Summary
Epidemics of novel or re-emerging infectious diseases have quickly spread globally via air travel, as highlighted by pandemic H1N1 influenza in 2009 (pH1N1). Federal, state, and local public health responders must be able to plan for and respond to these events at aviation points of entry.
The emergence of a novel influenza virus and its spread to the United States were simulated for February 2009 from 55 international metropolitan areas using three basic reproduction numbers (R0): 1.53, 1.70, and 1.90. Empirical data from the pH1N1 virus were used to validate our SEIR model.
Time to entry to the U.S. during the early stages of a prototypical novel communicable disease was predicted based on the aviation network patterns and the epidemiology of the disease. For example, approximately 96% of origins (R0 of 1.53) propagated a disease into the U.S. in under 75 days, 90% of these origins propagated a disease in under 50 days. An R0 of 1.53 reproduced the pH1NI observations.
The ability to anticipate the rate and location of disease introduction into the U.S. provides greater opportunity to plan responses based on the scenario as it is unfolding. This simulation tool can aid public health officials to assess risk and leverage resources efficiently.
Keywords: Influenza transmission, Susceptible-exposed-infectious-recovered (SEIR) disease-spread modeling, Public health aviation screening, Pandemic response, Points of entry
Introduction
As the world's population becomes ever more closely connected and as the numbers of international flights and air passengers continue to increase, so too has the spread of communicable diseases of public health concern via air travel.1 Novel infectious diseases have emerged and rapidly spread around the globe during the modern jet travel era. Examples include the Severe Acute Respiratory Syndrome (SARS) outbreak that started in southern China in 20022 and the pandemic influenza A (pH1N1) virus that was first reported in Mexico in 2009.3 Recent evidence, including analyses of the spread of pH1N1,4, 5, 6 has demonstrated how quickly transmissible diseases can be spread by air travelers.7, 8 Due to these health issues, the dramatic increase of international aviation travel and security concerns, the term “border” no longer denotes a static, fixed entity that begins and ends at political boundaries, but instead has been extended virtually to include pre- and post-travel geo-temporal space.9 Because of this global interconnectedness, adverse health consequences and economic and travel disruptions can result from the emergence of rapidly spreading novel communicable diseases anywhere in the world.1
Whenever global or regional public health threats emerge, countries predictably implement mitigation measures at their international points of entry.4, 10, 11, 12 Anticipating how such events will emerge and unfold is the essence of preparedness planning, and a critical practice for public health authorities who wish to mitigate the impact of such events.13 Previous aviation point of entry modeling studies have evaluated the potential effectiveness of some measures for mitigating global outbreaks of communicable disease, such as improving the timeliness of diagnostic testing for epidemic diseases,14, 15 and rapidly developing and distributing sufficient quantities of pharmaceutical countermeasures, such as vaccines and antimicrobial drugs.6, 16, 17, 18 Post-pH1N1 airport-based public health interventions including traveler screening have been described.19 While collectively these reports remain useful in analyzing the potential value of public health interventions at airports, to our knowledge no studies have used modeling to provide operational planning guidance assumptions for policy makers and public health authorities who would implement such measures.
For this paper, and as part of pre-pandemic preparedness planning, we estimated the geo-temporal components of disease spread between city pairs via air travel. More specifically, we examined the time-course for infectious air travelers to arrive in the United States (U.S.) from international cities with the highest U.S.-bound flight traffic for the month of February. Previous U.S. modeling and planning efforts for point of entry response to pandemic influenza assumed a system-wide “all or nothing” initiation of traveler screening, aligned with World Health Organization phases. Temporal and geographical risk-based response stratification was underrepresented in those analyses, while spatial considerations and disease epidemiological characteristics guided most airport response planning. This paper describes a method that would allow for a more flexible approach, which could be applied to threats other than pandemic influenza. We used a traditional Susceptible-Exposed-Infectious-Recovered (SEIR) model20, 21 and an illustrative scheduled-flight dataset, to demonstrate how public health authorities could prioritize the allocation of response-resources in the U.S. at point of entry in response to a novel disease that was spreading rapidly outside of North America.
Methods
Disease-spread model
To characterize possible patterns and rates of spread for an emerging infectious disease that could enter North America from various geographic points of origin, a prototypical novel pandemic influenza virus was simulated as an example of a human-to-human transmissible disease that is known to spread rapidly via air travel. The model propagates disease based on population sizes from metropolitan areas using calculations that estimate the dynamics of disease spread within each city and by air travel of individuals from one city to another, as previously described and calibrated by Epstein et al. and Bobashev et al.20, 21 The simulated population is mutually assigned into one of the following disease states: susceptible (S), exposed (E), symptomatically infectious (I_s), asymptomatically infectious (I_a), and recovered (R). These disease states are illustrated in Fig. 1 . A person can move from being susceptible, to exposed, then to either asymptomatically or symptomatically infectious prior to recovery. The model is implemented such that people in the infectious state have a 66.67% chance of being symptomatic and a 33.33% chance of being asymptomatic.18 People who transition to the recovered state would no longer be susceptible and remain in the recovered state for the duration of the simulation. Unlike prior implementation of this model, people in the infectious states were disallowed from entering a dead state in part to maximize person-to-person spread in the early stages of a pandemic and to create an upper boundary for this modeling study. As such the case-fatality proportion was intentionally set to zero.
Figure 1.
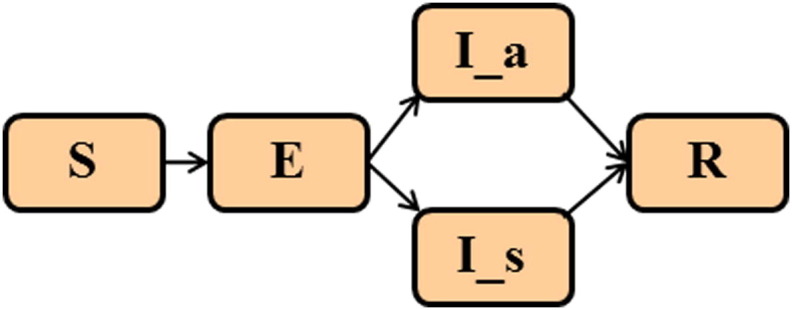
Mutually exclusive, allowable disease states of the model: susceptible (S), exposed (E), asymptomatically infectious (I_a), symptomatically infectious (I_s), and recovered (R).
The model was propagated so that a susceptible person could become exposed, on average for 1.2 days. The exposed person could then become symptomatically infectious or asymptomatically infectious on average for 4.1 days, with a maximum infectious duration of 6 days. Our SEIR model does not differentiate symptomatic and asymptomatic infection in the average infectious period calculation, nor does the model account for enhanced level of infectivity which could for example occur during early stages of influenza while a person is asymptomatic. The simulation evolved in discrete units of 1 day. Selected parameters used in this model are given in Supplemental Table S1. Unlike prior implementations of the model20, 21, 22 which permitted only asymptomatically infectious people to travel, the present implementation permitted both asymptomatically and symptomatically infectious people to travel.
In addition, the model includes 177 major metropolitan areas (see Supplemental Table S2) around the world, including the 100 largest airports, the 100 largest cities worldwide,20, 21 and 35 cities in the U.S. (Supplemental Tables S3 and S4). This approach led to some parts of the globe being sparsely represented, commensurate with the level of direct aviation traffic those regions send to the U.S. All estimates of population data were from sources released in 2007 or later.23, 24, 25 International metropolitan population data were taken from United Nations estimates and the World Gazetteer. U.S. metropolitan population data were based on US Census estimates for the 35 metropolitan statistical areas, totaling 126 million people.
Points of origin
Points of origin were selected on the basis of the numbers of actual flights between international metropolitan areas and the U.S. during February 9–13, 2009. The study used the top 55 points of origin, which accounted for approximately 94% of all international air passengers traveling to the U.S.
America's neighbor countries, Mexico and Canada, each have numerous ground border crossings with the U.S., many of them heavily trafficked; therefore it has been assumed that infectious diseases that originated in Mexico or Canada would quickly propagate to the U.S. through these ground channels. In fact, in the recent pH1N1 pandemic, the virus spread rapidly into the U.S. from Mexico through multiple points of entry.3 For this reason in the points of origin analysis presented here, Mexico, Canada and the U.S. have been treated as a single world region, and we have not calculated times required for an infectious disease to propagate to the U.S. from points of origin in Canada or Mexico.
In contrast, Honolulu was treated as an international point of origin because the state of Hawaii has limited non-aviation links to the rest of the world, including the U.S. In fact, public health authorities watching for the spread of disease to North America from East Asia consider Honolulu an important sentinel site for surveillance.26 Thus, Honolulu's role in the model closely resembles that of other points of origin outside North America.
Flight data
To populate the disease-spread model, weekly direct flight data were extracted from scheduled flights listed in a market intelligence source for aviation industry data (i.e., Diio® LLC27) for one week of February 2009, because the 1918 and H1N1 pandemics emerged in late winter. Database extraction included origins and destinations from 272 airports in 177 metropolitan areas (Supplemental Table S4). Weekly seats for flights between different airports in the same city pair were combined. If the summed seats between a city pair differed in directionality, the average seat counts were applied. Weekly seat counts were converted to daily seat counts and then multiplied by 70% to obtain enplanement estimates for all city pairs. The 70% load factor is consistent with information reported by the Bureau of Transportation and Statistics for the average of scheduled and non-scheduled flights (http://www.bts.gov). The flight matrix used in this study is available upon request. Since this study focused on forecasting and predictions, best available scheduled flight data were used, with the understanding that not all flights into the U.S. were recorded in scheduled flight databases (e.g., chartered flights and unscheduled flights were not represented).
Rationale for model, analysis assumptions, and limitations
Known assumptions for pandemic influenza are summarized in Supplemental Table S1. Model output included how quickly the disease reached the U.S. via the global airline transportation network and also which U.S. metropolitan areas the disease reached first. Time to reach the United States, referred to as “early disease arrival time,” was defined as the number of days it took for the tenth symptomatically infectious person from anywhere in the simulated worldwide network to appear in U.S. metropolitan areas. It is important to note that the “early disease arrival time” includes the period of time during which the virus is spreading person-to-person but unknown to public health authorities. The 10-person threshold was selected on the basis of a 2005 report by the U.S. Department of Homeland Security's National Infrastructure and Simulation Analysis Center, which showed that the arrival of as few as 10 infected people would be sufficient to propagate a disease in the U.S. for simulations seeded with an attack rate of 25% or greater.28 In this case, “attack rate” referred to the cumulative number of people infected at the peak of the pandemic, normalized by the population at the start of the pandemic. In a given simulation, the 10 symptomatically infectious people used to determine the early disease arrival time could be 10 infectious travelers arriving in the U.S., or a combination of infectious travelers and people within the U.S. who became infected following contact with an infectious traveler.
In addition to modeling an R 0 of 1.53 which best approximates the 2009 pH1N1, we selected two additional R 0's, 1.7 and 1.9, to simulate based on evaluations of R 0 values from all actual 20th century influenza pandemics.8, 17, 29, 30 Although R 0 values for some of these pandemics have been estimated as being greater than 1.9,6, 17, 20, 29 research has shown that mitigation measures such as point of entry interventions may be less effective at such high R 0 values.29, 31 For the purpose of pandemic response planning, we assumed that well over 100 exposed persons at each point of origin was a realistic scenario.32
The model accounted for seasonality by assuming that cities within the tropics have the same viral transmission year round, while cities outside the tropics exhibit transmission rates that vary sinusoidally with peak transmission occurring on January 1 in the northern hemisphere and July 2 in the southern hemisphere. Although the model can be implemented with flight data files reflecting true traveler patterns across city pairs for each simulated increment in time, we choose to use representative data from the month of Feb. 2009 for the purpose of this pre-pandemic baseline study. Clearly, the analysis presented in this paper would depend on the granularity of the actual data used by the model.
Visualization
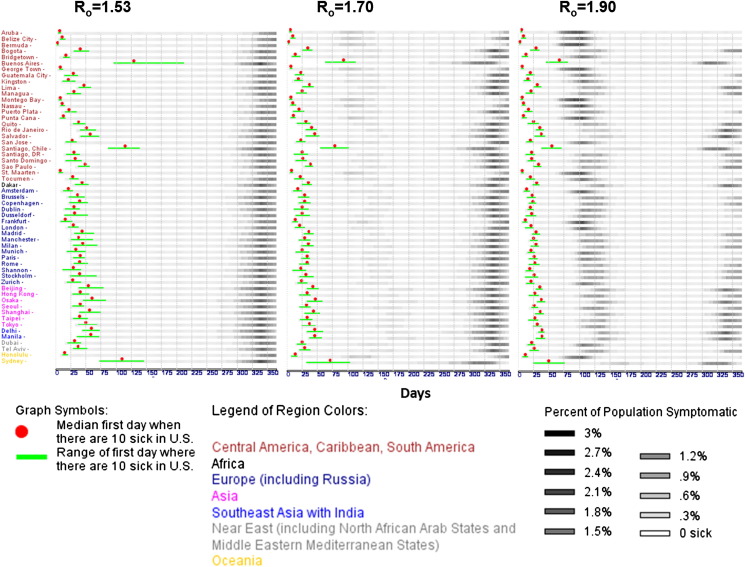
All simulation outputs were interactively accessed via a visualization tool, written in AnyLogic 6, XJ Technologies, St. Petersburg, Russian Federation. We vertically grouped points of origin by world regions, as follows: (1) Central America, Caribbean, South America, (2) Africa, (3) Europe including Russia, (4) Asia, (5) Southeast Asia with India, (6) Near East including North African Arab States and Middle East Mediterranean States, and (7) Oceania (see Supplemental Table S2 for world region classification). Results are shown in the left-, middle-, and right-hand panels of Fig. 2 respectively for the three reproduction numbers modeled: R 0 = 1.53, 1.7, and 1.9. Gray-scale raster plots represent the trial-average number of symptomatically infected people over time as a fraction of the aggregate simulated U.S. metropolitan populations. A solid red dot was superimposed onto each aggregate wave to indicate the median early disease arrival time across all trials for a given point of origin. A green bar illustrating the minimum and maximum range of the early disease arrival time was also superimposed on each aggregate wave. To determine which airports have the highest probability of being affected by disease spread via air travel, we computed the number of times a particular U.S. airport received any of the first ten symptomatically infectious passengers for all points of origin and for points of origin segregated by their respective world regions.
Figure 2.
Predicted disease spread time-course. Simulations were based on 100 exposed people from each international metropolitan point of origin across three reproduction numbers: 1.53, 1.7, and 1.9. The Y-axis displays origins that are first grouped by continent and then sorted alphabetically. The X-axis denotes time in days relative to the start of the disease-spread simulation. Gray-scale raster plots represent the number of infected people across time as a fraction of the aggregate U.S. metropolitan population size employed by the model. Each row shows the median disease spread for all suprathreshold trials (out of 40) in which at least 10 symptomatically infectious people appeared in the United States. In the R0 = 1.53 simulations, early disease arrival times varied from about 5 days to slightly under 225 days from disease emergence in a population. Approximately 95% of origins resulted in median early disease arrival under 75 days, with 85% of those origins resulting in median early disease arrival in less than 50 days. In the R0 = 1.7 simulations, early disease arrival time ranged from about 5 days to approximately 100 days. Approximately 95% of origins resulted in median early disease arrival in less than 50 days, with 37% of those origins resulting in median early disease arrival in less than 25 days. In the R0 = 1.9 simulations, early disease arrival time varied from about 5 days to slightly under 80 days. Approximately 96% of origins resulted in median early disease arrival under 50 days, with 53% of those origins showing median early disease arrival in less than 25 days.
Results
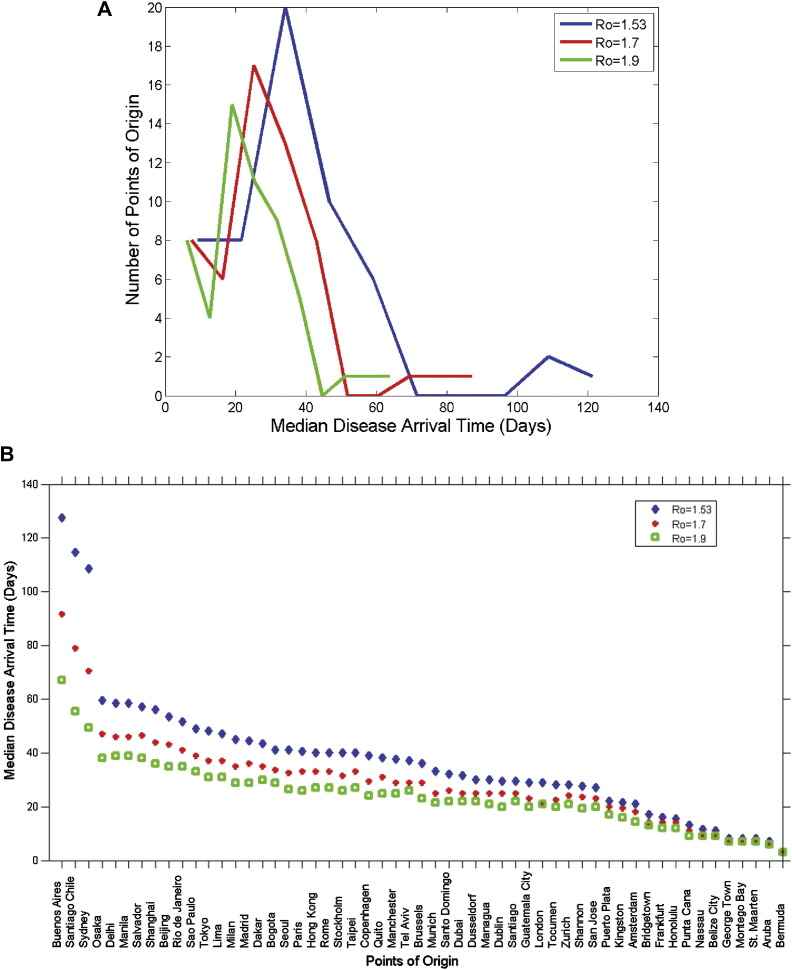
The time-course of disease entry into the U.S is presented in Fig. 2 as aggregate pandemic waves from all points of origin seeded for three R 0's. For response planning purposes, we are most interested in the time leading up to the day in which ten infectious people appear in the U.S. (red dot). It is apparent that at least two clusters of median early disease arrival times appeared for each panel of Fig. 2. The separation in clusters was most pronounced from the R 0 = 1.53 simulation, followed by a decrease in separation as the R 0 values increased (see Fig. 3 A). More specifically, median early disease arrival times in the 25th and 75th percentile were under and over 25 days, respectively (see Table 1 ). This observation suggests that response planning could be conducted differently for points of origin depending on their respective quartile and R 0 classifications.
Figure 3.
Predicted median early disease arrival times. A. Histogram of median early disease arrival time for all points of origin and R0. B. Median early disease arrival time sorted by speed of arrival into the U.S. for every point of origin and R0.
Table 1.
Distribution of medians based on early disease arrival times (days).
| R0 | 25th percentile | 50th percentile | 75th percentile |
|---|---|---|---|
| 1.53 | 24.5 | 36 | 44.75 |
| 1.7 | 21.75 | 29 | 35.5 |
| 1.9 | 18.25 | 23 | 29.5 |
Disclaimer: Table is included for illustrative purposes only. Medians can change from day to day depending on actual travel patterns.
When points of origin were grouped by their respective world regions (see Table 2 ), median early disease arrival times from Central America, the Caribbean, South America, Europe, the Near East, and Oceania (Honolulu) were shorter than those from Asia, Africa, Southeast Asia including India, and Oceania (Australia). Fig. 3B, in which median early disease arrival times into the U.S were plotted in decreasing magnitude for each point of origin and R 0, further elucidates this trend.
Table 2.
Median disease arrival time by world regions.
| World regions | Median early disease arrival time (days) |
||
|---|---|---|---|
| R0 = 1.53 | R0 = 1.7 | R0 = 1.9 | |
| Africa | 43.5 | 35 | 30 |
| Asia | 47 | 37 | 31 |
| Central America, Caribbean, South America | 26 | 22 | 19 |
| Europe | 33 | 27 | 23 |
| Near East (North African Arab States, Middle East Mediterranean States) | 34 | 27 | 24 |
| Oceania – Honolulu | 15.5 | 14 | 12 |
| Oceania – Sydney | 108.5 | 70.5 | 49.5 |
| Southeast Asia including India | 58.5 | 46 | 39 |
Disclaimer: Table is included for illustrative purposes only. Medians can change from day to day depending on true travel patterns.
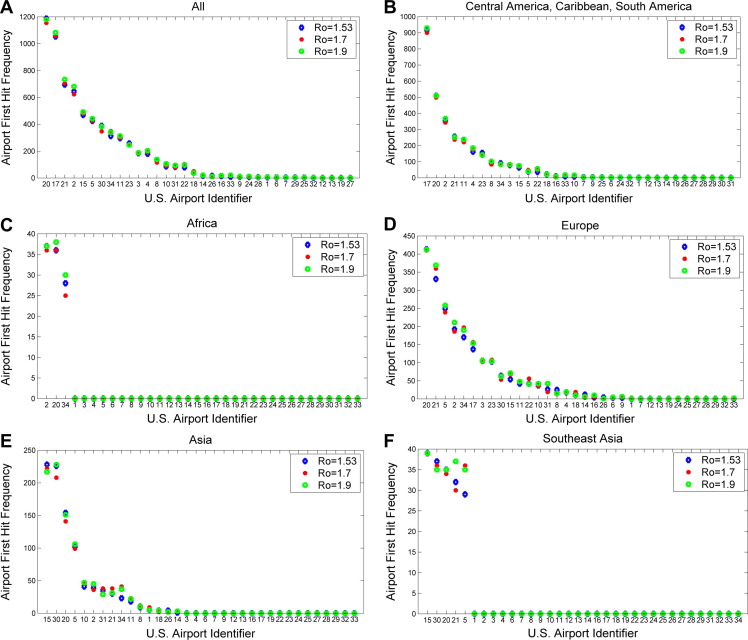
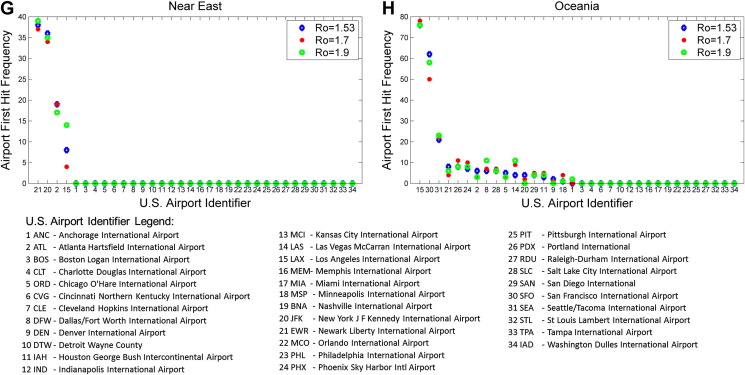
A summary examination of how U.S. airports would be affected by the first ten symptomatic people entering the U.S. revealed that New York, Miami, Newark, Atlanta, Los Angeles would experience the earliest impact (Fig. 4 A). Further, detailed examinations of how these airports would be affected by points of origin from specific world regions are also presented: Central America, Caribbean, or South America (Fig. 4B), Africa (Fig. 4C), Europe (Fig. 4D), Asia and Southeast Asia (Fig. 4E and F), Near East (Fig. 4G) and Oceania (Fig. 4H). Our analysis indicated that Los Angeles and San Francisco airports would experience the earliest impact for disease originating from Asia, Southeast Asia and Oceania, while New York and Atlanta airports would experience one of the earliest impacts for diseases originating from all other world regions.
Figure 4.
Airport first hit frequency. Calculations were based on the number of times a given airport was an entry point for any of the first 10 symptomatically infectious passengers over the course of all simulation trials. Analysis of simulation results for three reproduction numbers (1.53 in blue, 1.7 in red, and 1.9 in green) are presented according to world regions. (A) When considering all infectious disease origination points, analysis indicates that JFK (New York) would experience the earliest impact followed by MIA (Miami), EWR (Newark), ATL (Atlanta), LAX (Los Angeles), ORD (Chicago), SFO (San Francisco), and others. (B) For Central and South American disease origination points, MIA would experience the earliest impact followed by JFK, ATL, EWR, and IAH (Houston). (C) For African disease origination points, ATL and JFK would experience the earliest impact followed by IAD. (D) For European disease origination points, JFK and EWR would experience the earliest impact, followed by ORD, ATL, IAD (Washington), MIA and others. For (E) Asian and (F) Southeast Asian disease origination points, LAX and SFO would experience the earliest impact followed by JFK, ORD, EWR and others. For (G) Near East disease origination points, EWR and JFK would experience the earliest impact followed by ATL and LAX. For (H) Oceania disease origination points, LAX would experience the earliest impact followed by SFO, and others.
Unlike our points of origin analysis, when we validated our model based on data from the 2009 pH1N1, we did not treat the U.S. and Mexico as one mixing body. Rather, we used Mexico City as a proxy for the village of La Gloria, Veracruz, Mexico, from where some of the earliest pH1N1 cases were reported.3 In addition, we replicated the mean U.S. incidence rate of 0.997%, as reported by the CDC for pH1N1 cases in the U.S. between April and July 23, 200933 based on the U.S. cities used in the model (listed in Supplemental Table 2). In order to replicate the mean incidence rate of 0.997%, we evaluated two parameters: 1) The basic reproduction number, which represents a measure of the average number of people in a totally susceptible population to whom one infected individual transmits a disease, was allowed to vary between 1.4 and 1.7 in increments of 0.01. 2) The number of days it takes the model to predict the targeted mean incidence rate which best matches July 23, 2009, i.e., the 165th day after Feb 15th, 2009. We confirmed that our model generated a U.S. incidence rate of 1.0482% (standard deviation of 0.0067%) on the 165th day after disease onset for an R 0 of 1.53; these parameters are very close to the reported mean incidence rate of 0.997% by July 23, 2009. An R 0 of 1.53 is well within published estimates of R 0 for the 2009 pH1N1 which ranged from 1.4 to 1.6.3, 34
Discussion
Since the 1968 H3N2 (Hong Kong) influenza pandemic, global air travel has nearly increased by a factor of ten, from 261 million passengers35 worldwide to more than 2.5 billion passengers in 2010.36, 37 Air travel contributes greatly to the rapidity of communicable disease transfer across international borders and thus federal, state, and local public health authorities must understand and plan for aviation point of entry response to these threats.5, 9, 13 The model results presented here resemble those from related modeling studies in that they suggest an important influence of geographic origin of the outbreak on the timing and location of disease introduction into the U.S. during an emerging disease event.38 In this respect, the range of public health tools available for point of entry intervention (observation, health information distribution, health questionnaires, individual screening, illness response, contact investigations, isolation and quarantine, etc.) could be activated based on anticipated city-to-city spread of the disease in question, rather than being “turned on” system-wide unnecessarily, at great potential cost to taxpayers and need for use of government resources.
Our model output shows how quickly the disease reached the U.S. via the global airline transportation network and which U.S. metropolitan areas the disease reached first. We found variance in arrival times and locations of infectious passengers from origin points at different global locales. Model results indicated that a staggered “turn on” of point of entry responses according to strategic risk assessments, resources available, and time required to mount an effective response, would be feasible and should guide response planning.
The model, as well as the actual events during the SARS and pH1N1 outbreaks of the past decade, reinforced that a short time window would be available to implement measures at points of entry that could be effective in mitigating disease spread.13 As occurred during pH1N1, the initial outbreak and subsequent spread could go undetected for weeks or even months before the first cases were identified and laboratory-confirmed,32 drastically curtailing the time and options available to respond effectively. Since the U.S. essentially serves as a global hub for aviation travel with 88 international arrival and departure points for regularly scheduled commercial flights27; with an additional 113 U.S. airports that received passengers from international charter, private and/or air ambulance flights in 2010,39 it is not surprising that once a novel infectious disease emerges and begins person-to-person spread near international transportation hubs anywhere in the world, that novel disease will in all likelihood appear in the U.S. within days or, at most, a few weeks' time. Spurred by a rise in global traveler numbers and favorable changes in aviation regulations in many countries, the dramatic increase in the sheer number of aviation international arrival and departure points located within geographically and politically distinct entities,9 has resulted in the creation of new pathways for passengers – and diseases – to enter into local communities worldwide. Our model could prove useful for those countries with multiple entry points to consider when planning for a pandemic.
Although this paper describes new data for policy makers and planners to use for planning of public health interventions at point of entry, the effectiveness of point of entry interventions should be examined closely so that resources are not diverted from community infection control and prevention efforts, where they could have greater impact. Point of entry interventions for influenza have been questioned, for example, because of the many travelers who could be infected but not symptomatic at the time of entry (thus avoiding detection), greatly reducing the efficacy of any public health intervention. Computer simulations have demonstrated that even drastic travel restrictions (e.g., border closure) would achieve only limited delays in the introduction of a severe novel influenza virus, for example, into the U.S. during the early stages of an outbreak.17, 20, 30, 40, 41, 42 Indeed, the World Health Organization (WHO) discouraged point of entry screening measures both before and during pH1N1,43, 44, 45 and the WHO Director-General Margaret Chan asserted very early in the pandemic that travel restrictions would be counterproductive and “serve no purpose”.37, 46 However, prudence dictates planning for such a contingency as one possible component of a comprehensive and coordinated public health response to an emerging disease threat. There are many drivers behind the decision to screen aviation travelers during novel communicable disease outbreaks spreading regionally or globally; despite recommendations to the contrary, and known and potential limitations to aviation traveler screening activities for novel influenza, many countries mounted broad screening efforts at their borders during pH1N119 nonetheless.
In calibrating an effective public health response, knowing the “where” is just as important as knowing the “what” and the “when.” Thus, the unique geospatial characteristics of specific originating locations and regions need to be identified to accurately assess risk and develop effective response plans for point of entry interventions. For example, if a rapidly unfolding outbreak were taking place in Central America, it would make sense to anticipate higher need for public health resources focused on direct flights coming to the U.S. from high-volume contributors such as Managua and Guatemala City. Although other passengers from Central America may come to the U.S. via connecting flights, these travelers would be so low in number as to present minimal risk, and to expend resources to address them initially may divert from more effective efforts with the bulk of travelers arriving on direct flights at a few U.S. locations. Our analysis of 2009 flight data revealed that, by volume, the airports most affected by flights from Central America would be Miami (MIA), New York (JFK), Atlanta (ATL), Newark (EWR), and Houston (IAH). With this information, the U.S. can plan, prioritize, and position initial public health resources accordingly. For example, from December 21, 2010–March 31, 2011, the U.S. point of entry response to the cholera outbreak in Haiti included delivering Travelers' Health Alert Notices (T-HANs) with symptoms and medical information to arriving travelers from Haiti at the highest-volume receiving U.S. airports: Miami, New York (JFK), Fort Lauderdale, and San Juan, Puerto Rico. Out of 77,121 travelers who arrived at these ports from Haiti during this period, U.S. Customs and Border Protection and the U.S. Centers for Disease Control and Prevention provided T-HANs to about 73,474 (95%) travelers.
This approach seems intuitive, but others have observed that this risk-based, targeted approach to bolstering surveillance and response capacities at key airports has been the exception rather than the norm, and that careful planning for public health response at key points of entry can be more effective than the usual first response of implementing travel restrictions.13, 41, 47
In light of current global economic uncertainties and the overall decline of resources for public health in the U.S. and other countries, current planning and future point of entry interventions for global disease outbreaks must optimize the resources that are available, quickly and accurately assess risks and prioritize efforts to maximize impact of intervention efforts.48 The “node-to-node” approach embodied in our model will allow public health officials to preserve resources for longer-term, community-based mitigation and prevention activities by targeting response to those points of entry likely to be affected earliest.
Limitations
Although scientific modeling and simulation have proven to be effective tools for visualizing and planning responses to outbreaks of infectious diseases, in particular for influenza pandemics,28, 42, 49 these analyses should not be the only drivers for planners or policy makers when anticipating future outbreaks.50, 51, 52 As with other studies reported in the literature, this model shows that the speed with which a communicable disease spreads to the U.S. will depend on the R 0 of the disease in question.g This finding could represent a limitation of using the model for planning, since R 0 is quite difficult to determine for an emerging disease.
Further, the model employed in this study is not agent-based, and the attack rates reported by agent-based models cannot be computed identically using this equation-based model. This is because the total number of people who became infectious during a complete model run cannot be tallied without modeling each person as an agent.
Although we used the well-known and accepted assumptions of a prototypical pandemic influenza virus to examine early disease arrival times, the recent pH1N1 pandemic makes it clear that novel viruses do not always behave as expected. Some disease-specific assumptions and outcomes of the model may therefore not be useful in planning for future disease outbreaks.
Since this study focused on forecasting and predictions, future scheduled nonstop flights for a single month were utilized as the basis for estimating travel for a simulated period of one year. This approach simplifies the complex phenomenon of international travel. A rigorous comparison between scheduled seats and actual enplanements (which would include, among other things, chartered flights and actual load factors), was not conducted in part because such data for non-U.S. cities were not readily available. The spread of disease during travel from infectious passengers to susceptible passengers was not modeled separately from spread in the general population. Variation in travel based on seasonality was not included and thus the analysis presented in this study only applies to a disease that spread in the month of February 2009 based on travel in that month. Finally, this approach neglects travel between cities that does not take place via nonstop flights on the same day – in particular, connecting flights through gateway cities, or non-aviation means such as car or train for nearby cities such as Paris, France, and Amsterdam, The Netherlands. Thus, new analyses should be conducted periodically, and especially as an event is unfolding which may require point of entry intervention.
Future research
Policy makers and public health authorities must always consider aviation points of entry as potential foci for interrupting or slowing the global spread of disease. Therefore, future modeling should consider how to maximize effectiveness of public health interventions while minimizing passenger delays and travel disruptions, by analyzing the efficacy and cost of specific interventions such as dispensing antivirals at points of entry, health questionnaires with real-time data entry using handheld devices, and fever detection using technology such as thermal imaging. Further analysis and reporting of worldwide government responses to the emergence of pH1N1 and other regional disease outbreaks would help to fill in some of the information gaps regarding point of entry interventions.
Future models should also examine various pathogens (e.g., smallpox, pneumonic plague, a SARS-like virus) and their rates of spread. The rate of global spread from specific geographic locations around the world should also be studied using various known assumptions and pathogen characteristics. Those results could usefully be compared with the pH1N1 and SARS experiences.
Conclusion
Our findings indicate that time to disease entry to the U.S. during the early stages of an emerging pandemic would vary and can be predicted based on point of origin and point of entry into the U.S. This ability to anticipate the rate and location of disease introduction into the U.S. provides greater opportunity to plan responses based on the scenario as it is unfolding. This simulation tool can aid public health officials to assess risk and leverage resources efficiently via targeted and scalable border mitigation measures, especially at key U.S. airports that would be most expected to bear the initial brunt of an international outbreak.
Disclaimer
The findings and conclusions in this paper are those of the author(s) and do not necessarily represent the views of the Centers for Disease Control and Prevention, the Federal Aviation Administration (FAA), or the U.S. Department of Transportation (DOT). The United States Government, the FAA, and the DOT make no any warranty or guarantee, expressed or implied, concerning the content or accuracy of these views. Approved for Public Release; Distribution Unlimited. Case Numbers 11-2678, 11-2756.
Conflict of interest
All authors have confirmed that they have no conflict of interest either financially or via personal relationships as defined by Travel Medicine and Infectious Disease.
Acknowledgments
This research was funded by The MITRE Corporation's Bio-Threat Aircraft Warning System project and HHS/CDC/DGMQ Purchase Order 200-2010-F-37011. Dr. Goedecke of RTI International was partially funded by the Models of Infectious Disease Agent Study (MIDAS) Cooperative Agreement from NIGMS (U01 GM070698) and by The MITRE Corporation under Research Agreement # 82209. The authors thank Tonia Korves and Nicki Cohen for critically reviewing the manuscript. Dr. Hwang received financial salary supported from Rick Sciambi, John Piescik, and Alan Moore.
Footnotes
Although influenza was used, the model can be extrapolated for other communicable diseases of public health importance.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tmaid.2011.12.003.
Appendix. Supplementary material
References
- 1.Relman D., Choffnes E., Mack A. The National Academies Press; Washington, DC: 2010. Infectious disease movement in a borderless world: workshop summary. [PubMed] [Google Scholar]
- 2.Peiris J.S.M., Yuen K.Y., Osterhaus A.D.M.E., Stöhr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 3.Fraser C., Donnelly C.A., Cauchemez S., Hanage W.P., Van Kerkhove M.D., Hollingsworth T.D. Pandemic potential of a strain of influenza A (H1N1): early findings. Science. 2009;324:1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao B., Li X.-W., Mao Y., Wang J., Lu H.-Z., Chen Y.-S. Clinical features of the initial cases of 2009 pandemic influenza A (H1N1) virus infection in China. N Engl J Med. 2009;361:2507–2517. doi: 10.1056/NEJMoa0906612. [DOI] [PubMed] [Google Scholar]
- 5.Khan K., Arino J., Hu W., Raposo P., Sears J., Calderon F. Spread of a novel influenza A (H1N1) virus via global airline transportation. N Engl J Med. 2009;361:212–214. doi: 10.1056/NEJMc0904559. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y., Sugimoto J.D., Halloran M.E., Basta N.E., Chao D.L., Matrajt L. The transmissibility and control of pandemic influenza A (H1N1) virus. Science. 2009;326:729–733. doi: 10.1126/science.1177373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grais R.F., Ellis J.H., Glass G.E. Assessing the impact of airline travel on the geographic spread of pandemic influenza. Eur J Epidemiol. 2003;18:1065–1072. doi: 10.1023/a:1026140019146. [DOI] [PubMed] [Google Scholar]
- 8.Rvachev L.A., Longini I.M., Jr. A mathematical model for the global spread of influenza. Math Biosci. 1985;75:3–22. [Google Scholar]
- 9.Budd L., Bell M., Warren A. Maintaining the sanitary border: air transport liberalisation and health security practices at UK regional airports. Trans Inst Br Geogr. 2011;36:268–279. [Google Scholar]
- 10.http://www.who.int/csr/sars/travel/airtravel/en/index.html (accessed 31.05.11).
- 11.Nishiura H., Kamiya K. Fever screening during the influenza (H1N1-2009) pandemic at Narita International Airport, Japan. BMC Infect Dis. 2011;11:111. doi: 10.1186/1471-2334-11-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venkatesh S., Memish Z.A. SARS: the new challenge to international health and travel medicine. East Mediterr Health J. 2004;10:655–662. [PubMed] [Google Scholar]
- 13.Hosseini P., Sokolow S.H., Vandegrift K.J., Kilpatrick A.M., Daszak P. Predictive power of air travel and socio-economic data for early pandemic spread. PLoS ONE. 2010;5:e12763. doi: 10.1371/journal.pone.0012763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brigantic R., Malone J.D., Muller G.A., Lee R., Kulesz J., Delp W.W. Simulation to assess the efficacy of US airport entry screening of passengers for pandemic influenza. Int J Risk Assess Manag Int. 2009;12:290–310. [Google Scholar]
- 15.Malone J.D., Brigantic R., Muller G.A., Gadgil A., Delp W., McMahon B.H. U.S. airport entry screening in response to pandemic influenza: modeling and analysis. Travel Med Infect Dis. 2009;7(4):181–191. doi: 10.1016/j.tmaid.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balcan D., Hu H., Goncalves B., Bajardi P., Poletto C., Ramasco J.J. Seasonal transmission potential and activity peaks of the new influenza A (H1N1): a Monte Carlo likelihood analysis based on human mobility. BMC Med. 2009;7:45. doi: 10.1186/1741-7015-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper B.S., Pitman R.J., Edmunds W.J., Gay N.J. Delaying the international spread of pandemic influenza. PLoS Med. 2006;3:e212. doi: 10.1371/journal.pmed.0030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longini I.M., Jr., Nizam A., Xu S., Ungchusak K., Hanshaoworakul W., Cummings D.A.T. Containing pandemic influenza at the source. Science. 2005;309:1083–1087. doi: 10.1126/science.1115717. [DOI] [PubMed] [Google Scholar]
- 19.Public health measures taken at international borders during early stages of pandemic influenza A (H1N1) 2009: preliminary results. Wkly Epidemiol Rec. 2010;21:186–194. [PubMed] [Google Scholar]
- 20.Epstein J.M., Goedecke D.M., Yu F., Morris R.J., Wagener D.K., Bobashev G.V. Controlling pandemic flu: the value of international air travel restrictions. PLoS ONE. 2007;2:e401. doi: 10.1371/journal.pone.0000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bobashev G., Morris R.J., Goedecke D.M. Sampling for global epidemic models and the topology of an international airport network. PLoS ONE. 2008;3:e3154. doi: 10.1371/journal.pone.0003154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.https://www.epimodels.org/midas/globalmodel.do (accessed 07.11.10).
- 23.http://www.census.gov/popest/metro/CBSA (accessed 26.02.11).
- 24.http://www.world-gazetteer.com/ (accessed 26.02.11).
- 25.http://data.un.org/ (accessed 26.02.11).
- 26.Aiello A.E., Coulborn R.M., Aragon T.J., Baker M.G., Burrus B.B., Cowling B.J. Research findings from nonpharmaceutical intervention studies for pandemic influenza and current gaps in the research. Am J Infect Control. 2010;38:251–258. doi: 10.1016/j.ajic.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 27.https://diio.net/home.shtml (accessed 29.11.11).
- 28.Maclaren J., Brown T., Michelson R.E. The Department of Homeland Security's National Infrastructure Simulation & Analysis Center; 2005. Analysis of Avian Influenza virus issues for the catastrophic assessment task force (CATF) table-top exercise. LAUR-05-9254. [Google Scholar]
- 29.Colizza V., Barrat A., Barthelemy M., Valleron A.-J., Vespignani A. Modeling the worldwide spread of pandemic influenza: baseline case and containment interventions. PLoS Med. 2007;4:e13. doi: 10.1371/journal.pmed.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hollingsworth T.D., Ferguson N.M., Anderson R.M. Will travel restrictions control the international spread of pandemic influenza? Nat Med. 2006;12:497–499. doi: 10.1038/nm0506-497. [DOI] [PubMed] [Google Scholar]
- 31.Ferguson N.M., Cummings D.A.T., Cauchemez S., Fraser C., Riley S., Meeyai A. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 2005;437:209–214. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- 32.Lipsitch M., Lajous M., O'Hagan J.J., Cohen T., Miller J.C., Goldstein E. Use of cumulative incidence of novel influenza A/H1N1 in foreign travelers to estimate lower bounds on cumulative incidence in Mexico. PLoS ONE. 2009;4:e6895. doi: 10.1371/journal.pone.0006895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reed C., Angulo F.J., Swerdlow D.L., Lipsitch M., Meltzer M.I., Jernigan D. Estimates of the prevalence of pandemic (H1N1) 2009, United States, April–July 2009. Emerg Infect Dis. 2009;15:2004–2007. doi: 10.3201/eid1512.091413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chowell G., Echevarria-Zuno S., Viboud C., Simonsen L., Tamerius J., Miller M. Characterizing the epidemiology of the 2009 influenza A/H1N1 pandemic in Mexico. PLoS Med. 2011;8:e1000436. doi: 10.1371/journal.pmed.1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.International_Civil_Aviation_Organization 1968 statistics section. News release. Growth in civil aviation; 1968.
- 36.International_Civil_Aviation_Organization 2010 annual report of the council. ICAO; Montreal: 2010. http://www.icao.int/icaonet/dcs/9952/9952_en.pdf [Google Scholar]
- 37.Warren A., Bell M., Budd L. Airports, localities and disease: representations of global travel during the H1N1 pandemic. Health Place. 2010;16:727–735. doi: 10.1016/j.healthplace.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brownstein J.S., Wolfe C.J., Mandl K.D. Empirical evidence for the effect of airline travel on inter-regional influenza spread in the United States. PLoS Med. 2006;3:e401. doi: 10.1371/journal.pmed.0030401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.http://transtats.bts.gov/DL_SelectFields.asp?Table_ID=261&DB_Short_Name=AirCarriers. (accessed 29.11.11).
- 40.Valleron A.J., Cori A., Valtat S., Meurisse S., Carrat F., Boelle P.Y. Transmissibility and geographic spread of the 1889 influenza pandemic. Proc Natl Acad Sci U S A. 2010;107:8778–8781. doi: 10.1073/pnas.1000886107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferguson N.M., Cummings D.A.T., Fraser C., Cajka J.C., Cooley P.C., Burke D.S. Strategies for mitigating an influenza pandemic. Nature. 2006;442:448–452. doi: 10.1038/nature04795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Germann T.C., Kadau K., Longini I.M., Jr., Macken C.A. Mitigation strategies for pandemic influenza in the United States. Proc Natl Acad Sci U S A. 2006;103:5935–5940. doi: 10.1073/pnas.0601266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.WHO consultation on priority public health interventions before and during an influenza pandemic: working group two: public health interventions. 2011. http://www.who.int/csr/disease/avian_influenza/Public_health_interventions.pdf Available at: (accessed 26.02.11) [PubMed] [Google Scholar]
- 44.World Health Organization Swine influenza – update 3. 2011. http://www.who.int/csr/don/2009_04_27/en/index.html Available at: (accessed 26.02.11) [Google Scholar]
- 45.World_Health_Organization_Writing_Group Nonpharmaceutical interventions for pandemic influenza, national and community measures. Emerg Infect Dis. 2006;12:88–94. doi: 10.3201/eid1201.051371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.http://www.who.int/mediacentre/news/statements/2009/h1n1_20090427/en/index.html (accessed 26.02.11).
- 47.Hufnagel L., Brockmann D., Geisel T. Forecast and control of epidemics in a globalized world. Proc Natl Acad Sci U S A. 2004;101:15124–15129. doi: 10.1073/pnas.0308344101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frieden T.R. A framework for public health action: the health impact pyramid. Am J Public Health. 2010;100:590–595. doi: 10.2105/AJPH.2009.185652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Halloran M.E., Ferguson N.M., Eubank S., Longini I.M., Jr., Cummings D.A.T., Lewis B. Modeling targeted layered containment of an influenza pandemic in the United States. Proc Natl Acad Sci U S A. 2008;105:4639–4644. doi: 10.1073/pnas.0706849105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bauch C.T., Lloyd-Smith J.O., Coffee M.P., Galvani A.P. Dynamically modeling SARS and other newly emerging respiratory illnesses: past, present, and future. Epidemiology. 2005;16:791–801. doi: 10.1097/01.ede.0000181633.80269.4c. [DOI] [PubMed] [Google Scholar]
- 51.Timpka T., Eriksson H., Gursky E.A., Nyce J.M., Morin M., Jenvald J. Population-based simulations of influenza pandemics: validity and significance for public health policy. Bull World Health Organ. 2009;87:305–311. doi: 10.2471/BLT.07.050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riley S. Large-scale spatial-transmission models of infectious disease. Science. 2007;316:1298–1301. doi: 10.1126/science.1134695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.