Abstract
Coronaviral infection of New World camelids was first identified in 1998 in llamas and alpacas with severe diarrhea. In order to understand this infection, one of the coronavirus isolates was sequenced and analyzed. It has a genome of 31,076 nt including the poly A tail at the 3′ end. This virus designated as ACoV-00-1381 (ACoV) encodes all 10 open reading frames (ORFs) characteristic of Group 2 bovine coronavirus (BCoV). Phylogenetic analysis showed that the ACoV genome is clustered closely (> 99.5% identity) with two BCoV strains, ENT and LUN, and was also closely related to other BCoV strains (Mebus, Quebec, DB2), a human corona virus (strain 043) (> 96%), and porcine hemagglutinating encephalomyelitis virus (> 93% identity). A total of 145 point mutations and one nucleotide deletion were found relative to the BCoV ENT. Most of the ORFs were highly conserved; however, the predicted spike protein (S) has 9 and 12 amino acid differences from BCoV LUN and ENT, respectively, and shows a higher relative number of changes than the other proteins. Phylogenetic analysis suggests that ACoV shares the same ancestor as BCoV ENT and LUN.
Keywords: Coronavirus, Alpaca, Bovine coronavirus, Alpaca coronavirus
Introduction
Coronaviruses are a genus in the family Coronaviridae, which are enveloped viruses with large, positive sense RNA genomes of 29 to 32 kb. They are important causes of human and animal diseases that include respiratory infections, gastroenteritis, hepatic and neurological disorders, as well as immune-mediated diseases such as SARS and feline infectious peritonitis (reviewed in de Groot-Mijnes et al., 2005, Kahn, 2006, Spaan et al., 1988, Wege et al., 1982). The coronaviruses possess a characteristic genome composition. The 5′ two-thirds of the genome encodes two polyproteins (1a and 1ab) that contain proteins necessary for RNA replication. The 3′ one-third encodes two non-structural proteins (NS1 and 2) and several structural proteins, including a nucleocapsid protein (N) and three or four envelope proteins: the membrane (M), spike (S), hemagglutinin-esterase (HE), and/or a small membrane (E) proteins.
Originally, coronaviruses were classified into three groups on the basis of their antigenic cross reactivity (Cavanagh et al., 1993, Cavanagh and Davis, 1993, Tsunemitsu et al., 1995). When coronavirus genome sequence data became available, the original antigenic groups were converted into three genetic groups based on the similarity of their nucleotide sequences (Gonzalez et al., 2003, Lai, 2003, Vijgen et al., 2006). Group 2 coronaviruses include murine hepatitis virus (MHV), bovine coronaviruses (BCoV), human coronavirus OC43 (HCoV-OC43), rat sialodacryoadenitis virus, porcine hemagglutinating encephalomyelitis virus (PHEV), canine respiratory coronavirus and equine coronavirus. Fifteen strains of BCoV have been sequenced and have been implicated in a variety of diseases including respiratory and enteric infections (Chouljenko et al., 2001a, Dea et al., 1995, Han et al., 2006, King and Brian, 1982, Park et al., 2006, Woloszyn et al., 1990). For example, the ENT and LUN strains were isolated from animals with fatal shipping fever pneumonia (Chouljenko et al., 2001a). The former was associated with enteritis; whereas the latter is associated with respiratory infection (pneumonia). Mebus, Quebec and DB2 are virulent strains associated with neonatal calf diarrhea and winter dysentery in adult dairy cattle (Dea et al., 1995, Han et al., 2006, Park et al., 2006).
Coronavirus associated with outbreaks of diarrhea in all age groups of New World camelids (llamas and alpacas) was first identified in 1998, in Oregon. The symptoms were similar to cattle infected with bovine coronavirus. Sick camelids showed varying degrees of severity of clinical disease, with some animals dying and others requiring intensive medical care. Coronavirus-like viruses were frequently isolated from the diseased animals (Cebra et al., 2003). Because of the severity of the infection and the ability of coronaviruses to cross the species barriers, including human–animal barriers, we undertook an investigation to further characterize this virus. In this report we describe the sequence of the ACoV and compare it to other members of the coronavirus genus. We found that it is closely related to Group 2 bovine coronaviruses.
Results and discussion
Cloning and sequence analysis
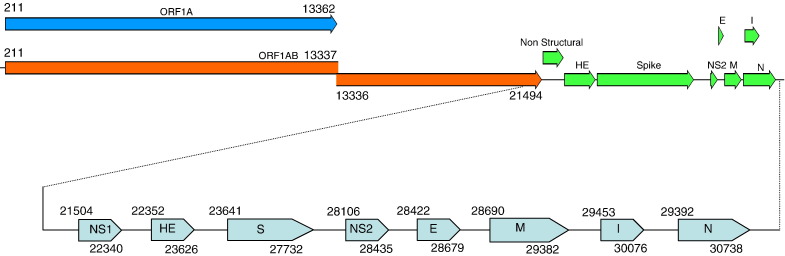
A total of 21 overlapping cDNA fragments were generated by RT-PCR cDNA cloning to encompass the entire RNA genome. The oligonucleotide primer sets used for these amplifications are listed in Table 1 . The 5′ and 3′ end of the genome were determined by 5′ and 3′RACE, respectively. This resulted in a genome sequence of 31,076 nt including a poly A tail of 38 nt. The genome encodes 10 ORFs characteristic of Group 2 BCoV and 5′ and 3′ untranslated sequences of 210 and 298 nts (which do not include the poly A tail), respectively (Fig. 1 ). The ORF coordinates are shown in Fig. 1. The predicted ORF 1a and ORF 1ab contained 13,152 and 21,282 nt, respectively (Fig. 1). ORF 1ab contains a 26 nt region that overlapped with ORF 1a and included a predicted ‘slippery’ sequence UUUAAAC. Based on evidence from other coronaviruses (Chouljenko et al., 2001b), this sequence causes a − 1 frameshift during the translation of ORF1a. This results in a portion of the proteins (called ORF 1ab) avoiding translation termination and containing an additional 2711 amino acids. Upstream of five of the genes, there is a repeated intergenic sequence, UCUAAAC, that is predicted to interact with the viral transcriptase along with cellular factors to ‘splice’ the leader sequence onto the start of each ORF. ORF-NS1, HE, S, M and N were all preceded by this sequence (Table 2 ), which would be predicted to give rise to a nested set of mRNAs characteristic of the order Nidovirales.
Table 1.
Oligonucleotide primer sets used for RT-PCR, 5′RACE and 3′RACE
| No. | Name | Primer sequences |
|---|---|---|
| 5′end | 5RACE1 | CGCAGTGGTGGAGCATACTA |
| 5RACE2 | GGCCACTGCCTAGGATACAA | |
| 1. | LC1F | GATTGCGAGC GATTTGCGTG |
| LC2016R | GCAGATTTTATCTGCGTAGTCA | |
| 2. | LC1112F | TGCGTGATCCACGTTATGTT |
| LC4092R | CATTAGCAGGATTTACAACGACT | |
| 3. | LC3779F | GTGCCATTTACAGCCCACTT |
| LC5138R | AGGCAAGCAATTCCTTCTGA | |
| 4. | LC4133F | GGTGGTGTTGCAAAGGCTAT |
| LC6993R | AACCCACATCCTGAATGGAA | |
| 5. | LC6921F | CGCACAGTGGATTAAGAGCA |
| LC8738R | ACAGCAACAACAATGGGACA | |
| 6. | LC8221F | CAGCTGATTTAGGTGTTCTGA |
| LC10021R | ATGTCTGGGACAGTAGACCT | |
| 7. | LC10000F | GGTCTACTGTCCCAGACA |
| LC12781R | CAAGGAGGATCTAACTCCCA | |
| 8. | LC12539F | GCAAATCGGCATAATGAGGT |
| LC13913R | ACCTCCACCAATTTGTCTGC | |
| 9. | LC13085F | TCATATGGTGGTGCGTCTGT |
| LC16485R | TGCAGCAAACAATTTCAAGC | |
| 10. | LC16484F | AGCGCTTGAAATTGTTTGCT |
| LC17313R | CAATTGAGCAGGATCACCAA | |
| 11. | LC17071F | CGTATTGTTCCTGCCAGGT |
| LC19948R | CTCCAACTTGTCCACCACT | |
| 12. | 19519F | ACAGGACAGGCTGGTGAAAT |
| 20125R | TTGCCAGAGCATCATTACCA | |
| 13. | LC20050F | AGTGGTGGACAAGGTTGGAG |
| LC22190R | TCATCATTCTCGGGAAGGTC | |
| 14. | 21908F | TCACTGATGCTGCACTTTCC |
| 23688R | ATAACAGCAAAAGCCGTTGG | |
| 15. | LC22190R | TCATCATTCTCGGGAAGGTC |
| LC27152F | GTTAGTCCCGGTCTGTGCAT | |
| 16. | 26132F | TGTGGTGATTATGCAGCATGT |
| 27183R | CTACCACCAGCAATGCACAG | |
| 17. | LC27152F | GTTAGTCCCGGTCTGTGCAT |
| LC29816R | GTCGGTGCCATACTGGTCTT | |
| 18. | LC29642F | GACAAGGTGTGCCTATTGCA |
| LC30662R | GCTGATGTCCTCTGCAGTCA | |
| 19. | LC304953F | TGAATAAACCCCGCCAGA |
| LC30782F | GAATGGATGTCTTGCTGCTA | |
| 3′end | 3RACE | GAATCTTGACGAACCCCAGA |
Fig. 1.
Map of the alpaca coronavirus genome (GenBank accession no. DQ915164) and ORFs of the viral coding sequence. NS1, non-structural protein gene 1; HE, hemagglutinin-esterase gene; S, spike gene; NS2, non-structural protein gene 2; E, envelope protein gene; M, membrane protein gene; N, nucleocapsid gene; I, internal ORF. The numbers above and below each ORF correspond to the nt coordinates of each gene.
Table 2.
Intergenic sequences upstream of ACoV genes
| Gene name | Coding sequence | Intergenic sequence upstream of the AUG |
|---|---|---|
| NS1 | 21,504–22,340 | UCUAAACUUUAAAAAUG |
| HE | 22,352–23,626 | ACUAAACUCAGUGAAAAUG |
| S | 23,641–27,732 | AAUCUAAACAUG |
| M | 28,690–29,382 | AAUCCAAACAUUAUG |
| N | 29,392–30,738 | AUCUAAACUUUAAGGAUG |
Sequence comparisons with bovine coronaviruses
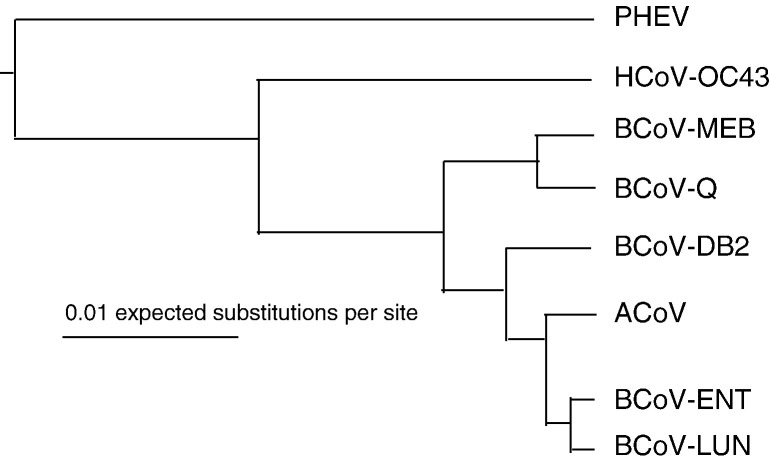
The sequence of ACoV was closely related to bovine coronaviruses (Table 3 ). It has 99.54% to 99.55% identity to the BCoV ENT and LUN strains, respectively. It was slightly less related to the Mebus and Quebec strains at about 98.5% (Table 3). ACoV is also closely related to a human corona virus (strain 0C43) (> 96%) and porcine hemagglutinating encephalomyelitis virus (> 93% identity) (Fig. 2 and Table 3). Phylogenetic analysis of the entire genome of ACoV and other selected coronaviruses was carried out and the results are summarized in Fig. 2 and Table 3.
Table 3.
Sequence comparisons of ACoV with selected Group 2 coronaviruses
| ACoV-alpaca | BCoV-DB2 | BCoV-ENT | BCoV-LUN | BCoV-MEB | BCoV-Q | HCoV-OC43 | PHEV-VW572 | |
|---|---|---|---|---|---|---|---|---|
| ACoV-alpaca | – | 99.18 | 99.54 | 99.55 | 98.53 | 98.48 | 96.52 | 93.4 |
| BCoV-DB2 | – | 99.33 | 99.33 | 99.05 | 98.97 | 96.81 | 93.67 | |
| BCoV-ENT | – | 99.66 | 98.73 | 98.66 | 96.62 | 93.56 | ||
| BCoV-LUN | – | 98.74 | 98.66 | 96.61 | 93.58 | |||
| BCoV-MEB | – | 99.72 | 96.94 | 93.67 | ||||
| BCoV-Q | – | 96.87 | 93.59 | |||||
| HCoV-OC43 | – | 93.5 |
BCoV: bovine coronavirus; HCoV: human coronavirus; PHEV: porcine hemagglutinating encephalomyelitis virus; Q: Quebec; MEB: Mebus.
Fig. 2.
A maximum-likelihood phylogenetic tree of selected coronavirus genomes. ACoV (GenBank accession no. DQ915164) was compared to other Group 2 Coronaviruses including porcine hemagglutinating encephalomyelitis virus (PHEV VW572, GenBank accession no. YP_459958); human coronavirus OC43 (HCoV-OC43, AY391777); bovine coronavirus Mebus strain (BCoV-Meb, U00735); bovine coronavirus Quebec strain (BCoV-Q, D00662); bovine coronavirus DB2 strain (BCoV-DB2, DQ811784); bovine coronavirus ENT strain (BCoV-ENT, Q91A22); bovine coronavirus LUN strain (BCoV-LUN, AF391542). The scale bar represents the genetic distance (nucleotide substitution per site).
Comparison of nine predicted BCoV and ACoV proteins
Analysis of the predicted ORF 1a and ORF 1b shows 14 aa and 1 aa changes, respectively, compared to the corresponding ORFs in the most closely related BCoV ENT strain. The ACoV HE protein is phylogenetically similar to those of BCoV ENT and LUN, with one and three amino acid differences, respectively (Fig. 3B). In addition, the following differences were noted between ACoV and the most closely related BCoV strains for the structural proteins: membrane (M), one difference out of 229 aa; internal orf (I), two differences out of 207 aa; and N, one to five differences out of 448 aa. The nucleocapsid (N) protein of coronavirus has been used as an early diagnostic marker for coronaviruses such as the SARS virus (Che et al., 2004). The highly conserved small membrane envelope (E) protein is unique to BCoV Group 2 and is identical between ACoV and the closely related BCoV strains. A similar pattern of relatedness was evident for the non-structural (NS) proteins. There were one to two aa differences in the predicted 32 kDa NS1 protein of ACoV compared to the most closely related BCoV ENT and LUN strains. This protein is not essential for virus replication in vitro, but has been implicated in virus pathogenicity (Vijgen et al., 2006). There were also one to two aa mutations between the predicted 12.7 kDa NS2 and those from the most closely related BCoV isolates.
Fig. 3.
A phylogenetic tree of selected Group 2 spike proteins and hemagglutinin-esterase amino acid sequences. (A) The S proteins from the selected strains were analyzed by the neighbor-joining method. (B) The hemagglutinin-esterase protein of the alpaca strain was compared to other selected Group 2 coronavirus hemagglutinin-esterase proteins. For information on the viruses analyzed, see legend to Fig. 2.
Comparison of the BCoV and ACoV predicted spike protein
Because the S protein has been implicated in tissue tropism (Gallagher and Buchmeier, 2001, Godet et al., 1994, Schultze et al., 1991, Schultze and Herrler, 1994), its affinity may be reflected in the type of diseases caused. The spike protein of ACoV and BCoV is 1363 aa long. The predicted proteolytic cleavage site (KRRSRR) was conserved between all strains. Cleavage at this site results in the production of two subunits (S1 and S2) in the well-characterized murine coronaviruses (de Haan et al., 2006). S1 and S2 of ACoV were predicted to be 760 and 603 amino acids, respectively. The neighbor-joining method of molecular evolutionary analysis revealed that the ACoV S protein appears to evolve at a somewhat accelerated rate compared to other proteins, e.g., the HE (Fig. 3). The predicted ACoV S protein had 9 and 12 amino acid differences from the predicted S proteins of BCoV strains LUN and ENT, respectively. Most of the mutations occurred in the S1 subunit. Three ACoV S amino acid changes are particularly striking, including: serine at aa #174; proline at 565; and serine at 702. In all the other S proteins analyzed (Fig. 3A and Table 4 ), these amino acids replace a Pro, Leu and Leu, respectively. The loss of one proline (#174) and the gain of another (#565) could significantly alter the structure of ACoV relative to the homologs from the closely related viruses. It has been shown that S2 is not directly involved in receptor binding, indicating that changes in S1 could be involved in host specificity (de Haan et al., 2006).
Table 4.
The amino acid differences of the S glycoprotein among ACoV and the two most related BCoV strains, ENT and LUN
| Predicted S protein residues | Amino acid |
||
|---|---|---|---|
| ACoV-00-1381 (ABI93999)a | LUN (AAL57308)a | ENT (NP_150077)a | |
| 54 | N | K | ⁎ |
| 174 | S | P | P |
| 179 | R | ⁎ | Q |
| 370 | D | ⁎ | Y |
| 483 | P | ⁎ | S |
| 531 | N | D | D |
| 565 | P | L | L |
| 571 | Y | H | H |
| 702 | S | L | L |
| 965 | D | E | E |
| 1052 | A | T | ⁎ |
| 1082 | D | E | E |
| 1180 | G | ⁎ | D |
| 1242 | D | ⁎ | Y |
Sequence accession number.
Same as ACoV strain.
Conclusions
The sequence analyses presented in this report demonstrate that ACoV-00-1381, which is associated with diarrhea in camelids (Cebra et al., 2003), is closely related to bovine coronaviruses. It appears to have been derived from the same ancestor as the LUN strain isolated from cattle with fatal shipping fever pneumonia and the ENT strain isolated from cattle with either pneumonia or enteritis (Chouljenko et al., 2001a, Storz et al., 1996). New World camelids have been in contact with cattle for over 500 years in South America, compared to their relatively recent and small-scale introduction to North America. Although the identification of coronaviral infection and epidemic diarrhea in all age groups of llamas and alpacas occurred only recently, it is possible that the virus crossed between species during earlier interspecies contact or is a BCoV strain that is pathogenic for both bovids and camelids, although BCoV infection in camelids had not been described previous to the Cebra et al. (2003) report.
The most significant difference that we observed between ACoV and BCoV strains was in the spike protein. The S protein forms distinctive surface projections on the virions and is responsible for the primary attachment of the virus to cell surface receptors (Schultze et al., 1991). It is a glycosylated and acetylated polypeptide with a molecular weight of 170 kDa to 220 kDa and is the major hemagglutinin of bovine coronaviruses (Schultze and Herrler, 1994). It has been suggested that the high degree of variation in host range and tissue tropism of coronaviruses is largely attributable to variations in the S glycoprotein (Gallagher, 2001). There are a number of distinct differences in S proteins between the respiratory isolates and diarrhea isolates (of bovine coronaviruses, human and porcine). Although the importance of such variability in the virulence and tropism of BCoV is unknown, some amino acid changes could have significant effects on the conformation, charge, hydrophobicity and antigenic regions of the protein. These mutations could change either the protein folding or physicochemical characteristics and could be involved in altering the host specificity of the virus.
Material and methods
Cells and virus
An alpaca coronavirus isolate designated ACoV-00-1381 was obtained from a diarrhea sample by the Veterinary Diagnostic Lab at Oregon State University (Cebra et al., 2003). The isolate was grown on human rectal tumor (HRT-18G) cells, which were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen), penicillin (100 U/ml) and streptomycin (100 μg/ml) (Sigma-Aldrich, Inc.) at 37 °C with 5% CO2 in a humidified incubator. The virus was propagated in Dulbecco's modified Eagle's medium supplemented with 2.5 μg/ml trypsin and 2.5 μg/ml pancreatin, 1× insulin–transferrin–selenium (Cat No. 51500-056, GIBCO).
Viral RNA preparation
Cells were infected at a multiplicity of infection of 1 to 3 and were incubated for 3 to 5 days. The supernatant was collected and clarified with a bench-top Beckman Coulter Allegra 64R centrifuged at 9000×g for 30 min. Viruses were isolated from the supernatant by centrifuging through a 30% sucrose cushion with an ultracentrifuge (Beckman model XL-70) at 25,000 rpm for 2 h in an SW28 rotor. The pellets were re-suspended in 100 μl TE buffer (10 mM Tris–HCl, 1 mM EDTA [pH 8.0]), and viral RNA was extracted with Trizol (Invitrogen) as described in the manufacturer's instructions.
RT-PCR amplification
A one-step reverse transcriptase (RT)-PCR kit (Invitrogen) was used to generate viral DNA sequences for analysis. Four microliters of the RNA extract (0.5 μg/μl) was added to the RT-PCR mixture (in total 20-μl reaction) containing final concentrations of 1.25 μM of each forward and reverse primer, 1× buffer for RT-PCR, 0.1 mM MgSO4 and 1 U of RT/Taq. The RT reaction was performed at 50 °C for 45 min, then the reverse transcriptase was inactivated and Taq was activated at 94 °C for 2 min. PCR was performed for 30 cycles as the following: 94 °C for 30 s, 50 °C for 45 s, 72 °C for 2.5 min, followed by a 7 min elongation reaction at 72 °C after the final cycle.
5′RACE
The 5′ end of the viral genome was amplified by 5′RACE kit (Invitrogen), following the manufacturer's instructions. Briefly, the first-strand cDNA was synthesized with gene-specific primer 5RACE1 (Table 1). Approximately 1 to 3 μg of total viral RNA was used as the template in a 20-μl RT reaction containing the 5RACE1 primer. After purification of the first-strand cDNA, the 5′ end was tailed with dCTP using terminal deoxynucleotidyltransferase. The oligo(dC) cDNA was then amplified with a second gene-specific primer (5RACE2) (Table 1) and the abridged anchor primer (AAP) specific for the 5′ dC tail. The primary PCR products were then reamplified with the hemi-nested gene-specific primer 5RACE2 and the abridged universal amplification primer (AUAP), under conditions recommended by the manufacturer. The 5′RACE products were cloned and sequenced as described for the RT-PCR amplimers.
3′RACE
The 3′ end of the viral genome was amplified by 3′RACE kit (Invitrogen) following the manufacturer's instructions. Briefly, the first-strand cDNA was synthesized with an oligo(dT)-containing adaptor primer (AP) to the end of the viral genome. Approximately 1 to 3 μg of total viral RNA was used as the template in a 20-μl RT reaction containing the oligo(dT)-AP primer. After the cDNA synthesis, 3 μl of the cDNA was used directly in PCR reaction with a gene-specific primer 3′RACE and abridged universal amplification primer (AUAP) in a reaction recommended by the manufacturer. The 3′RACE products were then cloned and sequenced as described for the RT-PCR amplimers.
Primers
The oligonucleotide primer sets used for RT-PCR, 5′RACE and 3′RACE, are listed in Table 1. Selection of the sequences used to design these primers for RT-PCR of the ACoV genome was based on the known genomic information of several strains of coronaviruses BCoV-LUN and BCoV-ENT (GenBank accession number AF391541 for ENT and AF391542 for LUN). They were selected by Primer 3 program available online. The walking primers (not shown) were selected from the ACoV sequence by the DNA sequence core facility at Oregon State University.
TOPO TA cloning
The RT-PCR, 5′RACE and 3′RACE products were cloned into the pCR 2.1-TOPO plasmid vector (vector) following the manufacturer's instructions (Invitrogen).
DNA sequencing and analysis
Nucleotide sequences were determined by sequencing cloned plasmid DNA with the universal primers first, then using primer walking to complete the sequence. In addition,three independently generated RT-PCR reaction products were combined and processed with a ChargeSwitch PCR Clean-Up kit (Invitrogen) before sequencing. Selected regions were reconfirmed by sequencing the RT-PCR products directly. All the cDNA clones and RT-PCR products were sequenced by the DNA sequence core facility at Oregon State University. The nucleotide sequences were assembled and analyzed with the EMBOSS software. Additional analyses were carried out using MacVector and programs described by Esteban et al. (2005).
Nucleotide sequence accession number
The sequences reported in this work have been deposited in the GenBank database under accession number DQ915164.
Acknowledgment
This study was supported by a grant (F0434A) from the Alpaca Research Foundation.
References
- Cavanagh D., Davis P.J. Sequence analysis of strains of avian infectious bronchitis coronavirus isolated during the 1960s in the U.K. Arch. Virol. 1993;130(3–4):471–476. doi: 10.1007/BF01309675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D., Brian D.A., Brinton M.A., Enjuanes L., Holmes K.V., Horzinek M.C., Lai M.M., Laude H., Plagemann P.G., Siddell S.G. The Coronaviridae now comprises two genera, coronavirus and torovirus: report of the Coronaviridae Study Group. Adv. Exp. Med. Biol. 1993;342:255–257. doi: 10.1007/978-1-4615-2996-5_39. [DOI] [PubMed] [Google Scholar]
- Cebra C.K., Mattson D.E., Baker R.J., Sonn R.J., Dearing P.L. Potential pathogens in feces from unweaned llamas and alpacas with diarrhea. J. Am. Vet. Med. Assoc. 2003;223(12):1806–1808. doi: 10.2460/javma.2003.223.1806. [DOI] [PubMed] [Google Scholar]
- Che X.Y., Qiu L.W., Pan Y.X., Wen K., Hao W., Zhang L.Y., Wang Y.D., Liao Z.Y., Hua X., Cheng V.C., Yuen K.Y. Sensitive and specific monoclonal antibody-based capture enzyme immunoassay for detection of nucleocapsid antigen in sera from patients with severe acute respiratory syndrome. J. Clin. Microbiol. 2004;42(6):2629–2635. doi: 10.1128/JCM.42.6.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouljenko V.N., Foster T.P., Lin X., Storz J., Kousoulas K.G. Elucidation of the genomic nucleotide sequence of bovine coronavirus and analysis of cryptic leader mRNA fusion sites. Adv. Exp. Med. Biol. 2001;494:49–55. doi: 10.1007/978-1-4615-1325-4_7. [DOI] [PubMed] [Google Scholar]
- Chouljenko V.N., Lin X.Q., Storz J., Kousoulas K.G., Gorbalenya A.E. Comparison of genomic and predicted amino acid sequences of respiratory and enteric bovine coronaviruses isolated from the same animal with fatal shipping pneumonia. J. Gen. Virol. 2001;82(Pt. 12):2927–2933. doi: 10.1099/0022-1317-82-12-2927. [DOI] [PubMed] [Google Scholar]
- Dea S., Michaud L., Milane G. Comparison of bovine coronavirus isolates associated with neonatal calf diarrhoea and winter dysentery in adult dairy cattle in Quebec. J. Gen. Virol. 1995;76(Pt. 5):1263–1270. doi: 10.1099/0022-1317-76-5-1263. [DOI] [PubMed] [Google Scholar]
- de Groot-Mijnes J.D., van Dun J.M., van der Most R.G., de Groot R.J. Natural history of a recurrent feline coronavirus infection and the role of cellular immunity in survival and disease. J. Virol. 2005;79(2):1036–1044. doi: 10.1128/JVI.79.2.1036-1044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Te Lintelo E., Li Z., Raaben M., Wurdinger T., Bosch B.J., Rottier P.J. Cooperative involvement of the S1 and S2 subunits of the murine coronavirus spike protein in receptor binding and extended host range. J. Virol. 2006;80(22):10909–10918. doi: 10.1128/JVI.00950-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban D.J., Da Silva M., Upton C. New bioinformatics tools for viral genome analyses at Viral Bioinformatics–Canada. Pharmacogenomics. 2005;6(3):271–280. doi: 10.1517/14622416.6.3.271. [DOI] [PubMed] [Google Scholar]
- Gallagher T.M. Murine coronavirus spike glycoprotein. Receptor binding and membrane fusion activities. Adv. Exp. Med. Biol. 2001;494:183–192. [PubMed] [Google Scholar]
- Gallagher T.M., Buchmeier M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279(2):371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet M., Grosclaude J., Delmas B., Laude H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 1994;68(12):8008–8016. doi: 10.1128/jvi.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J.M., Gomez-Puertas P., Cavanagh D., Gorbalenya A.E., Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 2003;148(11):2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.G., Cheon D.S., Zhang X., Saif L.J. Cross-protection against a human enteric coronavirus and a virulent bovine enteric coronavirus in gnotobiotic calves. J. Virol. 2006;80(24):12350–12356. doi: 10.1128/JVI.00402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn J.S. The widening scope of coronaviruses. Curr. Opin. Pediatr. 2006;18(1):42–47. doi: 10.1097/01.mop.0000192520.48411.fa. [DOI] [PubMed] [Google Scholar]
- King B., Brian D.A. Bovine coronavirus structural proteins. J. Virol. 1982;42(2):700–707. doi: 10.1128/jvi.42.2.700-707.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M. SARS virus: the beginning of the unraveling of a new coronavirus. J. Biomed. Sci. 2003;10(6 Pt. 2):664–675. doi: 10.1007/BF02256318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Jeong C., Yoon S.S., Choy H.E., Saif L.J., Park S.H., Kim Y.J., Jeong J.H., Park S.I., Kim H.H., Lee B.J., Cho H.S., Kim S.K., Kang M.I., Cho K.O. Detection and characterization of bovine coronaviruses in fecal specimens of adult cattle with diarrhea during the warmer seasons. J. Clin. Microbiol. 2006;44(9):3178–3188. doi: 10.1128/JCM.02667-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze B., Herrler G. Recognition of cellular receptors by bovine coronavirus. Arch. Virol., Suppl. 1994;9:451–459. doi: 10.1007/978-3-7091-9326-6_44. [DOI] [PubMed] [Google Scholar]
- Schultze B., Gross H.J., Brossmer R., Herrler G. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic acid as a receptor determinant. J. Virol. 1991;65(11):6232–6237. doi: 10.1128/jvi.65.11.6232-6237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W., Cavanagh D., Horzinek M.C. Coronaviruses: structure and genome expression. J. Gen. Virol. 1988;69(Pt. 12):2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- Storz J., Stine L., Liem A., Anderson G.A. Coronavirus isolation from nasal swab samples in cattle with signs of respiratory tract disease after shipping. J. Am. Vet. Med. Assoc. 1996;208(9):1452–1455. [PubMed] [Google Scholar]
- Tsunemitsu H., el-Kanawati Z.R., Smith D.R., Reed H.H., Saif L.J. Isolation of coronaviruses antigenically indistinguishable from bovine coronavirus from wild ruminants with diarrhea. J. Clin. Microbiol. 1995;33(12):3264–3269. doi: 10.1128/jcm.33.12.3264-3269.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Lemey P., Maes P., Van Reeth K., Nauwynck H., Pensaert M., Van Ranst M. Evolutionary history of the closely related Group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 2006;80(14):7270–7274. doi: 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege H., Siddell S., ter Meulen V. The biology and pathogenesis of coronaviruses. Curr. Top. Microbiol. Immunol. 1982;99:165–200. doi: 10.1007/978-3-642-68528-6_5. [DOI] [PubMed] [Google Scholar]
- Woloszyn N., Boireau P., Laporte J. Nucleotide sequence of the bovine enteric coronavirus BECV F15 mRNA 5 and mRNA 6 unique regions. Nucleic Acids Res. 1990;18(5):1303. doi: 10.1093/nar/18.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]