Abstract
Background
Rapid and specific molecular tests for identification of the recently identified pandemic influenza A/H1N1 2009 virus as well as rapid molecular tests to identify antiviral resistant strains are urgently needed.
Objectives
We have evaluated the performance of two novel reverse transcriptase polymerase chain reactions (RT-PCRs) targeting specifically hemagglutinin and neuraminidase of pandemic influenza A/H1N1 virus in combination with a conserved matrix PCR. In addition, we investigated the performance of a novel discrimination RT-PCR for detection of the H275Y resistance mutation in the neuraminidase gene.
Study design
Clinical performance of both subtype specific RT-PCR assays was evaluated through analysis of 684 throat swaps collected from individuals meeting the WHO case definition for the novel pandemic influenza virus. Analytical performance was analyzed through testing of 10-fold serial dilutions of RNA derived from the first Dutch sequenced and cultured confirmed case of novel pandemic influenza infection. Specificity and discriminative capacities of the H275Y discrimination assay were performed by testing wild type and recombinant H275Y pandemic influenza.
Results
121 throat swaps collected from April 2009 to July 2009 were positive by at least two out of three RT-PCRs, and negative for the seasonal H3/H1 subtype specific RT-PCR assays. 117 of these were tested positive for all three (Ct-values from 15.1 to 36.8). No oseltamivir resistance was detected.
Conclusions
We present a sensitive and specific approach for detection of pandemic influenza A/H1N1 2009 and a rapid RT-PCR assay detecting a primary oseltamivir resistance mutation which can be incorporated easily into clinical virology algorithms.
Abbreviations: RT-PCR(s), reverse transcriptase polymerase chain reaction(s); WHO, World Health Organisation; RNA, ribonucleic acid; HA, hemagglutinin; NA, neuraminidase; H1/3, hemagglutinin type 1 or 3; N1/2, neuraminidase type 1 or 2; NAI(s), neuraminidase inhibitor(s); Ct, cycle threshold; SNP, single nucleotide polymorphisms; LNA, locked-nucleic acid; TCID50, tissue culture infection dose 50; Ml, millilitre; Vp, viral particles; cDNA, complementary DNA; BHQ, black hole quencher; H275Y, histidine to tyrosine substitution at position 275; PDV, phocine distemper virus
Keywords: Pandemic, Influenza, Oseltamivir, Resistance, RT-PCR, H274Y
1. Background
The early identified outbreak of the new variant influenza A/H1N1 virus or ‘Mexican flu’ in April 2009, has recently been declared the new influenza pandemic.1 Many countries around the world have been stockpiling antiviral drugs for the treatment of infected patients and to minimize further spread of the virus during the first pandemic phase, the period when no appropriate vaccine against the pandemic influenza strain is available.2
Rapid detection of the pandemic influenza A/H1N1 variants, distinguishing these strains from circulating epidemic strains and identification of antiviral resistant variants is important for pandemic surveillance and patient management. Currently, the pandemic influenza variant is naturally resistant to amantadine and rimantidine, but is still susceptible to the neuraminidase inhibitors (NAIs) oseltamivir (Tamiflu) and zanamivir (Relenza).3
Since the beginning of the seasonal influenza epidemic of 2007–2008, resistance of influenza A/H1N1 viruses to the antiviral drug oseltamivir has been a cause of concern. Of the circulating viruses that year, 16% were found to be oseltamivir resistant and early data from the southern hemisphere suggested up to 100% oseltamivir resistance for the 2008–2009 epidemic.4 All of these viruses were found to be resistant by a histidine to tyrosine substitution in the viral neuraminidase at position 275 (residue 274 in the N2 gene). The same resistance mutation was found in viruses isolated from influenza A/H5N1 infected patients treated with oseltamivir.5 As a consequence of massive use of NAIs during the pandemic, appearances of influenza viruses resistant to these drugs are to be expected.6, 7
The gold standards for antiviral resistance screening are phenotypic resistance assays,8 but these are time consuming and would include viral culturing of a pandemic strain. To date, NAIs resistance patterns of the pandemic influenza A/H1N1 virus are unknown. However, potential resistance patterns may occur in a similar way, since a structural comparison between the neuraminidase of the epidemic and pandemic influenza A/H1N1 show great similarities between the active sites of both enzymes.3
A single nucleotide mutation (cytosine to thymine) at position 823 of the pandemic neuraminidase gene can result in a histidine to tyrosine substitution at position 275. Pyrosequencing,9 a method to detect single nucleotide polymorphisms (SNP), has become a well accepted method for sensitive screening of SNP resistant mutations as such. However, as a result of extensive washing procedures with reverse transcriptase polymerase chain reaction (RT-PCR) generated amplicons this technique may lead to false positive results. RT-PCR can be used for SNP analysis as well, by designing primers and probes that discrimination between wild type and mutant sequences. Excellent discrimination properties are achieved by incorporation of locked-nucleic acid (LNA) bases in these probes that increase melting temperatures, thus allowing the probe to be shorter with increased discriminative capacities.10
2. Objectives
We have developed and evaluated the performance of a novel RT-PCR assay targeting both the hemagglutinin and neuraminidase genes of the pandemic influenza A/H1N1 virus and a discrimination assay for detection of the H275Y oseltamivir resistance mutation in neuraminidase.
3. Study design
3.1. Laboratory strains
Laboratory cultured influenza H1N1 virus A/NL/602/2009(v), propagated on Madin–Darby canine kidney (MDCK) cells to a titer of 6.31 × 107 TCID50/ml, was 10-fold serial diluted for positive control samples. For conversion of RT-PCR threshold cycle (Ct) values into a semi-quantitative viral particles count, dilutions of an electron microscopic counted influenza virus A/PR/8/34 stock (Advanced biosciences, Columbia, USA) were run in parallel. To obtain a pandemic influenza A/H1N1 neuraminidase target with a H275Y substitution in the neuraminidase, the influenza virus A/NL/602/2009 (Genbank accession number CY039528 for neuraminidase gene) was rescued as described previously.11 In the neuraminidase gene of this clone a cytosine to thymine mutation was introduced at position 823 by site-directed mutagenesis (Quick-change, Stratagene, The Netherlands) and primer c823t_sense 5′-tcagtcgaaatgaatgcccctaattattactatgaggaatgc-3′.
3.2. Specimens
For the validation of both subtype specific RT-PCR assays, a total of 684 samples were used that were collected from suspected pandemic influenza infected individuals, according to the WHO case definition of a suspected novel pandemic influenza cases.12 All samples were tested for influenza A with a slightly modified version of a previously described RT-PCR targeting a conserved region in the influenza matrix gene.13, 14 The neuraminidase genes of clinical isolates with viral loads higher than 1.0 × 103 viral particles per milliliter (vp/ml) were sequenced by conventional Sanger sequencing. Cross-reactivity of the RT-PCR assays were checked by analyzing 53 throat swaps that were tested positive for seasonal influenza A/H1N1 (35) or A/H3N2 (18) virus (season 2008–2009), 87 supernatants of cultured seasonal influenza A/H1N1 virus (season 2007–2008) and 19 respiratory samples tested negative for influenza A virus but positive for rhino- (13), adeno- (3) or coronavirus 229E (3). The samples were collected in The Netherlands, Norway, France, Poland and in the United States.
3.3. Assay design
To design primers and probes for the pandemic influenza A/H1N1 RT-PCR assays, reference sequences of the hemagglutinin (HA) and neuraminidase (NA) that were deposited in the NCBI (National Center of Biotechnology Information: accessed May 2009) were aligned by clustalW. Primer Express 3.0 software (Applied Biosystems, Nieuwerkerk a/d Ijssel, The Netherlands) was used to design primers and probes. The primers and probes for the H275Y discrimination assay were designed around nucleotide position 823 of the neuraminidase gene. Five LNA bases were incorporated in the probes allowing them to be shorter with increased discriminative capacities. The primers and probes are listed in Table 1 (Eurogentec, Maastricht, The Netherlands).
Table 1.
Primers and probes used for the RT-PCR assays.
| Primers and probes | Sequence (5′–3′) | Label (5′-/-3′) | Position | References |
|---|---|---|---|---|
| Influenza A matrix RT-PCR | ||||
| InfA-sense-TM | aagaccaatcctgtcacctctga | 144–166 | Ward et al.13 | |
| InfA-antisense-TM | caaagcgtctacgctgcagtcc | 217–238 | Ward et al.13 | |
| InfA-probe-2 | tttgtgttcacgctcaccgtgcc | 6-FAM/BHQ-1 | 184–206 | Munster et al.14 |
| Pandemic influenza A/H1N1-H1 and A/H1N1-N1 2009 duplex RT-PCR | ||||
| panH1-forward | ggaaagaaatgctggatctggta | 822–844 | This study | |
| panH1-reverse | atgggaggctggtgtttatagc | 904–926 | This study | |
| panH1-probe | tgcaatacaacttgtcagacacccaaggg | Dragonfly/BHQ-2 | 874–902 | This study |
| panN1-sense | acatgtgtgtgcagggataactg | 865–887 | This study | |
| panN1-antisense | tccgaaaatcccactgcatat | 949–969 | This study | |
| panN1-probe | atcgaccgtgggtgtctttcaacca | 6-FAM/BHQ-1 | 899–923 | This study |
| Pandemic influenza A/H1N1-N1 H274Y | ||||
| panN1-H275-sense | cagtcgaaatgaatgcccctaa | 797–818 | This study | |
| panN1-H275-antisense | tgcacacacatgtgatttcactag | 854–877 | This study | |
| panN1-275H-probe | ttaTCActAtgAggaatga | 6-FAM/BHQ-1 | 819–837 | This study |
| panN1-275Y-probe | ttaTTActAtgAggaatga | Dragonfly/BHQ-2 | 819–837 | This study |
LNA nucleotides are denoted in upper case, DNA nucleotides are denoted in lower case and LNA nucleotide complementary to the predicted single nucleotide polymorphism (SNP) is underlined.
3.4. H275Y discrimination RT-PCR
For the evaluation of the primers and probes for the H275Y discrimination assay, two plasmids pJET1.2-NA-H275H and pJET1.2-NA-H275Y were constructed containing neuraminidase regions 797–836 of influenza virus strain A/NL/602/2009(v). At position 823 of pJET1.2-NA-H275Y, the cytosine was replaced by a thymine, resulting in the mutant sequence. Run-off RNA transcripts were produced by XbaI (New England Biolabs, Westburg, Leusden, The Netherlands) linearization of the constructs and the T7 Ribomax express large scale RNA production system (Promega, Leiden, The Netherlands) according to the manufactures protocol.
3.5. Protocol
Nucleic acids were extracted from 350 μl of respiratory specimen. Nucleic acids were eluted to a volume of 110 μl using the pathogen complex 200 protocol and the QiaSymphony machine (Qiagen, Venlo, The Netherlands) according to the instructions of the manufacturer. To monitor the whole process from isolation of nucleic acids up to RT-PCR, a universal internal control consisting of a seal distemper virus (PDV) preparation, was added to the original clinical sample.15 The rtTH-based EZ RT-PCR kit (Applied Biosystems, Nieuwerkerk a/d Ijssel, The Netherlands) was used for all RT-PCR assays, but with different cycle conditions. For the influenza A matrix, pandemic H1 and N1 RT-PCRs the following amplification conditions were used: UNG reaction at 50 °C for 2 min, cDNA reaction at 60 °C for 30 min and PCR amplification at 95 °C for 5 min, followed by 45 cycles at 95 °C for 20 s, 62 °C for 1 min. The matrix RT-PCR included 40 pmol forward, 30 pmol reverse primer and 5 pmol of the probe. For the pandemic H1 and N1 RT-PCR, 30 pmol of the primers and 10 pmol probes was used. The H275Y discrimination assay compromises 20 pmol for forward and reverse primer and 5 pmol for both probes and the thermal cycling profile was similar except for the elongation temperature which was 60 °C instead of 62 °C. All reactions were done in a total reaction volume of 50 μl, including 20 μl of nucleic acid. Amplification and detection was performed on a LightCycler 480 instrument using the FAM (465–510 nm) and Dragonfly (533–580 nm) filter (Roche, Almere, The Netherlands).
4. Results
4.1. Assay performances of subtype specific RT-PCR assay
4.1.1. Analytical sensitivity
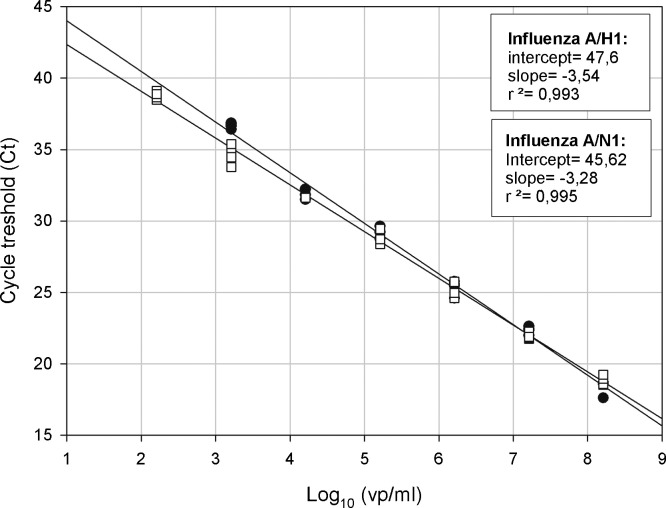
Serial 10-fold dilutions of RNA from the pandemic influenza A/NL/602/2009 strain in RNAse-free water were used for determination of the linear range of the RT-PCR assays targeting hemagglutinin and neuraminidase genes. The approximated sensitivity of RT-PCR assays was determined to be 1.6 × 103 viral particles per milliliter (vp/ml) for the H1 (4/4 reactions detected) and 1.6 × 102 vp/ml for the N1 (4/4 reactions detected). Both assays were linear over a wide and clinical significant range with slopes of −3.54 (R 2 = 0.99) and −3.28 (R 2 = 0.99) for the H1 and N1 RT-PCR, respectively (see Fig. 1 ).
Fig. 1.
Linearity of the H1 and N1 subtype specific RT-PCRs for detection of pandemic influenza A/H1N1 (2009). 10-Fold serial dilutions of target RNA obtained from A/NL/602/2009(v) were analyzed by both sub type RT-PCRs in fourplex and plotted as Ct value versus the log of the calculated viral particles per milliliter in each dilution.
4.1.2. Clinical sensitivity
Analysis of 684 throat swaps collected from individuals suspected for infection with pandemic influenza revealed 121 pandemic influenza samples tested positive by at least two out of three RT-PCR assays (see Table 2 ). 14% of these samples were follow-up samples. In total, 117 samples were tested positive by the influenza A matrix (Ct-values from 15.1 to 36.8) and both sub-typing RT-PCR assays (96.7%). Samples that were not detected by all RT-PCRs all had high Ct-values, indicating low viral loads in these samples.
Table 2.
Performances of the influenza A matrix, pandemic influenza A/H1 and A/N1 RT-PCR assay.
| RT-PCR target(s) | Positive | Mean (Ct) | Range (Ct) |
|---|---|---|---|
| Scores when influenza A matrix positive | |||
| H1 and N1 | 117 | 27.1 | 15.1–36.8 |
| H1 (N1 negative) | 1 | 37.3 | |
| N1 (H1 negative) | 1 | 37.7 | |
| Scores when influenza A matrix negative | |||
| H1 and N1 | 2 | 37.5 | 34.0–37.4 |
| N1 (H1 negative) | 7 | 37.1 | 34.5–39.0 |
| H1 (N1 negative) | 5 | 35.9 | 34.6–37.0 |
Ct-values from the influenza A matrix RT-PCR were taken when influenza A was positive. Samples were determined positive for novel pandemic influenza A/H1N1 2009 virus when either all three or two out of three RT-PCRs were positive.
4.1.3. Specificity
A total of 159 pandemic virus negative samples were tested for cross-reactivity. These samples were tested positive for seasonal influenza A virus H1N1 (122), H3N2 (18) or were confirmed to contain at least one other respiratory virus (19).
No cross-reactivity was observed when testing these samples.
4.2. Performance of pandemic influenza H275Y discrimination RT-PCR
4.2.1. Sensitivity
The assay sensitivity was tested using both RNA obtained from influenza A/NL/602/2009 virus as well as the H275Y recombinant. For both targets, the cut-off value was determined to be approximately 5.0 × 103 vp/ml (Ct ∼ 34).
4.2.2. Discrimination capacity
Serial 10-fold dilutions of in vitro transcribed RNA containing wild type or mutant sequence were analyzed using the H275Y discrimination RT-PCR assay. Two differently labeled LNA probes are used in this multiplex assay which allows data acquisition in both FAM (465–510 nm) and Dragonfly (533–580 nm) filter. Only when wild type RNA was present fluorescence was detected in the FAM-filter and, vice versa, when mutant RNA was present only fluorescence emission was detected in the Dragonfly filter. No wild type or mutant fluorescence was emitted in the negative controls and the samples that were used to check for cross-reactivity. In mixtures of wild type H275 and mutant H275Y RNA, up to 5% of mutant virus could be detected in a reaction with a total input of 1.0 × 105 vp/ml. In addition, when analyzing 61 clinical samples from oseltamivir naïve pandemic influenza A/H1N1 infected patients, only the wild type genotype was found (see Fig. 2 ).
Fig. 2.
H275Y endpoint fluorescence scatter plot. Mixtures of in vitro transcribed wild type and mutant RNA (total input 1.0 × 105 vp/ml) were analyzed using the H275Y discrimination assay (black dots). Relative H275 wild type (465–533 nm) and 275Y mutant (533–580 nm) fluorescence emissions were plotted on the x-axis and y-axis, respectively. In addition, 61 clinical isolates from naïve pandemic influenza A/H1N1 (2009) infected patients were analyzed (white squares) to determine a threshold for detecting mutant genotypes in mixed virus populations (5%).
5. Discussion
Within the first months of the novel pandemic influenza outbreak antiviral drugs are widely used for the treatment of infected patients and to temper viral transmission. Currently, no vaccines are available and antiviral drugs are the only line of defense. Recently, a pandemic influenza sequence was uploaded to the influenza sequence database that encoded for a H275Y variant (Genbank accession no. GQ365445). It is still unclear whether this virus is oseltamivir resistant and can be transmitted, but since the global spread of oseltamivir resistance in the seasonal influenza A/H1N1 virus population, focus on this mutation is needed. We have evaluated a novel RT-PCR assay for sub-typing the pandemic influenza A/H1N1 virus. Both RT-PCRs are sensitive, specific and can be incorporated directly into existing influenza detection and sub-typing algorithms. For validation of the H275Y discrimination assay, virus isolates were chosen that were derived from pandemic influenza A/H1N1 2009 infected patients before oseltamivir treatment. These patients respond well to oseltamivir treatment in general,16 therefore we assume that no H275Y minor variants were present in these samples. Taken these samples as a baseline, up to 5% for the H275Y oseltamivir resistant mutants could be detected in mixed virus populations using the H275Y discrimination assay (see Fig. 2). A drawback of genotypic screening for antiviral resistance mutations by RT-PCR is the potential appearances of unwanted mismatches in the primer and probe regions, due to antigenic drift. For diagnostic purposes, the discrimination RT-PCR assays should therefore be performed in parallel with influenza RT-PCR assays targeting more conserved regions of the influenza viral genome.
In conclusion, the sub-typing RT-PCR assays described appear sensitive and specific in this validation pilot. Using the H275Y discrimination RT-PCR, pandemic influenza A/H1N1 viruses can be screened for oseltamivir susceptibility within 4–6 h time, which makes it useful for surveillance and valuable in clinical patient management.
Conflict of interest
None declared.
Acknowledgment
Part of this study was funded by Hoffmann-La Roche Inc.
References
- 1.WHO. World now at the start of 2009 influenza pandemic, June 11, 2009 http://www.who.int/mediacentre/news/statements/2009/h1n1_pandemic_phase6_20090611/en/index.html; 2009.
- 2.Lackenby A., Thompson C.I., Democratis J. The potential impact of neuraminidase inhibitor resistant influenza. Curr Opin Infect Dis. 2008;21(December (6)):626–638. doi: 10.1097/QCO.0b013e3283199797. [DOI] [PubMed] [Google Scholar]
- 3.Rungrotmongkol T., Intharathep P., Malaisree M., Nunthaboot N., Kaiyawet N., Sompornpisut P. Susceptibility of antiviral drugs against 2009 influenza A (H1N1) virus. Biochem Biophys Res Commun. 2009;385(July (3)):390–394. doi: 10.1016/j.bbrc.2009.05.066. [DOI] [PubMed] [Google Scholar]
- 4.Hurt A.C., Ernest J., Deng Y.M., Iannello P., Besselaar T.G., Birch C. Emergence and spread of oseltamivir-resistant A(H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antiviral Res. 2009;83(July (1)):90–93. doi: 10.1016/j.antiviral.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 5.de Jong M.D., Tran T.T., Truong H.K., Vo M.H., Smith G.J., Nguyen V.C. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N Engl J Med. 2005;353(December (25)):2667–2672. doi: 10.1056/NEJMoa054512. [DOI] [PubMed] [Google Scholar]
- 6.Singer A.C., Nunn M.A., Gould E.A., Johnson A.C. Potential risks associated with the proposed widespread use of Tamiflu. Environ Health Perspect. 2007;115(January (1)):102–106. doi: 10.1289/ehp.9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poland G.A., Jacobson R.M., Ovsyannikova I.G. Influenza virus resistance to antiviral agents: a plea for rational use. Clin Infect Dis. 2009;48(May (9)):1254–1256. doi: 10.1086/598989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zambon M., Hayden F.G. Position statement: global neuraminidase inhibitor susceptibility network. Antiviral Res. 2001;49(March (3)):147–156. doi: 10.1016/s0166-3542(01)00124-3. [DOI] [PubMed] [Google Scholar]
- 9.Bright R.A., Shay D.K., Shu B., Cox N.J., Klimov A.I. Adamantane resistance among influenza A viruses isolated early during the 2005–2006 influenza season in the United States. JAMA. 2006;295(February (8)):891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- 10.You Y., Moreira B.G., Behlke M.A., Owczarzy R. Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res. 2006;34(8):e60. doi: 10.1093/nar/gkl175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Wit E., Spronken M.I.J., Vervaet G., Rimmelzwaan G.F., Osterhaus A., Fouchier R.A.M. A reverse-genetics system for influenza A virus using T7 RNA polymerase. J Gen Virol. 2007;88(April):1281–1287. doi: 10.1099/vir.0.82452-0. [DOI] [PubMed] [Google Scholar]
- 12.Hahné S., Donker T., Meijer A., Timen A., van Steenbergen J., Osterhaus A. Epidemiology and control of influenza A(H1N1)v in the Netherlands: the first 115 cases. Euro Surveillance. 2009;14(July (27)) doi: 10.2807/ese.14.27.19267-en. pii=19267. [DOI] [PubMed] [Google Scholar]
- 13.Ward C.L., Dempsey M.H., Ring C.J., Kempson R.E., Zhang L., Gor D. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J Clin Virol. 2004;29(March (3)):179–188. doi: 10.1016/S1386-6532(03)00122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munster V.J., Baas C., Lexmond P., Bestebroer T.M., Guldemeester J., Beyer W.E.P. Practical considerations for high-throughput influenza A virus surveillance studies of wild birds by use of molecular diagnostic tests. J Clin Microbiol. 2009;47(March (3)):666–673. doi: 10.1128/JCM.01625-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Doornum G.J.J., Schutten M., Voermans J., Guldemeester G.J.J., Niesters H.G.M. Development and implementation of real-time nucleic acid amplification for the detection of enterovirus infections in comparison to rapid culture of various clinical specimens. J Med Virol. 2007;79(October (12)):1868–1876. doi: 10.1002/jmv.21031. [DOI] [PubMed] [Google Scholar]
- 16.Koopmans MP, van Gageldonk M, Hahné S, Isken L, de Jong MD, Jonges M, et al. Clinical findings, viral loads, response to therapy, and resistance monitoring in patients with new influenza A(H1N1)v; submitted for publication.