Highlights
-
•
In vitro screens have very low success rate in follow up preclinical trials.
-
•
In vitro systems do not reflect the complexity of the host.
-
•
The main advantage of Drosophila is its significant similarity with humans.
-
•
Drug screens in Drosophila may increase the success of preclinical trials.
Abstract
Following an expansion in the antibiotic drug discovery in the previous century, we now face a bottleneck in the production of new anti-infective drugs. Traditionally, chemical libraries are screened either using in vitro culture systems or in silico to identify and chemically modify small molecules with antimicrobial properties. Nevertheless, almost all compounds passing through in vitro screening fail to pass preclinical trials. Drug screening in Drosophila offers to fill the gap between in vitro and mammalian model host testing by eliminating compounds that are toxic or have reduced bioavailability and by identifying others that may boost innate host defence or selectively reduce microbial virulence in a whole-organism setting. Such alternative screening methods in Drosophila, while low-throughput, may reduce the cost and increase the success rate of preclinical trials.
Current Opinion in Pharmacology 2013, 13:763–768
This review comes from a themed issue on Anti-infectives
Edited by George P Tegos and Michael R Hamblin
For a complete overview see the Issue and the Editorial
Available online 28th August 2013
1471-4892/$ – see front matter, © 2013 Elsevier Ltd. All rights reserved.
Chemicals related to this manuscript
Isoniazid, pyrazinamide, voriconazole, terbinafine, tarcolimus, posaconazone, 2-aminoacetophenone, deferasirox, methyl paraben and propionic acid.
Introduction
A common countermeasure to the ever-growing antibiotic drug resistance is the production of new effective drugs. Nevertheless, the rate of production of new antibiotics is steadily declining [1]. One reason for this might be the chemical screening methods that rely solely on in vitro culture systems. Traditionally, drug research is moving from in vitro small molecule screens to preclinical assessment in mammalian hosts. There are two problems with this approach: first, tests in mammals are costly and can usually be restricted to a few compounds at a time, and second, in vitro assays are inappropriate to capture the complexity of an infected host [2]. Live hosts are preferable because they enable drug toxicity and bioavailability assessment at the organismal level [3] and [4]. In addition, drugs that might interfere with the host microenvironment or microbial virulence per se can only be assessed upon the interaction of microbes with a host. Thus, quality anti-infective drug assessment in simple model hosts might be a more effective way to identify new drug leads. In this review, we aim to examine the suitability of Drosophila melanogaster as a model organism for anti-infective drug assessment due to its high degree of molecular, cellular and physiological conservation with humans, which allows the modelling of infections that recapitulate aspects of human disease [5•, 6]. In this respect, Drosophila might fill the gap between in vitro screens and preclinical trials or be used directly, instead of in vitro screens.
Advantages of Drosophila in terms of laboratory use
Drosophila has a short life cycle of ∼10 days from egg to sexually mature adult as compared to the ∼2.5 months of mice (Table 1 ). Large numbers of flies can be propagated quickly, since tens of females can produce hundreds of offspring within two weeks. The offspring become sexually mature very early in their adult life, enabling the life cycle to continue [2]. Due to its small size of 2 mm in length thousands of flies can be contained in a space that would normally fit less than 10 mice. In addition, fly food is usually made of grocery store ingredients such as cornmeal, yeast and sucrose, thus the cost of maintenance is quite low. Moreover, there are no ethical concerns or regulated protocols for its use in biomedical research.
Table 1.
Comparison of model organisms most commonly used in drug discovery
| C. elegans | Drosophila | Mouse | ||
|---|---|---|---|---|
| Practical aspects | Embryogenesis and sexual maturation | 3 days | ∼10 days | ∼2.5 months |
| Size | 1 mm | 2 mm | 10 cm | |
| Cost | Low | Low | Medium | |
| Similarity to humans | Number of genes | 21,187 | 15,867 | 34,293 |
| Disease homologs | ∼65% | ∼75% | ∼95% | |
| Physiology | Low | Medium | High | |
| Innate immunity | Low | Medium | High | |
| Genetic tools | Whole-genome RNAi | Yes | Yes | No |
| Tissue/time specific RNAi | No | Yes | No | |
| Gene knockouts | ∼50% | ∼50% | ∼10% | |
| Transgenesis | Easy | Easy | Laborious | |
| Drug testing | Drug delivery | Feeding | Feeding-injection | Feeding-injection |
| Drug quantity | μl | μl–nl | ml–μl | |
| Throughput | High | Low | Very low |
As an advantage over Caenorhabditis elegans, a popular invertebrate model host, drugs can not only be mixed in the fly food but also administered by injection (Table 1). Precise doses of 2–200 nl of drug solutions can customarily be injected in each fly [7] and less than 200 μl on a paper disc suffice to feed 20 flies for 24 hours [8•]. Hence, only small quantities of drugs are required during experiments; yet another reason why drug tests in flies are not expensive. In addition, Drosophila can be used for toxicological studies because the relative toxicity of chemicals in flies correlates well with that in mammals [9]. Finally, Drosophila infection and inflammation can easily be studied in relation to aging overcoming the barrier of long experimental time [10]. This is because Drosophila maximum life span ranges between 60 and 90 days, with 1 day of the fly roughly corresponding to 1 year of humans. That is, flies exhibit aging effects as early as 20 days post the onset of adulthood.
Advantages of Drosophila genetics
Drosophila has a long history as a model organism for genetics and a significant similarity with humans in terms of gene homologs. It has functional homologs for 75% of human disease related genes [11], more than any other invertebrate model host studied today (Table 1 and [12]). Its genome is fully sequenced and is one of the best-annotated among eukaryotes. Thus, many technologies have been developed and techniques are easily and commonly used, such as transgenesis, RNA interference (RNAi) technology and gene microarrays. Double-stranded RNAs have been synthesized for almost all genes and the tools are commercially available for the conditional inactivation of essentially any gene of interest in vivo or in cell culture [13]. For example, Drosophila cells have been used in genome-wide RNAi screens to rapidly identify genes required for replication of influenza and dengue viruses [14, 15]. Furthermore, there are large collections of mutants and transgenic Drosophila stocks maintained at Bloomington and other stock centers around the world (http://flybase.org). Moreover, the Drosophila genome contains fewer genes than humans, and consequently, presents less overall genetic redundancy. This allows for an easier target identification, although multiple or modified drugs might be needed in mammals to affect the multiple gene variants. Finally, a variety of genetic tools and markers are available today in order to study the role of microbial pathogenicity tissue-specifically using the GAL4/UAS system [2]. This is an advantage over other model hosts, because expression of any Drosophila gene can be controlled time and tissue-specifically (Table 1). For example, tissue-specific and temporal RNAi allowed the identification of the JAK/STAT signalling pathway as a regulator of the intestinal immune response and regeneration in the fruit fly [16]. In addition, intestinal damage and regeneration can be studied by flip-out clones of cells emanating from intestinal stem cells [17, 18], as well as mitotic clones using either the β-galactosidase marker or the “Mosaic Analysis with a Repressible Cell Marker” method [2].
Drosophila physiology and the immune system — conservation and significance for mammalian research
Several organs and specific cells fundamental to the immune response are highly conserved between flies and mammals. This is the most significant advantage over all other invertebrate model hosts studied today (Table 1). Flies have a defined brain that interacts with other organs, for example, the fat body and the intestine via cytokines and insulin peptides, respectively [19, 20]. The fat body is the equivalent of the mammalian liver, an innate immunity and a metabolic organ [19, 20]. The fly intestine bears many similarities with that of mammals in terms of cellular and molecular biology and epithelial architecture [2]. Plasmatocytes are the macrophage-like cells of Drosophila that detect and phagocytose microbes and secrete cytokines and antimicrobial peptides [21]. The muscle cells of the Drosophila flight muscle, heart and intestine are stratified or smooth similarly to those of humans and share a role in host response to infection [2, 22]. The Drosophila trachea is an air-transporting organ with similarities to the human vasculature [23]. Finally, the nephrocytes and the malpighian tubules are kidney-like cells with a role in host defence [24, 25].
The Drosophila epithelia that are attached to the cuticle, as well as those of the intestine and the trachea are physical barriers to pathogen invasion and the first to respond to external microbes. Should microbes invade these epithelia other local tissues, such as the Drosophila flight muscle, respond to wound infection eliciting a localized host defence response orchestrated by the highly conserved JNK pathway [22]. Importantly, muscle responses to wound infection appear to be conserved in mice and in humans [22, 26]. On the other hand, when bacteria enter and damage the intestine, they induce enterocyte regeneration, which serves as a defence response to protect the host [17]. Numerous conserved signalling pathways are involved in intestinal regeneration upon infection, including the Wnt/Wg, Notch, Hippo, JNK, INSR/InR, K-Ras/Ras1, JAK-STAT and the NF-κB pathways [27].
In case microbes pass through intestinal or other barrier epithelia, additional mechanisms of protection take place. These include phagocytosis by the plasmatocytes, which are analogous to the mammalian macrophages, and the production of antimicrobial peptides by the fat body [21]. Many bacteria and fungi induce the Toll and/or the immune deficiency (Imd) pathways, which are the two highly conserved NF-κB pathways of the systemic Drosophila immune response [27].
Viral infections elicit systemic immune responses via the universally conserved RNAi mechanism. The Drosophila small interfering RNA pathway is activated by double-stranded viral RNA or DNA [28]. Moreover, DExD/H box helicases, cell autophagy as well as the conserved JAK/STAT, Imd/TNF and/or the Toll/TLR innate immune cascades play a crucial role in responding to viral RNA in flies and mammals [6].
Human related microbes studied in Drosophila
Many human bacterial pathogens have been studied in Drosophila including the Gram-positive bacteria Enterococcus faecalis, Staphylococcus aureus, Steptococcus pneumoniae, Bacillus cereus and Listeria monocytogenes, and the Gram-negative bacteria Vibrio cholerae, Serratia marcescens, Pseudomonas aeruginosa, Salmonella typhimurium, Chlamydia spp., Burkholderia cepacia, Yersinia pseudotuberculosis, Francisella tularensis, Legionella pneumophila and Mycobacterium marinum [7]. Of those, P. aeruginosa and M. marinum may suppress the innate immune response as part of their virulence repertoire [22, 29]. Interestingly, the antibiotics rifampicin, dinitrobenzamide, amikacin and isoniazid show good bioavailability, because when fed to the flies they alleviate systemic M. marinum infection. Of special note, the success of the antituberculosis drugs isoniazid and pyrazinamide against the tuberculosis model microbe M. marinum is facilitated by a boost in host cell autophagy in flies and mammals [30••]. These data suggest that not only direct antibacterial efficacy but also innate immune induction share similarities between flies and mammals and can be exploited for pharmacological assessments in flies.
Intestinal P. aeruginosa induces damage and apoptosis of midgut enterocytes in Drosophila, which in turn induces intestinal stem cell proliferation, a process that is however reversible upon bacteria clearance by the common food preservatives methyl paraben and propionic acid [17]. Strikingly, K-Ras/Ras1 oncogene expressing Drosophila hindgut cells induce tumors and delaminate through the basal side of the epithelium upon P. aeruginosa infection, which is an additional process that can be inhibited by eliminating infection using food preservatives [31•, 32]. Furthermore, 2-aminoacetophenone, a small chemical produced by P. aeruginosa, has been shown to reduce P. aeruginosa virulence in Drosophila and mice [8•]. Finally, researchers have exploited phages as anti-infectives against P. aeruginosa using Drosophila. Fruit flies infected with P. aeruginosa can be treated with bacteriophages MPK1, MPK6 by feeding [33, 34•]. Such findings encourage future assessment of food preservatives and natural or biological products, including bacterial metabolites and bacteriophages, as anti-infectives.
Apart from bacteria, human fungal pathogens can also inflict disease in flies. Candida albicans, Aspergillus fumigatus, Aspergillus hyphae, Cryptococcus neoformans, Cunninghamella berthollethiae, Scedosporium spp. and Fusarium spp. have been studied in flies [13, 35]. Of those, the zygomycete C. berthollethiae has been meticulously studied in combination with chemical modifiers of iron in Drosophila. Enhancers of zygomycetes virulence traditionally used in humans, such as corticosteroids, increase iron supply, and iron availability through treatment with deferoxamine dramatically increases pathogenicity by zygomycetes. Accordingly, iron starvation induced by treatment with the iron chelator deferasirox significantly protects infected flies [36]. Another common antifungal, voriconazole is potent against F. moniliforme and S. apiopermum infection in flies [37]. Moreover, combinatorial drug assessment assays in Drosophila reveal a synergism between voriconazole and terbinafine against Aspergillus fumigatus, similar to that seen in mammals [38]. Recently, another synergy was shown between tarcolimus and posaconazone in flies and mice against Ryzopus oryzae [39•]. Because all of the aforementioned treatments were administered by feeding in flies, while infections were either superficial or systemic, many antifungal drugs appropriate for humans may have good bioavailability and efficacy in flies.
Human related viruses that have been studied in flies include, Dengue virus, Epstein-Barr virus, Hepatitis B virus, Human cytomegalovirus, HIV-1, Influenza A virus, SARS coronavirus, Simian valuolating virus 40, Vaccinia virus, Sindbis virus, Vesicular Stomatitis virus and West Nile virus [6]. The last three of those have also been studied in adult flies, thus allowing the assessment of treatments against them in a whole organism setting. Pertinent to the identification of gene target against these viruses, Drosophila NRAMP and its human homologue NRAMP2 have been identified as necessary for the entry of Sindbis virus into the host cells [40••]. In addition, Drosophila Toll-7 has been identified similar to its mammalian ortholog TLR-7 as important for host defence to infection against Vesicular Stomatitis virus via the induction of cell autophagy [41]. Finally, West Nile virus 3’-untranslated region-derived RNA molecule, known as subgenomic flavivirus RNA, suppresses the siRNA-induced and miRNA-induced RNAi pathways in both mammalian and insect cells [42], indicating that RNAi-based therapies might be a goal for the near future against insect-borne flaviviruses.
Obstacles and disadvantages of the model
Despite the numerous advantages of D. melanogaster as a model organism for the study of anti-infectives, there are also several shortcomings. That flies are infected and maintained at a temperature of 25–29°C can be a problem for the study of pathogens and virulence factors that require the mammalian body temperature, that is, 37°C [13]. Also, its inability to simulate human intestinal anaerobic microflora can be a disadvantage. While microaerophilic and aerotolerant bacteria might be used to infect flies, the presence of oxygen in the fly intestine prohibits fly infections with strict anaerobes, which are plentiful in the human gut [43]. Nevertheless, as with any microbe that is difficult to establish an infection with, specific virulence factors can be expressed or administered to flies to study their virulence. Moreover, pharmacokinetic analyses are still problematic in insects as there is not a precise method to measure the levels of administered drug tissue-specifically and insect xenobiotic metabolism might be very different from that of mammals. Furthermore, as opposed to mammals, Drosophila lacks an adaptive immune system and specialized immune response cells, such as dendritic cells (DC), B and T lymphocytes, which are responsible for immunological specificity and memory [44]. In addition, despite the significant conservation of the core of Drosophila signalling pathways, some of them might be activated differently between flies and mammals. For example, the mammalian Toll/TLR pathway that is directly activated by microbially associated molecular patterns, while the Drosophila Toll is activated indirectly through a cascade of proteases [45] and the mammalian TLR-7 that is localized in intracellular membranes versus the plasma membrane-localized Drosophila Toll-7 [41]. Finally, high-throughput screening for anti-infectives has not been developed in Drosophila and this is its major drawback as compared to other invertebrate hosts (Table 1).
Concluding remarks and Future perspectives
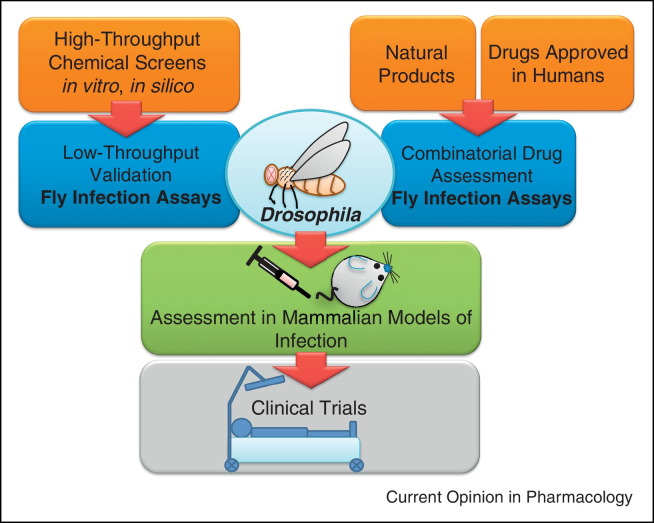
In recent years, the conventional methods used in most pharmacological studies for the discovery of new therapeutic drugs are based either on screening of small molecule libraries for the capacity to induce a specific phenotype in vitro or in silico [46, 47]. However, the efficacy of these methods is very low, because they lack the complex and dynamic host–pathogen interactions, which occur in vivo. Consequently, the use of mammalian hosts in such studies is needed and seems to be very widespread and prevalent nowadays. Even so, using a conventional animal model for this purpose can be time-consuming, laborious and expensive, not to mention the ethical concerns. Exploiting alternative strategies, D. melanogaster is a very promising and useful host, which may cover this gap between the computational or cellular testing studies and the tests in mammals (Figure 1 ). While low-throughput drug assessment in Drosophila has been proven meaningful, large-scale assessments might also be possible on the basis of protocols used for the identification of molecules that modify disease progression in Fragile X syndrome though a screen of 2000 compounds in Fmr1-mutant flies [48] and a screen of 1280 small molecules that identified reserpine as a sleep regulator [49]. In addition, the fly can be used to assess drugs already approved for human use (Figure 1). Indeed, the efficacy of a number of licensed anti-infective agents has been evaluated in Drosophila, demonstrating a significant correlation in drug efficacy between flies and mammals. Therefore, the use of Drosophila for anti-infective drug discovery may be a promising auxiliary tool for preclinical research.
Figure 1.
Drosophila can be used either to validate candidate drugs or in combinatorial drug assessment assays to identify synergistic drug combinations. Flies have significant similarities with humans enabling a facile and cost effective assessment of anti-infective drugs during the interaction of microbes with a host. Hits selected from in vitro or in silico chemical screens can be further screened in Drosophila survival or microbial colonization assays to select drug candidates that will have a higher success rate in preclinical trials. In addition, natural products, for example, microbial secondary metabolites and drugs approved in humans can be tested for the fist time combinatorially in flies to identify synergistic effects between two or more chemicals.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
Funding from Marie Curie (GIG-InfectionCancer) and Fontation Sante (YASante2013) to YA.
References
- 1.Butler M.S., Cooper M.A. Antibiotics in the clinical pipeline in 2011. J Antibiotics. 2011;64:413–425. doi: 10.1038/ja.2011.44. [DOI] [PubMed] [Google Scholar]
- 2.Apidianakis Y., Rahme L.G. Drosophila melanogaster as a model for human intestinal infection and pathology. Dis Models Mech. 2011;4:21–30. doi: 10.1242/dmm.003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomé S., Bizarro C.R., Lehmann M., de Abreu B.R., de Andrade H.H., Cunha K.S., Dihl R.R. Recombinagenic and mutagenic activities of fluoroquinolones in Drosophila melanogaster. Mutat Res. 2012;742:43–47. doi: 10.1016/j.mrgentox.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Kislukhin G., Murphy M.L., Jafari M., Long A.D. Chemotherapy-induced toxicity is highly heritable in Drosophila melanogaster. Pharmacogenet Genomics. 2012;22:285–289. doi: 10.1097/FPC.0b013e3283514395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5•.Pandey U.B., Nichols C.D. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol Rev. 2011;63:411–436. doi: 10.1124/pr.110.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]; An excellent review on the role of Drosophila as a pharmaceutically amenable model organism for human disease.
- 6.Hughes T.T., Allen A.L., Bardin J.E., Christian M.N., Daimon K., Dozier K.D., Hansen C.L., Holcomb L.M., Ahlander J. Drosophila as a genetic model for studying pathogenic human viruses. Virology. 2012;423:1–5. doi: 10.1016/j.virol.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Apidianakis Y., Rahme L.G. Drosophila melanogaster as a model host for studying Pseudomonas aeruginosa infection. Nat Protoc. 2009;4:1285–1294. doi: 10.1038/nprot.2009.124. [DOI] [PubMed] [Google Scholar]
- 8•.Kesarwani M., Hazan R., He J., Que Y., Apidianakis Y., Lesic B., Xiao G., Dekimpe V., Milot S., Deziel E., Lépine F., Rahme L.G. A quorum sensing regulated small volatile molecule reduces acute virulence and promotes chronic infection phenotypes. PLoS Pathog. 2011;7:e1002192. doi: 10.1371/journal.ppat.1002192. [DOI] [PMC free article] [PubMed] [Google Scholar]; Feeding or injecting a P. aeruginosa metabolite to flies and mice reduces virulence of its own species. An example of how bacterial metabolites from pathogens might help us fight infection with the same pathogens.
- 9.Inamdar A.A., Zaman T., Morath S.U., Pu D.C., Bennett J.W. Drosophila melanogaster as a model to characterize fungal volatile organic compounds. Environ Toxicol. 2012 doi: 10.1002/tox.21825. [DOI] [PubMed] [Google Scholar]
- 10.Rera M., Clark R.I., Walker D.W. Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc Natl Acad Sci U S A. 2012;109:21528–21533. doi: 10.1073/pnas.1215849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reiter L.T., Potocki L., Chien S., Gribskov M., Bier E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 2001;6:1114–1125. doi: 10.1101/gr.169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogel H., Altincicek B., Glöckner G., Vilcinskas A. A comprehensive transcriptome and immune-gene repertoire of the lepidopteran model host Galleria mellonella. BMC Genomics. 2011;12:308. doi: 10.1186/1471-2164-12-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lionakis M. Drosophila and Galleria insect model hosts. New tools for the study of fungal virulence, pharmacology and immunology. Virulence. 2011;2:521–527. doi: 10.4161/viru.2.6.18520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao L., Sakurai A., Watanabe T., Sorensen E., Nidom C.A., Newton M.A., Ahlquist P., Kawaoka Y. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sessions O.M., Barrows N.J., Souza-Neto J.A., Robinson T.J., Hershey C.L., Rodgers M.A., Ramirez J.L., Dimopoulos G., Yang P.L., Pearson J.L., Garcia-Blanco M.A. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cronin S.J., Nehme N.T., Limmer S., Liegeois S., Pospisilik J.A., Schramek D. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science. 2009;325:340–343. doi: 10.1126/science.1173164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apidianakis Y., Pitsouli C., Perrimon N., Rahme L. Synergy between bacterial infection and genetic predisposition in intestinal dysplasia. Proc Natl Acad Sci U S A. 2009;106:20883–20888. doi: 10.1073/pnas.0911797106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang H., Patel P.H., Kohlmaier A., Grenley M.O., McEwen D.G., Edgar B.A. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajan A., Perrimon N. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell. 2013;152:1197. doi: 10.1016/j.cell.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu S.C., Liao C.W., Pan R.L., Juang J.L. Infection-induced intestinal oxidative stress triggers organ-to-organ immunological communication in Drosophila. Cell Host Microbe. 2012;11:410–417. doi: 10.1016/j.chom.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 21.Ulvila J., Vanha-Aho L.M., Rämet M. Drosophila phagocytosis – still many unknowns under the surface. APMIS. 2011;119:651–662. doi: 10.1111/j.1600-0463.2011.02792.x. [DOI] [PubMed] [Google Scholar]
- 22.Apidianakis Y., Mindrinos M.N., Xiao W., Tegos G.P., Papisov M.I., Hamblin M.R., Davis R.W., Tompkins R.G., Rahme L.G. Involvement of skeletal muscle gene regulatory network in susceptibility to wound infection following trauma. PLoS ONE. 2007;2:e1356. doi: 10.1371/journal.pone.0001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner C., Isermann K., Fehrenbach H., Roeder T. Molecular architecture of the fruit fly's airway epithelial immune system. BMC Genomics. 2008;9:446. doi: 10.1186/1471-2164-9-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denholm B., Skaer H. Bringing together components of the fly renal system. Curr Opin Genet Dev. 2009;19:526–532. doi: 10.1016/j.gde.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verma P., Tapadia M.G. Immune response and anti-microbial peptides expression in Malpighian tubules of Drosophila melanogaster is under developmental regulation. PLoS ONE. 2012;7:e40714. doi: 10.1371/journal.pone.0040714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apidianakis Y., Que Y.A., Xu W., Tegos G.P., Zimniak P., Hamblin M.R., Tompkins R.G., Xiao W., Rahme L.G. Down-regulation of glutatione S-transferase α4 (hGSTA4) in the muscle of thermally injured patients is indicative of susceptibility to bacterial infection. FASEB J. 2012;26:730–737. doi: 10.1096/fj.11-192484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panayidou S., Apidianakis Y. Regenerative inflammation: lessons from Drosophila intestinal epithelium in health and disease. Pathogens. 2013;2:209–231. doi: 10.3390/pathogens2020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bronkhorst A.W., van Cleef K.W., Vodovar N., Ince I.A., Blanc H., Vlak J.M., Saleh M.C., van Rij R.P. The DNA virus invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc Natl Acad Sci U S A. 2012;109:E3604–E3613. doi: 10.1073/pnas.1207213109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh C.T., Moon C., Choi T.H., Kim B.S., Jang J. Mycobacterium marinum infection in Drosophila melanogaster for antimycobacterial activity assessment. J Antimicrob Chemother. 2013;68:601–609. doi: 10.1093/jac/dks425. [DOI] [PubMed] [Google Scholar]
- 30••.Kim J.J., Lee H.M., Shin D.M., Kim W., Yuk J.M., Jin H.S., Lee S.H., Cha G.H., Kim J.M., Lee Z.W., Shin S.J. Host cell autophagy activated by antibiotics is required for their effective antimycobacterial drug action. Cell Host Microbe. 2012;11:457–468. doi: 10.1016/j.chom.2012.03.008. [DOI] [PubMed] [Google Scholar]; An elegant demonstration of how two antimycobacterial drugs, apart from their direct antibiotic role, induce a conserved in flies and mammals autophagy-dependent host defence response.
- 31•.Bangi E., Pitsouli C., Rahme L.G., Cagan R., Apidianakis Y. Immune response to bacteria induces dissemination of Ras-activated Drosophila hindgut cells. EMBO Rep. 2012;13:569–576. doi: 10.1038/embor.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bacterial clearance or pharmacological inhibition of JNK eliminates innate immune-induced enterocyte invasion.
- 32.Christofi T., Apidianakis Y. Ras-oncogenic Drosophila hindgut but not midgut cells use an inflammation-like program to disseminate to distant sites. Gut Microbes. 2013;4:1–6. doi: 10.4161/gmic.22429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heo Y.J., Lee Y.R., Jung H.H., Lee J.E., Ko G.P., Cho Y.H. Antibacterial efficacy of phages against Pseudomonas aeruginosa infections in mice and Drosophila melanogaster. Antimicrob Agents Chemother. 2009;53:2469–2474. doi: 10.1128/AAC.01646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34•.Chung I.Y., Sim N., Cho Y.H. Antibacterial efficacy of temperate phage-mediated inhibition of bacterial group motilities. Antimicrob Agents Chemother. 2012;56:5612–5617. doi: 10.1128/AAC.00504-12. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bacteriophage treatments are simple to perform in flies. Their specificity and effectiveness in both flies and mice is a promising way to develop biological anti-infective therapies.
- 35.Lionakis M.S., Kontoyiannis D.P. Drosophila melanogaster as a model organism for invasive aspergillosis. Methods Mol Biol. 2012;845:455–468. doi: 10.1007/978-1-61779-539-8_32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chamilos G., Lewis R.E., Hu J., Xiao L., Zal T., Gilliet M., Halder G., Kontoyiannis D.P. Drosophila melanogaster as a model host to dissect the immunopathogenesis of zygomycosis. Proc Natl Acad Sci U S A. 2008;8:9367–9372. doi: 10.1073/pnas.0709578105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamaris G.A., Chamilos G., Lewis R.E., Kontoyiannis D.P. Virulence studies of Scedosporium and Fusarium species in Drosophila melanogaster. J Infect Dis. 2007;196:1860–1864. doi: 10.1086/523765. [DOI] [PubMed] [Google Scholar]
- 38.Lionakis M.S., Lewis R.E., May G.S., Wiederhold N.P., Albert N.D., Halder G., Kontoyiannis D.P. Toll-deficient Drosophila flies as a fast, high-throughput model for the study of antifungal drug efficacy against invasive aspergillosis and Aspergillus virulence. J Infect Dis. 2005;191:1188–1195. doi: 10.1086/428587. [DOI] [PubMed] [Google Scholar]
- 39•.Lewis R.E., Ben-Ami R., Best L., Albert N., Walsh T.J., Kontoyiannis D.P. Tacrolimus enhances the potency of posaconazole against Rhizopus oryzae in vitro and in an experimental model of mucormycosis. J Infect Dis. 2013;207:834–841. doi: 10.1093/infdis/jis767. [DOI] [PMC free article] [PubMed] [Google Scholar]; In vivo combinatorial drug assays reveal that antifungals may work synergistically and in a similar fashion between flies and mice.
- 40••.Rose P.P., Hanna S.L., Spiridigliozzi A., Wannissorn N., Beiting D.P., Ross S.R., Hardy R.W., Bambina S.A., Heise M.T., Cherry S. Natural resistance-associated macrophage protein is a cellular receptor for sindbis virus in both insect and mammalian hosts. Cell Host Microbe. 2011;10:97–104. doi: 10.1016/j.chom.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; Drosophila NRAMP and mammalian NRAMP2 are conserved receptors for Sindbis virus. This put Drosophila in an excellent position for screening for pharmacological inhibitors of Sindbis virus entry into the host cells.
- 41.Nakamoto M., Moy R.H., Xu J., Bambina S., Yasunaga A., Shelly S.S., Gold B., Cherry S. Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity. 2012;36:658–667. doi: 10.1016/j.immuni.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnettler E., Sterken M.G., Leung J.Y., Metz S.W., Geertsema C., Goldbach R.W., Vlak J.M., Kohl A., Khromykh A.A., Pijlman G.P. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and mammalian cells. J Virol. 2012;86:13486–13500. doi: 10.1128/JVI.01104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., Mende D.R. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christofi T., Apidianakis Y. Drosophila immune priming against Pseudomonas aeruginosa is short-lasting and depends on cellular and humoral immunity. F1000Research. 2013;2:76. doi: 10.12688/f1000research.2-76.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gangloff M., Weber A.N.R., Gibbard R.J., Gay N.J. Evolutionary relationships, but functional differences, between the Drosophila and human Toll-like receptor families. Biochem Soc Trans. 2003;31:659–663. doi: 10.1042/bst0310659. [DOI] [PubMed] [Google Scholar]
- 46.Schein C.H., Chen D., Ma L., Kanalas J.J., Gao J., Jimenez M.E., Sower L.E., Walter M.A., Gilbertson S.R., Peterson J.W. Pharmacophore selection and redesign of non-nucleotide inhibitors of anthrax edema factor. Toxins (Basel) 2012;4:1288–1300. doi: 10.3390/toxins4111288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kortagere S. Screening for small molecule inhibitors of Toxoplasma gondii. Expert Opin Drug Discov. 2012;7:1193–1206. doi: 10.1517/17460441.2012.729036. [DOI] [PubMed] [Google Scholar]
- 48.Chang S., Bray S.M., Li Z., Zarnescu D.C., He C., Jin P., Warren S.T. Identification of small molecules rescuing fragile X syndrome phenotypes in Drosophila. Nat Chem Biol. 2008;4:256–263. doi: 10.1038/nchembio.78. [DOI] [PubMed] [Google Scholar]
- 49.Nall A.H., Sehgal A. Small-molecule screen in adult Drosophila identifies VMAT as a regulator of sleep. J Neurosci. 2013;33:8534–8540. doi: 10.1523/JNEUROSCI.0253-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]