Abstract
A 1998 paper that delineated desirable characteristics, or desiderata for controlled medical terminologies attempted to summarize emerging consensus regarding structural issues of such terminologies. Among the Desiderata was a call for terminologies to be “concept oriented.” Since then, research has trended toward the extension of terminologies into ontologies. A paper by Smith, entitled “From Concepts to Clinical Reality: An Essay on the Benchmarking of Biomedical Terminologies” urges a realist approach that seeks terminologies composed of universals, rather than concepts. The current paper addresses issues raised by Smith and attempts to extend the Desiderata, not away from concepts, but towards recognition that concepts and universals must both be embraced and can coexist peaceably in controlled terminologies. To that end, additional Desiderata are defined that deal with the purpose, rather than the structure, of controlled medical terminologies.
Keywords: Controlled terminology, Ontology, Medical knowledge representation, Philosophy
1. Whence the Desiderata?
It would be a rare biomedical information system that did not require some means for representing data and/or knowledge in a formal, reproducible, and useful way. The uses can be quite varied but they all require some capability for symbolic manipulation. Otherwise, why bother with controlled representation when one could just store raw signals? At the simplest level, symbolic manipulation at least requires a set of symbols (often referred to as identifiers) that distinguish the various elements of data and knowledge. Human-understandable labels (often referred to as terms) for these elements are not always required, 1 but they are usually deemed convenient, if for no other reason than the need to map human-collected data into symbols and to map symbols into human-usable results. Sets of these symbols and labels (or terms and identifiers) are usually referred to as controlled terminologies.
The design and content of controlled terminologies are quite diverse but, ultimately, they serve some common purposes: they must support the capture, storage, manipulation, and retrieval of the information they represent in ways that faithfully preserve and communicate the original information. The construction of a controlled terminology does not always guarantee that successful representation will occur; some approaches will predictably work better than others. Biomedical informatics researchers have been studying and describing these approaches for decades. One attempt to synthesize and summarize that body of work, written in the late 1990s, identified 12 desiderata that reflected the ideas of many researchers of the time [1].
One of the key points in the Desiderata was that terminologies should focus not only on the names of the data and knowledge elements they intended to represent, but also on their underlying meanings. The term “meaning” has several definitions (or meanings!), including “the thing one intends to convey especially by language” [2]. How one conveys these “things” may be accomplished by various methods, depending on what that thing is. For example, if the thing is a particular object in reality (say, a particular person or automobile), its meaning might be conveyed by a proper noun or some unique identifier (e.g., the person’s name or the automobile’s registration number). If the thing is a generalization of some particular set of things in reality (that is, a universal that has some extension, or set of instances in reality), its meaning might be conveyed by a general term for the things or by some set of characteristics that all these things have in common (e.g., “person” or “passenger vehicle, self-propelled, four wheels …”—of course, this raises the need to define these additional terms). Finally, if the thing is an idea, we might refer to it with some description of the idea or by describing the particular instances on which the idea is based.
A particular term is not restricted to a single method for the conveyance of its meanings. Depending on how the thing to which the term corresponds is viewed, it might have multiple aspects that could be conveyed in multiple ways. For example, a controlled terminology for speaking about celestial bodies might contain a term with the name “solar planet,” the meaning of which might be expressed by referring to a list of known planets (“any of Mercury, Venus, Earth …”) or by a intentional definition (“a large body that orbits around the Sun”). 2
The names (terms) used for these things usually serve the purpose of conveying the meanings, when the reader has appropriate background knowledge. The Desiderata attempted to stress the fact that something more formal than the language-specific linguistic shorthand with which humans are comfortable was required to convey meanings in useful ways. Specifically, some formal representation to convey the intended meanings of the data and knowledge elements was requested. Formal representations could, themselves, be represented symbolically (that is, using a controlled terminology) and would be independent of the fashions of language. In the “solar planet” example above, this might be accomplished by including in the terminology elements corresponding to each of the planets, by including elements that correspond to the Sun and the notion of orbit, or by both.
While the formal representation of terms constitutes a type of knowledge, the Desiderata stopped short of using the word “ontology” to refer to this knowledge. This omission was due, in part, to the absence of the term in the reviewed informatics literature, although speakers at the time sometimes referred to it [3], usually as “the O Word” because of the perceived overuse of the term by the computer science community [4], [5]. Instead, the Desiderata attempted to describe this focus on meanings to be “concept oriented.” The use of “concept” was chosen as a way to refer to the meanings of the symbols, as understood by humans. But, the notion of “concept”—an abstract idea generalized from specific instances [2]—is only one aspect of the data and knowledge items that we might wish to represent. As a result, while the Desiderata may be interpreted narrowly, in fact, the work from which they were synthesized related to broader aspects of symbolic representation.
2. Aconceptual terminologies
In “From Concepts to Clinical Reality: An Essay on the Benchmarking of Biomedical Terminologies” [6], Smith holds that terminologies should not rely on the use of concepts. He is troubled by the fact that concepts can be viewed linguistically, psychologically, epistemologically, and ontologically. He sees concept-oriented terminologies, on the one hand, as collections of elements that may or may not correspond to things in reality and, on the other hand, as little more than groupings of the synonymous terms by which humans express ideas.
Smith provides many examples of ways in which terminologies composed of expressions of ideas lead to difficulties: in one, taken from Campbell et alia’s description of the National Library of Medicine’s (NLM) Unified Medical Language System (UMLS), “aspirin” and “Aspergum” refer to the same meaningful element in the UMLS’s Metathesaurus [7]. He holds this paradox to be an artifact of the “conceptualist” view.
He prefers to focus, instead, on terminologies composed of references to universals only, and to their corresponding instances in reality—what he calls the “realist” view. In this view—which assumes that there is only one, universal objective reality—only things in reality would be considered, such as pieces of Aspergum and portions of aspirin. Each of these objects would be instances of one of two, mutually exclusive universals (a universal of gum products made from aspirin, perhaps called “Aspergum,” and universal of portions of aspirin, perhaps called “aspirin”). More general universals would be considered (such as “aspirin-containing drugs” and “organic acids”) which would also extend to real world objects, including (respectively) pieces of Aspergum and portions of aspirin. All of this is done without the messiness of concepts and without any confusion of what is the chewable thing and what belongs on a chemist’s shelf.
3. What is on the side of the patient
Smith considers clinical terminologies, in particular, as domains where concept-orientation should be eschewed in deference to realism. It is natural to consider that a patient in reality has real attributes that can be described and used to support activities such as diagnostic and therapeutic decision-making. Each of these (the patient and the attributes) would be represented in a biomedical terminology as a universal. These universals might or might not have names associated with them as linguistic dressing. The important thing is that their meanings are determined solely by their extensions in reality, not by concepts spawned in human minds as ways of thinking about reality.
Strong arguments can be made for the ontological purity of this approach and for the clarity by which it can support reasoning. After all, the reality is what it is, and is not perverted by the fashionable views of society. A patient who behaves in the same particular manner from year to year might be labeled as having a psychological disease one year, as having a personality disorder the next year, and to be a normal variant the year after that, without any change in the actual patient. Recording the mannerisms themselves, rather than some momentary conceptual perspective, allows us to reconsider past information in the past (and future) contexts, as our understanding of the causes of the mannerisms becomes better understood. In fact, one of the novel ideas in Ledley and Lusted’s 1950 landmark paper was a call for the use of such primary patient attributes for automated medical diagnosis [8].
This approach is strengthened by including unique identifiers not just in the terminology of universals but “on the side of the patient”—that is, each attribute of the patient is, itself, a unique entity in reality and is assigned its own identifier. When a patient’s temperature is measured, the measurement is an instantaneous entity (a perdurant), while the polyps seen on colonoscopy are persisting entities (endurants); each is given an identifier by which they can be referenced, so that multiple temperature measurements can be related to each other and individual polyps can be followed over time.
4. Terminologies of universals
The notion of terminologies that are limited to well-behaved universals, each one clearly understood because of its extension in reality, is appealing and, if possible, would make the lives of clinical system developers much simpler. In fact, many patient records today do, to a limited extent, implicitly represent patient information as extensions in reality, as discussed below, through their data models. While a clinical record may appear to capture that, as Smith charges, a physician fits together a patient and a concept (e.g., a disease), the clinical record is usually capturing (with some unique identifier composed of time, patient, physician, and concept) that at some point in time, the physician is observing attributes of the patient that leads the physician to believe that the signs the patient is manifesting are evidence that the patient has some disease. Thus, while the concept may appear to be separate from the patient, there is a deeper connection that is implied by the design of the patient record [9].
The idea of unique identifiers for aspects “on the side of the patient” that are instances of universals in the terminology is not entirely new. Clinical laboratory systems have long included controlled terminologies for specimens. The terms in such terminologies arguably correspond to universals, since they have extensions in reality, in the form of actual instances of specimens collected from actual patients. When an actual specimen is collected from an actual patient, it is assigned a unique identifier (at New York Presbyterian Hospital, these are called “accession numbers”). Using such identifiers, it is possible to perform multiple measurements on the same specimen (to check for consistency), to perform different measurements on the same specimen (to correlate findings), and to report multiple results on the same specimen (such as preliminary and final results). It is even possible to invalidate a previous measurement, if some problem with an analysis is discovered.
The idea of uniquely enumerating each of the patient’s problems can be traced back to Weed’s landmark paper on problem-oriented medical records [10], later implemented in his PROMIS system. A more elaborate example of unique identifiers for patient attributes can be found in a paper by Barrows and Johnson [11] that describes the assignment of persistent identifiers to patient problems. As a particular problem (or the understanding of the problem) evolves over time, new interpretations can be assigned without losing the reference to the original problem. Smith promotes the extension of this approach (as proposed with Ceusters and Smith [12]) to permeate the patient record. Everything about the patient that is medically salient receives a unique identifier instance that relates it back to a universal in the terminology.
The tokenization of the entire patient record in this way is a bold proposal and, if technically feasible, would open up entire new pathways to patient care, epidemiology, quality assurance, and various other ways to use and reuse patient data. There is no question that such recording should be done more often than it is now, and that coded electronic record systems as they exist today actually do obliterate some of the instance-recording that occurs in patient care. Most notably, effort spent on encoding billable diagnoses and procedures divert valuable resources away from recording the “what it is on the side of the patient” in favor of data that are less-resusable [13].
I set aside here questions of whether the complete extension of this approach can actually be accomplished without paralyzing care providers with attribute-identification tasks (will endoscopists tattoo each polyp so that they can identify them later?) and how such an approach could be studied to gather evidence of its cost-effectiveness and safety. More immediately, we must consider whether a terminology that supports this approach can truly be composed of only universals and not include what have been traditionally understood as conceptual entities.
Laboratory specimen terminologies, as they exist today, typically do not include definitional information about the various specimens (collection methods, equipment used, body parts collected, etc.) beyond their name (e.g., “blue top blood specimen”). However, the meanings of these terms are generally understood, at least to the laboratory technicians, and these meanings are based on general attributes (e.g., “5 ml test tube with heparin and venous blood from the patient”) rather than a set of extensions in reality (e.g., “Specimens 22122, 37812, 45092, … ”). In fact, the terminology could include terms for specimens that don’t actually exist in reality (“blue top saliva specimen”). Such terms would not be included for reasons of pure fantasy (such as support for a pathologist-turned-fiction writer), but rather in anticipation of the eventual occurrence of such a specimen [14]. While there is no need to precoordinate every possible permutation of body part and collection method, there is a need to create terms in a terminology for specimens that will soon be needed, but for which no actual instance yet exists. Thus, if we assume that recording what is on the side of the patient will involve instances of laboratory specimens collected from the patient, and that the laboratory information system would benefit from having some controlled set of well-defined terms for universals to process the actual specimens, we cannot escape the practical need to include terms for things that do not yet actually exist—that is, terms that are, at least temporarily, purely conceptual. The alternative, to add a new specimen term to the laboratory system’s terminology when the first instance of specimen is created, is simply not practical: laboratory systems dictate to the clinicians which specimens are allowed, not the other way around.
Other attribute domains may prove equally messy when trying to limit a terminology to a set of terms for universals. For example, the clinical collection of patient temperature is a routine procedure that will continue regardless of how we represent the resulting data. Recording a number of degrees in the patient record, along with a unique identifier for that recording, is not sufficient for the information to be used in care of the patient; somehow, the notion that it is a body temperature must be conveyed. To really know the patient’s body temperature, we would need to know the kinetic energy of every one of the patient’s molecules. Any measurement process that attempted to detect this would (according to Heisenberg) render the result moot by destroying the patient. Instead, we use a device that detects, imperfectly, the average kinetic energy of a small subset of the patient’s molecules. From this, we make a guess at the patient’s actual temperature, taking into account that the reliability of our estimate is further influenced by the orifice containing the aforementioned molecules. I submit, then, that when we record a patient’s temperature, we can only do so through reference to concepts; we do not have the luxury of “real” universals. We might allow that since this is the best approximation we can make, that patient body temperature is, for all intents and purposes, a universal, or we can acknowledge that we use the instances of measurements to help us create a conceptual representation from which we can reason. The result—what we think we are dealing with and what we do about it—is the same. The advantages of an imperfect universal over an imperfect concept are not self-evident.
Patient temperatures are sometimes needed as primary parameters for patient management, such as hypo- and hyperthermia. Most of the time, however, the temperature is used as a proxy for detecting and monitoring underlying conditions. We speak of the term of “fever” and its more specific, yet less-well-defined term “low-grade fever.” If these terms correspond to the ways we think about various states of patient temperature, then they are concepts. Can we at least do away with these concepts and reason from first principles about our patients? This approach is appealing, until we consider that we don’t have much of a clue about the first principles that relate disease processes to particular values of body temperature. Is a patient with a temperature of 39 °C twice as sick as a patient with a temperature of 38 °C, or only 2.6% sicker? Of course, neither is the case, but we have no algorithm nor body of experience with which to characterize patient states based on temperature. Instead, we mentally convert temperature measurements into conceptual representations of the patient’s true body temperature (as above) and then further use concepts like “low-grade fever” to help us match patients tacitly to conceptual patterns that correspond to various disease states (such as mild upper respiratory tract infection—a cold).
The medical literature that is part of the foundation of clinical education, and the original studies on which that literature is based have been derived in part from patient data that were recorded conceptually, rather than realistically. Perhaps past behavior is no excuse: if we begin today to record patient data through the exclusive use of observations on the side of the patient, we might eventually reach some point at which we can compare a patient before us with our experience by matching patient attributes, rather than concepts. Homer Warner and others have argued that such data should be the basis for logical diagnostic reasoning, rather than reliance on abstractions that are the product of human experts, no matter how experienced [15].
5. What is on the side of the clinician
Even if we are to discard the past hundred years or so of clinical literature, we are still faced with the fact that human beings reason based, necessarily, on concepts; the best clinical reasoners rely on tacit knowledge that not only is conceptual in nature but is, by definition, inexpressible [16], [17]. Our eventual liberation from the vagaries of human expert reasoners may not relieve us of this reliance on conceptual representations. When Warner attempted to integrate his diagnostic expert system with his clinical information system, he found the mapping of patient data to clinical concepts to be a significant challenge [18].
Consider, for example, “severe acute respiratory syndrome” (SARS). When the condition first arose, we might have chosen to define this term based on a set of actual cases in reality that shared a set of particular attributes (i.e., certain clinical manifestations with particular geographic and chronological characteristics). While such characteristics were certainly true for each individual patient, we must also consider how clinicians dealt with this condition. Did they hold in their minds the unique identifiers of the individual cases or did they use some abstract representation, based on their understanding of the disease at the time? It is certainly the latter, for without the conceptual representation, they would have no way to consider cases that fit some of the pattern of the disease without having all of the characteristics of the initial cases—for example, they would have to treat cases arising in Canada as a different set of instances of a different universal, since they do not match the previously identified geographic characteristics. Smith does not say how we would know to relax those constraints to recognize these cases as being instances of the SARS universal. Humans, however, achieve such reclassification readily, even subconsciously.
Instead, clinicians made use of a SARS concept, which included conjectures, such as “probably viral.” This allowed them to consider aspects that would not be evoked solely by the then-known characteristics of the individual cases—for example, the recognition of cases that fit the pattern but not the definition based on known cases. Canadian SARS cases can be classified as such according to the conceptual representation, allowing us to relax the geographic constraints retroactively. It is fitting, then, that a controlled terminology contain terms corresponding to such concepts, to record, in the patient’s record, what diagnosis the clinician is considering at some point in time. If new information arises, the clinician might discard the previous diagnosis and consider a new one. The terminology can be employed to record both considerations.
6. If universals are insufficient, can we live with concepts?
We appear to be in a quandary: Smith would like us to work with patient information at the instance level, and reason with universals, thus avoiding the muddiness inherent in concept-level representation in reasoning. However, we are trapped into using concepts, as long as we deal with human reasoners (and their computer systems) and are not able to escape them when dealing with human patients (as per the fever example, above). Perhaps, though, things may not be as bleak as they seem.
An analysis of the great aspirin–Aspergum controversy leads to the conclusion that it is a problem not of semantics but of language. It is possible that someone, somewhere, refers to this chewable product of the Insight Pharmaceutical Corporation (Plymouth Meeting, PA) as “aspirin.” This is an example of the rhetorical device known as synecdoche, in which a general word or phrase is used to stand in for a more specific one (or vice versa). In this case, there is no conceptual dilemma—when the speaker says “aspirin,” he is actually attempting to communicate a meaning that is synonymous with the meaning generally associated with “Aspergum.” That he risks being misunderstood is merely the effect of the ambiguities that beset human language. Had he, instead, used some agreed upon identifier (whether a code or an agreed-upon name, recognized by speaker and listener as being a unique preferred term in a controlled terminology), there would be no conceptual dissonance.
Another possibility (and the one that is involved in this particular example) is that there was an error in judgment during the construction of the UMLS that led to the merging of two meanings, such that the same unique identifier was assigned to both meanings (and their corresponding terms)—an example of spurious synonymy—what the Desiderata called ambiguity. I believe that Smith would argue that this is precisely his point: attempting conceptual orientation inevitably leads to such muddiness. This may be as true of concept orientation as any other orientation, but in this case the error was a systematic one that concept-orientation itself actually helped to resolve. It seems that, at the time that terms for drug products were added to the UMLS, they were simply treated as instances of chemicals and were therefore indistinguishable from the chemicals from which they were composed (Nelson, personal communication). 3
Eventually, the NLM determined that two meanings were present—one that referred to an organic acid (a chemical) and one that referred to a drug (a manufactured object). The NLM determined that the chemicals and manufactured objects were mutually exclusive semantic types; therefore, the assignment of these two meanings to the same identifier could automatically be determined to represent ambiguity. Today, the UMLS contains two separate unique identifiers to which these terms are assigned.
I believe that other apparent contradictions in concept representation can be peaceably resolved when they are discovered, not by throwing them away and replacing them with something that extends to collections of real-world instances, but by doing the hard work of understanding their intended meaning(s) and purpose(s) to resolve the contradictions through improved representation. It is my experience that not only can concepts and universals coexist in the same controlled terminology, but that this is a desirable situation.
By way of example, consider the Medical Entities Dictionary (MED), the controlled terminology used at New York Presbyterian Hospital [19]. The MED needs identifiers for a wide variety of entities that are represented in patient records, including laboratory tests, laboratory specimens, radiological and cardiologic procedures, and diagnoses. Some of these can be considered to be universals (such as laboratory tests, medications, and the aforementioned specimens), while others are conceptual (such as diagnoses). There is certainly room for many more universals, should some method be developed for recording more instances on the side of the patient. Meanwhile, far too little is understood about the diseases represented by the diagnosis terms for us to represent them as universals, yet they are far too useful to discard.
The MED attempts to adhere to the Desiderata, including the use of formal definitions. While these definitions are present for a only subset of terms in the MED, and while the MED has been rightfully denied the characterization of “ontology” [20], it nevertheless contains ontological information and it provides an example of how concepts and universals can safely intermingle in the same terminology. Fig. 1 shows a small sample of entities from the MED, drawn from laboratory tests, procedures, medications, and clinical information system constructs (summary reports). Also shown is some of the ontological information contained in the MED, expressed as semantic relationships among the entities. These entities do more than share a common set of identifiers; their inclusion together in the MED supports automated reuse of patient data (for example, to aggregate comparable data into summary reports) [21], automated inferencing (for example, in decision support) [22], and automated translation [19], [23].
Fig. 1.
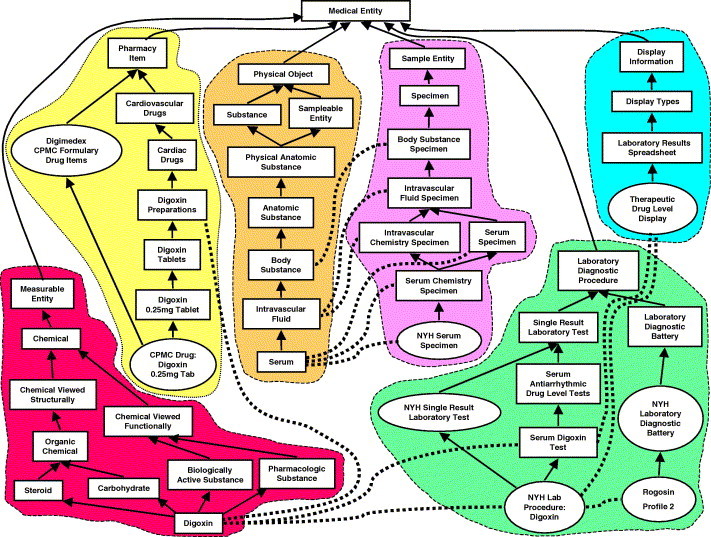
Sample contents of the Columbia University/New York Presbyterian Hospital Medical Entities Dictionary (MED). Rectangles correspond to terms that represent conceptual entities, while ovals correspond to terms that represent universals. The extensions of the universals are actual tests, procedures and medications that are related to patients in the electronic health record. Dashed lines delineate different areas in the MED, such as (from left to right) measurable entities (including chemicals), specimens, sampleable entities (including body substances), diagnostic procedures (including laboratory tests), and the data dictionary, (including display information, which is used by the clinical information system for organizing patient data) The solid arrows represent is-a links in the MED, while the dotted lines represent nonhierarchical semantic relations, such as “measures substance” (between tests and chemicals), “has specimen” (between tests and specimens), “samples” (between specimens and anatomic substances), “has pharmaceutic component” (between drugs and chemicals), “has display parameter” (between laboratory results displays and laboratory tests) and “has test part” (between laboratory diagnostic batteries and laboratory tests). Laboratory results displays, such as “Therapeutic Drug Level Display” refer to concepts used by the clinical information system for summary result reporting. The Therapeutic Lab Display shows results for digoxin tests, as well as other tests not shown here, such as theophylline tests, verapamil tests, etc.. A “Rogosin Profile 2” is an orderable battery of blood tests that includes the “NYH Lab Procedure: Digoxin,” as well as other tests not shown here such as “NYH Lab Procedure: Iron” and “NYH Lab Procedure: Magnesium.” For clarity, some high-level intermediate hierarchical terms between “Medical Entity” and “Laboratory Diagnostic Procedure” and between “Medical Entity” and “Pharmacy Concept” have been omitted, as has the is-a link between Chemical” and “Substance.” “NYH” stands for “New York Hospital”, a part of New York Presbyterian Hospital. More information about the MED, including a browser, is available at http://onto.cpmc.columbia.edu/medsite/med1.htm.
7. Whither the Desiderata?
Although the Desiderata might be construed to be some kind of commandments (“no false gods”—concept orientation, “thou shalt not kill”—concept permanence, “honor thy father and thy mother”—multiple hierarchies, and so on), they were only a synthesis of contemporary thought. At the time they were presented (at the 1997 IMIA Working Group 6 conference, in Jacksonville, Florida), the only objection raised was to the rejection of “not elsewhere classified” (NEC) terms [24]. Eight years later, at a subsequent meeting in Rome of the same working group, the NEC issue was not even raised, most likely because the general consensus is now that such terms are antithetical to ontologies. 4
It is a given that our knowledge expands and evolves. Our knowledge about knowledge is subject to this same evolutionary force. But rather than discard what we have learned to replace it with this alternative world view, I believe that we can expand our understanding of how controlled biomedical terminologies might be further developed to embrace both perspectives. Smith chooses a path he calls “realism.” One cynical definition of reality is “The dream of a mad philosopher” [25]. A more balanced definition is “the totality of real things and events; something that is neither derivative nor dependent but exists necessarily” [2]. I suggest a path that acknowledges the importance of representing reality, as best we can know it, but accepts the need for concepts to help us, among other things, reason under uncertainty. I consider this the realistic path.
In the realistic approach, terminologies contain terms that refer to universals and to concepts, along with various names and unique identifiers for these. Sometimes, a single term will refer to an entity that has both universal and conceptual characteristics. Terminologies also contain, to the fullest extent possible, ontological information to include what we know about the meanings of the terms and the entities that they represent. That ontological information for terms referring to concepts is, as Smith argues, problematic; I argue that it is no more problematic than ontological information for terms referring to universals. In any case, being problematic does not render such information valueless and should not dissuade us from including it where we can.
We therefore consider some desirable characteristics of controlled biomedical terminologies that address not the structural and content issues of the original Desiderata, but their purpose:
(1) Terminologies should support capturing what is known about the patient. This is at the level of what is actually observed, not just how we interpret our observations or what we infer. For example, when recording the medication given to a patient, we should record the specific product—“Aspergum,” for example, when it is known. However, when such details are not know, we will need the terminology to provide us with a more general term—“aspirin preparation,” for example.
(2) Terminologies should support retrieval. This has implications for the how the terminology is used at the time of data recording and at the time of querying. In both settings, we should strive to make the meanings of the terms universally understood; linguistic representation should support, rather than obfuscate, this understanding. For example, although “aspirin” is often a shorthand form of the term “aspirin preparation,” the terminology should make the distinction between the two terms clear to the person recording a patient’s medications, such that someone later encountering the information in the patient’s record will be able to determine the meaning intended by the recorder.
(3) Terminologies should allow storage, retrieval, and transfer of information with as little information loss as possible. This has implications for how terminologies evolve over time, while the data they are used to record remain as frozen artifacts. Changes in terminologies should not hamper our understanding of what was stored on the side of the patient. For example, many medical products that contained phenylpropanolamine—a drug that has been prohibited by the US Food and Drug Administration—have continued to be manufactured with a substitute ingredient (pseudoephedrine). While the names of these medications have not changed, the identifiers used to refer to them must change, so that we can know, from a patient’s medical record, which form of the medication the patient received.
(4) Terminologies should support aggregation of data. While we want our terminologies to support those who record data, we must recognize the legitimate needs for abstraction of data, perhaps from multiple perspectives. For example, if we want to know which patients are taking aspirin preparations, we will want to be able to identify those patients whose records contain “Aspergum” (or any of a large number of other specific products), as well as those whose records merely show that they are taking an “aspirin preparation.”
(5) Terminologies should support reuse of data. Users of data may wish to consider transformations other than simple aggregation, using what is known about the terms by which the data are recorded. For example, if we wish to know whether a patient is taking an antiplatelet agent, an antipyretic, an analgesic, or a nonsteroidal anti-inflammatory agent, we would want to be able to identify our Aspergum-taking patient as such.
(6) Terminologies should support inferencing. The knowledge underlying the terms used to record data should be compatible with knowledge used for conceptual representations for reasoning (by humans and computers), such that the transformation from the former to the latter can be accomplished. We need to be able to reach across from what is on the side of the patient to use it on the side of the clinician; terminologies can help. For example, we would like to be able to use the knowledge that the (conceptual) condition “aspirin allergy” is related to the chemical “aspirin” and, from that, infer that we should be concerned about aspirin-allergic patients (instances of a universal) who are given aspirin-containing products (instances of another universal).
If we can accept that the characteristics above are reasonable expectations for controlled biomedical terminologies, we can then proceed to determine how best to realize them. We must recognize that, after all, everything we say about the patient is, on some level, an abstraction of reality and that how we record what we say—that is, its context—is as important as what we say.
8. Conclusion
The original Desiderata paper discussed the entities represented by controlled terminologies without reference to ontologies, but it nevertheless reflected ontological principles. While it referred to the terminologic entities as “concepts,” it was describing desired characteristics of universals as well. As long as we consider that the purpose of terminologies is to support the recording and use of actual data, rather than primarily as a pure knowledge base of what is known in biomedicine, I believe that concepts and universals can coexist and commingle in controlled terminologies, to the advantage of those who seek to improve patient care through symbolic representation of patient information.
Acknowledgments
I thank IMIA Working Group 6 for the opportunity to present this work in an intellectually stimulating setting. In particular, I thank Dr. Barry Smith not only for his work on organizing the conference, but for his valuable comments on earlier drafts of this paper. I also thank Mark Musen and the anonymous JBI reviewers for their valuable comments on the later draft of this paper.
Footnotes
Consider, for example, the nodes in a hidden layer of a neural network. Each node represents coordination of values from input nodes and conveys some information to the output nodes, but we do not characterize these nodes as corresponding to any real or conceptual intermediate entity.
With the recent discovery of a new large body orbiting the Sun outside the orbit of Pluto, the extentional meaning will need to be updated with some reference to the new body, which will be awkward until it is named; meanwhile, the intentional meaning remains valid.
This error originated with the general assumption that, if a UMLS source terminology considers two terms to be synonymous, and no other source terminology treated them as distinct concepts, then the UMLS would perpetuate the synonymy. In the Medical Subject Headings (MeSH), “Aspergum” was listed as an Entry Term that pointed to the MeSH Heading “Aspirin” as the preferred term to use for indexing (i.e., “‘Aspergum’: see ‘Aspirin”’). Entry Terms may be synonyms of the MeSH Headings to which they refer, but they are not necessarily so, as in this case. But, because no other UMLS Source Terminology, at the time, contained the term “Aspergum” as an entity distinct form “Aspirin,” they were automatically assumed to be synonymous.
While the terms continue to exist, they are generally recognized to refer not to disease concepts or universals, but to instances of utterances made by clinicians when describing their beliefs and actions about a particular patient case.
References
- 1.Cimino J.J. Desiderata for controlled medical vocabularies in the Twenty-First Century. Methods Inf Med. 1998;37(4–5):394–403. [PMC free article] [PubMed] [Google Scholar]
- 2.Merriam-Webster, Incorporated. Merriam-Webster’s Collegiate Dictionary, 11th ed. Springfield, MA: Merriam-Webster, Incorporated; 2005.
- 3.Musen M.A., Wieckert K.E., Miller E.T., Campbell K.E., Fagan L.M. Development of a controlled medical terminology: knowledge acquisition and knowledge representation. Methods Inf Med. 1995;34(1–2):85–95. [PubMed] [Google Scholar]
- 4.Schreiber G., Wielinga B., Jansweijer W. Proceedings of IJCAI95 workshop on basic ontological issues in knowledge sharing. Montreal; Canada: 1995. The KAKTUS View on the ‘O’ Word. [Google Scholar]
- 5.Sim I. The “O” Word Revisited. <http://smi-web.stanford.edu/auslese/smi-web/archivedEventDetail.jsp?eventID=63>.
- 6.Smith B. From Concepts to Clinical Reality: An Essay on the Benchmarking of Biomedical Terminologies. J Biomed Inform. 2006;39(3):288–298. doi: 10.1016/j.jbi.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Campbell K.E., Oliver D.E., Spackman K.A., Shortliffe E.H. Representing thoughts, words, and things in the UMLS. J Am Med Inform Assoc. 1998;(5):421–431. doi: 10.1136/jamia.1998.0050421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ledley R.S., Lusted L.B. Reasoning foundations of medical diagnosis; symbolic logic, probability, and value theory aid our understanding of how physicians reason. Science. 1959;130(3366):9–21. doi: 10.1126/science.130.3366.9. [DOI] [PubMed] [Google Scholar]
- 9.Rector A.L., Nowlan W.A., Kay S. Foundations for an electronic medical record. Methods Inf Med. 1991;30(3):179–186. [PubMed] [Google Scholar]
- 10.Weed L.L. Medical records that guide and teach. N Engl J Med. 1968;278(11):593–600. doi: 10.1056/NEJM196803142781105. [DOI] [PubMed] [Google Scholar]
- 11.Barrows R.C., Johnson S.B. A data model that captures clinical reasoning about patient problems. In: Gardner R.M., editor. Proceedings of the nineteenth annual symposium on computer applications in medical care. Hanley & Belfus, Philadelphia; NewOrleans, LA: 1995. pp. 402–405. [PMC free article] [PubMed] [Google Scholar]
- 12.Ceusters W., Smith S. Strategies for referent tracking in electronic health records. J Biomed Inform. 2006;39(3):362–378. doi: 10.1016/j.jbi.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Jollis J.G., Ancukiewicz M., DeLong E.R., Pryor D.B., Muhlbaier L.H., Mark D.B. Discordance of databases designed for claims payment versus clinical information systems. Implications for outcomes research. Ann Intern Med. 1993;119(8):844–850. doi: 10.7326/0003-4819-119-8-199310150-00011. [DOI] [PubMed] [Google Scholar]
- 14.Wolff A., Begleiter A., Moskona D. A novel system of human submandibular/sublingual saliva collection. J Dent Res. 1997;76(11):1782–1786. doi: 10.1177/00220345970760111001. [DOI] [PubMed] [Google Scholar]
- 15.Lau L.M., Warner H.R. Performance of a diagnostic system (Iliad) as a tool for quality assurance. Comput Biomed Res. 1992;25(4):314–323. doi: 10.1016/0010-4809(92)90022-3. [DOI] [PubMed] [Google Scholar]
- 16.Patel V.L., Arocha J.F., Kaufman D.R. Expertise and tacit knowledge in medicine. In: Sternberg R.J., Horvath J.A., editors. Tacit knowledge in professional practice: researcher and practitioner perspectives. Lawrence Erlbaum Associates, Publishers; Mahwah, NJ: 1999. pp. 75–99. [Google Scholar]
- 17.Cimino J.J. Development of expertise in medical practice. In: Sternberg R.J., Horvath J.A., editors. Tacit knowledge in professional practice: researcher and practitioner perspectives. Lawrence Erlbaum Associates, Publishers; Mahwah, NJ: 1999. pp. 101–120. [Google Scholar]
- 18.Li Y.C., Haug P.J., Warner H.R. Automated transformation of probabilistic knowledge for a medical diagnostic system. In: Ozbolt J.G., editor. Proceedings of the eighteenth annual symposium on computer applications in medical care. Hanley & Belfus; Washington, DC Philadelphia: 1994. pp. 765–769. [PMC free article] [PubMed] [Google Scholar]
- 19.Cimino J.J. From data to knowledge through concept-oriented terminologies: experience with the Medical Entities Dictionary. J Am Med Inform Assoc. 2000;7(3):288–297. doi: 10.1136/jamia.2000.0070288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierre Zweigenbaum, Bruno Bachimont, Jacques Bouand, Jean Charlet, Jean-François Bosvieux. Le rôle du lexique sémantique et de l’ontologie dans le traitement automatique de la langue médicale. In: Le Beux P., Burgun A., editors. Actes du Colloque CRISTAL’S. Saint-Malo; 1996. [Google Scholar]
- 21.Elhanan G., Cimino J.J. Controlled vocabulary and design of laboratory results displays. J Am Med Inform Assoc. 1997;4(Suppl.):719–723. [PMC free article] [PubMed] [Google Scholar]
- 22.Hripcsak G., Clayton P.D., Jenders R.A., Cimino J.J., Johnson S.B. Design of a clinical event monitor. Comput Biomed Res. 1996;29(3):194–221. doi: 10.1006/cbmr.1996.0016. [DOI] [PubMed] [Google Scholar]
- 23.Cimino J.J., Barnett G.O. Automated translation between medical terminologies using semantic definitions. MD Computing. 1990;7(2):104–109. [PubMed] [Google Scholar]
- 24.Ingenerf J., Giere W. Concept-oriented standardization and statistics-oriented classification: continuing the classification versus nomenclature controversy. Methods Inf Med. 1998;37(4–5):527–539. [PubMed] [Google Scholar]
- 25.Bierce A. In: The unabridged devil’s dictionary. Schultz D.E., Joshi S.T., editors. University of Georgia Press; Athens, GA: 2000. [Google Scholar]


