Abstract
Emerging infectious diseases present a complex challenge to public health officials and governments; these challenges have been compounded by rapidly shifting patterns of human behaviour and globalisation. The increase in emerging infectious diseases has led to calls for new technologies and approaches for detection, tracking, reporting, and response. Internet-based surveillance systems offer a novel and developing means of monitoring conditions of public health concern, including emerging infectious diseases. We review studies that have exploited internet use and search trends to monitor two such diseases: influenza and dengue. Internet-based surveillance systems have good congruence with traditional surveillance approaches. Additionally, internet-based approaches are logistically and economically appealing. However, they do not have the capacity to replace traditional surveillance systems; they should not be viewed as an alternative, but rather an extension. Future research should focus on using data generated through internet-based surveillance and response systems to bolster the capacity of traditional surveillance systems for emerging infectious diseases.
Introduction
Emerging infectious diseases are of particular concern to public health. Emergence is driven by sociocultural, environmental, and ecological factors.1 The vulnerability of people to emerging infectious diseases has been shown by the emergence of AIDS in the late 1970s, severe acute respiratory syndrome (SARS) in 2003, pandemic influenza H1N1 in 2009, and multidrug-resistant nosocomial pathogens, as well as the re-emergence of dengue, chikungunya, and malaria. Traditionally, effective disease surveillance is expensive and needs a formal public health network.2 Such systems are maintained by most countries, to varying degrees. Data sources, surveillance methods, analytical approaches, and factors affecting these systems are varied and have been reviewed in detail elsewhere.3, 4 Traditional, passive surveillance systems typically rely on data submitted to the relevant public health authority by physicians, laboratories, and other health-care providers; they provide information crucial to the effective functioning of health systems.5 These systems can be complex and expensive. Time and resource constraints, as well as a lack of operational knowledge of reporting systems, adversely affect the completeness of reporting,6 resulting in an incomplete account of disease emergence. Furthermore, substantial lags between an event and its notification are common; a result of late or failed reporting and the hierarchical structure of these systems.7 The average delay from receipt to dissemination of data by traditional sentinel surveillance networks is roughly 2 weeks.8
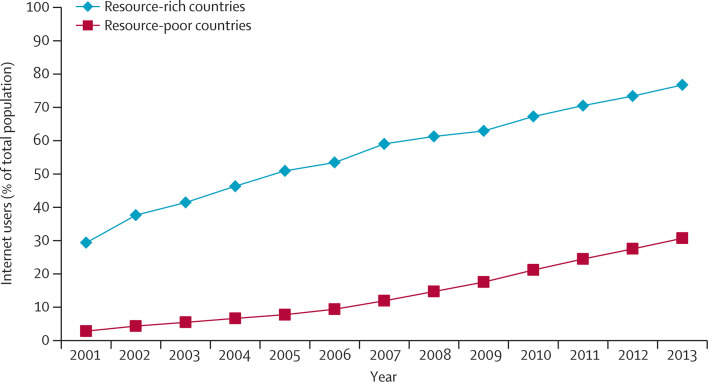
Internet availability and use has increased greatly in the past 10 years (figure 1 ).9, 10 The availability of health-related information on the internet (of varying quality and legitimacy) has also changed how people seek information about health.10, 11 These changes provide a new means to detect and monitor infectious diseases. The nature of emerging infectious diseases often limits the effectiveness of traditional surveillance systems.12 Digital surveillance could improve both the sensitivity and timeliness of detection of health events.13
Figure 1.
Internet access in resource-rich and resource-poor countries
Data taken from the International Telecommunications Union.9
We review recent studies that have exploited internet use and search trends to monitor two acute-onset viral illnesses of worldwide importance that have substantial seasonal and geographic variation: influenza and dengue. We critically analyse the effectiveness of monitoring internet data to track these diseases and discuss the advantages and limitations of this approach. Finally, we make recommendations for future research into these systems.
Digital surveillance
Digital surveillance attempts to provide knowledge of public health issues by analysis of health information stored digitally, as well as the distribution and patterns governing access to these data. Approaches to digital surveillance vary according to the media targeted. However, all exploit changes in behaviour related to information seeking, collection, storage, and communication pathways that have occurred with the development and increased availability of the internet and associated technologies.
Several surveillance systems use non-structured, event-based, digital data.14 The Global Public Health Intelligence Network (GPHIN)—developed by the Public Health Agency of Canada— automatically retrieves information about potential public health emergencies from news feed aggregators and distributes this information to public health agencies, including the WHO Global Outbreak Alert and Response Network.2, 15 The effectiveness of this system was shown during the SARS outbreak; GPHIN detected SARS more than 2 months before the first publications by the WHO.2 Other systems—eg, HealthMap16 and ProMED-mail17, 18—provide information about emerging public health problems by aggregating information about emerging diseases from various structured and non-structured data sources. These and other similar systems are reviewed elsewhere.13, 14, 19
Internet use has increased consistently in almost every country.20 Internet users in the USA alone generate 8 million queries for health-related information every day.21 The increase in worldwide internet availability and use over the past 10 years, combined with these changes in health-seeking behaviour, has created new possibilities for the development of innovative surveillance systems.9, 10, 22, 23 Although still very much in its infancy, analysis of digital data has been used to monitor communicable2, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33 and non-communicable diseases,34, 35 as well as mental health,36, 37 illegal drug use,38 health policy impact,39 and behaviours with potential health implications.40
Numerous studies have sought to exploit online health-seeking behaviour to monitor disease incidence. Although these studies use different data sources, they all rely on the premise that people who contract a disease will seek information about their condition from the internet and that incidence can be estimated by tracking changes in frequencies of searches for key terms. By monitoring search queries submitted to the search engine Yahoo!, Polgreen and colleagues41 predicted increases in positive influenza cultures1, 2, 3 weeks before their occurrence. Similarly, Hulth and co-workers42 developed a model for estimating intensity and peak incidence of influenza in Sweden by monitoring queries submitted to the medical web site Vårdguiden. This model correlated closely with both data for influenza-like illness (R2=0·89) and laboratory-confirmed cases of influenza (R2=0·90). A subsequent study showed this model to have good congruence with sentinel data over the course of the 2009 influenza H1N1 pandemic (r=0·88–0·90).43 More recently, influenza incidence in China was estimated by assessment of searches submitted to Baidu (the most commonly used search engine in China).44 This study reported a correlation of R2=0·96 between a composite search index (eight terms) and monthly Ministry of Health influenza reports. Furthermore, using a combination of Ministry of Health and Baidu data, the researchers produced accurate estimates (R2=0·95) of incidence 1–2 weeks before of Ministry of Health reports.
Google search queries also correlate highly with disease incidence. Historical logs of aggregated Google search queries—presented as normalised time series—are publically available through Google Trends from Jan 1, 2004. These data are available by country, state, and city in the USA, but only at country-level for many other regions (especially low-income countries). Previously, Google offered two user interfaces to access search reports: Google Trends and Google Insights for Search, which were merged in September, 2012.45 Carneiro and Mylonakis24 used Google Trends to analyse worldwide search frequency for the term “bird flu”. They reported an increase in search frequency between 2005, and 2006, coinciding with the spread of avian influenza from China to Turkey. Other studies reported the use of Google search data to monitor the frequency of searches for manually selected terms related to influenza in Chinese,46 Spanish,31 and French.28 These studies reported high degrees of correlation, which shows the potential application of this technology in languages other than English. Ginsberg and colleagues47 used an automated approach to select search terms from Google search logs with the greatest correlation with the US Influenza Sentinel Provider Surveillance Network of the US Centers for Disease Control and Prevention (CDC). The terms were then used to develop a model for monitoring influenza activity. Estimates from the model correlated highly with regional Centers for Disease Control and Prevention data (r=0·80–0·96, nine regions) and accurately estimate incidence of influenza-like illness 1–2 weeks before surveillance reports.47 An online influenza surveillance tool—Google Flu Trends—is based on the model of Ginsberg and coworkers and now includes 29 countries.
To date, two publications have reported the use of internet search data to estimate incidence of dengue. Chan and colleagues48 used a similar method to Ginsberg and coworkers47 to create models of dengue transmission for Bolivia, Brazil, India, Indonesia, and Singapore. Correlations between model estimates and holdout surveillance data (data excluded from the model, used for validation) were high for all countries (r=0·83–0·99). These models have been used to develop a free, publically available online resource for dengue surveillance: Google Dengue Trends. Althouse and colleagues49 used Google Insights for Search to monitor searches for dengue-related terms and applied these results to step-down linear regression models for Bangkok (Thailand) and Singapore. Both models showed a high degree of correlation with surveillance data (R2=0·95 for Bangkok and 0·94 for Singapore). Additionally, this study developed support vector machine50 and logistic regression models to predict periods of high dengue incidence. Area-under-the-receiver-operating-characteristic-curve, using the 75th percentile, was 0·960 for Bangkok and 0·906 for Singapore according to the support vector machine model compared with 0·960 and 0·896 respectively for the logistic regression model.
Google Flu Trends
Several studies have compared the performance of Google Flu Trends with national data for influenza-like illness. Google Flu Trends was visually compared with surveillance data for Australia51 and New Zealand52 over the course of the 2009 H1N1 influenza pandemic. Both studies reported good correlation. Another Australian study compared Google Flu Trends and emergency department presentation or hospital admissions for influenza-like illness in 2006–09 (r=0·35 for 2006, r=0·88 for 2007, r=0·91 for 2008, and r=0·76 for 2009).53 These findings accord with the results of Hulth and colleagues43 and Valdivia and colleagues54 who showed that Google Flu Trends correlated strongly with estimates of influenza incidence and peak incidences produced from data collected by sentinel physician networks throughout Europe. Finally, Google Flu Trends correlated highly with the Electronic Surveillance System for the Early Notification of Community-Based Epidemics (ESSENCE; a syndromic surveillance system run by the US Department of Defence; r=0·88)55 and with influenza-like illness estimates produced for Flanders, Belgium, by the Great Influenza Survey (a weekly, online influenza survey; r=0·62–0·94).56
Traditional influenza surveillance systems commonly monitor incidence with virological data, rather than influenza-like illness. Data from Google Flu Trends was highly correlated with data from the US Influenza Virologic Surveillance System (r=0·72);57 however, this correlation was lower than that reported for Google Flu Trends and CDC influenza-like illness data (r=0·94). Google Flu Trends estimates have been reported to correlate highly with laboratory-confirmed influenza at a provincial and city level. Malik and colleagues12 reported the correlation between weekly counts of laboratory-confirmed H1N1 influenza cases in Manitoba, Canada, and Google Flu Trends data during the 2009 influenza pandemic (R2=0·69; 2 week lag). Similarly, Google Flu Trends had a high level of congruence with virology data from a Baltimore hospital (adult r=0·88; paediatric r=0·72).58 Dugas and coworkers58 noted that Google Flu Trends correlated well with paediatric emergency department crowding measures, leading them to suggest that Google Flu Trends could be used for strategic management of emergency department resources. The potential applications of Google Flu Trends data to strategic allocation of resources and priority setting is further shown by Patwardhan and Bilkovski,59 who compared sales of four drugs commonly prescribed for treatment of influenza with Google Flu Trends and CDC ILINet data; aggregate correlation between Google Flu Trends and prescription sales was r=0·92.
Any changes to the status quo of internet search behaviour could alter how well Google Flu Trends models actual influenza incidence. Loss of resolution might occur as a result of media-driven interest or through other events that change search behaviour.43, 49, 57, 58, 60, 61 Google Flu Trends accounts for changing search behaviour by updating the model each year to best represent reference surveillance data.62 Despite this precaution, a loss of resolution was reported to have occurred during the 2009 H1N1 influenza pandemic.53, 58 Cook and coworkers62 updated the Google Flu Trends model with a larger pool of candidate queries, less common queries, and historical logs that included searches done during the initial months of the 2009 H1N1 influenza pandemic. The new model incorporated roughly 160 queries versus 45 used in the original model. Although the original Google Flu Trends model had a high degree of correlation with ILINet data before the 2009 pandemic (September, 2003, to March, 2009; r=0·91) and over the entire course of the outbreak (March–December, 2009; r=0·91), correlation during the initial wave of the pandemic was low (March–August, 2009; r=0·29). Correlations of the updated Google Flu Trends model were higher for all periods analysed, most notably during the initial wave (r=0·95). These results show the effect that changing search behaviours can have on surveillance systems based on internet search queries and the importance of continual assessment of the performance of such systems.
Predictive models and integration of surveillance systems based on internet search queries
Studies have predominantly focused on retrospective assessment of the performance of Google Flu Trends. However, almost real-time disease tracking can be done by application of Google Flu Trends data to a season-specific compartmental mathematical model.63 Google Flu Trends data can also be used for early detection of epidemics.64 Pervaiz and colleagues applied various algorithms to Google Flu Trends data to develop early epidemic detection systems capable of generating actionable alerts. Although this study did not identify a single best method, it showed the potential use of Google Flu Trends data in this manner. Zhou and coworkers65 have developed a system to predict epidemic alert levels from daily Google Trends data. Hidden Markov model-based methods predicted influenza alert levels in real-time with 97·7% accuracy and provided an indication of influenza activity up to 4 weeks ahead of the release of CDC reports. In another recent study, ensemble adjustment was used to assimilate Google Flu Trends data into a humidity-driven compartmental mathematical model, enabling real-time predictions of peaks to be made more than 7 weeks in advance of their occurrence.66 Finally, using a negative binomial generalised autoregressive moving average model—which included Google Flu Trends data as a secondary variable—Dugas and colleagues67 predicted weekly influenza cases at a medical centre with a high degree of accuracy (83% of estimates were within seven cases).
These models are promising and, overall, Google Flu Trends seems to provide timely and accurate estimates of influenza-like illness and laboratory-confirmed influenza. However, methods to integrate this information into existing surveillance systems need to be developed.55 Scarpino and colleagues68 postulated that the predictive power of the Texas ILINet could be improved by use of a smaller set of carefully chosen sentinel providers. Additionally, they investigated the potential of incorporating Google Flu Trends data into the network as a virtual provider. Google Flu Trends was reported to have a high degree of correlation with the ILINet in Texas (R2=0·77 at a 0 week lag). It was the most informative provider, matching the predictive performance of an optimised network of 44 sentinel providers.
Social media
The power of social media as both a source of information and as a means of disseminating information is increasingly recognised in public health.69, 70 Corley and colleagues71 have proposed that influenza incidence could be estimated by tracking use of key terms in web and social media. They analysed the frequency of English language blog posts that contained the terms “influenza” or “flu” and compared these with CDC ILINet data. Correlation was r=0·63 for this study and r=0·55 in a subsequent study with an extended dataset.72 Microblogs (such as Twitter) were not included in these studies. Collier and coworkers73 used supervised learning to categorise expressions from Twitter messages into five influenza-related categories and correlated these expressions with CDC data for positive influenza A H1N1 tests. Correlations in this study (r=0·58–0·67) were similar to those for Corley and colleagues. 71, 72 Chew and Eysenbach74 sorted Twitter posts containing terms related to influenza A H1N1 into groups describing “personal experiences” or “concern” and compared these with H1N1 incidence rates in the USA. Correlations were r=0·77 for “personal experiences” and r=0·66 for “concern”. These correlations are not as high as those reported for approaches based on internet search queries. However, Lampos and Cristianini75 reported correlations of up to r=0·933 for their analysis of influenza created with a supervised learning framework compared with influenza-like illnesses reported by the UK Health Protection Agency. They concluded that a supervised learning framework is a suitable method for selection of features for use in digital surveillance systems. Culotta76 reported that the accuracy of estimates could be improved by use of a document classification component; they reported correlations of up to r=0·97.
Advantages and limitations of web-based surveillance systems
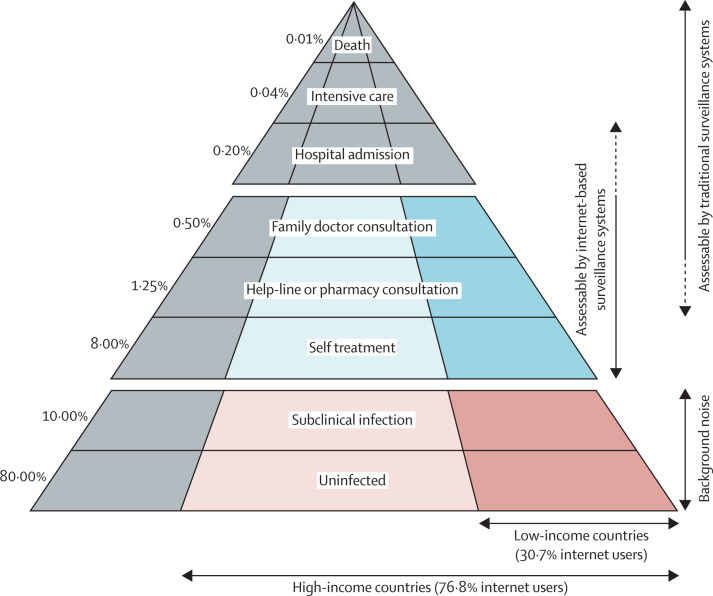
Google Flu Trends usually showed an increase of influenza incidence 0–2 weeks before traditional systems. Internet-based surveillance systems circumvent the bureaucratic structure of traditional systems. Furthermore, they target a different section of the community to traditional surveillance systems. Zeng and Wagner's77 model of patient behaviour during epidemics identifies four phases in health-care seeking: recognition of symptoms, interpretation of symptoms, representation of illness, and seeking treatment. Traditional surveillance systems only source data from people seeking treatment. Internet-based surveillance systems access people from not just the final phase, but also the earlier interpretation of symptoms and representation of illness phases.48 However, internet-based surveillance systems are limited to people who seek health-related information on the internet (or proxies, such as parents or carers of sick children). Despite this limitation, they can capture many cases. Attrition during disease pathogenesis or health-seeking pathways is both high and cumulative—results of a study done in rural Cambodia showed that 67% of cases of haemorrhagic fever were treated at home, rather than in a health facility; thus, a health-care-based surveillance system would miss 67% of information before it even becomes accessible.78 Systems that target points earlier in surveillance will produce more timely information. For an influenza epidemic with a 20% infection rate, 10% clinical attack rate, 2% case hospital admission rate, and 0·1% symptomatic case fatality rate,79 the fraction of the population assessable by an internet-based surveillance system (7488 people per 100 000 patients) would be nearly ten-times that of a traditional system (750 people per 100 000 patients), for a population with the internet use of an average high-income country (76·8%; figure 2 ).9
Figure 2.
Proportions of the population assessable by traditional and internet-based surveillance systems during an influenza epidemic in high-income and low-income countries
Light blue sections correspond with fractions visible to internet-based surveillance systems in high-income countries, dark blue corresponds to low-income countries. Red sections indicate background noise. Grey sections indicate fractions not visible to internet-based surveillance systems. Adapted from Watson and Pebody.79
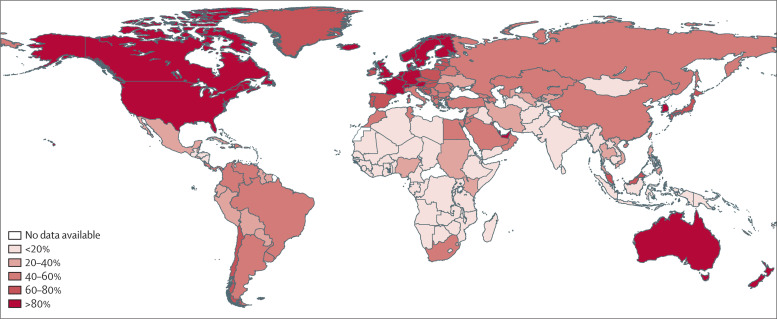
Internet-based surveillance systems work best for large populations24 and their use can be limited by national infrastructure (figure 3 ). Although the fraction of people assessable with an internet-based system in the average low-income country (30·7% internet access) is only 2993 people per 100 000 patients (figure 2), this fraction still exceeds that of a traditional surveillance system (750 people per 100 000 patients). The number of people who have access to the internet is not the only relevant factor. Internet use and health-seeking behaviour vary between different sectors of the community.23, 80 The accuracy of national Google Flu Trends estimates is positively correlated with the proportion of the population who use the internet to obtain health-related information.54 However, large discrepancies exist between availability and uptake of the internet, and seeking health-care information as a proxy for disease has biases stemming from unequal use and access.20
Figure 3.
Percentage of population who use the internet, by country
2012 data9 were used for all countries, except the British Virgin Islands (2010).
The spatial resolution of Google Flu Trends (and Google Trends) is improving. At present, Google Flu Trends offers some city-level estimates of influenza incidence in the USA but probably has neither the sensitivity nor spatial resolution necessary to detect small, localised outbreaks.12 Spatial resolution is limited by the level of data aggregation and search volume; resolution should improve over time as overall internet access increases and the internet becomes more widely accepted as a source of health-based information.54 Results of these systems should also be interpreted carefully. Although internet-based surveillance systems seem to have high correlation with traditional surveillance systems, overall correlations could hide short-term periods of high variance. 62
Translation of internet-based data into an accurate, meaningful, and useful format is a challenge. Bias introduced by self-reporting and media-driven interest might be the biggest confounder of internet-based surveillance systems. Targeting microblogs has the potential to track, not just disease activity, but also related community concerns and perceptions.73 However, the frequency of posts on social media is generally accepted to be a function of personal experience and perception of what an individual believes their friends and followers would find interesting, rather than a true reflection of the occurrence of an event.81 Similarly, the media drive search frequency. An increase in searches for “bird flu” occurred in the USA between 2005 and 2006, despite no avian influenza being detected; this trend was attributed to media-driven interest about the influenza outbreak affecting Asia at the time.24 A similar occurence was reported for dengue-related searches in India in 2006; an unusually large spike in searches was attributed to news that a member of the prime minister's family had been admitted to hosptial for dengue.48 To reduce the effect of media-driven searches, the Google Dengue Trends model replaces spikes that exceed the mean of the previous 4 weeks by five SDs with an imputed value.48 Media-driven behaviour does not exclusively affect internet-based surveillance systems. On April 26, 2009, the US CDC declared a national public-health emergency in response to the emerging H1N1 pandemic; the following week was termed fear week.61 Despite state-wide viral surveillance data showing little influenza activity, emergency department patient volumes increased substantially. A similar trend occurred in a Baltimore paediatric emergency department.58 Because changes in Google Flu Trends over this period correlated with the increase in emergency department patient volumes, the investigators suggested that Google Flu Trends could have a role in planning emergency department surge capacity;58 rather than representing influenza incidence, Google Flu Trends identified public perceptions of the threat of influenza and predicted the associated increase in health-care demand.
Unlike systems that rely on input from health-care practitioners or laboratories, internet-based surveillance instruments are unlikely to become overwhelmed during a pandemic and, because they are automated, are available year-round (contingent on sufficient search volume), whereas traditional networks might only operate seasonally.82 These internet-based systems could be of particular use in countries with poorly developed traditional surveillance systems.52 However, implementation of such systems in these countries is fraught with difficulties. Internet-based surveillance systems work on the premise that disease incidence correlates with frequency of information-seeking using specific terms. Textual information can be difficult to classify and interpret83 and accuracy might be heavily affected by cultural nuances, language shifts, and use of colloquialisms or even memes. The model of Collier and coworkers73 needed a filter to reduce the effect of terms such as “Bieber fever” (which refers to infatuation with Canadian pop musician Justin Bieber) on the keyword of interest, “fever”. Changes to search behaviours and information-seeking practices will affect the performance of these models;62 furthermore, such changes are unlikely to occur uniformly. The re-emergence of infectious diseases with similar clinical presentations—eg, chikungunya in dengue-endemic areas—also presents a difficulty.48 Models should be designed for a specific system (country or region) and be validated against reference data before they are used to guide health policy or action. As such, they cannot replace traditional surveillance.42
The problem of privacy has been raised by several researchers.2, 48, 83 For ethical reasons, data are de-identified or—in the case of data from Google—aggregated before public release, precluding identification of the source of specific posts or searches. Although not a problem in itself, this process could make interpretation difficult. Content cannot be connected with individuals and care should be taken not to commit an ecological fallacy—to make inferences about the characteristics of individuals based on aggregate data.36 Finally, the security of health information is an imperative.84 Google Flu Trends and Google Dengue Trends are operated by the philanthropic arm of Google, which is a publicly listed company. Although these services are freely available, Google does not release the search terms used in the algorithms; caution is urged in relying too heavily on closed-source data that are under the control of a multinational company.
Integration of internet-based surveillance technologies into existing surveillance systems
Few studies have explored how to translate internet-based surveillance systems into a public health response. Search queries submitted to Vårdguiden have been used to develop an automated system for generation of reports about epidemiological trends.85 GET WELL (Generating Epidemiological Trends from WEb Logs, Like) extracts search queries from Vårdguiden logs, aggregates the data (weekly), and produces time-series graphs. Additionally, the system enables custom statistical analyses to be integrated; this function is routinely used for norovirus and influenza. GET WELL is used by Swedish Institute for Infectious Disease Control in conjunction with traditional surveillance networks to identify emerging concerns and to focus epidemiological investigations.
The potential for internet-based surveillance systems to revolutionise emerging infectious disease surveillance was shown by Scarpino and colleagues. 68 They presented a method for optimisation of sentinel surveillance networks that enabled integration of Google Flu Trends into the network as a virtual provider (enabling it to function as a sentinal provider reporting influenza-like illness within the community). Google Flu Trends alone explained roughly 60% of influenza-associated hospital admissions in Texas; which is equivalent to the performance of an optimised sentinel network with 44 providers (R2=0·63). Furthermore, Google Flu Trends outperformed the 2008 Texas ILINet which drew information from 82 providers (R2=0·57). An optimised network of 82 providers outperformed Google Flu Trends (R2=0·77); however, the best predictive performance was achieved by optimised hybrid networks, which allowed use of Google Flu Trends as a virtual provider. Allowing Google Flu Trends as a virtual provider in a network of 82 providers increased predictive performance by a further 12·5% (R2=0·90).68 These studies show potential applications of internet-based surveillance systems in bolstering traditional surveillance system capacity and guiding public health action. However, the routine integration of non-traditional, unstructured, internet-based data into existing surveillance systems will necessitate a change in the structure and rhetoric of units responsible for surveillance if it is to be effectively translated into public health action.86
Future research
To date, most studies of internet-based surveillance systems are retrospective analyses of performance; the prospective performance of these systems needs to be assessed. Future studies should not only focus on development of new detection methods nor on application of these methods to new diseases, but they should also explore ways to integrate these approaches into existing systems.55 In doing so, care must be taken to ensure that new systems add to the capacity of old ones. The potential application of internet-based surveillance systems is not restricted to surveillance. They can also be strategic instruments for resource management and allocation,58, 59 which warrants further investigation. Finally, despite the potential of internet-based surveillance systems, they have not been applied with a global focus. Strategies for surveillance of infectious diseases have been criticised for focusing too heavily on high-income countries.87 New infectious diseases emerge all over the world and their emergence is affected by many sociocultural, economic, environmental, and ecological factors.1 The international nature of emerging infectious diseases, combined with the globalisation of travel and trade, have increased the interconnectedness of all countries. Strategies to detect, monitor, and control emerging infectious diseases should recognise this change—these diseases are a global concern. The potential to develop global surveillance systems for emerging infectious diseases that use internet-based data should be explored.
Assessment of internet queries for surveillance of emerging infectious diseases is a new concept that has been applied with promising results. These systems are appealing from a logistical, economical, and epidemiological standpoint. Internet-based systems are intuitive, adaptable, operate in almost real-time and, once established, are cheap to operate and maintain.12 Furthermore, these systems do not rely on the health-care system to provide and analyse data, or a government to disseminate information and advise the international community of emerging concerns—all limitations of traditional surveillance systems. However, internet-based surveillance does not provide an alternative to traditional surveillance systems. Rather, these systems are an extension of traditional systems. The societal effect and extent of spread of infectious diseases within a community cannot be measured by any one surveillance system.43 Surveillance systems should be flexible, built with models that incorporate several means of collecting information, and integrate information from other sources to create a comprehensive understanding of and approach to addressing emerging problems.86 Furthermore, addressing emerging infectious diseases is contingent on their recognition as global, rather than regional, issues. A global response requires concerted international approaches to strengthen the capacity of emerging infectious diseases surveillance systems worldwide. Future research needs to focus on how to use internet-based surveillance systems to complement existing systems.
Search strategy and selection criteria
We searched Medline (via PubMed) and Web of Science with the following search terms: “digital disease detection” “Google Flu Trends”, “Google Insights”, “Google Trends”, “infodemiology”, “infoveillance”, “real-time disease surveillance”, and “syndromic surveillance”. We also did searches with the terms “dengue”, “infectious”, OR “influenza” AND “early warning”, “Google”, “internet”, “search engine”, “social media”, “Twitter”, “Facebook”, OR “web”. Finally, we did searches for the terms “internet” OR “web” AND “disease surveillance” OR “disease detection”. To be eligible for inclusion, studies needed to be peer reviewed, describe the use of internet-search metrics or social media data for surveillance of influenza or dengue, and assess performance of this surveillance approach by comparing it with data from traditional surveillance approaches. Results were restricted to those published in English between Jan 1, 2008, and June 30, 2013. The appendix shows the publications that fit the inclusion criteria.
Acknowledgments
Acknowledgments
GJM's salary was provided through the National Health and Medical Research Council, Australia (grant #1002608).
Contributors
WH developed the original idea for this Review. The structure of the Review was developed by WH and GJM. GJM did the literature search, wrote the first draft, and created tables and figures. WH produced the map. WH, ACAC, and GMW provided editorial advice on the report and the final version was approved by all authors.
Conflicts of interest
We declare that we have no conflicts of interest.
Supplementary Material
References
- 1.Jones KE, Patel NG, Levy MA. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson K, Brownstein JS. Early detection of disease outbreaks using the internet. Can Med Assoc J. 2009;180:829–831. doi: 10.1503/cmaj.090215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brachman PS. Public health surveillance. In: Brachman PS, Abrutyn E, editors. Bacterial infections of humans. Springer; US: 2009. pp. 51–67. [Google Scholar]
- 4.Van Beneden CA, Lynfield R. Public health surveillance for infectious diseases. In: Lee LM, Teutsch SM, Thacker SB, St Louis ME, editors. Principles and practice of public health surveillance. 3rd edn. Oxford University Press; 2010. pp. 236–254. [Google Scholar]
- 5.O'Connell EK, Zhang GY, Leguen F, Llau A, Rico E. Innovative uses for syndromic surveillance. Emerg Infect Dis. 2010;16:669–671. doi: 10.3201/eid1604.090688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doyle TJ, Glynn MK, Groseclose SL. Completeness of notifiable infectious disease reporting in the United States: an analytical literature review. Am J Epidemiol. 2002;155:866–874. doi: 10.1093/aje/155.9.866. [DOI] [PubMed] [Google Scholar]
- 7.Madoff LC, Fisman DN, Kass-Hout T. A new approach to monitoring dengue activity. PLoS Negl Trop Dis. 2011;5:e1215. doi: 10.1371/journal.pntd.0001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng CK, Lau EH, Ip DK, Yeung AS, Ho LM, Cowling BJ. A profile of the online dissemination of national influenza surveillance data. BMC Public Health. 2009;9:339. doi: 10.1186/1471-2458-9-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International Telecommunications Union World Telecommunication/ICT Indicators Database 2013 (17th edition) 2013. http://www.itu.int/en/ITU-D/Statistics/Pages/publications/wtid.aspx (accessed Julu 19, 2013).
- 10.Rice RE. Influences, usage, and outcomes of internet health information searching: multivariate results from the Pew surveys. Int J Med Inform. 2006;75:8–28. doi: 10.1016/j.ijmedinf.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 11.Leung L. Internet embeddedness: links with online health information seeking, expectancy value/quality of health information websites, and internet usage patterns. Cyberpsychol Behav. 2008;11:565–569. doi: 10.1089/cpb.2007.0189. [DOI] [PubMed] [Google Scholar]
- 12.Malik MT, Gumel A, Thompson LH, Strome T, Mahmud SM. “Google flu trends” and emergency department triage data predicted the 2009 pandemic H1N1 waves in Manitoba. Can J Public Health. 2011;102:294–297. doi: 10.1007/BF03404053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morse SS. Public health surveillance and infectious disease detection. Biosecur Bioterror. 2012;10:6–16. doi: 10.1089/bsp.2011.0088. [DOI] [PubMed] [Google Scholar]
- 14.Keller M, Blench M, Tolentino H. Use of unstructured event-based reports for global infectious disease surveillance. Emerg Infect Dis. 2009;15:689–695. doi: 10.3201/eid1505.081114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mykhalovskiy E, Weir L. The Global Public Health Intelligence Network and early warning outbreak detection: a Canadian contribution to global public health. Can J Public Health. 2006;97:42–44. doi: 10.1007/BF03405213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freifeld CC, Mandl KD, Ras BY, Bronwnstein JS. HealthMap: global infectious disease monitoring through automated classification and visualization of Internet media reports. J Am Med Inform Assoc. 2008;15:150–157. doi: 10.1197/jamia.M2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madoff LC. ProMED-mail: an early warning system for emerging diseases. Clin Infect Dis. 2004;39:227–232. doi: 10.1086/422003. [DOI] [PubMed] [Google Scholar]
- 18.Pollack MP, Pringle C, Madoff LC, Memish ZA. Latest outbreak news from ProMED-mail: novel coronavirus—Middle East. Int J Infect Dis. 2013;17:e143–e144. doi: 10.1016/j.ijid.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brownstein JS, Freifeld CC, Madoff LC. Digital disease detection—harnessing the web for public health surveillance. N Engl J Med. 2009;360:2153–2155. doi: 10.1056/NEJMp0900702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The World Bank Internet users (per 100 people) 2012. http://data.worldbank.org/indicator/IT.NET.USER.P2/countries/1W?display=default (accessed Dec 10, 2012).
- 21.Fox S. Online health search 2006. Pew Internet and American Life Project; 2006. [Google Scholar]
- 22.Sadasivam RS, Kinney RL, Lemon SC, Shimada SL, Allison JJ, Houston TK. Internet health information seeking is a team sport: analysis of the Pew Internet Survey. Int J Med Inform. 2013;82:193–200. doi: 10.1016/j.ijmedinf.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Fox S. The social life of health information, 2011. Pew Internet and American Life Project; 2011. [Google Scholar]
- 24.Carneiro HA, Mylonakis E. Google trends: a web-based tool for real-time surveillance of disease outbreaks. Clin Infect Dis. 2009;49:1557–1564. doi: 10.1086/630200. [DOI] [PubMed] [Google Scholar]
- 25.Desai R, Hall AJ, Lopman BA. Norovirus disease surveillance using Google internet query share data. Clin Infect Dis. 2012;55:E75–E78. doi: 10.1093/cid/cis579. [DOI] [PubMed] [Google Scholar]
- 26.Desai R, Lopman BA, Shimshoni Y, Harris JP, Patel MM, Parashar UD. Use of internet search data to monitor impact of rotavirus vaccination in the United States. Clin Infect Dis. 2012;54:CP8–C11. doi: 10.1093/cid/cis121. [DOI] [PubMed] [Google Scholar]
- 27.Dukic VM, David MZ, Lauderdale DS. Internet queries and methicillin-resistant Staphylococcus aureus surveillance. Emerg Infect Dis. 2011;17:1068–1070. doi: 10.3201/eid1706.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelat C, Turbelin C, Bar-Hen A, Flahault A, Valleron A. More diseases tracked by using Google Trends. Emerg Infect Dis. 2009;15:1327–1328. doi: 10.3201/eid1508.090299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samaras L, Garcia-Barriocanal E, Sicilia MA. Syndromic surveillance models using web data: the case of scarlet fever in the UK. Inform Health Soc Care. 2012;37:106–124. doi: 10.3109/17538157.2011.647934. [DOI] [PubMed] [Google Scholar]
- 30.Seifter A, Schwarzwalder A, Geis K, Aucott J. The utility of “Google Trends” for epidemiological research: Lyme disease as an example. Geospatial Health. 2010;4:135–137. doi: 10.4081/gh.2010.195. [DOI] [PubMed] [Google Scholar]
- 31.Valdivia A, Monge-Corella S. Diseases tracked by using Google trends, Spain. Emerg Infect Dis. 2010;16:168. doi: 10.3201/eid1601.091308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou X, Ye J, Feng Y. Tuberculosis surveillance by analyzing Google Trends. IEEE Trans Biomed Eng. 2011;58:2247–2254. doi: 10.1109/TBME.2011.2132132. [DOI] [PubMed] [Google Scholar]
- 33.Zhou XC, Shen HB. Notifiable infectious disease surveillance with data collected by search engine. J Zhejiang Univ-SCI C. 2010;11:241–248. [Google Scholar]
- 34.Breyer BN, Sen S, Aaronson DS, Stoller ML, Erickson BA, Eisenberg ML. Use of Google Insights for search to track seasonal and geographic kidney stone incidence in the United States. Urology. 2011;78:267–271. doi: 10.1016/j.urology.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willard SD, Nguyen MM. Internet search trends analysis tools can provide real-time data on kidney stone disease in the United States. Urology. 2011;81:37–42. doi: 10.1016/j.urology.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 36.Reilly S, Richey S, Taylor JB. Using Google search data for state politics research: an empirical validity test using roll-off data. State Politics Policy Q. 2012;12:146–159. [Google Scholar]
- 37.Chen YD, Brown SA, Hu PJH, King CC, Chen HC. Managing emerging infectious diseases with information systems: reconceptualizing outbreak management through the lens of loose coupling. Info Sys Res. 2011;22:447–468. [Google Scholar]
- 38.Yin S, Ho M. Monitoring a toxicological outbreak using internet search query data. Clin Toxicol. 2012;50:818–822. doi: 10.3109/15563650.2012.729667. [DOI] [PubMed] [Google Scholar]
- 39.Reis BY, Brownstein JS. Measuring the impact of health policies using internet search patterns: the case of abortion. BMC Public Health. 2010;10:514. doi: 10.1186/1471-2458-10-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deutsch CM, Bronstein AC, Spyker DA. A spoonful of cinnamon: The “cinnamon challenge” — Google Trends and the National Poison Data System. Clin Toxicol. 2012;50:645. [Google Scholar]
- 41.Polgreen PM, Chen Y, Pennock DM, Nelson FD. Using internet searches for influenza surveillance. Clin Infect Dis. 2008;47:1443–1448. doi: 10.1086/593098. [DOI] [PubMed] [Google Scholar]
- 42.Hulth A, Rydevik G, Linde A. Web queries as a source for syndromic surveillance. PLoS One. 2009;4:e4378. doi: 10.1371/journal.pone.0004378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hulth A, Rydevik G. Web query-based surveillance in Sweden during the influenza A(H1N1)2009 pandemic, April 2009 to February 2010. Euro Surveill. 2011;16 PII:19856. [PubMed] [Google Scholar]
- 44.Yuan Q, Nsoesie EO, Lv B, Peng G, Chunara R, Brownstein JS. Monitoring influenza epidemics in China with search query from Baidu. PLoS One. 2013;8:e64323. doi: 10.1371/journal.pone.0064323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi HY, Varian H. Predicting the present with Google Trends. Economic Record. 2012;88:2–9. [Google Scholar]
- 46.Kang M, Zhong H, He J, Rutherford S, Yang F. Using Google Trends for influenza surveillance in South China. PLoS One. 2013;8:e55205. doi: 10.1371/journal.pone.0055205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ginsberg J, Mohebbi MH, Patel RS, Brammer L, Smolinski MS, Brilliant L. Detecting influenza epidemics using search engine query data. Nature. 2009;457:1012–1014. doi: 10.1038/nature07634. [DOI] [PubMed] [Google Scholar]
- 48.Chan EH, Sahai V, Conrad C, Brownstein JS. Using web search query data to monitor dengue epidemics: a new model for neglected tropical disease surveillance. PLoS Negl Trop Dis. 2011;5:e1206. doi: 10.1371/journal.pntd.0001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Althouse BM, Ng YY, Cummings DA. Prediction of dengue incidence using search query surveillance. PLoS Negl Trop Dis. 2011;5:e1258. doi: 10.1371/journal.pntd.0001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hastie T, Tibshirani R, Friedman J. The elements of statistical learning: data mining, inference, and prediction. 2nd edn. Springer; New York: 2009. [Google Scholar]
- 51.Kelly H, Grant K. Interim analysis of pandemic influenza (H1N1) 2009 in Australia: surveillance trends, age of infection and effectiveness of seasonal vaccination. Euro Surveill. 2009;14 doi: 10.2807/ese.14.31.19288-en. PII:19288. [DOI] [PubMed] [Google Scholar]
- 52.Wilson N, Mason K, Tobias M, Peacey M, Huang QS, Baker M. Interpreting Google Flu Trends data for pandemic H1N1 influenza: the New Zealand experience. Euro Surveill. 2009:14. [PubMed] [Google Scholar]
- 53.Boyle JR, Sparks RS, Keijzers GB, Crilly JL, Lind JF, Ryan LM. Prediction and surveillance of influenza epidemics. Med J Aust. 2011;194:S28–S33. doi: 10.5694/j.1326-5377.2011.tb02940.x. [DOI] [PubMed] [Google Scholar]
- 54.Valdivia A, Lopez-Alcalde J, Vicente M, Pichiule M, Ruiz M, Ordobas M. Monitoring influenza activity in Europe with Google Flu Trends: comparison with the findings of sentinel physician networks — results for 2009–10. Euro Surveill. 2010;15 doi: 10.2807/ese.15.29.19621-en. PII:19261. [DOI] [PubMed] [Google Scholar]
- 55.Pattie DC, Cox KL, Burkom HS, Lombardo JS, Gaydos JC. A public health role for internet search engine query data? Mil Med. 2009;174:11–12. [PubMed] [Google Scholar]
- 56.Vandendijck Y, Faes C, Hens N. Eight years of the great influenza survey to monitor influenza-like illness in Flanders. PLoS One. 2013;8:e64156. doi: 10.1371/journal.pone.0064156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ortiz JR, Zhou H, Shay DK, Neuzil KM, Fowlkes AL, Goss CH. Monitoring influenza activity in the United States: a comparison of traditional surveillance systems with Google Flu Trends. PLoS One. 2011;6:e18687. doi: 10.1371/journal.pone.0018687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dugas AF, Hsieh YH, Levin SR. Google Flu Trends: correlation with emergency department influenza rates and crowding metrics. Clin Infect Dis. 2012;54:463–469. doi: 10.1093/cid/cir883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patwardhan A, Bilkovski R. Comparison: flu prescription sales data from a retail pharmacy in the US with Google Flu Trends and US ILINet (CDC) data as flu activity indicator. PLoS One. 2012;7:e43611. doi: 10.1371/journal.pone.0043611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watts G. Google watches over flu. BMJ. 2008;337:a3076. doi: 10.1136/bmj.a3076. [DOI] [PubMed] [Google Scholar]
- 61.McDonnell WM, Nelson DS, Schunk JE. Should we fear “flu fear” itself? Effects of H1N1 influenza fear on ED use. Am J Emerg Med. 2012;30:275–282. doi: 10.1016/j.ajem.2010.11.027. [DOI] [PubMed] [Google Scholar]
- 62.Cook S, Conrad C, Fowlkes AL, Mohebbi MH. Assessing Google Flu Trends performance in the United States during the 2009 influenza virus A (H1N1) pandemic. PLoS One. 2011;6:e23610. doi: 10.1371/journal.pone.0023610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dukic V, Lopes HF, Polson NG. Tracking epidemics with Google Flu Trends data and a state-space SEIR model. J Am Stat Assoc. 2012;107:1410–1426. doi: 10.1080/01621459.2012.713876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pervaiz F, Pervaiz M, Abdur Rehman N, Saif U. FluBreaks: early epidemic detection from Google Flu Trends. J Med Internet Res. 2012;14:e125. doi: 10.2196/jmir.2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou XC, Li Q, Zhu ZL, Zhao H, Tang H, Feng YJ. Monitoring epidemic alert levels by analyzing internet search volume. IEEE Trans Biomed Eng. 2013;60:446–452. doi: 10.1109/TBME.2012.2228264. [DOI] [PubMed] [Google Scholar]
- 66.Shaman J, Karspeck A. Forecasting seasonal outbreaks of influenza. Proc Natl Acad Sci USA. 2012;109:20425–20430. doi: 10.1073/pnas.1208772109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dugas AF, Jalalpour M, Gel Y. Influenza forecasting with Google Flu Trends. PLoS One. 2013;8:e56176. doi: 10.1371/journal.pone.0056176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scarpino SV, Dimitrov NB, Meyers LA. Optimizing provider recruitment for influenza surveillance networks. PLoS Comp Biol. 2012;8:e1002472. doi: 10.1371/journal.pcbi.1002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.St Louis C, Zorlu G. Can Twitter predict disease outbreaks? BMJ. 2012;344:e2353. doi: 10.1136/bmj.e2353. [DOI] [PubMed] [Google Scholar]
- 70.Sofean M, Smith M. A real-time disease surveillance architecture using social networks. Stud Health Technol Inform. 2012;180:823–827. [PubMed] [Google Scholar]
- 71.Corley CD, Cook DJ, Mikler AR, Singh KP. Using web and social media for influenza surveillance. Adv Exp Med Biol. 2010;680:559–564. doi: 10.1007/978-1-4419-5913-3_61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corley CD, Cook DJ, Mikler AR, Singh KP. Text and structural data mining of influenza mentions in web and social media. Int J Environ Res Public Health. 2010;7:596–615. doi: 10.3390/ijerph7020596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Collier N, Son NT, Nguyen NM. OMG U got flu? Analysis of shared health messages for bio-surveillance. J Biomed Semantics. 2011;2(suppl 5):S9. doi: 10.1186/2041-1480-2-S5-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chew C, Eysenbach G. Pandemics in the age of Twitter: content analysis of tweets during the 2009 H1N1 outbreak. PLoS One. 2010;5:e14118. doi: 10.1371/journal.pone.0014118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lampos V, Cristianini N. Nowcasting events from the social web with statistical learning. ACM Trans Intell Syst Technol. 2012;3:72. [Google Scholar]
- 76.Culotta A. Lightweight methods to estimate influenza rates and alcohol sales volume from Twitter messages. Lang Resources Eval. 2013;47:217–238. [Google Scholar]
- 77.Zeng X, Wagner M. Modeling the effects of epidemics on routinely collected data. J Am Med Inform Assoc. 2002;9:S17–S22. [PMC free article] [PubMed] [Google Scholar]
- 78.Oum S, Chandramohan D, Cairncross S. Community-based surveillance: a pilot study from rural Cambodia. Trop Med Int Health. 2005;10:689–697. doi: 10.1111/j.1365-3156.2005.01445.x. [DOI] [PubMed] [Google Scholar]
- 79.Watson JM, Pebody RG. Influenza surveillance and pandemic requirements. In: Van-Tam J, Sellwood C, editors. Pandemic influenza. 2nd edn. CABI; 2013. pp. 9–18. [Google Scholar]
- 80.Hale TM, Cotten SR, Drentea P, Goldner M. Rural–urban differences in general and health-related internet use. Am Behav Sci. 2010;53:1304–1325. [Google Scholar]
- 81.Kiciman E. OMG, I have to tweet that! a study of factors that influence tweet rates. Proceedings of the Sixth International AAAI Conference on Weblogs and Social Media; Trinity College, Dublin, Ireland; 2012.
- 82.Mohebbi M, Vanderkam D, Kodysh J, Schonberger R, Choi H, Kumar S. Google correlate whitepaper. 2011. http://googleproof.org/trends/correlate/whitepaper.pdf (accessed Jan 1, 2013).
- 83.Eysenbach G. Infodemiology and infoveillance tracking online health information and cyberbehavior for public health. Am J Prev Med. 2011;40:S154–S158. doi: 10.1016/j.amepre.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 84.Chunara R, Freifeld CC, Brownstein JS. New technologies for reporting real-time emergent infections. Parasitology. 2012;139:1843–1851. doi: 10.1017/S0031182012000923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hulth A, Rydevik G. GET WELL: an automated surveillance system for gaining new epidemiological knowledge. BMC Public Health. 2011;11:252. doi: 10.1186/1471-2458-11-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khan AS, Fleischauer A, Casani J, Groseclose SL. The next public health revolution: public health information fusion and social networks. Am J Public Health. 2010;100:1237–1242. doi: 10.2105/AJPH.2009.180489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barclay E. Predicting the next pandemic. Lancet. 2008;372:1025–1026. doi: 10.1016/S0140-6736(08)61425-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.