Abstract
Background
Numerous viruses are responsible for respiratory infections; however, both their distribution and genetic diversity, in a limited area and a population subgroup, have been studied only rarely during a sustained period of time.
Methods
A 2-year surveillance program of children presenting with acute respiratory infections (ARIs) was carried out to characterize the viral etiology and to assess whether using gene amplification and sequencing could be a reliable approach to monitor virus introduction and spread in a population subgroup.
Results
Using multiplex RT-PCR, 15 different respiratory viruses were detected within the 486 nasopharyngeal positive samples collected among 817 children aged <9-year old who presented with ARI during October 2006 to September 2008. A single virus was detected in 373 patients (45.7%), and two to four viruses in 113 patients (13.8%). The most frequent causative viruses were respiratory syncytial virus (RSV) (24.7%), human bocavirus (24.5%), and human rhinovirus (HRV) (15%). RSV was more prevalent in winter and among young infants. Cases of seasonal influenza A and B viruses were reported mainly in January and August. An increase in adenovirus infection was observed during the spring of the second year of the study. Sequence analyses showed multiple introductions of different virus subtypes and identified a high prevalence of the newly defined HRV-C species. A higher viral incidence was observed during the winter of 2008, which was unusually cold.
Conclusions
This study supports the usefulness of multiplex RT-PCR for virus detection and co-infection, and for implementation of a molecular monitoring system for endemic and epidemic viral respiratory infections.
Keywords: Acute respiratory infection, Respiratory viruses, Genotypes, Co-infection, Pediatric
1. Background
Acute respiratory infections (ARIs) are the leading cause of pediatric morbidity worldwide.1, 2 Many viruses are associated with ARIs: influenza viruses A, B and C (IAV, IBV and ICV); respiratory syncytial virus (RSV); human metapneumovirus (HMPV); human coronaviruses (HCoV) NL63, 229E, OC43, and HKU1; parainfluenza viruses (PIV) 1–4; human rhinovirus (HRV); human enterovirus (HEV); and adenovirus (ADV).3 A new pathogen, human bocavirus (HBoV) has been shown to be associated with respiratory illnesses, mainly when it is present at a high viral load.4, 5, 6, 7
Since the epidemic of severe acute respiratory syndrome (SARS) in 2003, and the recent attention on possible influenza pandemics, sustained surveillance project was required to detect endemic, epidemic and newly emerging respiratory pathogens.
The diagnosis of respiratory viruses mainly relies on molecular techniques. Multiplex RT-PCR (mRT-PCR) techniques allow identification of a majority of respiratory viruses8, 9, 10 as well as co-infections.11, 12
2. Objectives
In the present 2-year study, we used a five-tube mRT-PCR assay we implemented in the Pasteur Institute network in the Asian region (http://www.pasteur-international.org/ip/easysite/pasteur-international/activites-scientifiques/projets/tous-les-projets/sisea), which covered 17 common respiratory viruses,10 to identify viruses in nasopharyngeal specimens in 817 children with ARI. Sequencing primers specific to other fragments of viral genes were employed to amplify the positive samples for sequencing and phylogenetic analysis. The sequences showed the genetic variation of viruses circulating in the region, and identified the virus evolution and introduction of new variants. This study allowed cartography of viral etiology of a large panel of viruses that were co-circulating in ARI in children in a district of Shanghai, with the aim of implementing a monitoring system for endemic or epidemic viral respiratory infections.
3. Study design
3.1. Patient population
Between October 1, 2006 and September 30, 2008, all subjects aged <9 years, who presented with an ARI syndrome and attended the outpatient ward of the pediatric department of Shanghai Nanxiang Hospital, China, were enrolled prospectively. The protocol was approved by the Ethical Committee of Shanghai Nanxiang Hospital and by the Biomedical Committee of Institut Pasteur in Paris. Written informed consent of a parent or a legal guardian was required. ARI was defined as the presence of fever (at least 38 °C) plus cough and/or sore throat (Table 1 ). Study enrollment was organized twice weekly (every Monday and Thursday); all patients who consulted on Monday and Thursday and presented with the above case definition were included in the study.
Table 1.
Criteria of patient enrollment.
| Criteria | |
|---|---|
| Inclusion | Exclusion |
| Children younger than 9-year old, first onset within 48 h, | Subjects already under antiviral treatment for any prophylactic or curative purpose |
| AND | |
| Fever (T ≥ 38 °C) plus cough and/or sore throat | |
| AND/OR | |
| Dyspnoea or tachypnea, cyanosis, cough, pleuritic chest pain, hypoxemia | |
| AND | |
| Signature of the patient consent agreement | |
3.2. Data and specimen collection
Upon enrollment, systematic recordings were made of the patients’ demographic characteristics and medical history using a standardized questionnaire. The questions included detailed signs and symptoms, laboratory and radiology examinations; the presence of a chronic underlying disease; and family smoking history. After a complete physical examination, the children were classified into three different disease groups on the basis of signs and/or symptoms indicating the inflammation site: upper respiratory tract infection, bronchitis and pneumonia. Patients and their parents were interviewed by the same doctor to obtain demographic data and information about their clinical presentation.
For each ARI case, nasopharyngeal swabs (NPSs) were obtained by the same nurse. Specimens were collected with sterile cotton-tipped swabs that were introduced into the nostril and the pharyngeal areas, and then placed in 2 ml viral transport medium. The NPSs were then transported at 4 °C to the Virology Department of the Institut Pasteur of Shanghai, where they were divided into aliquots, and stored at −80 °C.
RNA extraction—Total RNA from NPS aliquots was extracted using a QIAamp viral RNA Minikit (Qiagen, Hilden, Germany) in accordance with the manufacturer's protocol. Purified RNA was frozen at −80 °C in aliquots.
Multiplex RT-PCR—A mRT-PCR previously published was improved and employed in this study for virus detection. It was initially described by Bellau-Pujol et al. (2005) for 12 virus identification including IAV, IBV, ICV, RSV, HMPV, PIV1–4, HCoV-OC43 and 229E, and HRV in three tubes with hemi-nested PCR8 then later improved by Vabret et al. for 14 viruses plus with HCoV-HKU1 and HCoV-NL63 in four tubes.13 Besides, the primers used for HRV detection could also detect HEV, and the two viruses were then differentiated based on the sizes of the amplified products. The DNA band with the size of 550–574 bp corresponded to rhinovirus while the band with the size of 600–700 bp corresponded to enterovirus. Hence, the mRT-PCR could detect 15 viruses. The mRT-PCR multiplex 1 and the hemi-nested multiplex 1 detected 1 and 0.1 TCID50 of RSV A, respectively, and 0.01 and 0.001 TCID50 of influenza virus A/H3N2, respectively.8
We have improved the method to detect 17 viruses in a five-tube mRT-PCR assay by introducing specific primers to ADV and HBoV.14, 15 Moreover, we eliminated previous hemi-nested PCR step to avoid cross-contamination but introduced sequencing of amplification products. The assay was compared with commercialized Resplex II assay (Qiagen) in previous study and its sensitivity reached 0.01 TCID50 of RSV B, 0.3 TCID50 of influenza virus A/H1N1, and 0.001 TCID50 of IBV, respectively.12
Tube 1 targeted IAV, IBV, RSV, HMPV; tube 2, PIV1–4; tube 3, HRV and ICV; and tube 4, HCoV-229E, HCoV-OC43, HCoV-NL63 and HCoV-HKU1.8, 10, 12 Tube 5 targeted ADV and HBoV using previously published primers: HBoV (188F, 5′-GASCTCTGTAAGTACTATTAC-3′; 542R, 5′-CTCTGTGTTGACTGAATACAG-3′); ADV (ADHEX1F, 5′-CAACACCTAYGASTACATGAA-3′; ADHEX2R, 5′-ACATCCTTBCKGAAGTTCCA-3′).14, 15 RNA was amplified using a one-step RT-PCR kit (Qiagen) as previously described.12 In brief, 2.5 μl of extracted RNA was mixed with a 5× buffer and 0.2 mM dNTP, 0.2 μM of each primer, and l μl of enzyme mix, and DEPC-treated ultrapure water was added to a final volume of 25 μl. Amplification programs included reverse transcription at 50 °C for 30 min, inactivation at 95 °C for 15 min, followed by 40 cycles at 94 °C for 30 s, 50 °C (tubes 1, 2 and 5) or 55 °C (tubes 3 and 4) for 30 s, 72 °C for 45 s and final extension at 72 °C for 10 min. The amplified DNA products were detected by 0.5 g/l ethidium bromide/2% agarose gel electrophoresis.
Cloning and sequencing—Sequencing primers were designed based on the conserved region of each virus or on previous publications16, 17, 18, 19, 20, 21 (Table 2 ). Extracted RNA was amplified by specific monoplex RT-PCR following the same protocol as above, but with a hybridization temperature of 53 °C. The DNA products were purified from agarose gels using QIAquick Gel Extraction Kit (Qiagen), and were ligated into pMD20-T vector (Takara Biotechnology, Dalian, China). Recombinant plasmids were sequenced by Biosune Sequence Company and Invitrogen Biotechnology Company in Shanghai, China.
Table 2.
Sequencing primers.
| Virus | Primer name | Sequence 5′ → 3′ | Region | Position | Reference strain | Reference |
|---|---|---|---|---|---|---|
| 229E | 229E/SPIKE-F | CTACAAATGGGMTGAACACTAGTYACTC | Spike | 53–80 | AY386395 | This study |
| 229E/SPIKE-R | TACGTGGTTGAACAGCAATTATAG | 1679–1702 | This study | |||
| NL63 | NL63/SPIKE-F | GAGTTTGATTAAGAGTGGTAGGTTG | Spike | 20391–20415 | AY567487 | 16 |
| NL63/SPIKE-R | ACACGGCCATTATGTGTGGTGAC | 21049–21073 | 16 | |||
| HKU-1 | KHKU1-SPIKE-F | CYTCACCTCTTAATTGGGAACG | Spike | 23920–23939 | DQ415914 | This study |
| KHKU1-SPIKE-R | AAAACRCAACCAAGATAACTATC | 25067–25045 | This study | |||
| PIV1 | PIV1-HNF | CAACCTRYAAGGMAACARCATCYG | HN | 1–24 | M91648 | This study |
| PIV1-HNR | TCTATTTGTCATATAAATRTCTATTCATGC | 1835–1864 | This study | |||
| PIV3 | PIV3-HNF | AAATCCKAGGATCTCTCATAYTTTY | HN | 7522–7545 | EU424062 | This study |
| PIV3-HNR | GCCYTTGTCAACAACAATRATRG | 7946–7966 | This study | |||
| PIV4 | PIV4-HN-S | GGAACRCRCTTCTCAGCTC | HN | 65–81 | M34033-4A | This study |
| PIV4-HN-ANTI | GAGAAGTAAGTTATTGTGTATGAGTC | 1417–1439 | This study | |||
| ADV | ADHEX1F | CAACACCTAYGASTACATGAA | Hexon | 19002–19022 | FJ169625 | 15 |
| ADHEX2R | ACATCCTTBCKGAAGTTCCA | 19255–19274 | 15 | |||
| HMPV | F698 | ACATGCCAACATCTGCAGGACAAATAAAAC | Fusion | 698–727 | EU857610 | 17 |
| F1285 | ACATGCTGTTCACCTTCAACTTTGC | 1282–1307 | 17 | |||
| IBV | B/HA98 | ATAACATCGTCAAACTCACC | HA | 64–83 | EU852039 | 18 |
| B/HA836 | GCACCATGTAATCAACAACA | 780–799 | 18 | |||
| IBV | B/NA1 | GCTACCTTCAACTATACAAACG | NA | 3–24 | EU852040 | 19 |
| B/NA2 | AACGAGGGTATGTCCACTCC | 233–253 | 19 | |||
| IAV | 43f | GTCTGGTTTTCGCTCAAAAACTTCC | HA | 35–59 | EU716524 | This study |
| 1129r | GAATTTTGATGCCTGAAACCGTACC | 1097–1121 | This study | |||
| IAV | 32f | GATTGGCTCTGTTTCTCTCACC | NA | 27–48 | CY031565 | This study |
| 984r | CTGGGTGTGTCTCCAACAAGTCCTG | 956–980 | This study | |||
| HRV | P1-1 F | CAAGCACTTCTGTYWCCCC | 5′UTR | 163–181 | L24917 | 20 |
| P3-1 R | ACGGACACCCAAAGTAG | 536–552 | 20 | |||
| VP4/2 F | GGGACCAACTACTTTGGGTGTCCGTGT | VP2/4 | 528–554 | 21 | ||
| VP4/2 R | GCATCIGGYARYTTCCACCACCANCC | 1061–1086 | 21 | |||
| RSV | RSV-GLYCO-AF | ATCATATTCATAGCCTCRGCAAAC | Glycoprotein | 4833–4856 | NC_001803 | This study |
| RSV-GLYCO-AR | ACACTTCAAAGTGAAAATCATTAT | 5160–5132 | This study | |||
| RSV-GLYCO-BF | TATTCATCATCTCTGCGAATCAC | 4868–4890 | NC_001781 | This study | ||
| RSV-GLYCO-BR | AATCATCTTTGGGTTTTTTTGGTGG | 5176–5150 | This study |
Phylogenetic analysis—Multiple sequences were aligned using Clustal X (v1.83). The multiple-sequence alignment was subjected to phylogenetic analyses using programs in the PHYLIP package (v3.6). Bootstrap analysis was performed using SEQBOOT, in which replicate number was 1000. DNADIST and NEIGHBOR were used to obtain a distance matrix; in DNADIST, the transition/transversion ratio was 4. Consensus trees were computed by CONSENSE, and then rerooted with RETREE. The final tree was visualized and edited with MEGA version 4.
3.3. Statistical analysis
The patient data were analyzed using Statistical Package for the Social Sciences (SPSS) for Window version 17.0 (SPSS Inc. Chicago, IL, USA). A p value of <0.05 was considered statistically significant. Categorical variables were assessed for statistical significance by Pearson χ 2 test. Fisher's exact test was used if the expected values were <5.
4. Results
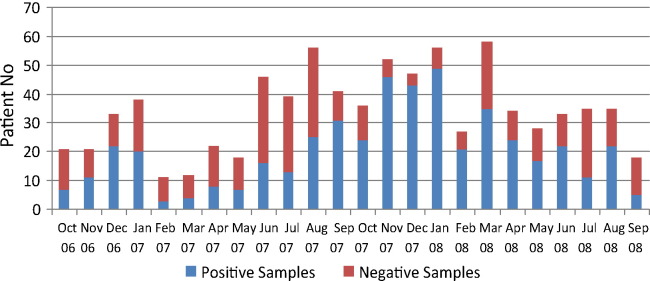
A viral etiology could be determined in 486 of the 817 (59.5%) patients. Multiple viral infections were detected in 113 (13.8%) patients (94 with two pathogens, 18 with three pathogens, and one with four pathogens) (Table 3 ). Among 486 virus-positive cases, 346 (71.2%) were diagnosed by clinicians as bronchitis, 135 (27.8%) as pneumonia, and 5 (1.0%) were diagnosed with upper respiratory tract infection.
Table 3.
Viral etiology of ARI in 817 outpatient children, Shanghai Nanxiang Hospital, October 2006 to September 2008.
| Virus | Virus detectiona |
Total virus strains | |
|---|---|---|---|
| Single infection | Co-infection | ||
| Human RSV | 71 | 49 | 120 |
| IV | 80 | 28 | 108 |
| IAV | 44 | 9 | 53 |
| IBV | 36 | 19 | 55 |
| HMPV | 33 | 11 | 44 |
| PIV | 52 | 25 | 77 |
| PIV1 | 27 | 18 | 45 |
| PIV3 | 23 | 6 | 29 |
| PIV4 | 2 | 1 | 3 |
| HRV | 43 | 30 | 73 |
| HEV | 5 | 6 | 11 |
| HCoV | 17 | 7 | 24 |
| HCoV-OC43 | 1 | 0 | 1 |
| HCoV-229E | 1 | 4 | 5 |
| HCoV-NL63 | 14 | 3 | 17 |
| HCoV-HKU1 | 1 | 0 | 1 |
| ADV | 18 | 25 | 43 |
| HBoV | 54 | 65 | 119 |
Case number.
Overall, 618 viral pathogens were detected: RSV was the most frequent pathogen (n = 120, 19.4%). The second to the fifth most frequent pathogens were HBoV (n = 119, 19.3%); IV (IAV and IBV) (n = 108, 17.5%); PIV1, 3 or 4 (n = 77, 12.5%), and HRV (n = 73, 11.8%). HMPV (n = 44, 7.1%), ADV (n = 43, 7.0%), HCoV-OC43, 229E, NL63 or HKU1 (n = 24, 3.9%). HEV (n = 11, 1.8%) were also detected occasionally.
Among the 113 patients diagnosed with a multiple viral infection, the most frequent pathogens were HBoV, RSV, HRV and ADV (data not shown).
4.1. Seasonality of viruses
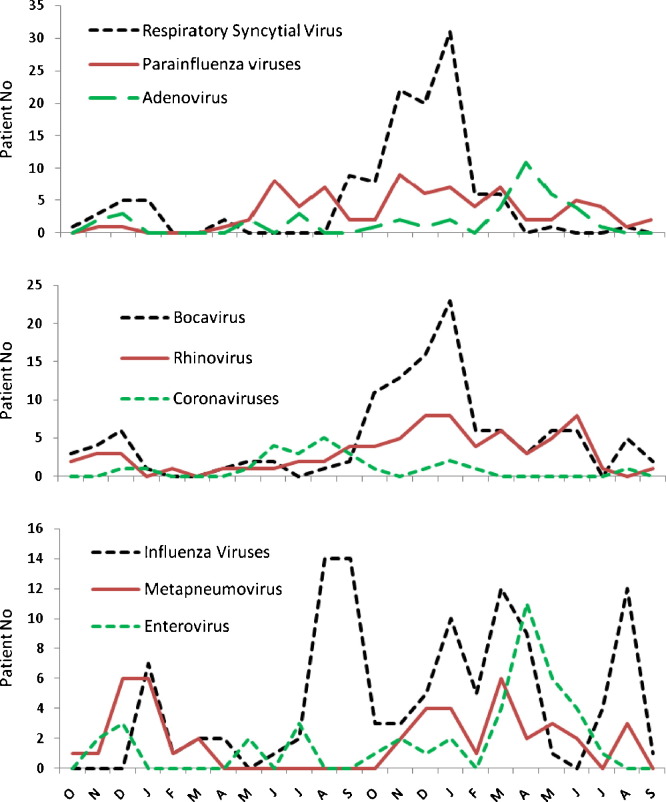
The monthly distribution of viruses is presented in Fig. 1, Fig. 2 . Viruses were detected significantly more often during fall or winter than during other seasons (71% and 49%, respectively, p < 0.01). RSV and HBoV identifications were most frequent in the fall and winter. IV showed a biannual distribution, one peak in winter and another in the summer. One important increase in ADV infection from March to June in 2008 was recorded. Twenty-two ADV strains among 118 samples were detected compared to five strains among 52 samples in the same period of 2007. A small number of HCoV-NL63 was detected during the summer of 2007. HRV, PIV and HMPV were detected continuously throughout the year.
Fig. 1.
Monthly distribution of ARI cases with mRT-PCR results, in 817 outpatient children, Shanghai Nanxiang Hospital, October 2006 to September 2008.
Fig. 2.
Monthly distribution of virus strains detected by mRT-PCR, in 817 outpatient children, Shanghai Nanxiang Hospital, October 2006 to September 2008.
4.2. Impact of age distribution
Patients included in this study were aged from 1 month to 9 years. The median age was 3 years. RSV was more frequent in younger children (p = 10−7) with 12 out of 23 virus-positive cases from 6 months to 2 years age group. No IAV or IBV infection was detected in children younger than 1-year old (Table 4 ). Only 8 patients from 1 to 6 months age group were enrolled in the study.
Table 4.
Frequencies of viral pathogens (per age group).
| ≤6 months |
6 months–1 year |
1–2 years |
2–4 years |
5–9 years |
All ages |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 8 | % | n = 31 | % | n = 121 | % | n = 449 | % | n = 208 | % | n = 817 | % | |
| Single infection | 4a | 50b | 11 | 35.5 | 37 | 30.6 | 236 | 52.6 | 85 | 40.9 | 373 | 45.7 |
| Co-infection | 0 | 12 | 38.7 | 36 | 29.8 | 38 | 8.5 | 27 | 13 | 113 | 13.8 | |
| PCR negative | 4 | 50 | 8 | 25.8 | 48 | 39.7 | 175 | 39 | 96 | 46.2 | 331 | 40.5 |
| RSV | 0 | 12 | 52.2 | 17 | 23.3 | 71 | 25.9 | 20 | 17.9 | 120 | 24.7 | |
| IV (any) | 0 | 3 | 13 | 11 | 15.1 | 56 | 20.4 | 38 | 33.9 | 108 | 22.2 | |
| IAV | 0 | 0 | 5 | 6.8 | 31 | 11.3 | 17 | 15.2 | 53 | 10.9 | ||
| IBV | 0 | 3 | 13 | 6 | 8.2 | 25 | 9.1 | 21 | 18.8 | 55 | 11.3 | |
| HMPV | 1c | 12.5d | 2 | 8.7 | 6 | 8.2 | 28 | 10.2 | 7 | 6.3 | 44 | 9.1 |
| PIV (any) | 0 | 5 | 21.7 | 10 | 13.7 | 49 | 17.9 | 13 | 11.6 | 77 | 15.8 | |
| Type 1 | 0 | 1 | 4.3 | 7 | 9.6 | 29 | 10.6 | 8 | 7.1 | 45 | 9.3 | |
| Type 3 | 0 | 2 | 8.7 | 3 | 4.1 | 19 | 6.9 | 5 | 4.5 | 29 | 6 | |
| Type 4 | 0 | 2 | 8.7 | 0 | 1 | 0.4 | 0 | 3 | 0.6 | |||
| HRV | 2 | 25 | 4 | 17.4 | 13 | 17.8 | 37 | 13.5 | 17 | 15.2 | 73 | 13 |
| HEV | 0 | 0 | 2 | 2.7 | 7 | 2.6 | 2 | 1.7 | 11 | 2.3 | ||
| HCoV (any) | 0 | 0 | 3 | 4.1 | 17 | 6.2 | 4 | 3.6 | 24 | 4.9 | ||
| OC43 | 0 | 0 | 0 | 1 | 0.4 | 0 | 1 | 0.2 | ||||
| 229E | 0 | 0 | 0 | 4 | 1.5 | 1 | 0.9 | 5 | 1 | |||
| NL63 | 0 | 0 | 2 | 2.7 | 12 | 4.4 | 3 | 2.7 | 17 | 3.5 | ||
| HKU1 | 0 | 1 | 4.3 | 0 | 0 | 0 | 1 | 0.2 | ||||
| ADV | 0 | 3 | 13 | 5 | 6.8 | 19 | 6.9 | 16 | 14.3 | 43 | 8.8 | |
| HBoV | 1 | 12.5 | 4 | 17.4 | 27 | 37 | 61 | 22.3 | 26 | 23.2 | 119 | 24.5 |
Case number.
Case percentage in group.
Detected virus.
Percentage of detected virus in virus infected cases of each group.
4.3. Prevalent types/subtypes of viruses
To identify the prevalent subtype of different viruses and the similarity of the virus strains, virus-positive samples were sequenced for target genes, when the amount of genetic material amplified was sufficient (Table 5 ), and phylogenetic trees were constructed (data not shown; dendrograms are available on request to the authors). Identity among isolates of each virus type or subtype was calculated by pairwise algorithm and their nearest reference strains are shown in Table 4.
Table 5.
Genetic variation of circulating viruses.
| Virus | Detected | Sequenced | Gene | Species | Genotype/type | Number | Nucleotide identity % among strains | Reference strain |
|---|---|---|---|---|---|---|---|---|
| HMPV | 44 | 39 | F | A1 | 7 | 98–99 | EU698012, EU179277 | |
| B1 | 16 | 97–100 | EU698017 | |||||
| B2 | 16 | 97–100 | EF694069 | |||||
| IAV | 53 | 37 | HA | H1 | 16 | 95–99 | CY031370 | |
| H3 | 21 | 92–99 | CY040098, CY044788 | |||||
| IBV | 55 | 36 | HA | 87–99 | CY015406, EU982188 | |||
| NA | 96–100 | CY040451, GQ423424 | ||||||
| HRV | 73 | 65 | VP4 | A | 25 | 73–100 | EU840871, EU840840 | |
| B | 5 | 75–82 | DQ473490, EF173424 | |||||
| C | 35 | 68–100 | EF582385, GQ223227, EF077280, EF582386, EF582387, EF077279, EF186077 | |||||
| ADV | 43 | 40 | Hexon | B | Type 2 | 37 | 94–100 | AY819918, AY819919 |
| C | 3 | 86–98 | AY819936 | |||||
| HCoV-NL63 | 17 | 13 | Spike | 94–100 | DQ445912 | |||
| HCoV-229E | 5 | 5 | Spike | 99 | AF304460 | |||
| HBoV | 119 | 17 | VP2 | Type 1 | 17 | 99.3 | DQ000496 | |
| RSV | 120 | 27 | Glycoprotein | A | 27 | 94–100 | AF512538 |
In 40 sequenced ADV strains, 37 were species B (92.5%) and three were species C (7.5%). Most of them were similar to serotype ADV-2 (data not shown). In 65 HRV strains, 25 were species A (38.5%), five were species B (7.7%), and 35 were species C (53.8%). HRV showed high variation in nucleotide identity (73–100% in HRV-A, 75–82% in HRV-B, and 68–100% in HRV-C) as previously described (Huang et al.).26 Among 37 IAV strains, 16 were seasonal H1N1 (95–99% nucleotide identity in the HA gene) and 21 were seasonal H3N2 (92–99% identity) strains. In 36 out of 55 IBV strains, HA and NA gene fragments were sequenced, and comparison showed that the IBV strains were more conserved in the NA gene (96–100% nucleotide identity) than HA gene (87–99% identity). Only 17 out of 119 HBoVs were sequenced but showed high nucleotide identity in the ST2 gene as previously described.7 The sequences of HCoVs were highly conserved (94–100% nucleotide identity in HCoV-NL63, 99% in HCoV-229E). In 39 HMPV strains, 16 were genotype B1 (41%), 16 were genotype B2 (41%), and seven were genotype A1 (17.9%) with high nucleotide identity (98–99% in A1 and 97–100% in B1 and B2). Only 27 out of 120 RSVs were sequenced and nucleotide sequence analysis of a glycoprotein gene fragment showed all of the strains were classified into RSV A subtype (data not shown). The results indicated a low variability of these viruses that circulated in the region during the 2-year period.
4.4. Clinical features
Among 486 virus-positive cases, 483 (99.4%) patients presented with high fever (>38 °C) and cough, 25 (5.1%) patients with dyspnoea or tachypnea and 46 (9.4%) patients with lymphopenia. Although 346 (71.2%) were clinically diagnosed as bronchitis, 135 (27.8%) were pneumonia and 5 (1.0%) were diagnosed with upper respiratory tract infection by the clinician, we considered that only 25 patients met the severe respiratory infection as showing dyspnoea or tachypnea symptoms. One hundred twenty patients (22.6%) were suggested to be hospitalized after their first consulting but none became inpatients. No correlation was observed between infection with any specific virus (single or co-infection) and clinical severity (Table 6A and B). In addition, the repartition of the different symptoms was statistically insignificant when compared with virus-negative diagnosed patients (Table 6A and B). Interestingly, 30 patients had polynucleosis associated to an inflammatory response may be linked to a bacterial infection. No specific virus was associated to polynucleosis.
Table 6.
Clinical features in 817 ARI cases.
| A | Single infection |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IAV | IBV | RSV | HMPV | PIV11 | PIV3 | PIV4 | RHV | HEV | OC43 | 229E | HKU-1 | NL63 | ADV | HBoV | |
| Total case | 44a | 36 | 71 | 33 | 27 | 23 | 2 | 43 | 5 | 1 | 1 | 1 | 14 | 18 | 54 |
| Fever (>38 °C) | 44 | 36 | 71 | 32 | 27 | 23 | 2 | 41 | 5 | 1 | 1 | 1 | 14 | 18 | 54 |
| Cough | 44 | 36 | 70 | 32 | 27 | 23 | 2 | 42 | 5 | 1 | 1 | 1 | 14 | 18 | 54 |
| Pleuritic pain | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Purulent expectoration | 5 | 8 | 16 | 8 | 5 | 3 | 2 | 11 | 0 | 1 | 0 | 0 | 3 | 3 | 10 |
| Dyspnoea or tachypnea | 1 (2.3) | 3 (8.3) | 2 (2.8) | 1 (3.0) | 1 (3.7) | 1 (4.3) | 1 (50) | 3 (7.0) | 0 | 0 | 0 | 0 | 0 | 1 (5.6) | 3 (5.6) |
| Hypoxemia | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Interstitial abnormality at chest X-ray | 25 (56.8)b | 25 (69.4) | 47 (66.2) | 17 (51.5) | 15 (55.6) | 14 (60.9) | 1 (50) | 29 (67.4) | 2 (40) | 1 | 1 | 0 | 9 (64.3) | 11 (61.1) | 34 (63.0) |
| Polynucleosis (>10,000 on absolute count) | 3 | 2 | 8 | 1 | 1 | 1 | 1 | 6 | 0 | 0 | 0 | 0 | 1 | 2 | 4 |
| Lymphopenia (<1500 on absolute count) | 12 (27.3) | 4 (11.1) | 4 (5.6) | 4 (12.1) | 0 | 1 (4.3) | 0 | 2 (9.3) | 0 | 0 | 1 | 0 | 1 (7.1) | 1 (5.6) | 4 (7.4) |
| Bronchitis | 31 (70.5) | 29 (80.6) | 46 (64.8) | 22 (66.7) | 20 (74.1) | 19 (82.6) | 0 | 35 (81.4) | 5 (100) | 1 | 0 | 1 | 10 (71.4) | 13 (72.2) | 38 (70.4) |
| Pneumonia | 13 (29.5) | 7 (19.4) | 24 (33.8) | 10 (30.3) | 6 (22.2) | 4 (17.4) | 1 (50) | 8 (18.6) | 0 | 0 | 1 | 0 | 4 (28.6) | 5 (27.8) | 16 (29.6) |
| Upper respiratory tract Infection | 0 | 0 | 1 (1.4) | 1 (3.0) | 1 (3.7) | 0 | 1 (50) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Suggested to be hospitalized | 8 (18.2) | 10 (27.8) | 21 (29.6) | 10 (30.3) | 5 (18.5) | 4 (17.4) | 0 | 8 (18.6) | 0 | 1 | 0 | 0 | 3 (21.4) | 5 (27.8) | 17 (31.5) |
| B | All viruses |
RT-PCR negative | |
|---|---|---|---|
| Single | Co-infection | ||
| Total case | 373 | 113 | 331 |
| Fever (>38 °C) | 370 | 113 | 329 |
| Cough | 370 | 113 | 330 |
| Pleuritic pain | 4 | 4 | 4 |
| Purulent expectoration | 75 | 25 | 50 |
| Dyspnoea or tachypnea | 17 (4.6) | 8 (7.1) | 18 (5.4) |
| Hypoxemia | 1 | 0 | 1 |
| Interstitial abnormality at chest X-ray | 201 (53.9) | 81 (71.7) | 174 (52.6) |
| Polynucleosis (>10,000 on absolute count) | 30 | 11 | 21 |
| Lymphopenia (<1500 on absolute count) | 34 (9.1) | 12 (10.6) | 21 (6.3) |
| Bronchitis | 270 (72.4) | 76 (67.3) | 208 (62.8) |
| Pneumonia | 99 (26.5) | 36 (31.9) | 91 (27.5) |
| Upper respiratory tract Infection | 4 (1.1) | 1 (0.9) | 32 (9.7) |
| Suggested to be hospitalized | 92 (24.7) | 28 (24.8) | 88 (26.6) |
A: each virus; B: all viruses including single infection and co-infection.
Case number.
Percentage (%).
5. Discussion
From October 2006 to September 2008, 817 outpatients aged from 1 month to 9 years were included in a surveillance program of viral etiology in ARI. Less than 1% of the children enrolled in this outpatient study were aged of less than 6 months, suggesting that very young children may show more severe symptoms and hospitalized. Fifteen different viruses were detected in 486 samples (59.5%). Thus, respiratory viruses were the major pathogens responsible for ARI in children in Shanghai and multi-infections of different viruses (13.8%) were frequently observed.
Although serotype identification is critical for epidemiological surveillance, the serotyping is time consuming and costly, and limited due to cross-reactivity of the tests. We sequenced the fragments of genes coding for virus antigenic proteins and analyzed by phylogenetic analysis the sequence diversity to monitor the molecular evolution of circulating virus.
During the outbreak of ADV from March to June in 2008, the majority of strains (37 out of 40) was ADV-B species and of serotype ADV-2. In ADV-B species, only serotype ADV-14 was reported to cause severe infection,22 which may partly explained that in our study ADV-infected patients showed only mild symptoms.
The IAV strains detected during the period from January 2007 to April 2008 were mainly H3N2 but the strains detected from July 2008 to September 2008 were mainly H1N1. This suggests that subtype H1N1 replaced the H3N2 subtype and predominated during the next year in the region. As vaccination for seasonal influenza (IAV H1N1, H3N2 and IBV) was not included in the children's routine vaccination program in Shanghai, and was usually based on the strains circulating worldwide in the precedent summer, the local population was not protected against the new subtype that emerged in the summer of 2008.
HRV is classified into three species: HRV-A, HRV-B and HRV-C by phylogenetic analysis based on sequences of VP4 gene and/or 5′UTR.23, 24, 25 A predominance of the newly identified species HRV-C (53.8%) and the recombinant strains were observed based on which two new subspecies of HRV-C were proposed as HRV-Ca and HRV-Cc.26 This suggests the emergence of new variant strains of HRV in future that might cause a new epidemic. Eleven HEV were detected in the study but were not analyzed further.
HMPV is another recently identified respiratory virus27 and has been found worldwide.28 It is grouped into four distinct genetic lineages based on the F gene: A1, A2, B1, and B2.29 In this study, two strains of HMPV-A1, 11 strains of HMPV-B1 and two strains of HMPV-B2 were detected in the first season, whereas one strain of HMPV-A1, three strains of HMPV-B1 and 12 strains of HMPV-B2 were identified in the second season. No HMPV-A2 lineage was found. Hence, a change of predominant lineage in the seasons was observed, but no association between the severity of infection and genetic drift of HMPV was found, as shown in other previous studies.30, 31 Besides, studies showed more sequence diversity in G and SH genes but not in F gene,30 which could explain the constant incidence of HMPV infection in the population.
HCoV-HKU1 (in group II with HCoV-OC43) and HCoV-NL63 (in group I with HCoV-229E) are two novel coronaviruses.32, 33 During the present 2-year study, HCoV-NL63 and HCoV-229E were the major HCoV circulating in Shanghai, whereas only one strain of HCoV-OC43 and one of HCoV-HKU1 were detected, indicating a sporadic introduction of group II HCoV to the region. In context that the recent emerged HCoV, like HKU-1 and SARS whose sequence is more homologous to group II virus, could cause severe respiratory infection, the surveillance for emergence of new species of HCoV is necessary.
The co-infection of RSV and HBoV was frequently detected among the samples, whereas these two viruses co-dominated in cold season. HBoV was the second most prevalent virus (24.5%), and the co-infection rate of HBoV with other respiratory viruses was 54.6%, compared to 14% in non-HBoV-infected patients. This was lower than the co-infection rate reported previously, which ranged up to 71%.34, 35, 36, 37 A previous study showed that HBoV increased the severity of bronchiolitis in children less than 1-year-old co-infected with RSV, and that it is not an occasional virus.38 However, no correlation of HBoV infection with clinical severity was observed in this and its related study.7
One study carried out in Wuhan, China analyzed peripheral blood samples by indirect immunofluorescence to detect RSV, IAV, IBV, ADV, PIV1–3, Chlamydia pneumonia and Mycoplasma pneumonia in children ARI inpatients and used viremia as sign of severe infection.39 It showed that 36% of cases were co-infected by multiple agents and IAV, IBV and PIV1 were associated with co-infection. In addition, studies showed up to 30% of co-infection in hospitalized children40 and RSV co-infection was associated with clinical severity.38, 39 However, no such correlation was found in this study. This may be due to differences in the criteria of patient enrollment and in the lower severity of clinical signs observed. Bacteria-virus co-infection was commonly found in inpatients. In our study, only polynucleosis (>10,000 on absolute count) was considered as a sign of bacterial infection and was observed in 30 patients infected with a respiratory virus. Hence, future studies should focus on severe respiratory infection to identify viral determinants of disease severity and should introduce bacteriological test.
Up to 331 specimens were negative in mRT-PCR, despite all of them matched well with the inclusion criteria for ARI. Negative results could have resulted from the low load of viral material in samples, or to infection with bacteria instead of virus. New sensitive tools such as Mass-Tag41 or high-throughput sequencing42 have been developed recently to identify new viruses and bacterial pathogens.43 Implementation of these new molecular techniques for samples that are negative in mRT-PCR might be considered in the future.
This is believed to be the first study in China to characterize 17 common respiratory viruses in pediatric ARI during a 2-year consecutive period in a limited community with an important immigrant population. Using mRT-PCR followed by sequencing and phylogenetic analysis, we could identify a wide variety of agents and differentiate highly pathogenic viruses from less virulent seasonal respiratory viruses. The sequence analysis result could be useful to improve the primer design for RT-PCR and to identify new subtype virus, for example HRV-C. It monitored sustaining virus circulation in the community, which could serve as a baseline of the annual distribution of viruses for surveillance of unusual prevalence of one specific virus.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
The study was supported by the Li Ka-Shing Foundation (RESPARI network) for discovery of new emerging viruses, the French Agency for the Development for Surveillance and Investigation of Epidemic Situations in Southeast Asia (SISEA project), the National Science and Technology Major Project (2009ZX10004-105) of the Chinese Ministry of Health for the establishment of pathogen and immuno-response detection platforms for respiratory and central nervous system viral infections, the AREVA Foundation which supports the salary of Dr. Wei Wang. We thank Drs. François Freymuth (Virology Laboratory, CHU Caen, France) and Sylvie van der Werf (National Reference Center for Influenza virus in Northern France, Institut Pasteur Paris), and Dr. Jean-Claude Manuguerra (Urgent Response to Biological Threats) for having facilitated this work. We also thank Y.M. Zheng for manuscript preparation.
References
- 1.Shay D.K., Holman R.C., Newman R.D., Liu L.L., Stout J.W., Anderson L.J. Bronchiolitis-associated hospitalizations among US children, 1980–1996. JAMA. 1999;282:1440–1446. doi: 10.1001/jama.282.15.1440. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong G.L., Conn L.A., Pinner R.W. Trends in infectious disease mortality in the United States during the 20th century. JAMA. 1999;281:61–66. doi: 10.1001/jama.281.1.61. [DOI] [PubMed] [Google Scholar]
- 3.Mackay I.M. Human rhinoviruses: the cold wars resume. J Clin Virol. 2008;42:297–320. doi: 10.1016/j.jcv.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allander T., Jartti T., Gupta S., Niesters H.G., Lehtinen P., Osterback R. Human bocavirus and acute wheezing in children. Clin Infect Dis. 2007;44:904–910. doi: 10.1086/512196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fry A.M., Lu X., Chittaganpitch M., Peret T., Fischer J., Dowell S.F. Human bocavirus: a novel parvovirus epidemiologically associated with pneumonia requiring hospitalization in Thailand. J Infect Dis. 2007;195:1038–1045. doi: 10.1086/512163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kantola K., Hedman L., Allander T., Jartti T., Lehtinen P., Ruuskanen O. Serodiagnosis of human bocavirus infection. Clin Infect Dis. 2008;46:540–546. doi: 10.1086/526532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K., Wang W., Yan H., Ren P., Zhang J., Shen J. Correlation between bocavirus infection and humoral response, and co-infection with other respiratory viruses in children with acute respiratory infection. J Clin Virol. 2009 doi: 10.1016/j.jcv.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellau-Pujol S., Vabret A., Legrand L., Dina J., Gouarin S., Petitjean-Lecherbonnier J. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J Virol Methods. 2005;126:53–63. doi: 10.1016/j.jviromet.2005.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunstein J., Thomas E. Direct screening of clinical specimens for multiple respiratory pathogens using the Genaco Respiratory Panels 1 and 2. Diagn Mol Pathol. 2006;15:169–173. doi: 10.1097/01.pdm.0000210430.35340.53. [DOI] [PubMed] [Google Scholar]
- 10.Freymuth F., Vabret A., Cuvillon-Nimal D., Simon S., Dina J., Legrand L. Comparison of multiplex PCR assays and conventional techniques for the diagnostic of respiratory virus infections in children admitted to hospital with an acute respiratory illness. J Med Virol. 2006;78:1498–1504. doi: 10.1002/jmv.20725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunstein J.D., Cline C.L., McKinney S., Thomas E. Evidence from multiplex molecular assays for complex multipathogen interactions in acute respiratory infections. J Clin Microbiol. 2008;46:97–102. doi: 10.1128/JCM.01117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W., Ren P., Sheng J., Mardy S., Yan H., Zhang J. Simultaneous detection of respiratory viruses in children with acute respiratory infection using two different multiplex reverse transcription-PCR assays. J Virol Methods. 2009;162:40–45. doi: 10.1016/j.jviromet.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vabret A., Dina J., Gouarin S., Petitjean J., Tripey V., Brouard J. Human (non-severe acute respiratory syndrome) coronavirus infections in hospitalised children in France. J Paediatr Child Health. 2008;44:176–181. doi: 10.1111/j.1440-1754.2007.01246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casas I., Avellon A., Mosquera M., Jabado O., Echevarria J.E., Campos R.H. Molecular identification of adenoviruses in clinical samples by analyzing a partial hexon genomic region. J Clin Microbiol. 2005;43:6176–6182. doi: 10.1128/JCM.43.12.6176-6182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moes E., Vijgen L., Keyaerts E., Zlateva K., Li S., Maes P. A novel pancoronavirus RT-PCR assay: frequent detection of human coronavirus NL63 in children hospitalized with respiratory tract infections in Belgium. BMC Infect Dis. 2005;5:6. doi: 10.1186/1471-2334-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung J.Y., Han T.H., Kim B.E., Kim C.K., Kim S.W., Hwang E.S. Human metapneumovirus infection in hospitalized children with acute respiratory disease in Korea. J Korean Med Sci. 2006;21:838–842. doi: 10.3346/jkms.2006.21.5.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chi X.S., Hu A., Bolar T.V., Al-Rimawi W., Zhao P., Tam J.S. Detection and characterization of new influenza B virus variants in 2002. J Clin Microbiol. 2005;43:2345–2349. doi: 10.1128/JCM.43.5.2345-2349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chi X.S., Bolar T.V., Zhao P., Rappaport R., Cheng S.M. Cocirculation and evolution of two lineages of influenza B viruses in europe and Israel in the 2001–2002 season. J Clin Microbiol. 2003;41:5770–5773. doi: 10.1128/JCM.41.12.5770-5773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee W.M., Kiesner C., Pappas T., Lee I., Grindle K., Jartti T. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS One. 2007;2:e966. doi: 10.1371/journal.pone.0000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savolainen C., Blomqvist S., Mulders M.N., Hovi T. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J Gen Virol. 2002;83:333–340. doi: 10.1099/0022-1317-83-2-333. [DOI] [PubMed] [Google Scholar]
- 22.Acute respiratory disease associated with adenovirus serotype 14—four states, 2006–2007. MMWR Morb Mortal Wkly Rep. 2007;56:1181–1184. [PubMed] [Google Scholar]
- 23.Lau S.K., Yip C.C., Tsoi H.W., Lee R.A., So L.Y., Lau Y.L. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J Clin Microbiol. 2007;45:3655–3664. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McErlean P., Shackelton L.A., Andrews E., Webster D.R., Lambert S.B., Nissen M.D. Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C) PLoS One. 2008;3:e1847. doi: 10.1371/journal.pone.0001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominguez S.R., Briese T., Palacios G., Hui J., Villari J., Kapoor V. Multiplex MassTag-PCR for respiratory pathogens in pediatric nasopharyngeal washes negative by conventional diagnostic testing shows a high prevalence of viruses belonging to a newly recognized rhinovirus clade. J Clin Virol. 2008;43:219–222. doi: 10.1016/j.jcv.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang T., Wang W., Bessaud M., Ren P., Sheng J., Yan H. Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection. PLoS One. 2009;4:e6355. doi: 10.1371/journal.pone.0006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van den Hoogen B.G., de Jong J.C., Groen J., Kuiken T., de Groot R., Fouchier R.A. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahn J.S. Human metapneumovirus: a newly emerging respiratory pathogen. Curr Opin Infect Dis. 2003;16:255–258. doi: 10.1097/00001432-200306000-00012. [DOI] [PubMed] [Google Scholar]
- 29.van den Hoogen B.G., Herfst S., Sprong L., Cane P.A., Forleo-Neto E., de Swart R.L. Antigenic and genetic variability of human metapneumoviruses. Emerg Infect Dis. 2004;10:658–666. doi: 10.3201/eid1004.030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agapov E., Sumino K.C., Gaudreault-Keener M., Storch G.A., Holtzman M.J. Genetic variability of human metapneumovirus infection: evidence of a shift in viral genotype without a change in illness. J Infect Dis. 2006;193:396–403. doi: 10.1086/499310. [DOI] [PubMed] [Google Scholar]
- 31.Williams J.V., Wang C.K., Yang C.F., Tollefson S.J., House F.S., Heck J.M. The role of human metapneumovirus in upper respiratory tract infections in children: a 20-year experience. J Infect Dis. 2006;193:387–395. doi: 10.1086/499274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C. Identification of a new human coronavirus. Nat Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allander T. Human bocavirus. J Clin Virol. 2008;41:29–33. doi: 10.1016/j.jcv.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 35.Dina J., Vabret A., Gouarin S., Petitjean J., Lecoq J., Brouard J. Detection of human bocavirus in hospitalised children. J Paediatr Child Health. 2009;45:149–153. doi: 10.1111/j.1440-1754.2008.01442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Longtin J., Bastien M., Gilca R., Leblanc E., de Serres G., Bergeron M.G. Human bocavirus infections in hospitalized children and adults. Emerg Infect Dis. 2008;14:217–221. doi: 10.3201/eid1402.070851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pozo F., Garcia-Garcia M.L., Calvo C., Cuesta I., Perez-Brena P., Casas I. High incidence of human bocavirus infection in children in Spain. J Clin Virol. 2007;40:224–228. doi: 10.1016/j.jcv.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Midulla F., Scagnolari C., Bonci E., Pierangeli A., Antonelli G., De Angelis D. Respiratory syncytial virus, human bocavirus and rhinovirus bronchiolitis in infants. Arch Dis Child. 2010;95:35–41. doi: 10.1136/adc.2008.153361. [DOI] [PubMed] [Google Scholar]
- 39.Peng D., Zhao D., Liu J., Wang X., Yang K., Xicheng H. Multipathogen infections in hospitalized children with acute respiratory infections. Virol J. 2009;6:155. doi: 10.1186/1743-422X-6-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miron D., Srugo I., Kra-Oz Z., Keness Y., Wolf D., Amirav I. Sole pathogen in acute bronchiolitis: is there a role for other organisms apart from respiratory syncytial virus? Pediatr Infect Dis J. 2010;29:e7–e10. doi: 10.1097/INF.0b013e3181c2a212. [DOI] [PubMed] [Google Scholar]
- 41.Briese T., Palacios G., Kokoris M., Jabado O., Liu Z., Renwick N. Diagnostic system for rapid and sensitive differential detection of pathogens. Emerg Infect Dis. 2005;11:310–313. doi: 10.3201/eid1102.040492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cox-Foster D.L., Conlan S., Holmes E.C., Palacios G., Evans J.D., Moran N.A. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 43.Palacios G., Druce J., Du L., Tran T., Birch C., Briese T. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med. 2008;358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]