Abstract
Endophytic bacteria, which are common in plant tissues, may help to control plant pathogens and enhance plant growth. Camellia oleifera, an oil-producing plant, is widely grown in warm, subtropical, hilly regions in China. However, C. oleifera is strongly negatively affected by C. oleifera anthracnose, which is caused by Colletetrichum fructicola. To find a suitable biocontrol agent for C. oleifera anthracnose, 41 endophytes were isolated from the stems, leaves, and roots of C. oleifera. Bacterial cultures were identified based on analyses of 16S rDNA sequences; most strains belonged to the genus Bacillus. The antagonistic effects of these strains on C. fructicola were tested in vitro. In total, 16 strains inhibited C. fructicola growth, with B. subtilis strain 1-L-29 being the most efficient. Strain 1-L-29 demonstrated antagonistic activity against C. siamense, C. asianum, Fusarium proliferatum, Agaricodochium camellia, and Pseudomonas syringae. In addition, this strain produced indole acetic acid, solubilized phosphate, grew on N-free media, and produced siderophores. To facilitate further microecological studies of this strain, a rifampicin-resistant, green fluorescent protein (GFP)-labeled strain, 1-L-29gfpr, was created using protoplast transformation. This plasmid had good segregational stability. Strain 1-L-29gfpr was re-introduced into C. oleifera and successfully colonized root, stem, and leaf tissues. This strain remained at a stable concentration in the root more than 20 d after inoculation. Fluorescence microscopic analysis showed that strain 1-L-29gfpr thoroughly colonized the root surfaces of C. fructicola as well as the root vascular tissues of Arabidopsis thaliana.
Introduction
Oil extracted from the seeds of the tea-oil camellia (Camellia oleifera Abel), which is rich in unsaturated fatty acids, vitamins, and various antioxidants, is commonly used in China for cooking [1]. Camellia oil is also used as a remedy for bowel, stomach, and burn-associated ailments in traditional Chinese medicine [2]. Thus, C. oleifera is widely commercially cultivated in many parts of China. C. oleifera anthracnose, a fungal infection caused by the Colletetrichum gloeosporioides species complex (CGSC) [3,4], is one of the most serious diseases affecting the tea-oil camellia [5–7]. This disease has severe detrimental effects, such as fruit drop, seed loss, or branch death, and may even lead to plant mortality [4].
Although chemical pesticides are a powerful and cost-effective method of anthracnose prevention, overuse of chemical pesticides might stimulate the development of pesticide-resistant fungal strains, and might have negative effects on human and environmental health [8]. Biocontrol methods, which utilize plant extracts and other biological agents, may be a promising alternative of anthracnose control. For example, endophytic bacteria may act as biocontrol agents, as these bacteria compete with bacterial pathogens.
Endophytes are plant-associated microorganisms that live in plant tissues without negatively affecting the plant host [9,10]. It has been shown that endophytic microorganisms may control plant pathogens [11–13], enhance plant growth [14], and improve phytoremediation. However, previous studies of endophytic bacteria have generally focused on model plants, including Arabidopsis thaliana [15], potatoes [16,17], and soybeans [18], as well as oil-producing trees, including olive trees [19,20], oil palms [21], and Vernicia fordii [22]. For example, the endophytic biocontrol strain Pseudomonas fluorescens PICF7 was extracted from olive trees; this bacterial strain was shown to interact with pathogens to induce a systemic defense response in the host tree; P. fluorescens PICF7 also colonizes and persists on or in wheat and barley root tissues [23–25]. Although endophytic bacteria in C. oleifera have been investigated, previous studies have focused on the identification of antagonistic bacteria. The community structures of endophytic bacteria in C. oleifera have not yet been studied.
Here, we aimed to address this knowledge gap by (i) characterizing the structure of the culturable endophytic microbial community in C. oleifera; (ii) evaluating the potential biocontrol applications of the endophytic bacteria based on their anti-pathogenic behaviors; and (iii) determining the growth-promotion potential and colonization capacities of these endophytic bacteria.
Materials and methods
Sample collection and isolation of endophytic bacteria
Leaf, stem, and root samples were collected from healthy tea-oil camellias growing in the Tianjilin Mountain, Experimental Base of Central South University of Forestry and Technology, Hunan Province, China (28°06′–28°07′ N, 113°02′–113°03′ E). Samples were surface-sterilized using a stepwise washing procedure [ethanol, 0.1% (w/v) mercuric chloride (HgCl2), and water] following Schulz et al. [26]. The sterilized samples were crushed in 5 ml of sterile distilled water for 30 min. Then, 50 μL of each suspension was plated onto nutrient agar (NA). In addition, 100 μL aliquots of the water from the final wash were plated onto NA to check the efficiency of sterilization. All plates, including the control, were incubated at 26–28°C for 7 d. Morphologically distinct colonies were selected and purified. Bacteria were grown in NA for 12 h at 28°C. Sterile glycerol was then added to the bacterial culture to a final concentration of 15%, and the bacterial-glycerol suspension was stored at −80°C until further analysis.
DNA extraction, amplification, and sequencing
The endophytic bacterial isolates were grown in Luria-Bertani medium for 24 h at 28°C. Genomic DNA was extracted using Tiangen Bacterial Genomic DNA Extraction Kits (Tiangen, Beijing, China). To extract DNA, cells were lysed in three cycles of −80°C for 15 min and 37°C for 5 min. We PCR amplified the 16S rRNA gene in 25 μL reaction volumes, each containing 10 μL GoTaq Master mix (2X) (Promega, Wisconsin, USA), 1 μL of DNA template, and 0.2 μM of each primer (27F: 5’-AGAGTTTGATCCTGGCTCAG-3’ and 1492R: 5’-TACTTGTTACGACTT-3’). The PCR cycling condition were 5 min at 95°C; 35 cycles of 15 s at 94°C, 15 s at 50°C, and 90 s at 72°C; and 5 min at 72°C. PCR products were purified using Tiangen PCR Cleanup Kits, and sequenced on an ABI 3730 DNA Analyzer (ABI, CA, USA). Generated sequences were compared with public databases using NCBI BLASTN online (http://www.ncbi.nlm.nih.gov/). A phylogenetic tree was constructed based on this alignment using the neighbor-joining algorithm in MEGA 6.
Identification of a highly antifungal bacterial endophyte
To quantify the antifungal activity levels of the endophytic bacterial taxa isolated from C. oleifera against Colletetrichum fructicola in vitro, the dual culture method was used [27]. We selected C. fructicola to represent the CGSC complex, as this species has a high isolation rate and is extremely virulent. In brief, for each endophyte strain, C. fructicola was inoculated in the middle of a potato dextrose agar (PDA) plate (d = 90 mm), and one endophyte was inoculated in an equilateral triangle approximately 3 cm from the pathogen. Control plates, inoculated with the pathogen but not the endophytic bacteria, were prepared in parallel. Plates were incubated for 7 d at 28°C, and then the diameters of the pathogen colonies were measured. The inhibition rate was then calculated as [(colony radius of the control group—colony radius of the test group) / colony radius of the control group] × 100%.
Antagonistic activity of strain 1-L-29
Preliminary results showed that the endophyte identified as B. subtilis strain 1-L-29 had the highest level of antagonistic activity against C. fructicola. We repeated the dual culture method (following the methods described above) to measure the antifungal activity of strain 1-L-29 against two other members of the C. gloeosporioides complex, C. siamense and C. asianum, as well as three common fungal pathogens of C. oleifera: Athelia rolfsii, Fusarium proliferatum, and Agaricodochium camellia [28]. The antagonistic activity of strain 1-L-29 against a pathogenic plant bacterium, Pseudomonas syringae pv. Tomato D3000, was measured following the methods of Ghorbani [29].
The effects of strain 1-L-29 on the mycelial morphology of C. fructicola were observed under a microscope. Then, 10 μL of spore suspension (106 spores/mL) was mixed with 20 μL of PDA medium containing 1 μL 1-L-29 (OD 0.8) on a sterile glass microscope slide. Each slide was placed under a sterile plastic petri dish with water-soaked filter paper. Plates were incubated at 28°C for 18 h.
Physiological properties of strain 1-L-29
The nitrogen fixation capacity of strain 1-L-29 was estimated on nitrogen-free bromothymol blue (NFb) medium following Swamy et al. [30]; the inorganic phosphorus fixation capacity of this strain was estimated on Pikovskaia’s (PKO) medium following Li et al. [31]; and the organic phosphorus fixation capacity of this strain was estimated on Mongina organic culture medium following Schwyn and Neilands [32]. The siderophore production of strain 1-L-29 was measured on Chrom-Azurol Siderophore (CAS) agar medium following Rahman et al. [33]. Finally, the indole-3-acetic acid (IAA) production of strain 1-L-29 was estimated as previously described [34].
Preparation of protoplasts
Strain 1-L-29 was incubated in LB liquid medium for 12 h at 37°C with shaking at Strain 1-L-29 was incubated in LB liquid medium by shaking at 200 rpm for 12 h at 37°C, and then 5 mL of this bacterial suspension was centrifuged at 4,000 rpm for 10 min at room temperature. The bacterial pellet was resuspended in 0.5 mL SMMP and lysozyme was added. SMMP medium was prepared by mixing equal volumes of 4X Penassay broth and 2X SMM (0.5 M sucrose, 0.02 M Maleate, and 0.02 M MgC12, pH 6.5 adjusted with NaOH). The cells were treated with 0.2, 5, 10, 20, 30, or 40 mg/mL lysozyme at 37°C with shaking at 120 rpm, and protoplast formation was determined after 0.5, 1, 1.5, and 2 h. Lysozyme was removed by centrifugation at 4,000 rpm for 5 min, followed by washing once in 2 mL of SMMP. The protoplasts were centrifuged again at 4,000 rpm for 5 min, resuspended in 0.5 mL of SMMP, diluted, and spread onto DM3 medium. Each liter of DM3 medium contained 200 mL of 4% agar, 500 mL of 1 M sodium succinate (pH 7.3), 100 mL of 5% casamino acids, 50 mL of 10% yeast extract, 100 mL of 3.5% K2HPO4 and 1.5% KH2PO4, 25 mL of 20% glucose, 20 mL of 1 M NaCl, and 5 mL of filter-sterilized 2% bovine serum albumin (added when the temperature of the mixture was ~55°C). Protoplasts were cultured on DM3 for 2–3 d at 37°C. The rate of protoplast formation was then calculated as (A − B) / A × 100%, where A and B were the number of colonies on the LB medium before and after lysozyme treatment, respectively.
GFP labeling of strain 1-L-29
Strain 1-L-29 protoplasts were transformed following Guo [35]. Plasmid pGFP22 was constructed following Yao [36]. In brief, approximately 20 μL of pGFP22 and 20 μL of SMMP were thoroughly mixed into 0.5 mL of the protoplast SMMP suspension (protoplasts were produced under the optimal conditions determined above). The suspension was then incubated with 1.5 mL of 40% polyethylene glycol (PEG) 6000 in a 37°C water bath for 2 min. After incubation, 0.5 mL of SMMP was added to stop the reaction. The mixture was centrifuged at 4,000 rpm for 5 min, re-suspended in 1 mL of SMMP, and shaken at 60 rpm at 37°C for 2 h. Then, 0.1 mL of the suspension was spread onto DM3 medium containing 100 μg/mL ampicillin and cultured for 2–3 d at 37°C. Successful GFP labeling was confirmed with PCR. Plasmid pGFP22 DNA was purified using the Plasmid Mini Purification Kit. We PCR amplified the plasmid pGFP22 in 25 μL reaction volumes, each containing 10 μL GoTaq Master mix (2X) (Promega, WI, USA), 1 μL of DNA template, and 0.2 μM of each primer (gfpF: 5’-TAA GGG GGA AAT CAC ATG AGT AAA GGA GAA GAA-3’ and gfpR: 5’-GGG GTA CCA TTA TTT TTG ACA CCA GA-3’). The PCR cycling condition were 5 min at 95°C; 30 cycles of 30 s at 94°C, 30 s at 56°C, and 30 s at 72°C; and 5 min at 72°C. GFP-expressing bacteria were visualized using a Carl Zeiss AxioObserver A1 (Carl Zeiss, Jena, Germany) fluorescence microscope.
Biological characteristics of strain 1-L-29gfp
To analyze growth curves, colonies of strain 1-L-29gfp and1-L-29 were inoculated into 100 mL of LB liquid medium, 1-L-29gfp medium supplemented with ampicillin, and cultured overnight at 37°C in a shaking incubator at 60 rpm. After incubation, 1 mL of each overnight culture was inoculated into 100 mL of ampicillin-containing LB, and cultured at 37°C in a shaking incubator at 150 rpm. The OD600 values of the two cultures were measured every 2 h to compare the growth curve of strain 1-L-29 to that of strain 1-L-29gfp.
The antifungal activity levels of strain 1-L-29gfp against Colletetrichum fructicola, Colletetrichum siamense, and Colletetrichum asianum were measured as described above.
The segregational stability of the plasmid was determined with a serial dilution culture method, using successive incubations. Specifically, strain 1-L-29gfp was diluted 1:1,000 in 5 mL antibiotic-free LB liquid medium and cultured for 60 h. Continual dilutions were performed during incubation, and 100 μL aliquots of the bacterial cultures were transferred every 5 h before dilution. Each aliquot was spread onto an antibiotic-free LB plate and cultured at 37°C overnight. Total colonies and GFP-positive colonies were counted under ultraviolet light. The segregational stability of the plasmid was represented by the number of GFP-positive colonies, as a percentage of the total number of colonies.
The growth-promotion potential of 1-L-29gfpr
Strain 1-L-29gfpr, which was used in the inoculation, was grown in LB liquid medium for 24 h at 28°C. The cells were harvested by centrifugation at 4,000 rpm, and bacterial cell suspensions were prepared in sterile water (108 CFU /mL). C. oleifera seeds were soaked in the bacterial suspension for 30 min, and then put on moistened absorbent cotton. Control seeds were soaked in distilled water. All treated C. oleifera seeds were maintained in a plant incubator (Panasonic, Ehime-ken, Japan) at 25°C with a 16-h light/8-h dark cycle. Various plant growth parameters, including fresh weight (FW), dry weight (DW), and root length were measured after 30 d of treatment. DW was measured after drying the samples in a hot air oven at 70°C for 72 h. This experiment was repeated three times.
Colonization of C. oleifera by strain 1-L-29gfpr
Rifampicin-resistant strain 1-L-29gfp (designated strain 1-L-29gfpr) was produced following Glandorf [37]. In brief, rifampicin-resistant mutants of 1-L-29gfp were obtained by transferring colonies of this strain to LB medium agar plates containing increasing concentrations (5, 10, 20, 30, 50, 70, 100 μg/mL) of rifampicin. Strain 1-L-29gfpr, which showed the same antagonistic properties against C. fructicola as strain 1-L-29gfp, was selected.
We next tested whether 1-L-29gfpr was able to colonize the roots of C. oleifera. In brief, strain 1-L-29gfpr was diluted to 108 CFU/mL using sterile water. The roots of two-year-old C. oleifera plants were soaked in the bacterial suspension for 20 min, and then transplanted into sterile soil. Positive controls and negative controls were soaked in the 1-L-29 bacterial suspension and in distilled water, respectively, before planting. Treated C. oleifera plants were kept at 25°C and 70% relative humidity with a 16-h light/8-h dark cycle. Endophytic bacteria were isolated from the roots, stems, and leaves of the treated plants at 0, 1, 3, 5, 7, 10, 15, 20, 25, and 30 d after treatment. Bacteria were isolated as described above, except that the nutrient agar medium was supplemented with 100 μg/mL rifampicin. This experiment was repeated three times.
In planta visualization of strain 1-L-29gfpr
Strain 1-L-29gfpr cells were washed twice in phosphate-buffered saline (1.44 g/L Na2HPO4, 0.24 g/L KH2PO4, 0.20 g/L KCl, 8.00 g/L NaCl; pH 7.4) (PBS) and resuspended in PBS (107 cells/mL) prior to use. The roots of C. oleifera seedlings were then soaked in the 1-L-29gfpr bacterial suspension for 20 min at 28°C. Control plants were soaked in sterilized PBS buffer under the same conditions. All seedlings were transplanted into sterile soil and maintained at 28°C with a 16-h light/8-h dark cycle. Plants were carefully uprooted from pots, and root systems were washed by dipping in tap water. Root samples were exposed to 10 cycles of 30 sec ultrasound treatments. Plant roots were sampled at 0, 24, 48, and 72 h after bacterial inoculation, and GFP fluorescence was visualized using a Carl Zeiss AxioObserver A1 fluorescence microscope with a 450–490 nm excitation filter. To observe whether the colonization mechanisms of strain 1-L-29 in C. oleifera differed from those in other plants, we studied the colonization of Arabidopsis thaliana roots by strain 1-L-29gfpr as described above for C. oleifera.
Data analysis
We performed analyses of variance (ANOVAs) on all data. Multiple comparisons of test data were implemented using Duncan’s new multiple range test at a 5% probability level, using SPSS version 16.0 (SPSS Inc., IL, USA).
Results
Identification of endophytic bacterial isolates
We isolated, purified, and cultured 41 bacterial species from healthy C. oleifera leaves (10 species; 24.4%), roots (23 species; 56.1%), and stems (8 species; 19.5%). The lack of bacterial colonies on control plates indicated that the isolates obtained were endophytic. Based on our 16S rRNA phylogeny, all isolated bacterial taxa fell into the genera Bacillus, Sporosarcina, Paenibacillus, Achromobacter, or Enterobacter (Table 1; S1 Fig).
Table 1. Endophytic bacteria isolated from surface-sterilized C. oleifera.
| Isolates | GenBank Accession No. a | Closest phylogenetic relative (GenBank Accession No.) | Identity (%) |
|---|---|---|---|
| Isolates from leaves | |||
| 1-L-28 | MK133136 | B. amyloliquefaciens (KY685067.1) | 99 |
| 1-L-26 | MK133133 | 99 | |
| 1-L-24 | MK133122 | 99 | |
| 1-L-32 | MK133119 | 99 | |
| 1-L-1 | MK133120 | 99 | |
| 1-L-28 | MK133123 | 99 | |
| 3-L-2 | MK133125 | 99 | |
| 1-L-29 | MK133134 | B. subtilis (MF957285.1) | 99 |
| 1-L-27 | MK133132 | 99 | |
| 1-L-21 | MK133128 | 99 | |
| Isolates from stems | |||
| 1-S-25 | MK133126 | B. amyloliquefaciens (KY685067.1) | 99 |
| 2-S-2 | MK133124 | 99 | |
| 1-S-15 | MK133121 | 99 | |
| 1-S-12 | MK133130 | 99 | |
| 2-S-1 | MK133127 | 99 | |
| 1-S-22 | MK133131 | A. xylosoxidans (LC125142.1) | 99 |
| 1-S-2 | MK133129 | 99 | |
| 2-S-3 | MK133102 | Paenibacillus sp. (JN617220.1) | 99 |
| Isolates from roots | |||
| 1-R-2 | MK133101 | B. cereus (KJ524505.1) | 99 |
| 1-R-4 | MK133103 | 99 | |
| 1-R-6 | MK133105 | 99 | |
| 1-R-7 | MK133106 | 99 | |
| 2-R-1 | MK133107 | 99 | |
| 2-R-2 | MK133108 | 98 | |
| 2-R-3 | MK133109 | 96 | |
| 2-R-4 | MK133110 | 96 | |
| 2-R-7 | MK133113 | 99 | |
| 1-R-9 | MK133117 | 98 | |
| 1-R-10 | MK133118 | 99 | |
| 1-R-1 | MK133100 | Enterobacter sp. (MH725605.1) | 99 |
| 1-R-5 | MK133104 | 99 | |
| 2-R-5 | MK133111 | 99 | |
| 2-R-6 | MK133112 | 97 | |
| 2-R-8 | MK133114 | 99 | |
| 2-R-9 | MK133115 | 97 | |
| 1-R-11 | MK133137 | E. cloacae (CP022148.1) | 99 |
| 1-R-12 | MK133138 | 99 | |
| 1-R-13 | MK133139 | 99 | |
| 1-R-14 | MK133140 | 99 | |
| 2-R-10 | MK133116 | S. luteola (KP100329.1) | 99 |
| 1-R-8 | MK133135 | B. amyloliquefaciens (KY685067.1) | 99 |
Anti-pathogenic and physiological properties of the endophytic bacteria
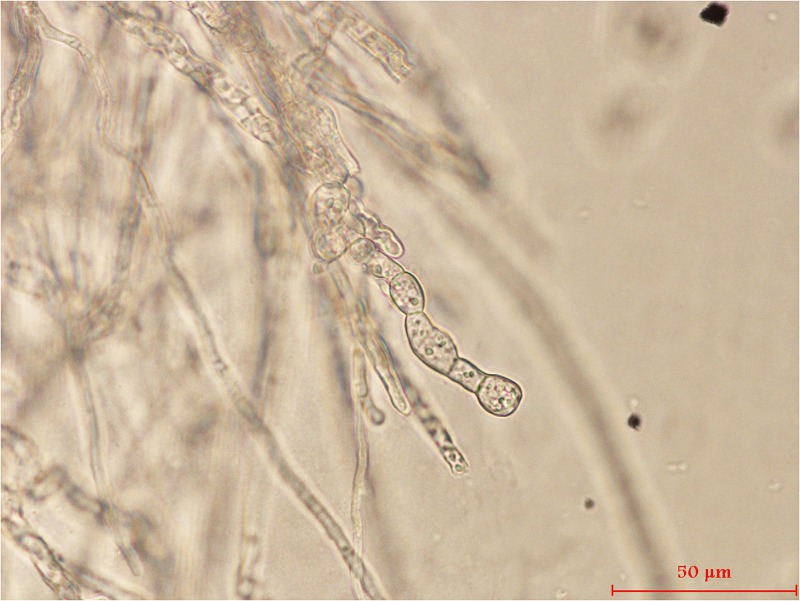
Our dual culture analyses showed that 16 endophytic bacterial strains (of 41 total) exhibited antifungal properties (Table 2). Each of these 16 strains competed with C. fructicola for nutrients or space, or otherwise inhibited the mycelial growth of this fungus. Strain 1-L-29 had the highest inhibition rate against C. fructicola (although not significantly higher than several other strains; Table 2). In addition, micrographs showed that exposure to strain 1-L-29 led to hyphal deformation in C. fructicola, as well as the enlargement of the cytoplasmic vacuoles (Fig 1). For these reasons, strain 1-L-29 was chosen for further analyses. We found that strain 1-L-29 inhibited C. siamense, C. asianum, F. proliferatum, A. camellia, and P. syringae, but not A. rolfsii (Table 3).
Table 2. Inhibition of C. fructicola mycelial growth by endophytic bacteria from C. oleifera.
| Endophyte strains | Pathogenic colony radius (mm) | Pathogen growth inhibition rate (%) |
|---|---|---|
| 1-L-29 | 22.33 ± 4.51 ab | (68.1 ± 6.44) a |
| 1-L-27 | 26 ± 4 a | 62.86 ± 5.71 ab |
| 1-R-8 | 28.67 ± 3.51 a | (59.05 ± 5.02) b |
| 2-S-3 | 27.67 ± 1.53 ab | (60.48 ± 2.18) ab |
| 1-S-22 | 28.33 ± 3.21 ab | (59.52 ± 4.59) b |
| 1-L-26 | 27.47 ± 2.2 ab | (60.76 ± 3.15) ab |
| 1-L-1 | 23.33 ± 3.06 ab | (66.67 ± 4.36) ab |
| 1-L-32 | 25.2 ± 2.43 ab | (64 ± 3.48) ab |
| 1-S-15 | 27.33 ± 2.31 ab | (60.95 ± 3.3) ab |
| 1-S-12 | 25.67 ± 3.21 ab | (63.33 ± 4.59) ab |
| 2-S-2 | 26.33 ± 4.04 ab | (62.38 ± 5.77) ab |
| 1-L-21 | 22.67 ± 4.62 ab | (67.62 ± 6.6) ab |
| 2-S-1 | 24.67 ± 1.15 ab | (64.76 ± 1.65) ab |
| 1-L-28 | 24.67 ± 4.62 ab | (64.76 ± 6.6) ab |
| 1-L-24 | 26.8 ± 1.39 b | (61.71 ± 1.98) ab |
| 1-S-25 | 26 ± 2 b | (62.86 ± 2.86) ab |
| CK | 70.31± 1.71 | 0 |
Data shown are mean ± standard error of three replicates. Different lowercase letters within the same column indicate significant differences (p < 0.05). CK: Control C. fructicola plates, without endophytic strains.
Fig 1. Mycelial morphology of C. fructicola after exposure to B. subtilis strain 1-L-29.
Table 3. Antagonistic activity of strain 1-L-29 against various pathogens.
| Pathogen | Antagonistic effect |
|---|---|
| C. siamense | Positive |
| C. asianum | Positive |
| A. rolfsii | Negative |
| F. proliferatum | Positive |
| A. camellia | Positive |
| P. syringae | Positive |
Strain 1-L-29 produced IAA, grew on N-free media, and solubilized both organic and inorganic phosphorus. When plated on CAS medium, strain 1-L-29 produced a zone of yellowish-orange color, indicating the production of siderophores.
Construction and verification of GFP-labeled strain 1-L-29gfp
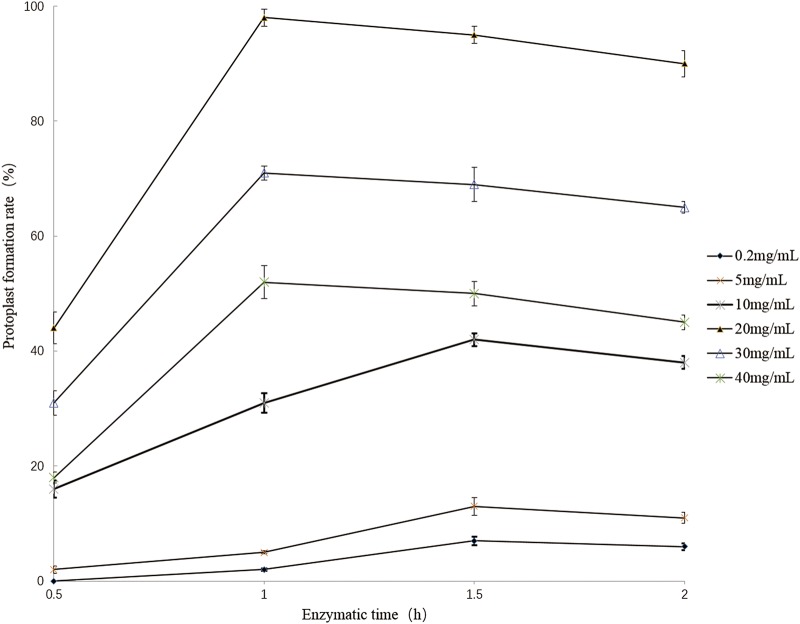
The protoplast formation rate for strain 1-L-29 was greatest (98%) after 1 h of digestion with 20 mg/mL lysozyme (Fig 2). Therefore, these conditions were used to produce protoplasts from strain 1-L-29.
Fig 2. Effects of lysozyme concentration and digestion time on the protoplast formation of Bacillus subtilis strain 1-L-29.
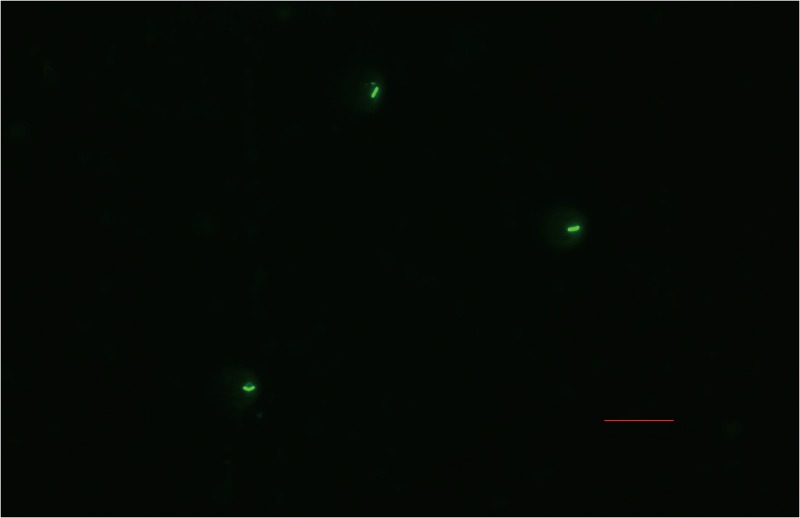
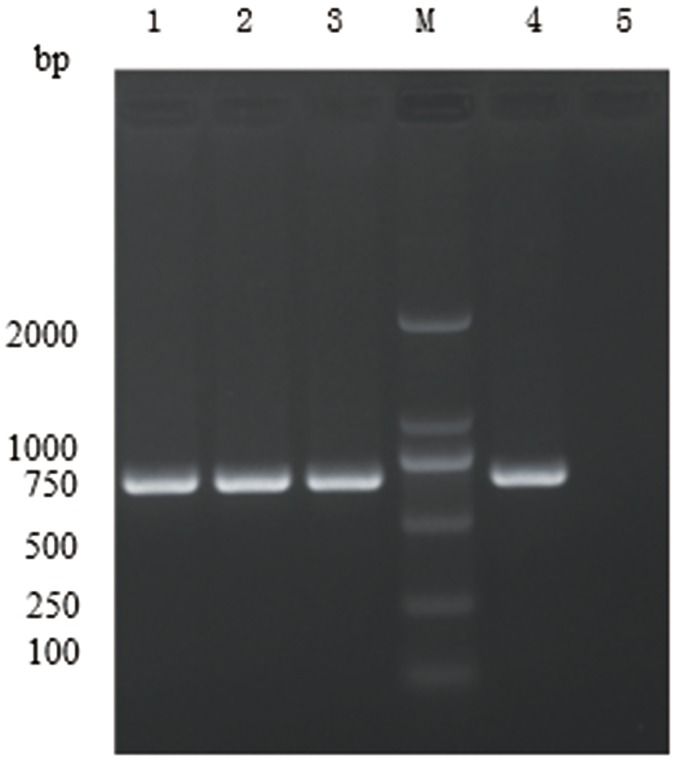
Strain 1-L-29 transformed with the recombinant expression vector pGFP22 fluoresced green (Fig 3), and the expected 750 bp PCR product was amplified using GFP-specific primers (Fig 4). This indicated that the GFP-labeled strain 1-L-29 (strain 1-L-29gfp) had been successfully constructed.
Fig 3. Cells of Bacillus subtilis strain 1-L-29gfp under a fluorescence microscope (40 × magnification).
Fig 4. Electrophoresis of PCR products.
Lane M: DNA marker; lanes 1–3: genomic DNA of strain 1-L-29gfp; 4: plasmid pGFP22; 5: genomic DNA of strain 1-L-29.
The growth curves of wild-type strain 1-L-29 and GFP-labeled strain 1-L-29gfp were similar (S2 Fig): both strains reached the logarithmic growth phase after 2 h of culture, the late stage of logarithmic growth after 10 h, and then entered the stationary phase. Thus, the presence of the plasmid pGFP22 and the expression of GFP did not noticeably affect the growth of 1-L-29. The inhibition rates of 1-L-29gfp against C. fructicola, C. siamense and C. asianum did not differ significantly from those of 1-L-29 (S1 Table). After 10 generations, the plasmid segregational stability of 1-L-29gfp was 88% in continual dilution culture, and 91% after successive incubations (S3 Fig).
The growth-promotion potential of 1-L-29gfpr
Root length, fresh weight, and dry weight were significantly greater in 1-L-29gfpr-inoculated C. oleifera plants than in control plants (Table 4). In 1-L-29gfpr-inoculated seedlings, root length increased by 49.3%, root fresh weight increased by 62.9%, and root dry weight increased by 77.7% as compared to control seedlings.
Table 4. Effects of 1-L-29gfpr on root length, fresh weight, and dry weight.
| Treatment | Root length (cm) | Root fresh weight (g) | Root dry weight (g) |
|---|---|---|---|
| 1-L-29gfpr | 7.27±0.41 a | 0.589±0.045 a | 0.192±0.014 a |
| CK | 4.87±0.33 b | 0.365±0.019 b | 0.108±0.014 b |
Data shown are mean ± standard error of three replicates. Different lowercase letters within the same column indicate significant differences (p < 0.05).
Colonization of C. oleifera by strain 1-L-29gfpr
The mutant strain 1-L-29gfpr was resistant to 100 μg/mL rifampicin. There were no obvious differences in morphology or antifungal activity between strain 1-L-29gfp and strain 1-L-29gfpr. Colonization of C. oleifera by strain 1-L-29gfpr peaked on day 0 after inoculation in the root, and on day 1 after inoculation in the stem and leaf (Table 5). Colonization steadily decreased in all plant tissues after this point, plateauing after day 20.
Table 5. Colonization of C. oleifera after a single inoculation with B. subtilis strain 1-L-29gfpr.
| Treatments | Day post-inoculation | Colonization (103 CFU/g) | ||
|---|---|---|---|---|
| Root | Stem | Leaf | ||
| 1-L-29gfpr | 0 | 17.8 ± 0.97 a | 0 ± 0 f | 0 ± 0 h |
| 1 | 11.97 ± 0.39 b | 6.01 ± 0.17 a | 3.06 ± 0.09 a | |
| 3 | 8.62 ± 0.37 c | 2.33 ± 0.13 b | 1.32 ± 0.06 b | |
| 5 | 5 ± 0.73 d | 1.3 ± 0.19 c | 0.83 ± 0.07 c | |
| 7 | 2.16 ± 0.31 e | 0.66 ± 0.08 d | 0.5 ± 0.03 d | |
| 10 | 1.9 ± 0.04 ef | 0.35± 0.06 df | 0.34 ± 0.06 e | |
| 15 | 1.42 ± 0.09 efg | 0.21± 0.03 df | 0.25 ± 0.05 f | |
| 20 | 1.18 ± 0.03 fg | 0.13± 0.05 df | 0.16 ± 0.02 g | |
| 25 | 0.73 ± 0.03 g | 0.11± 0.05 df | 0.14 ± 0.02 g | |
| 30 | 0.64 ± 0.04 g | 0.1 ± 0.05 df | 0.13 ± 0.02 g | |
| Positive control | 0–30 | - | - | - |
| Negative control | 0–30 | - | - | - |
Positive controls were soaked in the 1-L-29 bacterial suspension; Negative controls were soaked in distilled water. -, no bacteria. Data shown are mean ± standard error of three replicates. Different lowercase letters within the same column indicate significant differences (p < 0.05).
In planta visualization of strain 1-L-29gfpr
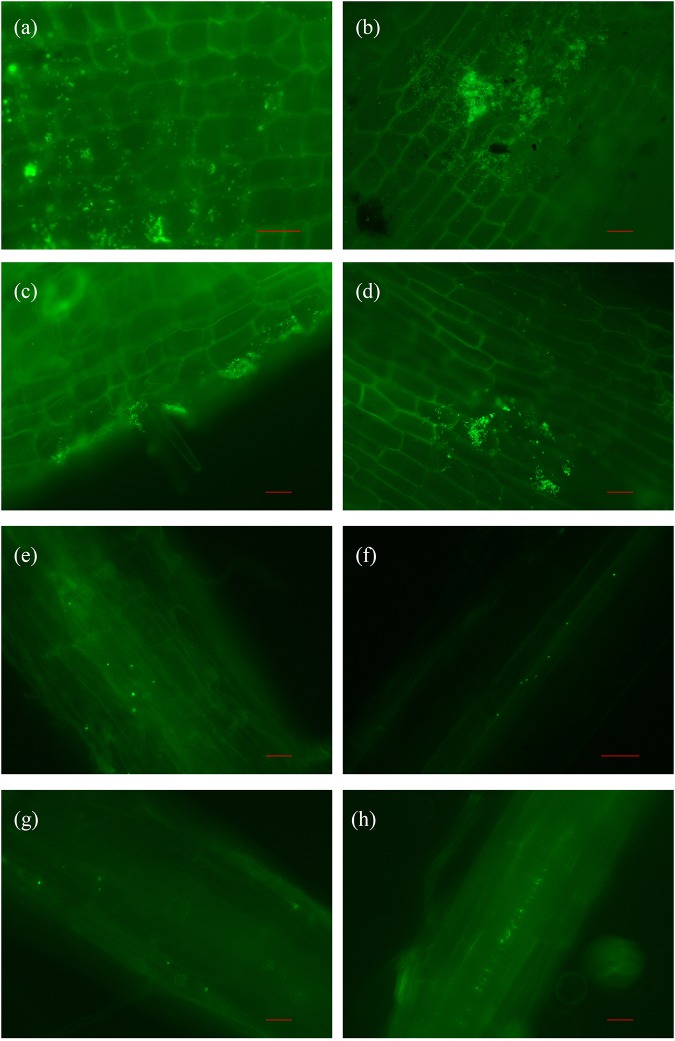
GFP-expressing cells were visible at high densities in the roots of C. oleifera and A. thaliana, clearly demonstrating successful colonization (Fig 5). At 24 h post-inoculation, many 1-L-29gfpr cells were distributed uniformly on the surfaces of the C. oleifera roots (Fig 5a). However, bacterial numbers decreased between 24 and 72 h post-inoculation, and few bacteria were observed on the root surfaces of C. oleifera at 72 h post-inoculation (Fig 5g). Colonies of 1-L-29gfpr cells were also observed on the root surfaces and in the vascular tissues of A. thaliana. In contrast to C. oleifera, few bacteria were observed on the root surfaces of A. thaliana. In the vascular tissues of A. thaliana, more 1-L-29gfpr cells were observed after 72 h (Fig 5h) then after 24 h (Fig 5f).
Fig 5. Colonization of the roots of (a–d) C. oleifera and (e–h) A. thaliana by B. subtilis strain 1-L-29gfpr (300 × magnification).
Colonization after (a) 0 h; (b, e, f) 24 h; (c) 48 h; and (d, g, h) 72 h is indicated by green fluorescence. Scale bars = 50 μm.
Discussion
Although C. oleifera is currently of major agricultural importance in southern China as the source of tea oil, farmers do not manage this plant intensively, and may apply fertilizer only once or not at all. Therefore, it is important to identify a microorganism that can effectively colonize C. oleifera, prevent plant disease, and improve soil fertility. Endophytic bacteria are widely used as biocontrol agents as they can successfully colonize target plants. The endophytic bacteria of C. oleifera have been studied to some extent. For example, He isolated and screened antagonistic endophytic bacteria, aiming to control C. oleifera anthracnose [38]. However, the pathogen identified in this previous study, C. gloeosporioides, was later shown not to be the primary pathogen causing C. oleifera anthracnose [4]. Here, we identified microbes that inhibited the primary pathogen causing C. oleifera anthracnose, C. fructicola. The identified bacterial strains also solubilized phosphate, grew on N-free media, and produced siderophores. These properties rendered the identified strains suitable for the soil environment of Hunan, China, and for the management of C. oleifera.
The endophytic bacteria of C. oleifera were studied using cultivation methods. Most of these bacteria were obtained from the root. This suggested that roots are appropriate habitats for endophytic bacteria. The 41 endophytic bacteria identified in C. oleifera fell into the classes Bacilli, γ-Proteobacteria, and β-Proteobacteria. Of the identified strains, B. subtilis strain 1-L-29 strongly inhibited C. fructicola, as well as several other plant pathogens. Culturable endophytic Bacillus species are frequently isolated from soils and plant tissues, including rhizospheres and leaves [39–42]. Due to the spore-forming abilities of these bacteria, they are highly resistant to adverse ecological conditions. Many Bacillus strains promote plant growth and/or produce a wide variety of antibiotic metabolites; as such, Bacillus strains are often used for the biocontrol of plant diseases [43].
Here, strain 1-L-29 negatively affected the mycelial growth and morphology of C. fructicola without direct contact, possibly because this strain produces antifungal chemicals or emits antifungal volatile organic compounds. Similarly, exposure to the crude culture filtrate extract of Streptomyces sp. MJM5763 inhibited spore germination in C. gloeosporioides and greatly altered mycelial morphology, causing swelling, excessive branching with large vesicles, and stunted hyphal growth [44]. In addition, treatment with B. amylolicefaciens reduced C. lindemuthianum spore numbers and inhibited mycelial growth [45]. B. subtilis 1-L-29 inhibited C. fructicola, C. siamense, C. asianum, F. proliferatum, A. camellia, and P. syringae, but not A. rolfsii (Table 3). Bacillus species, which are considered good sources of molecules with antimicrobial activity, produce well-known substances such as bacitracin, bacteriocins, and antimicrobial lipopeptides. B. subtilis strains also produce volatile organic compounds (VOCs), some of which promote plant growth and/or activate plant defense mechanisms by triggering systemic resistance. Interestingly, our preliminary tests indicated that volatile substances have inhibitory effects on C. fructicola mycelium growth and pigmentation (results not shown).
Strain 1-L-29 might also improve plant growth. That is, the acidic soils in which C. oleifera is grown in southern China commonly have low levels of available phosphorus; this may limit tea-oil yield [46]. As our results indicated that strain 1-L-29 solubilized organic and inorganic phosphorus, treatment with this strain might increase the amount of phosphorus available to C. oleifera, enhancing plant growth. Endophytic bacteria can also promote plant growth by producing the phytohormone IAA [47]. IAA increases root size and distribution, resulting in greater nutrient absorption from the soil. Although we found that strain 1-L-29 produced IAA, we did not quantify the concentration of IAA produced. To test whether strain 1-L-29 improved growth, C. oleifera seeds were soaked in a bacterial suspension. Subsequent phenotypic evaluation showed that bacterial exposure increased root length, fresh weight, and dry weight. This suggested that strain 1-L-29 might promote growth when used to treat C. oleifera seeds.
Strain 1-L-29 might be useful for biocontrol and growth-promotion applications if this strain successfully colonizes C. oleifera. To test this, it was necessary to tag the strain. Although previous studies have primarily used antibiotic markers, such as rifampicin, ampicillin, and kanamycin, to label endophytic bacteria, this method cannot distinguish the labeled strain from microorganisms with natural antibiotic resistance [48]. Labeling with GFP overcomes this problem. Typically, marker genes are introduced into B. subtilis strains using natural competence, protoplast transformation, or electroporation [49]. Electroporation, which is efficient, simple, and widely applicable, is the most commonly-used transformation method [49]. However, a higher voltage is required to penetrate Bacillus cell walls as compared to the cell walls of Escherichia coli, and this higher voltage often kills the target cell. Therefore, the preparation of electrocompetent Bacillus cells is complicated, and protoplast transformation efficiency is low. In this study, we attempted electroporation with strain 1-L-29 under a variety of conditions but were unsuccessful. We thus used the protoplast method, and successfully constructed a GFP-tagged strain.
Strains 1-L-29gfp and 1-L-29 had similar rates of growth and levels of antipathogenic activity, suggesting that exogenous plasmids had no significant effects on bacterial growth and antagonistic activity.
Strain 1-L-29 was labeled with the gfp-containing plasmid. The segregational stability of this the plasmid is generally low. However, we found that 91% of all 1-L-29gfp cells contained the recombinant plasmid after 60 h of serial dilution culture. If we assume that the doubling time of 1-L-29gfp is 20 min in a serial dilution culture, then the 1-L-29gfp cells would have divided approximately 180 times in 60 h, implying a plasmid loss frequency of about 5 × 10−4/generation. The plasmid segregational stability of strain 1-L-29gfp under continuous culture conditions was higher than that of this strain under continuous dilution culture conditions. This might be because the engineered strain was prone to spore formation due to nutrient limitation in the continuous culture, and plasmids were easily lost during spore formation. This suggested that 1-L-29gfp required additional nutrients. To obtain high stability and to prevent genetic burden from affecting growth of the labeled strain, a single copy of the gfp+ gene could be integrated into the bacterial chromosome. However, the expression of the single gfp+ copy in the bacterial genome would result in a relatively low intensity of green fluorescence; this is especially problematic in G+ bacteria with cell envelopes consisting of multiple peptidoglycan layers [50]. Therefore, bacteria with a low intensity of green fluorescence might be barely distinguishable from auto-fluorescent plant tissues. Therefore, a plasmid was chosen to carry the gfp gene.
Our colonization experiments showed that the density of 1-L-29gfpr inside C. oleifera roots and leaves was greater than 0.13 × 103 CFU/g (Table 5). In addition, 1-L-29gfpr colonized the root epidermal surfaces (Fig 5), suggesting endophytic colonization. Strain 1-L-29gfpr colonized the roots, stems, and leaves of C. oleifera, but 1-L-29gfpr density was typically higher in the root. In general, populations of introduced endophytic bacteria remain stable at 103–105 CFU/g in the roots of most plant species investigated to date [51]. However, due to the long treatment time (30 d), the densities of 1-L-29gfpr were lower than have been reported previously. Although bacteria were isolated from the leaves of C. oleifera, the densities of the 1-L-29gfpr colonies in the roots were greater than the densities of these colonies in the leaves. The explanation for this is unclear, but it is possible that the leaf bacteria we isolated entered the plant through the roots and were then transported to the leaves.
The fluorescence microscope images showed that strain 1-L-29gfpr successfully colonized the roots of C. oleifera, which is a woody plant. In the vascular tissues of A. thaliana, more 1-L-29gfpr cells were observed after 72 h (Fig 5h) then after 24 h. This indicated that the strain 1-L-29gfpr also successfully colonized, and was transported throughout, the herbaceous plant A. thaliana. Thus, strain 1-L-29 might be useful as a biocontrol agent in herbaceous plants, as well as in woody plants.
In C. oleifera, strain 1-L-29gfp extensively colonized the root surfaces, forming aggregates. These aggregations were not present in A. thaliana, possibly because plants selectively recruit beneficial rhizobacteria [52].
In summary, our results showed that the endophytic B. subtilis strain 1-L-29, isolated from C. oleifera, might have broad applications as a biocontrol and growth-promotion agent in both woody and herbaceous plants. Our results may be particularly useful for the biological control of C. oleifera anthracnose.
Supporting information
(TIF)
(TIF)
(TIF)
(XLSX)
(PDF)
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Science Foundation for Young Scientists of China (Grant No. 31600515, http://www.nsfc.gov.cn/) and the Natural Science Foundation of Hunan Province, China (Grant No. 2019JJ50999, http://kjt.hunan.gov.cn/zxgz/zkjj/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ma J, Ye H, Rui Y, Chen G, Zhang N (2011) Fatty acid composition of Camellia oleifera oil. Journal Fur Verbraucherschutz Und Lebensmittelsicherheit-Journal of Consumer Protection and Food Safety 6: 9–12. [Google Scholar]
- 2.Cheng YT, Wu SL, Ho CY, Huang SM, Cheng CL, et al. (2014) Beneficial Effects of Camellia Oil (Camellia oleifera Abel.) on Ketoprofen-Induced Gastrointestinal Mucosal Damage through Upregulation of HO-1 and VEGF. Journal of Agricultural and Food Chemistry 62: 642–650. 10.1021/jf404614k [DOI] [PubMed] [Google Scholar]
- 3.Weir BS, Johnston PR, Damm U (2012) The Colletotrichum gloeosporioides species complex. Studies in Mycology: 115–180. 10.3114/sim0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Zhou G-Y, Liu J-A, Xu J (2016) Population Genetic Analyses of the Fungal Pathogen Colletotrichum fructicola on Tea-Oil Trees in China. Plos One 11: e0156841 10.1371/journal.pone.0156841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Chen J, Li D-W, Zheng L, Huang J (2017) CglCUT1 gene required for cutinase activity and pathogenicity of Colletotrichum gloeosporioides causing anthracnose of Camellia oleifera. European Journal of Plant Pathology 147: 103–114. [Google Scholar]
- 6.Xu M, He R, Peng Y, Zeng C-B, Liu Y, et al. (2017) Isolation and Molecular Identification of Colletotrichum gloeosporioides Causing Brown Spot Disease of Camellia oleifera in Hainan of China. Journal of Phytopathology 165: 380–386. [Google Scholar]
- 7.He L, Zhou G, Lu L, Liu J (2009) Isolation and identification of endophytic bacteria antagonistic to Camellia oleifera anthracnose. African Journal of Microbiology Research 3: 315–318. [Google Scholar]
- 8.Abass K, Reponen P, Pelkonen O (2009) Metabolic and interaction properties of selected fungicides, In Fungicides: Chemistry, Environmental, Impact and Health. pp. 25–62. [Google Scholar]
- 9.Ryan RP, Germaine K, Franks A, Ryan DJ, Dowling DN (2008) Bacterial endophytes: recent developments and applications. FEMS Microbiol Lett 278: 1–9. 10.1111/j.1574-6968.2007.00918.x [DOI] [PubMed] [Google Scholar]
- 10.Schulz BJE, Boyle CJC, Sieber TN (2006) Microbial root endophytes. Springer; Berlin: 9. [Google Scholar]
- 11.Innerebner G, Knief C, Vorholt JA (2011) Protection of Arabidopsis thaliana against Leaf-Pathogenic Pseudomonas syringae by Sphingomonas Strains in a Controlled Model System. Applied and Environmental Microbiology 77: 3202–3210. 10.1128/AEM.00133-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forchetti G, Masciarelli O, Alemano S, Alvarez D, Abdala G (2007) Endophytic bacteria in sunflower (Helianthus annuus L.): isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Applied microbiology and biotechnology 76: 1145–1152. 10.1007/s00253-007-1077-7 [DOI] [PubMed] [Google Scholar]
- 13.Kim N, Jeon HW, Mannaa M, Jeong SI, Kim J, et al. (2019) Induction of resistance against pine wilt disease caused by Bursaphelenchus xylophilus using selected pine endophytic bacteria. Plant Pathology 68: 434–444. [Google Scholar]
- 14.Li J, Zheng B, Hu R, Liu Y, Jing Y, et al. (2019) Pseudomonas species isolated from tobacco seed promote root growth and reduce lead contents in Nicotiana tobacum K326. Canadian Journal of Microbiology 65: 214–223. 10.1139/cjm-2018-0434 [DOI] [PubMed] [Google Scholar]
- 15.van der Meij A, Willemse J, Schneijderberg MA, Geurts R, Raaijmakers JM, et al. (2018) Inter- and intracellular colonization of Arabidopsis roots by endophytic actinobacteria and the impact of plant hormones on their antimicrobial activity. Antonie Van Leeuwenhoek International Journal of General and Molecular Microbiology 111: 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen Thanh H, Tran Quang M, Pham Xuan H, Nguyen Thi Thanh T, Furuya N, et al. (2018) Biological control of potato tuber soft rot using N-acyl-L-homoserine lactone-degrading endophytic bacteria. Current Science 115: 1921–1927. [Google Scholar]
- 17.Ardanov P, Sessitsch A, Haggman H, Kozyrovska N, Pirttila AM (2012) Methylobacterium-Induced Endophyte Community Changes Correspond with Protection of Plants against Pathogen Attack. Plos One 7: e46802 10.1371/journal.pone.0046802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Almeida Lopes KB, Carpentieri-Pipolo V, Fira D, Alberto Balatti P, Yanil Lopez SM, et al. (2018) Screening of bacterial endophytes as potential biocontrol agents against soybean diseases. Journal of Applied Microbiology 125: 1466–1481. 10.1111/jam.14041 [DOI] [PubMed] [Google Scholar]
- 19.Mercedes Maldonado-Gonzalez M, Bakker PAHM, Prieto P, Mercado-Blanco J (2015) Arabidopsis thaliana as a tool to identify traits involved in Verticillium dahliae biocontrol by the olive root endophyte Pseudomonas fluorescens PICF7. Frontiers in Microbiology 6: 266 10.3389/fmicb.2015.00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mercedes Maldonado-Gonzalez M, Schiliro E, Prieto P, Mercado-Blanco J (2015) Endophytic colonization and biocontrol performance of Pseudomonas fluorescens PICF7 in olive (Olea europaea L.) are determined neither by pyoverdine production nor swimming motility. Environmental Microbiology 17: 3139–3153. 10.1111/1462-2920.12725 [DOI] [PubMed] [Google Scholar]
- 21.Sunpapao A, Chairin T, Ito S-i (2018) The biocontrol by Streptomyces and Trichoderma of leaf spot disease caused by Curvularia oryzae in oil palm seedlings. Biological Control 123: 36–42. [Google Scholar]
- 22.Qin S, Xing K, Fei SM, Lin QA, Chen XM, et al. (2011) Pseudonocardia sichuanensis sp nov., a novel endophytic actinomycete isolated from the root of Jatropha curcas L. Antonie Van Leeuwenhoek International Journal of General and Molecular Microbiology 99: 395–401. [DOI] [PubMed] [Google Scholar]
- 23.Schiliro E, Ferrara M, Nigro F, Mercado-Blanco J (2012) Genetic Responses Induced in Olive Roots upon Colonization by the Biocontrol Endophytic Bacterium Pseudomonas fluorescens PICF7. Plos One 7: e48646 10.1371/journal.pone.0048646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prieto P, Mercado-Blanco J (2008) Endophytic colonization of olive roots by the biocontrol strain Pseudomonas fluorescens PICF7. FEMS microbiology ecology 64: 297–306. 10.1111/j.1574-6941.2008.00450.x [DOI] [PubMed] [Google Scholar]
- 25.Gomez-Lama Cabanas C, Schiliro E, Valverde-Corredor A, Mercado-Blanco J (2014) The biocontrol endophytic bacterium Pseudomonas fluorescens PICF7 induces systemic defense responses in aerial tissues upon colonization of olive roots. Frontiers in Microbiology 5: 427 10.3389/fmicb.2014.00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulz B, Wanke U, Draeger S, Aust HJ (1993) Endophytes from herbaceous plants and shrubs: effectiveness of surface sterilization methods. Mycol Res 97: 1447–1450. [Google Scholar]
- 27.Lang J-F, Tian X-L, Shi M-W, Ran L-X (2018) Identification of endophytes with biocontrol potential from Ziziphus jujuba and its inhibition effects on Alternaria alternata, the pathogen of jujube shrunken-fruit disease. Plos One 13: e0199466 10.1371/journal.pone.0199466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Junang, Pan Huaping, Wu Nan, Li H (2010) Spatial distribution pattern of major diseases in Camellia oleifera. Forest Pest and Disease 29: 7–10. [Google Scholar]
- 29.Ghorbani S, Harighi B (2018) Characterization of endophytic bacteria with plant growth promotion and biological control potential isolated from walnut trees. Forest Pathology 48: 9. [Google Scholar]
- 30.Swamy CT, Gayathri D, Devaraja TN, Bandekar M, D’Souza SE, et al. (2016) Plant growth promoting potential and phylogenetic characteristics of a lichenized nitrogen fixing bacterium, Enterobacter cloacae. Journal of Basic Microbiology 56: 1369–1379. 10.1002/jobm.201600197 [DOI] [PubMed] [Google Scholar]
- 31.Li H, Jiang Y, Yao T, Hou D, Ma Y, et al. (2018) Isolation, screening, identification and growth promoting characteristics of plant growth promoting rhizobacteria of vegetable crops. Journal of Plant Protection 45: 836–845. [Google Scholar]
- 32.Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Analytical biochemistry 160: 47–56. 10.1016/0003-2697(87)90612-9 [DOI] [PubMed] [Google Scholar]
- 33.Rahman A, Sitepu IR, Tang S-Y, Hashidoko Y (2010) Salkowski’s Reagent Test as a Primary Screening Index for Functionalities of Rhizobacteria Isolated from Wild Dipterocarp Saplings Growing Naturally on Medium-Strongly Acidic Tropical Peat Soil. Bioscience Biotechnology and Biochemistry 74: 2202–2208. [DOI] [PubMed] [Google Scholar]
- 34.Yao T (2002) Characteristics and Biofertilizer of Plant Growth Promoting Rhizobacteria Isolated from Oat and Wheat in Northwest China. Gansu Agriculture University.
- 35.Guo XH, Jia SF, Chen NY, Guo SD, Men DP (1982) Bacillus Protoplasts as Recipients for Plasmid DNA Transformation. Acta Microbiologica Sinica 22: 263–268. [Google Scholar]
- 36.Yao ZS, Chen ZY, Chen ZY, Zheng XB, Zhang J, et al. (2003) Genetically Marking of Natural Biocontrol Bacterium Bacillus subtilis Strains with Green Fluorescent Protein Gene. Chinese Journal of Biotechnology 19: 551–555. [PubMed] [Google Scholar]
- 37.Glandorf DCM, Brand I, Bakker PAHM, Schippers B (1992) Stability of rifampicin resistance as a marker for root colonization studies of Pseudomonas putida in the field. Plant and Soil 147: 135–142. [Google Scholar]
- 38.Li H, Zhou GY, Lu LL, Ang LJ (2009) Isolation and identification of endophytic bacteria antagonistic to Camellia oleifera anthracnose. African Journal of Microbiology Research 3: 315–318. [Google Scholar]
- 39.Hall ME, Wilcox WF (2019) Identification and Frequencies of Endophytic Microbes within Healthy Grape Berries. American Journal of Enology and Viticulture 70: 212–219. [Google Scholar]
- 40.Manjunatha BS, Paul S, Aggarwal C, Bandeppa S, Govindasamy V, et al. (2019) Diversity and Tissue Preference of Osmotolerant Bacterial Endophytes Associated with Pearl Millet Genotypes Having Differential Drought Susceptibilities. Microbial Ecology 77: 676–688. 10.1007/s00248-018-1257-2 [DOI] [PubMed] [Google Scholar]
- 41.Etminani F, Harighi B (2018) Isolation and Identification of Endophytic Bacteria with Plant Growth Promoting Activity and Biocontrol Potential from Wild Pistachio Trees. Plant Pathology Journal 34: 208–217. 10.5423/PPJ.OA.07.2017.0158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Compant S, Mitter B, Colli-Mull JG, Gangl H, Sessitsch A (2011) Endophytes of Grapevine Flowers, Berries, and Seeds: Identification of Cultivable Bacteria, Comparison with Other Plant Parts, and Visualization of Niches of Colonization. Microbial Ecology 62: 188–197. 10.1007/s00248-011-9883-y [DOI] [PubMed] [Google Scholar]
- 43.Wu L, Wu HJ, Qiao J, Gao X, Borriss R (2015) Novel Routes for Improving Biocontrol Activity of Bacillus Based Bioinoculants. Front Microbiol 6: 1395 10.3389/fmicb.2015.01395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palaniyandi SA, Yang SH, Cheng JH, Meng L, S J-W (2011) Biological control of anthracnose (Colletotrichum gloeosporioides) in yam by Streptomyces sp.MJM5763. Journal of Applied Microbiology 111: 443–455. 10.1111/j.1365-2672.2011.05048.x [DOI] [PubMed] [Google Scholar]
- 45.Martins SJ, Faria AF, Pedroso MP, Cunha MG, Rocha MR, et al. (2019) Microbial volatiles organic compounds control anthracnose (Colletotrichum lindemuthianum) in common bean (Phaseolus vulgaris L.). Biological Control 131: 36–42. [Google Scholar]
- 46.Jie L, Wu L, Dong C, Yu Z, Wei C (2018) Development of a soil quality index for Camellia oleifera forestland yield under three different parent materials in Southern China. Soil & Tillage Research 176: 45–50. [Google Scholar]
- 47.Dias ACF, Costa FEC, Andreote FD, Lacava PT, Teixeira MA, et al. (2009) Isolation of micropropagated strawberry endophytic bacteria and assessment of their potential for plant growth promotion. World Journal of Microbiology & Biotechnology 25: 189–195. [Google Scholar]
- 48.Liu Youzhou, Liang Xuejie, Qiao Junqing, Zhang Rongsheng, Chen Z (2014) Bacillus subtilis PTS-394 labeled by green fluorescent protein and its colonization. Acta Phytophylacica Sinica 41: 416–422. [Google Scholar]
- 49.Lu YP, Zhang C, Lv FX, Bie XM, Lu ZX (2012) Study on the electro-transformation conditions of improving transformation efficiency for Bacillus subtilis. Letters in Applied Microbiology 55: 9–14. 10.1111/j.1472-765X.2012.03249.x [DOI] [PubMed] [Google Scholar]
- 50.Fan B, Chen XH, Budiharjo A, Bleiss W, Vater J, et al. (2011) Efficient colonization of plant roots by the plant growth promoting bacterium Bacillus amyloliquefaciens FZB42, engineered to express green fluorescent protein. Journal of Biotechnology 151: 303–311. 10.1016/j.jbiotec.2010.12.022 [DOI] [PubMed] [Google Scholar]
- 51.Lee SW, Ahn I-P, Sim S-Y, Lee S-Y, Seo M-W, et al. (2010) Pseudomonassp. LSW25R, antagonistic to plant pathogens, promoted plant growth, and reduced blossom-end rot of tomato fruits in a hydroponic system. European Journal of Plant Pathology 126: 1–11. [Google Scholar]
- 52.Lakshmanan V, Castaneda R, Rudrappa T, Bais HP (2013) Root transcriptome analysis of Arabidopsis thaliana exposed to beneficial Bacillus subtilis FB17 rhizobacteria revealed genes for bacterial recruitment and plant defense independent of malate efflux. Planta 238: 657–668. 10.1007/s00425-013-1920-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(XLSX)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.