Highlights
-
•
IFITMs are interferon inducible membrane proteins that restrict viral infection.
-
•
Influenza infection is exacerbated in Ifitm3 −/− mice.
-
•
A SNP in IFITM3 may increase the severity of influenza infection in humans.
-
•
Cellular distributions of IFITMs may determine the range of viruses they restrict.
-
•
IFITM expression modulates membrane fluidity.
Abstract
Interferon inducible transmembrane (IFITM) proteins are a recently discovered family of cellular anti-viral proteins that restrict the replication of a number of enveloped and non-enveloped viruses. IFITM proteins are located in the plasma membrane and endosomal membranes, the main portals of entry for many viruses. Biochemical and membrane fusion studies suggest IFITM proteins have the ability to inhibit viral entry, possibly by modulating the fluidity of cellular membranes. Here we discuss the IFITM proteins, recent work on their mode of action, and future directions for research.
Current Opinion in Virology 2014, 4:71–77
This review comes from a themed issue on Virus entry
Edited by Mark Marsh and Jane A McKeating
For a complete overview see the Issue and the Editorial
Available online 28th January 2014
1879-6257/$ – see front matter, © 2014 Elsevier B.V. All rights reserved.
Introduction
In recent years, studies of innate defence mechanisms have identified a number of cellular proteins that interfere with the replication of human and animal viruses. Many of these so-called ‘restriction factors’ have been most intensively studied for human immunodeficiency virus (HIV-1). For example, tripartite motif-containing protein 5 (TRIM5) [1], APOBEC3G [2], 2′,3′-cyclic-nucleotide 3′-phosphodiesterase [3] and tetherin [4] have been found to affect uncoating, reverse transcription, virus assembly and virus release, respectively. A new addition to this antiviral repertoire is myxoma resistance protein B (MxB/Mx2) [5] that inhibits HIV-1 at a late post-entry step. However, restriction factors for other viruses have also been identified, including: RNA-activated protein kinase (PKR), that restricts Hepatitis C and other viruses [6]; MX1, that restricts influenza A virus (IAV) and measles virus [7]; and 2′-5′-oligoadenylate synthase/RNase L, that restricts Hepatitis C and other viruses [8]. Many of these factors are components of the broad antiviral response induced by interferons, collectively known as interferon stimulated genes (ISGs: for review see [9]).
Although recognised to act at different stages in viral replication cycles, most of the well-characterised restriction factors affect steps following virus entry. Recently, a new family of proteins has been identified that appears to act specifically on virus entry, the interferon inducible transmembrane (IFITM) proteins. Here we review the antiviral capacity of three of these proteins, IFITM1–3.
Identification of IFITMs
The IFITM gene family was initially identified more than 20 years ago [10], with particular interest in the interferon-stimulated response elements (ISREs) they contained. The IFITM transcripts were originally named 9-27, 1-8D and 1-8U, however, the antiviral properties of the encoded proteins were only identified in 2009 in an RNAi screen for host factors that influence IAV replication. Knock-down of IFITM3 led to enhanced viral replication. Conversely, overexpression of IFITM1, 2, or 3 inhibited early viral replication [11•].
Subsequent genome analyses have indicated that the IFITM genes are likely to have arisen by gene duplication very early in vertebrate evolution [12], since ‘lower’ vertebrates, such as lampreys, possess at least one IFITM-like gene [13]. To date, five IFITM genes have been identified in humans, of which IFITM1, 2, 3 and 5 are clustered within a 26 kb region towards the telomere on the short arm of chromosome 11. IFITM5 is not IFN inducible and is involved in bone mineralisation [14]. The fifth gene, IFITM10, is located 1.4 Mb towards the centromere, but little is known about its function. IFITM4 is not present in humans, but is located close to Ifitm1, 2, 3, and 5 in the mouse genome [15], in which the locus has expanded to encode seven Ifitm genes. Analogous genes have also been found in other mammals [12], including marsupials [13], and avian species [16].
Although the molecular function of these proteins has been largely studied in cell culture systems, studies in mice and humans suggest IFITM proteins, and IFITM3 in particular, restrict IAV infection in vivo. Ifitm3 −/− mice fail to control infection by mildly-pathogenic strains of IAV compared to their wild type littermates, developing fatal fulminant viral pneumonia [17•, 18]. Everitt et al. also found that the minor C allele of human IFITM3 (synonymous single nucleotide polymorphism (SNP) rs12252) was enriched in a cohort of Caucasian patients hospitalised with either IAV H1N1/09 or influenza B in the 2009 pandemic [17•]. Although the C allele is rare in Caucasians, replication of this genetic association was shown in a cohort of Han Chinese patients (the SNP is more prevalent in this population) with severe symptoms following influenza infection. The minority CC genotype was found in 69% of patients with severe disease compared to only 25% with mild symptoms [19], further suggesting that deleterious changes in the IFITM3 gene can influence the severity of influenza infection. It is currently unclear how this allele impacts IAV pathogenesis, but the alteration of a splice acceptor site may lead to the synthesis of a truncated IFITM3 protein that lacks the N-terminal 21 amino acids, and is expressed primarily on the cell surface rather than in endosomes (see below) [17•, 20]. Aside from rs12252, little investigation has been carried out into other SNPs reported for IFITM3. One study carried out by John et al. [21] made alterations of non-synonymous SNPs H3Q/rs1136853, D56G/rs55794999, H57D/rs1553883, N69D/rs12778, and G95R/rs61744108, with only G95R showing a small reduction in IAV restriction compared to wild type.
Broad-spectrum antiviral function
Using cell culture systems, and often pseudotype viruses, several groups demonstrated that, in addition to IAV, entry and infection by representatives of multiple virus families (including filoviruses, rhabdoviruses and flaviviruses) [22•, 23, 24] were also inhibited by overexpression of IFITMs, particularly IFITM3 (see Table 1 ). Interestingly, these restricted viruses are all enveloped, with ssRNA genomes, and considered to enter cells by membrane fusion following endocytosis. However, some retroviruses (e.g. Moloney leukaemia virus (MLV)) and several arenaviruses were apparently not restricted. Although restriction of HIV-1 infection was not initially detected [11•], several more recent studies have reported some restriction of cell infection [20, 25, 26]. Most recently, evidence that IFITM3 can also restrict a non-enveloped reovirus has been published [27], suggesting the range of viruses influenced by the IFITM proteins is not limited to those with an envelope.
Table 1.
Summary of viruses IFITM proteins have been tested against
| Family | Virus | pH dependent | Restricts infectivity | Prevents cell–cell fusion | Pseudotyped virions (P) or live virus (L) | Restriction status | Reference |
|---|---|---|---|---|---|---|---|
| Enveloped | |||||||
| Orthomyxoviridae | Influenza A virus | ✓✓ | ✓ | ✓ | P L | M1–3 | Brass et al. [11•], Smith et al. [16] |
| Influenza B virus | ✓✓ | ✓ | L | M1–3 | Everitt et al. [17•] | ||
| Flaviviridae | West Nile virus | ✓ | ✓ | P | M1–3 | Brass et al. [11•] | |
| Dengue virus | ✓✓ | ✓ | P | M1–3 | Brass et al. [11•] | ||
| Hepatitis C virus | ✓ | ✓/× | P L | M3 — no, M1 — yes | Brass et al. [11•], Wilkins et al. [24] | ||
| Rhabdoviridae | Vesicular stomatitis virus | ✓ | ✓ | ✓ | P L | M1–3 | Weidner et al. [23] |
| Rabies virus | ✓✓ | ✓ | P | M2–3 | Smith et al. [16] | ||
| Lagos Bat virus | ✓✓ | ✓ | P | M2–3 | Smith et al. [16] | ||
| Filoviridae | Marburg virus | Δ | ✓ | P L | M1–3 | Huang et al. [22•] | |
| Ebola virus | Δ | ✓ | P L | M1–3 | Huang et al. [22•] | ||
| Coronaviridae | SARS coronavirus | Δ | ✓ | P L | M1–3 | Huang et al. [22•] | |
| Retroviridae | HIV-1 | × | ✓/× | P L | Mixed results | Brass et al. [11•], Lu et al. [26], Jia et al. [20] | |
| Moloney leukaemia virus | × | × | P L | No | Brass et al. [11•], Huang et al. [22•] | ||
| Jaagsiekte sheep retrovirus | ✓ | ✓ | ✓ | P | M1 best | Li et al. [28•] | |
| Arenaviridae | Lassa virus | ✓ | × | P | No | Brass et al. [11•] | |
| Machupo virus | ✓ | × | P | No | Brass et al. [11•] | ||
| Lymphocytic choriomeningitis virus | ✓ | × | P | No | Brass et al. [11•] | ||
| Alphaviridae | Semliki Forest virus | ✓ | ✓ | ✓ | L | M2 and M3 best | Li et al. [28•] |
| Bunyaviridae | La Crosse virus | ✓✓ | ✓ | L | M1-3 | Mudhasani et al. [30] | |
| Hantaan virus | ✓✓ | ✓ | L | M1-3 | Mudhasani et al. [30] | ||
| Andes virus | ✓✓ | ✓ | L | M1-3 | Mudhasani et al. [30] | ||
| Rift Valley fever virus | ✓✓ | ✓ | L-attenuated | M2 and M3 | Mudhasani et al. [30] | ||
| Crimean–Congo haemorrhagic fever virus | ✓✓ | × | L | No | Mudhasani et al. [30] | ||
| Non-enveloped | |||||||
| Reoviridae | Reovirus | ✓✓ | ✓ | L | M3 | Anafu et al. [27] | |
✓, fuses at pH >6; ✓✓, fuses at pH <6; ×, does not require fusion; Δ, requires cathespin L in lysosome.
Using a pseudotype virus carrying the Jaagsiekte sheep retrovirus (JSRV) envelope protein (Env), for which fusion requires initial Env priming by receptor binding and subsequent exposure to pH 6.3, IFITM1 seems to restrict replication more potently than IFITM2 and 3 [28•]. As IFITM1 appears to be located earlier in the endocytic pathway, where the pH is higher [29], these data suggest that the cellular location of different IFITM proteins determines the range of viruses that each restricts. Although not strictly pH-related, restriction correlates with the cellular compartment where fusion or penetration occurs. Differential restriction of viruses in the vector-borne Bunyaviridae family has also been found [30] (Table 1); only IFITM 2 and 3 were capable of restricting Rift Valley fever virus (RVFV) and none of the IFITM proteins prevented replication of Crimean–Congo haemorrhagic fever virus (CCHFV). The reason(s) underlying this difference in susceptibility are unclear as the bunyaviruses share similar morphologies and glycoproteins (GN and GC) on their envelopes.
IFITMs: protein structure and cellular distribution
IFITM3 is expressed constitutively in cells of the upper airway and visceral pleura [18], but otherwise its expression, and that of IFITM1 and 2, in vivo is poorly understood. In both cell lines and primary cells ex vivo, IFITM protein expression is upregulated by IFNs, though the relative levels of each protein in a given cell type or tissue has yet to be determined. All three IFN-inducible human IFITM proteins show high amino acid sequence similarity and all are membrane located, though their topologies remain to be clearly established [31]. Initially proposed as transmembrane proteins (Figure 1 ), with both the N-termini and C-termini located externally, subsequent studies suggested both the N-termini and C-termini, as well as the so-called conserved intracellular loop (CIL), are located cytoplasmically, with the hydrophobic domains interacting with the membrane but not spanning it [31]. More recently, a model for IFITM3 in which the N-terminal and CIL domains are located in the cytoplasm and the C-terminal domain is extracellular, suggested a type II transmembrane topology for the second hydrophobic domain [32]. Our own work suggests a similar topology for human IFITM1 (S Weston et al., unpublished data). This topology is consistent with observations that all three IFN-inducible IFITM proteins contain conserved cysteine residues at the junctions of the CIL domain, and the putative membrane interacting domains. These cysteines (C71, 72 and 105 in human IFITM3) are palmitoylated, and this modification is required for full viral restriction [33]. Substitution of the cysteines for alanines in IFITM3 inhibits protein clustering in membranes and reduces its antiviral function [21]. IFITM3 can also be ubiquitinated on any of four lysines in the N-terminal and CIL domains. Ubiquitination enhances IFITM3 turnover [31], thus substitution of the lysines with alanines slows the protein's degradation and increases its antiviral activity [25].
Figure 1.
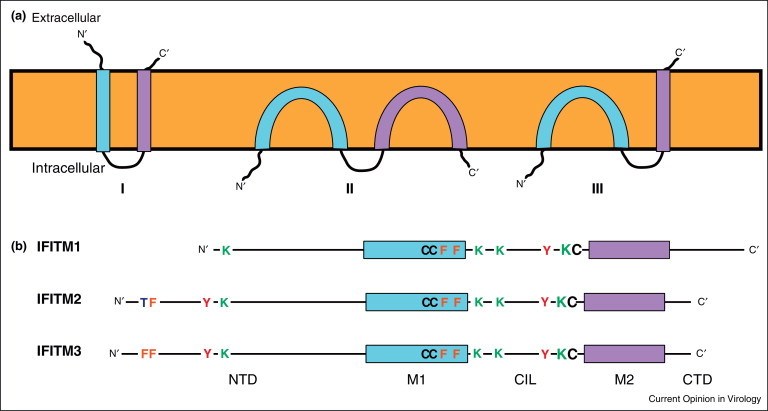
IFITM protein topology and domain organisation. Panel (a) Topological models for IFITM proteins. (I) Represents an initial model for the proteins as transmembrane molecules with both the N-terminal and C-terminal domains (NTD and CTD) extracellular and the conserved intracellular loop (CIL) facing the cytoplasm [33]. Subsequently, an alternative model (II) was proposed with the NTD, CTD and CIL all positioned intracellularly, and neither membrane domain (M1 and M2, blue and purple respectively) spanning the bilayer [31]. The most recent model (III) combines models I and II, positioning the NTD and CIL in the cytoplasm and the CTD extracellularly. Currently, the topology represented by III is only established for murine IFITM3 [32]. Panel (b) Linear representation of human IFITM1, 2 and 3 showing key amino acids. In all cases, modifications and functional activities have only been established with IFITM3, but conserved residues in IFITM1 and 2 are shown.
The N-terminal domains of IFITM2 and 3 are 20 and 21 amino acids longer, than IFITM1, respectively (Figure 1). These N-terminal extensions include a key tyrosine (Y20 in IFITM3 that appears to control the cellular distributions of the two longer IFITMs) [17•, 20, 21]. Thus IFITM1 is predominantly at the plasma membrane, while IFITM2 and 3 are located mainly in intracellular compartments. IFITM3 is reported to reside primarily in endosomal organelles, identified by co-labelling with endosomal markers, including Lamp1, Rab7 and CD63 [21, 34, 35•], but the location of IFITM2 remains to be clearly established (Figure 2 ). Therefore, Y20 may be a component of a YxxØ-type sorting signal for clathrin-mediated trafficking [20]. Significantly, Y20 has also been identified as a target for Fyn-mediated phosphorylation, suggesting that perhaps the activity of this motif as a trafficking signal can be regulated [20, 36].
Figure 2.
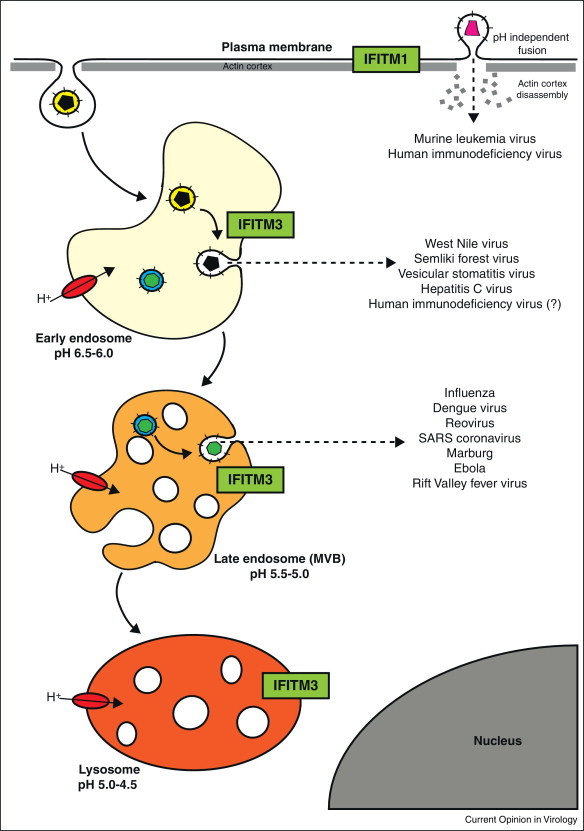
IFITM proteins inhibit virus entry at different stages of cell trafficking. Viruses enter cells by fusing with or penetrating a limiting cellular membrane. For most enveloped viruses fusion occurs either at the cell surface or, following uptake by endocytosis, from within endosomes. Acid-dependent viruses require acidification of the endosomal lumen by the membrane-associate vacuolar proton ATPase for fusion (shown in red). Trafficking through the endocytic system, from early to late endosomes, exposes virions to increasingly acidic environments. IFITM proteins (green) can inhibit entry and infection by a number of viruses that fuse/penetrate at the cell surface or from within endosomes. IFITM1 is expressed primarily at the cell surface, while IFITM2 and 3 are primarily intracellular. IFITM3 has been localised to endosomal compartments, but the distribution of IFITM2 still needs to be clearly established.
It is important to note that studies of the subcellular location of the IFITMs to date have for the most part used epitope-tagged proteins, where tagging and/or overexpression (in transient systems) may have an impact on protein localisation and/or detection. Recently John et al. [21] showed that IFITM3 can interact with itself, as well as IFITM1 and 2, and that phenylalanine residues (F75 and F78) are required for this interaction. Although the significance of this association is unclear, the formation of homo-oligomers and/or hetero-oligomers might also influence the distribution and functional activities of these proteins.
Mode of action
Reovirus subvirus particles (ISVPs), in contrast to replication competent reovirus, do not require endosomal acidification for entry and are not inhibited by IFITM3 expression, suggesting that IFITM3 may perturb endosomal acidification [27]. However, studies with various enveloped viruses suggest a different mode of action. Morphological analysis of IFITM3-restricted IAV in cells showed the accumulation of viral particles in acidified endosomal compartments, suggesting there is no effect on receptor-binding, endocytosis or acidification [22•, 34].
Studies using cell-cell fusion assays suggest that IFITM3 blocks enveloped virus entry by preventing fusion of the viral membrane with a limiting membrane of the host cell, either the plasma membrane and/or endosomal membranes [28•]. Fusion is an essential step in enveloped virus entry, and results in the transfer of viral capsids into the cytoplasm of a target cell. This process is extremely well characterised for a number of viruses, in particular IAV. Low pH in the endosomal lumen triggers conformational changes in one of the viral envelope proteins, haemagglutinin (HA). This change results in fusion of the outer leaflet of the viral membrane with the luminal leaflet of endosomal membranes forming a short-lived hemifusion intermediate. Resolution of the hemifusion intermediate allows fusion of the viral membrane inner leaflet with the cytoplasmic leaflet of endosomal membranes and the opening of a stable fusion pore [37]. Although often not a reflection of the pathway of infectious virus entry, a commonly used approach to studying viral fusion mechanisms is the formation of syncytia by cell-cell fusion. This requires the presence of viral fusion proteins in the plasma membrane of cells and appropriate signals, such as receptor-bearing cells and/or a transient change in the pH of the medium. Using the JSRV Env discussed previously, the IFITMs had no effect on either priming or pH-induced conformational changes [28•]. Moreover, syncytia formation induced by representatives of all three classes of viral fusion proteins [38] could be blocked by IFITM1. Using cold to arrest fusion at the hemifusion state, and chlorpromazine to resolve this, IFITM proteins were found to inhibit the early stages of viral envelope fusion with cellular membranes [28•].
The mechanism(s) through which the IFITMs inhibit the early stages of fusion is unclear. Two-photon laser scanning and fluorescence lifetime imaging (FLIM) of Laurdan-labelled cells, together with the effects of oleic acid treatment on cell-cell fusion, suggest that IFITM proteins may reduce membrane fluidity and increase spontaneous positive curvature in the outer leaflet of membranes [28•]. Such changes might be expected to impact on fusion, but how IFITMs affect membrane fluidity, and whether this has consequences for other membrane functions in the absence of infection, is unclear. One mechanism, however, has been suggested from experiments on IFITM3. Amini-Bavil-Olyaee et al. show IFITM3 interacts with vesicle membrane protein associated protein A (VAPA) and disrupts its association with an oxysterol binding protein that regulates the cholesterol content of endosomal membranes. Overexpression of IFITM3 increases endosomal cholesterol, which may impact on viral fusion through a corresponding decrease in endosomal membrane fluidity [35•].
Although other mechanisms may contribute to IFITM inhibition of virus entry [39], analysis of changes in the physical properties of cellular membranes induced by IFITM expression is likely to shed light on the processes underlying the broad anti-viral effects of these proteins, as well as the mechanisms involved in the fusion/penetration and entry of a number of viruses.
Conclusions and outlook
Knock-down of IFITM proteins in cell culture can increase infection by a range of enveloped and non-enveloped viruses, and knock-out of IFITM genes in vivo can influence pathogenesis. By contrast, overexpression of the proteins in human cells can inhibit infection at an early stage of the replication cycle. Although the mechanism of IFITM antiviral activity remains unclear, possible IFITM induced decreases in membrane fluidity, or increased outward curvature, may inhibit the initial stages of enveloped virus fusion. Why each protein can exhibit variable restriction on viruses of the same family, for instance the Bunyaviridae, and how a virus that does not require fusion for entry (reovirus) is restricted under the proposed mechanisms remains unclear.
In many cases viruses have evolved mechanisms to antagonise the activity of a number of cellular restriction factors (e.g. see [2, 4]). As yet no mechanisms to inhibit the activity of the IFITM proteins have been identified, though the indication that the IFITM proteins do not inhibit the entry of some viruses suggests that these agents are either refractory to the effects of the IFITM proteins, or have indeed evolved mechanisms to antagonise their function.
Although questions about the topology and intracellular distribution of the IFN-induced IFITMs remain, when taken together a model emerges suggesting that these proteins may have evolved to provide coverage of the main cellular membrane systems that have been implicated in virus entry, that is, the plasma membrane and compartments of the endocytic pathway. The implication is that perhaps IFITM1 primarily restricts viruses that fuse or penetrate at the cell surface, whereas IFITM2 and 3 primarily restrict viruses entering from endocytic organelles. That two IFITM proteins appear to function intracellularly may reflect the fact that endocytosis has been implicated in the entry of an increasing number of both pH-dependent and independent vertebrate viruses [40]. While there is some suggestion that this pattern may be true (see Table 1), there are exceptions. It is clear that there is much to be learned about the mechanisms through which these newly identified cellular proteins inhibit virus entry, and their full potential to restrict viral transmission in vivo.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
Acknowledgements
This work was supported by the Wellcome Trust grant (098051), and funding from the Medical Research Council grant (G1000413) as well as core funding to the MRC Laboratory for Molecular Cell Biology. We thank Joe Grove for help with Figure 2 and Jason Mercer for critical comments on the manuscript.
References
- 1.Stremlau M., Owens C.M., Perron M.J., Kiessling M., Autissier P., Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 2.Sheehy A.M., Gaddis N.C., Choi J.D., Malim M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 3.Wilson S.J., Schoggins J.W., Zang T., Kutluay S.B., Jouvenet N., Alim M.A., Bitzegeio J., Rice C.M., Bieniasz P.D. Inhibition of HIV-1 particle assembly by 22,32-cyclic-nucleotide 32-phosphodiesterase. Cell Host Microbe. 2012;12:585–597. doi: 10.1016/j.chom.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neil S.J., Zang T., Bieniasz P.D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 5.Goujon C., Moncorge O., Bauby H., Doyle T., Ward C.C., Schaller T., Hue S., Barclay W.S., Schulz R., Malim M.H. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. 2013;502:559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.García M.A., Meurs E.F., Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Haller O., Staeheli P., Kochs G. Interferon-induced Mx proteins in antiviral host defense. Biochimie. 2007;89:812–818. doi: 10.1016/j.biochi.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Silverman R.H. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duggal N.K., Emerman M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol. 2012;12:687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewin A.R., Reid L.E., McMahon M., Stark G.R., Kerr I.M. Molecular analysis of a human interferon-inducible gene family. Eur J Biochem. 1991;199:417–423. doi: 10.1111/j.1432-1033.1991.tb16139.x. [DOI] [PubMed] [Google Scholar]
- 11•.Brass A.L., Huang I.C., Benita Y., John S.P., Krishnan M.N., Feeley E.M., Ryan B.J., Weyer J.L., van der Weyden L., Fikrig E., Adams D.J. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and Dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study used a genome-wide siRNA knock-down screen and identified IFITM proteins as potent inhibitors of several viruses at the early stages of the virus life cycle.
- 12.Siegrist F., Ebeling M., Certa U. Phylogenetic analysis of interferon inducible transmembrane gene family and functional aspects of IFITM3. Cytokine. 2009;48:87–89. [Google Scholar]
- 13.Hickford D.E., Frankenberg S.R., Shaw G., Renfree M.B. Evolution of vertebrate interferon inducible transmembrane proteins. BMC Genomics. 2012;155:1–7. doi: 10.1186/1471-2164-13-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moffatt P., Gaumond M.H., Salois P., Sellin K., Bessette M.C., Godin E., de Oliveira P.T., Atkins G.J., Nanci A., Thomas G. Bril: a novel bone-specific modulator of mineralization. J Bone Miner Res. 2008;23:1497–1508. doi: 10.1359/jbmr.080412. [DOI] [PubMed] [Google Scholar]
- 15.Sällman Almén M., Bringeland N., Fredriksson R., Schiöth H.B. The dispanins: a novel gene family of ancient origin that contains 14 human members. PLOS ONE. 2012;7:e31961. doi: 10.1371/journal.pone.0031961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith S.E., Gibson M.S., Wash R.S., Ferrara F., Wright E., Temperton N., Kellam P., Fife M. Chicken IFITM3 restricts Influenza viruses and Lyssaviruses in vitro. J Virol. 2013;87:12957–12966. doi: 10.1128/JVI.01443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Everitt A.R., Clare S., Pertel T., John S.P., Wash R.S., Smith S.E., Chin C.R., Feeley E.M., Sims J.S., Adams D.J., Wise H.M. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work was the first to report that IFITM3 was required for the in vivo control of IAV infection in a mouse model and that the IFITM3 SNP rs12252-C was strongly associated with worse clinical outcomes for patients infected with 2009 pandemic IAV.
- 18.Bailey C.C., Huang I.C., Kam C., Farzan M. Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog. 2012;8:e90991002. doi: 10.1371/journal.ppat.1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y.H., Zhao Y., Li N., Peng Y.C., Giannoulatou E., Jin R.H., Yan H.P., Wu H., Liu J.H., Liu N., Wang D.Y. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat Commun. 2013;8:1–5. doi: 10.1038/ncomms2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia R., Pan Q., Ding S., Rong L., Liu S.L., Geng Y., Qiao W., Liang C. The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J Virol. 2012;86:13697–13707. doi: 10.1128/JVI.01828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.John S.P., Chin C.R., Perreira J.M., Feeley E.M., Aker A.M., Savidis G., Smith S.E., Elia A.E., Everitt A.R., Vora M., Pertel T. The CD225 domain of IFITM3 is required for both IFITM protein association and inhibition of influenza A virus and dengue virus replication. J Virol. 2013;87:7837–7852. doi: 10.1128/JVI.00481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Huang I.C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J., Brass A.L., Ahmed A.A., Chi X., Dong L., Longobardi L.E. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]; An extensive cell based study which showed that IFITM1-3 were able to restrict more viruses of the Filoviridae and Coronaviridae families.
- 23.Weidner J.M., Jiang D., Pan X.-B., Chang J., Block T.M., Guo J.-T. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J Virol. 2010;84:12646–12657. doi: 10.1128/JVI.01328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilkins C., Woodward J., Lau D.T., Barnes A., Joyce M., McFarlane N., McKeating J.A., Tyrrell D.L., Gale M., Jr. IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology. 2013;57:461–469. doi: 10.1002/hep.26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chutiwitoonchai N., Hiyoshi M., Hiyoshi-Yoshidomi Y., Hashimoto M., Tokunaga K., Suzu S. Characteristics of IFITM, the newly identified IFN-inducible anti-HIV-1 family proteins. Microbes Infect. 2013;15:280–290. doi: 10.1016/j.micinf.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu J., Pan Q., Rong L., Liu S.-L., Liang C. The IFITM proteins inhibit HIV-1 infection. J Virol. 2011;85:2126–2137. doi: 10.1128/JVI.01531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anafu A.A., Bowen C.H., Chin C.R., Brass A.L., Holm G.H. Interferon inducible transmembrane protein 3 IFITM3 restricts reovirus cell entry. J Biol Chem. 2013;24:17261–17271. doi: 10.1074/jbc.M112.438515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Li K., Markosyan R.M., Zheng Y.-M., Golfetto O., Bungart B., Li M., Ding S., He Y., Liang C., Lee J.C., Gratton E. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013;9:e1003124. doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]; Use cell–cell fusion assays to show that IFITM proteins block the fusion lipid membrane bilayers and increase the rigidity of the membrane, further decreasing the fusion potential.
- 29.Mellman I., Fuchs R., Helenius A. Acidification of the endocytic and exocytic pathways. Annu Rev Biochem. 1986;55:663–700. doi: 10.1146/annurev.bi.55.070186.003311. [DOI] [PubMed] [Google Scholar]
- 30.Mudhasani R., Tran J.P., Retterer C., Radoshitzky S.R., Kota K., Altamura L.A., Smith J.M., Packard B.Z., Kuhn J.H., Costantino J., Garrison A.R. Ifitm-2 and Ifitm-3 but not Ifitm-1 restrict Rift Valley fever virus. J Virol. 2013;87:8451–8464. doi: 10.1128/JVI.03382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yount J.S., Karssemeijer R.A., Hang H.C. S-palmitoylation and ubiquitination differentially regulate IFITM3-mediated resistance to influenza virus. J Biol Chem. 2012;287:19631–19641. doi: 10.1074/jbc.M112.362095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey C.C., Kondur H.R., Huang I.-C., Farzan M. Interferon-induced transmembrane protein 3 is a type II transmembrane protein. J Biol Chem. 2013;288:32184–32193. doi: 10.1074/jbc.M113.514356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yount J.S., Moltedo B., Yang Y.-Y., Charron G., Moran T.M., Lopez C.B., Hang H.C. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat Chem Biol. 2010;6:610–614. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feeley E.M., Sims J.S., John S.P., Chin C.R., Pertel T., Chen L.-M., Gaiha G.D., Ryan B.J., Donis R.O., Elledge S.J., Brass A.L. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7:e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Amini-Bavil-Olyaee S., Choi Y.J., Lee J.H., Shi M., Huang I.C., Farzan M., Jung J.U. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe. 2013;13:452–464. doi: 10.1016/j.chom.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; IFITM3 associates with VAPA, increasing the levels of intracellular cholesterol and thereby blocking viral replication.
- 36.Shiratori T., Miyatake S., Ohno H., Nakaseko C., Isono K., Bonifacino J.S., Saito T. Tyrosine phosphorylation controls internalization of CTLA-4 by regulating its interaction with clathrin-associated adaptor complex AP-2. Immunity. 1997;6:583–589. doi: 10.1016/s1074-7613(00)80346-5. [DOI] [PubMed] [Google Scholar]
- 37.Sieczkarski S.B., Whittaker G.R. Viral entry. Curr Top Microbiol Immunol. 2005;285:1–23. doi: 10.1007/3-540-26764-6_1. [DOI] [PubMed] [Google Scholar]
- 38.Igonet S., Rey F.A. SnapShot: viral and eukaryotic protein fusogens. Cell. 2012;151:1634. doi: 10.1016/j.cell.2012.11.041. [DOI] [PubMed] [Google Scholar]
- 39.Wee Y.S., Roundy K.M., Weis J.J., Weis J.H. Interferon-inducible transmembrane proteins of the innate immune response act as membrane organizers by influencing clathrin and v-ATPase localization and function. Innate Immun. 2012;30:30. doi: 10.1177/1753425912443392. [DOI] [PubMed] [Google Scholar]
- 40.Mercer J., Helenius A. Gulping rather than sipping: macropinocytosis as a way of virus entry. Curr Opin Microbiol. 2012;15:490–499. doi: 10.1016/j.mib.2012.05.016. [DOI] [PubMed] [Google Scholar]


