Highlights
► We described requirements for global animal disease surveillance. ► We explored the use of tools for spatio-temporal analysis on animal disease surveillance. ► Highly pathogenic avian influenza in Denmark and Sweden was used as an example.
Keywords: Global animal disease surveillance, Disease BioPortal, Avian influenza
Abstract
Development and implementation of global animal disease surveillance has been limited by the lack of information systems that enable near real-time data capturing, sharing, analysis, and related decision- and policy-making. The objective of this paper is to describe requirements for global animal disease surveillance, including design and functionality of tools and methods for visualization and analysis of animal disease data. The paper also explores the potential application of techniques for spatial and spatio-temporal analysis on global animal disease surveillance, including for example, landscape genetics, social network analysis, and Bayesian modeling. Finally, highly pathogenic avian influenza data from Denmark and Sweden are used to illustrate the potential application of a novel system (Disease BioPortal) for data sharing, visualization, and analysis for regional and global surveillance efforts.
1. Introduction
Animal disease surveillance may be defined as an active, ongoing, formal, and systemic process of collection, collation, and interpretation of information, aimed at early detection of a specific disease or agent in a population or early prediction of an elevated risk, with a pre-specified action that follows the detection (Meah and Lewis, 1999, Doherr and Audige, 2001, Thurmond, 2003). The World Organization for Animal Health (OIE) has defined animal disease surveillance as “the process of demonstrating the absence of disease or infection, determining the occurrence or distribution of disease or infection, while also detecting as early as possible exotic or emerging diseases” (OIE, 2010a).
Animal disease surveillance is one of the most important components of an effective and efficient program for disease prevention and control. For that reason, veterinary services dedicate many of their available financial and human resources to surveillance planning and implementation (Doherr and Audige, 2001, Thurmond, 2003). Animal disease surveillance has been traditionally been implemented at sub-national and national scales. The need for establishing surveillance systems at continental and global scales has become evident over the last few decades as a consequence of major animal disease epidemics caused by the transboundary spread of infectious agencies. Some of the best documented examples include highly pathogenic avian influenza (HPAI), classical and African swine fever, foot-and-mouth disease (FMD), bluetongue, Rift Valley fever (RVF), and West Nile fever (OIE, 2010b). In response to this need, international agencies and research groups have developed information systems to facilitate data collection and sharing at a global scale. Examples of information systems developed by international agencies include the OIE’s WAHIS (http://www.oie.int/wahis/public.php?page=weekly_report_index&admin=0) and the FAO’s EMPRES-i (http://www.empres-i.fao.org/empres-i/home). The OIE’s WAHIS includes official information submitted by OIE member countries in the form of (a) immediate and follow-up notifications on exceptional disease events, or (b) semester and annual reports on the situation of OIE-listed diseases and background information on animal health and control programs. The FAO’s EMPRES-i includes official and unofficial reports on disease events collected from different sources, such as country or regional project reports, field mission reports, partner and cooperating institutions and organizations, national governments, the FAO or other United Nations agency’s local and regional representations, public domains, the media, and web-based health surveillance systems. WAHIS and EMPRES-i represent a step forward in the implementation of surveillance systems at the global scale; however, there are critical components required for global surveillance that they are still missing.
A global surveillance system may be regarded as a diagnostic test applied at the global scale for the detection of particular agents or diseases. As such, it’s ability to determine infection status or risk for a population may be evaluated, for example in terms of its efficiency, accuracy, precision, rapidity and cost-effectiveness (Thurmond, 2003). Information systems implemented by international organizations are exclusively based on the voluntary reporting of cases of disease by member countries. Therefore, it is reasonable to assume that the specificity of those reporting systems is high i.e. it is unlikely or rare for countries to falsely report cases of disease. Conversely, the sensitivity of international disease reporting systems is often unknown (specific metrics of the probability of detection of disease cases are typically unavailable) or probably low in many regions of the world, and that sensitivity likely varies in time and space.
The purpose of this paper is to describe the desirable features and attributes of a global animal disease surveillance system, the requirements that are necessary to develop such a system, and the potential contribution that spatial analytical techniques, methods and tools may have in the development of a global surveillance system. A prototype web-based system (the Disease BioPortal) is also described, using the retrospective analysis of surveillance data for the detection and spread of avian influenza in Denmark and Sweden in 2006 as an example of its application.
2. Limitations and requirements of global animal disease surveillance systems
The ability of a system to manage and organize disparate streams of information is a critical component of a global surveillance system. National and international organizations, private veterinary practices, abattoirs, poultry slaughterhouses, veterinary hospital’s registries, pharmaceutical and agricultural commercial institutes, zoological gardens, agricultural organizations, commercial livestock enterprises, non-veterinary government departments, farm records, veterinary schools, feral animal organizations, research and reference laboratories, pet-food manufacturers, certification schemes, pet shops, breeding societies, serum banks sentinel units, field observations, human surveillance data, and gene banks are potential sources of data and information in a surveillance system (Doherr and Audige, 2001, Thrusfield, 2007, OIE, 2010a, OIE, 2010b). Data structure, language, semantics and relations differ among those potential sources of information. These need to be combined and organized into one single system in order to build a global surveillance system. The most important component of a surveillance system is the data. The successful process of early detection or risk prediction of an infectious agent or disease in the population largely depends on the quality of collected data, as well as methods of collection, sharing, and communication (Thurmond, 2003). Unfortunately, data reported and collected in many information systems are rarely verified and they are susceptible to multiple, often unknown, biases.
There have been several initiatives to improve the data collection and organization process. For example, hierarchical coding systems allow data of varying degrees of refinement or granularity to be recorded, therefore assisting in the recording of disease profiles according to the available diagnostic detail (Stark et al., 1996, Thrusfield, 2007). The human medical community has initiated major programs that recognized the shortcomings in their disease surveillance systems with regards to the use of standard terminologies, thus the consolidated health informatics initiatives seeks to standardize human medical terminology (Case, 2005). However, standardization of terminology may be difficult to achieve because laboratories, agencies, and organizations often have different objectives for their information systems, organizational structure, and type and quality of data collected. Alternatively, adaptors (grossly defined as IT applications for converting attributes of a given system into attributes that are compatible with those from another system) may be used to relate disparate databases through the identification and relation of fields that contain equivalent information in each database.
Data are the most important component of a surveillance system. The establishment of global surveillance systems has been greatly impaired by the quality and quality of the collected data. Many factors have contributed to this situation, including limited willingness of some research groups, agencies and laboratories to share disease data freely and transparently. For example, sequencing information is typically made available by laboratories only after publication of results in scientific journals, a process which can typically take several years since the initial collection of the samples. Issues related to data confidentiality have also limited the development of surveillance systems (Thrusfield, 2007). Some countries are simply not interested in reporting animal disease cases to the international community because they do not perceive any benefit. Moreover, some countries may perceive disease reporting to have a negative outcome because sanctions (such as trade restrictions) are typically imposed on countries that report the occurrence of particular diseases. Finally, lack of resources in certain regions of the world limits the development of global surveillance systems. Limited resources may affect, for example, the ability to collect data in the field, the quality of laboratory diagnosis, and the ability of veterinary services to prevent and control disease spread.
Issues related to data control and data quality represents, in many cases, a critical limitation to the development of surveillance systems at the global scale. However, development of the IT components of a surveillance system, has progressed rapidly over recent decades. Web-based formal and informal information systems have become popular and accessible to the general public. They provide valuable information for early detection of infectious disease outbreaks, especially in areas invisible to the global public efforts (Woodall, 1997, Sturtevant et al., 2007). For example, during the epidemic of severe acute respiratory syndrome (SARS) in Guangdong Province, China, a number of public and private initiatives aggregated unstructured data from internet-based discussion forums, news outlets and blogs. This helped in the early dissemination of data and information (Grein et al., 2000, Sturtevant et al., 2007). Increased uptake of unstructured internet data, by international organizations, such as the OIE and the World Health Organization (WHO), strongly suggests that the public has an increasingly important role in global disease surveillance (Keystone et al., 2001, Sturtevant et al., 2007). However, it is still recognized that developing countries have a limited ability to participate in internet disease reporting due to the lack of financial resources, education, and limited internet access (Sturtevant et al., 2007). This has an adverse impact on the efficiency of new introduced global surveillance systems that depend largely on internet-based technologies. Moreover, there have not been any international initiatives that focus on the verification and maintenance of the quality of shared disease data derived from internet-based reporting.
A detailed description of the attributes and features of surveillance systems is available elsewhere (Thurmond, 2003). Briefly, in terms of evaluation of the attributes of a surveillance system, novel analytical methods have been recently developed to evaluate the sensitivity of the system. For example, some have proposed the application of models that use disparate data sources to describe each component of a surveillance system and to estimate the sensitivity of the composite surveillance system to support claims of freedom from disease (Martin et al., 2007a, Martin et al., 2007b). However, those methods are applicable to limited geographical areas and diseases for which eradication has been achieved, as documented through accumulated surveillance data (Alban et al., 2008).
3. Application of spatial analysis to animal disease surveillance
It is expected that over the coming decades, the repertoire of analytical techniques available for data analysis at a global scale will increase. Such growth will likely drive countries and organizations to improve the quality of data collection and sharing. Here we review the potential application of methodological approaches in the field of veterinary epidemiology, namely disease mapping, social network analysis (SNA), Bayesian techniques, and landscape genetics and phylogenetic analysis.
3.1. Disease mapping
Disease mapping is one of the most commonly used applications of geographical information systems (GIS) in veterinary epidemiology, because it provides an intuitive way to represent surveillance data in form of maps that facilitate interpretation, synthesis and recognition of frequency and cluster phenomena, and decision making (Rinaldi et al., 2006, Thrusfield, 2007). Application of GIS and remote sensing (RS; the process of acquiring data using sensing devices that are remotely located from the data being collected) is rapidly advancing in veterinary medicine. In addition, during the past decade many research articles, reviews and conferences in veterinary medicine have placed a special emphasis on GIS or RS (Rinaldi et al., 2006). Several types of spatial analyses are now integrated into GIS interfaces and software and are routinely used. These have become a useful complement for disease surveillance systems. Such tools include, for example, cluster and clustering detection techniques, generation of buffers, overlay analysis, network analysis, smoothing and prediction techniques, area and distance calculations, and three dimensional surface modeling (Ward and Carpenter, 2000, Carpenter, 2001, Rinaldi et al., 2006, Ward, 2007a, Ward, 2007b). RS data are increasingly used for surveillance and monitoring, particularly for human vector borne diseases (Beck et al., 2000, Saxena et al., 2009). Because disease vectors have specific requirements regarding climate, vegetation, soil and other edaphic factors, and are sensitive to changes in these factors that can be only captured and recorded by the current satellite technology, RS can be used to predict their distributions (Rinaldi et al., 2006, Saxena et al., 2009). RS technology has been used in a very limited way in veterinary surveillance (Ward et al., 2005). However, there are some important applications of RS technology to animal disease surveillance, for example, to survey and monitor herds and their movements and surrounding landscapes to predict the risk for spread of transboundary infectious animal diseases.
Gao et al. (2008) proposed 4 major challenges facing the development of disease mapping for surveillance systems, namely (i) disease data heterogeneities, which is mainly caused by the lack of an unified procedure for data collection by different international organizations, which may also lead to the duplication of reports; (ii) difficulties in integration and reusability; (iii) lack of interoperability between different disease services; and (vi) concerns over appropriate cartographical representation and dissemination of sensitive disease information. As noted above, these four challenges are also limitations for the implementation of surveillance systems at a global scale.
3.2. Social network analysis (SNA)
SNA has been defined as a group of elements and the nature and extent of the connections, relationships or interactions between them (Martinez-Lopez et al., 2009). One of the most important advantages of SNA is its inherent ability to incorporate relationships that are bi-directional, such as contacts between individuals, trade or animal movements (Martinez-Lopez et al., 2009). Mathematical methods and applications of SNA has been discussed and illustrated extensively elsewhere (Christley et al., 2005, Garabed et al., 2008, Martinez-Lopez et al., 2009). Application of SNA has emerged only recently in the field of veterinary preventive medicine (Christley et al., 2005). Most of the initial studies focused on the 2001 FMD epidemic in the UK, and were mostly related to the movements of FMD-susceptible species (Webb, 2005, Webb, 2006, Woolhouse et al., 2005, Kiss et al., 2006, Robinson and Christley, 2007, Robinson et al., 2007). SNA had shown to be a useful technique to identify individuals, populations and regions that are important in terms of the risk for animal disease introduction, spread, dissemination and maintenance. Furthermore, SNA can identify the common attributes, forces, factors and characteristics of these important nodes so that they can be selectively targeted as a part of a prevention or control scheme. SNA can also be used as a support tool for epidemiological modeling, risk analysis, space-time analysis or hazard analysis and critical control points. In addition, SNA can facilitate the identification of individuals (network nodes) in populations that are critical for the maintenance of the network structure. This in turn can help formulate prevention, control, surveillance, and eradication programs (Christley et al., 2005). SNA can be used to identify locations where disease is more likely to be maintained and spread in the event of its introduction into a region. This makes the technique suitable for integration into a surveillance system, in which selected locations could be targeted for surveillance, thus enhancing the sensitivity and reducing the time-to-detection of the surveillance system. Therefore, SNA can improve the efficiency of animal tracing systems – an increasing requirement for trade within and among countries. For these reasons, SNA may substantially support the decision- and policy-making process (Stevenson et al., 2007, Sturtevant et al., 2007, Martinez-Lopez et al., 2009).
3.3. Bayesian techniques
Rapid advancement in the field of Bayesian modeling and Markov chain Monte Carlo (MCMC) methods for statistical estimation and inference was possible due to the development of fast and powerful computing hardware and software (Lawson, 2009, Macnab, 2010). In recent years, the use of Bayesian disease mapping has been progressively expanding spatial statistical methodology for studies on spatial and spatiotemporal variations, trends and clustering of disease and health outcome risks in populations, as well as associated ecological risk factors (Macnab, 2010). Common methods in Bayesian disease mapping include the classical Bayesian hierarchical models for spatial disease mapping, spatiotemporal disease mapping and more recently bivariate and multivariate disease mapping (Biggeri et al., 2006, Lawson, 2006, Lawson, 2009, Zheng et al., 2008, Macnab, 2010). The concept of global and local smoothing –pooling data and borrowing-strength for small area mapping of rare events – is at the core of the Bayesian disease mapping approach (Lawson, 2009, Macnab, 2010). This makes the technique an ideal analytical tool to be integrated into a future global surveillance system. In veterinary medicine for example, Biggeri et al. (2006) used Bayesian Gaussian spatial exponential models and Bayesian kriging on canine parasitic disease data in Italy to demonstrate the accuracy of such approaches in identifying areas at risk of zoonotic parasitic diseases. Branscum et al. (2007) used a flexible Bayesian spatiotemporal regression approach to quantify the association between FMD outbreaks and selected epidemiological factors in Turkey from 1996 to 2003; this type of models could be scaled to manage international and global data. The model identified provinces with high and low risk for FMD occurrence in space and time. Branscum et al. (2007) demonstrated that the model can be useful when integrated into strategic targeting within a surveillance system, whereby surveillance sampling could be intensified in areas and at times when FMD was predicted to increase, which aids in adjusting control and prevention resources accordingly (Branscum et al., 2007). Recently Chan et al. (2010) constructed a timely influenza syndromic surveillance system using a Bayesian hierarchical model which incorporates the impact of spatial and temporal dependence, accounts for meteorological data, and performs probabilistic prediction on influenza. Their study concluded that Bayesian hierarchical models can be applied in a dynamic surveillance system to provide early warning for disease emergence, an advantage to previous classical syndromic surveillance approaches (Chan et al., 2010). Integration of such spatial and space-time Bayesian cluster detection models in a global surveillance system can provide a broad, unbiased and flexible range of diagnostic outputs that could be simplified and made widely available (Lawson, 2006, Hossain and Lawson, 2010). However, application of Bayesian methods to quantification of risk at the global scale has been limited. For example, Garabed et al. (2008) used Bayesian model averaging procedures and governance and economic health data to estimate the national probability of FMD outbreaks at a global scale from 1997 to 2005. That model identified significant risk factors that can be targeted to selectively allocate resources for disease control at regional and global scales. Recently, the use of Bayesian formulation in infectious disease models has been proposed for understanding spatial spread and identifying infected, undetected premises over the FMD epidemic that affected the UK in 2001 (Jewell et al., 2009a) and a hypothetical AI epidemic in the same country (Jewell et al., 2009b).
3.4. Landscape genetics and phylogeography
Landscape epidemiology has the potential to become a very important element in the establishment of a global animal disease surveillance system. Clements and Pfeiffer (2009) defined landscape epidemiology as the study of relationships between ecology and disease aimed at predicting and identifying the spatial distribution of diseases and their vectors or hosts. Landscape epidemiology is a modern epidemiological approach which integrates surveillance data, RS-derived climatic, topographical and other information relating to the environment (such as land cover and the normalized difference vegetation index) within a GIS environment. In addition to statistical analysis of the relationships between the epidemiological and environmental data, the application can lead to prediction of disease outcomes at non-sampled locations (Clements and Pfeiffer, 2009). The integration of landscape epidemiology into a global surveillance system may substantially improve its performance, sensitivity and specificity, since the ecology of most animal diseases differs substantially from country to regional to continental levels. An extension of landscape epidemiology that incorporates molecular data into the analysis has been referred to as landscape genetics.
It has recently been suggested that transboundary animal disease emergence is simultaneously influenced by both genetic and ecological factors and some studies have recently proposed general frameworks for linking molecular changes of infectious agents with the ecological dynamics of the diseases they cause (Real et al., 2005, Lemey et al., 2009). The recently developed field of phylogeography has been defined as the spatial analysis of genetic variation that aims to determine the genetic evolution of diseases in space and time, supplementing epidemiological knowledge of disease emergence (Real et al., 2005, Clements and Pfeiffer, 2009). Real et al. (2005) illustrated the linkage between ecological and evolutionary dynamics in rabies virus during its epidemic expansion in eastern and southern Ontario, Canada (Real et al., 2005). Chen et al. (2006) conducted a large scale phylogeographical study using HPAI H5N1 surveillance data from Asia. They found regionally-distinct sublineages of the virus that have become established in poultry populations in different geographical regions of Southeast Asia, and confirmed that Southern China was the focal point for multiple introductions into neighboring countries (Chen et al., 2006). FMD outbreaks reported in Israel and Palestine in 2007 were likely a consequence of different epidemics associated with circulation and spread of FMD virus strains from different regions of the Middle East, as suggested by a study that combined the application of molecular and cluster detection techniques (Alkhamis et al., 2009). Molecular and cluster detection techniques were also jointly used to identify areas in which vesicular stomatitis virus was likely to over-winter in the United States (Perez et al., 2010).
Most of the recent applications of phylogeography have been either descriptive or exploratory. Future integration of phylogeography with ecological methods, landscape genetics, spatial methods, and other mathematical modeling approaches would maximize its potential for quantifying and predicting the disease distribution at a global scale (Clements and Pfeiffer, 2009). Consequently, this will facilitate the integration of phylogeography in the establishment of a global surveillance system for transboundary animal diseases (Chen et al., 2006). In that context, Gallego et al. (2009) have demonstrated the potential of an innovative space-time cluster genotyping technology to enhance temporal sensitivity of any surveillance system by aiding the detection of moderate and small epidemics. For these reasons, real-time genotyping will also be an essential component of future real-time or near real-time surveillance systems, as sequencing and analysis of samples are expected to become more affordable and rapid, which will contribute to the broad application of such techniques.
4. The Disease BioPortal
Disease BioPortal (previously the FMD BioPortal) is a web-based system that facilitates near real-time animal disease data and information sharing, visualization and analysis. For those reasons, the Disease BioPortal provides an example of an IT system that could be used to support international or global animal disease surveillance efforts. Here, some of the attributes of the system are illustrated.
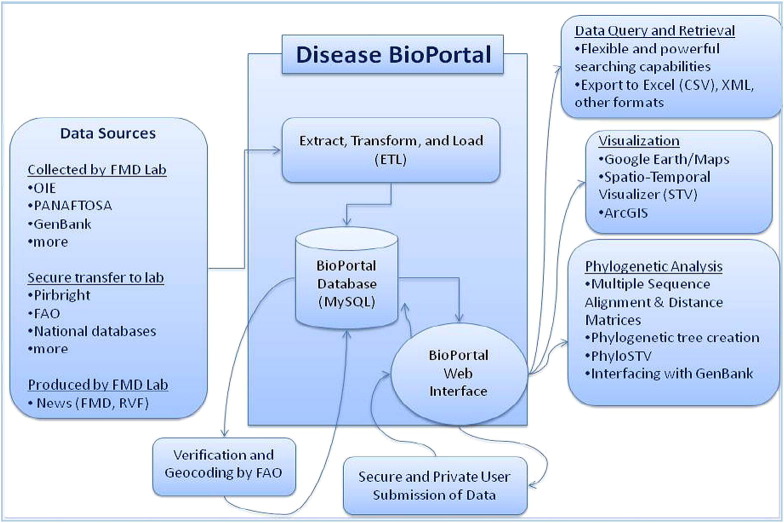
The Disease BioPortal is an unrestricted, public web site created and maintained by the Center for Animal Disease Modeling and Surveillance (CADMS) at the University of California, Davis (http://www.fmdbioportal.ucdavis.edu/). Information and data on >40 animal diseases and syndromes originally collected and stored at various databases around the world are compiled and organized in the Disease BioPortal central data repository. The Disease BioPortal makes available to the user a set of techniques for cluster detection and phylogenetic analysis of sequences. Users can also depict and analyze data using a novel visualization environment which supports multiple displays including those for illustration of spatial and temporal patterns using the spatio-temporal-visualized (STV) and the Phylo-STV (tutorial video available at http://www.fmd.ucdavis.edu/ma/rcm/Japan_2010.wmv) tools. Disease BioPortal functionalities are accessible through the web and data confidentiality is secured through user access control and computer network security techniques such as Secure Sockets Layer (SSL). Disease BioPortal operates with multiple streams of information (Fig. 1 ). Databases are actively collected by, securely transferred to, or produced at CADMS. Databases available in the Disease BioPortal include OIE’s WAHID, FAO’s Empres-i, GenBank, the World Reference Laboratory’s (Pirbright) FMD serological database, PANASFTOSA’s weekly reports on vesicular-like diseases, and the FMD News and RVF News produced by the CADMS. By subscribing to FMD News, daily or weekly FMD-related news items captured by different media sources in several languages and in near-real-time can be received (link of FMD News: http://www.fmd.ucdavis.edu/news.php).
Fig. 1.
Flow diagram depicting the procedure used to collect, store and share databases in the Disease BioPortal (http://www.fmdbioportal.ucdavis.edu/).
More than 459 users from >50 countries have registered to the Disease BioPortal. Since the inception of the project in 2004, more than 11,000 FMD news items from 128 countries have been delivered to subscribers in near real-time. The use of systems like the Disease BioPortal will enhance the ability of countries and agencies to prepare for and respond to transboundary animal disease epidemics (Perez et al., 2009).
5. Retrospective analysis of avian influenza surveillance data using the Disease BioPortal
Potential applications and the impact that web-based surveillance systems can have on regional and global surveillance of animal diseases are illustrated here through the use of the Disease BioPortal for the retrospective analysis and visualization of joint datasets of Swedish and Danish records from the HPAI H5N1 epidemic that affected mainly wild birds around the Baltic Sea during the winter and spring of 2006. Use of the Disease BioPortal to analyze the Danish dataset has been described elsewhere (Willeberg et al., 2010). The Swedish dataset was recently added to the newest version of the Disease BioPortal system and provides illustrates how disparate databases may be combined for joint analysis as part of a hierarchical surveillance system. The Danish epidemic has previously been documented in official reports from the Danish Veterinary and Food Administration (DVFA, 2007) and from the European Commission (Hesterberg et al., 2007). Spatial distribution of infection within the country (Vigre et al., 2007, Willeberg et al., 2010), its phylogenetic distribution (Bragstad et al., 2007), and its potential public health implications have been described elsewhere (Molbak et al., 2006). The official Danish wild bird surveillance data from 2006 includes >6700 records of tested samples, of which >5,500 were fecal samples obtained from migrating wild birds during resting (i.e, surveillance with active collection of data, referred to as “active surveillance data” here); approximately 1200 samples collected from dead wild birds (i.e., surveillance with passive data collection, referred to as “passive surveillance” here) were tested (Willeberg et al., 2010). From February 15 through May 31, approximately 1000 dead birds were tested. The total number of H5N1-positive wild bird samples was 45, of which only one positive sample was found through active surveillance. The first positive bird was found on March 13, 2006(DVFA, 2007). In addition, a mixed backyard poultry flock was found positive at the end of the epidemic in May (DVFA, 2007). A total of 22 HPAI H5N1 viruses were isolated, of which 8 have been sequenced and were found to be closely related. The remaining positive birds were positive to real-time polymerase chain reaction (RT-PCR) (Bragstad et al., 2007). Spatio-temporal analyses identified an early cluster of high HPAI H5N1 virus prevalence among Tufted Ducks (Aythya fuligula), which appeared to spread quickly to other wild bird species in the close vicinity (Willeberg et al., 2010). The Swedish 2006 surveillance dataset contains >4800 sample records, with approximately 1000 samples tested during the risk period of February 15–May 31. During this period 518 diseased or dead wild birds were tested in the passive surveillance, of which 65 were HPAI H5N1-positive to RT-PCR. The first positive Swedish bird was identified on February 24, 2010. No cases were detected in the active surveillance program (live birds trapped or hunted). Published sequence data from positive birds revealed two distinct sub-lineages (Kiss et al., 2008, Zohari et al., 2008). In a hunting preserve with mallards (Anas platyrhynchos), located within the active surveillance zone around the index case, one of the birds was found positive on March 20. Although we failed to identify published epidemiological studies from Sweden, some data were available elsewhere (Sabirovic et al., 2006, Hesterberg et al., 2007, Komar and Olsen, 2008).
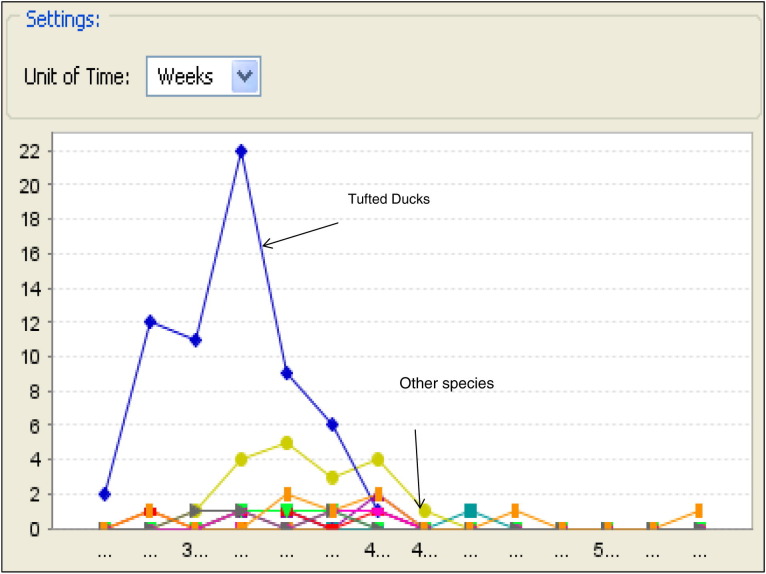
The epidemics in both countries had several characteristics in common. Most PCR-positive birds were found close to the shore along the shallow waters of the Baltic Sea and of its associated straits (Fig. 2 ). The duration of the epidemic (the time period between the first and the last detection of a positive case) was 56 days for Sweden (February 24–April 21) and 70 days for Denmark (March 13–May 22). In each country there was one positive poultry farm, both with a mix of species including ducks. Table 1 summarizes the numerical composition of the joint database by country and species (Tufted Ducks and other species, respectively), as well as the resulting relative risk estimated for species (Tufted Ducks, other species) stratified by country, and for country (Denmark, Sweden) stratified by species. The proportion of Tufted Ducks that tested positive was high in both countries (Table 1) and the proportion of positive Tufted Ducks was high during the first period of the epidemics (Fig. 3 ). Disease prevalence was higher in Sweden than in Denmark, except among Tufted Ducks where the reverse was true (Table 1). Such differences may have been influenced by various selection biases because of the passive collection of surveillance data (Breed et al., 2010). Spatial (Fig. 2), temporal (Fig. 3), and phylogenetic relationships (Fig. 4 ) are simultaneously displayed (in color coding) on the monitor during the on-line session in the Disease BioPortal.
Fig. 2.
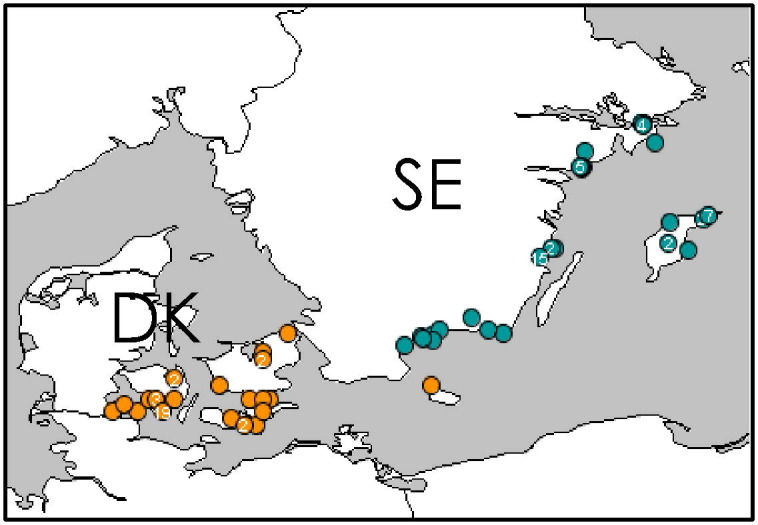
Cases of highly pathogenic avian influenza in Sweden (blue dots) and Denmark (orange dots) illustrated using the Disease BioPortal (http://www.fmdbioportal.ucdavis.edu/) spatio-temporal visualizer (STV) tool; note location of collected positive birds along the shorelines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Number of tested and PCR-positive birds by country and species, and relative risk stratified by species and by country for highly pathogenic avian influenza H5N1, 2006.
| Country | Species | PCR-positive | Tested | % Positive | Relative risk |
|
|---|---|---|---|---|---|---|
| Species | Country | |||||
| Sweden | All | 65 | 518 | 12.5 | - | 1 |
| Tufted D. | 36 | 83 | 43.4 | 6.5 | 1 | |
| Other sp. | 29 | 435 | 6.7 | 1 | 1 | |
| Denmark | All | 44 | 951 | 4.6 | - | 0.4 |
| tufted D. | 26 | 36 | 72.2 | 36.7 | 1.7 | |
| Other sp. | 18 | 915 | 2 | 1 | 0.3 | |
| Total | All | 109 | 1,469 | 7.4 | - | - |
Fig. 3.
Cases of highly pathogenic avian influenza in Sweden and Denmark stratified by species and week depicted using the Disease BioPortal (http://www.fmdbioportal.ucdavis.edu/) spatio-temporal visualizer (STV) tool; note that Tufted Ducks dominate early in the epidemic.
Fig. 4.
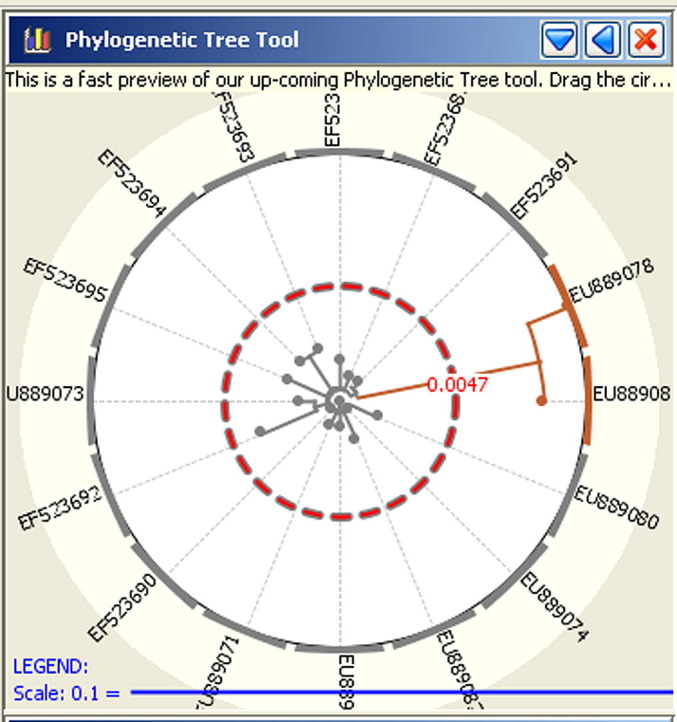
Phylogenetic tool from the Disease BioPortal (http://www.fmdbioportal.ucdavis.edu/) applied to the combined pool of 8 Danish and 8 Swedish H5N1 highly pathogenic avian influenza viruses. Users may change the value of genetic distance that is used as a threshold (red circle) to categorize isolates as “similar” and “different”, which are depicted in the tree and in the spatial visualizer using colors specific for each category (orange and gray in the example hear). Here, the tree was built using a neighboring joining algorithm; users may also implement maximum likelihood and maximum parsimony algorithms. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
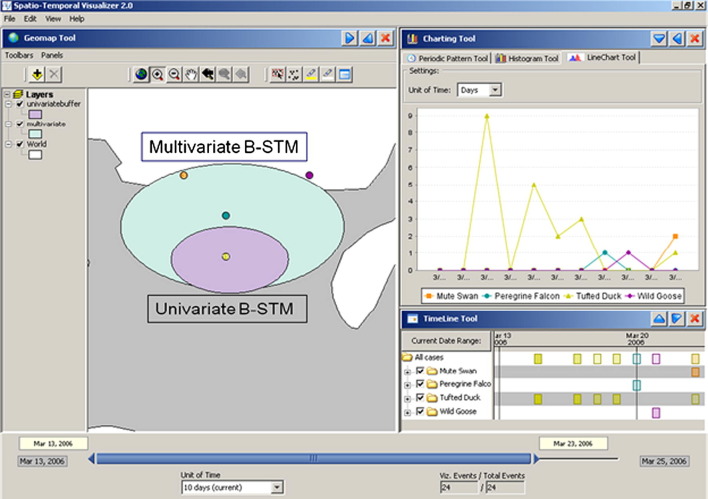
Spatio-temporal cluster detection in the Danish dataset was performed using a scan algorithm implemented in the SaTScan software (Kulldorff and Information Management Services, 2007), with both uni- and multivariate methodology (Kulldorff et al., 2007) to relate clustering among Tufted Ducks to that among other species (Fig. 5, Fig. 6 ). Both the univariate and the multivariate models were statistically significant (P < 0.001) (Willeberg et al., 2010).
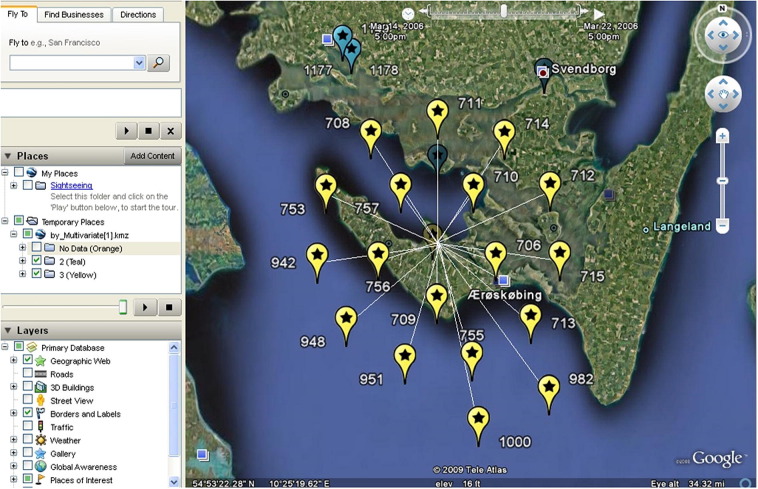
Fig. 5.
Location of cases included in the multivariate cluster. The temporal window for the cluster is March 15–23, 2006, and it is illustrated using a Google Earth map available through the Disease BioPortal (http://www.fmdbioportal.ucdavis.edu/).
Fig. 6.
Univariate and multivariate clusters around H5N1 highly pathogenic avian influenza cases in Denmark, March 15–23, 2006, depicted using the Disease BioPortal (http://www.fmdbioportal.ucdavis.edu/) spatio-temporal visualizer tool.
6. Discussion
Emergence and spread of diseases at a global scale have created unprecedented new challenges to solving local, regional, and global problems of animal and human health and to mitigating their social, economic, and political consequences. Global efforts to maintain animal and human health need knowledge of disease incidence and changing risk factors – such as environmental, economic and political factors and the international movement of humans and animals. A prerequisite to understanding local, regional or global distributions of disease or disease risks is rapid access to current information on these distributions.
Most existing animal disease surveillance systems are limited in their scope and do not completely address the need for real-time surveillance of emerging animal and zoonotic infections (Martin et al., 2007c). As an example, the Disease BioPortal was developed to fill this gap as a generic, publicly accessible tool providing flexible input and output solutions (Thurmond et al., 2007). Focusing on a restricted geographical region the present study applied some of its visualization tools, such as the STV (Fig. 2, Fig. 3, Fig. 6), Phylo-STV (Fig. 4), and Google Earth (Fig. 5). The most recent Disease BioPortal version accommodates techniques for detection of spatio-temporal clusters as an integral part of the on-line functionality.
Here, we demonstrated the use of the Disease BioPortal system through the retrospective analysis of HPAI data collected by Sweden and Denmark Real-time use of the Disease BioPortal would have certainly improved the repertoire of tools and techniques available to veterinary services to impose efficient control measures. For example, when the epidemics ended in the affected regions of Sweden and Denmark it was feared that the infection was moving north – following the natural migratory patterns of birds – and that northern regions of Denmark and Sweden and neighboring countries such as Norway and Finland may become infected. Consequently, there was a mobilization of manpower and technical resources by the national veterinary services and other government authorities to prevent further spread of infection. Control measures were quite costly for the Danish and Swedish economies and included, for example, the imposition to keep poultry indoors, temporary zoning around all wild-bird cases, limiting the movement of poultry and poultry products, and screening of poultry and back-yard flocks inside the control zones before restrictions could be lifted (DVFA, 2007). However, no cases were detected in neighboring countries and in retrospect, Denmark was the last European country to become infected in the European spring 2006 epidemic. Because only the south-eastern part of Denmark became affected (Fig. 2), it has been hypothesized that the wild-bird infection died out in that specific region, possibly due to increasing air and water temperatures (Ottaviani et al., 2010, Reperant et al., 2010). Arguably, early appreciation of the temporal-spatial regional evolution of the epidemic in Denmark and Sweden, which could have been through the real time analysis of data using the tools shown here, might have reduced the costs associated with a continued high alert level in the region as well as the fear that the infection might return with the fall migration of wild birds. Similarly, systems with the attributes and features such as those available in the Disease BioPortal would provide support for decision-makers in the face of an infectious disease epidemic. Because there is an urgent need for the implementation of a near-real-time animal disease surveillance system at a global scale, and considering that analytical techniques are rapidly evolving, it is expected that over the next few years there will be substantial progress in the establishment of a global surveillance effort. However, many issues that have prevented the development of such a system still need to be solved including, for example, the role that international and national organizations will play in maintenance and operation of the system, quality of the data, temporal sensitivity of the system, and financial and logistic support. Because of the recent development of new technology, once those issues are solved, it is expected that such development will represent a substantial step forward in the prevention and control of transboundary animal diseases at regional and global scales.
Acknowledgments
This study was funded in part by grants from the U.S. National Center for Medical Intelligence, the U.S. Department of Agriculture, the University of California in Davis, the Kansas Bioscience Authority, and the U.S. Foreign Animal Disease Center – Department of Homeland Security under Grant Award Number 2010-ST-061-AG0002.
References
- Alban L., Boes J., Kreiner H., Petersen J.V., Willeberg P. Towards a risk-based surveillance for Trichinella spp. In Danish pig production. Prev Vet Med. 2008;87:340–357. doi: 10.1016/j.prevetmed.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Alkhamis M.A., Perez A.M., Yadin H., Knowles N.J. Temporospatial clustering of foot-and-mouth disease outbreaks in Israel and Palestine, 2006–2007. Transbound Emerg Dis. 2009;56:99–107. doi: 10.1111/j.1865-1682.2009.01066.x. [DOI] [PubMed] [Google Scholar]
- Beck L.R., Lobitz B.M., Wood B.L. Remote sensing and human health: new sensors and new opportunities. Emerg Infect Dis. 2000;6:217–227. doi: 10.3201/eid0603.000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggeri A., Dreassi E., Catelan D., Rinaldi L., Lagazio C., Cringoli G. Disease mapping in veterinary epidemiology: a Bayesian geostatistical approach. Stat Methods Med Res. 2006;15:337–352. doi: 10.1191/0962280206sm455oa. [DOI] [PubMed] [Google Scholar]
- Bragstad K., Jorgensen P.H., Handberg K., Hammer A.S., Kabell S., Fomsgaard A. First introduction of highly pathogenic H5N1 avian influenza A viruses in wild and domestic birds in Denmark, Northern Europe. Virol J. 2007;4:43. doi: 10.1186/1743-422X-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branscum A.J., Perez A.M., Johnson W.O., Thurmond M.C. Bayesian spatiotemporal analysis of foot-and-mouth disease data from the Republic of Turkey. Epidemiol Infect. 2007;1:10. doi: 10.1017/S0950268807009065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breed A.C., Harris K., Hesterberg U., Gould G., Londt B.Z., Brown I.H., Cook A.J. Surveillance for avian influenza in wild birds in the European Union in 2007. Avian Dis. 2010;54:399–404. doi: 10.1637/8950-053109-Reg.1. [DOI] [PubMed] [Google Scholar]
- Carpenter T.E. Methods to investigate spatial and temporal clustering in veterinary epidemiology. Prev Vet Med. 2001;48:303–320. doi: 10.1016/s0167-5877(00)00199-9. [DOI] [PubMed] [Google Scholar]
- Case J.T. Are we really communicating? Standard terminology versus terminology standards in veterinary clinical pathology. Vet Clin Pathol. 2005;34:5–6. doi: 10.1111/j.1939-165x.2005.tb00001.x. [DOI] [PubMed] [Google Scholar]
- Chan T.C., King C.C., Yen M.Y., Chiang P.H., Huang C.S., Hsiao C.K. Probabilistic daily ILI syndromic surveillance with a spatio-temporal Bayesian hierarchical model. PLoS One. 2010;5:e11626. doi: 10.1371/journal.pone.0011626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Smith G.J., Li K.S., Wang J., Fan X.H., Rayner J.M., Vijaykrishna D., Zhang J.X., Zhang L.J., Guo C.T., Cheung C.L., Xu K.M., Duan L., Huang K., Qin K., Leung Y.H., Wu W.L., Lu H.R., Chen Y., Xia N.S., Naipospos T.S., Yuen K.Y., Hassan S.S., Bahri S., Nguyen T.D., Webster R.G., Peiris J.S., Guan Y. Establishment of multiple sublineages of H5N1 influenza virus in Asia: implications for pandemic control. Proc Natl Acad Sci USA. 2006;103:2845–2850. doi: 10.1073/pnas.0511120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christley R.M., Pinchbeck G.L., Bowers R.G., Clancy D., French N.P., Bennett R., Turner J. Infection in social networks: using network analysis to identify high-risk individuals. Am J Epidemiol. 2005;162:1024–1031. doi: 10.1093/aje/kwi308. [DOI] [PubMed] [Google Scholar]
- Clements A.C., Pfeiffer D.U. Emerging viral zoonoses: frameworks for spatial and spatiotemporal risk assessment and resource planning. Vet J. 2009;182:21–30. doi: 10.1016/j.tvjl.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherr M.G., Audige L. Monitoring and surveillance for rare health-related events: a review from the veterinary perspective. Philos Trans R Soc Lond B Biol Sci. 2001;356:1097–1106. doi: 10.1098/rstb.2001.0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DVFA, 2007. Highly Pathogenic Avian Influenza H5N1, Denmark, Spring 2006. Danish Veterinary and Food Administration.
- Gallego B., Sintchenko V., Wang Q., Hiley L., Gilbert G.L., Coiera E. Biosurveillance of emerging biothreats using scalable genotype clustering. J Biomed Inform. 2009;42:66–73. doi: 10.1016/j.jbi.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Gao S., Mioc D., Anton F., Yi X., Coleman D.J. Online GIS services for mapping and sharing disease information. Int J Health Geogr. 2008;7:8. doi: 10.1186/1476-072X-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garabed R.B.J., Johnson W.O., Gill J., Perez A.M., Thurmond M.C. Exploration of associations between governance and economics and country level foot-and-mouth disease status by using Bayesian model averaging. J R Statis soc A. 2008;171:699–722. [Google Scholar]
- Grein T.W., Kamara K.B., Rodier G., Plant A.J., Bovier P., Ryan M.J., Ohyama T., Heymann D.L. Rumors of disease in the global village: outbreak verification. Emerg Infect Dis. 2000;6:97–102. doi: 10.3201/eid0602.000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesterberg U., Harris K., Cook A., Brown I. Annual Report of the EU Avian Influenza Surveillance in Wild Birds 2006. In: Disease C.R.L.f.a.i.a.N., editor. Surveillance for Avian Influenza in Wild Birds carrIed out By Member States, February–December 2006. European Commission, Health & Consumer Protection directorate-General; 2007. [Google Scholar]
- Hossain M.M., Lawson A.B. Space-time Bayesian small area disease risk models: development and evaluation with a focus on cluster detection. Environ Ecol Stat. 2010;17:73–95. doi: 10.1007/s10651-008-0102-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell C.P., Keeling M.J., Roberts G.O. Predicting undetected infections during the 2007 foot-and-mouth disease outbreak. J R Soc Interface. 2009;6:1145–1151. doi: 10.1098/rsif.2008.0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell C.P., Kypraios T., Christley R.M., Roberts G.O. A novel approach to real-time risk prediction for emerging infectious diseases: a case study in Avian Influenza H5N1. Prev Vet Med. 2009;91:19–28. doi: 10.1016/j.prevetmed.2009.05.019. [DOI] [PubMed] [Google Scholar]
- Keystone J.S., Kozarsky P.E., Freedman D.O. Internet and computer-based resources for travel medicine practitioners. Clin Infect Dis. 2001;32:757–765. doi: 10.1086/319234. [DOI] [PubMed] [Google Scholar]
- Kiss I.Z., Green D.M., Kao R.R. The network of sheep movements within Great Britain: network properties and their implications for infectious disease spread. J R Soc Interface. 2006;3:669–677. doi: 10.1098/rsif.2006.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss I., Gyarmati P., Zohari S., Ramsay K.W., Metreveli G., Weiss E., Brytting M., Stivers M., Lindstrom S., Lundkvist A., Nemirov K., Thoren P., Berg M., Czifra G., Belak S. Molecular characterization of highly pathogenic H5N1 avian influenza viruses isolated in Sweden in 2006. Virol J. 2008;5:113. doi: 10.1186/1743-422X-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar N., Olsen B. Avian influenza virus (H5N1) mortality surveillance. Emerg Infect Dis. 2008;14:1176–1178. doi: 10.3201/eid1407.080161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulldorff M., Mostashari F., Duczmal L., Katherine Yih W., Kleinman K., Platt R. Multivariate scan statistics for disease surveillance. Stat Med. 2007;26:1824–1833. doi: 10.1002/sim.2818. [DOI] [PubMed] [Google Scholar]
- Kulldorff M, Information Management Services I, 2007. SatScan v7.0.3 Software for the spatial and space-time scan statistics. <http://www.satscan.org>.
- Lawson A. CRC Press; Boca Raton: 2009. Introduction, Bayesian disease mapping. Bayesian disease mapping hierarchical modeling in spatial epidemiology. [Google Scholar]
- Lawson A.B. Disease cluster detection: a critique and a Bayesian proposal. Stat Med. 2006;25:897–916. doi: 10.1002/sim.2417. [DOI] [PubMed] [Google Scholar]
- Lemey P., Rambaut A., Drummond A.J., Suchard M.A. Bayesian phylogeography finds its roots. PLoS Comput Biol. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab YC. On Gaussian Markov random fields and Bayesian disease mapping. Stat Methods Med Res; 2010. [DOI] [PubMed]
- Martin P.A., Cameron A.R., Barfod K., Sergeant E.S., Greiner M. Demonstrating freedom from disease using multiple complex data sources 2: case study – classical swine fever in Denmark. Prev Vet Med. 2007;79:98–115. doi: 10.1016/j.prevetmed.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Martin P.A., Cameron A.R., Greiner M. Demonstrating freedom from disease using multiple complex data sources 1: a new methodology based on scenario trees. Prev Vet Med. 2007;79:71–97. doi: 10.1016/j.prevetmed.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Martin V., Von Dobschuetz S., Lemenach A., Rass N., Schoustra W., Desimone L. Early warning, database, and information systems for avian influenza surveillance. J Wildlife Dis. 2007;43:S71–S76. [Google Scholar]
- Martinez-Lopez B., Perez A.M., Sanchez-Vizcaino J.M. Social network analysis. Review of general concepts and use in preventive veterinary medicine. Transbound Emerg Dis. 2009;56:109–120. doi: 10.1111/j.1865-1682.2009.01073.x. [DOI] [PubMed] [Google Scholar]
- Meah M.N., Lewis G.A. Ministry of Agriculture, Fisheries and Food (MAFF); London: 1999. A review of veterinary surveillance in England and Wales with special reference to work supported by the ministry of Agriculture, Fisheries and Food. Veterinary surveillance in England and Wales – a review. [Google Scholar]
- Molbak K, Trykker H, Mellergaard S, Glismann S. Avian influenza in Denmark, March–June 2006: public health aspects. Euro Surveill 2006; 11, E060615 060613. [DOI] [PubMed]
- OIE, 2010a. Animal health Surveillance In: Health WOfA, editor. Terrestrial Animal Health Code. Chapter 1.4.
- OIE W. World animal health information database (WAHID) Interface; 2010b. Weekly disease information.
- Ottaviani D., de la Rocque S., Khomenko S., Gilbert M., Newman S.H., Roche B., Schwabenbauer K., Pinto J., Robinson T.P., Slingenbergh J. The cold European winter of 2005–2006 assisted the spread and persistence of H5N1 influenza virus in wild birds. Ecohealth. 2010;7:226–236. doi: 10.1007/s10393-010-0316-z. [DOI] [PubMed] [Google Scholar]
- Perez A.M., Pauszek S.J., Jimenez D., Kelley W.N., Whedbee Z., Rodriguez L.L. Spatial and phylogenetic analysis of vesicular stomatitis virus over-wintering in the United States. Prev Vet Med. 2010;93:258–264. doi: 10.1016/j.prevetmed.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Perez A.M., Zeng D., Tseng C.J., Chen H., Whedbee Z., Paton D., Thurmond M.C. A web-based system for near real-time surveillance and space-time cluster analysis of foot-and-mouth disease and other animal diseases. Prev Vet Med. 2009;91:39–45. doi: 10.1016/j.prevetmed.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Real L.A., Henderson J.C., Biek R., Snaman J., Jack T.L., Childs J.E., Stahl E., Waller L., Tinline R., Nadin-Davis S. Unifying the spatial population dynamics and molecular evolution of epidemic rabies virus. Proc Natl Acad Sci USA. 2005;102:12107–12111. doi: 10.1073/pnas.0500057102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reperant L.A., Fuckar N.S., Osterhaus A.D., Dobson A.P., Kuiken T. Spatial and temporal association of outbreaks of H5N1 influenza virus infection in wild birds with the 0°C isotherm. PLoS Pathog. 2010;6:e1000854. doi: 10.1371/journal.ppat.1000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi L., Musella V., Biggeri A., Cringoli G. New insights into the application of geographical information systems and remote sensing in veterinary parasitology. Geospat Health. 2006;1:33–47. doi: 10.4081/gh.2006.279. [DOI] [PubMed] [Google Scholar]
- Robinson S.E., Christley R.M. Exploring the role of auction markets in cattle movements within Great Britain. Prev Vet Med. 2007;81:21–37. doi: 10.1016/j.prevetmed.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Robinson S.E., Everett M.G., Christley R.M. Recent network evolution increases the potential for large epidemics in the British cattle population. J R Soc Interface. 2007;4:669–674. doi: 10.1098/rsif.2007.0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabirovic M., Wilesmith J., Hall S., Coulson N., Landeg F. Department for Environment, Food and Rural Affairs; 2006. Outbreaks of HPAI H5N1 virus in Europe during 2005/2006. [Google Scholar]
- Saxena R., Nagpal B.N., Srivastava A., Gupta S.K., Dash A.P. Application of spatial technology in malaria research & control: some new insights. Indian J Med Res. 2009;130:125–132. [PubMed] [Google Scholar]
- Stark KDC, Frei PP, Frie-Staheli C, Pfeiffer DU, Audige L. A health profile of swiss dairy cow. In: Thrusfield MV, Goodall EA, editors. Society for Veterinary Epidemiology and Preventive Medicine, Glasgow; 1996. p. 86–93.
- Stevenson M.A., Sanson R.L., Miranda A.O., Lawrence K.A., Morris R.S. Decision support systems for monitoring and maintaining health in food animal populations. N Z Vet J. 2007;55:264–272. doi: 10.1080/00480169.2007.36780. [DOI] [PubMed] [Google Scholar]
- Sturtevant J.L., Anema A., Brownstein J.S. The new International Health Regulations: considerations for global public health surveillance. Disaster Med Public Health Prep. 2007;1:117–121. doi: 10.1097/DMP.0b013e318159cbae. [DOI] [PubMed] [Google Scholar]
- Thrusfield M.V. Veterinary epidemiology. Blackwell; Oxford/Ames: 2007. Surveillance. pp. 168–187. [Google Scholar]
- Thurmond M., Perez A., Tseng C., Chen H., Zeng D. Springer-Verlag; New Brunswick, NJ, USA: 2007. Global foot-and-mouth disease surveillance using bioportal. Proceedings of the 2nd NSF conference on Intelligence and security informatics: BioSurveillance. pp. 169–179. [Google Scholar]
- Thurmond M.C. Conceptual foundations for infectious disease surveillance. J Vet Diagn Invest. 2003;15:501–514. doi: 10.1177/104063870301500601. [DOI] [PubMed] [Google Scholar]
- Vigre H., Jensen Handberg K., Therkildsen O.R., Mortenesen S., Hammer A.-S., Kebell S., Michel M., Elvested K., Jørgens P.H. GISVET; Copenhagen: 2007. Spatial pattern of high pathogenic H5N1 Avian influenza infection in dead wild birds in Denmark 2006. [Google Scholar]
- Ward M.P. Geographic information system-based avian influenza surveillance systems for village poultry in Romania. Vet Ital. 2007;43:483–489. [PubMed] [Google Scholar]
- Ward M.P. Spatio-temporal analysis of infectious disease outbreaks in veterinary medicine: clusters, hotspots and foci. Vet Ital. 2007;43:559–570. [PubMed] [Google Scholar]
- Ward M.P., Carpenter T.E. Techniques for analysis of disease clustering in space and in time in veterinary epidemiology. Prev Vet Med. 2000;45:257–284. doi: 10.1016/s0167-5877(00)00133-1. [DOI] [PubMed] [Google Scholar]
- Ward M.P., Ramsay B.H., Gallo K. Rural cases of equine West Nile virus encephalomyelitis and the normalized difference vegetation index. Vector Borne Zoonotic Dis. 2005;5:181–188. doi: 10.1089/vbz.2005.5.181. [DOI] [PubMed] [Google Scholar]
- Webb C.R. Farm animal networks: unraveling the contact structure of the British sheep population. Prev Vet Med. 2005;68:3–17. doi: 10.1016/j.prevetmed.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Webb C.R. Investigating the potential spread of infectious diseases of sheep via agricultural shows in Great Britain. Epidemiol Infect. 2006;134:31–40. doi: 10.1017/S095026880500467X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willeberg P., Perez A., Thurmond M., Ascher M., Carpenter T., AlKhamis M. Visualization and analysis of the Danish 2006 highly pathogenic avian influenza virus H5N1 wild bird surveillance data by a prototype avian influenza BioPortal. Avian Dis. 2010;54:433–439. doi: 10.1637/8820-040209-Reg.1. [DOI] [PubMed] [Google Scholar]
- Woodall J. Official versus unofficial outbreak reporting through the Internet. Int J Med Inform. 1997;47:31–34. doi: 10.1016/s1386-5056(97)00079-8. [DOI] [PubMed] [Google Scholar]
- Woolhouse M.E., Shaw D.J., Matthews L., Liu W.C., Mellor D.J., Thomas M.R. Epidemiological implications of the contact network structure for cattle farms and the 20–80 rule. Biol Lett. 2005;1:350–352. doi: 10.1098/rsbl.2005.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W.J., Li X.Y., Chen K. Bayesian statistics in spatial epidemiology. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2008;37:642–647. doi: 10.3785/j.issn.1008-9292.2008.06.017. [DOI] [PubMed] [Google Scholar]
- Zohari S., Gyarmati P., Thoren P., Czifra G., Brojer C., Belak S., Berg M. Genetic characterization of the NS gene indicates co-circulation of two sub-lineages of highly pathogenic avian influenza virus of H5N1 subtype in Northern Europe in 2006. Virus Genes. 2008;36:117–125. doi: 10.1007/s11262-007-0188-7. [DOI] [PubMed] [Google Scholar]