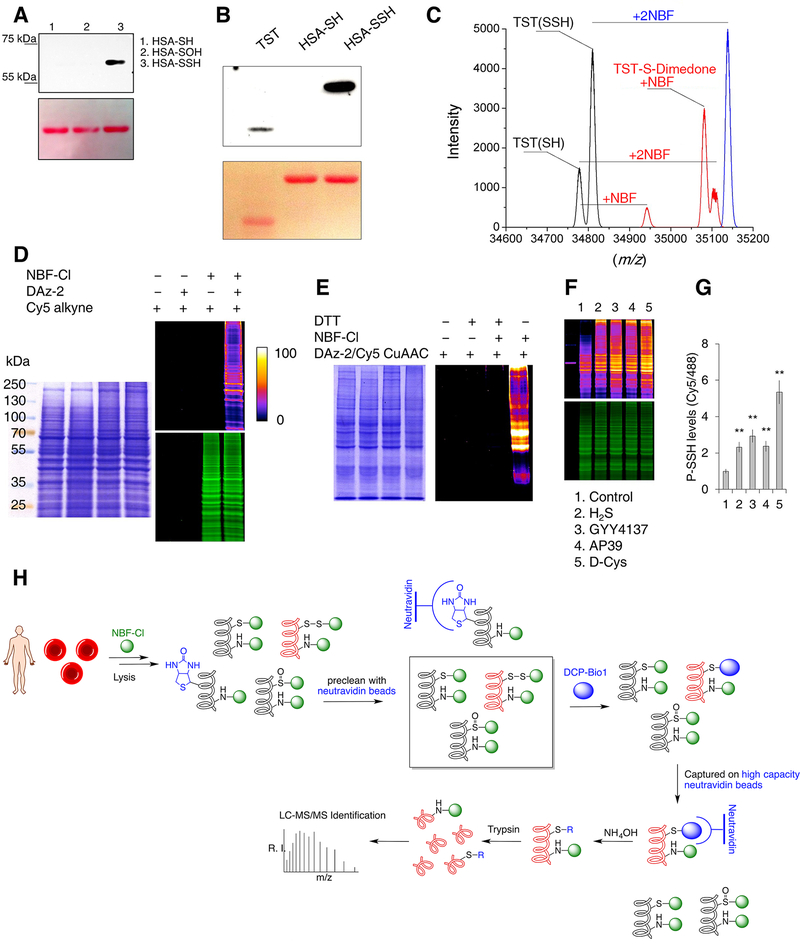
We then evaluated the selectivity of the dimedone switch method by using human serum albumin (HSA) as a model (Cuevasanta et al., 2015). HSA has 17 disulfides and 1 free thiol, and is therefore both a good control and an example of a protein with oxidized cysteines. HSA-SH, its sulfenylated (HSA-SOH) and persulfidated (HSA-SSH) forms reacted with NBF-Cl to generate products with distinct absorbance maxima that fit well to those reported in the literature (Ellis and Poole, 1997) (Figure S1G, I, K). The subsequent addition of dimedone caused tag-switching only in the HSA-SSH sample (Figure 1F,G, Figure S1H,J) resulting in the loss of HSA-SS-NBF absorbance and formation of both HSA-S-dimedone and NBF-SH products. Separation of treated samples by electrophoresis and subsequent immunoblotting with anti-dimedone antibody (Seo and Carroll, 2009) gave a positive signal for HSA-SSH only (Figure 2A). To confirm the labeling of naturally occurring persulfides, we used Thiosulfate Sulfur Transferase (TST, also known as Rhodanese), an enzyme that forms a persulfide during its catalytic cycle, as a model (Filipovic et al., 2018). Both immunoblotting with anti-dimedone antibody (Figure 2B) and ESI-TOF-MS (Figure 2C) revealed TST labeled with dimedone.
Figure 2. Protein persulfide labeling and identification.
(A-B) Selectivity of dimedone-switch method for protein persulfides. Human serum albumin (HSA, A) and TST (B) were used as models. Dimedone labeling was visualized by rabbit polyclonal anti-dimedone antibody. Ponceau S staining was used for the protein load.
(C) Deconvoluted mass spectra 20 μM rhodanese (black), rhodanese treated with 100 μM NBF-Cl (blue) and rhodanese treated first with 100 μM NBF-Cl then with 500 μM dimedone (red).
(D) In-gel detection of cellular PSSH levels. HeLa cells were lysed with or without supplementation of 10 mM NBF-Cl, and probed for persulfide labeling with or without DAz-2, followed by Cy5-alkyne using CuAAC. Gels were also stained with Coomassie Brilliant Blue. Fire pseudo-colouring was used to visually enhance the signal. Green fluorescence corresponds to the total protein load (NBF-protein adducts).
(E) MEF cells lysed with or without 20 mM NBF-Cl samples and then treated with or without 20 mM DTT and labeled with DAz-2/Cy5-alkyne using CuAAC.
(F-G) Protein persulfidation levels in HeLa cells treated with different H2S donors: 200 μM Na2S (H2S) for 45 min, 200 μM GYY4137 for 2 hr, 200 nM AP39 for 2 hr and 2 mM D-cysteine (D-Cys) for 1 hr. Ratio of Cy5/488 signals is used for the quantification (G). Data shown as a mean ± SD. of 3 individual experiments. ** p < 0.01 vs. control.
(H) Schematic depiction of the protocol used for the proteomic analysis of endogenous persulfidation in RBC.
With these data in hand, we envisioned that the dimedone switch method could enable the installation of various payloads onto a protein of interest that would allow specific identification/visualization. We first used DCP-Bio1, a biotinylated form of dimedone (Figure 1A). Persulfides of HSA and GAPDH showed positive staining (Figure S2A, B), as detected by Cy5-streptavidin. Separation of samples by streptavidin magnetic beads and subsequent in-gel detection of green fluorescence originating from amino groups labeled with NBF-Cl (Figure 1B), showed the selective labeling of PSSH but not other oxPTM (Figure S2A, B).
Next, we tested if the method can be used for proteomic analysis. TST persulfide was tag-switched with DCP-Bio1 and subjected to either trypsin or chymotrypsin digestion. MS/MS sequence coverage was 95% of the structure (Figure S2C, Supporting Dataset S1, S2) and only C248, present as a persulfide in the active site of the enzyme, was found to be labelled by DCP-Bio1 (Figure S2C). Other cysteine residues and several lysine residues were labeled with NBF alone (Supporting Data S1, S2), according to the reaction scheme in Figure 1B.
We also tested the possibility of detecting PSSH directly in gel, by installing a Cy5-fluorescent moiety through Copper(I)-catalyzed Azide-Alkyne Cycloaddition (CuAAC, click chemistry). In addition to labeling cysteines, NBF-Cl also reacts with amino groups giving a characteristic fluorescence with λex at 488 nm (Bernal-Perez et al., 2012). Commercially available TST (already partially present as a persulfide) was incubated with either thiosulfate (TS) or dithiothreitol (DTT) to form a fully persulfidated or reduced TST, respectively. 20 μM reduced or persulfidated TST was mixed with 50 μM NBF-Cl and the persulfide was visualized using DAz-2/Cy5 CuAAC. On the one hand, while both untreated (lane 1, Figure S2D) and thiosulfate treated (lane 3, Figure S2D) TST showed Cy5 signal, the green fluorescence signal was significantly reduced in the fully persulfidated enzyme, despite the same load. On the other hand, the green fluorescence signal was much stronger in the fully reduced enzyme (lane 2, Figure S2D), suggesting that at low NBF-Cl/protein ratio, switching caused by the dimedone-based probe could affect the intensity of green fluorescence (Figure S2D), so we opted for using at least 10-fold excess of NBF-Cl (Figure S2E). This use of NBF-Cl in excess additionally offered the opportunity to use the green fluorescence as a measure of the total protein load and to therefore quantify the persulfidation levels by measuring the Cy5/488 fluorescence signal ratio. Different components of click chemistry did not cause any unspecific labeling in samples blocked with NBF-Cl (Figure S2F).
To be able to label cell extracts, we had to ensure that the method shows sufficient selectivity. The selectivity of dimedone probes for sulfenic acids have been demonstrated previously and no cross-reactivity with disulfides, S-nitrosothiols, HNE-modified cysteines or any other nucelophilic centers could be observed (Charles et al., 2007; Klomsiri et al., 2010; Yang et al., 2014). In fact, a blind proteomic search for off-targets of dimedone did not identify any unwanted labeling (Yang et al., 2014). However, in the dimedone-switch labeling (i) incomplete blocking of sulfenic acids by NBF-Cl and (ii) potential interference of sulfenamides could represent possible caveats (Figure S2G).
To ensure efficient and selective labeling we decided to perform the cell lysis under denaturing conditions, which will permit fast unfolding and exposure of all cysteine and amine residues to NBF-Cl. Large excess of NBF-Cl and incubation at 37 °C should provide an efficient kinetic push for fast and complete labeling and blocking. Protein unfolding will also expose cyclic sulfenamides to more water, and since they exist in the equilibrium with sulfenic acids this equilibrium will be shifted towards sulfenic acids (Gupta and Carroll, 2016). Furthermore, NBF-Cl has been demonstrated to efficiently react with cyclic sulfenamides (Figure S2G) (Gupta and Carroll, 2016). Nonetheless, we used protein tyrosine phosphatase 1B (PTP1B), which forms a stable cyclic sulfenamide, as a model system (Paulsen and Carroll, 2013). PTP1B treated with H2O2 (to form a mixture of sulfenic acid and cyclic sulfenamide) was labeled with DAz-2 (that bears a biorthogonal azide group) and then subsequently coupled to Cy5-alkyne via a CuAAC reaction, as expected, but not if it first reacted with NBF-Cl (lanes 1 and 2, Figure S2H). PTP1B persulfide however, could be labeled only if it first reacted with NBF-Cl and then with DAz-2/Cy5 CuAAC (lane 3, Figure S2H). Labeling was absent when NBF-Cl step was omitted (lane 4, Figure S2H).
The method’s selectivity was further confirmed in cell lysates. HeLa cells lysed with NBF-Cl and tag switched with DAz-2/Cy5 CuAAC resulted in the labeling and in-gel detection of a red fluorescence signal only when all reagents were used (Figure 2D, Figure S2I). Barely any signal could be detected in a control sample where NBF-Cl was absent (Figure 2D), confirming that lysis, incubation and protein precipitation (Figure S2I) would already be sufficient to get rid of reactive sulfenic acids and cyclic sulfenamides that could have remained uncapped by NBF-Cl. We found that lysis with 5 mM NBF-Cl is already sufficient to give maximal persulfide signal (Figure S2J). Treatment of cell lysates with DTT to reduce the disulfide bond formed in the reaction of NBF-Cl and protein persulfides, abolished the detected Cy5 fluorescence (Figure 2E), confirming the chemical mechanism of the dimedone switch approach proposed in Figure 1B. Lysis with dimedone to trap all sulfenic acids (Figure S2K) and subsequent labeling with NBF-Cl and then DCP-Bio1 (Figure S2L) or DAz-2/Cy5 CuAAC (Figure S2M) to switch tag persulfides, showed that the removal of sulfenic acids prior to persulfide labeling did not affect the detected signal, further confirming that our dimedone switch approach shows no cross-reactivity with sulfenic acids and/or cyclic sulfenamides (Figures S2K–M). Additionally, treatment of HeLa cells with different sources of H2S increased the intracellular persulfidation levels several-fold (Figure 2F, G). 200 nM mitochondria-targeted H2S donor, AP39, induced comparable increase as 200 μM Na2S, confirming the strong pharmacological potential of this compound (Figure 2F, G).
We used DCP-Bio1 as a switch label to identify the endogenously persulfidated proteins in human erythrocytes (Figure 2H, Table S1). Out of 56 identified proteins, more than half were previously identified to bear oxidized cysteine residues. These proteins were either found to be prone to cysteine oxidation in red blood cells (RBC) depleted of haemoglobin (Delobel et al., 2016) or treated with diamide (Zaccarin et al., 2014), in RBC from peroxiredoxin II deficient mice (Yang et al., 2012) or found to be directly persulfidated (Valentine et al., 1987). More importantly, both enzymes involved in H2S production in erythrocytes were also found to be persulfidated: 3-mercaptopyruvate sulfur transferase (MPST or MST) and methanethiol oxidase. The former is known to form a persulfide during the catalytic cycle (Yadav et al., 2013), while the latter produces both H2O2 and H2S (Pol et al., 2018), facilitating the oxidation of a cysteine residue to a sulfenic acid first and then to form a persulfide. In addition, peroxiredoxins, known to form a sulfenic acid during their catalytic cycle (Wood et al., 2003), were found to be persulfidated as well. It is also worth noting that all identified peptides, even those not falling within selection criteria (at least 2 reliable peptides and −10logP>50), originated from cysteine containing proteins, indicative of the very high selectivity of this approach.
Persulfidation is evolutionarily conserved and controlled by H2S generation from transsulfuration pathway and cysteine catabolism
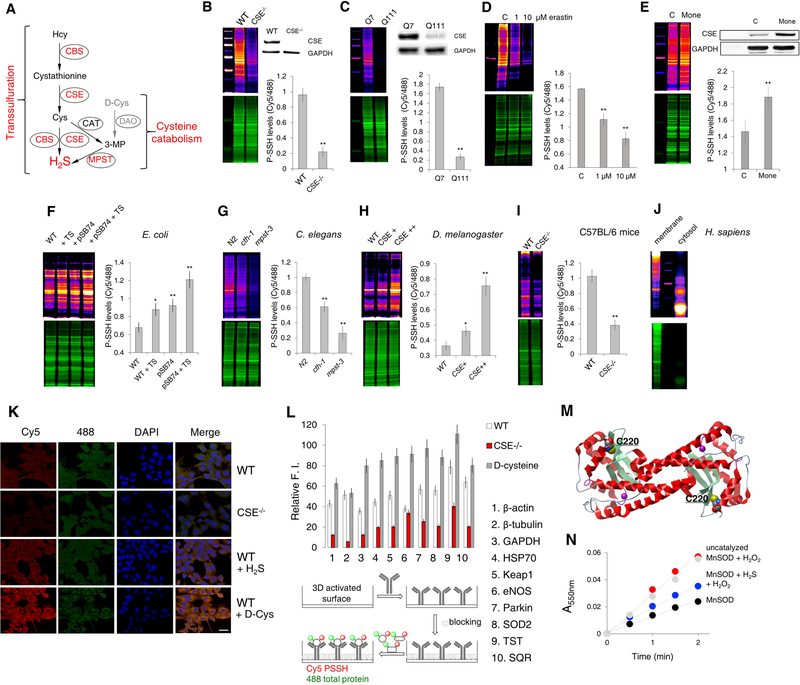
Although initial studies suggested that the main source of intracellular persulfides is H2S, produced predominantly by CSE (Figure 3A) (Filipovic et al., 2018; Mustafa et al., 2009; Paul and Snyder, 2012), recent findings questioned this by claiming that persulfides are synthesized during protein translation and are not related to the transsulfuration pathway or cysteine catabolism (Figure 3A) (Akaike et al., 2017). Persulfidation levels were significantly reduced in mouse embryonic fibroblasts (MEFs) originating from CSE−/− animals (Figure 3B). Interestingly, repeated cell splitting of the same cell line leads to less pronounced differences in these levels due to compensatory overexpression of cystathionine-β-synthase (CBS, Figure S3A, B). CSE is profoundly diminished in Huntington’s Disease (HD), a neurodegenerative disease triggered by the expansion of polyglutamine repeats in the huntingtin protein (Paul et al., 2014). In striatal cell-line models of HD (STHdhQ7/Q7 and STHdhQ111/Q111) harbouring 7 and 111 polyglutamine repeats, we now show that the lack of CSE results in the barely detectable PSSH levels in the STHdhQ111/Q111 cells (Figure 3C). CSE is known to be the predominant source of H2S in this cell type (Paul et al., 2014; Sbodio et al., 2016). Furthermore, the inhibition of cystine transporter, system xc− with erastin also resulted in the loss of protein persulfidation (Figure 3D). Additionally, pharmacologically induced overexpression of CSE by Golgi stressor, monensin (Sbodio et al., 2018), on the other hand, resulted in an increase of intracellular persulfidation (Figure 3E).
Figure 3. Intracellular persulfidation is evolutionarily conserved and controlled by H2S producing enzymes.
(A) Intracellular H2S production is catalyzed by cystathionine γ-lyase (CSE) and cystathionine-β-synthase (CBS), via the reverse transsulfuration pathway, and by 3-mercaptopyruvate sulfur transferase (MPST) in the cysteine catabolism pathway. Hcy: homocysteine; Cys: cysteine; 3MP: 3-mercaptopyruvate; CAT: cysteine aminotransferase; DAO: D-amino acid oxidase.
(B) PSSH levels in MEF cells from wild type (WT) and CSE−/− mice. Ratio of Cy5/488 signals is used for the quantification. n = 4. ** p < 0.01 vs. WT. Data are shown as mean ± SD. Inset: Western blot analysis of CSE levels. n = 3.
(C) PSSH levels in STHdhQ7/Q7 and STHdhQ111/Q111 cells. n = 4. ** p < 0.01 vs. Q7. Inset: Representative Western blot of CSE protein expression levels. n = 3.
(D) The effect of 1 and 10 μM Erastin (18.5 hr) on PSSH levels in WT MEF cells. n = 4. ** p<0.01 vs. control.
(E) PSSH levels in WT MEF cells for control, C, and treated with 1 μM Monensin, Mone (18 hr). n = 3. ** p<0.01 vs. control. Inset: Representative Western blot of CSE protein expression levels. n = 3.
(F) PSSH levels in E. coli without (WT) or with phsABC operon (pSB74 plasmid) that encodes thiosulfate reductase and results in H2S production. Both strains were treated with or without thiosulfate (TS, 4 hr at 37°C). n = 3. * p < 0.05, ** p < 0.01 vs. control.
(G) PSSH levels in wild type (N2), cth-1 and mpst-3 C. elegans mutants. ~ 16000 worms per sample. Ratio of Cy5/488 signals is used for the quantification. n = 3. ** p < 0.01 vs. control.
(H) PSSH levels in wild type (y1w118) Drosophila melanogaster and flies with different levels of CSE overexpression. 3–4 flies per samples. n = 3. * p<0.05, ** p<0.01 vs. WT.
(I) PSSH levels in kidney extracts form wild type (C57BL/6J) and CSE−/− mice. n = 3 animals. ** p < 0.01 vs. WT.
(J) Protein persulfidation in RBC membrane and cytosol from a healthy human donor.
(K) Confocal microscopy images of intracellular protein persulfide levels of WT and CSE−/− MEFs treated or not with 200 μM Na2S (H2S) or 2 mM D-Cys for 1 hr. Cy5 signal corresponds to protein persulfides, 488 nm signal corresponds to NBF-adducts. Nuclei stained with DAPI. Scale bar 20 μm.
(L) Antibody microarray-like approach to study persulfidation status of specific proteins. Schematic depiction of the method (lower part) and the actual readout (upper part) for the ten listed proteins. Cell lysates from WT, CSE−/− and WT MEFs treated with D-Cys (2 mM, 1 hr) were compared. Results are presented as a mean ± SD from 3 independent experiments.
(M) Ribbon structure of two subunits from human MnSOD (PDB: 1pl4), highlighting the cysteine residues and manganese containing active site.
(N) Persulfidation of MnSOD protects it from the H2O2-induced inactivation. SOD activity was measured using cytochrome c as a reporting molecule which is reduced by the superoxide generated from the xanthine/xanthine oxide system. Results are presented as a mean ± SD. from 3 independent experiments.
We expanded the screening to different phyla and regna, and in all of them endogenous persulfidation was found to be controlled by H2S produced via the transsulfuration pathway or cysteine catabolism (Figure 3A). We used an E. coli strain that is transformed with the phsABC operon (pSB74 plasmid) encoding for thiosulfate reductase, causing increased H2S production. Treatment of these bacteria with thiosulfate resulted in a two-fold increase of bacterial protein persulfidation when compared to control (Figure 3F). On the other hand, cth-1 and mpst-3 mutants of C. elegans (lacking CSE and MPST, respectively, Figure 3G) showed lower PSSH levels. Drosophila melanogaster flies overexpressing CSE (Snijder et al., 2015) showed increased PSSH levels (Figure 3H), while kidneys from CSE−/− mice showed reduced persulfidation levels (Figure 3I). Finally, endogenous persulfidation could be observed in human RBC, in both membrane and cytoplasm, confirming the proteomic data (Figure 3J, Table S1).
The dimedone switch method was also successfully applied for the visualization of intracellular persulfides by confocal microscopy (Figure 3K, Figure S3C). MEFs lacking CSE showed barely any detectable intracellular levels of PSSH, while both H2S and D-cysteine treatments increased those levels several-fold. Independent of CSE, both sources of H2S increased the PSSH levels highlighting the essential role of H2S in protein persulfide formation (Figure 3K, Figure S3C). Furthermore, wide-field fluorescence deconvolution microscopy provided the first high-resolution images of intracellular persulfidation (Figure S3D–F). The PSSH signal is dispersed throughout the cell, with some of it being observed even in the nucleus. The PSSH signal in D-cysteine treated cells seems to be predominantly localized in mitochondria, in accordance with the fact that D-cysteine is a substrate for cysteine catabolism path via MPST (Shibuya et al., 2013) (Figure S3F).
Broad applicability of dimedone switch method: antibody microarray
To further showcase the applicability of the dimedone switch method, we used an antibody microarray-like approach, where antibodies for specific proteins were immobilized on an NHS-activated surface (Figure 3L). As the samples carry both green and red fluorescence, reflecting the total load and PSSH levels respectively, proteins of interest could be analyzed by this approach and their PSSH levels assessed. We selected antibodies against a series of proteins (Figure 3L) for which persulfidation has been shown, or which form persulfides in their catalytic cycles. In general, the lack of CSE reduced PSSH levels of target proteins while D-cysteine treatment increased it, albeit with different efficiencies. The selectivity of the method is once more demonstrated, as enzymes reported to form persulfides at their active sites during the H2S oxidation, such as sulfide:quinone oxidoreductase (SQOR or SQR) and TST, showed high endogenous PSSH levels with minimal changes upon further D-cysteine treatment. However, a significant decrease of steady-state persulfide levels of those enzymes was observed in cells lacking CSE. Aside from the proteins whose persulfidation has already been demonstrated (GAPDH, HSP70, Keap 1, β-actin, Parkin) (Mustafa et al., 2009; Vandiver et al., 2013; Yang et al., 2013; Zhang et al., 2014), this approach led to the observation that manganese superoxide dismutase (MnSOD) could be persulfidated as well (Figure 3L). Unlike prokaryotes, most eukaryotic MnSOD have at least one cysteine residue (Figure 3M, Figure S3G) and exhibit strong product inhibition by H2O2 (Hearn et al., 2001). Our experiments with human recombinant MnSOD showed that a 15 min exposure to a 3-fold excess of H2O2, inhibited MnSOD activity (0.15 ± 0.06 × 103 U/mg vs. 2.91 ± 0.07 × 103 U/mg in the control) while the co-treatment with 5-fold excess of H2S rescued the enzymatic activity (1.92 ± 0.07 × 103 U/mg) (Figure 3N). MS/MS analysis of human recombinant MnSOD treated with H2O2 and H2S and labeled by the dimedone switch method (using DCP-Bio1 as a switching agent) confirmed that C193 was indeed persulfidated (Figure S3H, I, Dataset S3, S4). Other studies have pointed out that the same cysteine residue is redox sensitive (Matsuda et al., 1990). Furthermore, persulfidated MnSOD was more resilient to tyrosine nitration by peroxynitrite (yield of nitration per subunit 3±2%) when compared to the control (15±4%/subunit), suggesting that persulfidation of MnSOD might serve as a protective mechanism against detrimental enzyme nitration found in many disease states (Szabó et al., 2007).
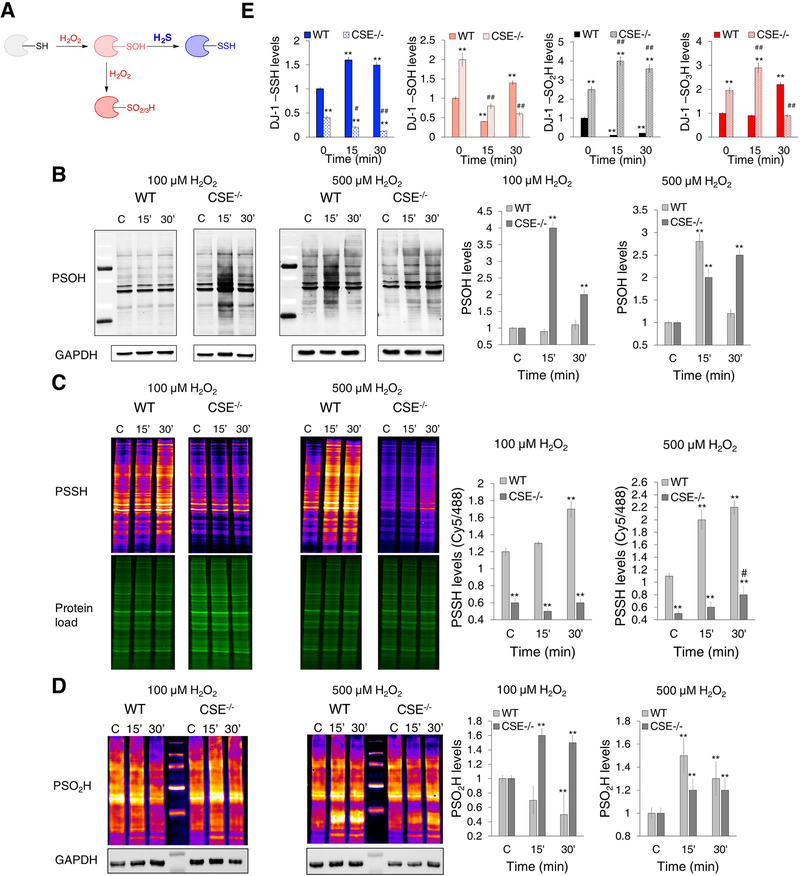
Persulfidation is intrinsically linked to H2O2
For H2S to be able to modify cysteine residues, an oxidant is required - a role that could be played by H2O2. Protein sulfenylation, as a consequence of H2O2 production, represents an important signaling event (Paulsen and Carroll, 2013; Poole et al., 2004). However, PSOH formation should be controlled in order to prevent overoxidation of cysteine residues to sulfinic (PSO2H) and sulfonic acids (PSO3H) that results in a loss of protein function (Figure 4A). Previous studies showed that protein PSOH react two orders of magnitude faster with H2S, than with glutathione at pH 7.4 (Cuevasanta et al., 2015) and our proteomic analysis of persulfidated proteins in RBC showed a significant overlap with proteins known to be sulfenylated (Table S1). We hypothesized that the reaction of H2S with PSOH could represent an integral part of H2O2-induced redox signaling and the main way for resolving PSOH back to thiols. To test this, we first exposed CSE+/+ wild type (WT) and CSE−/− MEFs to H2O2. While 100 μM H2O2 induced no detectable increase in PSOH levels in WT cells, a massive increase in sulfenylation was detected in CSE−/− that decreased as exposure time increased (Figure 4B). 500 μM H2O2 was required to cause the same magnitude of PSOH formation in WT cells (Figure 4B). This effect could be completely abolished by pre-incubating the cells with 100 μM H2S donor, GYY4137 (Figure S4A). Conversely, the PSSH levels in WT cells increased time-dependently when treated with 100 and 500 μM H2O2 but remained very low in CSE−/− cells (Figure 4C). Recent development of selective probes for sulfinic acids (PSO2H) (Akter et al., 2018) allowed us to test how sulfinylation changes in cells lacking endogenous H2S. A strong overall increase of sulfinylation was observed in CSE−/− cells treated with 100 μM H2O2 for 15 min, but this PSO2H dropped back to normal after 30 min suggesting that those cysteines either became hyperoxidized or reduced back by sulfiredoxin (Akter et al., 2018) (Figure 4D). 500 μM H2O2 caused increase of sulfinylation only on a selected group of proteins in WT cells. Sulfinylation of CSE−/− cells, caused by 500 μM H2O2, seems to be somewhat lower than with 100 μM dose, presumably due to their higher sensitivity and a stronger cysteine hyeproxidation to sulfonic acid.
Figure 4. Endogenous H2S controls cysteine oxidation caused by H2O2.
(A) The proposed mechanism for the redox switching between H2O2-induced thiol oxidation and persulfidation.
(B-D) Cysteine oxPTM levels in WT and CSE−/− MEF cells treated with 100 or 500 μM H2O2 for 15 and 30 min. (B) Protein sulfenylation (PSOH) (labeled with DCP-Bio1 and visualized with streptavidin-488). GAPDH was used as a loading control. n = 4. (C) Protein persulfidation (PSSH) (labeled with DAz-2:Cy5 as a switching agent). Ratio of Cy5/488 signals is used for the quantification. n = 3. (D) Protein sulfinylation (PSO2H) (labeled with BioDiaAlk and visualized with streptavidin-Cy5). GAPDH was used as a loading control. n = 5. PSOH and PSSH values were normalized to the levels found in untreated cells. ** p < 0.01 compared to the untreated WT cells; # p < 0.05 compared to the untreated CSE−/− cells.
(E) Persulfidation, sulfenylation, sulfinylation and sulfonylation of DJ-1. WT and CSE−/− MEF cells were treated with 100 μM H2O2 for 15 or 30 min, labeled for PSSH, PSOH and PSO2H, immunoprecipitated with anti-DJ-1 antibody immobilized to agarose beads and immunoblotted with anti-biotin antibody. For sulfonylated DJ-1 (DJ-1-SO3H), antibody selective for C106 sulfonic acid of DJ-1 was used. n = 4. ** p < 0.01 vs. untreated WT. # p < 0.05, ## p < 0.01 vs. untreated CSE−/− cells.
To address how endogenous H2S controls H2O2-induced cysteine oxidation on a molecular level we monitored the cysteine oxidation status in the redox sensitive protein, DJ-1 (also known as PARK7). The C106 in DJ-1 is known to undergo oxidation to a sulfinic (Akter et al., 2018) and even sulfonic acid (Fernandez-Caggiano et al., 2016), while our proteomic analysis identified DJ-1 as a target for persulfidation as well (Supporting Table S1). WT and CSE−/− MEF cells were treated with 100 μM H2O2 for 15 and 30 min, labeled for PSOH (using DCP-Bio1), PSSH (using DCP-Bio1 as a switching reagent) and PSO2H (using BioDiaAlk) and immunoprecipitated. In parallel, using an antibody selective for the C106 sulfonic acid of DJ-1, we assessed the DJ-1-SO3H levels in those samples (Figure 4E, Figure S4B). H2O2 treatment of WT cells resulted in increased persulfidation of DJ-1 (in 15 and 30 min) and increased sulfenylation and sulfonylation (PSO3H) after 30 min. However, in CSE−/− cells already low PSSH levels continued to decrease in cells treated with H2O2. The basal levels of DJ-1-SOH, DJ-1-SO2H and DJ-1-SO3H were much higher in untreated CSE−/− cells when compared to WT. While the DJ-1-SOH decreased with H2O2 treatment in CSE−/− cells, DJ-1-SO2H and DJ-1-SO3H levels continued to further increase (Figure 4E, Figure S4B) confirming that the lack of H2S to trap PSOH results in cysteine hyperoxidation. Taken together these data confirmed that persulfidation, controlled by endogenous H2S production, is an integral part of H2O2-induced redox changes in proteins.
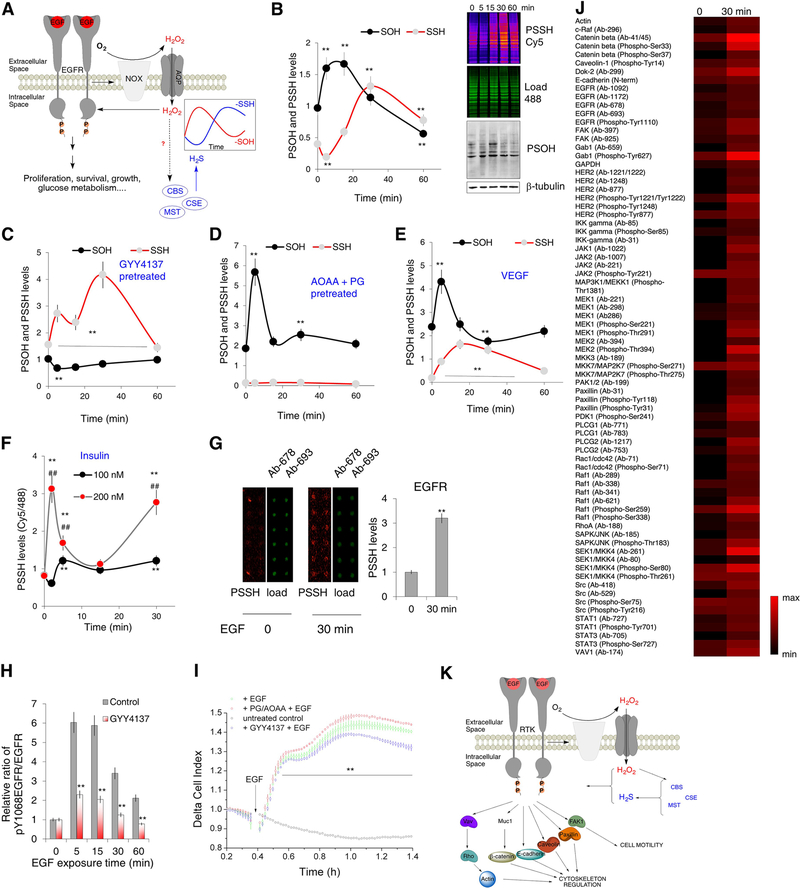
PSSH waves follow PSOH formation: implications for RTK-H2O2 signaling
The importance of PSOH signaling is best exemplified by the receptor tyrosine kinase (RTK) activation (Finkel, 2011; Paulsen et al., 2011; Sundaresan et al., 1995), thus we looked for the temporal correlation between PSOH and PSSH (Figure 5A). HeLa cells treated with 100 ng/ml of epithelial growth factor (EGF) responded by a sharp rise in PSOH within the first 5–15 min that dropped back to basal values by 30 min (Figure 5B). PSSH levels however, followed a phase shifted curve, with the levels initially dropping at 5 min and then reaching a maximum at 30 min (Figure 5B). This correlated well with the increase in expression of all three H2S producing enzymes with EGF (Figure S4C).
Figure 5. Waves of protein persulfidation in RTK signaling.
(A) Schematic representation of the signaling events triggered by the epidermal growth factor receptor (EGFR) activation. Nox: NADPH oxidase; AQP: aquaporin.
(B) HeLa cells treated with 100 ng/mL EGF for 5, 15, 30 or 60 min were analyzed for protein sulfenylation (labeled using DCP-Bio1 and visualized with streptavidin-488, levels calculated using β-tubulin as a loading control) and protein persulfidation (using dimedone switch method with Cy5 as a reporting molecule, levels calculated as a ratio of Cy5/488 fluorescence readouts). (Top) In-gel fluorescence of PSSH levels and Western blots for PSOH levels. (Bottom) Temporal dynamics of PSSH and PSOH changes upon EGF exposure. n ≥ 3. Values are presented as a mean ± SD. ** p < 0.01 vs. control (0).
(C) Quantification of PSSH and PSOH changes as a function of time upon EGF exposure in HeLa cells, pretreated with GYY4137 (100 μM) for 30 min, prior the EGF treatment. n ≥ 3. Values are presented as a mean ± SD. ** p < 0.01 vs. control.
(D) Quantification of PSSH and PSOH changes as a function of time upon EGF exposure in HeLa cells, pretreated with 2 mM mixture of inhibitors, aminooxyacetic acid (AOAA) and propargylglycine (PG) (1:1, 30 min), prior the EGF treatment. n ≥ 3. Values are presented as a mean ± SD. ** p < 0.01 vs. control.
(E) Quantification of PSSH and PSOH changes in HUVEC as a function of time upon VEGF (40 ng/mL) exposure. n ≥ 3. Values are presented as a mean ± SD. ** p < 0.01 vs. control.
(F) The effect of different insulin concentrations on PSSH levels in neuroblastoma (SHSY5Y) cells as a function of time of insulin exposure. n ≥ 3. Values are presented as a mean ± SD. ** p < 0.01 vs. untreated, ## p < 0.01 100 nM vs. 200 nM.
(G) Persulfidation of EGF receptor of HeLa cells treated with 100 ng/mL EGF for 30 min, detected by two different antibodies using antibody microarray slides. Each antibody was spotted in pentaplicated. 2 technical replicates were performed. Values are presented as a mean ± SD. ** p < 0.01 vs. untreated (0).
(H) Time-dependent phosphorylation of EGF receptor tyrosine 1068 (Y1068) as a response to EGF. HeLa cells were pretreated or not with GYY4137 (100 μM) for 2 hr prior to exposure to EGF (100 ng/mL). n = 3. ** p < 0.01 GYY4137 treated vs untreated.
(I) Real-time measurement of EGF receptor activation in living cells recorded with xCELLigence RTCA DP system. HeLa cells were also pretreated with GYY4137 (100 μM, 30 min) or with 2 mM mixture of AOAA and PG (1:1, 30 min). EGF receptor activation was initiated by the addition of 150 ng/mL EGF. n = 4. Values are presented as a mean ± SD. ** p < 0.01 vs. untreated control.
(J) Antibody microarray analysis of persulfidation of different kinases involved in the EGF signaling. HeLa cells were treated with 100 ng/mL EGF for 30 min. Each antibody was spotted in pentaplicated. 2 technical replicates were performed.
(K) Schematic presentation of protein targets involved in actin remodeling, cytoskeleton regulation and cell motility, found to be persulfidated in cells treated with 100 ng/mL EGF for 30 min.
To confirm the interplay between of PSOH and PSSH, we pretreated HeLa cells with GYY4137 (Figure 5C, Figure S4D) or with a mix of CSE and CBS inhibitors (propargylglycine and aminooxyacetic acid, Figure 5D, Figure S4E) for 30 min to either increase or decrease, respectively, the intracellular H2S and PSSH levels. Pretreatment with GYY4137 indeed induced an increase of PSSH, and upon EGF stimulation these levels continued to rise over 30 min, while PSOH initially dropped and remained low and unchanged (Figure 5C, Figure S4D). In contrast, the pharmacological inhibition of endogenous H2S production resulted in a sharp rise of PSOH levels, peaking at 5 min, and being fully resolved at 15 min, presumably due to cysteine overoxidation, as PSSH levels remained very low and unchanged (Figure 5D, Figure S4E). These results strongly suggested that persulfidation represents the innate mechanism that cells use to resolve signaling by sulfenylation. At the same time, as both labeling approaches use dimedone-based probes (for PSSH using at a switching agent), these data confirmed once more that the dimedone switch method can distinguish PSSH from PSOH.
We then tested other RTK pathways. The treatment of human umbilical endothelial vein cells (HUVEC) with 40 ng/ml of vascular endothelial growth factor (VEGF) showed similar phase-shift curves for PSOH and PSSH, with PSOH peaking at 5 min and reverting back to basal levels already after 15 min, while PSSH levels peaked at 15 min and stayed high even after 30 min (Figure 5E, Figure S4F). PSSH increased ~ 9 times in cells exposed to VEGF for 15 min, in accordance with the very high H2S production rate that these cells possess (Filipovic et al., 2018; Lin et al., 2013). The treatment of neuroblastoma cells (SH-SY5Y) with insulin, produced again a distinct peak of PSSH levels, with the peaking time and the intensity of change correlating directly with the dose of insulin (Figure 5F, Figure S4G). A sharp peak of sulfenylation was observed after 2 min of exposure to 100 ng/mL insulin and it was followed by a wave of persulfidation that peaked at 5 min (Figure 5F, Figure S4G,H). When 200 ng/mL was used, the PSSH wave of greater amplitude was detected, with peaking time being 2 min, presumably due to a stronger and faster H2O2 flux produced by higher insulin dose (Figure 5F, Figure S4G). The kinetics of intracellular sulfenylation preceded the PSSH wave in a phase shifted manner (Figure S4H). Finally, we used WT and CSE−/− MEFs and treated them with 100 ng/ml EGF. The temporal profile of PSSH in WT was quite similar to that observed for HeLa cells and was inhibited in CSE−/− cells (Figure S4I,J). On the other hand, sulfenylation was much stronger in CSE−/− cells (Figure S4I,J).
Next, we turned our attention to understanding the biological relevance of these waves of persulfidation. EGF receptor activation is regulated by sulfenylation (Paulsen et al., 2011) so we focused on understanding if and how persulfidation could control the duration of EGF signaling. We first looked for the persulfidation of EGFR using commercially available antibody microarray plates for the EGFR pathway. Pentaplicates of two different antibodies for EGFR showed a strong increase in EGFR persulfidation in HeLa cells treated with EGF for 30 min (Figure 5G). Persulfidation of EGFR had a functional effect on the downstream signaling. Phosphorylation of Y1068, activated by cysteine sulfenylation, was strongly impaired in GYY4137 pretreated HeLa cells (Figure 5H). Correspondingly, the activation of the EGF receptor monitored in live cells also revealed a stronger receptor activation in cells pretreated with CSE and CBS inhibitors (PG and AOAA), an effect that could be reduced by GYY4137 pretreatment (Figure 5I). Furthermore, the inhibition of H2S production to increase the half-life of EGFR sulfenylation caused a significant increase of extracellular signal-regulated kinase (ERK) phosphorylation even without EGF stimulation (Figure S4K).
Many of the cysteine containing phosphatases important for EGFR signaling have been already shown to be sulfenylated (Paulsen et al., 2011). This is particularly true for PTEN, PTP1B and SHPTP2, which we now find to be persulfidated as well (Figure S4L). In addition, using the EGFR pathway antibody microarray, we also assessed the persulfidation status of kinases downstream of EGFR (Figure 5J, Dataset S5). Numerous protein targets were identified with an increased persulfidation status upon exposure to EGF. It is interesting that besides actin, kinases involved in the regulation of cytoskeleton rearrangements and cell motility are particularly affected (Figure 5J, K, Figures S4M–R). Taken together these data suggest that the formation of protein persulfides represents a redox switch for controlling cellular signaling initiated by H2O2 and PSOH formation.
Protein persulfidation as a rescuing path from cysteine hyperoxidation
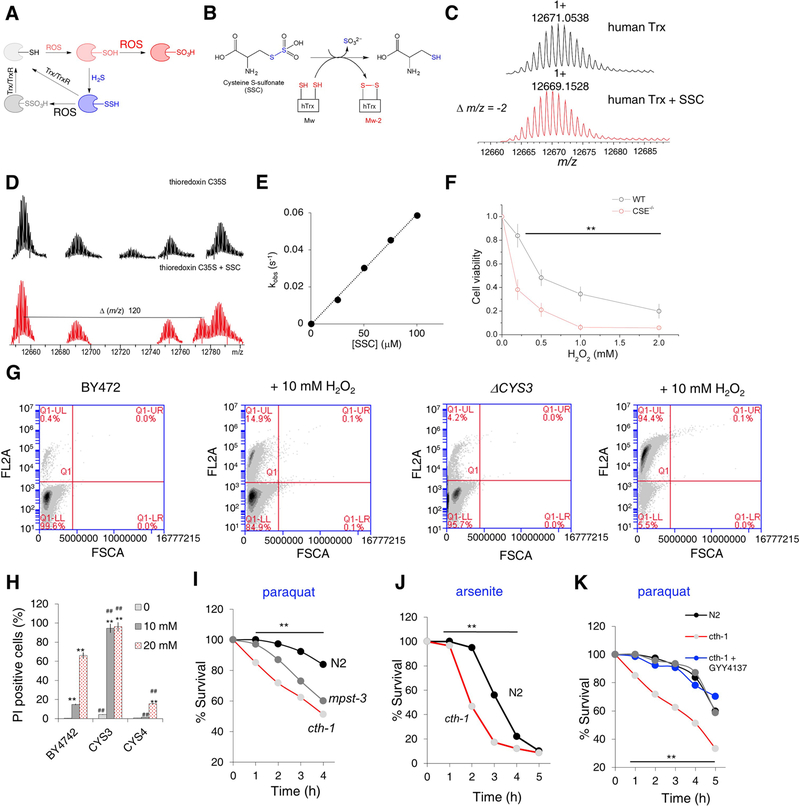
While protein sulfenylation (and even sulfinylation for some proteins) represents a signaling event, uncontrollable production of H2O2 or any of the ROS would result in cysteine hyperoxidation and loss of function (Figure 6A). Our initial hypothesis was that due to its small size, diffusibility and high production flux (Cuevasanta et al., 2012; Filipovic et al., 2018; Vitvitsky et al., 2012), H2S could react as the first line of cellular defence against cysteine hyperoxidation (Figure 6A). Formed persulfides would be much better nucleophiles than cysteines alone, and could scavenge (Cuevasanta et al., 2015) more of the damaging oxidants. Owing to their reducible S-S bond, the ensuing S-sulfocysteine should be readily reduced by disulfide reductases, such as thioredoxin (Trx) (Figure 6A) creating a reduction pathway which would recycle it back to its thiol form. It has been already demonstrated that Trx could reduce S-sulfocysteine formed in the active site of 3′-phosphoadenosine-5′-phosphosulfate reductase (Palde and Carroll, 2015).
Figure 6. Cytoprotective effects of protein persulfidation.
(A) The proposed mechanism for the protective effects of protein persulfidation. Trx-thioredoxin, TrxR-thioredoxin reductase.
(B) Model reaction of S-sulfocysteine (SSC) with human thioredoxin (hTrx).
(C) Deconvoluted MS spectrum of 10 μM human recombinant Trx (black) and Trx treated with 10 μM S-sulfocysteine (SSC) (red).
(D) Deconvolution of MS of 10 μM C35S Trx before (black) and after (red) the reaction with 10 μM SSC showing appearance of TrxS-S-Cys adduct in sample treated with SSC.
(E) Plot of kobs vs. concentration of SSC for the reaction with human recombinant Trx. Reaction was followed fluorometrically by measuring conformational changes induced in Trx due to the cysteine oxidation. Values presented as a mean ± SD. n = 3.
(F) Toxicity of H2O2 in WT and CSE−/− MEFs. Values presented as a mean ± SD. n = 3, ** p < 0.01 vs. control.
(G-H) Flow cytometry analysis of cell death using propidium iodide (FL2A channel). Different S. cerevisiae strains were cultured overnight, adjusted to OD600 = 2, and grown for 27 hr without or with 10 mM and 20 mM H2O2. Upper left quadrant was used as a measure of dead cells. 150000 cells were analysed per measurement. n=2. ** p < 0.01 vs. untreated cells in the same group, ## p < 0.01 vs. corresponding treatment of BY4742 cells.
(I-J) Survival curves of N2, cth-1 and mpst-3 C. elegans strains exposed to 60 mM paraquat (I) and 5 mM sodium arsenite (J). N>80 worms. Experiments were performed in triplicate. ** p < 0.01 vs. control.
(K) The effect of short-term (3 hr) pre-exposure to GYY4137 (500 μM) or AP39 (100 nM) on survival rate of cth-1 C. elegans mutants treated with 60 mM paraquat. N>80 worms. Experiments were performed in triplicate. ** p < 0.01 vs. control.
To address the reversibility of persulfide oxidation, we used S-sulfocysteine (SSC) as a model system (Figure 6B). Incubation of human recombinant Trx with SSC at an equimolar ratio resulted in the complete oxidation of Trx, as observed by the leftward mass shift (Δm/z = −2), corresponding to the formation of a disulfide bond (Figure 6C). When catalytically inactive C32S mutant was incubated with the same amount of SSC, no spectral change could be observed (Figure S5A); whilst incubation with the C35S mutant, which initially reacts with the substrate but cannot complete the catalytic cycle, resulted in the characterization of the disulfide adduct of Trx with cysteine (Figure 6D, Figure S5B). Estimated rate constant of 6.1 ± 0.2 × 104 M−1 s−1 (Figure 6E, S5C) is one order of magnitude higher than that for the reaction of Trx with cysteine persulfide and two orders of magnitude higher than for the Trx reaction with cystine (Wedmann et al., 2016). Coupled with thioredoxin reductase (TrxR), the reaction was also faster than with cystine (Figure S5D). Thioredoxin-related protein (TRP14) however, was not as efficient in cleaving SSC as human Trx (Figure S5E).
We then assessed how persulfidation levels of different living systems correlate with their ability to resist stress. CSE−/− MEFs showed slower growth and were much more sensitive to H2O2 when compared to the WT cells (Figure 6F); S. cerevisiae proved to be no exception either. Despite a slightly different cysteine and H2S metabolism (Figure S5F), the S. cerevisiae mutant Δcys3 (CSE) showed growth retardation (Figure S5G) and had lower PSSH and H2S levels (Figure S5H,I). This mutant was also found to be more sensitive to H2O2 than the wild type (Figure 6G, H). Δcys4 (CBS), on the other hand, exhibited somewhat higher H2S and PSSH levels (Figure SH,I) and was seen to be more resistant to H2O2 (Figure 6H).
We also tested the sensitivity of C. elegans to different ROS-inducing stressors. cth-1 (CSE) and mpst-3 C. elegans mutants showed enhanced sensitivity to paraquat (Figure 6I) with ~ 50% of cth-1 animals dead within 4 hr of exposure (compared to ~80% alive for N2). Another ROS stressor, sodium arsenite, also proved to be more toxic to cth-1 mutants compared to N2 (Figure 6J). Enhanced sensitivity of cth-1 to paraquat and arsenite could be rescued by pretreatment with H2S donors to increase the intracellular PSSH levels (Figure 6K, Figure S5J, K). Even the N2 showed better survival after exposure to paraquat or arsenite, just by a 3 hr pretreatment with either GYY4137 or AP39 (Figure S5J, K).
Protein persulfidation decreases with aging
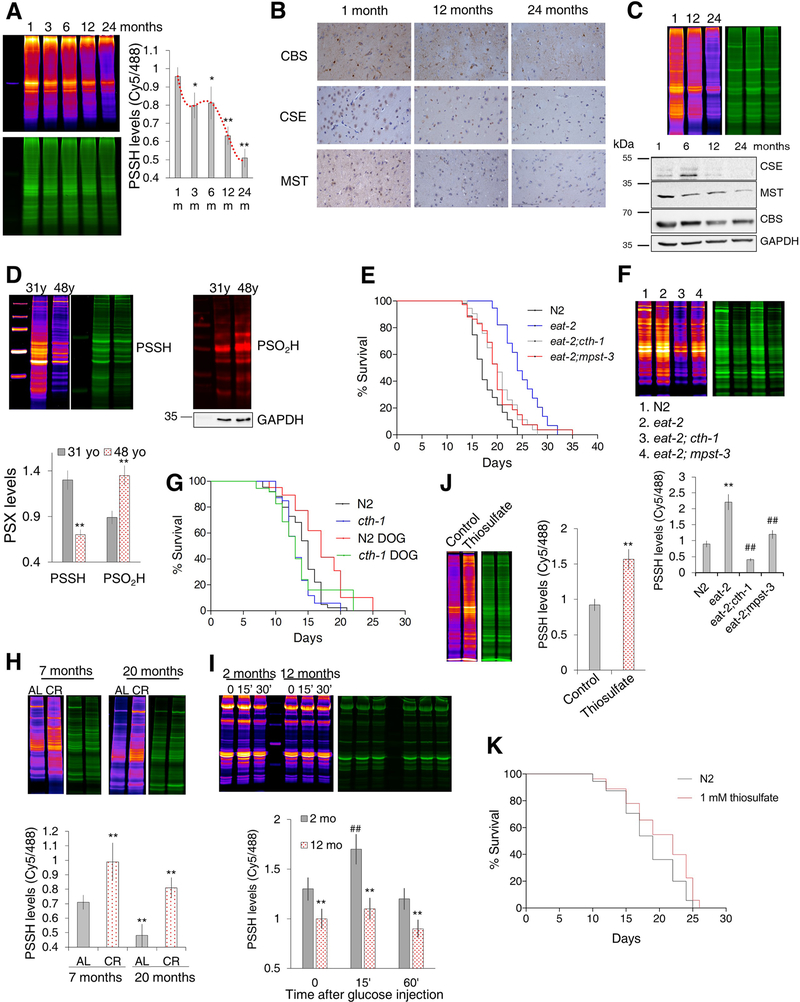
With aging there is an imbalance between ROS production and removal, resulting in an increase in oxidative damage (Balaban et al., 2005; Finkel and Holbrook, 2000; Liochev, 2013; Redman et al., 2018). Moreover, two independent quantitative proteomics studies found that in C. elegans, CSE is decreased during aging (Aging et al., 2015; Narayan et al., 2016). Here we tested the hypothesis that protective pools of intracellular PSSH decline with age and correlate with the lifespan of individuals within a given species. While the cth-1 C. elegans mutants did not display an overall significantly shorter median/maximal life span, they initially exhibited a much higher death rate (Figure S6A). Mpst-3 mutants, which had an even lower PSSH pool (Figure 3G), lived significantly shorter (Figure S6A). Additionally, the persulfidation levels in wild type N2 worms gradually decreased from day 1 to day 7 of adulthood (Figure S6B). To confirm the evolutionary conservation of this phenomenon, we looked at Wistar rats of 1, 3, 6, 12, and 24 months of age. In brain extracts, reduced protein persulfidation levels were observed beginning at 6 months of age, with an approximately 50% lower persulfidation in 24 -month-old rats relative to 1-month-old rats (Figure 7A). The reduction of PSSH correlated with the loss of protein expression of all three H2S producing enzymes (Figure 7B, Figure S6C). We also observed a profound decrease in protein persulfidation and H2S-generating enzyme levels (CSE, MPST) in rat hearts as a function of age (Figure 7C), while in liver PSSH and CSE and CBS levels trended lower but were not significantly different between 12- and 24-month-old rats (Figure S6D).
Figure 7. Anti-aging properties of protein persulfidation.
(A) Changes in the persulfidation levels in brain extracts of male Wistar rats 1, 3, 6, 12 and 24 months of age, calculated as a ratio of Cy5/488 signal. Values are presented as a mean ± SD. n = 3/age group. * p < 0.05, ** p < 0.01 vs 1 month (1m).
(B) Immunohistochemical analysis of CSE, CBS and MST expression levels in the cortex of 1-, 12- and 24-month-old male Wistar rats. Images are representative of 3 animals/experimental group, magnification 20x.
(C) Protein persulfidation levels of 1-, 12- and 24-month-old hearts of male Wistar rats (top). Expression levels of CSE, CBS and MPST in hearts of 1-, 6-, 12- and 24-month-old male Wistar rats (bottom). Images are representative of 3 animals/experimental group.
(D) PSSH and PSO2H levels in human fibroblasts originating from the same donor but collected at the age of 31 and 48. Quantification of thiol modifications, marked on y axis as PSX, represents average ± SD of n = 3. ** p < 0.01 vs. 31 yr.
(E) Survival curves for N2, eat-2, eat-2;cth-1 and eat-2; mpst-3 double mutants. n > 100 per line. N2 = 17.8±0.5 days; eat-2 = 24.5±0.9 days; eat-2;cth-1 = 20.3±0.6 days; eat-2;mpst-3 = 20.2±0.7 days. For eat-2;cth-1 vs. eat-2 and eat-2;mpst-3 vs. eat-2 p < 0.001.
(F) Persulfidation levels in N2, eat-2, eat-2;cth-1 and eat-2; mpst-3 C. elegans mutants. Values are presented as average ± SD. Protein extracts from ~16000 worms were used for each lane. n = 3. ** p < 0.01 vs. N2, ## p < 0.01 vs. eat-2.
(G) Survival curves for N2 and cth-1 mutants grown in the absence or presence of 5 mM 2-deoxy-D-glucose (DOG). n = 110 per each line. N2 = 14.2±0.4 days, N2 5 mM DOG = 17.2 ± 1.0 days; cth-1 = 13.3 ± 0.4 days; cth-1 5 mM DOG = 13.7 ± 1.0 days. For N2 vs. N2 5 mM DOG p= 0.005; for cth-1 vs. cth-1 5 mM DOG p = n.s, for N2 vs. cth-1 p = 0.0565.
(H) Age-induced PSSH changes in 7- and 20- month-old mice fed ad libitum (AL) and mice fed with calorie restriction diet (CR). n = 5 animals per group. ** p < 0.01 vs. 7-month AL mice.
(I) Time-dependent PSSH changes in the muscle tissue of 2- and 12- month old male mice injected i.p. with D-glucose (2 g/kg body weight). n ≥ 3 animals per group. ** p < 0.01 control vs. 2-month old mice, ## p<0.01 2-month vs. 12month old mice.
(J) Persulfidation levels in N2 worms with and without treatment of 1 mM thiosulfate. n = 3. ** p < 0.01 vs. control.
(K) Survival curves for N2 C. elegans, and N2 treated with 1 mM thiosulfate. n > 160 per group. N2 = 18.5 ± 0.3 days, 1 mM thiosulfate = 20.3 ± 0.4 days. p < 0.0001
Finally, as a proof-of-concept experiment we analyzed the persulfidation and sulfinylation levels in human fibroblasts originating from the same individual but collected at different ages (31 and 48 years of age). The results displayed a decrease in PSSH levels and an increase in PSO2H (Figure 7D) in accordance with our hypothesis (Figure 6A).
Life-span extension by dietary restriction is caused by increased protein persulfidation
Recent studies suggest that dietary restriction (DR) increases endogenous H2S production (Hine et al., 2015; Mitchell et al., 2016) and that this increase is associated with multiple benefits including extended longevity in different species. However, the mechanism by which H2S contributes to DR benefits remains poorly characterized. We tested the hypothesis that DR increases longevity in part by increasing protein persulfidation. We started with C. elegans by using the eat-2 genetic model of DR which eats less and lives longer than N2 controls. An additional deletion of either cth-1 or mpst-3 reduced the lifespan of the eat-2 mutant back to control levels (Figure 7E). Interestingly, this was in excellent concordance with the total PSSH pool in these worms; eat-2 mutants had approximately 3 times higher PSSH levels than N2, while double mutants had PSSH levels close to or even lower than N2 (Figure 7F). Furthermore, lifespan extension induced by 2-deoxy-D-glucose (DOG) treatment in wildtype N2 worms was completely reversed in cth-1 and mpst-3 mutants (Figure 7G).
We next looked at 7- and 20-month-old C57BL/6J mice fed ad libitum (AL) or subject to daily 30% calorie restricted (CR) from the age of 2 months. Liver persulfidation levels declined with age, but were higher in both CR groups compared to AL (Figure 7H).
Considering that glucose and insulin tolerance are affected with aging (Fink et al., 1983), and that we already observed that persulfidation is an integral part of insulin signaling (Figure 5F) we monitored PSSH changes in young and old male mice (2- vs 12-month-old) challenged with i.p. glucose injection. A lower persulfidation in old mice, as well as a decrease of the H2S producing enzymes (Figure S6E) were obvious. The peak of persulfidation was observed in the muscle tissue of young mice treated with glucose but was blunted in the 12-month-old animals (Figure 7I), suggesting that the loss of persulfidation capacity with aging is also affecting the insulin signaling and that the beneficial effects of CR to a glucose load (Fontana et al., 2010) could partially be due to the increased persulfidation.
Finally, we tested whether pharmacological interventions to increase persulfidation levels could extend lifespan. We opted for testing an established FDA approved therapeutic drug, sodium thiosulfate, which does not release H2S itself but exhibits beneficial effects mimicking those of H2S (Snijder et al., 2015). We observed that treatment of cell lysates with either thiosulfate, TST or both increased the persulfidation levels several fold (Figure S6F). C. elegans grown on medium supplemented with 1 mM thiosulfate showed higher persulfidation levels (Figure 7J) as such, we decided to test its effect on lifespan. Indeed, worms grown on medium supplemented with 1 mM thiosulfate had a significant increase (~ 15%) in median longevity (Figure 7K).
DISCUSSION
Versatility and selectivity of dimedone switch method
By combining commercially available and well-characterized chemicals, our novel dimedone switch method is a simple, versatile and robust approach for selectively labeling protein persulfides, that enables the installation of various groups to proteins and the use of a global of detection methods. Moreover, our chemical method allows for a wide scale analysis of metabolic pathways that could be controlled by persulfidation and the identification of new redox switches. The versatility of dimedone based probes also permits site-centric identification and quantitation of persulfides by MS, as done for protein sulfenylation (Yang et al., 2014). The installation of different fluorophores through CuAAC and almost undetectable unselective background in microscopic analysis also carries a potential for further exploration of protein persulfidation in tissue sections. Finally, the use of simple in-gel detection prevents all the problems associated with column separation and Western blot transfer, commonly used in other persulfide detection methods.
Persulfidation as integral part of H2O2 signaling or/and as evolutionary conserved route to rescue cysteines
As life emerged in an H2S-enriched environment (Olson and Straub, 2016) and under conditions of extensive radiation where ROS were formed by photolysis of water (Liang et al., 2006), protection of cysteine residues was necessitated. The formation of persulfides poses as the simplest answer. In persulfides, sulfur atoms are oxidized, however concurrently sulfur is deprotonated and highly nucleophilic, reducing ROS at least one order of magnitude faster than its corresponding thiolate (Cuevasanta et al., 2015; Filipovic et al., 2018). In other words, while a cysteine gets oxidized to form a persulfide, it forms a strong reducing agent at the same time.
H2O2 is now widely accepted as a signaling molecule (D’Autréaux and Toledano, 2007; Holmström and Finkel, 2014; Yang et al., 2014). Approximately 1000 cysteine sites have been found to be sulfenylated, raising the question of how these sites are rescued. In our study, waves of persulfidation are observed following sulfenylation, as a response to RTK activation, confirming that persulfidation is an integral part of RTK signaling. A recent study demonstrated that a large number of proteins undergo further oxidation to sulfinic acid as well (Akter et al., 2018). Our results demonstrate that without H2S/persulfidation proteins undergo a substantial thiol oxidation (sulfenylation, sulfinylation and sulfonylation) even with H2O2 concentrations that cause no change in normal cells. This overlooked phenomenon questions whether and in which concentration range H2O2 acts as a signaling molecule without H2S.
We demonstrate that as cells age their persulfidation levels decrease due to the loss of H2S producing enzymes. Reacting with ROS, a persulfide will form a S-sulfonate which we show could be readily reduced back to a thiolate by thioredoxin (Figure 6A). This rescue loop for preserving cysteine residues in proteins and preventing their loss of function is probably an evolutionary remnant used as a general protective mechanism in all life forms. This is best documented by the enhanced sensitivity of different life forms with low PSSH levels to oxidant stressors and their increased resistance once the intracellular PSSH pool is increased even by a short-term pretreatment with H2S donors. This unifying mechanism explains the beneficial effects of DR, already known to result in H2S overproduction (Hine et al., 2015), as well as lifespan extension caused by pharmacological increase of protein persulfidation. Thus, our results further strengthen the ROS theory of aging (Redman et al., 2018).
It is worth mentioning that beside a general protection, persulfidation of specific proteins could result in an alteration of their function (Filipovic et al., 2018; Paul and Snyder, 2015; Vandiver et al., 2013), as we observe in the case of MnSOD, whose activity is preserved after persulfidation. It is possible that DR-induced decrease of ROS (Redman et al., 2018) could also be partially related to better activity of ROS removing enzymes.
Beside aging, general protective effects of protein persulfidation could be translated to many other disease states. HD and neurodegeneration are strongly linked to CSE expression and pharmacological interventions to increase CSE levels, such as monensin treatment, show promising therapeutic effects (Paul et al., 2014; Sbodio et al., 2016, 2018). Decreased persulfidation in HD cells and increased PSSH levels by upregulating CSE expression reported in our study could provide a general explanation for this. Taken together, our data represent a good starting point for the fine tuning of targeted therapeutic approaches to improve healthspan and lifespan.
Limitations of study
In this study we report a new chemoselective approach to label protein persulfidation and its broad applicability. We prove that persulfidation has a protective role, rescuing cysteines from hyperoxidation. However, the extent to which this modification is just protective or also regulatory remains unclear. Quantitative, site-centric proteomic analysis combined with metabolomic and targeted structural analysis of the persulfidated proteins would shed light on the functional impact that this posttranslational modification has.
Moreover, the intertwined nature of H2O2 and H2S signalling, described in our work, warrants further investigation. We show this to be the case for receptor tyrosine kinase signalling, but H2O2 signaling has been implicated in many other physiological processes. The role of H2S through persulfidation in these pathways remains to be further investigated, in order to position this PSOH/PSSH switch globally in the signalling scheme.
Finally, we demonstrate that persulfidation decreases with aging and that pharmacological or dietary interventions to increase persulfidation extend lifespan. How and why H2S producing enzymes decrease with aging remains to be understood, as well as the exact mechanism by which increased persulfidation correlates with lifespan, or even healthspan extension. However, the dimedone switch method that we describe herein allows us to dig further into these processes that appear to be just the beginning of a clearer picture.
STAR METHODS
LEAD CONTACT AND METERIALS AVAILABILITY
This study did not generate any new reagents. Further information and requests for resources and reagents should be directed to, and will be fulfilled by the Lead Contact, Milos Filipovic (milos.filipovic@ibgc.cnrs.fr)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines
HeLa cells and Neuroblastoma cells (SH-SY5Y) were both obtained from ECACC. HeLa cells were cultured in Dulbecco’s modified Eagle’s media (DMEM, high glucose and sodium pyruvate) supplemented with 2 mM L-glutamine, 1% penicillin-streptomycin and 10% calf serum at 37 °C and 5% CO2.
SH-SY5Y cells were cultured in Ham’s F12 : DMEM (1 : 1) media, supplemented with 2 mM glutamine, 1% penicillin-streptomycin and 10% calf serum at 37 °C and 5% CO2.
C-pooled human umbilical vein endothelial cells (HUVEC) were obtained from PromoCell, Germany and cultured in Endothelial Cell Growth Medium Kit (C-22110, PromoCell, Germany) at 37 °C and 5% CO2.
Mouse Embryonic Fibroblasts (MEF) were generated from male wild type (WT) and CSE−/− mice and immortalized using SV40T antigen (Sbodio et al., 2016). MEF cells were cultured in DMEM (high glucose), supplemented with 2 mM L-glutamine, 1% penicillin-streptomycin and 10% calf serum at 37 °C and 5% CO2.
The striatal progenitor cell line STHdhQ7/Q7, expressing wild-type huntingtin, and STHdhQ111/Q111, expressing mutant huntingtin, harbouring 111 glutamine repeats (referred to as Q7 and Q111 cells, respectively), were from M. MacDonald (Massachusetts General Hospital, Boston, MA). The cells were maintained in DMEM (low glucose, no pyruvate) supplemented with 2 mM L-glutamine, 1% penicillin-streptomycin and 10% calf serum at 33 °C and 5% CO2.
Human Fibroblasts cell lines AG08790 (31 years old) and AG14245 (48 years old - a culture initiated from a biopsy taken 17 years earlier from the same male donor is AG08790) were obtained from Coriell Institute for Medical Research. Fibroblasts were cultured in DMEM supplemented with 2 mM L-glutamine and 10% calf serum at 37 °C and 5% CO2.
C. elegans
The following strains were used in this study: Wild-type Bristol N2, cth-1(ok3319)V, mpst-3(tm4387)V, eat-2(tm5786)II, eat-2(tm5786)II;cth-1(ok3319)V, eat-2(tm5786)II;mpst-3(tm4387)V. The original isolate VC2569 cth-1(ok3319)V was provided by the Caenorhabditis Genetics Center whereas the original isolates FX04387 mpst-3(tm4387)V and FX19451 eat-2(tm5786)II were provided by the MITANI Lab through the National Bio-Resource Project of the MEXT, Japan. All mutant strains were outcrossed at least 4 times before use. Double-mutant strains were constructed by using standard techniques (Brenner, 1974; Sulston and Hodgkin, 1988) and the presence of both mutations were checked by PCR. Worms were grown at 20 °C on standard nematode growth medium (NGM) plates, unless otherwise stated, using standard C. elegans techniques (Stiernagle, 2006) with sufficient food (E. coli OP50–1) for at least two generations prior to use.
S. cerevisiae
Yeast cells were grown in liquid YPD media (1% yeast extract, 1% bactopeptone, and 2% glucose), unless otherwise stated.
Wistar rats
Male Wistar rats used in the experiment were bred and housed at the Institute for Biological Research “Siniša Stanković”, Belgrade, Serbia, under constant laboratory conditions (22±2˚C, 12–12 hr light-dark cycle). Food and water were available ad libitum. Healthy control animals were decapitated at the ages of 1, 3, 6, 12 and 24 month(s). All animal procedures were in compliance with the EEC Directive (86/609/EEC) on the protection of animals used for experimental and other scientific purposes and were approved by the Ethical Committee for the Use of Laboratory Animals of the Institute for Biological Research “Siniša Stanković”, University of Belgrade.
Male C57BL/6J mice (AL and CR experiments)
All experiments were performed with the approval of the Harvard Medical Area Institutional Animal Care and Use Committee (IACUC). Male C57BL/6J mice were obtained from the NIA Aging Colony Resource at Charles River Laboratories (CRL) at 6 months (young) and 19 months (old) of age. Mice were allowed to acclimatize to the facility for one month to ensure weight stabilization after shipment, so at sacrifice were 7mo old (young) and 20 months (old). Initiation of caloric restriction (CR) was performed as previously described (Turturro et al., 1999) (starting at 14 weeks of age, CR is initiated in a step-down fashion, where it its increased to 25% restriction at 15 weeks, and the full 40% restriction imitated at 16 weeks where it is maintained throughout the life of the animal. Ad libitum (AL) animals were fed NIH-31 diet, while CR animals are fed NIH-31 fortified diet (Turturro et al., 1999). Other details about the husbandry conditions at CRL can be found here: https://www.nia.nih.gov/research/dab/aged-rodent-colonies-handbook/barrier-environmental-information.
Mice were single housed for the duration of their life in standard mouse cages. Upon arrival at Harvard, mice were housed in the barrier facility at HSPH in microisolator cages with corncob bedding, a nestlet and one shepherd shack. Mice receive free access to water, and either AL NIH-31, or fortified NIH-31 (1 pellet per day for the CR mice). CR mice were fed daily between the hours of 7:00–8:00am by feeding the ration of food onto the floor of the cage. Any remaining food was removed the next day prior to giving the new food ration. Mice were housed in a room on a 12/12 hr light/dark cycle (7am-7pm) with temperature between 20–23 °C with 30%–70% relative humidity. Cages were changed weekly using full sterile technique and performed in Class II laminar flow change stations with spot changes as required.
Male C57BL/6J mice (Glucose Tolerance Test)
Animals were housed on a 12-h light–dark schedule and received food and water ad libitum, except for the glucose tolerance experiment, where the mice were starved overnight before injection with glucose. All animals were treated in compliance with the recommendations of the National Institutes of Health and approved by the Johns Hopkins University Committee on Animal Care.
METHOD DETAILS
All chemicals were purchased from Sigma Aldrich, unless otherwise mentioned. All buffers were prepared with nanopure water and treated with Chelex-100 resins to remove traces of metal ions. Na2S solutions were prepared and handled as recommended (Wedmann et al., 2014). Nmc-penicillamine was synthesized, as previously described (Artaud and Galardon, 2014). Morpholin-4-ium 4 methoxyphenyl(morpholino) phosphinodithioate (GYY4137) and (10-oxo-10-(4-(3-thioxo-3H-1,2-dithiol-5-yl)phenoxy)decyl)triphenylphosphonium bromide (AP39) were synthesized in house according to (Alexander et al., 2015; Le Trionnaire et al., 2014).
Preparation of Specific oxPTM of Proteins
Preparation of HSA-SH, HSA-SOH and HSA-SSH
Reduced HSA (HSA-SH)
1 g HSA was dissolved in 10 ml dH2O and adjusted to pH 3 using concentrated HCl. Charcoal (0.5 g) was then added to the solution and stirred on ice for 1 h. The solution was centrifuged (16000 x g, 30 min, 4 °C), the supernatant was collected and centrifuged (26000 x g, 30 min, 4 °C) again and then filtered on ice, using a 22 μm filter. This was then reduced by adding 5 mM β-mercaptoethanol and incubating for 1 hr at RT or overnight at 4 °C. The resulting solution was filtered using a Sephadex 10-based gel filtration chromatography with 30 mM phosphate buffer. The fractions containing HSA were collected, pooled and the protein concentration was determined using UV/Vis spectrophotometry, using the molar extinction coefficient of ε280 = 35279 M−1 cm−1.
Sulfenylated HSA (HSA-SOH)
Reduced HSA-SH in phosphate buffer was incubated with 10-fold excess H2O2 at 37 °C for 15 min and then cleaned using a biospin column (BioRad), as instructed by the manufacturer.
Persulfidated HSA (HSA-SSH via HSA-TNB)
Reduced HSA-SH in phosphate buffer was incubated with 20-fold excess of Ellman’s Reagent at RT for 1 h, with constant mixing. The resulting solution was cleaned on Sephadex G10-column with 30 mM phosphate buffer. 1.5 equivalents of H2S were added to 1 equivalent of HSA-TNB and incubated for 15 min at 37 °C or 1 hour on ice. The solution was immediately cleaned through a biospin column to obtain HAS-SSH. Once made, HSA-SSH was kept on ice and used immediately.
Preparation of GAPDH persulfide
GAPDH persulfide was prepared as described for HSA derivatives.
Preparation of Rhodanese (TST) modifications
Reduced TST (TST-SH)
Bovine TST (Sigma Aldrich) was treated with 100-fold excess of DTT for 30 min at 37 °C and then cleaned by gel filtration using a mini biospin column (BioRad) as instructed by manufacturer.
Persulfidated TST (TST-SSH)
Bovine TST (already largely present as a persulfide) was incubated with a 10-fold excess of sodium thiosulfate in 10 mM Tris buffer (pH 8) at 37 °C for 30 min and then cleaned with a mini biospin column (Bio-Rad) as instructed by manufacturer.
Dimedone Switch Method for Purified Proteins
Protein was incubated with 5 or 10 mM NBF-Cl in 50 mM PBS (40mM Na2HPO4, 10 mM NaH2PO4 and 135 mM NaCl, pH 7.4) supplemented with SDS (final conc. 2%) for 30 min at 37 °C. The solution was then precipitated by methanol/chloroform precipitation; Sample/MeOH/CHCl3, 4/4/1 (v/v/v) was added and centrifuged (14000 x g, 15 mins, 4 °C). The protein pellet obtained will be between the organic and aqueous layers, both layers was aspirated and H2O/MeOH/CHCl3, 4/4/1 (v/v/v) was added to the protein pellet and centrifuged. Supernatant was aspirated again and the pellet was subsequently washed with MeOH 2–3 times. Pellet was resuspended in 50 mM PBS containing 2% SDS, incubated with either 50 μM dimedone or DCP-Bio1 for 1 hr at 37 °C, precipitated with methanol/chloroform as previously mentioned, and re-suspended in PBS containing 2% SDS. 1 equivalent of Laemmli (4X) buffer (BioRad) supplemented with 10% β-mercaptoethanol, was then added to 3 equivalents of sample for SDS-PAGE and boiled at 95 °C for 5 min protected from light.
For some experiments, the DCP-Bio1-labeled samples were redissolved in 50 mM PBS only, incubated with neutravidin beads (Thermo Fischer Scientific) for 2 hr at room temperature (RT) with continuous mixing. Beads were then washed with 10 volumes of PBS supplemented with 0.01% Tween-20 and bound proteins were eluted by boiling the beads in a minimum volume of Laemmli buffer supplemented with 10% β-mercaptoethanol (1X with PBS) buffer for SDS-PAGE for 5 min. Samples were resolved by SDS-PAGE and gels were fixed in fixation buffer for 30 min, protected from light. The gel was recorded, at 635 nm for the Cy5 signal and 473 nm for NBF-Cl signal, on the Typhoon FLA 9500 (GE Healthcare).
ESI-TOF MS of the dimedone switch reaction with LMW persulfides
Mass spectrometry was performed on maXis 5G, Bruker Daltonic (Bremen, Germany), an ESI-TOF MS capable of resolution of at least 40,000 FWHM. Detection was in positive-ion mode. 100 μM nmc-penicillamine persulfide was mixed with 100 μM NBF-Cl in ammonium bicarbonate buffer, (pH 7.4, 23 °C) and the reaction monitored for 15 min. 500 μM dimedone was then added and the reaction monitored for an additional 15 min.
MS of Protein Persulfide Models Labeled with DCP-Bio1
Protein persulfides switch labeled with DCP-Bio1 as previously described and resolved by SDS-PAGE, and protein bands excised and digested with either trypsin or chymotrypsin following previously described protocol (Crouzet et al., 2017). Digested peptides were analyzed by LC-MS/MS on a Thermo Scientific Q Exactive Orbitrap mass spectrometer in conjunction with a Proxeon Easy-nLC II HPLC (Thermo Fisher Scientific) and Proxeon nanospray source at Bordeaux Proteomic Platform. The digested peptides were loaded a 100 micron x 25 mm Magic C18 100Å 5U reverse phase trap where they were desalted online before being separated with a 75 micron x 150 mm Magic C18 200Å 3U reverse phase column. Peptides were eluted using a 120 min gradient with a flow rate of 300 nL/min. An MS survey scan was obtained for the m/z range of 350–1600; MS/MS spectra were acquired using a top 12 method, where the top 12 ions in the MS spectra were subjected to High Energy Collisional Dissociation (HCD). An isolation mass window of 2 m/z was used for the precursor ion selection, and normalized collision energy of 27% was used for fragmentation. Five second duration was used for the dynamic exclusion. Peptide identification was performed using PEAKS Studio (BSI, Canada) (Zhang et al., 2012). The search settings were: precursor Δm tolerance = 10 ppm, fragment Δm tolerance = 0.2 Da, missed cleavages = 2, modifications of lysine: NBF (163.0012), modifications of cysteine: NBF (163.0012), DCP-Bio1 (394.1557), or hydrolyzed DCP-Bio1 (168.0786).
Proteomic Analysis of Persulfidated Proteins in Red Blood Cell Lysates
9 mL of peripheral whole blood from participants of this study, who provided informed consent in accordance with the Declaration of Helsinki, was collected in citrate and processed immediately following previously described protocol (Pasini et al., 2006) with the modification that the lysis buffer contained 5 mM NBF-Cl. After 30 min of incubation with lysis buffer at 4 °C, additional NBF-Cl was added (to final concentration of 15 mM NBF-Cl) with SDS (to final concentration of 2%) and incubated for 30 min. Methanol/chloroform precipitation was performed as previously described, and the protein pellet obtained was redissolved in 50 mM PBS supplemented with 0.1% SDS. Endogenously biotinylated proteins were precleared by incubating with Pierce™ NeutrAvidin™ Agarose (Thermo Fisher Scientific) at RT for 2 hr with agitation. The resins were subsequently removed on a Pierce™ Disposable Column (Thermo Fisher Scientific) and the solution obtained was precipitated by methanol/chloroform. The resulting protein pellet was redissolved in 50 mM PBS supplemented with 2% SDS and was incubated with 50 μM DCP-Bio1 at 37 °C for 1.5 hr. Solution was precipitated with methanol/chloroform and redissolved in 50 mM PBS supplemented with 0.1% SDS. The protein solution was incubated with Pierce™ High Capacity NeutrAvidin™ Agarose (Thermo Fisher Scientific) at 4 °C overnight with agitation. Samples were then brought to RT and loaded on a column. The resins were washed with 8 column volumes of 50 mM PBS supplemented with 0.001% Tween-20, 2 column volumes of 50 mM PBS and finally with 1 column volume of H2O. After washing, the resins were collected from the column and incubated with 2.25 M ammonium hydroxide at RT, overnight with agitation. The sample was then neutralized with formic acid and protein concentration was determined. 1 equivalent of enzyme digestion buffer was added (for trypsin digestion: 100 mM ammonium bicarbonate buffer; for chymotrypsin digestion: 100 mM Tris and 10 mM CaCl2, pH 7.8). Digestion was performed at an enzyme-to-substrate ratio of 1:50 (wt:wt) and incubated at 37 °C overnight with agitation. Enzyme was added again to solution at an enzyme-to-substrate ratio of 1:20 (wt:wt) and reincubated at 37 °C for 5 hr with agitation. The resulting peptide sample was then quenched by bringing to pH 3, with conc. HCl, and analyzed at Bordeaux Proteomic Platform. Trypsin digested peptides were analyzed by LC-MS/MS on a Thermo Scientific Q Exactive Orbitrap mass spectrometer in conjunction with a Proxeon Easy-nLC II HPLC (Thermo Fisher Scientific) and Proxeon nanospray source. Samples were separated on 300-μm ID x 5-mm C18 PepMapTM precolumn and 75 μm ID x 25 cm nanoViper C18, 2 μm, 100 Å – Acclaim® PepMap RSLC column using 4–40% gradient of B (A: H2O/MeCN/HCOOH, 95/05/0.1, B: H2O/MeCN/HCOOH, 20/80/0.1). Peptides were eluted using a 120 min gradient with a flow rate of 300 nL/min. An MS survey scan was obtained for the m/z range of 3501600; MS/MS spectra were acquired using a top 12 method, where the top 12 ions in the MS spectra were subjected to High Energy Collisional Dissociation (HCD). An isolation mass window of 2 m/z was used for the precursor ion selection, and normalized collision energy of 27% was used for fragmentation. Five second duration was used for the dynamic exclusion. Peptide identification was performed using PEAKS Studio (BSI, Canada). The search settings were: precursor Δm tolerance = 10 ppm, fragment Δm tolerance = 0.2 Da, missed cleavages = 2, −10logP>50, modifications of lysine: NBF (163.0012), modifications of cysteine: NBF (163.0012), DCP-Bio1 (394.1557), or hydrolyzed DCP-Bio1 (168.0786).
In Gel Detection of Persulfidation (Dimedone Switch Method)
Preparation of DAz-2:Cy-5 Click Mix
Final concentrations of 1 mM DAz-2 (Cayman Chemical), 1 mM Cyanine5 alkyne (Lumiprobe), 2 mM copper(II)-TBTA complex (Lumiprobe) and 4 mM ascorbic acid made in situ, were added sequentially in 15 mM PBS buffer mixed with 30% (vol/vol) acetonitrile. The solution was mixed at RT overnight and then quenched with 20 mM ethylenediaminetetraacetic acid (EDTA) and mixed at RT for 2 hr.
Persulfidation of PTP1B
PTP1B, human recombinant, was purchased from Abcam at 1 mg/mL. Protein was desalted on biospin columns (BioRad) and concentration adjusted to 10 μM. Samples were either left untreated, exposed to 50 μM H2O2, or combination of 50 μM H2O2 and H2S for 15 min at 37 °C. Samples were then desalted and treated with or without 1 mM NBF-Cl in HEPES buffer supplemented with 2 % SDS for 1 h at 37 °C. Following a desalting step on biospin column, the same samples were treated with 20 μM DAz-2:Cy-5 Click Mix for 30 min at at 37 °C. Unreacted reagent was removed by desalting on biospin columns and samples mixed with Laemmli buffer supplemented with 10% β-mercaptoethanol.
Persulfide Detection in Cell Lysates
Cells were grown to 80–90% confluency in a 10 cm Petri dish, and following respective treatments, were gently washed twice with cold 15 mM PBS. 1 ml of cold HEN lysis buffer (50 mM Hepes, 1 mM EDTA, 0.1 mM Neocuproine, 1% IGEPAL and 2% SDS, adjusted to pH 7.4, 1% protease inhibitor) supplemented with 5 mM 4-chloro-7-nitrobenzofurazan (NBF-Cl) was then added onto the cells. Cells were gently scrapped, lysates were collected, homogenized with a syringe and needle and immediately placed for incubation at 37 °C for 30 min, protected from light. A methanol/chloroform precipitation was performed, as previously described, protected from light. Protein pellets obtained were then redissolved in 50 mM Hepes (Euromedex, adjusted pH 7.4) supplemented with SDS (2% final conc.). Once fully dissolved, protein concentration was determined using a DC Assay (BioRad) and adjusted to approx. 3 mg/ml. DAz-2:Cy-5 click mix (final. conc. 25 μM), prepared as described above, was added to the adjusted samples and incubated at 37 °C for 30 min, protected from light. The sample was then precipitated by methanol/chloroform and the protein pellet obtained was redissolved in Hepes with SDS (final conc. 2%). The protein concentration was adjusted to 2–2.5 mg/ml and sample was prepared for SDS-PAGE and recorded, as previously described.
For H2S donor treatments were performed with 200 μM Na2S for 45 min, 200 μM GYY4137 for 2 hr, 200 nM AP39 for 2 hr and 2 mM D-Cysteine for 1 hr. Erastin treatments were performed by treating cells with 1 μM and 10 μM erastin for 18.5 hr. Monensin treatments were performed with 1 μM monensin for 18 hr.
For PSSH labeling in thiosulfate and TST treated HeLa lysates, cells were lysed with RIPA lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 2 mM EDTA, 1% IGEPAL and 2% SDS, adjusted to pH 7.4) supplemented with 1% protease inhibitor, as previously described. Lysates were precipitated by TCA precipitation; 100% TCA stock solution was added to sample to obtain a final mixture of 20% TCA. The mixture was incubated on ice for 10 min followed by centrifugation (30000 x g, 10 min, 4 °C) and supernatant was aspirated. Protein pellet was then washed with cold acetone and centrifuged (30000 x g, 10 min, 4 °C) twice, and left to dry. Protein pellet was redissolved in 50 mM Hepes and treated with or without 500 μM thiosulfate, 5μM thiosulfate sulfur transferase (TST) or a co-treatment of both, for 1 hr at 37 °C. Next, 10 mM NBF-Cl was added to the mixtures, incubated for 1 hr at 37 °C and cleaned by methanol/chloroform precipitation. Samples were then switch labeled with DAz-2:Cy5 preclick mix and processed for SDS PAGE as previously described.
Persulfide Detection in Escherichia coli Lysates
The laboratory strain E. coli MG1655 was transformed with the pSB74 plasmid that contains the phsABC operon of S. Enterica serovar Typhimurium for H2S production. As a negative control, E. coli was transformed with an empty vector (pTrc99a). Both strains were streaked on TSA plates supplemented with ampicillin 50 mg/ml, and incubated overnight at 37 °C. The next day, one colony of each strain was inoculated into TSB medium supplemented with 50 mg/ml of ampicillin and grown at 37 °C with agitation (100 rpm), as a starter inoculum. 10 μl of each of these overnight-grown cultures were transferred into 10 ml of fresh TSB medium supplemented with the appropriate antibiotic. Both strains were treated with or without 20 mM sodium thiosulfate for 4 hr at 37 °C with agitation (120 rpm). After the incubation time, samples were harvested by centrifugation at 5000 rpm for 4 min, washed with ice-cold PBS and resuspended in HEN lysis buffer supplemented with 1% protease inhibitor and 25 mM NBF-Cl. Cells were disrupted on ice by sonication, 20 seconds at 190 MHz 2 times with a 2 min pause, and incubated at 37 °C for 1 hr, protected from light. Samples were then precipitated and protein pellets were processed as previously described for persulfide labeling, with 50 μM DAz-2:Cy5 preclick mix, for 1 hr at 37 °C protected from light.
Persulfide Detection in Saccharomyces cerevisiae Lysates
200 μL of yeast cell strains WT, Δcys3, Δcys4 and Δtum1 were mixed with 1 mL HEN lysis buffer supplemented with 1% protease inhibitor for yeast and 20 mM NBF-Cl. Mixture was added dropwise into liquid nitrogen and grinded using a mortar and pestle together with glass beads, reaching a fine powder consistency. Samples were transferred in 2 ml tubes and centrifuged (1500 x g, 15 min at 4 °C). Supernatants were collected and incubated at 37 °C for 1 hr, protected from light. After precipitation, protein pellets were processed as previously described with 50 μM DAz-2:Cy5 preclick mix for 45 min at 37 °C.
Persulfide Detection in Caenorhabditis elegans Lysates
Synchronous populations of embryos were obtained by lysing gravid hermaphrodites in alkaline bleach as previously described (Emmons et al., 1979). Once washed free of the alkaline bleach by centrifugation, the embryos were put on standard NGM agar plates seeded with E. coli OP50–1, ~4000 embryos/plate. At Day-1 adult stage worms of different strains (N2, cth-1, mpst-3, eat-2, eat-2;cth-1, eat-2;mpst-3) were collected from the NGM plates, 4 plates/strain, into 15 ml falcons using M9 buffer and washed three times. Worm pellets were frozen in liquid nitrogen and 500 μl of glass beads was added in every tube. Samples were put in the bead beater (FastPrep-24, MP Biomedicals, California, USA) for 35 seconds at speed 6.5 m/s, followed by an additional cycle at the same speed for 20 seconds. HEN lysis buffer supplemented with 1% protease inhibitor and 20 mM NBF-Cl was added to each tube, and centrifuged for 15 min at 13000 rpm at 4°C. Supernatants were collected and incubated at 37 °C for 45 min. Samples were then precipitated and protein pellets were switch labeled for persulfides and processed as previously described. Synchronous populations of embryos were obtained as described above and put on standard NGM agar or NGM agar plates supplemented with 1 mM sodium thiosulfate seeded with E. coli OP50–1. The plates were then incubated at 20 °C until worms reached Day-1 adult stage. Worms were collected from the plates and processed for persulfide labeling, as previously described. For the detection of persulfide levels in aging N2 worms (Day-1, Day-3 and Day-7) synchronous populations of embryos were placed on standard NGM-agar plates seeded with E. coli OP50–1. L4 staged worms were transferred to NGM agar plates containing 25 μM 5-Fluoro-2’-deoxyuridine (FUdR). One day after transfer, worms were collected with M9 buffer and filtered (Cell Strainer 40 μm, ClearLine) in order to remove the eggs. Worms were either directly lysed and processed for persulfide labeling as previously described (for Day-1 of adulthood) or placed back onto FUdR containing plates and grown for respective times (Day-3 and −7 of adulthood), whilst filtering and transferring to fresh FUdR plates every 2–3 days.
Persulfide Detection in Drosophila melanogaster Lysates
As wild-type control, the y1w1118 Drosophila line was used. Eip55E (Drosophila CSE)-overexpressing lines were a kind gift from Professor Ody Sibon (University of Groningen). 3–4 whole flies were grinded in dounce homogenizer with 50 μL of HEN lysis buffer supplemented with 1% protease inhibitor and 20 mM NBF-Cl on ice. Homogenates were centrifuged at 30000 x g for 20 min and the supernatant was incubated for 30 min at 37 °C, protected from light. Samples were then precipitated and protein pellets were switch labeled for persulfides and processed as previously described.
Persulfide Detection in Mouse Kidney Lysates
CSE +/+ and CSE −/− C57BL/6 mice were generated and previously characterized (Markó et al., 2016). CSE+/− males and females were bred to obtain WT and CSE−/− littermates. Mice were allowed free access to standard chow and water. The mice were kept in a 12:12-hr light-dark cycle. The kidneys from these mice were kind gift from Professor Maik Gollasch (Charité Medical Faculty). 5–10 mg of kidney tissue was homogenized with 500 μL of HEN lysis buffer supplemented with 1% protease inhibitor and 20 mM NBF-Cl on ice in dounce homogenizer. Homogenates were centrifuged at 30 000 x g for 20 min and the supernatant was incubated for 30 min at 37 °C, protected from light. Samples were then precipitated and protein pellets were switch labeled for persulfides and processed as previously described.
Persulfide Detection in Rat Tissue Lysates
Immediately after decapitation, the brain, heart and liver were quickly removed and snap frozen in liquid nitrogen. The organs were shredded and homogenized in HEN lysis buffer supplemented with 1% protease inhibitor and 20 mM NBF-Cl using a dispersion system (Ultra-Turax T25, Janke & Kunkel, IKA-Labortechnik, Germany) at 8000 rpm on ice. After 1 hr incubation at 37 °C protected from light, samples were precipitated, processed for persulfidation labeling (with 50 μM DAz-2:Cy5 preclick mix) as previously described.
Persulfide Detection in Liver of Aging Mice following dietary restriction (AL and CR)
On the day of the experiment, mice were brought to the procedure room (6.30 am) and placed in clean cages. Food for AL mice was transferred to the hopper, and CR mice were fed per usual at 7am with one pellet per mouse on the floor of the cage. They were allowed to eat for 2 hr and then starting at 9am, mice were anesthetized with isoflurane (2–5% in oxygen) and a cardiac puncture was performed to withdraw blood. Cervical dislocation was performed to ensure euthanasia and tissues were excised and snap frozen in liquid nitrogen. From the time of cervical dislocation to excision and snap freezing of the liver, this period did not exceed 30 seconds (mice were not fasted for this experiment). 20 mg of liver was cut into small pieces with a scalpel placed in 1 ml of HEN buffer supplemented with 1% protease inhibitor and 20 mM NBF-Cl, which were then homogenized using a dispersion system on ice and switch labeled with 50 μM DAz-2:Cy5 preclick mix and processed as previously above.
Persulfide Detection in Skeletal Muscle of Mice following a Glucose Tolerance Test
A glucose tolerance test (GTT) was performed on 2-month old and 12-month-old mice which were fasted for 16 hr prior to being injected with D-glucose (i.p. 2 g/kg body weight). Blood glucose level was recorded by tail vein bleeding immediately before and at indicated time points (15 min and 60 min) after injection using an Ascensia Contour blood glucose meter and test strips. At the indicated time points, the mice were euthanized by cervical decapitation and the skeletal muscle was isolated and snap frozen in liquid nitrogen. 20 mg of frozen muscle was cut in to small pieces with a scalpel and placed in 2 ml of HEN buffer supplemented with 1% protease inhibitor and 20 mM NBF-Cl. The suspension was then homogenized using a dispersion system on ice and processed for persulfidation labeling (50 μM DAz-2:Cy5 preclick mix), as previously described.
Persulfide Detection in Human Erythrocyte Lysates
Erythrocyte lysates were prepared in two ways, to obtain membranes and cytosol. Packed erythrocytes were lysed with 5 volumes of 10 mM phosphate buffer containing 5 mM NBF-Cl for 30 min at 4 °C with frequent vortexing. After 20 min centrifugation (30,000 x g) supernatant was separated from cell membrane pellet. SDS (for a final conc. of 2%) and 10 mM NBF-Cl (final conc.) were added to the supernatant and incubated for an additional 60 min at 4 °C. Membrane pellets were washed 3 times with PBS containing 5 mM NBF-Cl, and then resuspended in PBS containing 5 mM NBF-Cl and SDS (final conc. 2%) and incubated for 30 min at 37 °C. Both membrane and cytosol proteins were precipitated and processed for persulfidation labeling (50 μM DAz-2:Cy5 preclick mix) as previously described.
Persulfidation Detection by Confocal Microscopy and Epifluorescence Deconvolution Microscopy
WT and CSE−/− MEF cells were grown in μ-Dish (35 mm, high Glass Bottom, 81158) obtained from Ibidi® (Martinsried, Germany) following manufacturer’s instructions. The treatments with 2 mM D-cysteine and 200 μM Na2S (H2S) were performed over 1 hr at 37 °C. After treatments, the cells were washed twice with warm sterile PBS, and incubated with 1 mM NBF-Cl in PBS for 30 min at 37 °C. Fixation was carried out by incubation with ice-cold methanol at −20 °C for 20 min and subsequent permeabilization with ice-cold acetone at −20 °C for 5 min. The dishes were washed with PBS and incubated with an additional 1 mM (final conc.) NBF-Cl in 2 ml PBS for 1 hr at 37 °C. Cells were washed with PBS overnight with agitation, and incubated with 10 μM (final conc.) DAz-2:Cy-5 click mix in 2 ml PBS for 1 hr at 37 °C. For the negative control cells were incubated with 10 μM DAz-2:Cy-5 click mix prepared without DAz-2. After overnight washing with PBS, cells were washed with methanol 3 × 10 min, followed by an additional washing with PBS. DAPI staining was performed by incubating cells with 300 nM DAPI (final conc.) in 2 ml 15 mM PBS, for 5 min with agitation, protected from light, and then cells were washed 5 times gently with 15 mM PBS. Images were obtained using a Confocal Leica TCS SP5 microscope equipped with an Argon laser (458, 476, 488, 514 nm), a diode laser (405 nm) and Helium-Neon laser (633 nm). A x40 oil objective lens was used. For examination of co-localization of immunofluorescence, single optical sections at the same focus plane were taken separately and the 3 corresponding channels (405 nm (DAPI), 488 nm (NBF-adducts) and 633 nm (Cy5 for PSSH)) were merged into a 8-bit RGB tiff-file using ImageJ. Z-stack images were taken on Olympus IX81 inverted fluorescence microscope using x 100 oil objective lens, used for image deconvolution.
Antibody Array-like Approach for Detection of Persulfidation
Each antibody was added into the appropriate wells of a 96 well plate with 3D-NHS Surface (PolyAn, Berlin) at a final volume of 50 μl in PBS buffer (150 mM Na2HPO4 / NaHPO4 and 50 mM NaCl, pH 8.5). Additionally, in the negative control wells 50 μl of 5% BSA in TBST (137 mM NaCl and 20 mM Trizma base, pH 7.4, supplemented with 0.1% Tween) and 0.002% NaN3 was added. The plate was covered and incubated at 4 °C overnight with agitation. The solutions were discarded and the wells were washed 5 times with 15 mM PBS buffer supplement with 0.01% Tween, using a multi-channel and inverting the clean plate against paper towels for complete removal of liquid. All wells were then blocked with 50 mM ethanolamine in 100 mM Tris at pH 9 for 2 hr at RT with agitation. The blocking solution was discarded and wells were re-blocked with 5% BSA in TBS with 0.01% Tween-20 for a further 1 hr at RT with agitation. Wells were washed again as described above. Following complete removal of liquid, 100 μl of treated samples were added to appropriate wells and incubated at 4 °C, overnight with agitation. After washing, the plate was recorded on Typhoon FL9500 at 473 nm and 635 nm.
For experiments where WT, CSE−/− and WT treated with D-cysteine MEF cells, lysates were obtained and labeled for persulfides, as previously described and redissolved in 50 mM Hepes at a concentration of 0.3 mg/ml. The following antibodies were used: 1. β-actin (0.04 mg/ml, sc-47778, Santa Cruz Biotechnology); 2. β-tubulin (0.04 mg/ml, T0198, Sigma Aldrich); 3. GAPDH (0.04 mg/ml, G8795, Sigma Aldrich); 4. HSP70 (0.04 mg/ml, ab5439, Abcam); 5. KEAP1 P586 (0.0062 mg/ml, 4678, Cell Signalling); 6. eNOS (0.0025 mg/ml, 32027, Cell Signalling); 7. Parkin (0.04 mg/ml, sc-136989, Santa Cruz Biotechnology); 8. SOD-2 (0.04 mg/ml, sc-137254, Santa Cruz Biotechnology); 9. Anti-TST (0.04 mg/ml, GTX114858, GeneTex); 10. Anti-SQR (0.04 mg/ml, HPA017079, Sigma Aldrich).
For experiments with lysates of HeLa cells treated in a time-dependent manner with EGF and labeled as described above. The following antibodies were used: PTEN (1:1000, sc-7974, Santa Cruz Biotechnology,); 2. PTP1B (1:1000, sc-133259, Santa Cruz Biotechnology); 3. SH-PTP2 (0.04 mg/ml, sc-7384, Santa Cruz Biotechnology); 4. EGFR (0.04 mg/ml, sc-03-G, Santa Cruz Biotechnology).
MnSOD Persulfidation and Activity Experiments
Human recombinant MnSOD was purchased from Creative BioMart. SOD activity was measured using cytochrome c assay, as described previously (Liu et al., 2007). SOD activity of 13 μM MnSOD, MnSOD pretreated with 3-fold excess of H2O2 (15 min, 37 °C) and MnSOD pretreated with both 3-fold excess of H2O2 and 5-fold excess of H2S was measured. Peroxynitrite was prepared following well established protocol (Filipovic et al., 2012) and tyrosine nitration assessed using characteristic spectral properties: an increase in the absorbance at 430 nm was attributed to nitrotyrosine formation due to the characteristic shift (from 430 to 357 nm) observed with decreasing pH (Filipovic et al., 2012).
Detection of Protein Sulfenylation
Following respective treatments, cells were lysed in cold HEN lysis buffer supplemented with 1% protease inhibitor and 100 μM DCP-Bio1. Cells were gently scrapped, lysate was collected, homogenized with a syringe and needle and immediately placed for incubation at 37 °C for 1 hr. A methanol/chloroform precipitation was performed as previously described, and protein pellet obtained was redissolved in 50 mM Hepes with SDS (final conc. 2%). Protein pellets were prepared for SDS-PAGE, as previously explained. After resolving samples using SDS-PAGE, protein transfer was performed on nitrocellulose membrane, followed by blocking in 5% dry-milk in PBS supplemented with 0.1% Tween (PBST) and incubated with Streptavidin Protein DyLight 488 in 50 mM PBS (1:10000, 21832, Thermo Fisher Scientific) for 1 hr protected from light. Nitrocellulose membrane was recorded at 473 nm, using a Typhoon FLA 9500 (GE Healthcare). Membrane was then striped using 0.04M NaOH, incubated with agitation for 5 mins twice, and then washed with PBST (5 times, 5 min), blocked as previously described and incubated with GAPDH or β-tubulin as a loading control. Membranes were washed and incubated with respective secondary horseradish-conjugated antibodies for 2 hr at RT. Membranes were then washed and visualized using Clarity™ Western ECL Substrate (BioRad) on a G:Box Chemi-XT4 (Syngene).
Detection of Protein Sulfinylation
Following respective treatments, cells were lysed in cold HEN lysis buffer supplemented with 1% protease inhibitor and 5 mM NBF-Cl or 20 mM NEM. Cells were gently scrapped, lysate was collected, homogenized with a syringe and needle and immediately placed for incubation at 37 °C for 30 min or 1 hr respectively. A methanol/chloroform precipitation was performed and protein pellet obtained was redissolved and protein concentration adjusted as previously described. Samples were treated with 1 mM BioDiaAlk at RT for 1 hr, protected from light. Reaction was quenched by the addition of 1 mM DTT incubated overnight at 4 °C or by a methanol/chloroform precipitation. 3 equivalents of sample was then mixed with 1 equivalent of Laemmli buffer (4X) supplemented with 10% β-mercaptoethanol and incubated for 20 min at 55 °C. After resolving samples using SDS-PAGE, protein transfer was performed, followed by blocking in 5% dry-milk in PBST and incubated with Cy5-Streptavidin (1:4000, 21832, Thermo Fisher Scientific) for 1 hr protected from light. Nitrocellulose membrane was recorded at 635 nm, using a Typhoon FLA 9500 (GE Healthcare). GAPDH was used as a loading control as described above.
Receptor Tyrosine Kinase Activation of Cells
Activation Conditions
EGF (PromoKine) treatments in HeLa and MEF cells, and insulin treatments in SH-SY5Y cells, were performed with cells cultured in 100 mm cell culture dishes at a 80–90% confluency. VEGF (PromoCell) treatments were performed with HUVECs cultured in T25 flasks (Greiner) at a 70–80% confluency. For pretreatments, media was replaced with complete media supplemented with 100 μM GYY4137 or inhibitor mix (PG and AOAA, 1 mM of each), incubated at 37 °C and 5% CO2 for 30 min and then washed with warm sterile PBS. Subsequently, media was replaced with complete media (as described above) for control and media supplemented with respective treatments (100 or 200 ng/ml EGF; 100 nM or 200 nM insulin; 40 ng/ml VEGF). Following treatments, cells were washed with cold PBS twice and lysed for persulfide or sulfenic acid labeling, as previously explained.
Real-time activity of EGFR in HeLa cells
Cells were seeded at 1 × 104 cell/well in an equilibrated E-Plate VIEW 16 PET (ACEA Biosciences, San Diego, USA) and grown overnight at 37 °C with 5% CO2. Next, cells were incubated in serum-free complete medium for 4 hr prior to experiments and pretreated with 100 μM of GYY4137 or inhibitor mix (PG and AOAA, 1 mM of each), as indicated. Basal receptor tyrosine kinase activity in cells was recorded for 20 min and upon the addition of 150 ng/ml EGF, cells were further recorded in 2 hr with the integration time of 1 minute using xCELLigence RTCA DP system (ACEA Biosciences, San Diego, USA).
Antibody Microarray Detection of Persulfidation of EGFR Pathway kinases
The persulfidation of the proteins associated to the EGFR pathways were assessed using an EGF pathway phospho antibody array comprising of 214 antibodies related to the EGF pathway (Full Moon Biosystems, CA), performed in duplicates. The glass strips (barcode: 4000026018, 4000026019, 4000026022 and 4000026023) were equilibrated to RT for 1 hr and dried for 45 min. They were then blocked with 5% BSA in TBST by rocking at 55 rpm at RT for 45 mins. The slides were then washed with TBST (twice), TBS (twice) and then with H2O (5 times). Lysates of HeLa cells treated with or without 100 ng of EGF for 30 mins and labeled for persulfides, as previously explained. Samples were then dissolved in 50 mM Hepes with SDS (final conc. 2%) and further diluted 60 times with 1% BSA in TBS up to 6 ml. The glass strips were then submerged in the samples and incubated by rocking at 55 rpm for 2 hr. The slides were washed as done previously, left to dry and recorded at 473 nm and 635 nm, using a Typhoon FLA 9500 (GE Healthcare). Persulfide levels of each dot for each type of antibody (in pentaplicates on glass strip) were averaged. The intensity of their cy5 (635 nm) signal was adjusted to the 473 nm signal for the internal standards, GAPDH and β-actin.
Immunoblotting
Untreated or specifically treated cells were washed twice with cold PBS and harvested in RIPA lysis buffer supplemented with 1% protease inhibitor. Cells were scrapped, lysates were collected, homogenized with syringe and needle, and centrifuged at 30,000 x g for 5 min at 4 °C. Clear lysate was transferred to a new tube. Protein concentration of lysate was determined using the DC assay (BioRad). Protein samples were resolved by SDS-PAGE and transferred to a nitrocellulose membrane (GE Healthcare).
Primary antibodies used: CBS (1:1000, sc-133154, Santa Cruz Biotechnology), MPST (1:4000, HPA001240, Sigma Aldrich), CTH (MEF cells, 1:4000), CTH (HeLa cells, 1:1000, sc-374249, Santa Cruz Biotechnology), p-ERK (1:1000, sc-7383, Santa Cruz Biotechnology), total-ERK (1:1000, sc-271269, Santa Cruz Biotechnology), DJ-1 (1:250, sc-55572, Santa Cruz Biotechnology), DJ-1 Oxidized At C106 (1:1000, HCA024, BioRad) and β-tubulin (1:5000, T0198, Sigma Aldrich). Species-specific horseradish-conjugated secondary antibodies (1:5000, Santa Cruz Biotechnology) were used for antigen detection and visualized using Clarity™ Western ECL Substrate (BioRad) on a G:Box Chemi-XT4 (Syngene).
Immunoprecipitation and detection of DJ-1 Persulfidation, Sulfinylation and Sulfenylation Following H2O2 Treatment
MEF cells (WT and CSE−/−) were treated with 100 μM H2O2 for 15 or 30 min. Samples were labeled for oxPTMs as previously described; for the detection of persulfides NBF-Cl was used and switched with DCP-Bio1; for sulfenic acid labeling DCP-Bio1 was used; for sulfinic acid labeling NEM was used and then labeled with BioDiaAlk. Proteins were then precipitated and resuspended in 50 mM Hepes with SDS (final conc. 0.01%) at 1 mg/ml. Samples were incubated overnight at 4 °C with anti-DJ-1 agarose coupled antibody (Santa Cruz Biotechnology, sc-55572 AC). After incubation agarose resins were collected by centrifugation (2,000 x g, 10 min) and washed with 10 mM TBS supplemented with 0.001% Tween (3 times) and with 10 mM TBS (twice). DJ-1 protein was eluted from the resins for 10 min at 95 °C in 10 mM TBS with 3.5% SDS. This was then mixed with Laemmli buffer (4X) supplemented with 10% β-mercaptoethanol (3 eq. sample : 1 eq. buffer) and incubated overnight at 55 °C. Elution fractions were collected by centrifugation (30,000 x g, 20 min) and supernatants were resolved by SDS-PAGE. Protein transfer was performed, followed by blocking in 1% BSA in TBST. Monoclonal anti-Biotin-Peroxidase-conjugated antibody (1:1000, Sigma Aldrich) was used for detection and visualized using Clarity™ Western ECL Substrate (BioRad) on a G:Box Chemi-XT4 (Syngene).
Trx-catalyzed reduction of S-sulfocysteine (SSC)
Cloning and Mutagenesis of Human TRP14 and Trx1
Genetic sequence of human TRP14 was obtained by generating cDNA from mRNA isolated from human fibroblasts. The target sequence was amplified by PCR using forward primer 5’-ACCATCACGGATCCATGGCCCGCTATGAGGAGG-3’ containing a BamHI restriction site (underlined) and a start codon (boldface type), in combination with reverse primer 5’-CCGGGGTACCGTTAATCTTCAGAGAACAACATTTCCACCAG-3’ 3’containing a KpnI restriction site (underlined) and a stop codon (boldface type). The sequence of the target band was verified by commercial sequencing (GATC Biotech). The PCR product was ligated into a pQE80-L expression vector (Qiagen). Human Thioredoxin 1 (Trx1) was expressed in E. coli using pET28a expression vector. Mutagenesis of the C32S & C35S variants were conducted using PCR-mediated site-directed mutagenesis, and the Trx1-pET28a expression plasmid was used as a scaffold.
Expression and Purification of Human TRP14 and Trx1
E. coli BL21 (DE3) Rosetta cells were transformed with the respective expression plasmid and cultured at 37 °C, 140 rpm in LB medium supplemented with the necessary antibiotics and grown until OD600 = 0.5. Protein expression was induced using isopropyl-β-ᴅ-thiogalactopyranosid (IPTG) at a final concentration of 250 μM. The expression temperature and rotation was changed to 30 °C, 120 rpm for TRP14, and 18 °C, 120 rpm for Trx1 variants; expression was conducted for ~16 hr. Cells were harvested and lysed in lysis buffer (50 mM potassium phosphate, pH 7.4, 250 mM NaCl, 10 mM imidazole) supplemented with complete EDTA-free protease inhibitor (Roche). Cells were lysed using egg white lysozyme (VWR) and homogenized in an Emulsiflex. Residual nucleic acids were disrupted using a sonication rod at 25% amplitude, 10 seconds. The cell lysate was centrifuged at 48000 x g for 45 min, and His-tagged proteins were isolated using His-Pur Ni-NTA Superflow Agarose (Thermo Fisher Scientific) in accordance with the manufacturer’s protocol. Elution was performed using lysis buffer supplemented with 250 mM imidazole. Elution fractions were analyzed using SDS-PAGE and subsequent Coomassie Brilliant Blue staining, and fractions containing increased amounts of target protein were further purified using anion exchange chromatography (Source 15Q, GE Healthcare; low salt buffer: 50 mM sodium phosphate, pH 7.4; high salt buffer: 50 mM sodium phosphate, pH 7.4, 1 M NaCl). Elution was performed in a gradient over 5 column volumes. Elution fractions were again analyzed using SDS-PAGE and Coomassie Brilliant Blue staining. Relevant fractions were buffer exchanged into storage buffer (50 mM sodium phosphate, pH 7.4, 150 mM NaCl, 100 μM DTT), flash-frozen in liquid nitrogen and stored at −80 °C. Proteins were again buffer exchanged after thawing into the relevant experimental buffer.
Coupled Assay of Trx1 / TRP14 and TrxR with Cystine / SSC
Activities of hTrx and TRP14 with cystine and SSC were recorded measuring NADPH oxidation on a Tecan Infinite M200 plate reader, set to record absorption at 340 nm. 3 μM hTrx1 or TRP14, 30 nM TrxR from rat liver (Sigma Aldrich) and 250 μM NADPH were used in all measurements. The concentration of cystine or SSC were set at 1 mM. Absorption at 340 nm were recorded in 10 seconds intervals over 900 seconds. The initial rate of A340 decrease was fitted linearly, and consumption of NAPDH over time was determined using an NADPH extinction coefficient ε340 of 6020 M−1 cm−1.
Kinetics of Direct Reaction of Human Trx with SSC
Kinetics of the reaction of hTrx with cysteine persulfide was monitored on a FP-8200 spectrofluorometer (Carry Eclipse, Agilent) using an excitation wavelength of 280 nm and a maximal emission of 345 nm. Concentration of the enzyme was kept at 4 μM while substrate concentrations ranged between 25 μM and 100 μM. Given pseudo first-order conditions, observed rate constants kobs were obtained by fitting the decrease in emission at 345 nm at a given SSC concentration using a first order exponential decay fit in Origin® analysis software.
Ultra High Resolution ESI-TOF Mass Spectrometry of Reaction Between Trx and SSC
10 μM of human recombinant Trx, Trx C35S or Trx C32S were incubated with 10 μM SSC in 20 mM ammonium carbonate buffer pH 7.8 for 5 min and recorded on maXis 5G (Bruker Daltonics) ESI-TOF MS capable of resolution of at least 40,000 FWHM, following previously described protocol (Wedmann et al., 2016).
MEF cells Stress Assays
MEF cells (WT and CSE−/−) were plated in 96-well plates at 5 × 104 cells/well. Cells were treated with H2O2, as indicated, for 24 hr and the cell survival was assessed by an MTT assay as previously described (Liu et al., 2015).
C. elegans Stress Assays
Worms were initially synchronised by picking approximately 100 young adult worms per strain onto NGM-plates, and allowed to lay eggs over 4 hr at 20 °C. The young adults were then removed, and the remaining embryos were washed off and collected from the NGM-plates using M9 buffer, and centrifuged (850 x g, 1 min) and washed with M9 buffer 3 times. The resulting pellet of embryos was re-suspended in 12 ml of M9 buffer, and incubated with agitation (100 rpm) for 24 hr at 20 °C. The synchronised L1 worms were collected, centrifuged (850 x g, 1 min) and re-suspended in S-basal buffer. The number and synchronicity of L1 worms was determined and the worms were transferred to an Erlenmeyer, diluted in the respective amount of HB101 in S-Basal buffer for a final conc. of 60 worms/10 mg of HB101/ml of S-basal buffer. They were then incubated with agitation (115 rpm) for 48 hr at 20 °C. The worms were then collected, centrifuged (400 rpm, 1 min), washed with M9 buffer 3 times and re-suspended in M9 buffer.
For experiments with the pretreatment of worms, synchronised young adults (as described above) were transferred in an Erlenmeyer with 12 ml of M9 buffer in which GYY4137 (final conc. 500 μM) or AP39 (final conc. 100 nM) was added. Worms were then incubated for 3 hr at 20 °C with agitation (115 rpm). Worms were centrifuged (400 rpm, 1 min) and re-suspended in M9 buffer. To assess the effect of CTH on stress resistance, paraquat dichloride hydrate (final conc. 60 mM) or sodium (meta) arsenite (final conc. 5 mM) was added to the synchronised young adult worm suspension and plated on a 96 well plate with approximately 5 – 10 worms per well. Viability was monitored by counting dead worms over 5 – 6 hr, whilst incubating the plate at 20 °C, with agitation (110 rpm).
Yeast stress Assays and H2S production
Spot assays were carried out by spotting 5 μl of early exponential phase cultures (OD600=0.5) of different strains of S. cerevisiae, sequentially diluted (approximately 3.5 × 104 to 3.5 cells) on plates with YPD media supplemented with different concentrations of H2O2 (0, 1 mM, 2 mM and 5 mM). Growth was recorded after incubation of 24 and 48 hr, at 30 °C. Survival assays were done by preparing overnight cultures in YPD media (cell in stationary phase), from which the experimental cultures were set, by diluting to OD600=2 in a 5 ml final volume, with or without respective concentrations of H2O2 (0, 10 mM and 20 mM). The cultures were subsequently grown in culture tubes for 27 hr at 30 °C with agitation (180 rpm). Yeast cells were then washed once in PBS and collected by centrifugation (5000 x g, 3 min). Cells were then resuspended in PBS supplemented with 2 μM propidium iodide, at 1×106 cells/ml and incubated for 5 min in the dark. Analysis was performed by flow cytometry using 150,000 cells per condition, on a BD Accuri™ C6 (BD Biosciences) and results were analysed using the CFlow Plus Software. For the quantification of H2S levels, overnight cultures were washed with PBS and diluted as described for survival assays. Cell suspensions were incubated with 20 μM MeRho-Az sensor for 45 mins at 30 °C and analysed by flow cytometry, on a BD Accuri™ C6 (BD Biosciences).
C. elegans Lifespans
For the lifespan experiments a synchronous population of worms was obtained by transferring 5–6 young adults on medium plates and allowing them to lay eggs over 3 hr at 20 °C. Lifespan measurements were conducted at 20 °C, worms were transferred daily during the reproductive period. Death was scored by failure of the animal to move in response to gentle prodding with a platinum wire. For lifespan analysis with 1 mM sodium thiosulfate treatment or 5 mM 2-deoxy-D-glucose (DOG), new plates containing treatments in NGM agar were prepared every second day. Lifespan measurements were repeated at least twice unless otherwise stated.
Immunohistochemistry on Male Rat Brains
Brains were fixed in 4% paraformaldehyde for 24 hr, dehydrated in a series of increasing concentrations of ethanol (30%–100%), enlightened in xylene and embedded in Histowax® (Histolab Product AB, Göteborg, Sweden). Sagittal plane of each brain was sectioned at 5 μm thickness on a rotary microtome (RM 2125RT Leica Microsystems, Wetzlar, Germany). Sections were placed on Superfrost Ultra Plus® manufactured slides and used for immunohistochemical staining. After tissue deparaffinization, brain sections were exposed to heat-induced antigen retrieval to demask target antigens. Slides were placed in a container and covered with 0.01 mol/l sodium citrate buffer pH 6.0, and then heated at 750 W in microwave oven for 3 × 7 min. Next, sections were incubated with 0.3% H2O2 in MeOH for 15 min to block endogenous peroxidase. Slides were washed in PBS (pH 7.4) and reduction of non-specific background staining was achieved by incubation with normal swine (1:10, X0901, Dako) and donkey serum (1:10, X0903, Abcam) for 45 min at RT. Next, sections were incubated with primary antibodies; CBS (1:200, sc-67154, Santa Cruz Biotechnology), MPST (1:500, HPA001240, Sigma Aldrich), or with CTH/CSE antibody (1:200, sc-365382, Santa Cruz Biotechnology), overnight at 4 °C. For the negative control, the primary antibody was substituted with PBS. After washing for 5 min in PBS, brain tissue sections were incubated with swine-anti-rabbit (1:100, P0399, Dako) and donkey-anti-mouse IgG-HRP (1:100, ab6820, Abcam) for 1 hr at RT. Slides were washed in PBS and visualization was performed using Dako liquid 3,3′-diaminobenzidine tetrahydrochloride (DAB) substrate chromogen system (Dako) at concentrations suggested by the manufacturer. Hematoxylin was used as counterstain and slides were mounted in DPX medium (Sigma Aldrich).
QUANTIFICATION AND STATISTICAL ANALYSIS
The experiments were performed in at least in triplicates by at least 2 different researchers. Key methodological experiments were verified by at least three different researchers in three different laboratories and all data are presented as mean ±SD. Protein expression levels, persulfidation, sulfenylation and sulfinylation levels were compared with an unpaired t test using the asterisk (*) or the number sign (#). No results/data points were excluded. For both signs, the single symbol represents a p value of < 0.05 and two symbols of p values of < 0.01, throughout the study. Data of Mass Spectroscopy was analyzed using PEAKS Studio. Lifespan data were analyzed using Kaplan-Meier survival analysis to detect statistical differences. For lifespan experiments at least 100 worms per experiment were used. Plotting of the data were performed using GraphPad Prism 5.0, Origin 8 and Microsoft Excel.
DATA AND CODE AVAILABILITY
The raw data corresponding to the Antibody Microarray Detection of Persulfidation of EGFR Pathway kinases represent entirely new application of the dimedone switch method and are therefore available in more detail from the corresponding author on request. Proteomic data used for Figure S2C are stored in Data S1, S2 and for Figure S3H, I in Data S3, S4. All in-gel persulfidation, as well as Western blot sufenylation and sulfynilation data are reported in main and supporting Figures. Other raw data are available at DOI: 10.17632/pw2wz39tsk.1.
Supplementary Material
Dataset S1: MS/MS data of the persulfidated TST digested with trypsin. Related to Figure 2.
Dataset S2: MS/MS data of the persulfidated TST digested with chymotrypsin. Related to Figure 2.
Dataset S3: MS/MS data of the persulfidated MnSOD digested with trypsin. Related to Figure 3.
Dataset S5: Antibody microarray analysis of protein persulfidation. Related to Figure 5.
Dataset S4: MS/MS data of the persulfidated MnSOD digested with chymotrypsin. Related to Figure 3.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-β-actin, mouse monoclonal | Santa Cruz Biotechnology | Cat# sc-47778 |
| Anti-β-tubulin, mouse monoclonal | Sigma Aldrich | Cat# T0198 |
| Anti-GAPDH, mouse monoclonal | Sigma Aldrich | Cat# G8795 |
| Anti-HSP70, mouse monoclonal | Abcam | Cat# Ab5439 |
| Anti-KEAP1 P586, rabbit polyclonal | Cell Signalling | Cat# 4678 |
| Anti-eNOS, rabbit monoclonal | Cell Signalling | Cat# 32027 |
| Anti-Parkin, mouse monoclonal | Santa Cruz Biotechnology | Cat# sc-136989 |
| Anti-SOD-2, mouse monoclonal | Santa Cruz Biotechnology | Cat# sc-137254 |
| Anti-TST, rabbit polyclonal | GeneTex | Cat# GTX114858 |
| Anti-SQRDL, rabbit polyclonal | Sigma Aldrich | Cat# HPA017079 |
| Anti-PTEN, mouse monoclonal | Santa Cruz Biotechnology | Cat# sc-7974 |
| Anti-PTP1B, mouse monoclonal | Santa Cruz Biotechnology | Cat# sc-133259 |
| Anti-SH-PTP2, mouse monoclonal | Santa Cruz Biotechnology | Cat# sc-7384 |
| Anti-EGFR, rabbit polyclonal | Santa Cruz Biotechnology | Cat# sc-03-G |
| Anti-CBS, mouse monoclonal | Santa Cruz Biotechnology | Cat# sc-133154 |
| Anti-CBS, rabbit polyclonal | Santa Cruz Biotechnology | Cat# sc-67154 |
| Anti-MPST, rabbit polyclonal | Sigma Aldrich | Cat# HPA001240 |
| Anti-CTH, rabbit | From Prof. Snyder’s lab (Johns Hopkins University School of Medicine) | (Paul et al., 2014) |
| Anti-CTH, mouse monoclonal | Santa Cruz Biotechnology | Cat# sc-365382 |
| Anti- p-ERK, mouse monoclonal | Santa Cruz Biotechnology | Cat# sc-7383 |
| Anti ERK 1, mouse monoclonal | Santa Cruz Biotechnology | Cat# sc-271269 |
| Anti-DJ-1, mouse monoclonal | Santa Cruz Biotechnology | Cat# sc-55572 |
| Anti-DJ-1 AC, mouse monoclonal | Santa Cruz Biotechnology | Cat# sc-55572 AC |
| Anti DJ-1 (Oxidized At C106), mouse monoclonal | BioRad | Cat# HCA024 |
| Anti-Biotin-Peroxidase, mouse monoclonal | Sigma Aldrich | Cat# A0185 |
| Anti-Dimedone, rabbit polyclonal | From Dr. Carroll’s lab (The Scripps Research Institute) | (Seo and Carroll, 2009) |
| Mouse IgGκ BP-HRP | Santa Cruz Biotechnology | Cat# sc-516102 |
| Mouse anti-rabbit IgG-HRP | Santa Cruz Biotechnology | Cat# sc-2357 |
| Swine anti-rabbit IgG-HRP, polyclonal | Dako, Denmark | Cat# P0399 |
| Donkey anti-mouse IgG-HRP, polyclonal | Abcam, Cambridge, UK | Cat# ab6820 |
| Streptavidin Protein, DyLight 488 | Thermo Fisher Scientific | Cat# 21832 |
| Cy®5-Streptavidin | Sigma Aldrich | Cat# GEPA45001 |
| EGF Pathway Phospho Antibody Array | Full Moon Biosystems | Cat# PEG214 |
| Bacterial and Virus Strains | ||
| OP50-1 | Caenorhabditis Genetics Center | N/A |
| HB101 | Caenorhabditis Genetics Center | N/A |
| E.coli MG1655 (pTrc99a) | From the laboratory of Gonzales-Zorn, Spain | N/A |
| E.coli MG1655 (pSB74) | From the laboratory of Gonzales-Zorn, Spain | N/A |
| E. coli BL21 DE3 Rosetta | Novagen | Cat# 70954-3 |
| Biological Samples | ||
| Kidneys from C57BL/6 WT and CSE−/− mice | From Professor Maik Gollasch (Charité Medical Faculty) | (Markó et al., 2016) |
| D. melanogaster: y1w1118 | From Professor Ody Sibon (University of Groningen) | (Snijder et al., 2015) |
| D. melanogaster: Eip55E | From Professor Ody Sibon (University of Groningen) | (Snijder et al., 2015) |
| Chemicals, Peptides and Recombinant Proteins | ||
| Chelex-100 | Sigma Aldrich | Cat# C7901 |
| Sodium Sulfide | Sigma Aldrich | Cat# 407410 |
| GYY4137 | (Alexander et al., 2015) | N/A |
| AP39 | (Le Trionnaire et al., 2014) | N/A |
| D-Cystine | Sigma Aldrich | Cat# 30095 |
| DL-propargylglycine | Sigma Aldrich | Cat# P7888 |
| O-(Carboxymethyl)hydroxylamine hemihydrochloride (AOAA) | Sigma Aldrich | Cat# C13408 |
| Hydrogen peroxide solution | Sigma Aldrich | Cat# 216763 |
| Monensin sodium salt | Sigma Aldrich | Cat# M5273-1G |
| Erastin | Sigma Aldrich | Cat# E7781 |
| S-Sulfocysteine | Sigma Aldrich | Cat# C2196 |
| Cystine | Sigma Aldrich | Cat# 30200 |
| Imidazole | Carl Roth Gmbh | Cat# 3899.4 |
| Isopropyl β-D-1-thiogalactopyranoside | Inalco Pharmaceuticals | Cat# 1758-1400 |
| Dithiothreitol | GE Healthcare | Cat# 17-1318-02 |
| 4-Chloro-7-nitrobenzofurazan | Sigma Aldrich | Cat# 163260 |
| Dimedone | Sigma Aldrich | Cat# D153303 |
| DCP-Bio1 | Kerafast | Cat# EE0028 |
| DAz-2 | Cayman Chemicals | Cat# 13382 |
| BioDiaAlk | (Akter et al., 2018) | N/A |
| DiaAlk | (Akter et al., 2018) | N/A |
| Cyanine5 alkyne | Lumiprobe | Cat# FP-OO5590 |
| Copper(II)-TBTA | Lumiprobe | Cat# 21050 |
| L-Ascorbic acid | Sigma Aldrich | Cat# 795437 |
| 5-Fluoro-2′-deoxyuridine | Sigma Aldrich | #F0503 |
| Nmc-penicillamine | (Artaud and Galardon, 2014) | N/A |
| EGF human recombinant | PromoKine | Cat# 60170 |
| VEGF-165, human, recombinant | PromoCell | Cat# C-64422 |
| Insulin human | Sigma Aldrich | Cat# I0908 |
| Paraformaldehyde | Sigma Aldrich | Cat# P6148 |
| Sodium thiosulfate | PROLABO | Cat# 27 910.291 |
| Paraquat hydrochloride hydrate | Sigma Aldrich | Cat# 36541 |
| Sodium (meta)arsenite | Sigma Aldrich | Cat# S7400 |
| 2-Deoxy-D-glucose | Sigma Aldrich | Cat# D8375 |
| Thiazolyl Blue Tetrazolium Bromide | Sigma Aldrich | Cat# M5655 |
| Protease Inhibitor | Sigma Aldrich | Cat# P8340 |
| His-Pur Ni-NTA Superflow Agarose | Thermo Fisher Scientific | Cat# 25216 |
| Streptavidin Magnetic Beads | Sigma Aldrich | Cat# 11 641 778 001 |
| NeutrAvidin Agarose Resin | Thermo Fisher Scientific | Cat# 29201 |
| High Capacity NeutrAvidin Agarose Resin | Thermo Fisher Scientific | Cat# 29202 |
| Trypsin from porcine pancreas | Sigma Aldrich | Cat# T6567 |
| Chymotrypsin Sequencing Grade | Sigma Aldrich | Cat# 000000011418467001 |
| Human Serum Albumin (HSA) | Sigma Aldrich | Cat# A1887 |
| GAPDH | Sigma Aldrich | Cat# G2267 |
| Rhodanese from bovine liver (TST) | Sigma Aldrich | Cat# R1756 |
| MnSOD | Creative BioMart | Cat# SOD2-1039H |
| Cytochrome c | Sigma Aldrich | Cat# 30398 |
| Xanthine | Sigma Aldrich | Cat# X0626 |
| PTP1B human recombinant | Abcam, Cambridge, UK | Cat# ab51277 |
| Xanthine Oxidase | Sigma Aldrich | Cat# X1875 |
| Thioredoxin Reductase (TrxR) from rat liver | Sigma Aldrich | Cat# T9698 |
| TRP14 (From human fibroblast cDNA) | For this paper | N/A |
| hsTrx C32S | For this paper | N/A |
| hsTrx C35S | For this paper | N/A |
| hsTrx1 | For this paper | N/A |
| NADPH | Sigma Aldrich | Cat# N5130 |
| DAPI | Euromedex | Cat# 1050-A |
| MeRho-Az | From Michael D. Pluth’s lab (Dept. of Chemistry and Biochemistry, Eugene, OR) | N/A |
| Propidium Iodide | Sigma Aldrich | Cat# P4864 |
| Histowax® | Histolab Product AB, Sweden | Cat# 00405 |
| Normal swine serum | Dako, Denmark | Cat# X0901 |
| Normal donkey serum | Abcam, Cambridge, UK | Cat# X0903 |
| 3,3′-Diaminobenzidine tetrahydrochloride (DAB) | Dako North America, Inc. Carpintería, CA, USA | Cat# K3468 |
| DPX medium | Sigma-Aldrich | Cat# 06522 |
| Experimental Models: Cell Lines | ||
| Human: HeLa | ECACC | Cat# 93021013 |
| Human: HUVEC | PromoCell | Cat# C-12203 |
| Human: SH-SY5Y | ECACC | Cat# 94030304 |
| Human: Fibroblasts (male) | Coriell Institute | AG08790 and AG14245 |
| Mouse: MEF WT (WT) (male) | From Prof. Snyder’s lab (Johns Hopkins University School of Medicine) | (Sbodio et al., 2016) |
| Mouse: MEF CSE−/− (male) | From Prof. Snyder’s lab (Johns Hopkins University School of Medicine) | (Sbodio et al., 2016) |
| Mouse: Striatal progenitor cells STHdhQ7/Q7 (male) | M. MacDonald (Massachusetts General Hospital, Boston, MA) | N/A |
| Mouse: Striatal progenitor cells STHdhQ111/Q111 (male) | M. MacDonald (Massachusetts General Hospital, Boston, MA) | N/A |
| Experimental Models: C.elegans mutants | ||
| Wild-type Bristol N2 | Caenorhabditis Genetic Center | N/A |
| cth-1(ok3319)V | Caenorhabditis Genetic Center | VC2569 |
| mpst-3(tm4387)V | MITANI Lab, National Bio-Resource Project of the MEXT, Japan | FX04387 |
| eat-2(tm5786)II | MITANI Lab, National BioResource Project of the MEXT, Japan | FX19451 |
| eat-2(tm5786)II;cth-1(ok3319)V | This paper | N/A |
| eat-2(tm5786)II;mpst-3(tm4387)V | This paper | N/A |
| Experimental Models: Strains/Organisms | ||
| S. cerevisiae: BY4742 (WT) | Euroscarf | N/A |
| S. cerevisiae: Δcys4 | Euroscarf | ACCNO Y16696 |
| S. cerevisiae: Δcys3 | Euroscarf | ACCNO Y16865 |
| S. cerevisiae: Δtum1 | Euroscarf | ACCNO Y12507 |
| Male Wistar Rat | Institute for Biological Research “Siniša Stanković”, Belgrade, Serbia | N/A |
| Male C57BL/6J mice | NIA Aging Colony Resource at Charles River Laboratories (CRL) | N/A |
| Male C57BL/6 mice | Department of Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, MD | N/A |
| Oligonucleotides | ||
| 5’-GAAGGAGATATACCATGGTGAAGC-3’ (fwd) | Sigma Aldrich | hsTrx in pET28a |
| 5’-CGGATCTCAGTGGTGGTG-3’ (rev) | Sigma Aldrich | hsTrx in pET28a |
| 5’-CACGTGGTCTGGGCCTTG-3’ (fwd) | Sigma Aldrich | hsTrx C32S mutagenesis |
| 5’-CAAGGCCCAGACCACGTG-3’ (rev) | Sigma Aldrich | hsTrx C32S mutagenesis |
| 5’-GTGTGGGCCTTCCAAAATGATCAAG-3’ (fwd) | Sigma Aldrich | hsTrx C35S mutagenesis |
| 5’-CTTGATCATTTTGGAAGGCCCACAC-3’ (rev) | Sigma Aldrich | hsTrx C35S mutagenesis |
| 5’-ACCATCACGGATCCATGGCCCGCTATGAGGAGG-3’ (fwd) | Sigma Aldrich | hsTRP14 in pQE-80L |
| 5’-CCGGGGTACCGTTAATCTTCAGAGAACAACATTTCCACCAG-3’ (rev) | Sigma Aldrich | hsTRP14 in pQE-80L |
| Recombinant DNA | ||
| Plasmid: pET-28a(+) | Novagen | Cat# 69864-3 |
| Plasmid: pQE-80L | Qiagen | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/download.html |
| GraphPad Prism 5.0 | GraphPad Software | https://www.graphpad.com |
| PEAKS Studio | Bioinformatics Solutions Inc. | http://www.bioinfor.com/ |
| OriginPro 8 | OriginLab | https://www.originlab.com/ |
| RTCA Software Version 2.0 | ACEA Biosciences, Inc. | https://www.aceabio.com/products/rtca-dp/ |
| CFLow Plus Version 1.0.202.1 | BD Biosciences | http://www.bdbiosciences.com |
Acknowledgments
The authors would like to thank Dr. Michel Toledano for careful reading of the manuscript and helpful discussions, and Dr. Bertrand Daignan-Fornier for his help with yeast experiments.
Funding: This work was supported by the ATIP-Avenir grant, by the French State in the frame of the ‘Investments for the future’ Programme IdEx Bordeaux (ANR-10-IDEX-03-02) and by FRM (DEQ20180339181). MW acknowledges support from MRC, UK (MR/M022706/1), The Brian Ridge Scholarship (to RT) and the Northcott Devon Medical Research Foundation, UK. MM and VM were supported by the Ministry of Education, Science and Technology Development of the Republic of Serbia (173009). KSC acknowledges support by NIH (GM102187) and GS and JBK support by the DFG, Germany (SFB1218 TP B08). Project 19PABH134580006 from the AHA/Allen Initiative in Brain Health and Cognitive Impairment team (SHS and associates) is acknowledged.
Footnotes
Declaration of Interests: MW and the University of Exeter have patents on the therapeutic and agricultural use of mitochondria-targeted, and other, hydrogen sulfide delivery molecules. All other authors declare no competing interests.
REFERENCES
- Aging C, Walther DM, Kasturi P, Mann M, Hartl FU, Walther DM, Kasturi P, Zheng M, Pinkert S, Vecchi G, et al. (2015). Article Widespread Proteome Remodeling and Aggregation in Aging C. elegans. Cell 161, 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaike T, Ida T, Wei FY, Nishida M, Kumagai Y, Alam MM, Ihara H, Sawa T, Matsunaga T, Kasamatsu S, et al. (2017). Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat. Commun 8, 1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akter S, Fu L, Jung Y, Conte M. Lo, Lawson JR, Lowther WT, Sun R, Liu K, Yang J, and Carroll KS (2018). Chemical proteomics reveals new targets of cysteine sulfinic acid reductase. Nat. Chem. Biol 14, 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander BE, Coles SJ, Fox BC, Khan TF, Maliszewski J, Perry A, Pitak MB, Whiteman M, Wood ME, Nakashima I, et al. (2015). Investigating the generation of hydrogen sulfide from the phosphonamidodithioate slow-release donor GYY4137. Med. Chem. Commun 6, 1649–1655. [Google Scholar]
- Artaud I, and Galardon E (2014). A persulfide analogue of the nitrosothiol SNAP: formation, characterization and reactivity. Chembiochem 15, 2361–2364. [DOI] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, and Finkel T (2005). Mitochondria, Oxidants, and Aging. Cell 120, 483–495. [DOI] [PubMed] [Google Scholar]
- Bernal-Perez LF, Prokai L, and Ryu Y (2012). Selective N-terminal fluorescent labeling of proteins using 4-chloro-7-nitrobenzofurazan: A method to distinguish protein N-terminal acetylation. Anal. Biochem 428, 13–15. [DOI] [PubMed] [Google Scholar]
- Blackstone E (2005). H2S Induces a Suspended Animation-Like State in Mice. Science (80-. ). 308, 518–518. [DOI] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles RL, Schroder E, May G, Free P, Gaffney PRJ, Wait R, Begum S, Heads RJ, and Eaton P (2007). Protein Sulfenation as a Redox Sensor: Proteomics Studies Using a Novel Biotinylated Dimedone Analogue. Mol. Cell. Proteomics 6, 1473–1484. [DOI] [PubMed] [Google Scholar]
- Crouzet M, Claverol S, Lomenech AM, Le Sénéchal C, Costaglioli P, Barthe C, Garbay B, Bonneu M, and Vilain S (2017). Pseudomonas aeruginosa cells attached to a surface display a typical proteome early as 20 minutes of incubation. PLoS One 12, e0180341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevasanta E, Denicola A, Alvarez B, and Möller MN (2012). Solubility and Permeation of Hydrogen Sulfide in Lipid Membranes. PLoS One 7, e34562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevasanta E, Lange M, Bonanata J, Coitiño EL, Ferrer-Sueta G, Filipovic MR, and Alvarez B (2015). Reaction of Hydrogen Sulfide with Disulfide and Sulfenic Acid to Form the Strongly Nucleophilic Persulfide. J. Biol. Chem 290, 26866–26880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Autréaux B, and Toledano MB (2007). ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol 8, 813–824. [DOI] [PubMed] [Google Scholar]
- Delobel J, Prudent M, Crettaz D, ElHajj Z, Riederer BM, Tissot JD, and Lion N (2016). Cysteine redox proteomics of the hemoglobin-depleted cytosolic fraction of stored red blood cells. Proteomics - Clin. Appl 10, 883–893. [DOI] [PubMed] [Google Scholar]
- Dóka É, Pader I, Bíró A, Johansson K, Cheng Q, Ballagó K, Prigge JR, Pastor-Flores D, Dick TP, Schmidt EE, et al. (2016). A novel persulfide detection method reveals protein persulfide- and polysulfide-reducing functions of thioredoxin and glutathione systems. Sci. Adv 2, e1500968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis HR, and Poole LB (1997). Novel application of 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole to identify cysteine sulfenic acid in the AhpC component of alkyl hydroperoxide reductase. Biochemistry 36, 15013–15018. [DOI] [PubMed] [Google Scholar]
- Emmons SW, Klass MR, and Hirsh D (1979). Analysis of the constancy of DNA sequences during development and evolution of the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci 76, 1333–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Caggiano M, Schröder E, Cho HJ, Burgoyne J, Barallobre-Barreiro J, Mayr M, and Eaton P (2016). Oxidant-induced interprotein disulfide formation in cardiac protein DJ-1 occurs via an interaction with peroxiredoxin 2. J. Biol. Chem 291, 10399–10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovic MR, Miljkovic J, Allgäuer A, Chaurio R, Shubina T, Herrmann M, and Ivanovic-Burmazovic I (2012). Biochemical insight into physiological effects of H₂S: reaction with peroxynitrite and formation of a new nitric oxide donor, sulfinyl nitrite. Biochem. J 441, 609–621. [DOI] [PubMed] [Google Scholar]
- Filipovic MR, Zivanovic J, Alvarez B, and Banerjee R (2018). Chemical Biology of H2S Signaling through Persulfidation. Chem. Rev 118, 1253–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink RI, Kolterman OG, Griffin J, and Olefsky JM (1983). Mechanisms of insulin resistance in aging. J. Clin. Invest [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T (2011). Signal transduction by reactive oxygen species. J. Cell Biol 194, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, and Holbrook NJ (2000). Oxidants, oxidative stress and the biology of ageing. Nature 408, 239–247. [DOI] [PubMed] [Google Scholar]
- Fontana L, Klein S, and Holloszy JO (2010). Effects of long-term calorie restriction and endurance exercise on glucose tolerance, insulin action, and adipokine production. Age (Omaha). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster MW, Hess DT, and Stamler JS (2009). Protein S-nitrosylation in health and disease: a current perspective. Trends Mol. Med 15, 391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furdui CM, and Poole LB (2014). Chemical approaches to detect and analyze protein sulfenic acids. Mass Spectrom. Rev 33, 126–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, and Carroll KS (2016). Profiling the reactivity of cyclic C-nucleophiles towards electrophilic sulfur in cysteine sulfenic acid. Chem. Sci [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearn AS, Stroupe ME, Cabelli DE, Lepock JR, Tainer JA, Nick HS, and Silverman DN (2001). Kinetic analysis of product inhibition in human manganese superoxide dismutase. Biochemistry 40, 12051–12058. [DOI] [PubMed] [Google Scholar]
- Hine C, Harputlugil E, Zhang Y, Ruckenstuhl C, Lee BC, Brace L, Longchamp A, Treviño-Villarreal JH, Mejia P, Ozaki CK, et al. (2015). Endogenous Hydrogen Sulfide Production Is Essential for Dietary Restriction Benefits. Cell 160, 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmström KM, and Finkel T (2014). Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol 15, 411–421. [DOI] [PubMed] [Google Scholar]
- Klomsiri C, Nelson KJ, Bechtold E, Soito L, Johnson LC, Lowther WT, Ryu S-E, King SB, Furdui CM, and Poole LB (2010). Use of dimedone-based chemical probes for sulfenic acid detection evaluation of conditions affecting probe incorporation into redox-sensitive proteins. Methods Enzymol. 473, 77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M-C, Hartman H, Kopp RE, Kirschvink JL, and Yung YL (2006). Production of hydrogen peroxide in the atmosphere of a Snowball Earth and the origin of oxygenic photosynthesis. Proc. Natl. Acad. Sci 18896–18899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin VS, Lippert AR, and Chang CJ (2013). Cell-trappable fluorescent probes for endogenous hydrogen sulfide signaling and imaging H2O2-dependent H2S production. Proc. Natl. Acad. Sci 110, 7131–7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liochev SI (2013). Reactive oxygen species and the free radical theory of aging. Free Radic. Biol. Med 60, 1–4. [DOI] [PubMed] [Google Scholar]
- Liu G-F, Filipović M, Heinemann FW, and Ivanović-Burmazović I (2007). Seven-Coordinate Iron and Manganese Complexes with Acyclic and Rigid Pentadentate Chelates and Their Superoxide Dismutase Activity. Inorg. Chem 46, 8825–8835. [DOI] [PubMed] [Google Scholar]
- Liu HC, Chen WS, Chiang CM, Shia BC, and Ju JM (2015). Extending Liu’s ordering theory for cognitive diagnosis and remedial instruction. ICIC Express Lett. Part B Appl 6, 491–496. [Google Scholar]
- Marino SM, and Gladyshev VN (2010). Cysteine Function Governs Its Conservation and Degeneration and Restricts Its Utilization on Protein Surfaces. J. Mol. Biol 404, 902–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markó L, Szijártó IA, Filipovic MR, Kaβmann M, Balogh A, Park J-K, Przybyl L, N’diaye G, Krämer S, Anders J, et al. (2016). Role of Cystathionine Gamma-Lyase in Immediate Renal Impairment and Inflammatory Response in Acute Ischemic Kidney Injury. Sci. Rep 6, 27517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Higashiyama S, Kijima Y, Suzuki K, Kawano K, Akiyama M, Kawata S, Tarui S, Deutsch HF, and Taniguchi N (1990). Human liver manganese superoxide dismutase: Purification and crystallization, subunit association and sulfhydryl reactivity. Eur. J. Biochem 194, 713–720. [DOI] [PubMed] [Google Scholar]
- Mitchell SJ, Madrigal-Matute J, Scheibye-Knudsen M, Fang E, Aon M, González-Reyes JA, Cortassa S, Kaushik S, Gonzalez-Freire M, Patel B, et al. (2016). Effects of Sex, Strain, and Energy Intake on Hallmarks of Aging in Mice. Cell Metab. 23, 1093–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, and Snyder SH (2009). H2S Signals Through Protein S-Sulfhydration. Sci. Signal 2, ra72–ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan V, Ly T, Pourkarimi E, Murillo AB, Gartner A, Lamond AI, and Kenyon C (2016). Deep Proteome Analysis Identifies Age-Related Processes in C. elegans. Cell Syst. 3, 144–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson KR, and Straub KD (2016). The Role of Hydrogen Sulfide in Evolution and the Evolution of Hydrogen Sulfide in Metabolism and Signaling. Physiology 31, 60–72. [DOI] [PubMed] [Google Scholar]
- Ono K, Akaike T, Sawa T, Kumagai Y, Wink DA, Tantillo DJ, Hobbs AJ, Nagy P, Xian M, Lin J, et al. (2014). Redox chemistry and chemical biology of H2S, hydropersulfides, and derived species: implications of their possible biological activity and utility. Free Radic. Biol. Med 77, 82–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palde PB, and Carroll KS (2015). A universal entropy-driven mechanism for thioredoxin–target recognition. Proc. Natl. Acad. Sci 112, 7960–7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, and Carroll KS (2013). Persulfide reactivity in the detection of protein S-sulfhydration. ACS Chem. Biol 8, 1110–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini EM, Kirkegaard M, Mortensen P, Lutz HU, Thomas AW, and Mann M (2006). In-depth analysis of the membrane and cytosolic proteome of red blood cells. Blood 108, 791–801. [DOI] [PubMed] [Google Scholar]
- Patel BH, Percivalle C, Ritson DJ, Duffy CD, and Sutherland JD (2015). Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nat. Chem 7, 301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BD, and Snyder SH (2012). H2S signalling through protein sulfhydration and beyond. Nat. Rev. Mol. Cell Biol 13, 499–507. [DOI] [PubMed] [Google Scholar]
- Paul BD, and Snyder SH (2015). H2S: A Novel Gasotransmitter that Signals by Sulfhydration. Trends Biochem. Sci 40, 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BD, Sbodio JI, Xu R, Vandiver MS, Cha JY, Snowman AM, and Snyder SH (2014). Cystathionine γ-lyase deficiency mediates neurodegeneration in Huntington’s disease. Nature 509, 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen CE, and Carroll KS (2013). Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem. Rev 113, 4633–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, and Carroll KS (2011). Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat. Chem. Biol 8, 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol A, Renkema GH, Tangerman A, Winkel EG, Engelke UF, de Brouwer APM, Lloyd KC, Araiza RS, van den Heuvel L, Omran H, et al. (2018). Mutations in SELENBP1, encoding a novel human methanethiol oxidase, cause extraoral halitosis. Nat. Genet 50, 120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole LB, Karplus PA, and Claiborne A (2004). Protein sulfenic acids in redox signaling. Annu. Rev. Pharmacol. Toxicol 44, 325–347. [DOI] [PubMed] [Google Scholar]
- Redman LM, Smith SR, Burton JH, Martin CK, Il’yasova D, and Ravussin E (2018). Metabolic Slowing and Reduced Oxidative Damage with Sustained Caloric Restriction Support the Rate of Living and Oxidative Damage Theories of Aging. Cell Metab. 27, 805–815.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisz JA, Bechtold E, King SB, Poole LB, and Furdui CM (2013). Thiol-blocking electrophiles interfere with labeling and detection of protein sulfenic acids. FEBS J. 280, 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbodio JI, Snyder SH, and Paul BD (2016). Transcriptional control of amino acid homeostasis is disrupted in Huntington’s disease. Proc. Natl. Acad. Sci 113, 8843–8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbodio JI, Snyder SH, and Paul BD (2018). Golgi stress response reprograms cysteine metabolism to confer cytoprotection in Huntington’s disease. Proc. Natl. Acad. Sci. U S A 115, 780–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YH, and Carroll KS (2009). Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc. Natl. Acad. Sci. U. S. A 106, 16163–16168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya N, Koike S, Tanaka M, Ishigami-Yuasa M, Kimura Y, Ogasawara Y, Fukui K, Nagahara N, and Kimura H (2013). A novel pathway for the production of hydrogen sulfide from D-cysteine in mammalian cells. Nat. Commun 4, 1366. [DOI] [PubMed] [Google Scholar]
- Snijder PM, Baratashvili M, Grzeschik NA, Leuvenink HGD, Kuijpers L, Huitema S, Schaap O, Giepmans BNG, Kuipers J, Miljkovic JL, et al. (2015). Overexpression of cystathionine γ-lyase suppresses detrimental effects of spinocerebellar ataxia type 3. Mol. Med 21, 758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T (2006). Maintenance of C. elegans. WormBook 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J, and Hodgkin J (1988). Methods, The Nematode Caenorhabditis elegans. In Cold Spring Harbor Monograph Archive, Wood W, ed. pp. 587–606. [Google Scholar]
- Sundaresan M, Yu Z-X, Ferrans VJ, Irani K, and Finkel T (1995). Requirement for Generation of H2O2 for Platelet-Derived Growth Factor Signal Tran sduction. Science (80- ). 270, 296–299. [DOI] [PubMed] [Google Scholar]
- Szabó C (2007). Hydrogen sulphide and its therapeutic potential. Nat. Rev. Drug Discov 6, 917–935. [DOI] [PubMed] [Google Scholar]
- Szabó C, Ischiropoulos H, and Radi R (2007). Peroxynitrite: Biochemistry, pathophysiology and development of therapeutics. Nat. Rev. Drug Discov 6, 662–680. [DOI] [PubMed] [Google Scholar]
- Le Trionnaire S, Perry A, Szczesny B, Szabo C, Winyard PG, Whatmore JL, Wood ME, and Whiteman M (2014). The synthesis and functional evaluation of a mitochondria-targeted hydrogen sulfide donor, (10-oxo-10-(4-(3-thioxo-3H-1,2-dithiol-5-yl)phenoxy)decyl)triphenylphosphonium bromide (AP39). Med. Chem. Commun 5, 728–736. [Google Scholar]
- Turturro A, Witt WW, Lewis S, Hass BS, Lipman RD, and Hart RW (1999). Growth curves and survival characteristics of the animals used in the Biomarkers of Aging Program. J. Gerontol. A. Biol. Sci. Med. Sci 54, B492–501. [DOI] [PubMed] [Google Scholar]
- Valentine WN, Toohey JI, Paglia DE, Nakatani M, and Brockway RA (1987). Modification of erythrocyte enzyme activities by persulfides and methanethiol: possible regulatory role. Proc. Natl. Acad. Sci. U. S. A 84, 1394–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandiver MS, Paul BD, Xu R, Karuppagounder S, Rao F, Snowman AM, Seok Ko H, Il Lee Y, Dawson VL, Dawson TM, et al. (2013). Sulfhydration Mediates Neuroprotective Actions of Parkin. Nat. Commun 4, 1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitvitsky V, Kabil O, and Banerjee R (2012). High Turnover Rates for Hydrogen Sulfide Allow for Rapid Regulation of Its Tissue Concentrations. Antioxid. Redox Signal 17, 22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JL, and Wang R (2015). Hydrogen sulfide-based therapeutics: exploiting a unique but ubiquitous gasotransmitter. Nat. Rev. Drug Discov 14, 329–345. [DOI] [PubMed] [Google Scholar]
- Wang R (2012). Physiological Implications of Hydrogen Sulfide: A Whiff Exploration That Blossomed. Physiol. Rev 92, 791–896. [DOI] [PubMed] [Google Scholar]
- Wedmann R, Bertlein S, Macinkovic I, Böltz S, Miljkovic JL, Muñoz LE, Herrmann M, and Filipovic MR (2014). Working with “H2S”: Facts and apparent artifacts. Nitric Oxide 41, 85–96. [DOI] [PubMed] [Google Scholar]
- Wedmann R, Onderka C, Wei S, Szijártó IA, Miljkovic JL, Mitrovic A, Lange M, Savitsky S, Yadav PK, Torregrossa R, et al. (2016). Improved tag-switch method reveals that thioredoxin acts as depersulfidase and controls the intracellular levels of protein persulfidation. Chem. Sci 7, 3414–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood ZA, Poole LB, and Karplus PA (2003). Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science (80-. ). 300, 650–653. [DOI] [PubMed] [Google Scholar]
- Yadav PK, Yamada K, Chiku T, Koutmos M, and Banerjee R (2013). Structure and Kinetic Analysis of H2S Production by Human Mercaptopyruvate Sulfurtransferase. J. Biol. Chem 288, 20002–20013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Wu L, Jiang B, Yang W, Qi J, Cao K, Meng Q, Mustafa AK, Mu W, Zhang S, et al. (2008). H2S as a Physiologic Vasorelaxant: Hypertension in Mice with Deletion of Cystathionine -Lyase. Science (80-. ). 322, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Zhao K, Ju Y, Mani S, Cao Q, Puukila S, Khaper N, Wu L, and Wang R (2013). Hydrogen sulfide protects against cellular senescence via S-sulfhydration of Keap1 and activation of Nrf2. Antioxid. Redox Signal 18, 1906–1919. [DOI] [PubMed] [Google Scholar]
- Yang HY, Kwon J, Choi HI, Park SH, Yang U, Park HR, Ren L, Chung KJ, Kim YU, Park BJ, et al. (2012). In-depth analysis of cysteine oxidation by the RBC proteome: Advantage of peroxiredoxin II knockout mice. Proteomics 12, 101–112. [DOI] [PubMed] [Google Scholar]
- Yang J, Gupta V, Carroll KS, and Liebler DC (2014). Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat. Commun 5, 4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccarin M, Falda M, Roveri A, Bosello-Travain V, Bordin L, Maiorino M, Ursini F, and Toppo S (2014). Quantitative label-free redox proteomics of reversible cysteine oxidation in red blood cell membranes. Free Radic. Biol. Med 71, 90–98.24642086 [Google Scholar]
- Zhang D, Macinkovic I, Devarie-Baez NO, Pan J, Park C-M, Carroll KS, Filipovic MR, and Xian M (2014). Detection of Protein S-Sulfhydration by a Tag-Switch Technique. Angew. Chemie Int. Ed 53, 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Xin L, Shan B, Chen W, Xie M, Yuen D, Zhang W, Zhang Z, Lajoie GA, and Ma B (2012). PEAKS DB: De Novo Sequencing Assisted Database Search for Sensitive and Accurate Peptide Identification. Mol. Cell. Proteomics 11, M111.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset S1: MS/MS data of the persulfidated TST digested with trypsin. Related to Figure 2.
Dataset S2: MS/MS data of the persulfidated TST digested with chymotrypsin. Related to Figure 2.
Dataset S3: MS/MS data of the persulfidated MnSOD digested with trypsin. Related to Figure 3.
Dataset S5: Antibody microarray analysis of protein persulfidation. Related to Figure 5.
Dataset S4: MS/MS data of the persulfidated MnSOD digested with chymotrypsin. Related to Figure 3.
Data Availability Statement
The raw data corresponding to the Antibody Microarray Detection of Persulfidation of EGFR Pathway kinases represent entirely new application of the dimedone switch method and are therefore available in more detail from the corresponding author on request. Proteomic data used for Figure S2C are stored in Data S1, S2 and for Figure S3H, I in Data S3, S4. All in-gel persulfidation, as well as Western blot sufenylation and sulfynilation data are reported in main and supporting Figures. Other raw data are available at DOI: 10.17632/pw2wz39tsk.1.