Highlights
-
•
RNAs with three phosphates at the 5′-end are made by several RNA viruses.
-
•
Most RNAs found in the cytosol of uninfected cells do not bear 5′-triphosphates.
-
•
RIG-I, a sensor for virus infection, detects 5′-triphosphorylated viral RNA genomes.
-
•
RIG-I then induces type I interferons that trigger expression of IFIT proteins.
-
•
IFITs mediate anti-viral effects by sequestration of 5′-triphosphate viral RNAs.
Abstract
Some RNA virus genomes bear 5′-triphosphates, which can be recognized in the cytoplasm of infected cells by host proteins that mediate anti-viral immunity. Both the innate sensor RIG-I and the interferon-induced IFIT proteins bind to 5′-triphosphate viral RNAs. RIG-I signals for induction of interferons during RNA virus infection while IFITs sequester viral RNAs to exert an anti-viral effect. Notably, the structures of these proteins reveal both similarities and differences, which are suggestive of independent evolution towards ligand binding. 5′-triphosphates, which are absent from most RNAs in the cytosol of uninfected cells, are thus a marker of virus infection that is targeted by the innate immune system for both induction and execution of the anti-viral response.
Current Opinion in Microbiology 2013, 16:485–492
This review comes from a themed issue on Host–microbe interactions: viruses
Edited by Carlos F Arias
For a complete overview see the Issue and the Editorial
Available online 23rd May 2013
1369-5274/$ – see front matter, © 2013 Elsevier Ltd. All rights reserved.
Introduction
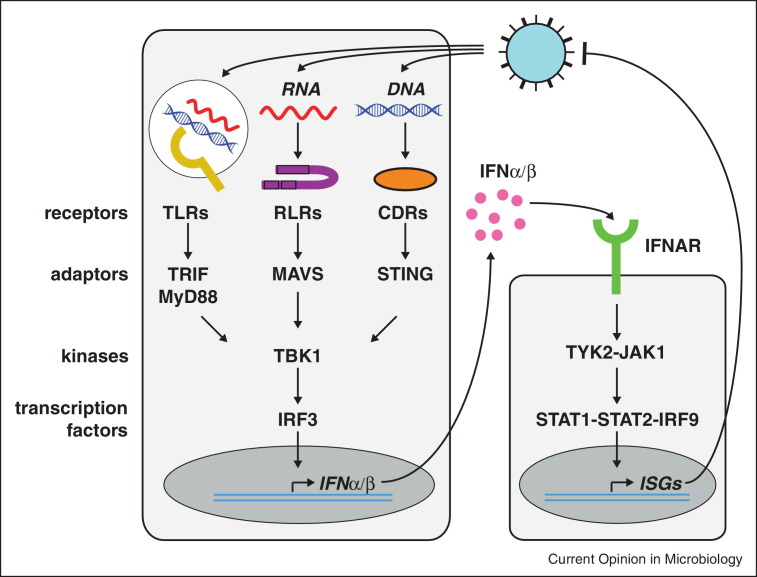
The interferon system is one of the cornerstones of innate immunity in mammals (Figure 1 ). Interferons were discovered as soluble factors secreted by virally infected cells that protected uninfected cells from subsequent viral challenge [1]. The type I interferons (referred to as IFN in this review) include interferon-β and multiple interferon-α subtypes, all of which signal through a common receptor IFNAR [2] (Figure 1). All nucleated cells can produce IFN in response to nucleic acids generated during virus infection. Detection of these nucleic acids is carried out within the infected cell by host proteins residing in the cytosol, such as RIG-I-like receptors (RLRs) and other virus-sensing receptors [3] (Figure 1). In addition, some cells possess the ability to detect viruses in the extracellular milieu using members of the toll-like receptor (TLR) family. This generally involves endocytic uptake of viruses or remnants of virally infected cells into vesicular compartments surveyed by TLR3, 7, 8 and 9, all of which bind nucleic acids (Figure 1). Either RLR or TLR signalling then induces expression of the IFN genes (Figure 1). Given the abundance of nucleic acids in uninfected healthy cells and the potential of IFN to contribute to autoinflammatory and autoimmune diseases [4], it is not surprising that sophisticated mechanisms ensure that activation of RLRs and TLRs occurs only during infection. One of these mechanisms is the recognition of viral RNA genomes bearing a 5′-triphosphate (5PPP) moiety by RIG-I, a member of the RLR family. 5PPP groups are present on the genomes of many RNA viruses (Table 1 ), but are not found on most cellular RNAs in the cytosol. As such, the presence of 5PPP RNA in the cytoplasm is a molecular signature of virus infection.
Figure 1.
Induction and effects of IFN during virus infection. Virus infection delivers nucleic acids into the cytosol or endosomal compartment. Innate nucleic acid sensors including TLRs, RLRs and the poorly characterized cytosolic DNA receptors (CDRs) detect these DNAs and RNAs and then trigger a signal transduction cascade that induces IFN. Adaptor proteins, kinases and transcription factors mediate signalling. Note that additional proteins have been implicated and that the figure only shows some selected key components. IFN signals via IFNAR resulting in the induction of ISGs that have direct and indirect anti-viral effects.
Table 1.
RIG-I and IFITs target viruses with 5PPP RNA genomes. For selected RNA viruses, recognition by RIG-I and restriction by IFITs is indicated.
| Virus (family) | 5′-end of genome | RIG-I recognition | IFIT restriction | References |
|---|---|---|---|---|
| Influenza A virus (Orthomyxoviridae) | PPP | Yes | Yes | [9••, 12, 13, 38] |
| Vesicular stomatitis virus (Rhabdoviridae) | PPP | Yes | Yes | [9••, 38, 46] |
| Rift Valley fever virus (Bunyaviridae) | PPP | Yes | Yes | [9••, 14•, 52] |
| Encephalomyocarditis virus (Picornaviridae) | VPg protein | No | No | [9••, 38] |
| Sendai virus (Paramyxoviridae) | PPP | Yes | Not testeda | [12, 13, 38] |
| Hantaan virus; Crimean-Congo haemorrhagic fever virus (Bunyaviridae) | P | No | Not tested | [52] |
| Borna disease virus (Bornaviridae) | P | No | Not tested | [52] |
IFIT1 inhibits viral mRNA translation of parainfluenza virus type 5, another member of the paramyxovirus family [54]. The mechanism of inhibition is unknown but is unlikely to involve 5PPP RNA binding as viral mRNAs are capped.
Secreted IFN acts in autocrine and paracrine fashion to turn on the transcription of several hundred IFN-stimulated genes (ISGs) (Figure 1) [5]. Examples include the genes encoding (i) RIG-I and other virus sensing receptors (providing a positive-feedback loop), (ii) proteins involved in cell-intrinsic anti-viral defence such as RNaseL, MxA or tetherin and (iii) molecules facilitating adaptive immune responses such as CD80 and CD86. One family of ISGs encodes the IFIT proteins that are characterized by tetratricopeptide repeats [6, 7]. IFITs control translation initiation, cell proliferation and cell migration, and exert anti-viral effects against a variety of RNA viruses and human papillomavirus [6, 7]. Interestingly, it has recently been found that some IFITs bind to 5PPP RNAs and antagonize viruses by sequestering viral RNA [8••, 9••]. Here, we review recent progress on the recognition of 5PPP RNA by RIG-I and IFITs and its subsequent impact on cell-intrinsic immunity to virus infection.
RIG-I detects 5PPP RNA
Studies on the types of RNA that can activate RIG-I have attracted much attention but have mostly been based on transfection of synthetic RNAs into reporter cells. Such experiments revealed that the most potent RIG-I agonists are RNAs that contain a triphosphate moiety and are base-paired at the 5′-end. RNA sequence is largely irrelevant so long as it does not impact on the base-pairing. A blunt end without overhangs is most effective and can be provided in trans by hybridization between two RNA molecules or in cis by complementarity between the 5′-end and 3′-end of a single RNA molecule (Figure 2a). These findings have been reviewed in detail elsewhere [10, 11] and are well supported by structural data (see below). Less is known about the types of RNA that activate RIG-I during virus infection. However, work from us and others showed that RIG-I recognizes the 5PPP RNA genome of influenza A virus (IAV) and Sendai virus in infected cells, and that both the 5PPP and the secondary structure are required for RIG-I-dependent IFN induction [12, 13]. Thus, these two studies in virus infection models nicely validate the predictions made by analysis of the RIG-I-mediated response to synthetic RNAs [12, 13]. Recent work shows that RIG-I is recruited to and activated by bunyavirus nucleocapsids, native complexes containing a 5PPP RNA genome and viral proteins [14•]. This demonstrates that RIG-I can indeed gain access to viral RNAs under physiological conditions. Like viral RNA, bacterial RNAs can possess 5PPP termini and secondary structures that are predicted to make them RIG-I agonists. Consistent with this notion, RIG-I can act as a sensor of bacterial RNA and may help maintain homeostasis to gut microbiota [15, 16].
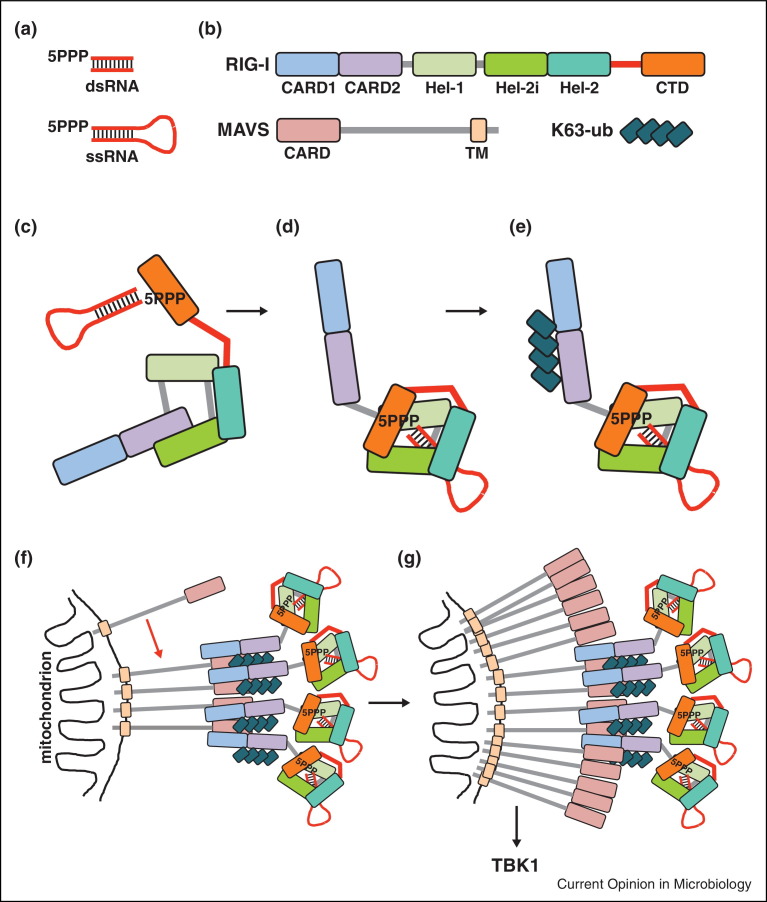
Figure 2.
Model of RIG-I activation and signalling. (a) The most potent RIG-I agonists are characterized by a 5PPP moiety and by base-pairing to a complementary stretch of RNA. This base-pairing can either be provided by a second molecule of RNA or by complementarity of the 5′-end and 3′-end of the 5PPP bearing RNA. As such, RIG-I agonists can be double-stranded (ds) and single-stranded (ss) RNAs. (b) The domain architecture of RIG-I and MAVS is shown schematically. CARD, caspase recruitment domain; CTD, C-terminal domain; TM, transmembrane domain; Hel-1, Hel-2i and Hel-2, subdomains of the RIG-I helicase domain; K63-ub, K63-linked polyubiquitin. The RIG-I pincer domain (also called bridging helices) is shown as a red line connecting the helicase domain and CTD. (c) In the autorepressed conformation, the CTD is flexibly connected to the helicase domain and this allows for binding of 5PPP groups of viral RNA genomes to the CTD. (d) Upon RNA binding to the CTD, the helicase domain makes contacts with the RNA and RIG-I undergoes a conformational change that exposes the CARDs. (e) RIG-I is then ubiquitylated or binds to free ubiquitin chains. (f and g) This facilitates RIG-I tetramerization and interaction with MAVS. Upon initial oligomerization of MAVS, a prion-like mechanism recruits additional MAVS molecules into the complex (red arrow) and signal transduction is initiated via TBK1 activation.
Interestingly, some RNAs lacking 5PPPs also trigger RIG-I upon transfection into cells. These RNAs include short double-stranded RNAs made by chemical synthesis [17, 18], certain forms of the enzymatically prepared RNA poly I:C [19] and dephosphorylated double-stranded RNAs made by in vitro transcription [20]. Further supporting the notion that non-5PPP RNAs can activate RIG-I is the observation that poly I:C triggers RIG-I signalling in a cell free reconstitution assay [21]. However, it remains unclear whether 5PPP-independent RIG-I signalling occurs in infected cells. It will be important to identify natural RNAs that activate RIG-I in cells infected with different viruses, including positive sense RNA viruses, to validate the extent to which RIG-I can be activated independently of 5PPPs.
Structural insights into RIG-I recognition
RIG-I features two caspase recruitment domains (CARDs) at its N-terminus, a central DECH-box type RNA helicase domain and a C-terminal domain (CTD) (Figure 2b) [22, 23]. The helicase domain and CTD are involved in RNA recognition while the CARDs initiate downstream signalling. Initial studies using nuclear magnetic resonance and X-ray crystallography elucidated the structure of the CTD in the absence [17, 24] and presence of RNA [25, 26, 27]. They showed that the CTD contains a positively charged pocket that can accommodate 5PPP ends of RNA molecules and thus provided a structural explanation for detection of this type of RNA [25, 26]. Much progress has been made in the last two years determining the structure of the other RIG-I domains and of the full-length protein [22, 23]. Without an RNA ligand, full-length RIG-I adopts an autorepressed state. The helicase domain is in an open conformation, flexibly linked to the CTD and bound to the second CARD [28••, 29, 30] (Figure 2c). For downstream signalling to occur, the CARDs need to be free (see below). However, the CARD2-helicase interaction observed in RNA-free RIG-I constrains this freedom and thus explains autorepression [28••, 30] (Figure 2c). Crystal structures of RIG-I bound to short double-stranded RNAs [28••, 31•, 32•] or to a 5PPP hairpin single-stranded RNA [33•] (Figure 3b) reveal how the autorepressed state is dismantled by structural rearrangement upon RNA agonist binding. The helicase adopts a closed conformation and completely surrounds the base-paired RNA along its length, while the CTD caps the 5′-end by directly interacting with the 5PPP and the first few nucleotides [28••, 31•, 32•, 33•] (Figure 2, Figure 3). The binding site for CARD2 on the helicase domain is now involved in RNA binding, thus allowing the CARDs to be released [28••, 31•, 32•, 33•]. Furthermore, in the RNA-bound conformation, a pincer domain (also called bridging helices) places the CTD in a position overlapping with that previously occupied by CARD2 [28••, 31•, 32•, 33•] (Figure 2d). RNA binding is therefore predicted to push away the CARDs. This is likely to happen in two steps: first, the CTD recognizes the 5PPP group and, second, the helicase domain binds to the base-paired region (Figure 2c,d) [22, 23]. The hydrolysis of ATP is thought to tighten the helicase domain around the RNA, further favouring CARD release [33•].
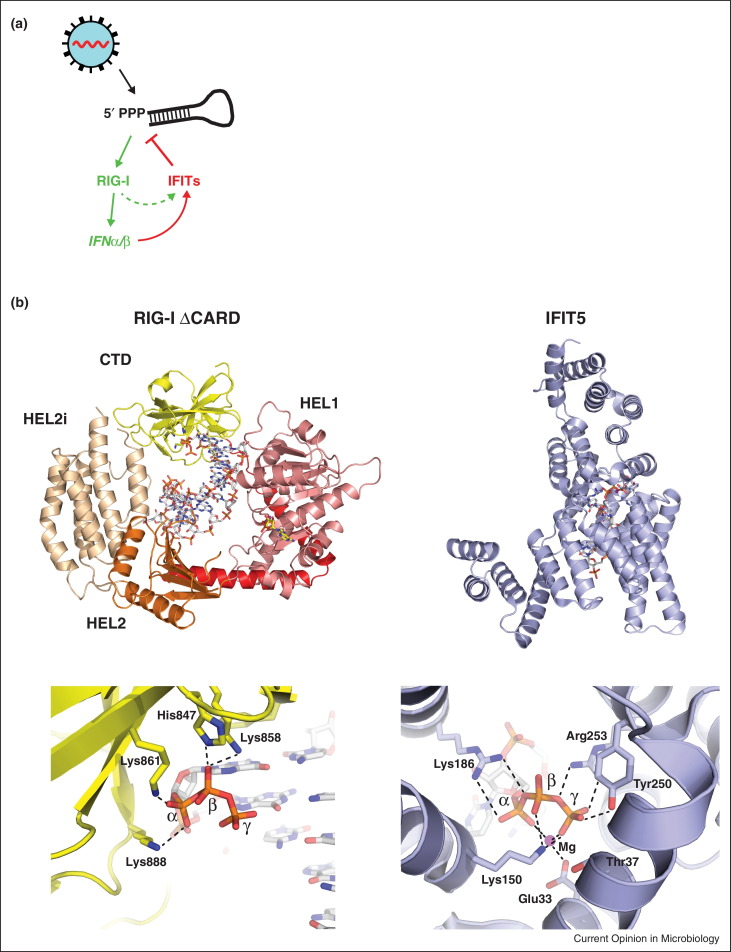
Figure 3.
RIG-I and IFITs recognize 5PPP RNA. (a) Several RNA viruses generate 5PPP RNAs, such as viral genomic RNAs. These RNAs are recognized by RIG-I that induces IFN production (green). IFNs then induce the expression of IFITs that bind and sequester 5PPP viral RNAs, preventing their translation, replication and/or packaging (red). Note that IFITs can also be induced directly by RIG-I/MAVS (dashed arrow) [43]. (b) The top row shows overall structures of RIG-I and IFIT5 in complex with 5PPP RNA; the bottom row zooms in on the 5PPP binding sites. The RIG-I structure is the RIG-I ΔCARD plus 5PPP RNA hairpin complex described in [33•] (PDB code 4AY2). IFIT5 is shown in complex with 5PPP-oligo-C [8••] (PDB code 4HOR). Amino acid residues contacting the 5PPP are shown in stick format and dashed lines indicate hydrogen-bonding interactions.
MDA5 is another RLR. A recent crystal structure of the MDA5 helicase domain and CTD in complex with double-stranded RNA shows that these domains form a ring around the RNA, leaving both ends free to extend [34••]. This is in contrast to the equivalent domains in RIG-I that cap one end of 5PPP base-paired RNA, due to tilting of the RIG-I CTD towards the RNA [34••]. This structural difference, together with the absence of a 5PPP-binding pocket in the CTD of MDA5 [25, 34••, 35, 36], may explain why the two RLRs recognize different types of RNA and mediate IFN responses to different viruses [37, 38].
RIG-I signalling via MAVS
Upon RNA agonist binding and CARD exposure, RIG-I initiates a signal transduction cascade that ultimately results in IFN gene transcription [3]. MAVS is the RIG-I and MDA5 proximal adaptor protein. It mediates activation of kinases such as TBK1, which phosphorylate transcription factors including IRF3 that then translocate to the cell nucleus to trigger transcription of IFN genes (Figure 1). MAVS has a C-terminal transmembrane domain and a single N-terminal CARD, which engages in CARD-CARD interactions with RIG-I. This interaction requires binding to K63-linked polyubiquitin chains generated by the ubiquitin ligase TRIM25 (Figure 2e) [21, 39, 40•]. One study suggests that TRIM25 mediates covalent ubiquitylation of RIG-I at lysine 172 in the second RIG-I CARD [39]. A different study indicates that the same lysine binds non-covalently to TRIM25-generated free ubiquitin chains [21]. CARD-associated ubiquitin chains are key for signalling as they trigger formation of RIG-I tetramers (Figure 2f) [40•]. The resulting CARD clusters recruit multiple MAVS molecules [40•, 41••]. Surprisingly, the stoichiometry of this reaction reveals signal amplification through self-propagation (Figure 2f,g). MAVS forms fibrils that convert additional MAVS molecules into the fibrillar conformation, in a self-amplification mode reminiscent of prions [41••]. The MAVS fibrils in turn mediate activation of TBK1 [41••] (Figure 2g). A subsequent study confirmed the prion-like behaviour of MAVS and showed that it can also be triggered by the CARDs of MDA5 [34••]. Self-propagation allows for sensitive detection of minute amounts of viral RNA: it has been estimated that 20 molecules of 5PPP RNA are sufficient to activate IRF3 via the RIG-I/MAVS pathway [21]. An important question for future studies is how signalling is terminated and how the self-propagating prion conformation of MAVS is cleared. Another area for future research is the spatiotemporal regulation of RLR signalling and how it is governed by the localization of MAVS [42]. Interestingly, MAVS has been found on mitochondria, peroxisomes and so-called MAMs (mitochondrial-associated endoplasmic reticulum membranes) [43, 44]. Much remains to be learned about how MAVS localization is regulated and how this impacts on RLR signalling during infection with different viruses.
IFIT proteins sequester viral 5PPP RNA
The human genome encodes four IFITs (IFIT1-3, IFIT5) whereas mice have three (IFIT1-3) [6, 7]. The expression of these proteins is potently induced by IFN and their anti-viral function has been reviewed elsewhere [6, 7]. Recent functional and structural data indicate that the anti-viral effect of IFIT1 and IFIT5 is due to their ability to recognize 5PPP RNA. A proteomics study identified all four human IFITs in a pull-down with synthetic 5PPP RNA as bait [9••]. IFIT1 and IFIT5 directly bound to the RNA, whereas IFIT2 and IFIT3 did so indirectly, through association with IFIT1 [9••]. Three viruses that produce 5PPP RNA (vesicular stomatitis virus (VSV), rift valley fever virus and IAV) showed enhanced replication in human or mouse cells depleted of IFIT1 whereas replication of encephalomyocarditis virus (EMCV) that lacks 5PPPs was normal [9••]. What is the molecular basis of IFIT1 mediated inhibition of virus replication? In vitro assays showed that recombinant IFIT1 blocks translation of 5PPP containing reporter RNAs, while having little effect on translation of the corresponding dephosphorylated RNA [9••]. IFIT1 also precipitated viral RNAs from cells infected with VSV or IAV [9••]. Together, these observations suggest that IFIT1 recognizes and sequesters viral 5PPP RNAs and thereby prevents their replication, packaging and/or translation.
Structural data demonstrate that human IFIT5 has a positively charged cavity accommodating 5PPP RNA [8••] (Figure 3b). Structure-guided mutation of residues involved in 5PPP RNA binding diminishes the anti-viral effects of IFIT5 and IFIT1 against VSV and IAV [8••, 9••]. Similar to 5PPP RNA recognition by RIG-I, the interaction between IFIT5 and RNA is not sequence specific [8••, 22, 23]. However, while RIG-I is most strongly activated by blunt ended 5PPP base-paired RNA [11], a 5′-overhang is required for RNA to be accommodated in IFIT5 [8••]. Whether this reflects binding of different types of RNA to IFITs and RIG-I or whether base-paired RIG-I agonists are unwound for IFIT recognition remains to be determined.
Genetically modified mice provide further evidence for a role of IFITs in host defence against viruses [9••, 45, 46, 47]. IFIT2-deficient animals are highly susceptible to intranasal VSV challenge [46], and similar data have been reported for Ifit1 −/− mice [9••]. It is unclear why another study using IFIT1-deficient mice independently derived from the same ES cell line failed to reproduce these results [9••, 46]. Interestingly, Ifit1 −/− mice and cells are more susceptible to infection with viruses lacking 2′-O-methyltransferases, including mutants of West Nile virus, coronavirus and poxvirus [45, 47, 48]. The 2′-O-methyltransferases encoded by the wild-type counterparts of these mutant strains modify the cap structure of viral messenger RNA by 2′-O-methylation. Cellular messenger RNAs are methylated at this position by a host-encoded methyltransferase, suggesting that some viruses mimic this to circumvent recognition by IFIT1 and, possibly, MDA5 [45, 47, 48]. This indicates that IFIT1 may interact not only with 5PPP RNA, but also with capped RNA lacking 2′-O-methyl groups, although the latter interaction could be indirect, perhaps via other IFITs. Indeed, a large spectrum of different RNAs may be targeted by IFITs: IFIT5 not only binds to 5PPP RNA but also to transfer RNAs that bear a single 5′-phosphate [49•] and IFIT2 interacts with AU-rich RNAs independently of 5′-phosphorylation [50]. It will be important to identify and characterize RNAs bound by IFITs in virally infected cells to further understand the role of these proteins in immune responses to virus infection.
Conclusions
The IFN system operates in two phases: first, virus presence is detected, resulting in the expression of IFN; second, IFN induces ISGs that exert anti-viral effects (Figure 1). Recent work shows that the IFN system targets 5PPP RNAs during both phases: both RIG-I, a virus sensor that induces IFN expression, and IFITs, effector molecules that execute anti-viral activities, can specifically recognize 5PPP RNAs. As such, 5PPP RNAs appear to be Achilles’ heel of many RNA viruses in their interaction with the innate immune system (Figure 3a). Why is this the case? Many RNA viruses use primer-independent means of replication. 5PPP moieties on viral RNA genomes, antigenomes and some viral transcripts are an inevitable consequence of primer-independent initiation of RNA synthesis by a single ribonucleoside-triphosphate [51]. The fundamental importance of RNA replication to the life cycle of RNA viruses may explain why viruses that use primer-independent strategies to initiate RNA synthesis have been forced to maintain 5PPP groups during evolution — despite the selective pressure exerted by the IFN system that detects these moieties. It is noteworthy that other mechanisms to initiate RNA synthesis and the removal of 5PPP groups have evolved in some virus families. For example, EMCV uses as primer for RNA replication a protein known as VPg, which becomes covalently attached to the 5′-end of the viral genome [51]. Hantaan virus, Crimean-Congo haemorrhagic fever virus and Borna disease virus post-transcriptionally process the 5′-end of their genomes to leave a mono-phosphate [52]. Consistent with these facts, none of these viruses trigger IFN induction via RIG-I and EMCV is not restricted by IFIT1 [9••, 38, 52] (Table 1). Notably, viruses that maintain 5PPP RNA and are recognized by RIG-I often encode proteins that specifically target and inhibit RLR signalling [53]. Studies of the sensors that detect RNA viruses and the identification of ISGs that restrict them remain challenges for the future.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We thank Andreas Pichlmair, Delphine Goubau and Safia Deddouche for their helpful comments, and Martin Jinek for his help in preparing Figure 3. We apologize to our colleagues whose work could not be cited due to space limitations. CRS is funded by Cancer Research UK, ERC and Foundation Bettencourt-Schueller. JR is funded by the UK Medical Research Council.
Contributor Information
Jan Rehwinkel, Email: jan.rehwinkel@imm.ox.ac.uk.
Caetano Reis e Sousa, Email: caetano@cancer.org.uk.
References
- 1.Isaacs A., Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- 2.Pestka S., Krause C.D., Walter M.R. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 3.Pichlmair A., Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Crow Y.J. Type I interferonopathies: a novel set of inborn errors of immunity. Ann N Y Acad Sci. 2011;1238:91–98. doi: 10.1111/j.1749-6632.2011.06220.x. [DOI] [PubMed] [Google Scholar]
- 5.Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., Rice C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fensterl V., Sen G.C. The ISG56/IFIT1 gene family. J Interferon Cytokine Res. 2011;31:71–78. doi: 10.1089/jir.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diamond M.S., Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8••.Abbas Y.M., Pichlmair A., Gorna M.W., Superti-Furga G., Nagar B. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature. 2013;494:60–64. doi: 10.1038/nature11783. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reveals the structural basis of how IFITs interact with 5PPP RNA.
- 9••.Pichlmair A., Lassnig C., Eberle C.A., Gorna M.W., Baumann C.L., Burkard T.R., Burckstummer T., Stefanovic A., Krieger S., Bennett K.L. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]; This paper reports the discovery that IFIT1 binds viral 5PPP RNAs to exert an anti-viral effect.
- 10.Rehwinkel J., Reis e Sousa C. RIGorous detection: exposing virus through RNA sensing. Science. 2010;327:284–286. doi: 10.1126/science.1185068. [DOI] [PubMed] [Google Scholar]
- 11.Schlee M., Hartmann G. The chase for the RIG-I ligand — recent advances. Mol Ther. 2010;18:1254–1262. doi: 10.1038/mt.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baum A., Sachidanandam R., Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A. 2010;107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rehwinkel J., Tan C.P., Goubau D., Schulz O., Pichlmair A., Bier K., Robb N., Vreede F., Barclay W., Fodor E. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 14•.Weber M., Gawanbacht A., Habjan M., Rang A., Borner C., Schmidt Anna M., Veitinger S., Jacob R., Devignot S., Kochs G. Incoming RNA virus nucleocapsids containing a 5′-triphosphorylated genome activate RIG-I and antiviral signaling. Cell Host Microbe. 2013;13:336–346. doi: 10.1016/j.chom.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reports that RIG-I is activated by bunyavirus 5PPP RNA genomes contained in nucleocapsids, which demonstrates that this innate receptor can sense viral RNA in its native complex with viral proteins.
- 15.Li X.D., Chiu Y.H., Ismail A.S., Behrendt C.L., Wight-Carter M., Hooper L.V., Chen Z.J. Mitochondrial antiviral signaling protein (MAVS) monitors commensal bacteria and induces an immune response that prevents experimental colitis. Proc Natl Acad Sci U S A. 2011;108:17390–17395. doi: 10.1073/pnas.1107114108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdullah Z., Schlee M., Roth S., Mraheil M.A., Barchet W., Bottcher J., Hain T., Geiger S., Hayakawa Y., Fritz J.H. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J. 2012;31:4153–4164. doi: 10.1038/emboj.2012.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahasi K., Yoneyama M., Nishihori T., Hirai R., Kumeta H., Narita R., Gale M., Jr., Inagaki F., Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 18.Marques J.T., Devosse T., Wang D., Zamanian-Daryoush M., Serbinowski P., Hartmann R., Fujita T., Behlke M.A., Williams B.R. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 19.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T.S., Fujita T., Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binder M., Eberle F., Seitz S., Mucke N., Huber C.M., Kiani N., Kaderali L., Lohmann V., Dalpke A., Bartenschlager R. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid-inducible gene-I (RIG-I) J Biol Chem. 2011;286:27278–27287. doi: 10.1074/jbc.M111.256974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A., Xu M., Chen Z.J. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolakofsky D., Kowalinski E., Cusack S. A structure-based model of RIG-I activation. RNA. 2012;18:2118–2127. doi: 10.1261/rna.035949.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Q.X., Chen Z.J. Structural insights into the activation of RIG-I, a nanosensor for viral RNAs. EMBO Rep. 2012;13:7–8. doi: 10.1038/embor.2011.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui S., Eisenacher K., Kirchhofer A., Brzozka K., Lammens A., Lammens K., Fujita T., Conzelmann K.K., Krug A., Hopfner K.P. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H., Juranek S., Sheng G., Micura R., Tuschl T. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu C., Xu H., Ranjith-Kumar C.T., Brooks M.T., Hou T.Y., Hu F., Herr A.B., Strong R.K., Kao C.C., Li P. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu C., Ranjith-Kumar C.T., Hao L., Kao C.C., Li P. Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5′ triphosphate. Nucleic Acids Res. 2011;39:1565–1575. doi: 10.1093/nar/gkq974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Kowalinski E., Lunardi T., McCarthy A.A., Louber J., Brunel J., Grigorov B., Gerlier D., Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]; This paper describes the structure of full-length RIG-I and reveals an important interaction between CARD2 and the helicase domain that explains RIG-I autorepression.
- 29.Civril F., Bennett M., Moldt M., Deimling T., Witte G., Schiesser S., Carell T., Hopfner K.P. The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling. EMBO Rep. 2011;12:1127–1134. doi: 10.1038/embor.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferrage F., Dutta K., Nistal-Villan E., Patel J.R., Sanchez-Aparicio M.T., De Ioannes P., Buku A., Aseguinolaza G.G., Garcia-Sastre A., Aggarwal A.K. Structure and dynamics of the second CARD of human RIG-I provide mechanistic insights into regulation of RIG-I activation. Structure. 2012;20:2048–2061. doi: 10.1016/j.str.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31•.Jiang F., Ramanathan A., Miller M.T., Tang G.Q., Gale M., Jr., Patel S.S., Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]; These studies report the crystal structure of the RIG-I helicase domain and CTD in complex with RNA and provide insights into the conformation of the activated receptor.
- 32•.Luo D., Ding S.C., Vela A., Kohlway A., Lindenbach B.D., Pyle A.M. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref. [31•].
- 33•.Luo D., Kohlway A., Vela A., Pyle A.M. Visualizing the determinants of viral RNA recognition by innate immune sensor RIG-I. Structure. 2012;20:1983–1988. doi: 10.1016/j.str.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref. [31•].
- 34••.Wu B., Peisley A., Richards C., Yao H., Zeng X., Lin C., Chu F., Walz T., Hur S. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]; This work on the structure of the MDA5 helicase domain and CTD highlights similarities and differences between RIG-I and MDA5 that may explain why these RLRs recognize different types of RNA.
- 35.Li X., Lu C., Stewart M., Xu H., Strong R.K., Igumenova T., Li P. Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5. Arch Biochem Biophys. 2009;488:23–33. doi: 10.1016/j.abb.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Takahasi K., Kumeta H., Tsuduki N., Narita R., Shigemoto T., Hirai R., Yoneyama M., Horiuchi M., Ogura K., Fujita T. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J Biol Chem. 2009;284:17465–17474. doi: 10.1074/jbc.M109.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rehwinkel J. Exposing viruses: RNA patterns sensed by RIG-I-like receptors. J Clin Immunol. 2010;30:491–495. doi: 10.1007/s10875-010-9384-7. [DOI] [PubMed] [Google Scholar]
- 38.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 39.Gack M.U., Shin Y.C., Joo C.H., Urano T., Liang C., Sun L., Takeuchi O., Akira S., Chen Z., Inoue S. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 40•.Jiang X., Kinch L.N., Brautigam C.A., Chen X., Du F., Grishin N.V., Chen Z.J. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that activated RIG-I forms tetramers and that non-covalent binding of RIG-I and MDA5 to K63-linked ubiquitin chains is required for signaling to MAVS.
- 41••.Hou F., Sun L., Zheng H., Skaug B., Jiang Q.X., Chen Z.J. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that MAVS forms prion-like aggregates that activate TBK1 and thus reveals an unexpected mechanism of signal amplification in the RIG-I pathway.
- 42.Kagan J.C. Defining the subcellular sites of innate immune signal transduction. Trends Immunol. 2012;33:442–448. doi: 10.1016/j.it.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dixit E., Boulant S., Zhang Y., Lee A.S., Odendall C., Shum B., Hacohen N., Chen Z.J., Whelan S.P., Fransen M. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horner S.M., Liu H.M., Park H.S., Briley J., Gale M., Jr. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.Y., Schneller S., Zust R., Dong H. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fensterl V., Wetzel J.L., Ramachandran S., Ogino T., Stohlman S.A., Bergmann C.C., Diamond M.S., Virgin H.W., Sen G.C. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog. 2012;8:e1002712. doi: 10.1371/journal.ppat.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szretter K.J., Daniels B.P., Cho H., Gainey M.D., Yokoyama W.M., Gale M., Jr., Virgin H.W., Klein R.S., Sen G.C., Diamond M.S. 2′-O methylation of the viral mRNA cap by West Nile virus evades ifit1-dependent and -independent mechanisms of host restriction in vivo. PLoS Pathog. 2012;8:e1002698. doi: 10.1371/journal.ppat.1002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zust R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49•.Katibah G.E., Lee H.J., Huizar J.P., Vogan J.M., Alber T., Collins K. tRNA binding, structure, and localization of the human interferon-induced protein IFIT5. Mol Cell. 2013;49:743–750. doi: 10.1016/j.molcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using biochemical and structural analysis, this work shows that IFIT5 also binds transfer RNAs bearing 5′-monophosphates, in addition to viral 5PPP RNAs [8••, 9••].
- 50.Yang Z., Liang H., Zhou Q., Li Y., Chen H., Ye W., Chen D., Fleming J., Shu H., Liu Y. Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms. Cell Res. 2012;22:1328–1338. doi: 10.1038/cr.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knipe D.M., Howley P.M. edn 5. Lippincott Williams & Wilkins; Philadelphia: 2007. Fields Virology. [Google Scholar]
- 52.Habjan M., Andersson I., Klingstrom J., Schumann M., Martin A., Zimmermann P., Wagner V., Pichlmair A., Schneider U., Muhlberger E. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS One. 2008;3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Versteeg G.A., Garcia-Sastre A. Viral tricks to grid-lock the type I interferon system. Curr Opin Microbiol. 2010;13:508–516. doi: 10.1016/j.mib.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andrejeva J., Norsted H., Habjan M., Thiel V., Goodbourn S., Randall R.E. ISG56/IFIT1 is primarily responsible for interferon-induced changes to patterns of parainfluenza virus type 5 transcription and protein synthesis. J Gen Virol. 2013;94:59–68. doi: 10.1099/vir.0.046797-0. [DOI] [PMC free article] [PubMed] [Google Scholar]