Abstract
Bovine toroviruses (BToVs), belong to the subfamily Toroviridae within the family Coronaviridae, and are pathogens, causing enteric disease in cattle. In Japan, BToVs are distributed throughout the country and cause gastrointestinal infection of calves and cows. In the present study, complete genome sequences of two Japanese BToVs and partial genome sequences of two Japanese BToVs and one porcine torovirus (PToV) from distant regions in Japan were determined and genetic analyses were performed. Pairwise nucleotide comparison and phylogenetic analyses revealed that Japanese BToVs shared high identity with each other and showed high similarities with BToV Breda1 strain in S, M, and HE coding regions. Japanese BToVs showed high similarities with porcine toroviruses in ORF1a, ORF1b, and N coding regions and the 5′ and 3′ untranslated regions, suggestive of a natural recombination event. Recombination analyses mapped the putative recombinant breakpoints to the 3′ ends of the ORF1b and HE regions. These findings suggest that the interspecies recombinant nature of Japanese BToVs resulted in a closer relationship between BToV Breda1 and PToVs.
Keywords: Bovine torovirus, Japan, Natural recombination, Porcine torovirus
Highlights
-
•
Recombination events between porcine and bovine torovirus were identified.
-
•
Recombinant breakpoints were mapped at ORF1b and HE coding regions.
-
•
These recombinant viruses are prevalent throughout Japan.
1. Introduction
Toroviruses (ToVs), belonging to the subfamily Toroviridae, are members of the family Coronaviridae, order Nidovirales (Cavanagh and Horzinek, 1993). ToVs have a single-stranded positive-sense RNA genome of nearly 28 kb, containing two large open reading frames (ORFs) encoding nonstructural proteins, ORF1a and ORF1ab, and four structural proteins, spike glycoprotein (S), membrane glycoprotein (M), hemagglutinin esterase (HE), and nucleocapsid phosphoprotein (N) (Draker et al., 2006, Sun et al., 2014).
ToVs are identified in various animals and humans and are thought to cause diarrheic and respiratory diseases (Duckmanton et al., 1997, Ito et al., 2009, Kroneman et al., 1998, Uziel et al., 1999, Vanopdenbosch et al., 1991, Woode et al., 1982). Berne virus (EToV Berne), the first isolated ToV, was isolated from a horse with diarrhea in 1972 in Switzerland (Weiss et al., 1983). In 1979, a bovine ToV (BToV), named Breda virus, was detected in calves with diarrhea in the United States (US). BToV Breda causes enteric disease in gnotobiotic reared calves (Woode et al., 1982). BToVs are found throughout the world including North America (Hoet et al., 2002, Hoet et al., 2003), Central America (Pérez et al., 1998), Europe (Haschek et al., 2006, Koopmans et al., 1991, Matiz et al., 2002), Asia (Aita et al., 2012, Ito et al., 2010, Park et al., 2008) and South Africa (Penrith and Gerdes, 1992). BToVs have been detected in 2.9–36.4% of fecal samples obtained from cattle with diarrhea (Duckmanton et al., 1998, Hoet et al., 2003, Ito et al., 2007, Kirisawa et al., 2007, Nogueira et al., 2013, Park et al., 2008). Porcine ToV (PToV) is also prevalent in piglets worldwide; however their pathogenicity in swine remains unclear (Anbalagan et al., 2014, Shin et al., 2010, Sun et al., 2014).
So far, only three whole genome sequences of ToV, namely, BToV Breda1 (AY427798) (Draker et al., 2006) and PToV NPL/2014 (KM403390), which was identified in the US in 2014 (Anbalagan et al., 2014) and PToV SH1 (JQ860350), which was identified in China in 2010 (Sun et al., 2014), are available on the database. To gain more information about genetic diversity, relationship, and evolution of ToVs, we performed whole genome analysis of Japanese BToVs and PToV. Our data showed natural interspecies recombination events of Japanese BToVs, which originated from genetic recombination of BToV Breda1 and PToV strains.
2. Material and methods
2.1. Viruses
Four BToVs and one PToV were studied. BToV Ishikawa/2010 (BToV Ishi) was isolated using HRT-18-Aich cells (Aita et al., 2012, Kuwabara et al., 2007) from a fecal sample of a cow with diarrhea in Ishikawa Prefecture in 2010 (Ito et al., 2012). BToV Kagoshima/2014 (BToV Kago), BToV Tochigi/2013 (BToV Tochi), and BToV Tokyo/2014 (BToV Tokyo) were detected in the course of metagenomics of fecal samples obtained from 18-, 12-, and 16-day-old calves with diarrhea in Kagoshima, Tochigi, and Tokyo Prefecture in 2014, 2013, and 2014, respectively. PToV Tottori/2015 (PToV Tottori) was identified in the course of metagenomics in fecal samples collected from a healthy two-month-old pig in 2015 in Tottori Prefecture.
2.2. Whole genome sequencing
Since viruses could not be isolated from samples using HRT-18-Aich cells by repeating passage thrice, fecal suspensions (20% v/v in sterile phosphate-buffered saline) of BToV Kago, BToV Tochi, BToV Tokyo, and PToV Tottori were used for RNA extraction. Viral RNA was extracted from 0.25 mL supernatant of BToV Ishi culture (105.3 TCID50/mL) or 0.25 mL fecal suspensions by using TRIzol® LS Reagent (Life Technologies, Carlsbad, CA, USA), followed by treatment of the RNA with DNase I (TaKaRa Bio Inc., Shiga, Japan). cDNA library was constructed using NEBNext® Ultra RNA Library Prep Kit for Illumina version 2.0 (New England Biolabs, Ipswich, MA, USA), as described previously (Nagai et al., 2015). The libraries obtained were loaded onto a MiSeq cartridge (MiSeq Reagent Kit V2 (300 cycles); Illumina, San Diego, CA, USA) and sequenced using a MiSeq bench-top sequencer (Illumina) with 151 paired-end reads. To obtain complete genome sequence of BToV Ishi and BToV Kago, rapid amplification of cDNA end method (RACE) (5′-Full RACE Core Set and 3′-Full RACE Core Set; TaKaRa Bio, Otsu, Japan) was employed.
2.3. Genome analysis
The sequence data were collected using the MiSeq Reporter program (Illumina) to generate reads in FASTQ format. Collected reads were trimmed and assembled into contigs by de novo assembly using CLC Genomics Workbench 6.5.1 (CLC bio, Aarhus, Denmark). The sequences were aligned using ClustalW in MEGA5.22 (Tamura et al., 2011). Pairwise sequence identity calculations were performed using CLC Genomics Workbench 6.5.1 (CLC bio). The phylogenetic tree was constructed by the maximum likelihood method statistically supported by bootstrapping with 1000 replicates by using MEGA5.22. Recombination analysis was performed using SimPlot software v. 3.5.1 (Lole et al., 1999) and the Recombination Detection Program (RDP) v. 4.58 (Martin and Rybicki, 2000, Martin et al., 2005).
3. Results
3.1. Deep sequencing and determination of whole genome sequences
We performed deep sequencing using the Illumina MiSeq sequencing system. The total ToV sequence read counts (percentage of ToV sequence reads: ToV sequence reads/total reads) of BToV Ishi, BToV Kago, BToV Tochi, BToV Tokyo, and PToV Tottori samples were 198,526 (13.2%), 31,685 (2.4%), 492 (0.04%), 1527 (0.4%) and 1805 (0.05%), respectively. An approximately 28-kb contig was obtained from BToV Ishi and BToV Kago samples with sufficient average sequence read depth of 1007 (maximum depth: 2384) and 162 (maximum depth: 320), respectively; however, large contigs were not obtained from BToV Tochi, BToV Tokyo, and PToV Tottori samples. Since nearly complete sequences of contigs were obtained, complete genome length of BToV Ishi and BToV Kago were determined using the 5′ and 3′ RACE method. The complete genome length of BToV Ishi and BToV Kago excluding poly(A) were 28,309 nucleotides (nt) and 28,301 nt, respectively, which were similar to the length of published sequences of ToVs; BToV Breda1 (28,475 nt), PToV NPL/2014 (28,305 nt), and PToV SH1 (28,301 nt). The nucleotide sequence identities among complete genome sequences of BToV Ishi and BToV Breda1, PToV NPL/2014, PToV SH1, and BToV Kago were 82.3%, 86.3%, 85.3%, and 97.6%, respectively. The sequences of BToV Ishi and BToV Kago were deposited in the DNA Data Bank of Japan, DDBJ/EMBL/GenBank database under the accession numbers LC088094 and LC088095, respectively.
3.2. Pairwise nucleotide sequence identity comparison
Pairwise alignment for comparing nucleotide sequences of BToV Ishi and BToV Kago to other BToV and PToVs was performed using the whole genomic region of the 5′ untranslated region (UTR), ORF1a, ORF1b, S, M, HE, N, and the 3′UTR (Table 1 ). BToV Ishi shared high sequence identities (96.8–99.4%) with BToV Kago for all genomic regions analyzed. Japanese BToVs showed high identities with PToVs in 5′UTR, ORF1a, ORF1b, N and 3′UTR (86.9–95.1%) and showed low identities with PToVs in S, M, and HE (70.2–80.3%). On the other hand, Japanese BToVs showed high identities with BToV Breda1 in S, M, and HE (87.9–95.7%) and showed low identities with BToV Breda1 in 5′UTR, ORF1a, ORF1b, N and 3′UTR (68.9–83.0%). These results suggest the occurrence of recombination events between BToV Breda1 and PToVs.
Table 1.
Pairwise nucleotide sequence comparison of BToV Ishikawa/2010 and BToV Kagoshima/2014 to the other BToV and PToVs.
| Pairwise nucleotide identity (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| BToV Ishikawa/2010 |
BToV Kagoshima/2014 |
||||||||
| PToV-NPL/2014 | PToV SH1 | BToV Breda | EToV Berne | BToV Kagoshima/2015 | PToV-NPL/2014 | PToV SH1 | BToV Breda | EToV Berne | |
| 5′UTR | 91.7 | 92.0 | 75.4 | 63.3 | 99.4 | 91.7 | 92.3 | 75.6 | 63.4 |
| ORF1a | 88.7 | 86.9 | 77.0 | 67.3 | 97.3 | 88.5 | 86.9 | 76.9 | 67.2 |
| ORF1b | 92.7 | 91.9 | 83.0 | 81.1 | 98.2 | 92.6 | 91.8 | 82.8 | 81.2 |
| S | 73.4 | 73.2 | 95.7 | 75.7 | 96.9 | 73.6 | 73.4 | 95.3 | 75.6 |
| M | 79.6 | 80.3 | 94.3 | 84.3 | 98.6 | 78.8 | 79.5 | 94.0 | 84.5 |
| HE | 70.8 | 71.7 | 88.3 | –a | 96.8 | 70.2 | 71.2 | 87.9 | –a |
| N | 92.9 | 90.2 | 69.1 | 67.1 | 98.2 | 93.1 | 90.2 | 69.1 | 67.3 |
| 3′UTR | 95.1 | 90.8 | 68.9 | 66.9 | 98.8 | 93.9 | 90.8 | 68.9 | 66.9 |
The complete HE sequence of EToV Berne strain is not available.
3.3. Recombination analysis
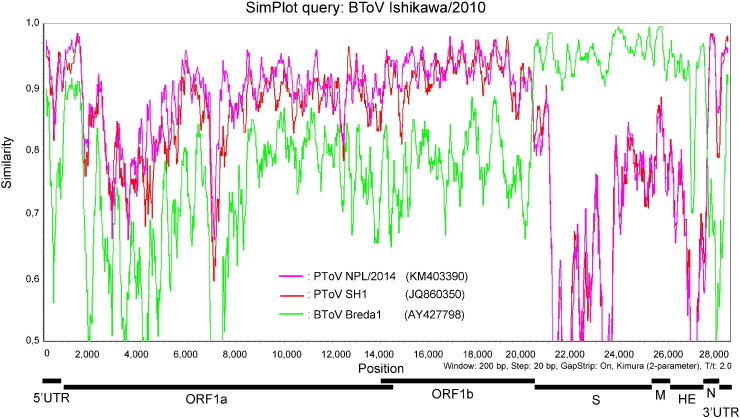
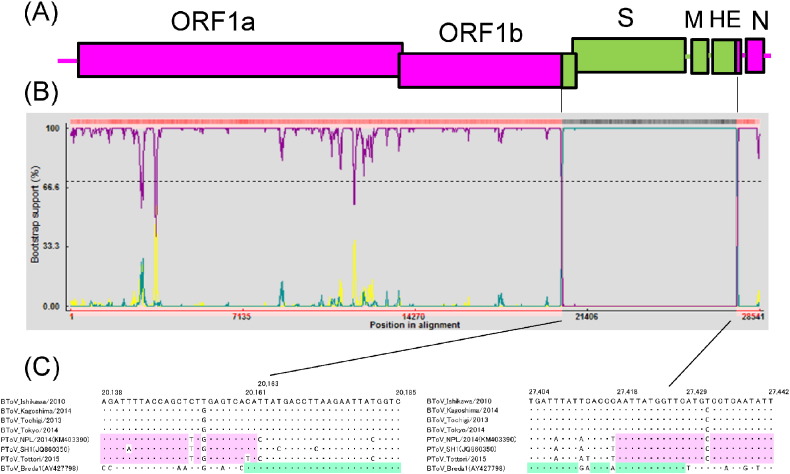
To investigate the recombination events, the complete genomes of BToV Ishi, BToV Kago, PToV NPL/2014, PToV SH1, and BToV Breda1 were aligned using ClustalW program in MEGA5.22 and standard similarity plot analysis was performed using SimPlot software v. 3.5.1 with BToV Ishi (Fig. 1 ) and BToV Kagoshima/2014 sequences as separate queries. Both SimPlot graph indicated that the sequences of Japanese BToV had high similarity with those of PToVs except in the 3′ end of ORF1b, S, M, and most of the HE coding regions, which had high similarity with that of BToV Breda1. The bootscanning analysis using RDP v. 4.58 was performed to identify the presumed recombinant breakpoints. Schema of ToV genome structure is shown in Fig. 2 . A. Beginning and end breakpoint positions were mapped near the 3′ of ORF1b and HE regions, respectively (Fig. 2B). At the starting putative recombination breakpoint, 25-nt sequences of Japanese BToVs corresponding to nt 20,138–20,162 of BToV Ishi showed low identities (72–76%) with that of BToV Breda1, whereas 25 nt sequences of Japanese BToVs corresponding to nt 20,161–20,185 showed high identities (100%) with that of BToV Breda1. At the end putative recombination breakpoint, 25 nt sequences of Japanese BToV corresponding to nt 27,404–27,428 of BToV Ishi showed low identities (84–88%) with those of PToVs; however, the 25-nt Japanese BToV sequences corresponding to nt 27,418 to 27,442 showed high identities (96–100%) with those of PToVs (Fig. 2C).
Fig. 1.
Similarity plot analyses of the entire genome length of PToV NPL/2013 (pink line), PToV SH1 (red line), BToV Breda1 (green line), and BToV Ishikawa/2010 (A), BToV Kagoshima/2014 (B), and BToV Tochigi/2–13 (C) as query sequences, using a sliding window of 200 nt and a moving step size of 20 nt.
Fig. 2.
Detection of recombination breakpoint in Japanese BToV genomes. (A) Genome organization of ToV. Presumed recombinant breakpoints were mapped to the 3′ end of ORF1b and of HE coding regions. Pink boxes indicate sequence regions originating from PToV. Green boxes indicate sequence regions originating from BToV Breda1 strain.(B) Bootscan analysis of BToV Ishikawa/2010 vs PToV NPL/2013 (purple line), BToV Ishikawa/2010 vs BToV Breda1 (green line) and PToV NPL/2013 vs BToV Breda1 (yellow line). Cut-off of the bootstrapping test (> 70%) is indicated by the break-point. (C) Sequence of recombination junctions of Japanese BToVs. Dots indicate sequences similar to that of BToV Ishikawa/2010. Identities between Japanese BToVs and PToV (NPL/2013 and SH1) are highlighted in pink; identities between Japanese BToV and BToV Breda1 are highlighted in green.
3.4. Phylogenetic analysis
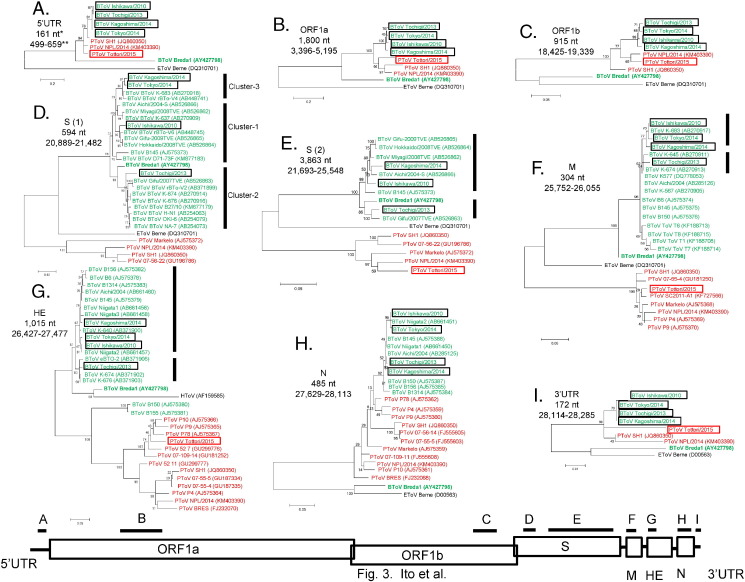
For further confirmation of the recombination event, phylogenetic trees of nine genomic regions were constructed using nucleotide sequences of BToVs and PToV determined in this study together with ToV and PToV genome sequences available in GenBank. Although the genome sequences of BToV Tochi, BToV Tokyo, and PToV Tottori had gaps owing to insufficient sequence read counts, sequence regions with no gap and corresponding to BToV Ishi were used: 5′UTR (161 nt, 499–659), ORF1a (1800 nt, 3396–5195), ORF1b (915 nt, 18,425–19,339), S (594 nt, 20,889–21,482 and 3863 nt, 21,693–25,548), M (304 nt, 25,752–26,055), HE (1015 nt, 26,427–27,477), N (485 nt, 27,629–28,113), and 3′UTR (172 nt, 28,114–28,285). However, we could not obtain the sequences of regions S (20,889–21,482) of PToV Tottori, S (21,693–25,548) of BToV Tokyo, and N (27,629–28,113) of PToV Tottori. The phylogenetic trees of S and N coding regions were constructed without these strains. The four Japanese BToVs branched separately from BToV Breda1, and clustered with PToV strains in the 5′UTR, ORF1a, ORF1b, N, and 3′UTR phylogenetic trees (Fig. 3 A-C, H and I). In addition, Japanese BToVs in this study together with other BToV strains from Japan and other countries selected from GenBank, clustered with BToV Breda1 but not with PToVs in the S, M, and HE phylogenetic trees (Fig. 3D-G). Ito et al. classified Japanese BToVs using 5′ portion of S coding region into three genotypes, Clusters 1–3 (Ito et al., 2007, Ito et al., 2009, and 2010). In the phylogenetic tree, the 5′ portion of S coding region of BToVs in this study showed that BToV Ishi and BToV Tochi clustered with Cluster-1 and Cluster-2 strains, respectively, while BToV Kago and BToV Tokyo clustered with Cluster-3 strain, though enough bootstrap support could not be obtained. (Fig. 3D). BToV Tochi separately clustered with BToV Ishi, BToV Kago, and BToV Tokyo in the S (21,693–25,548), M, and HE phylogenetic trees (Fig. 3E-G).
Fig. 3.
Phylogenetic trees based on sequences of 5′UTR (A), ORF1a (B), ORF1b (C), S (D, E), M (F), HE (G), N (H), and 3′UTR (I). Phylogenetic trees were constructed using the maximum likelihood method in MEGA5.22 with bootstrap values (1000 replicates). Scale bar indicates nucleotide substitutions per site. The BToV strains are represented by green (BToV Breda1 is indicated by boldface), whereas the PToV strains are represented by red. The BToV strains analyzed in this study are shown by black open square. *: Length of nucleotide sequences using analysis. **: Nucleotide position of BToV Ishikawa/2010.
4. Discussion
In the present study, we performed whole genome sequencing of Japanese BToVs and PToV by deep sequencing using supernatants of virus cultures and fecal suspensions. We could obtain a sufficient number of sequence reads and contigs of nearly 28 kb from 0.25 ml of 105.3 TCID50/mL BToV Ishi cell culture supernatant. Only one sample among three 0.25-mL fecal suspension samples gave a sufficient number of sequence reads and large contigs, though the number of sequence reads from this sample was lower than that from the viral culture supernatant. Unfortunately, we could not isolate BToV from fecal sample of BToV Kago by using HRT-18-Aich cells, which are useful for BToV isolation (Aita et al., 2012, Ito et al., 2012, Kuwabara et al., 2007), possibly because the samples could not be preserved at − 20 °C at the veterinary clinic. Although numerous sequence reads originating from bacterial species and hosts might interfere and reduce the number of sequence reads from fecal suspensions, the whole genome sequence of BToV could be obtained from this sample by using our deep-sequencing strategy.
The results of sequence comparison through pairwise alignment, similarity plot, bootscanning, and phylogenetic tree analyses of Japanese BToVs with BToVs and PToVs sequences from database were indicative of interspecies recombination between BToV Breda1 and PToV at the 3′ end of ORF1b and the end of HE coding region. Smits et al. provided evidence for recombination of ToV sequences between BToV Breda1 and PToV in the HE coding region (Smits et al., 2003). Italian and Dutch BToVs identified before the year 2000 were reported to possess a single recombination breakpoint located approximately 1100 nucleotides from the ATG initiation codon of the HE coding region. The putative recombinant breakpoint in the HE coding region of Japanese BToVs was quite identical to that of European BToVs. In the phylogenetic tree analyses, Japanese BToVs were found to be closely related to European BToVs in S, M, HE, and N coding regions. These findings suggest that Japanese BToVs and European BToVs have a common ancestor. However, owing to the lack of sequence information of 5′UTR, ORF1a and ORF 1b of European BToVs, we could not further compare Japanese BToVs with European BToVs. The HE coding region of ToV shares sequence homology to that of coronavirus and influenza C virus, however it is suggested that HE region of these viruses were introduced through independent heterologous recombination events (Smits et al., 2005). Homologous recombination of HE coding region was also reported in PToVs (Cong et al., 2013, Smits et al., 2003), suggesting that the HE coding region may be a hot spot for recombination events.
To our present knowledge, this is the first report to present the identification of a natural recombination event between BToV Breda1 and PToV at ORF1b. Approximately 76% of the genomes of BToV Ishi and BToV Kago are similar to that of PToV. Natural recombination events are common in coronaviruses, resulting in evolution and emergence of new variants that are pathogenic to poultry, cats, pigs, and humans (Graham and Baric, 2010, Herrewegh et al., 1998, Hewson et al., 2014, Lau et al., 2015, Song et al., 2013, Terada et al., 2014, Tian et al., 2014, Wang et al., 2015, Zhang et al., 2015a, Zhang et al., 2015b, Zhao et al., 2013). Co-infection of certain cells in bovine or swine with BToV Breda1 type virus and PToV is a pre-requisite for the generation of new recombinant viruses by double-template switch. The generated new recombinant virus, which has tropism to bovine cells, might adapt to bovine hosts and spread among cattle.
Although the four Japanese BToVs evaluated in this study were collected from distinct regions, the sequences of genomic regions apart from the S, M, and HE coding regions were closely related to each other and showed high identity (97.3–99.4%). Based on the sequences of the S, M, and HE coding regions, these BToVs were divided into at least two groups (Fig. 3D-G). The four Japanese BToVs showed genetic diversity, they all possessed a recombinant genome, suggesting that Japanese BToVs evolved after recombination events after or before being introduced in Japan. The Japanese and foreign BToV strains retrieved from the database clustered closely together in the phylogenetic tree, suggesting that all BToVs investigated in this study, except BToV Breda1, underwent recombination at the same genomic regions. Strains genetically closely related to Japanese BToVs are distributed worldwide.
In conclusion, we identified a natural recombination event between BToV Breda1 and PToV of Japanese BToVs. The four Japanese BToVs possessed two recombination breakpoints mapped to the 3′ end of ORF1b coding region and to the 3′ end of HE coding region and nearly 76% of the genome is similar to the PToV genome. Homologous recombination events of the virus genome are crucial for their evolution, and are significant factors for changes in host range and virulence of viruses. Our current data may provide important evidence to evaluate the epidemiological basis of ToV in cattle and swine population and to understand the mechanisms underlying the evolution of BToV.
Acknowledgements
This work was supported by the Grants from the Ministry of Health, Labour and Welfare of Japan and JSPS KAKENHI Grant Number 15K07718.
References
- Aita T., Kuwabara M., Murayama K., Sasagawa Y., Yabe S., Higuchi R., Tamura T., Miyazaki A., Tsunemitsu H. Characterization of epidemic diarrhea outbreaks associated with bovine torovirus in adult cows. Arch. Virol. 2012;157:423–431. doi: 10.1007/s00705-011-1183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbalagan S., Peterson J., Wassman B., Elston J., Schwartz K. Genome sequence of torovirus identified from a pig with porcine epidemic diarrhea virus from the United States. Genome Announc. 2014;2 doi: 10.1128/genomeA.01291-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D., Horzinek M.C. Genus torovirus assigned to the coronaviridae. Arch. Virol. 1993;128:395–396. doi: 10.1007/BF01309450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Zarlenga D.S., Richt J.A., Wang X., Wang Y., Suo S., Wang J., Ren Y., Ren X. Evolution and homologous recombination of the hemagglutinin-esterase gene sequences from porcine torovirus. Virus Genes. 2013;47:66–74. doi: 10.1007/s11262-013-0926-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draker R., Roper R.L., Petric M., Tellier R. The complete sequence of the bovine torovirus genome. Virus Res. 2006;115:56–68. doi: 10.1016/j.virusres.2005.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckmanton L., Luan B., Devenish J., Tellier R., Petric M. Characterization of torovirus from human fecal specimens. Virology. 1997;239:158–168. doi: 10.1006/viro.1997.8879. [DOI] [PubMed] [Google Scholar]
- Duckmanton L., Carman S., Nagy E., Petric M. Detection of bovine torovirus in fecal specimens of calves with diarrhea from Ontario farms. J. Clin. Microbiol. 1998;36:1266–1270. doi: 10.1128/jcm.36.5.1266-1270.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Baric R.S. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haschek B., Klein D., Benetka V., Herrera C., Sommerfeld-Stur I., Vilcek S., Moestl K., Baumgartner W. Detection of bovine torovirus in neonatal calf diarrhoea in Lower Austria and Styria (Austria) J. Vet. Med. B Infect. Dis Vet. Public Health. 2006;53:160–165. doi: 10.1111/j.1439-0450.2006.00936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh A.A., Smeenk I., Horzinek M.C., Rottier P.J., de Groot R.J. Feline coronavirus type II strains 79–1683 and 79–1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 1998;72:4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewson K.A., Noormohammadi A.H., Devlin J.M., Browning G.F., Schultz B.K., Ignjatovic J. Evaluation of a novel strain of infectious bronchitis virus emerged as a result of spike gene recombination between two highly diverged parent strains. Avian Pathol. 2014;43:249–257. doi: 10.1080/03079457.2014.914624. [DOI] [PubMed] [Google Scholar]
- Hoet A.E., Cho K.O., Chang K.O., Loerch S.C., Wittum T.E., Saif L.J. Enteric and nasal shedding of bovine torovirus (Breda virus) in feedlot cattle. Am. J. Vet. Res. 2002;63:342–348. doi: 10.2460/ajvr.2002.63.342. [DOI] [PubMed] [Google Scholar]
- Hoet A.E., Nielsen P.R., Hasoksuz M., Thomas C., Wittum T.E., Saif L.J. Detection of bovine torovirus and other enteric pathogens in feces from diarrhea cases in cattle. J. Vet. Diagn. Investig. 2003;15:205–212. doi: 10.1177/104063870301500301. [DOI] [PubMed] [Google Scholar]
- Ito T., Okada N., Fukuyama S. Epidemiological analysis of bovine torovirus in Japan. Virus Res. 2007;126:32–37. doi: 10.1016/j.virusres.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Okada N., Okawa M., Fukuyama S., Shimizu M. Detection and characterization of bovine torovirus from the respiratory tract in Japanese cattle. Vet. Microbiol. 2009;136:366–371. doi: 10.1016/j.vetmic.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Katayama S., Okada N., Masubuchi K., Fukuyama S., Shimizu M. Genetic and antigenic characterization of newly isolated bovine toroviruses from Japanese cattle. J. Clin. Microbiol. 2010;48:1795–1800. doi: 10.1128/JCM.02339-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Ichikawa Y., Shimoike K., Matsuda T., Uechi M., Shintani E. Outbreak of bovine torovirus diarrhea in Ishikawa Prefecture. J. Jpn. Vet. Med. Assoc. 2012;65:350–354. (In Japanese) [Google Scholar]
- Kirisawa R., Takeyama A., Koiwa M., IWAI H. Detection of bovine torovirus in fecal specimens of calves with diarrhea in Japan. J. Vet. Med. Sci. 2007;69:471–476. doi: 10.1292/jvms.69.471. [DOI] [PubMed] [Google Scholar]
- Koopmans M., van Wuijckhuise-Sjouke L., Schukken Y.H., Cremers H., Horzinek M.C. Association of diarrhea in cattle with torovirus infections on farms. Am. J. Vet. Res. 1991;52:1769–1773. [PubMed] [Google Scholar]
- Kroneman A., Cornelissen L.A., Horzinek M.C., de Groot R.J., Egberink H.F. Identification and characterization of a porcine torovirus. J. Virol. 1998;72:3507–3511. doi: 10.1128/jvi.72.5.3507-3511.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara M., Wada K., Maeda Y., Miyazaki A., Tsunemitsu H. First isolation of cytopathogenic bovine torovirus in cell culture from a calf with diarrhea. Clin. Vaccine Immunol. 2007;14:998–1004. doi: 10.1128/CVI.00475-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Feng Y., Chen H., Luk H.K., Yang W.H., Li K.S., Zhang Y.Z., Huang Y., Song Z.Z., Chow W.N., Fan R.Y., Ahmed S.S., Yeung H.C., Lam C.S., Cai J.P., Wong S.S., Chan J.F., Yuen K.Y., Zhang H.L., Woo P.C. Severe acute respiratory syndrome (SARS) coronavirus ORF8 protein is acquired from SARS-related coronavirus from greater horseshoe bats through recombination. J. Virol. 2015;89:10532–10547. doi: 10.1128/JVI.01048-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkani S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C. Fulllength human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D., Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- Martin D.P., Posada D., Crandall K.A., Williamson C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retrovir. 2005;21:98–102. doi: 10.1089/aid.2005.21.98. [DOI] [PubMed] [Google Scholar]
- Matiz K., Kecskeméti S., Kiss I., Adám Z., Tanyi J., Nagy B. Torovirus detection in faecal specimens of calves and pigs in Hungary: short communication. Acta Vet. Hung. 2002;50:293–296. doi: 10.1556/AVet.50.2002.3.5. [DOI] [PubMed] [Google Scholar]
- Nagai M., Omatsu T., Aoki H., Otomaru K., Uto T., Koizumi M., Minami-Fukuda F., Takai H., Murakami T., Masuda T., Yamasato H., Shiokawa M., Tsuchiaka S., Naoi Y., Sano K., Okazaki S., Katayama Y., Oba M., Furuya T., Shirai J., Mizutani T. Full genome analysis of bovine astrovirus from fecal samples of cattle in Japan: identification of possible interspecies transmission of bovine astrovirus. Arch. Virol. 2015;160:2491–2501. doi: 10.1007/s00705-015-2543-7. [DOI] [PubMed] [Google Scholar]
- Nogueira J.S., Asano K.M., de Souza S.P., Brandaão P.E., Richtzenhain L.J. First detection and molecular diversity of Brazilian bovine torovirus (BToV) strains from young and adult cattle. Res. Vet. Sci. 2013;95:799–801. doi: 10.1016/j.rvsc.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Oh E.H., Park S.I., Kim H.H., Jeong Y.J., Lim G.K., Hyun B.H., Cho K.O. Molecular epidemiology of bovine toroviruses circulating in South Korea. Vet. Microbiol. 2008;126:364–371. doi: 10.1016/j.vetmic.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penrith M.L., Gerdes G.H. Breda virus-like particles in pigs in South Africa. J. S. Afr. Vet. Assoc. 1992;63:102. [PubMed] [Google Scholar]
- Pérez E., Kummeling A., Janssen M.M., Jiménez C., Alvarado R., Caballero M., Donado P., Dwinger R.H. Infectious agents associated with diarrhoea of calves in the canton of Tilarán, Costa Rica. Prev. Vet. Med. 1998;33:195–205. doi: 10.1016/S0167-5877(97)00038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D.J., Park S.I., Jeong Y.J., Hosmillo M., Kim H.H., Kim H.J., Kwon H.J., Kang M.I., Park S.J., Cho K.O. Detection and molecular characterization of porcine toroviruses in Korea. Arch. Virol. 2010;155:417–422. doi: 10.1007/s00705-010-0595-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.L., Lavazza A., Matiz K., Horzinek M.C., Koopmans M.P., de Groot R.J. Phylogenetic and evolutionary relationships among torovirus field variants: evidence for multiple intertypic recombination events. J. Virol. 2003;77:9567–9577. doi: 10.1128/JVI.77.17.9567-9577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.L., Gerwig G.J., van Vliet A.L., Lissenberg A., Briza P., Kamerling J.P., Vlasak R., de Groot R.J. Nidovirus sialate-O-acetylesterases: evolution and substrate specificity of coronaviral and toroviral receptor-destroying enzymes. J. Biol. Chem. 2005;280:6933–6941. doi: 10.1074/jbc.M409683200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.E., Jeong W.G., Sung H.W., Kwon H.M. Sequencing, phylogenetic analysis, and potential recombination events of infectious bronchitis viruses isolated in Korea. Virus Genes. 2013;46:371–374. doi: 10.1007/s11262-012-0856-0. [DOI] [PubMed] [Google Scholar]
- Sun H., Lan D., Lu L., Chen M., Wang C., Hua X. Molecular characterization and phylogenetic analysis of the genome of porcine torovirus. Arch. Virol. 2014;159:773–778. doi: 10.1007/s00705-013-1861-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y., Matsui N., Noguchi K., Kuwata R., Shimoda H., Soma T., Mochizuki M., Maeda K. Emergence of pathogenic coronaviruses in cats by homologous recombination between feline and canine coronaviruses. PLoS One. 2014;9 doi: 10.1371/journal.pone.0106534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian P.F., Jin Y.L., Xing G., Qv L.L., Huang Y.W., Zhou J.Y. Evidence of recombinant strains of porcine epidemic diarrhea virus, United States. 2013. Emerg. Infect. Dis. 2014;20:1735–1738. doi: 10.3201/eid2010.140338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel Y., Laxer R.M., Petric M. Torovirus gastroenteritis presenting as acute abdomen. Clin. Infect. Dis. 1999;28:925–926. doi: 10.1086/517251. [DOI] [PubMed] [Google Scholar]
- Vanopdenbosch E., Wellemans G., Petroff K. Breda virus associated with respiratory disease in calves. Vet. Rec. 1991;129:203. doi: 10.1136/vr.129.9.203-b. [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu D., Shi W., Lu R., Wang W., Zhao Y., Deng Y., Zhou W., Ren H., Wu J., Wang Y., Wu G., Gao G.F., Tan W. Origin and possible genetic recombination of the Middle East respiratory syndrome coronavirus from the first imported case in China: phylogenetics and coalescence analysis. mBio. 2015;6 doi: 10.1128/mBio.01280-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M., Steck F., Horzinek M.C. Purification and partial characterization of a new enveloped RNA virus (Berne virus) J. Gen. Virol. 1983;64:1849–1858. doi: 10.1099/0022-1317-64-9-1849. [DOI] [PubMed] [Google Scholar]
- Woode G.N., Reed D.E., Runnels P.L., Herrig M.A., Hill H.T. Studies with an unclassified virus isolated from diarrheic calves. Vet. Microbiol. 1982;7:221–240. doi: 10.1016/0378-1135(82)90036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Han Z., Xu Q., Wang Q., Gao M., Wu W., Shao Y., Li H., Kong X., Liu S. Serotype shift of a 793/B genotype infectious bronchitis coronavirus by natural recombination. Infect. Genet. Evol. 2015;32:377–387. doi: 10.1016/j.meegid.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li J., Xiao Y., Zhang J., Wang Y., Chen L., Paranhos-Baccalà G., Ren L., Wang J. Genotype shift in human coronavirus OC43 and emergence of a novel genotype by natural recombination. J. Infect. 2015;70:641–650. doi: 10.1016/j.jinf.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Zou N., Wang F., Guo M., Liu P., Wen X., Cao S., Huang Y. Analysis of a QX-like avian infectious bronchitis virus genome identified recombination in the region containing the ORF 5a, ORF 5b, and nucleocapsid protein gene sequences. Virus Genes. 2013;46:454–464. doi: 10.1007/s11262-013-0884-4. [DOI] [PMC free article] [PubMed] [Google Scholar]