Abstract
Feline kobuvirus (FeKoV), a novel picornavirus of the genus kobuvirus, was initially identified in the feces of cats with diarrhea in South Korea in 2013. To date, there is only one report of the circulation of kobuvirus in cats in southern China. To investigate the presence and genetic variability of FeKoV in northeast China, 197 fecal samples were collected from 105 cats with obvious diarrhea and 92 asymptomatic cats in Shenyang, Jinzhou, Changchun, Jilin and Harbin regions, Northeast China, and viruses were detected by RT-PCR with universal primers targeting all kobuviruses. Kobuvirus was identified in 28 fecal samples with an overall prevalence of 14.2% (28/197) of which 20 samples were co-infected with feline parvovirus (FPV) and/or feline bocavirus (FBoV). Diarrhoeic cats had a higher kobuvirus prevalence (19.1%, 20/105) than asymptomatic cats (8.7%, 8/92). By genetic analysis based on partial 3D gene, all kobuvirus-positive samples were more closely related to previous FeKoV strains with high identities of 90.5%–97.8% and 96.6%–100% at the nucleotide and amino acid levels. Additionally, phylogenetic analysis based on the complete VP1 gene indicated that all FeKoV strains identified in this study were placed into a cluster, which separated from other reference strains previously reported, and three identical amino acid substitutions were present at the C-terminal of the VP1 protein for these FeKoV strains. Furthermore, two complete FeKoV polyprotein genomes were successfully obtained from two positive samples and designated 16JZ0605 and 17CC0811, respectively. The two strains shared 92.9%–94.9% nucleotide identities and 96.8%–98.4% amino acid identities to FeKoV prototype strains. Phylogenetic analysis indicated that FeKoVs were clustered according to their geographical regions, albeit with limited sequences support. This study provides the first molecular evidence that FeKoV circulates in cats in northeast China, and these FeKoVs exhibit genetic diversity and unique evolutionary trend.
Keywords: Kobuvirus, Cat, Genetic characterization, Phylogenetic analysis
Abbreviations: FeKoV, Feline kobuvirus; FPV, Feline parvovirus; FBoV, Feline bocavirus
Highlights
-
•
This study first provides the molecular evidence for the circulation of feline kobuvirus in cats in northeast China.
-
•
Feline kobuvirus infection is closely related to viral diarrhea in cats.
-
•
All the strains identified in this study displayed genetic diversity and unique evolutionary trend.
-
•
Feline kobuvirus strains were clustered according to their geographical regions.
1. Introduction
Kobuvirus (KoV), which belongs to a recently classified genus (Kobuvirus) of the family Picornaviridae, is a small, non-enveloped, spherical virus approximately 27–30 nm in diameter (Zell 2018). Kobuvirus has a single-stranded, positive-sense RNA genome of 8.2–8.4 kb consisting of 5′ untranslated region (UTR), one single open reading frame (ORF), 3′UTR and poly A tail (Han et al. 2011). This ORF encodes a large polyprotein which is cleaved to yield a non-structural protein L, three structural proteins (VP0, VP1 and VP3) and seven other non-structural proteins (2A-2C and 3A-3D) (Reuter et al. 2011). Of these, VP1 is the most important viral capsid protein determining the antigenicity and pathogenicity for kobuvirus. 3D gene encodes the RNA-dependent RNA polymerase (RdRp), which plays a critical role in viral replication (Lescar and Canard 2009). Based on the function of encoded proteins, the kobuvirus genome is generally divided into three functional regions: P1 (encoding structural protein VP0, VP1 and VP3), P2 and P3 (encoding non-structural protein 2A-2C and 3A-3D, respectively) (Han et al. 2011; Reuter et al. 2011).
Since human Aichi virus (AiV) was first recognized in 1989 as the cause of oyster-associated nonbacterial gastroenteritis in humans in Aichi Prefecture, Japan (Yamashita et al. 1993), the novel kobuviruses have been identified in many mammalian animals. In 2003, bovine kobuvirus (BKV) was first identified from a contaminant of HeLa cells in Japan (Yamashita et al. 2003). Subsequently, porcine kobuvirus (PKoV) was found in the stools of domestic pigs in Hungary in 2008 (Reuter et al. 2008). Canine kobuvirus (CaKoV) that was genetically related to human AiV was first identified in a domestic dog with acute gastroenteritis in USA in 2011 (Kapoor et al. 2011), and was subsequently found in healthy domestic dogs (Oem et al. 2014a) and wild carnivores, including wolves (Melegari et al. 2018), red foxes (Di Martino et al. 2014), golden jackals, side-striped jackal and spotted hyena (Olarte-Castillo et al. 2015). To date, kobuviruses have been reported in human (Yamashita et al. 1993), cattle (Yamashita et al. 2003), sheep (Reuter et al. 2010), pig (Reuter et al. 2008), rodents (Phan et al. 2011), goat (Oem et al. 2014b), wild boars (Reuter et al. 2013), roe deer (Di Martino et al. 2015a), rabbits (Pankovics et al. 2016), bats (Wu et al. 2016), ferrets (Smits et al. 2013), domestic and wild carnivores (Olarte-Castillo et al. 2015) and cats (Chung et al. 2013). According to the recent report of International Committee on Taxonomy of Viruses (ICTV) in 2017 (https://talk.ictvonline.org/taxonomy/), the genus Kobuvirus was classified into six officially recognized species, namely Aichivirus A (formerly Aichi virus), Aichivirus B (formerly bovine kobuvirus), Aichivirus C (porcine kobuvirus), Aichivirus D (kagovirus 1), Aichivirus E (rabbit kobuvirus) and Aichivirus F (bat kobuvirus), respectively (Adams et al. 2013; Adams et al. 2017). Aichivirus A includes six types: human AiV, CaKoV, murine kobuvirus, roller kobuvirus, Kathmandu sewage kobuvirus and feline kobuvirus (FeKoV).
Feline kobuvirus (FeKoV), a member of the species Aichivirus A, was first identified in feces of cats with diarrhea in South Korea in 2013 (Chung et al. 2013). The genetic analysis based on the partial RdRp gene indicated that FeKoV strains shared higher nucleotide (81.2%–82.1%) and amino acid identities (91.4%–92.1%) with CaKoV strains previously reported (Kapoor et al. 2011). In a study by Cho, et al., it was demonstrated that kobuvirus widely circulated in domestic cats and was associated with viral diarrhea (Cho et al. 2014). Recently, FeKoV infection in cats was reported in Italy (Di Martino et al. 2015b). In 2018, Lu et al. first reported the circulation of FeKoV in diarrhoeic cats in southern China. FeKoV RNA was found in 8 domestic cats with diarrhea, but was undetected in healthy cats (Lu et al. 2018). However, only four complete genomes of FeKoV strains, including FK-13 (Cho et al. 2014), 12D240 (Choi et al. 2015), TE/52/IT/2013 (Di Martino et al. 2015b) and WHJ-1 (Lu et al. 2018), have been sequenced until now. Furthermore, there is no data of kobuvirus infections in cats in northeast China. In this study, we provide the first molecular evidence for the circulation of FeKoV in northeast China, and investigate the prevalent levels, as well as genetic characteristics.
2. Materials and methods
2.1. Sample collection
In total, 197 fresh fecal samples were collected from 105 cats with diarrhea and 97 asymptomatic cats from five different regions in northeast China, including Shenyang, Jinzhou, Changchun, Jilin and Harbin, during January 2016 to November 2017. Individual fresh feces were immediately placed in RNase-free tubes and were stored at −70 °C until further use.
2.2. Nucleic acid extraction
Fecal samples were suspended in phosphate-buffered saline (PBS, pH = 7.4) at a concentration of approximately 0.5 g/ml, and then the suspension was centrifuged at 8000 ×g for 10 min at 4 °C to collect the supernatant. Total RNA was extracted from 250 μl of supernatant using AxyPrep Body Viral RNA Miniprep kit (CORNING, China), and reverse transcribed to synthesize cDNA using the RevertAid first strand cDNA synthesis kit (Invitrogen, USA) according to the manufacturer's instructions. Viral DNA of fecal samples was extracted using Viral DNA extraction kit I (OMEGA, China) according to the manufacturer's instruction.
2.3. Detection of FeKoV and other feline enteric viruses
The detection of kobuvirus was performed by RT-PCR using a pair of universal primer previously described (the primer sequences are shown in Table 1 ), UNIV-kobu-F/UNIV-kobu-R targeting 217-bp of partial 3D gene for all kobuviruses (Reuter et al. 2009). These samples were also examined for other feline enteric viruses, including feline parvovirus (FPV) and feline bocavirus (FBoV), using PCR assays previously described (Takano et al. 2016; Yang et al. 2010). The amplified products were separated after electrophoresis on 1.5% agarose gels at 160 V for 20 min, and were visualized using a gel documentation system (Wealtec, USA).
Table 1.
Sequences of oligonucleotide primers used in this study.
| Targate | Primer name | Sequence (5′-3′) | Product (bp) | Reference | |
|---|---|---|---|---|---|
| Partial 3D gene | UNIV-Kobu-F | TGGAYTACAAGTGTTTTGATGC | 216 | Reuter et al. 2009 | |
| UNIV-Kobu-R | ATGTTGTTRATGATGGTGTTGA | ||||
| FeKoV-3D-F | CTCCGCCCCACCGCTAAGG | 530 | This study | ||
| FeKoV-3D-R | GGGGGTTCCGTTGCGTAGATGA | ||||
| Complete VP1 gene | FeKoV-VP1-F | GCCCCCGCCTCTGCCATTGTG | 843 | This study | |
| FeKoV-VP1-R | CTTGATGACGGCGACGGACTTTTC | ||||
| Complete polyprotein genea | 5′UTR-VP0 | FeKoV-535-F | GTTCGTCCGGCTGTCCTTTGGTAA | 1164 | This study |
| FeKoV-1698-R | AGTAGCRGTGGGGTAGGCRAGRAA | ||||
| VP0-VP3 | FeKoV-1601-F | GCCTGGCYGCYCTCAATCCTTCA | 1127 | ||
| FeKoV-2727-R | GGGGGCAGCGGGGGTGTAGC | ||||
| VP3-2B | FeKoV-2567-F | TCCCCGTCTCCCCCAGTGCYATTG | 1654 | ||
| FeKoV-4220-R | GTGTCGGCGTCGGCGGTCAG | ||||
| 2B-2C | FeKoV-3753-F | AAGTCCGTCGCCGTCATCAAGA | 1330 | ||
| FeKoV-5087-R | GTCCCGGGCGGTCCATAGAAAT | ||||
| 2C-3D | FeKoV-4948-F | AGGATGCAAAATCTGGCCCACACTCTG | 1788 | ||
| FeKoV-6735-R | GGGGGCAGGTTCCTTCTTGATGG | ||||
| 3D-3′UTR | FeKoV-6503-F | TCCCCGCTTGTGACAGATGACC | 1672 | ||
| FeKoV-8155-R | TACAACCATGGCTTAGGGGCTCAC | ||||
The primers' positions are referred to the full-length genome of FeKoV reference strain 12D240 (Genbank accession number: KJ958930).
2.4. Sequencing of partial 3D gene and the complete VP1 gene
To further investigate the genetic diversity of FeKoV detected in the present study, two pairs of primers for the amplification of partial 3D gene and the complete VP1 gene were designed. The primer sequences are shown in Table 1. The PCR conditions were as follows: initial denaturation at 94 °C for 5 min, followed by 35 cycles of 94 °C for 1 min, 62 °C for 45 s and 72 °C for 1 min, and a final extension at 72 °C for 10 min. All PCR products were purified using AxyPrep DNA gel Extraction kit (CORNING, China), and then were cloned into PMD-18 T vector (TAKARA, China). Plasmid DNA was extracted using AxyPrep Plasmid Miniprep kit (CORNING, China), and positive DH-5α clones (three clones per sample) were sent to Sangon Biotech (Shanghai, China) for Sanger sequencing.
2.5. Amplification of the full-length polyprotein gene
Two FeKoV-positive samples were randomly selected to amplify the complete polyprotein gene sequences using six primer sets designed in the present study (primer sequences are shown in Table 1). The reaction conditions were described as follows: pre-denaturation at 94 °C for 5 min followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 60 °C for 1 min, extension at 72 °C for 30 s, and a final extension at 72 °C for 10 min. Purification of PCR products, clone of purified fragment, plasmid extraction and sequencing were performed with the same methods as previously described. The nucleotide sequences were assembled using SeqMan program, and the complete FeKoV polyprotein gene sequences were deposited in GenBank under accession numbers MH159813 (7621 nt) and MH159814 (7618 nt).
2.6. Genetic and phylogenetic analysis
Pairwise alignments among different kobuviruses based on the partial 3D gene and the complete VP1 gene were performed using online BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The nucleotide and amino acid identities among all sequences were calculated using Bioedit. For the obtained genome sequences, the ORF was predicted using ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/), and the potential cleavage sites of polyprotein were predicted using NetPicoRNA 1.0 and were further verified via the nucleotide and amino acid alignments with reference kobuviruses. Similarity plot analysis of the complete polyprotein gene was performed using SimPlot 3.5 software. Phylogenetic tree was constructed using the neighbour-joining method with 1000 bootstrap replicates in MEGA 7.0 software.
3. Results
3.1. Screening of FeKoV in fecal samples
We tested a total of 197 fecal samples from Shenyang (n = 36), Jinzhou (n = 17), Changchun (n = 85), Jilin (n = 33) and Harbin (n = 26) in northeast China. Background data was available for all cats of which 105 were diarrhoeic and 92 were healthy; 71 were collected from private veterinary clinics and 126 were collected from animal shelter centers. The screening results showed that 28 of the 197 samples (14.2%) were positive for kobuvirus. Out of these kobuvirus-positive samples, 8 were positive for FPV, 6 were positive for FBoV and 6 were co-infected with FPV and FBoV. Detailed information about kobuvirus-positive samples was shown in Table 2 . The diarrhoeic cats had a higher kobuvirus prevalence (19.1%, 20/105) than asymptomatic cats (8.7%, 8/92) and the positive rate of cats from animal shelter centers (16.7%, 21/126) were also higher than that of cats from private veterinary clinics (9.9%, 7/71). Moreover, there was no significant difference in the prevalence of samples from different regions (from 11.1% to 16.5%).
Table 2.
Detailed information of FeKoV-positive samples identified in this study.
| Number | ID | Year | Region | Source | Health status | Mixed infection |
|---|---|---|---|---|---|---|
| 1 | 16SY0707 | 2016 | Shengyang | ASC | Diarrhoeic | FBoV |
| 2 | 16SY0720 | 2016 | Shengyang | PVC | Diarrhoeic | FBoV |
| 3 | 16JZ0613 | 2016 | Jinzhou | ASC | Diarrhoeic | FBoV |
| 4 | 16JZ0605 | 2016 | Jinzhou | PVC | Diarrhoeic | FPV |
| 5 | 16CC0802 | 2016 | Changchun | ASC | Diarrhoeic | – |
| 6 | 16CC0805 | 2016 | Changchun | PVC | Diarrhoeic | FBoV |
| 7 | 16CC1104 | 2016 | Changchun | ASC | Diarrhoeic | – |
| 8 | 16CC1105 | 2016 | Changchun | ASC | Healthy | FBoV+FPV |
| 9 | 16JL0802 | 2016 | Jilin | ASC | Healthy | – |
| 10 | 17SY1202 | 2017 | Shengyang | ASC | Healthy | – |
| 11 | 17SY1207 | 2017 | Shengyang | ASC | Healthy | – |
| 12 | 17CC0302 | 2017 | Changchun | ASC | Diarrhoeic | FBoV+FPV |
| 13 | 17CC0305 | 2017 | Changchun | ASC | Healthy | – |
| 14 | 17CC0306 | 2017 | Changchun | ASC | Diarrhoeic | FPV |
| 15 | 17CC0308 | 2017 | Changchun | ASC | Diarrhoeic | FBoV+FPV |
| 16 | 17CC0311 | 2017 | Changchun | ASC | Diarrhoeic | FBoV+FPV |
| 17 | 17CC0702 | 2017 | Changchun | ASC | Healthy | – |
| 18 | 17CC0809 | 2017 | Changchun | PVC | Healthy | FBoV |
| 19 | 17CC0811 | 2017 | Changchun | ASC | Diarrhoeic | FPV |
| 20 | 17CC1101 | 2017 | Changchun | ASC | Diarrhoeic | FPV |
| 21 | 17CC1106 | 2017 | Changchun | ASC | Diarrhoeic | FPV |
| 22 | 17JL0303 | 2017 | Jilin | PVC | Diarrhoeic | FPV |
| 23 | 17JL0308 | 2017 | Jilin | ASC | Diarrhoeic | FPV |
| 24 | 17JL0311 | 2017 | Jilin | ASC | Diarrhoeic | FBoV+FPV |
| 25 | 17JL0312 | 2017 | Jilin | ASC | Diarrhoeic | FBoV+FPV |
| 26 | 17HRB0505 | 2017 | Harbin | PVC | Diarrhoeic | FPV |
| 27 | 17HRB0509 | 2017 | Harbin | PVC | Healthy | – |
| 28 | 17HRB0903 | 2017 | Harbin | ASC | Diarrhoeic | FBoV |
ASC, animal shelter centers; PVC, private veterinary clinics; FBoV, feline bocavirus; FPV, feline parvovirus.
3.2. Phylogenetic analysis of partial 3D gene
Partial 3D genes of 28 kobuvirus-positive samples were sequenced in this study, and the nucleotide sequences were deposited in GenBank under accession numbers MH159777-MH159804. The 28 sequences shared 90.8%–100% nucleotide identities and 97.5%–100% amino acid identities with each other. These sequences had the highest nucleotide (90.5%–97.8%) and amino acid identities (96.6%–100%) with FeKoV reference sequences deposited in GenBank, suggesting that all twenty-eight fecal samples were FeKoV-positive. Furthermore, the 28 sequences were 80.7%–85.8%, 80.2%–82.7% and 80.4%–83.2% similar to CaKoVs, human AiVs and murine kobuvirus at the nucleotide level, respectively.
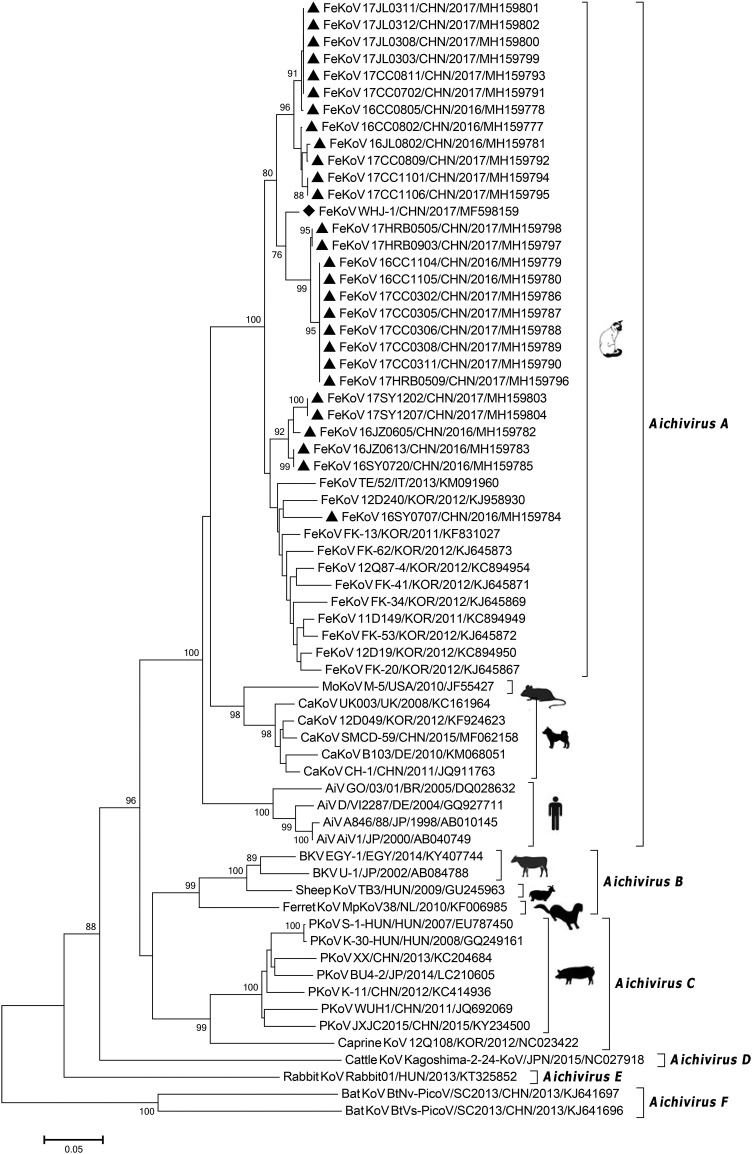
Phylogenetic analysis based on the partial 3D gene showed that the 28 sequences were more closely related to FeKoVs, clustering in the Aichivirus A, which included also human AiVs, CaKoVs and murine kobuvirus. The 28 FeKoV sequences were divided into two major groups: 22 sequences clustered with the Chinese FeKoV strain, WHJ-1 (Lu et al. 2018), and formed a major group, while the other 6 sequences and FeKoV reference strains identified in South Korea and Italy formed another group. Interestingly, only one sequence identified in this study appeared more closely related to the Korean FeKoV strain, 12D240 (Choi et al. 2015), while other sequences were separated from these reference sequences (Fig. 1 ).
Fig. 1.
Phylogenetic analysis based on the nucleotide sequences (363 nt) of partial 3D genes for kobuviruses. The phylogenetic tree was conducted using the neighbour-joining method with 1, 000 bootstrap replicates using MEGA 7.0 software. Black triangles indicate sequences identified in the present study, and black diamond indicates the Chinese FeKoV strain, WHJ-1. The silhouettes of hosts for different kobuviruses were observed on the branches. AiV, Aichi virus; BKV, bovine kobuvirus; CaKoV, canine kobuvirus; FeKoV, feline kobuvirus; MoKoV, murine kobuvirus; PKoV, porcine kobuvirus. BR, Brazil; CHN, China; DE, Germany; EGY, Egypt; HUN, Hungary; IT, Italy; JP, Japan; NL, Nederland; KOR, South Korea; UK, the United Kingdom; USA, the United States.
3.3. Complete VP1 gene sequence analysis
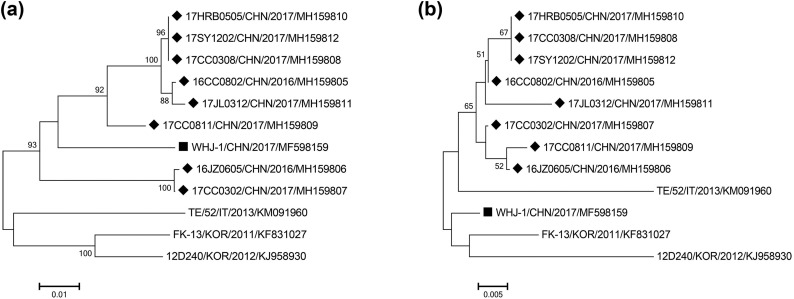
Eight samples were randomly selected from 28 FeKoV-positive samples, and their complete VP1 gene sequences were obtained in the present study (GenBank accession numbers MH159805-MH159812). The eight sequences shared nucleotide and deduced amino acid identities of 92.9%–100% and 98.4%–100% with each other and the highest nucleotide identities of 92.6%–94.5% with the Chinese FeKoV strain, WHJ-1 (Lu et al. 2018), when compared with FeKoV reference strains. The neighbour-joining tree based on the VP1 nucleotide sequences showed that our sequences were more closely related to FeKoV strain WHJ-1 and clustered within a major group, while other FeKoV reference strains identified in South Korea and Italy formed another group (Fig. 2a). Interestingly, the eight sequences and FeKoV strain WHJ-1 placed in different branches in the phylogenetic tree based on the deduced amino acid sequences (Fig. 2b). Then, we analyzed the amino acid mutation sites in the VP1 gene between the eight sequences and other FeKoV sequences previously described, and discovered that three identical amino acid substitutions at amino acid positions 182 (T → A), 235 (P → S) and 241(S → T) were exhibited in all sequences identified in this study (Table 3 ).
Fig. 2.
Phylogenetic trees based on the nucleotide (a) and deduced amino acid (b) sequences of the complete VP1 gene of FeKoV strains using the neighbour-joining method with 1, 000 bootstrap replicates. Black diamond indicate FeKoV sequences identified in the current study, and black squares indicate the Chinese FeKoV strain, WHJ-1. CHN, China; KOR, South Korea; IT, Italy.
Table 3.
Amino acid mutations in the VP1 protein of FeKoV strains identified in this study.
| Strains | Regions | Amino acid substitutions at position |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 22 (N) | 108 (I) | 149 (T) | 182 (T) | 186 (I) | 235 (P) | 238 (T) | 241 (S) | ||
| Reference strains | |||||||||
| FK-13/2011/KF831027 | South Korea | – | – | – | – | – | – | – | – |
| 12D240/2012/KJ958930 | South Korea | – | – | – | – | V | – | – | – |
| TE/52/2013/KM091960 | Italy | Y | – | A | – | V | A | – | T |
| WHJ-1 | China | – | – | – | – | V | – | – | T |
| This study | |||||||||
| 16JZ0605/2016 | China | – | – | – | A | A | S | – | T |
| 16CC0802/2016 | China | – | – | – | A | V | S | – | T |
| 17JL0312/2017 | China | – | T | – | A | V | S | – | T |
| 17CC0302/2017 | China | – | – | – | A | V | S | – | T |
| 17CC0308/2017 | China | – | – | – | A | V | S | I | T |
| 17CC0811/2017 | China | – | T | – | A | A | S | – | T |
| 17SY1202/2017 | China | – | – | – | A | V | S | I | T |
| 17HRB0505/2017 | China | – | – | – | A | V | S | I | T |
The amino acid positions are referred to the complete VP1 gene. Bold text indicates the identical amino acid substitutions for FeKoV sequences that identified in the present study.
3.4. Genomic and phylogenetic analyses of the complete polyprotein gene
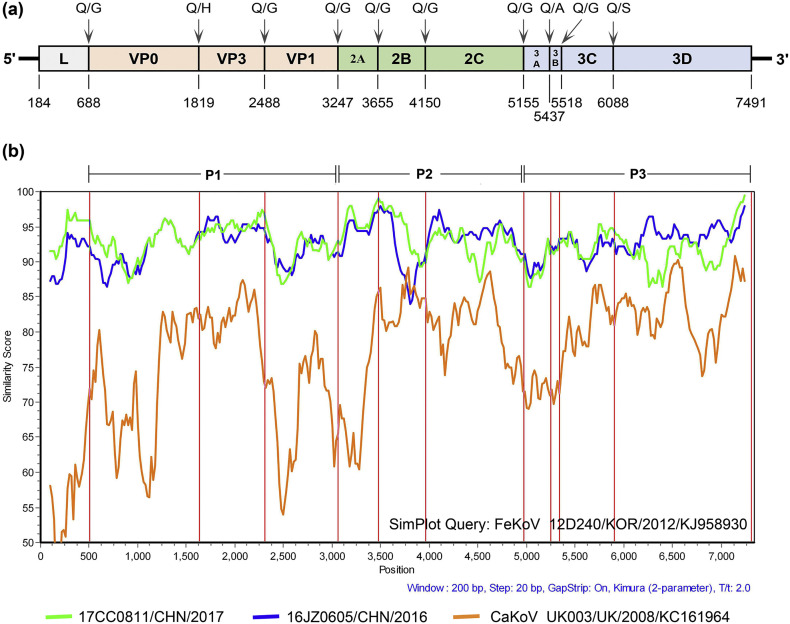
Two complete FeKoV polyprotein genomes were successfully sequenced from two kobuvirus-positive samples in the present study using six pairs of primers, and designated 16JZ0605 and 17CC0811, respectively. The obtained genome sequence of 16JZ0605 was 7621 nt in length and contained one ORF (7311 nt) encoding the polyprotein of 2436 aa, while the 17CC0811 was 7618 nt long. The ORF of 17CC0811 was 7308 nt in length with one-amino-acid deletion in VP0 gene. Moreover, we predicted and verified the genomic organization and potential cleavage sites for the complete polyprotein gene of 16JZ0605 and 17CC0811, which were identical to other FeKoV strains previously described (Fig. 3a). The 16JZ0605 shared 94.8% nucleotide identity and 98.5% amino acid identity with 17CC0811. Compared with other FeKoV reference strains, 16JZ0605 and 17CC0811 had 92.9%–93.4% nucleotide identities and 96.8%–97.9% amino acid identities with the Korean strains, FK-13 and 12D240, and the Italian strain, TE/52/IT/13, and shared a higher sequences homologies with the Chinese FeKoV strain, WHJ-1, at the nucleotide (94.0%–94.9%) and deduced amino acid (98.3%–98.4%) levels. We next compared the similarities of each functional region among 16JZ0605, 17CC0811 and other kobuviruses. For 16JZ0605, the highest nucleotide and amino acid identities were found in the WHJ-1 P2 region, with values of 95.2% and 98.6%, while the highest nucleotide and amino acid divergences were found in the 12D240 L region, with values of 91.3% and 91.5%. 17CC0811 shared amino acid identities of 93.9%–100%, 97.2%–98.0%, 97.3%–99.2% and 97.0%–98.5% with FeKoV reference strains at the L, P1, P2 and P3 regions, respectively. Furthermore, the nucleotide and amino acid homologies between 17CC0811 and WHJ-1 were higher than that between 16JZ0605 and WHJ-1 at each region of the polyprotein gene (Table 4 ). In order to further analyze the genetic characteristics of the complete polyprotein gene, the similarity plot analysis which compared polyprotein nucleotide sequences of 16JZ0605, 17CC0811 and one CaKoV sequence (used as a out-group sequence) to FeKoV reference strain 12D240/KJ958930 (used as a query sequence) was performed in this study. The analytical results showed that 16JZ0605 and 17CC0811 shared similar similarities with reference strain 12D240 in the VP0, VP3, VP1, 2A and from 3A to 3C regions, but different similarities in the L, 2B, 2C and 3D regions. In the L, 2A and 2B regions, 17CC0811 was more similar to reference strain than 16JZ0605, while 17CC0811 presented considerably lower similarities in other regions of polyprotein gene than 16JZ0605. Moreover, higher range of genetic variability was observed in P2 and P3 regions of polyprotein gene (Fig. 3b).
Fig. 3.
Genome organization and genetic characterization of 16JZ0605 and 17CC0811 identified in this study. (a) Graphical depiction of the complete polyprotein genome organization of FeKoV: the predicted cleavage sites (above bar) and nucleotide positions (below bar) are shown and the nucleotide positions are referred to obtained sequence of 16JZ0605. (b) Similarity plot analysis of the complete polyprotein gene of 16JZ0605 (blue line), 17CC0811 (green line), CaKoV strain UK003 as a out-group sequence (orange line) and FeKoV reference strain 12D240 as a query sequence, using a Kimura (2-parameter) model with a sliding window of 200 nt and a moving step size of 20 nt. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 4.
Nucleotide and deduced amino acid sequence identities of the complete polyprotein gene and L, P1-P3 regions between two Chinese FeKoV strains, 16JZ0605 and 17CC0811, and other kobuviruses.
| FeKoV |
CaKoV |
AiV |
MoKoV |
|||||
|---|---|---|---|---|---|---|---|---|
| 17CC0811 |
12D240 |
FK-13 |
TE/52 |
WHJ-1 |
UK003 |
AiV1 |
M-5 |
|
| MH159813 | KJ958930 | KF831027 | KM091960 | MF598159 | KC161964 | AB040749 | JF755427 | |
| 16JZ0605/MH159814 (percentage of nucleotide identity/percentage of amino acid identity) | ||||||||
| Polyprotein | 94.8/98.5 | 93.0/97.0 | 92.9/97.9 | 93.4/97.3 | 94.0/98.3 | 80.6/85.8 | 75.3/78.8 | 77.1/81.5 |
| L | 93.5/97.0 | 91.3/91.5 | 91.9/94.6 | 93.7/96.4 | 93.5/97.0 | 68.5/67.7 | 62.9/56.4 | 64.7/58.1 |
| P1 | 95.9/99.4 | 92.6/97.5 | 92.5/97.9 | 92.8/97.4 | 93.2/98.1 | 79.3/83.5 | 73.9/76.6 | 75.0/79.7 |
| P2 | 94.8/98.4 | 93.7/99.1 | 94.2/98.9 | 94.2/98.1 | 95.2/98.6 | 82.3/87.7 | 76.2/81.4 | 78.7/84.1 |
| P3 | 93.8/97.8 | 93.5/97.9 | 92.7/97.8 | 93.4/96.5 | 94.0/98.1 | 83.2/90.8 | 78.8/83.7 | 80.7/86.4 |
| 17CC0811/MH159813 (percentage of nucleotide identity/percentage of amino acid identity) | ||||||||
| Polyprotein | 100/100 | 93.2/96.8 | 92.9/97.9 | 93.1/97.3 | 94.9/98.4 | 81.0/85.5 | 75.2/78.4 | 76.9/81.3 |
| L | 100/100 | 94.4/93.9 | 93.5/97.6 | 94.2/98.8 | 94.8/100 | 68.5/67.7 | 62.2/55.2 | 63.5/57.5 |
| P1 | 100/100 | 93.0/97.3 | 92.7/97.5 | 92.8/97.2 | 94.4/98.0 | 79.4/83.3 | 74.2/76.7 | 75.2/79.7 |
| P2 | 100/100 | 94.0/98.4 | 94.1/98.6 | 94.1/97.3 | 95.5/99.2 | 83.3/87.3 | 75.7/80.7 | 78.5/83.6 |
| P3 | 100/100 | 92.3/97.6 | 92.0/97.9 | 92.3/97.0 | 95.2/98.5 | 83.6/90.4 | 78.7/83.3 | 80.2/86.3 |
FeKoV, Feline Kobuvirus; CaKoV, Canine kobuvirus; AiV, Aichi Virus; MoKoV, Murine Kobuvirus.
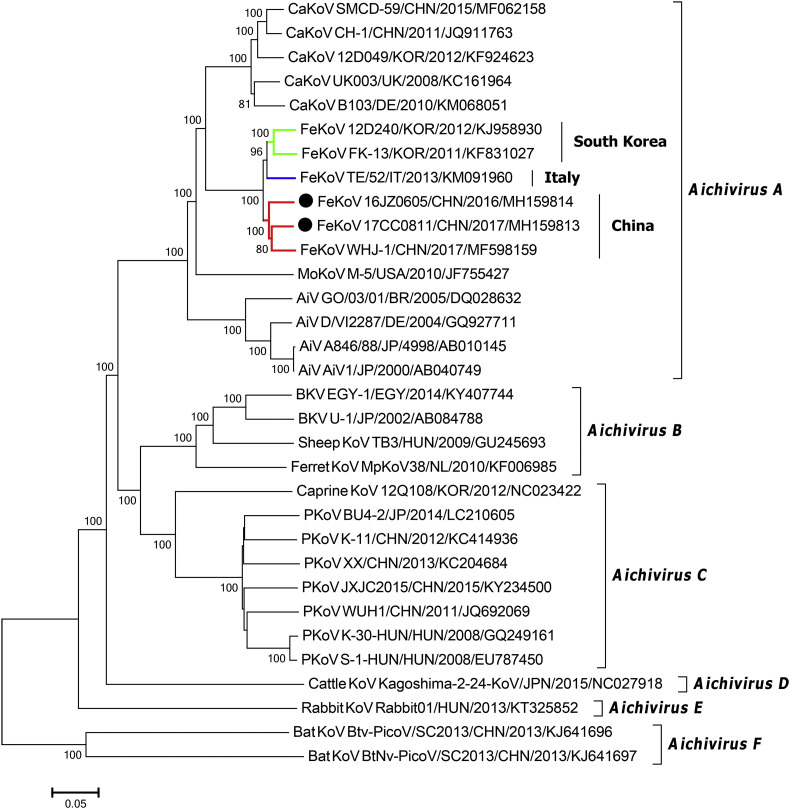
Phylogenetic analysis based on the complete polyprotein nucleotide sequences indicated that 16JZ0605 and 17CC0811 were more closely related to FeKoV reference strains than other kobuvirus strains and these FeKoV strains formed a group distinct from CaKoVs, human AiVs and murine kobuvirus, within the Aichivirus A. In the group of FeKoV, 16JZ0605 and 17CC0811 clustered with the Chinese FeKoV strain, WHJ-1, and formed a tight branch, while other FeKoV strains also formed different branches according to their geographical regions (Fig. 4 ). These results suggested that genetic diversity of FeKoV was presented in different geographical regions, albeit with limited sequences support.
Fig. 4.
Phylogenetic tree of kobuviruses based on the complete polyprotein nucleotide sequences. The tree is generated using the neighbour-joining method with 1000 bootstrap replicates, and only bootstrap values >70% are displayed above the tree branches. Black circles indicate FeKoV strains identified in the present study. All FeKoV strains are divided into different groups according their geographical regions: the Korean strains with green lines, Italian strain with blue line and Chinese strains with red lines. AiV, Aichi virus; BKV, bovine kobuvirus; CaKoV, canine kobuvirus; FeKoV, feline kobuvirus; MoKoV, murine kobuvirus; PKoV, porcine kobuvirus. BR, Brazil; CHN, China; DE, Germany; EGY, Egypt; HUN, Hungary; IT, Italy; JP, Japan; KOR, South Korea; UK, the United Kingdom; USA, the United States. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
In the past few years, the circulations of kobuvirus in cats had been reported in South Korea, Italy and southern China (Chung et al. 2013; Di Martino et al. 2015b; Lu et al. 2018). However, the related data of FeKoV in other countries and regions is lacking. This study presents the first identification and genetic characterization of kobuvirus in cats in northeast China. We investigated 197 fecal samples of which 28 (14.2%) were positive for kobuvirus. The prevalence rate is similar to previous reports in South Korea (15.4%, 6/39), Italy (13.5%, 5/37) and southern China (9.9%, 8/81) (Chung et al. 2013; Di Martino et al. 2015b; Lu et al. 2018), suggesting that kobuvirus widely circulates in domestic cats in northeast China. The prevalence of kobuvirus in diarrhoeic cats (19.1%, 20/105) is significantly higher than that in healthy cats (8.7%, 8/92), similar to a previous study by Cho, et al. (Cho et al. 2014). Moreover, we also tested other enteric viruses (FPV and FBoV) for kobuvirus-positive samples of which 20 samples were positive for FPV and/or FBoV. In previous investigations, the co-infection of FeKoV, FPV and feline enteric coronavirus (FECV) in diarrheal cats with a higher prevalence had been reported (Di Martino et al. 2015b), and it was also determined that human AiVs and other animal kobuvirus were associated with gastroenteritis (Yang et al. 2009; Zhai et al. 2017). These reveal that FeKoV, as a potential enteric virus, may be associated with viral diarrhea in cats.
Phylogenetic analysis based on partial 3D gene indicates that the 28 FeKoV sequences identified in this study clustered into two large groups, majority had a higher nucleotide identity with the Chinese FeKoV strain, WHJ-1 (Lu et al. 2018), and formed a novel group, while only 6 sequences are divided into another group which formed with other FeKoV strains identified in South Korea and Italy (Fig. 1). Moreover, two unique amino acid replacements (amino acid positions 281 and 282 in the complete 3D gene) were present in most sequences (excluding 16JZ0605, 16JZ0613, 16SY0707, 16SY0720, 17SY1202 and 17SY1707) identified in the present study and WHJ-1. These results suggest that a unique evolutionary trend is present in FeKoV strains circulating in China, when compared with FeKoV strains in other countries.
The VP1 protein of picornaviruses is not only an important capsid protein determining the antigenicity and pathogenicity for kobuvirus, but also is the most variable structural protein in the kobuvirus (Reuter et al. 2011). Phylogenetic analyses based on the complete VP1 sequences indicated that FeKoV strains identified in this study clustered together and formed a separate cluster compared to other FeKoV strains, and the identical amino acid mutations were present in the C-terminal of VP1 protein. Additionally, the different amino acid substitutions of the VP1 protein were observed in FeKoV strains identified in different regions (Table 3). These results suggest that the VP1 protein may be used as a considerable indication for the geographical distribution of FeKoVs. Furthermore, a recent study indicated that a polyproline helix structure, as integrin binding motifs, is present at the C-terminal of VP1 protein on the outer surface of human AiV, and predicted this polyproline motif may be associated with signal transduction, antigen recognition, and viral infectivity and pathogenicity (Zhu et al. 2016). Subsequently, the identical proline-rich motif was also reported in CaKoVs (Li et al. 2018). A polyproline fragment is present in amino acid positions 227–243 of the VP1 protein for all FeKoV strains, similar to human AiVs and CaKoVs. Interestingly, one amino acid substitution is observed in this polyproline motif for FeKoV strains identified in this study (one substitution from proline to serine at position 235) and that identified in Italy (one substitution from proline to alanine at position 235). The impact on viral infectivity for this amino acid mutation needs to be further investigated via structure prediction and viral isolation.
The complete polyprotein gene sequences of 16JZ0605 (7311 nt) and 17CC0811 (7308 nt) were successfully obtained in this study. Genomic analysis showed that the polyprotein of the two strains were all cleaved into 11 viral proteins, L, VP0, VP3, VP1, 2A, 2B, 2C, 3A, 3B, 3C and 3D. The predicted cleavage sites were Q/G, Q/H, Q/A and Q/S, in accordance with the Korean FeKoV strains, 12D240 and FK-13, and the Chinese strain, WHJ-1 (Cho et al. 2014; Choi et al. 2015; Lu et al. 2018). One of the important finding is that one-amino-acid deletion in VP0 gene of 17CC0811. In a previous study, thirty-amino-acid deletion was presented in 2B gene of PKoV from healthy piglets, and these deletions were possibly related to the pathogenicity of PKoV (Jin et al. 2015). However, in this study, 16JZ0605 and 17CC0811 were all identified from diarrhoeic cats, the pathogenicity of FeKoV seemed unaffected by this amino-acid deletion. Consequently, more complete polyprotein gene sequencing of Chinese FeKoV strains is needed to determine whether this amino-acid deletion is widely existent in FeKoV strains circulating in China, and further targated research is also needed to demonstrate the effect of this deletion in FeKoV. Phylogenetic analysis based on the complete polyprotein sequences showed that 16JZ0605 and 17CC0811 shared higher nucleotide and amino acid identities with the Chinese strain, WHJ-1, compared to other FeKoV strains, and all FeKoV strains were clustered according to their geographical regions, albeit with limited sequences support (Fig. 4). Moreover, 16JZ0605 and 17CC0811 shared high nucleotide and amino acid identities to each other in the VP1, 3A and 3C regions, but low identities to FeKoV reference strains. Recent research indicates that the 3A protein plays a vital role in hijacking host acyl-CoA-binding domain-containing protein-3 (ACBD3), which provides a site for the replication of kobuviruses (Klima et al. 2017). 3C protein of most picornaviruses is also important for viral replication (Fujita et al. 2007). Therefore, these mutations in VP1, 3A and 3B regions of 16JZ0605 and 17CC0811 may affect viral replication and antigenicity, more targeted researches are needed to further determine. Taken toghter, the considerable genetic diversity is existent in Chinese FeKoV strains.
Furthermore, these FeKoV strains shared high amino acid identity with CaKoVs and human AiVs in the complete polyprotein gene and different functional regions, especially in P3 region. Considering the close genetic relationship of these kobuviruses, and frequent contact among their own host, the cross-species transmission of kobuviruses is worth investigating. In previous studies, several findings had provided considerable evidences for the potential risks of cross-species transmission for FeKoV, including frequent genetic variation in VP1 gene of FeKoV with 1.29 × 10−2 substitutions/site/year substitution rates (Cho et al. 2014), high nucleotide and amino acid identities between FeKoVs and CaKoVs (Chung et al. 2013; Di Martino et al. 2015b; Lu et al. 2018), and the detection of IgG antibodies specific for AiV in cats (Carmona-Vicente et al. 2013). Moreover, it is also important to mention about conducting serological studies to investigate kobuvirus pathogenicity and whether the genetic diversity in FeKoV affects pathogenicity. Thus, periodic genetic and serological investigations of FeKoVs will be helpful for the assessment of cross-species transmission and pathogenicity for FeKoVs.
5. Conclusions
In conclusion, we provide the first molecular evidence for the circulation of feline kobuvirus in cats in northeast China. Our findings indicate that the circulation of FeKoV in domestic cats with diarrhea was more prevalent, suggesting FeKoV infections may be related to viral diarrhea in cats. Phylogenetic analyses based on partial 3D gene and complete VP1 gene indicate that the considerable genetic diversity is exhibited in Chinese FeKoV strains, and novel FeKoV strains with unique evolutionary trends are circulating in China. Moreover, the complete polyprotein genes of FeKoV strains 16JZ0605 and 17CC0811 are successfully sequenced in this study. These findings will help us to understand the epidemics and genetics of kobuvirus in cats in China. Further epidemiological and molecular investigations are also required to demonstrate the distribution, genetic diversity and potential risk of cross-species transmission of feline kobuvirus.
Author contributions
JTN, SSY and HLW conceived and designed the experiments; JTN, SSY, HLW and YBG performed the experiments; JTN, SSY, YLZ and HD analyzed the data; YBG, KW, SZ and GXH contributed reagents/materials/analysis tools; JTN, SSY, HD and GXH drafted the manuscript; JTN, SSY, HD, XW and YLZ revised the manuscript; HD and GXH supervised and approved the message for publication.
Conflict of interest
All authors declare that they have no competing interests.
Acknowledgements
This study was supported by the Research Project of the National Key Research and Development Plan of China (grant no. 2016YFD0501002). This study was also funded by National Key R&D Program of China (grant no. 2017YFD0501703).
References
- Adams M.J., King A.M., Carstens E.B. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2013) Arch. Virol. 2013;158:2023–2030. doi: 10.1007/s00705-013-1688-5. [DOI] [PubMed] [Google Scholar]
- Adams M.J., Lefkowitz E.J., King A.M.Q., Harrach B., Harrison R.L., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Mushegian A.R., Nibert M., Sabanadzovic S., Sanfacon H., Siddell S.G., Simmonds P., Varsani A., Zerbini F.M., Gorbalenya A.E., Davison A.J. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017) Arch. Virol. 2017;162:2505–2538. doi: 10.1007/s00705-017-3358-5. [DOI] [PubMed] [Google Scholar]
- Carmona-Vicente N., Buesa J., Brown P.A., Merga J.Y., Darby A.C., Stavisky J., Sadler L., Gaskell R.M., Dawson S., Radford A.D. Phylogeny and prevalence of kobuviruses in dogs and cats in the UK. Vet. Microbiol. 2013;164:246–252. doi: 10.1016/j.vetmic.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.Y., Lim S.I., Kim Y.K., Song J.Y., Lee J.B., An D.J. Molecular characterization of the full kobuvirus genome in a cat. Genome Announc. 2014;2 doi: 10.1128/genomeA.00420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.W., Lee M.H., Lee K.K., Oem J.K. Genetic characteristics of the complete feline kobuvirus genome. Virus Genes. 2015;50:52–57. doi: 10.1007/s11262-014-1144-y. [DOI] [PubMed] [Google Scholar]
- Chung J.Y., Kim S.H., Kim Y.H., Lee M.H., Lee K.K., Oem J.K. Detection and genetic characterization of feline kobuviruses. Virus Genes. 2013;47:559–562. doi: 10.1007/s11262-013-0953-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino B., Di Profio F., Melegari I., Robetto S., Di Felice E., Orusa R., Marsilio F. Molecular evidence of kobuviruses in free-ranging red foxes (Vulpes vulpes) Arch. Virol. 2014;159:1803–1806. doi: 10.1007/s00705-014-1975-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino B., Di Profio F., Melegari I., Di Felice E., Robetto S., Guidetti C., Orusa R., Martella V., Marsilio F. Molecular detection of kobuviruses in European roe deer (Capreolus capreolus) in Italy. Arch. Virol. 2015;160:2083–2086. doi: 10.1007/s00705-015-2464-5. [DOI] [PubMed] [Google Scholar]
- Di Martino B., Di Profio F., Melegari I., Marsilio F., Martella V. Detection of feline kobuviruses in diarrhoeic cats, Italy. Vet. Microbiol. 2015;176:186–189. doi: 10.1016/j.vetmic.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K., Krishnakumar S.S., Franco D., Paul A.V., London E., Wimmer E. Membrane topography of the hydrophobic anchor sequence of poliovirus 3A and 3AB proteins and the functional effect of 3A/3AB membrane association upon RNA replication. Biochemistry. 2007;46:5185–5199. doi: 10.1021/bi6024758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Zhang W., Xue Y., Shao S. Sequence analysis reveals mosaic genome of Aichi virus. Virol. J. 2011;8:390. doi: 10.1186/1743-422X-8-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W.J., Yang Z., Zhao Z.P., Wang W.Y., Yang J., Qin A.J., Yang H.C. Genetic characterization of porcine kobuvirus variants identified from healthy piglets in China. Infect. Genet. Evol. 2015;35:89–95. doi: 10.1016/j.meegid.2015.07.035. [DOI] [PubMed] [Google Scholar]
- Kapoor A., Simmonds P., Dubovi E.J., Qaisar N., Henriquez J.A., Medina J., Shields S., Lipkin W.I. Characterization of a canine homolog of human Aichivirus. J. Virol. 2011;85:11520–11525. doi: 10.1128/JVI.05317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klima M., Chalupska D., Rozycki B., Humpolickova J., Rezabkova L., Silhan J., Baumlova A., Dubankova A., Boura E. Kobuviral Non-structural 3A proteins act as molecular harnesses to Hijack the host ACBD3 protein. Structure. 2017;25:219–230. doi: 10.1016/j.str.2016.11.021. [DOI] [PubMed] [Google Scholar]
- Lescar J., Canard B. RNA-dependent RNA polymerases from flaviviruses and Picornaviridae. Curr. Opin. Struct. Biol. 2009;19:759–767. doi: 10.1016/j.sbi.2009.10.011. [DOI] [PubMed] [Google Scholar]
- Li M., Yan N., Wang M., Zhang B., Yue H., Tang C. Prevalence and genomic characteristics of canine kobuvirus in southwest China. Arch. Virol. 2018;163:459–466. doi: 10.1007/s00705-017-3648-y. [DOI] [PubMed] [Google Scholar]
- Lu G., Zhang X., Luo J., Sun Y., Xu H., Huang J., Ou J., Li S. First report and genetic characterization of feline kobuvirus in diarrhoeic cats in China. Transbound. Emerg. Dis. 2018;65:1357–1363. doi: 10.1111/tbed.12916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melegari I., Sarchese V., Di Profio F., Robetto S., Carella E., Bermudez Sanchez S., Orusa R., Martella V., Marsilio F., Di Martino B. First molecular identification of kobuviruses in wolves (Canis lupus) in Italy. Arch. Virol. 2018;163:509–513. doi: 10.1007/s00705-017-3637-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oem J.K., Choi J.W., Lee M.H., Lee K.K., Choi K.S. Canine kobuvirus infections in Korean dogs. Arch. Virol. 2014;159:2751–2755. doi: 10.1007/s00705-014-2136-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oem J.K., Lee M.H., Lee K.K., An D.J. Novel Kobuvirus species identified from black goat with diarrhea. Vet. Microbiol. 2014;172:563–567. doi: 10.1016/j.vetmic.2014.06.009. [DOI] [PubMed] [Google Scholar]
- Olarte-Castillo X.A., Heeger F., Mazzoni C.J., Greenwood A.D., Fyumagwa R., Moehlman P.D., Hofer H., East M.L. Molecular characterization of canine kobuvirus in wild carnivores and the domestic dog in Africa. Virology. 2015;477:89–97. doi: 10.1016/j.virol.2015.01.010. [DOI] [PubMed] [Google Scholar]
- Pankovics P., Boros A., Biro H., Horvath K.B., Phan T.G., Delwart E., Reuter G. Novel picornavirus in domestic rabbits (Oryctolagus cuniculus var. domestica) Infect. Genet. Evol. 2016;37:117–122. doi: 10.1016/j.meegid.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T.G., Kapusinszky B., Wang C., Rose R.K., Lipton H.L., Delwart E.L. The fecal viral flora of wild rodents. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G., Boldizsar A., Kiss I., Pankovics P. Candidate new species of Kobuvirus in porcine hosts. Emerg. Infect. Dis. 2008;14:1968–1970. doi: 10.3201/eid1412.080797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G., Boldizsar A., Pankovics P. Complete nucleotide and amino acid sequences and genetic organization of porcine kobuvirus, a member of a new species in the genus Kobuvirus, family Picornaviridae. Arch. Virol. 2009;154:101–108. doi: 10.1007/s00705-008-0288-2. [DOI] [PubMed] [Google Scholar]
- Reuter G., Boros A., Pankovics P., Egyed L. Kobuvirus in domestic sheep, Hungary. Emerg. Infect. Dis. 2010;16:869–870. doi: 10.3201/eid1605.091934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G., Boros A., Pankovics P. Kobuviruses - a comprehensive review. Rev. Med. Virol. 2011;21:32–41. doi: 10.1002/rmv.677. [DOI] [PubMed] [Google Scholar]
- Reuter G., Nemes C., Boros A., Kapusinszky B., Delwart E., Pankovics P. Porcine kobuvirus in wild boars (Sus scrofa) Arch. Virol. 2013;158:281–282. doi: 10.1007/s00705-012-1456-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.L., Raj V.S., Oduber M.D., Schapendonk C.M., Bodewes R., Provacia L., Stittelaar K.J., Osterhaus A.D., Haagmans B.L. Metagenomic analysis of the ferret fecal viral flora. PLoS One. 2013;8 doi: 10.1371/journal.pone.0071595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T., Takadate Y., Doki T., Hohdatsu T. Genetic characterization of feline bocavirus detected in cats in Japan. Arch. Virol. 2016;161:2825–2828. doi: 10.1007/s00705-016-2972-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Yang L., Ren X., He G., Zhang J., Yang J., Qian Z., Dong J., Sun L., Zhu Y., Du J., Yang F., Zhang S., Jin Q. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J. 2016;10:609–620. doi: 10.1038/ismej.2015.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T., Sakae K., Ishihara Y., Isomura S., Utagawa E. Prevalence of newly isolated, cytopathic small round virus (Aichi strain) in Japan. J. Clin. Microbiol. 1993;31:2938–2943. doi: 10.1128/jcm.31.11.2938-2943.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T., Ito M., Kabashima Y., Tsuzuki H., Fujiura A., Sakae K. Isolation and characterization of a new species of kobuvirus associated with cattle. J Gen Virol. 2003;84:3069–3077. doi: 10.1099/vir.0.19266-0. [DOI] [PubMed] [Google Scholar]
- Yang S., Zhang W., Shen Q., Yang Z., Zhu J., Cui L., Hua X. Aichi virus strains in children with gastroenteritis, China. Emerg. Infect. Dis. 2009;15:1703–1705. doi: 10.3201/eid1510.090522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Wang S., Feng H., Zeng L., Xia Z., Zhang R., Zou X., Wang C., Liu Q., Xia X. Isolation and characterization of feline panleukopenia virus from a diarrheic monkey. Vet. Microbiol. 2010;143:155–159. doi: 10.1016/j.vetmic.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell R. Picornaviridae-the ever-growing virus family. Arch. Virol. 2018;163:299–317. doi: 10.1007/s00705-017-3614-8. [DOI] [PubMed] [Google Scholar]
- Zhai S.L., Zhang H., Lin T., Chen S.N., Zhou X., Chen Q.L., Lv D.H., Wen X.H., Zhou X.R., Jia C.L., Wei W.K. A novel porcine kobuvirus emerged in piglets with severe diarrhoea in China. Transbound. Emerg. Dis. 2017;64:1030–1036. doi: 10.1111/tbed.12663. [DOI] [PubMed] [Google Scholar]
- Zhu L., Wang X., Ren J., Kotecha A., Walter T.S., Yuan S., Yamashita T., Tuthill T.J., Fry E.E., Rao Z., Stuart D.I. Structure of human Aichi virus and implications for receptor binding. Nat Microbiol. 2016;1(16150) doi: 10.1038/nmicrobiol.2016.150. [DOI] [PubMed] [Google Scholar]