Highlights
-
•
Review of the present status of the development of antimicrobial peptides (AMPs).
-
•
Recent advances in designing new anti-infective agents with enhanced activity.
-
•
Transient expression of AMPs in tobacco to boost the biosynthesis of pharmaceutics.
-
•
CRISPRs, an impressive genome editing tool to enhance the biosynthesis of new AMPs.
Abstract
Anti-infective drugs have had a key role in the contemporary world, contributing to dramatically decrease mortality rates caused by infectious diseases worldwide. Antimicrobial peptides (AMPs) are multifunctional effectors of the innate immune system of mucosal surfaces and present antimicrobial activity against a range of pathogenic viruses, bacteria, and fungi. However, the discovery and development of new antibacterial drugs is a crucial step to overcome the great challenge posed by the emergence of antibiotic resistance. In this review, we outline recent advances in the development of novel AMPs with improved antimicrobial activities that were achieved through characteristic structural design. In addition, we describe recent progress made to overcome some of the major limitations that have hindered peptide biosynthesis.
This review provides an integrated scenario of the most relevant aspects in the development of useful drugs provided by natural sources and recent advances in the biosynthesis and structural design of antimicrobial peptides.
Introduction
Most drugs currently used therapeutically were obtained as naturally occurring molecules purified from microorganisms, plants, and animals [1]. It is estimated that, from 1981 to 2006, anti-infective agents based on natural product scaffolds accounted for up to 75% of all approved antibacterial new chemical entities (NCEs), thus attesting for the importance of natural products as the main source of therapeutics against pathogenic bacteria [2]. In this context, microorganisms, notably members of the Gram-positive phylum Actinomycetes, have been the workhouse source of clinically approved antibacterial agents. The high diversity of candidate molecules extracted from microorganisms frequently leads to the discovery of new compounds with distinct mechanisms of action compared with drugs currently used for clinical purposes. However, in recent years, the traditional screening of drug candidates belonging to completely new antibacterial classes from microbial sources has suffered a considerable decline, mostly because of the presence of numerous, already well-characterized, molecules [3].
This scenario is particularly worrisome because of the increasing emergence of bacterial resistance to antibiotics, notably in developing countries, where there is a widespread and indiscriminate use of antibiotics for clinical and veterinary purposes. The search for new therapeutic molecules for commercial applications is an ongoing in the pharmaceutical industry, which now is focusing on not only drug prospection, but also the modification of existing antibiotics in a timely fashion that meets the customer's needs [3].
Among the diverse naturally occurring anti-infective agents that have been discovered to date, AMPs are particularly important [4]. These molecules are versatile, highly specific antimicrobial compounds that constitute promising candidates for commercial and clinical uses. AMPs are essential components of the innate immune system of vertebrates and the nonspecific host defense system of plants, fungi, and invertebrates that evolved over 2.6 billion years ago as potent anti-infective agents against different viruses, bacteria, fungi, and parasites. These small peptides usually comprise 10–50 amino acid residues and are 2–9 kDa in size [1]. Commonly, they show a cationic structure rich in positively charged arginine and lysine residues, which favors the interaction between these peptides and microbial cytoplasmic membranes. However, although less common, there are also anionic AMPs, notably in plants. AMPs usually kill pathogens by interacting with membrane phospholipids, resulting in membrane permeabilization and subsequent disruption. These peptides also show different secondary structures, such as β-sheets stabilized by two or three disulfide bridges and often display a helical amphipathic structure 1, 2, 3.
AMPs can target a variety of essential metabolic processes in the plasma membrane or at extra- and intracellular sites, and frequently exhibit immunomodulatory properties that stimulate cytokine production while repressing inflammation, can kill cancer cells, and promote wound healing [5]. In eukaryotes, most natural AMPs are encoded by specific genes, which are constitutively expressed at basal levels and rapidly transcribed after induction by contact or exposition to invading pathogens. Thereby, a variety of different AMPs can be found simultaneously in organisms, such as plants, in response to pathogen stimuli at different organs, such as roots, seeds, flowers, stems, and leaves [6].
Recent advances in in silico drug design and high-throughput screening of compound libraries based on protein–protein interactions have triggered a growing interest in the discovery of new antibacterial drugs from microbial, plant, and animal origins. Such efforts have contributed to uncover a variety of molecules with novel structures and improvement of the currently available classes of antibiotic, such as β-lactams, ketolides, macrolides, glycopeptides, aminoglycosides, oxazolidinones, and other anti-infective agents [7].
The use of viral vectors based on the tobacco mosaic virus (TMV) and potato virus X (PVX) for cloning and massively expressing AMP genes in Nicotiana benthamiana appears to be an interesting new approach to considerably enhance recombinant peptide production before structural and functional characterization. The rapid, high-throughput staggered biosynthesis of peptides using agroinfiltrated leaves of N. benthamiana constitutes the most promising strategy for delivering new antigenic peptides and vaccine candidates, addressing the interest of many private and state health institutes [8].
Finally, gene editing using the clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein (Cas) system is another molecular tool that could revolutionize the recombinant biosynthesis of AMPs [9]. This approach specifically targets DNA sequences by adding, removing, or replacing DNA segments mediated by cellular DNA repair mechanisms of site-specific double-stranded breaks. It allows the edition of specific genes within the host genome to integrate the fragments containing the coding sequence of a particular AMP in a hotspot site to enable increased AMP biosynthesis. Working on a case-to-case scenario, this expression strategy could overcome major drawbacks in the production of AMPs, notably the low biosynthesis levels presented by plant and animal AMPs [9].
Mechanisms of action of AMPs
The antimicrobial activity of a given AMP is specifically related to its amino acid composition and physical chemical properties, such as positive net charge, flexibility, size, hydrophobicity, and amphipathicity 5, 10. Marginal changes in peptide residue sequence are normally followed by major changes in antimicrobial activity [10].
AMPs present different mechanisms of action, many of them well described in the literature. The molecular interactions of some AMPs and microorganisms rely on the variety of targets presented by microorganisms, particularly Gram-positive and Gram-negative bacteria. [11].
Generally, the mechanisms of action of AMPs can be classified into two basic groups: (i) the disruptive mechanisms, which are associated with membrane lysis; and (ii) the membrane undisruptive mechanisms, which focus on neutralizing intracellular targets [10]. Independently of the proposed group, the first step of any mechanism is the molecular interaction between the AMP and the cytoplasmic membrane. The driving force of such interaction is the electrostatic force presented by the AMP, which is normally cationic, and the polyanionic surface of bacteria 5, 11. Differences in the cell wall composition among bacterial groups affect directly the mode of action of AMPs. Gram-negative bacteria present three major layers in the cell envelope: (i) the outer membrane (OM), comprising a lipid bilayer, mostly of lipopolysaccharide (LPS) interlaced by teichoic acid, which has a major role in protection against the environment; (ii) the peptidoglycan cell wall, comprising repeated units of a disaccharide (N-acetyl glucosamine-N-acetylmuramic acid) linked by pentapeptide side chains; and (iii) the cytoplasmic membrane–phospholipid bilayer. By contrast, in Gram-positive bacteria, there is no OM, but the peptidoglycan layer is thicker than in Gram-negative bacteria [5].
The disruptive mechanisms are presented in four classical models: (i) the Toroidal model; (ii) the Carpet model; (iii) the Aggregate model; and (iv) the Barrel model. Recently, new disruptive models or models indirectly associated with membrane disruption were described: (i) the Disordered toroidal model; (ii) the Membrane thinning/thickening model; (iii) the Charged lipid clustering model; (iv) the Non-bilayer intermediate model; (v) the Oxidized lipid targeting model; (vi) the Anion carrier model; (vii) the Non-lytic membrane depolarization mode; and (viii) the Electroporation model. All the models are non-mutually exclusive, allowing that a single AMP might present a multiple-hit strategy based on two or more simultaneous mechanisms. In the Toroidal model, the AMP binds the membrane and forms a ‘flip-flop’ translocation channel that opens the membrane vertically, with the AMP remaining closely associated to the lipid head-groups throughout the process 5, 12.
The Carpet model provides the disruption of the membrane without the internalization of the AMP. By contrast, the AMP remains associated with the membrane until a crucial concentration of the peptide contributes to weaken the hydrophobic interactions of structural phospholipids and AMPs form a carpet structure that increase membrane disruption [12].
In the Aggregate model, the AMPs have a detergent-like role by interweaving the phospholipids and disaggregating them, similar to a true detergent. Depending on the AMP and the membrane composition, this mode of action does not provide the membrane rupture. Instead, it just forms a thin channel for the crossing of AMP and other molecules into the cell [10].
The Barrel model provides the formation of a regularly organized aggregate of AMPs that interacts and associate with the membrane. The AMP oligomer leads to the formation of pores in the lipid bilayer by intimately interacting its hydrophobic side chains with hydrophobic parts of the membrane. The transmembrane pores allow the internalization of the hydrophilic part of the AMP that faces the internal region of the membrane [5].
Lastly, there is the model that shares the most similarities with the four classic models: the Disordered toroidal model. This mode of action provides a stochastic pore formation after inward lipid distortion that allows the aggregation of a maximum of two peptides in the center of the pore. However, in the external peripheral region of the pore, many peptides are set up before translocation 5, 12.
The undisruptive mechanisms are based on the AMP crossing the membrane because of the combined features of AMP sequence and membrane composition, and the inhibition of some reactions of cellular metabolism, causing cell death. There are two different ways that an AMP enters the cell. The first is an obscure spontaneous translocation across the membrane; the other is mainly the result of the presence of a secondary structure in the AMP that causes membrane permeabilization. In this method, named the Shai–Matsuzaki–Huang method, a α-helical AMP binds parallel to the membrane. Hydrophobic residues facing the membrane permit the internalization of part of the AMPs and the change of their organization to a transversal mode by forming toroidal pores 5, 10, 11.
The other model suggests that the β-sheet AMPs are organized into a flat-aggregate form that allows the insertion of some aromatic residues in the membrane and the opening of thin translocational spaces in which the AMPs can cross the membrane [9].
Once across the membrane, the AMPs can target a variety of intracellular sites, such as gene promoters and coding sequences, mRNA-binding sites, enzyme regulatory sites, and protein prefolding sites. Such inhibitory interactions involve blocking both DNA transcription and/or RNA translation, or incorrect protein folding, triggering the failure of metabolic pathways and cell death [12].
Recently, a new undisruptive mechanism was discovered in AMP-sensitive bacteria. Species such as Escherichia coli and Salmonella spp. have evolved the ability to detect and prevent AMP antibacterial activities by triggering pathways involved in sensing and amplifying resistance to cationic AMPS. Resistance is provided by the PhoQ/PhoP system of E. coli, which comprises the activation of the AMP detector kinase PhoQ by Mg2+ and Ca2+, which phosphorylates and activates the transcription factor PhoP [13]. The active PhoP activates pagP, which encodes a resistance enzyme against AMPs; mgtA, encoding a transporter of Mg2+; and hdeA, encoding a chaperone responsive to acidic conditions. Phosphorylated PhoP activates the transcription of the QueE gene, encoding an enzyme that controls the biosynthesis of hypermodified guanosine found in rare tRNAs and, once overexpressed, inhibits cell division by downregulating the bacterial divisome 12, 13.
Sublethal concentrations of the C18G AMP, a highly cationic, amphiphilic peptide derived from the C-terminal sequence of human protein platelet factor 4, can trigger the PhoQ-dependent filamentation of wildtype E. coli by the overexpression of QueE. The bacterial filamentation (tens to hundreds of microns in length) prevented fully bacterial growth and was detectable in genetically engineered strains with a PhoP-regulated promoter driving the transcription of a yellow fluorescent protein [13].
Another good example is Buforin II, a potent AMP from Chinese toad Bufo gargarizans, with antimicrobial activity against a range of microorganisms. Once inside the cell, Buforin II inhibits gene expression by associating with DNA and mRNA during transcription and translation. The same occurs with melittin, a bee venom peptide active against both Gram-positive and Gram-negative bacteria, as well as some pathogenic fungi, which is a typical example of nondisruptive AMP, discovered during the late 1980s, that supported the Carpet model of microorganism inhibition detailed in 2011 5, 12, 13.
Antimicrobial resistance to AMPs
The development of antibiotics has had a major impact on modern medicine. However, the increasing emergence of antibiotic resistance and the limited development of novel classes of antibiotic over the past four decades has led to a scenario in which some infections are no longer treatable with available antibiotics [14].
Antimicrobial resistance (AMR) is defined as the resistance of microorganisms to an antimicrobial against which they were once sensitive 3, 13. AMR is an inevitable evolutionary outcome once all organisms develop genetic mutations that can improve fitness and lead to selection as a response to the selective pressure of the environment. In fact, more than 70% of pathogenic bacteria are resistant to at least one type of antibiotic [1]. Undesired selection of microbial cells with resistance-conferring mutations or other resistant elements represents the main drawback in long-term treatment efficiency of patients, leading to intensive medical research to discover new targets to overcome multidrug resistance (MDR) 4, 6, 12.
The current classes of antibiotic face a constant threat represented by the diverse bacterial resistance mechanisms. The ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) represent the most dangerous pathogens to immunocompromised patients because they are commonly isolated as drug-resistant (DR) or MDR microorganisms [7].
There are two major ways in which common bacteria evolve to become resistant: by intrinsic or acquired resistance against AMPs. Intrinsic resistance occurs as a natural consequence of the presence of AMPs in the natural environment of bacteria, which develop mechanisms to resist antibiotic action [13]. This can happen via passive or inducible mechanisms. Passive resistance is always associated with less tight interactions between bacteria and AMPs because of the inherent accumulation of more positive charges in lipid A, and is more common in genera such as Proteus, Providencia, Burkholderia, Morganella, and Serratia [13]. Inducible resistance is a consequence of perennial, reversible modifications at the molecular level in both Gram-positive and Gram-negative bacteria [14].
Acquired resistance is a product of high-fitness mutants that often contain more than one mutation that is unrelated to the AMP selection. Bacterial mutated genes provide altered genetic systems that allow bacterial growth in the presence of AMPs and can be identified by experimental procedures with and without AMP in the culture media [15].
Membrane modifications related to AMP resistance
The bacterial OM and inner membrane architectures can be altered and sites for the ligation of AMPS can be protected in response to reduced levels of Ca2+, Mg2+, and specific proteins, and changes in lipid composition. This considerably reduces membrane fluidity and permeability to polymyxins, defensins, and cathelicidins 3, 16.
The bacterial membrane phospholipid content, such as phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL), reflects a global state of protection against pore formation [17]. The biosynthesis, turnover, and translocation of phospholipids to target sites in the membrane can be modulated by the expression of proteins closely related to resistance against AMPs 15, 16. Profiles of cationic AMP resistance can be shown by the 10% increase in the total content of CL in liposome model membranes [17]. Strains of S. aureus resistant to methicillin, for example, can increase lipid biosynthesis and translocation to the membrane by simply activating genes that encode multiple peptide resistance factor (mprF), cardiolipin synthase (cls), and phosphatidylglycerol synthase (pgsA) 18, 19.
The affinity between the cell membrane and AMPs can be considerably reduced by minimizing the negative charge of the phospholipid bilayer (i.e. the lipid composition). In addition, two-component signal regulatory systems (TCS), such as the PhoQ/PhoP system in P. aeruginosa and the ApsR/ApsS in Staphylococcus epidermidis, can act together with lipid modifications to enhance resistance against cationic AMPs [13].
Resistant bacteria with modifications in the cell wall
The cell wall is the outer barrier that acts as a secondary physical protection structure against pore formation in bacteria. It gives strength to the bacterial cell and influences the final cell format. AMPs often establish ionic and/or hydrophobic interactions with the cell wall. Modifications of the polysaccharide bilayer of peptidoglycan and teichoic acids in the cell wall are particularly interesting in Gram-positive bacteria. The AMP–cell wall interactions can be avoided in S. aureus and Staphylococcus xylosus, which present multiple copies of the dlt operon, a regulatory sequence that, when active, promotes d-alanylation of teichoic acids in the cell wall, reducing their anionic charges [20].
In some Gram-negative bacteria, the affinity of lipopolysaccharides (LPS) in the OM by cationic AMPs can be reduced by the increase of positively charged lipid A with substituents such as palmitate, phosphoethanolamine, and 4-amino-4-deoxy-l-arabinose at 1- or 4′-phosphate groups [21]. These modifications in lipid A are also induced by the PhoQ/PhoP system and appear to be closely related to cationic AMP resistance, mainly against polymyxin B, in Gram-negative bacteria. Glycosylation of lipid A was recently reported as another lipid modification that results in polymyxin resistance in EI Tor Vibrio cholerae [22]. The addition of glycine to lipid A is regulated by the almEFG operon, which controls the expression of several proteins (AImE, AlmF, and AlmG) related to glycine activation and transfer to lipid A. Glycine is activated by adenylation by AlmE, which transfers active glycine to the 4′-phosphopantetheine group of AlmF. Once active, AlmF donates the glycine to the hydroxylauryl chain of lipid A, a transfer reaction performed by AlmG [22].
In bacteria, is commonly stated that the thicker the cell wall, the less efficient an antibiotic is, and, by extension, an AMP will be. Improving the cell wall thickness is a strategy adopted by S. aureus against erythromycin, vancomycin, acriflavine, and many AMPs. The cell wall peptidoglycan layer can increase in thickness by the upregulation of glutamine synthase in E. coli strains that are resistant to magainin II, a characteristic absent in susceptible strains [22].
Resistance associated with changes in metabolism
Some bacteria have an arsenal of molecules that prevent cellular metabolism from suffering stress caused by AMPs. Several pathways can be up- or downregulated to increase the biosynthesis of proteases, modification of membrane sites recognizable by AMPs, overproduction of biofilms, and suppression of superficial elements related to pore formation [13].
An inherent mechanism of AMP resistance by bacteria is the partial or total proteolytic cleavage of AMPs. Strains of S. aureus overproducing the metalloprotease aureolysin are resistant to cathelicidin. Proteus mirabilis producing high amounts of the metalloproteases ZapA and LL-37 can avoid the antimicrobial activity of β-defensin 1 (hBD1) simply by breaking down the protein into six or nine innocuous peptides [23].
Other systems for the upregulation of the biosynthesis of bacteria proteases have been discussed elsewhere. The nisin resistance gene (nrs) of resistant strains of Lactococcus lactis is a central element of nisin-controlled gene expression systems (NICE). A protease encoded by the nrs gene cleaves the C terminus of nisin, conferring in vitro resistance for non-nisin-producing L. lactis [24]. By analogy, the SpeB cysteine protease of Streptococcus pyogenes catalyzes the proteolysis of LL-37 in vitro and in patients infected with resistant bacteria. Under stress caused by AMPs, S. pyogenes synthesizes a G-related α2-macroglobulin-binding (GRAB) protein that acts as a potent inhibitor of the protease inhibitor α2-macroglobulin, forming a cluster named the ‘GRAB-α2-macroglobulin complex’ that retains an active SpeB on the bacterial surface for posterior LL-37 cleavage, causing bacterial resistance [25].
Molecular traps that capture AMPs and biofilm production are also important resistance mechanisms evolved by different species of bacteria. For example, L. lactis expressing PilB, a pilus backbone protein in Gram-positive bacteria, can trap cathelicidin along with the cell wall, avoiding contact with the cytoplasmic membrane. Biofilm-producing bacteria can resist many different antibiotics. Biofilm-mediated resistance results mainly from extracellular polymeric substance (EPS), a liquid comprising mainly amyloid and adhesive fimbriae, extracellular DNA, and exopolysaccharides that embeds multiple cells throughout the biofilm matrix [26]. The EPS extracellular DNA of P. aeruginosa can induce resistance against polymyxin B and colistin in response to the chelating of environmental cations. The decrease in the concentration of biofilm cations modulates the upregulation of LPS modification genes associated with resistance, a typical mechanism in resistant strains of P. aeruginosa [26].
Therapeutic peptides: new drug candidates for the treatment of diseases
In addition to their natural antimicrobial activities, AMPs also have other potential applications in the therapy and treatment of disease. For example, some AMPs have antitumoral and immunomodulatory activities and these peptides are important drug candidates known as anticancer peptides (ACP) and host defense peptides (HDP), respectively. Several ACPs show improved absorption and higher specific cytotoxicity to tumor cells and fewer adverse effects compared with chemical agents. The high number of interactions between ACPs and tumor receptors could result from the presence of abundant anionic sites dispersed on the tumoral cell, resulting in rapid and selective binding and cell death [27].
By contrast, HDPs frequently show weak antimicrobial activity in mammals, but are potent triggers of the immune response through a variety of mechanisms that affect the innate immunity of hosts. These peptides have diverse structures and sequences because of constant interactions with different microbial cells presenting multiple infective strategies 2, 27. The innate immune systems of mammals show characteristics of nonspecific, quick antimicrobial therapies mediated by the activation of elements of resistance against pathogens [28]. HDPs act as triggers of immune responsive elements by several mechanisms, such as the upregulation of the expression of hundreds of genes in monocytes and epithelial cells, the induction of differentiation responses and chemokine synthesis, and promotion of angiogenesis, and inflammatory and wound-healing responses [29].
Some therapeutic peptides have already reached market status, with more than 60 peptides currently available on the market in the USA. It is estimated that, in 2015, approximately 500 therapeutic peptides reached preclinical trials and 140 were included clinical trials in the USA [27]. Despite the development of many potential pharmaceutical peptides, the road to market is restrictive because drug candidates must meet several requirements, such as similar or higher efficacy and tolerability compared with already existing analogous drugs, improved pharmacodynamics and pharmacokinetics, low toxicity, and safe use 2, 27. Economic issues must also be satisfied, mainly relating to market competition, scalable production, and intellectual property. For these reasons, more than 90% of novel therapeutic candidates fail to achieve marketable status [29].
Regulations surrounding the use of such molecules are also determined based on the physicochemical properties and manufacturing of each peptide. The US Food and Drug Administration (FDA) usually ranks peptides as conventional drugs mostly because their chemical structures exceed 100 residues, although exceptions are made mostly in the case of vaccines, which are ranked as biological products [2]. In Europe, the European Medicines Agency (EMA) evaluates the source of the peptide [2]. If the molecule was screened from a natural biological source, it is treated as a biological entity. By contrast, chemical entities are those that were chemically synthesized in vitro. In addition, manufacturing must guarantee the identity, purity, potency, and individual lot consistency 28, 30.
Examples of drug candidate failure are perhaps more common than might expected. The most famous case was the peptide magainin (pexiganan), a potent AMP isolated from the African clawed frog Xenopus laevis. After completion of Phase 3 clinical trials in 1999, the FDA did not approve its commercialization because the drug was not proved to be more effective compared with the antibiotics utilized during the trials [31].
Notable expansion in the peptide-based drug discovery field over the past 10 years, encouraged the screening and testing of new drug candidates. The approval rate since 2012 for peptides is around 20%. This reflects an increasing number of peptides entering annual clinical trials, one in 1970 compared with 20 in 2013. Most candidates that enter Phase 1 clinical trials are painkillers (>30%), or anticancer and anticardiovascular disease agents. ACPs dominate Phase 2 (15%) and 3 (40%) clinical trials, followed by painkillers, anti-infectious disease and antiallergen agents [32].
Tailored peptides: the design of engineered AMPs against MDR bacteria
Historically, the search for new efficient AMPs was based on the high-throughput screening (HTS) of biologically active molecules. This concept relies on the discovery of naturally occurring peptides using classic purification and in vitro and in vivo techniques for checking antimicrobial activity. Many AMPs have been identified and tested against clinical and natural strains of pathogens using this approach [15].
Bioactive peptides obtained from natural sources have been under evolutionary pressure for millennia, and consistently show high stability and target affinity and/or specificity. However, naturally occurring AMPs are normally synthesized at low rates by their biological sources, many are susceptible to protease degradation, and have low bioavailability (i.e. the presence of bioactive molecules at usual low levels). Despite the recent advances in HTS techniques, this approach is laborious and it is difficult to produce high yields of peptides in a scalable fashion 15, 33.
New AMPs with potent antimicrobial activities and lower propensity to select for drug resistance have been intensely investigated. In silico methodologies for the rational design of peptides aim to improve the biological activities and increase production efficiency, speeding up biosynthesis and decreasing production costs. These rational tailored peptides represent a new generation of designer drugs to simultaneously overcome pathogen resistance and enhance microbial killing 33, 34, 35.
AMP design is based primarily on structure–function relations of host-derived synthetic AMPs and computational analysis of surface interactions between the peptide and pathogen structures. There are three main ways to enhance peptide activities through computational design: (i) epitope and net charge engineering by de novo sequence optimization of AMP motifs; (ii) changes in post-translational patterns of glycosylated peptides by amino acid substitution in glycosylation sites; and (iii) engineering different peptides as chimeric molecules and/or biomaterial surfaces with AMP properties 7, 33, 36.
Sequence optimization of motifs is mainly applied for the design of cationic α-helical AMPs with improved and specific antimicrobial activities and low toxicity to mammalian cells. Diverse engineered cationic antimicrobial peptides (eCAPs) have been synthetically produced in laboratories worldwide, and show a range of in vitro and in vivo antimicrobial activities. Examination of structure–function relations is primarily performed for the prediction of protein interfaces to infer protein–protein and protein–lipid interaction networks 7, 37. Interface prediction is based on the physicochemical properties of residues in interfaces of protein complexes and by overlapping interface and non-interface segments of protein motifs. The most prominent characteristics of AMPs for peptide design are the sequence conservation of interface residues, proportion of the 20 types of amino acid residue, relative presence of secondary structures, solvent accessibility, and side-chain conformational entropy [38].
Some AMPs are synthesized as glycoconjugates, peptides carrying a variety of N- or O-linked glycans that are crucial for peptide recognition, binding, and protein–protein interactions. Most surface glycans are post-translationally added in the Golgi after the peptide has passed through the secretory pathway [39]. Plant-derived AMPs are the main primary targets for glycosylation engineering that aims to improve biological activity against phytopathogens and to humanize glycosylation for clinical use in humans. Plant N-glycans differ considerably from those in mammals. Typically, mammalian α1,6 fucose (N-acetylglucosamine of the core), β1,4N-acetylglucosamine (β-mannose of the core), and β1,4 galactose combined with sialic acid and linked to the terminal N-acetylglucosamine are substituted in plants, by an α1,3 fucose, a bisecting β1,2 xylose, and a β1,3 galactose and fucose α1,4-linked to the terminal N-acetylglucosamine, respectively 40, 41. Another important issue concerning the humanization of AMP glycosylation is avoiding the addition of allergenic glycoepitopes to the peptide surface, because humans are frequently allergic to plant α1,3 fucose and β2 xylose. Currently, the main efforts to minimize undesirable plant glycosylation of AMPs rely on avoiding the complete transit throughout the secretory pathway by confining peptides inside the endoplasmic reticulum, using N- or C-terminal retention signals, such as KDEL. Advances in the humanization of the glycosylation of proteins (i.e. the full substitution of non-mammalian host N-linked glycans for typical human glycans by knocking down plant xylosyl and fucosyltransferases and yeast mannosidases and expression of human glycosidases) is already a reality in the production of plant and yeast antibodies and interferons, but has not yet been fully applied to AMPs 41, 42.
Chimeric eCAPs are another major group of engineered AMPs. They are synthesized chemically or recombinantly as fusion peptides or as antimicrobial surface-coating agents, based on the potential synergistic effect of multiple active epitopes that considerably enhances their antimicrobial activities [43]. These features reveal an interesting aspect of chimeric eCAPs, namely the potential to prevent bacterial colonization and biofilm formation, a promising approach to eliminate implant infections. After synthesis, the individual domains of chimeric eCAPs must present solid-binding kinetics to nanoformulated surfaces or substrates without the loss of antimicrobial properties. These peptides remain one of the most promising engineered anti-infective agents to be popularized against opportunistic pathogens in postoperative care settings [44].
Naturally occurring AMPs are typically subject to proteolysis, because they comprise l-amino acids recognizable by proteases. To minimize peptide degradation, the rational design of sequences comprising analogous d-amino acids substituted for l-amino acids can consistently increase the peptide post-translational stability without altering biological function. Given that the interactions between AMPs and the bacterial membrane are not strictly dependent on interactions mediated by specific receptors, the D-enantiomers of a peptide often retain the antimicrobial activity [45].
Another interesting modification of eCAPs is the addition of polyalanine tails in the N or C terminus of the peptide. The amino acid alanine is moderately hydrophobic and, when polymerized as a repetitive peptide tail, can exceed the hydrophobic limit for insertion mechanisms in the cytoplasmic membrane. Polyalanine peptides can not only be inserted into the membrane of microbes without causing significant phospholipid displacement, maintaining the bilayer integrity, but also be internalized within the cell for further modifications in metabolic pathways. Therefore, polyalanine tails can induce undesirable peptide configurations that lead to the formation of peptide clusters that are unable to anchor lipid bilayers 46, 47.
One of the most promising modifications of the eCAP structure is PEGlyation: that is, the covalent addition of polyethylene glycol (PEG) chains to peptides. PEGlyation provides improved structural stability and higher bioavailability of modified peptides and proteins. PEGylated synthetic eCAPs can retain their antimicrobial activity with higher target specificity, although superfluous covalently attached PEG moieties can reduce the interactions between eCAP and the target sites of the cytoplasmic membrane of bacteria 48, 49.
When the peptide shows therapeutic functions, it must be injected into the bloodstream either in its pure form or nanoencapsulated. In such cases, the stability of the peptide must be preserved to maintain a high level of bioactivity. Peptide cyclization is a major strategy to improve the serum stability of synthetic peptides. The joining of the N and C terminus backbone or the formation of internal cross disulfide bridges cyclizes the peptide, hiding proteolytic cleavage sites from specific cellular aminopeptidases 50, 51.
The amidated C terminus of peptides appears to generally improve antimicrobial activity and stability. In general, amidated peptides exhibit higher antimicrobial activity. Amidation of the C terminus affects the hydrophobic moment of synthetic peptides, corroborating to enhance interactions with the membrane. Although this has been an important strategy to improve microbial death, it also appears to improve hemolytic activity, requiring case-by-case studies to evaluate the pros and cons of such modifications 52, 53.
Following the accurate prediction of structural changes, the engineered peptides can be chemically synthesized or routinely produced by many genetic engineering strategies (Fig. 1 ). In this context, in silico interaction databases are the most valuable tools to predict the sites where peptides physically interact with the microbial cell and the optimization of motif architecture and net charge by amino acid replacements.
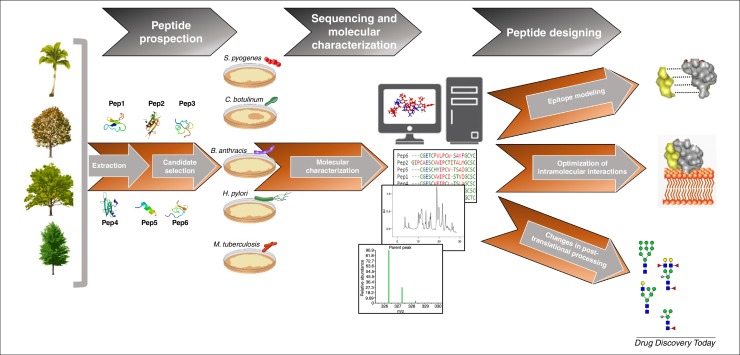
Figure 1.
Schematic diagram of the development of designed peptide. After prospection, natural occurring AMPs are tested against pathogen microorganisms and putative candidates are selected based upon their biological activities. Peptide sequencing and structural characterization are key points to understand the structure-functional relationships of the AMP and to determine the surface interactions between the peptide and pathogen structures. Structural changed AMPs are chemically synthesized or recombinantly produced and their antimicrobial activities are evaluated based in optimized molecular interactions with the target pathogen.
Many examples of rationally designed AMPs have already been described in the literature, most of which show true potential for future use in clinical settings. Often, these eCAPs present not only enhanced biological activities against MDR microorganisms, but also lower propensity to select for resistant bacteria in vitro compared with native analogs. Table 1 lists promising eCAPs for therapeutic use against MDR microbes 7, 33.
Table 1.
Promising characterized eCAPs against MDR microbes
| Name | Structural modification | Enhanced trait | Target | Refs |
|---|---|---|---|---|
| Arabidopsis thalianacyclin-dependent kinase | Recombinant scFv antigen produced in Nicotinia tabacum with no plant glycosylation | Biologically active | Immunomodulation against plant pathogens | [74] |
| Chimeric JEV E protein | Recombinant Escherichia coli expressed fusion of 27-amino acid JEV peptide with Mycobacterium tuberculosis hsp70 antigen | Chimeric protein elicited stronger immune response in mice than the single JEV antigen | Vaccine against Japanese encephalitis virus | [75] |
| Clavanin MO | Amino acid substitution | Improved antibacterial activity | Pseudomonas aeruginosa (immunomodulatory) | [76] |
| Cm-p5 | Peptide fragmentation, de novo sequence determination, amino acid substitution | Improved antibacterial activity | Candida albicans, Cryptococcus neoformans, Trichophyton rubrum | [77] |
| D5 and D6 decamers | Systematic Arg and Trp substitution | Improved antibacterial activity | Gram-positive and Gram-negative bacteria | [78] |
| Escherichia coliheat-labile toxin | Recombinant antigen produced in Zea mays with no plant glycosylation. Retention in endoplasmic reticulum | Biologically active | Vaccine against pathogenic E. coli | [79] |
| Hepatitis B surface antigen | Recombinant antigen produced in N. tabacum with no plant glycosylation. Retention in endoplasmic reticulum | Biologically active | Vaccine against Hepatitis B virus | [80] |
| Human carcinoembryonic antigen | Recombinant antigen produced transiently in N. tabacum | Biologically active | Activity against colon and breast cancer | [81] |
| Japanese cedar pollen allergens | Recombinant antigen produced in Oryza sativa with no plant glycosylation. Retention in endoplasmic reticulum | Biologically active | Vaccine against pollen allergy | [82] |
| Lytic base unit (LBU) | eCAPs with 12–48 residues. Optimized amphipathic helices with only Arg and Trp residues | Maximum antibacterial selectivity at 24 residues; increased activity at 12 residues in length | P. aeruginosa and Staphylococcus aureus | [83] |
| P307SQ-8C | C-terminal amino acid substitution | High in vitro activity against Gram-negative bacteria biofilms. Synergistic activity with polymyxin B. Did not lyse human red blood cells or B cells | Acinetobacter baumannii | [36] |
| Peptide derived from human lysosomal cathepsin G (cat G) | Substitution of residues 117–136 | Enhancement of activity against Gram-positive and Gram-negative bacteria | S. aureus, P. aeruginosa | [84] |
| Synthetic cxc cfcfc peptides | Dengue fever virus and Japanese encephalitis virus synthetic peptides with motifs to fit human leukocyte antigen (HLA) | Enhanced cellular immune response in the lymph nodes | Vaccine against Dengue virus for populations in developing countries | [58] |
| TiBP1 | Chimeric peptides with solid-binding kinetics to titanium substrate | Enhanced activity | Activity against bacteria commonly found in oral and orthopedic implants, such as Streptococcus mutans, Staphylococcus epidermidis, and E. coli | [85] |
| WLBU2 and WR12 | Idealized amphipathic helices with three and two amino acid substitution, respectively | WLBU2 eradicated lethal P. aeruginosa septicemia in mice | 142 isolates of ESKAPE pathogens | [86] |
Magnifection: a technology for the rapid and massive production of peptides in tobacco
After prospection or improvements in silico, a selected AMP must be produced on a large scale at a consistently high quality and under good manufacturing practice rules (GMP). The size and chemical properties of a given peptide will directly influence the choice of production strategy [54]. There are at least four major strategies to achieve satisfactory yields of high-quality products: (i) solid-state chemical synthesis; (ii) recombinant microbe platforms; (iii) transgenic plants and animals; and (iv) cell-free expression systems. Regardless of the method used, downstream processing is the crucial step before the commercialization of any peptide. This is the most expensive, time-consuming and testing phase of the production pipeline 55, 56.
Specially designed strains of bacteria or fungal accumulating induced mutations are typically utilized as reactors of naturally occurring AMPs. These strains can enhance protein synthesis and secretions, as well as upgrade peptide folding. Under these circumstances, peptide yields can increase by three orders of magnitude, but in many cases, the production levels will still be below the standard production levels required [54].
As an alternative to natural sources, the production technology can result in the chemical synthesis of partial or full peptide chains. There are three types of chemical synthesis: (i) the solution phase; (ii) the solid phase; and (iii) hybrid approaches [57]. Most of the commercially approved peptides, which are frequently small to medium in size, are synthesized by solution phase approaches 54, 58. This methodology provides standard protocols for the isolation, characterization, and purification of peptides. Solid-phase synthesis provides platforms for the production of large and structurally complex peptides on a large scale. The hybrid method combines characteristics of the two previous methodologies. Although efficient for the production of active peptides, the three approaches are expensive because they enable prolonged times for amino acid polymerization, a drawback that is particularly important during early clinical studies because it can invalidate economically the manufacturing of the peptides 2, 54, 55, 56.
To maximize peptide biosynthesis, genetically engineered bacteria and yeast cells are frequently explored as vehicles for the recombinant production of bioactive AMPs [59]. Many different AMPs have been synthesized in E. coli and Pichia pastoris [60]. Despite the high therapeutic potential of recombinant AMPs, limited investment of companies and drawbacks in terms of poor yield, low quality, and unsatisfactory in vivo activity have restricted commercial development to only a few promising AMPs. Regardless of these production limitations, some of these therapeutic peptides have reached advanced clinical trials before commercialization, as detailed in Table 2 .
Table 2.
Recombinant AMPs that reached advanced preclinical stages or are undergoing trials
| Name | Developer | Natural source | Target (prevented disease) | Status | Refs |
|---|---|---|---|---|---|
| Arenicin | Adenium Biotech (Copenhagen, Denmark) | Lugworm Arenicola marina | Multiresistant Gram-positive bacteria | Preclinical | [31] |
| Avidocin and purocin | AvidBiotics (San Francisco, CA, USA) | Pseudomonas aeruginosa | Gram-negative bacteria | Preclinical | [31] |
| IMX924 | Iminex (Coquitlam, BC, Canada) | Mammalian | Gram-negative and Gram-positive bacteria | Preclinical | [31] |
| Iseganan (IB-367) | Intrabiotics Pharmaceuticals, Inc. (Mountainview, CA, USA) | Pig leucocytes | Bacteria and fungi/chronic respiratory infections | Abandoned after Phase 2 clinical trials | [87] |
| MBI 594AN | Microbiologix Biotech (Vancouver, BC, Canada) | Chemically modified mammalian peptide | Propionibacterium acnes (acne) | Phase 2b clinical trial | [88] |
| Neuprex (rBPI21) | Xoma (US) LLC (Berkeley, CA, USA) | Vaccine against Hepatitis B virus | Neisseria meningitidis (severe meningococcemia) | Phase 3 clinical trial | [31] |
| Omiganen (MBI-226) | Microbiologix Biotech | Bovine neutrophils | Bacteria and fungi/bloodstream infections | Phase 3 clinical trial | [89] |
| P113 | Demegen (Pittsburgh, PA, USA) | Mammalian | Candida albicans (oral candidiasis) | Phase 1/2 clinical trial | [90] |
| Pexiganan (MSI-78) (magainin) | Magainin Pharmaceutical Inc., since renamed Genaera (Plymouth Meeting, PA, USA) | African clawed frog Xenopus laevis | Broad-spectrum activity against 3109 bacterial clinical isolates (diabetic foot ulcers) | Discontinued by showing same efficiency as other antibiotics | [91] |
| Plectasin | Novozymes (Bagsvaerd, Denmark) | Fungal (Pseudoplectania nigrella) | Streptococcus pneumoniae (pneumonia) | Preclinical | [92] |
| XMP.629 | Xoma (US) LLC | Mammalian | P. acnes (acne) | Phase 2 clinical trial | [61] |
Among the most important factors that limit the recombinant production of AMPs in microbial systems is the inner toxicity of the peptide toward host cells; however, this is not typically a limitation because many AMPs kill bacteria at very low, nontoxic concentrations. Another concern is the low quality of the peptide product following post-translational modifications. Under such circumstances, plants appear to be an interesting and promising alternative host system for the production of recombinant AMPs 61, 62, 63.
Although plants perform a range of post-translational modifications, low levels of recombinant biosynthesis of peptides are common, resulting in low quantities of purified products. However, a new transformative technology, called Magnifection, has emerged as a platform for the fast production of large numbers of plant-derived recombinant proteins and peptides 8, 63, 64.
Developed by Gleba and collaborators at the German biotech company Icon Genetics, Magnifection is a transient expression platform that utilizes Nicotinia tabacum or Nicotinia benthamiana plants as efficient reactors for the production of massive yields of recombinant proteins, in a rapid, scalable fashion 63, 65. The process is based on the infiltration of whole plants with a suspension of transgenic Agrobacterium tumefaciens cells carrying plasmids that encode viral RNAs replicons (Fig. 2 ). These Gram-negative soil bacteria have key roles in infection and movement throughout plant tissues because they systemically spread through the plant to eventually reach most leaves and stems 8, 63. Infiltrated plants contain viral vectors based on tobacco mosaic virus (TMV) or potato virus X (PVX) carrying AMP-coding sequences, delivered by bacteria to be transiently expressed and amplified [64]. These potent machines of transcript production use the viral machinery to enhance the production of viral proteins along with the selected AMP, without stable transgene integration, resulting in massive yields of AMPs [65].
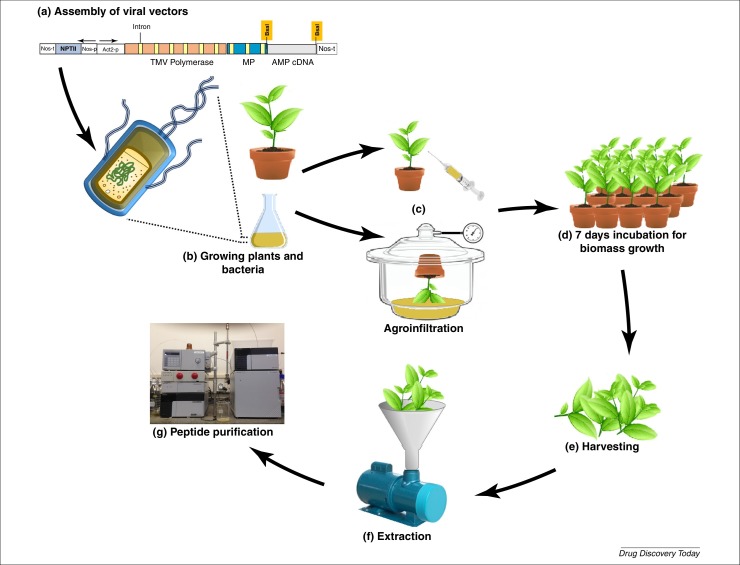
Figure 2.
Transient biosynthesis of AMPs using the Magnifection platform. (a) Assembled viral vector cassettes harboring the coding sequence of an AMP is introduced in cells of A. tumefaciens. After growth and selection of transgenic bacteria (b), leaves of N. benthamiana are agroinfiltrated using a syringe or vacuum (c) and the kinetics of transgene transient expression reaches its peak between 4 and 7 days after transfection (d). After harvesting the plant biomass (e) and peptide extraction using appropriate buffers (f), the AMPs are purified and evaluated in bio-assays (g).
Speed is one of the main advantages of Magnifection, because it provides expression kinetics that frequently reach the peak of peptide production 3–4 days after infiltration. Such conditions allow the scale-up of plant infiltration by vacuum and peptide biosynthesis, combining elements of three biological systems (i.e. viral potent transcription, bacterial systemic spread, and plant accurate post-translational modifications) in a single expression strategy 8, 64.
Therefore, Magnifection significantly reduces AMP production costs because of a rapid and straightforward approach that results in the production of the first milligrams of AMPs in just 4 weeks and up to 100 kg in 1 year, using a much-diluted A. tumefaciens suspension to infiltrate completely an entire green house. The high biomass of tobacco (>100.00 kg ha−1) also contributes to the scaling up of peptide production, while reducing overall costs 64, 65.
Using Magnifection, protein and peptide amounts frequently are 10–100-fold higher than those observed for stable genetically transformed plants, yielding up to 80% of the total soluble protein (TSP). Using this technology, amounts of up to 5 g of peptides per kilogram of fresh agroinfiltrated leaves have been obtained routinely 8, 63, 64.
The Magnifection platform has experienced considerable success in the production of a variety of proteins and peptides, notably vaccines [65]. The small size of antigen peptides and relatively simple chemical structures of some AMPs appear to fulfill the requirements of the platform. Since 2010, the Magnifection system has been explored by the Canadian biotech company Medicago (http://www.medicago.com/) for the industrial production of a vaccine against H1N1 flu in the USA. A US$21 million budget financial deal was signed between Medicago and the US Department of Defense Agency Defense Advanced Research Projects Agency (DARPA) to develop 10 million doses per month to avoid the risk of epidemic flu outbreaks 8, 63, 64, 65. The vaccine is currently in Phase 2 clinical trials, as is another vaccine against the variant virus H5N1, synthesized using the same platform [64]. Table 3 details other vaccines already synthesized in tobacco using the Magnifection system.
Table 3.
Examples of antigens transiently expressed using the Magnifection system
| Antigen | Disease/target | Status | Refs |
|---|---|---|---|
| Der p 1 | Allergy | In vitro | [93] |
| F1-V | Plague | Animal preclinical trial | [94] |
| Hepatitis B/C | HBsAg (Hep B) | Animal preclinical trial | [95] |
| HIV p24 capsid protein HIV | AIDS | In vitro | [96] |
| HSP-A | Helicobacter pylori | Phase 1/2 clinical trial | [97] |
| L1 major capsid protein | Cervical cancer | Animal preclinical trial | [98] |
| Protective antigen Der p 2 | Anthrax | Animal preclinical trial | [99] |
| SARS-CoV-S1 | SARS | Animal preclinical trial | [100] |
| Tet-C | Tetanus | Animal preclinical trial | [101] |
| Type 1 diabetes mellitus | GAD65 | Animal preclinical trial | [102] |
| VCA antigen | Epstein–Barr virus | In vitro | [103] |
| VP1 | Foot and mouth disease | Animal preclinical trial | [104] |
Although efficient for the transient biosynthesis of peptides, the addition of plant-specific post-translational modifications, notably the addition of N-glycans to peptides, is a potential limitation of the system and could lead to nonfunctional products or highly immunogenic vaccines. The threshold of economically accepted expression levels using Magnifection is a restrictive factor similar to other previously explored recombinant platforms. Although a good producer of full antibodies, the size of multimeric proteins constitutes another important challenge for the Magnifection system, requiring the manipulation of two viral vectors to express two or more assembled polypeptides 63, 64.
Genome editing by CRISPRs and AMP biosynthesis: advances, implications, and challenges
The development of new technologies to improve recombinant peptide biosynthesis has contributed to considerably increase expression levels and improve product quality. However, frequent gene silencing at the transcriptional level and instability of genes cloned in vectors for stable or transient expression remain major challenges limiting the efficient production of AMPs. Another persistent issue is the poor quality of the peptides endogenously synthesized in bacteria, yeast, and plants, particularly resulting from undesirable post-translational modifications 2, 33.
Improved targeted genome engineering represents a sophisticated approach that could help minimize such limitations. Over the past decade, alternative genome-editing tools, such as artificial engineered enzymes, zinc-finger nucleases (ZFNs), and transcription-activator-like effector nucleases (TALENs), have been successfully utilized to modify the genome of microbes, plants, and animals by adding, removing, or replacing segments of DNA [66].
A more efficient and less time-consuming technology for genome engineering was developed more recently, and appears to considerably expand the possible modifications of target sites in almost any sequenced genome: the clustered regularly interspaced short palindromic repeats system (CRISPRs) [66] (Box 1 ).
Box 1. Genome editing tools.
Strategic question
How do CRISPRs revolutionize the currently available molecular tools for genome editing and AMP biosynthesis? Here, we describe previous molecular tools based on the repair of artificially generated DNA DSBs.
Zinc finger nucleases
Zinc finger nucleases (ZNFs) are genetically engineered DNA-binding proteins utilized for the easy editing of the genome. They create DSBs at user-specified locations. ZFNs present two functional domains. The first is a DNA-binding domain with two-finger modules for DNA hexamer (6-bp) recognition. These two-fingered modules stitch together to produce the zinc finger proteins, which are capable of recognizing and specifically binding to a ≥24-bp DNA extension. The second domain is a nuclease FokI DNA-cleavage domain that, once fused to the DNA-binding domain, forms a molecular scissor that catalyzes phosphodiester disruption. When used together, two ZFNs can produce DSBs that are targets for precise genomic edits by natural DNA repair processes, such as homologous recombination and non-homologous end joining (NHEJ).
Pros of ZFNs for genome editing
-
•
Provide rapid DNA disruption of, and/or integration into, any genomic loci.
-
•
Functional in a range of mammalian somatic cell types.
-
•
Edit genomes through a single transfection round of DNA repair.
-
•
Screening dispenses antibiotics.
Cons of ZFNs for genomic editing
-
•
Screening and assembly is technically challenging.
-
•
Typically binds to short 9–18-bp sequences.
-
•
Replacement of fragments longer than 1 kb is difficult.
-
•
Off-target effects.
-
•
Target events in animals require screening.
Transcription activator-like effector nucleases
Transcription activator-like effectors (TALEs) are a class of proteins recent found exclusively in the plant pathogenic bacterium Xanthomonas. TALEs comprises a DNA-binding domain of 33–35 conserved amino acid repeat motifs organized in tandem arrays that individually recognizes a specific nucleotide of target DNA. TALE repeats provide two adjacent amino acids, named repeat-variable-di-residue (RVD), that specifically bind to a nucleotide, and can be arranged in different combinations according to the sequence that must be recognized in the target DNA. A new type of engineered TALE repeats presents a C-terminal FokI endonuclease domain, constituting a TALE nuclease (TALEN). TALEN are similar to ZFNs, providing DSBs when grouped in pairs after binding to DNA with high affinity.
Pros of TALENs for genome editing
-
•
Easier to design compared with ZFNs.
-
•
Bind 18-bp or longer sequences.
-
•
Fewer constraints on site location.
Cons of TALENs for genome editing
-
•
Larger than ZFNs (coded usually by 3-kb DNA).
-
•
Evidence that larger TALENs can lead to less specificity.
-
•
Off-target effects.
CRISPRs
CRISPRs are recently discovered bacterial adaptive immune systems that use a combination of short RNAs and associated proteins to target specific sequences of DNA for the generation of DSBs. CRISPRs revolutionized genome editing standards by presenting several technical advantages compared with previous systems, although they still have some disadvantages.
Pros of CRISPRs for genome editing
-
•
Easily adapted system to target and modify any genomic sequence.
-
•
The Cas9 protein, the main protein for DNA recognition and cleavage, remains unchanged for any CRISPRs.
-
•
Easy to use to target numerous sites or across genomic libraries.
-
•
Use of multiplex-based guide RNAs to simultaneously edit multiple sites.
Cons of CRISPRs for genome editing
-
•
Size of Cas9 (cDNA approximately 4.2 kb).
Conventional genome-editing systems use synthetic nucleases to induce genomic double-stranded breaks (DSBs) at target sites. DSBs are targets of the imprecise cellular repair machineries either mediated by non-homologous end-joining (NHEJ) or homology directed repair (HDR), which require a donor DNA template [98]. CRISPRs constitute an incredibly versatile genome-editing platform derived from the S. pyogenes CRISPR-associated protein 9 (Cas9) 67, 68. There are three types of CRISPRs. Type II is the simplest, presented only by bacteria and comprises four proteins (i.e. Cas1, Cas2, Cas9, and Cas4); it is the most widely used system for gene engineering. The most popular CRISPR–Cas system is constituted by Cas9 endonuclease proteins and CRISPR RNAs [68]. The nuclease Cas9 can catalyze the precise cleavage of target sites assisted by two short helper RNAs: cRNA and tracRNA. The fusion of both RNAs forms a hybrid single guide RNA (sgRNA) that binds to Cas9 to form a supramolecular complex named RNA-guided endonuclease, a sequence-specific recognizer complex that cleaves at specific sites in the genome, resulting in DSBs that are then repaired preferentially by HDR (in the case of bacterial genomes), thus altering the genome in a precise manner by site-specific modifications [69].
The CRISPR–Cas9 system has been used to introduce point mutations, modify gene function, generate gene knockouts, integrate foreign genes, repress and/or activate specific genes, deliver epigenetic modifications, and aid genomic loci accession by proteins. Although limited studies report the utilization of the type II CRISPRS–Cas system to edit plant genomes, it has been exploited for editing other organisms 66, 67, 68.
In addition genome editing, the CRISPR–Cas system can be utilized for other purposes within the cell. For instance, gene silencing by CRISPR interference (CRISPRi) has proved to be a powerful tool to complement (and, in many cases, improve) previously described RNAi, by the double formation of Cas9 bound to sgRNA and attached to the nontemplate strand of DNA, blocking transcription [69]. This approach can also be utilized to simultaneously silence multiple target genes (a process called multiplexing), thus overcoming one of the limitations of RNAi in terms of its gene silencing potential. Another interesting application of CRISPR–Cas is to block transcription initiation in a specific and reversible manner [70].
There is immense potential for the utilization of CRISPRs to improve the recombinant biosynthesis of AMPs in almost any organism. Tailored genomes modified by CRISPRs can harbor AMP coding sequences within the donor DNA fragment inserted in a genomic expression hot spot, potentially boosting heterologous production of the peptide to unprecedented levels 69, 71 (Fig. 3 ).
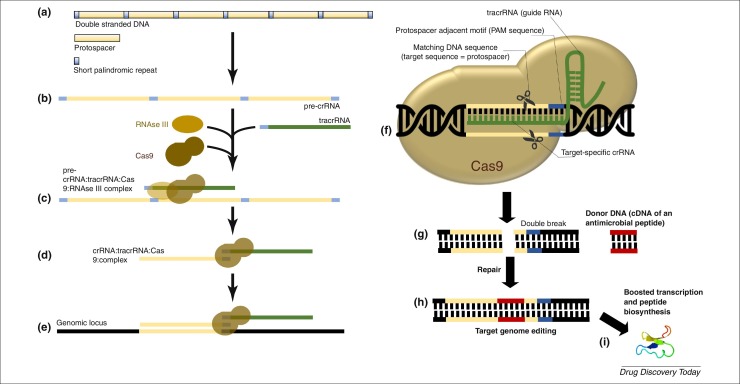
Figure 3.
CRISPR/Cas system II technology description for AMP biosynthesis mediated by genome edition. Spacer acquisition: (a) Formation of CRISPRs array by recognition and integration of foreign DNA as spacer within the CRISPR locus, or fully synthesized by genetic engineering. The protospacers are non-coding region inserted in the bacterial DNA with 24–48 bp. Adjacent to each protospacer are found 3–5 bp short DNA sequences termed protospacer adjacent motifs (PAM) crRNA processing: (b) The CRISPR array is transcribed as a long RNA (pre-crRNA) that is cleaved into crRNAs with the help of Cas proteins. An extra small RNA (tracrRNA) complementary to the repeat sequence is also synthesized. (c) The tracrRNA pairs with the repeated region of crRNA and helps in the processing of pre-crRNA into crRNA with the help of RNAse III for cleavage. Interference stage: crRNAS binds to Ca proteins (d) to form a complex that recognizes foreign DNA (e). A single multifunctional protein, Cas9, recruits crRNA and tracrRNA to cleave the recognized foreign DNA using internal endonuclease domains (f). All the process is based in the recognition and pairing of the PAM and the foreign DNA. Double strand breaks (DSBs) are generated (g). A donor DNA containing the coding sequence of an AMP of interest is integrated in the site of the DSBs by homologous recombination (i), and after gene expression, the AMP is extracted and purified (h).
To reduce undesirable post-translational modifications of AMPs, it might be possible to knockout host glycosylase genes or substitute them with coding sequences of human glycosylases to humanize glycopeptides. This could contribute to enhance antimicrobial activity and therapeutic peptide quality (purity, lack of undesirable post-translational modifications, high levels, among others). In addition glycosylation, other post-translational modifications and peptide-processing pathways could be manipulated with CRISPR–Cas. CRISPR–Cas represents a promising technology to improve peptide engineering and biosynthesis, pushing the boundaries of the development of clinical drugs to new levels of sophistication 68, 70, 71.
The major concern for the utilization of CRISPRs as a genome-editing tool is its potential secondary mutations or off-targets effects. This is a common scenario in human edited cells (however, on-target efficiency has been recently improved), but still rare in plants. To avoid off-target effects, it is imperative to properly design sgRNAs that direct Cas9 to an exact target in the genome, and efforts along these lines have been published recently. In addition, high levels of the nuclease Cas9 relative to sgRNA helps reduce the incidence of off-target effects [72]. The Cas9 nuclease can be further engineered into a nickase, which only cleaves a single strand of DNA and the same strategy on the other strand enhances the specificity of site recognition and consistently minimizes off-target effects 67, 70, 71, 72, 73.
Concluding remarks and future perspectives
The prospect of novel, efficient AMPs is a crucial starting point to combat antibiotic-resistant microbial pathogens. The emergence of MDR bacteria is a tremendous global health problem that has been predicted to lead to the death of 10 million humans per year by 2050 [73]. Microbes, plants, and animals are natural sources of therapeutic drugs and could also be a source of novel, biologically inspired, previously unknown antibiotics for human applications. In addition to their obvious importance, novel AMPs with high anti-infective properties are difficult to prospect and are naturally synthesized only at low levels in their respective host organisms. Therefore, there is an urgent need to increase the efficiency and final yield of peptides during their production to allow their economic exploitation.
New insights into the rational design of peptides are required to maximize AMP variability and increase specificity and antimicrobial activity. Currently, examples of synthetic and recombinant AMPs with tailored domains are now facing advanced clinical trials, with promising results. These rationally designed molecules present modified epitopes with improved net charge and enhanced antimicrobial activity, and represent a new generation of anti-infective agents with the potential to overcome MDR.
The transient expression of AMP genes in tobacco is a new technology with immense potential to considerably boost the heterologous production of new antibiotics and vaccines, in a fast, cheap, and efficient way. The use of potent viral vectors associated with the bacterial delivery of transcriptional units provides a scalable platform for the massive production of diversified AMPs, with potential desirable improvements in terms of processing and production costs.
Some drawbacks presented by plant expression systems are solved by the CRISPR system, a revolutionary genome-editing technology that presents myriad possibilities for genetic manipulation at the genomic level and provides unprecedented tools (CRISPRi and CRISPRa) to precisely control gene expression and the structural modification of AMPs. Despite its current minor limitations (e.g. off-target effects), CRISPR could have a key role in the future development of clinical drugs as biotechnological anti-infective agents, including the rational biosynthesis of next-generation antimicrobials.
Biographies

Dr Nicolau Brito da Cunha is a Biochemistry professor at the Catholic University of Brasilia (UCB) and fellow researcher at the Center of Proteomic and Biochemical Analysis (CAPB) of the same university. He participates in different research projects focusing the recombinant biosynthesis of different antimicrobial and therapeutic peptides in microbes and plants. He earned his PhD in Molecular Biology from University of Brasilia in 2012.

Nicole Berwanger Cobacho is a Master degree student at the Genomics and Biotechnology Post Graduation Program, at the Catholic University of Brasilia (UCB). The main theme of her research is the stable recombinant expression of antimicrobial peptides in prokaryotic systems and the transient expression of peptides in Nicotiana benthamiana leaves, aiming the prospection and study of potential drug candidates against pathogens like Klebsiella pneumoniae.

Simoni Campos Dias is a professor at the Genomics and Biotechnology Post Graduation Program, at the Catholic University of Brasilia (UCB) and fellow researcher at the Center of Proteomic and Biochemical Analysis (CAPB) of the same university. She participates in several projects in the area biochemistry, proteomics and molecular biology researching on the recombinant biosynthesis of different antimicrobials peptides and their use as biopharmaceuticals and plant defense.
References
- 1.Watkins R.R., Bonomo R.A. Overview: global and local impact of antibiotic resistance. Infect. Dis. Clin. N. Am. 2016;30:313–322. doi: 10.1016/j.idc.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Uhlig T. The emergence of peptides in the pharmaceutical business: from exploration to exploitation. EuPA Open Proteomics. 2014;4:58–69. [Google Scholar]
- 3.Andersson D.I. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist. Updates. 2016;26:43–57. doi: 10.1016/j.drup.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Karam G. Antibiotic strategies in the era of multidrug resistance. Crit. Care. 2016;20:136. doi: 10.1186/s13054-016-1320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen L.T. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011;29:464–472. doi: 10.1016/j.tibtech.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Mith O. The antifungal plant defensin AhPDF1.1b is a beneficial factor involved in adaptive response to zinc overload when it is expressed in yeast cells. MicrobiologyOpen. 2015;4:409–422. doi: 10.1002/mbo3.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deslouches B. Engineered cationic antimicrobial peptides to overcome multidrug resistance by ESKAPE pathogens. Antimicrob. Agents Chemother. 2015;59:1329–1333. doi: 10.1128/AAC.03937-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gleba Y. Magnifection: a new platform for expressing recombinant vaccines in plants. Vaccine. 2005;23:2042–2048. doi: 10.1016/j.vaccine.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Zhang F. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum. Mol. Genet. 2014;23:R40–R46. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- 10.Malanovic N., Lohner K. Antimicrobial peptides targeting Gram-positive bacteria. Pharmaceuticals. 2016;9:59. doi: 10.3390/ph9030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malanovic N., Lohner K. Gram-positive bacterial cell envelopes: the impact on the activity of antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2016;1858:936–946. doi: 10.1016/j.bbamem.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Shruti S. Antimicrobial peptides and their pore/ion channel properties in neutralization of pathogenic microbes. Curr. Top. Med. Chem. 2016;16:46–53. doi: 10.2174/1568026615666150703115454. [DOI] [PubMed] [Google Scholar]
- 13.Maria-Neto S. Understanding bacterial resistance to antimicrobial peptides: from the surface to deep inside. Biochim. Biophys. Acta Biomembr. 2015;1848:3078–3088. doi: 10.1016/j.bbamem.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 14.Fleitas O., Franco O.L. Induced bacterial cross-resistance toward host antimicrobial peptides: a worrying phenomenon. Front. Microbiol. 2016;7:381. doi: 10.3389/fmicb.2016.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parachin N.S., Franco O.L. New edge of antibiotic development: antimicrobial peptides and corresponding resistance. Front. Microbiol. 2014;5:147. doi: 10.3389/fmicb.2014.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mishra N.N., Bayer A.S. Correlation of cell membrane lipid profiles with daptomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2013;57:1082–1085. doi: 10.1128/AAC.02182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang T. Cardiolipin prevents membrane translocation and permeabilization by daptomycin. J. Biol. Chem. 2014;289:11584–11591. doi: 10.1074/jbc.M114.554444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayer A.S. Heterogeneity of mprF sequences in methicillin-resistant Staphylococcus aureus clinical isolates: role in cross-resistance between daptomycin and host defense antimicrobial peptides. Antimicrob. Agents Chemother. 2014;58:7462–7467. doi: 10.1128/AAC.03422-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra N.N. Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content. PLoS ONE. 2012;7:e43958. doi: 10.1371/journal.pone.0043958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kristian S.A. d-Alanylation of teichoic acids promotes group A Streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J. Bacteriol. 2005;187:6719–6725. doi: 10.1128/JB.187.19.6719-6725.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raetz C.R.H., Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson J.C. Antimicrobial peptide resistance of Vibrio cholerae results from an LPS modification pathway related to nonribosomal peptide synthetases. ACS Chem. Biol. 2014;9:2382–2392. doi: 10.1021/cb500438x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jusko M. A metalloproteinase karilysin present in the majority of Tannerella forsythia isolates inhibits all pathways of the complement system. J. Immunol. 2012;188:2338–2349. doi: 10.4049/jimmunol.1101240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maisey H.C. A group B streptococcal pilus protein promotes phagocyte resistance and systemic virulence. FASEB J. 2008;22:1715–1724. doi: 10.1096/fj.07-093963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen M. Protein GRAB of Streptococcus pyogenes regulates proteolysis at the bacterial surface by binding α2-macroglobulin. J. Biol. Chem. 1999;274:15336–15344. doi: 10.1074/jbc.274.22.15336. [DOI] [PubMed] [Google Scholar]
- 26.Dufour D. Bacterial biofilm: structure, function, and antimicrobial resistance. Endod. Top. 2010;22:2–16. [Google Scholar]
- 27.Fosgerau K., Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discov. Today. 2015;20:122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Hancock R.E.W., Sahl H-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 29.Kosikowska P., Lesner A. Antimicrobial peptides (AMPs) as drug candidates: a patent review (2003–2015) Expert Opin. Ther. Pat. 2016;26:689–702. doi: 10.1080/13543776.2016.1176149. [DOI] [PubMed] [Google Scholar]
- 30.Vergote V. Quality specifications for peptide drugs: a regulatory-pharmaceutical approach. J. Pept. Sci. 2009;15:697–710. doi: 10.1002/psc.1167. [DOI] [PubMed] [Google Scholar]
- 31.Fox J.L. Antimicrobial peptides stage a comeback. Nat. Biotechnol. 2013;31:379–382. doi: 10.1038/nbt.2572. [DOI] [PubMed] [Google Scholar]
- 32.Kaspar A.A., Reichert J.M. Future directions for peptide therapeutics development. Drug Discov. Today. 2013;18:807–817. doi: 10.1016/j.drudis.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 33.Deslouches B. Rational design of engineered cationic antimicrobial peptides consisting exclusively of arginine and tryptophan, and their activity against multidrug-resistant pathogens. Antimicrob. Agents Chemother. 2013;57:2511–2521. doi: 10.1128/AAC.02218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brogden K.A. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 35.Mihajlovic M., Lazaridis T. Antimicrobial peptides in toroidal and cylindrical pores. Biochim. Biophys. Acta. 2010;1798:1485–1493. doi: 10.1016/j.bbamem.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thandar M. Novel engineered peptides of a phage lysin as effective antimicrobials against multidrug resistant, Acinetobacter baumannii. Antimicrob. Agents Chemother. 2016;60:2671–2679. doi: 10.1128/AAC.02972-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pushpanathan M. Antimicrobial peptides: versatile biological properties. Int. J. Pept. 2013;2013:15. doi: 10.1155/2013/675391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou H-X., Qin S. Interaction-site prediction for protein complexes: a critical assessment. Bioinformatics. 2007;23:2203–2209. doi: 10.1093/bioinformatics/btm323. [DOI] [PubMed] [Google Scholar]
- 39.Gomord V. Biopharmaceutical production in plants: problems, solutions and opportunities. Trends Biotechnol. 2005;23:559–565. doi: 10.1016/j.tibtech.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 40.Bardor M. N-glycosylation of plant recombinant pharmaceuticals. In: Faye L., Gomord V., editors. Recombinant Proteins from Plants: Methods and Protocols. Humana Press; 2009. pp. 239–264. [Google Scholar]
- 41.Gomord V. Production and glycosylation of plant-made pharmaceuticals: the antibodies as a challenge. Plant Biotechnol. J. 2004;2:83–100. doi: 10.1111/j.1467-7652.2004.00062.x. [DOI] [PubMed] [Google Scholar]
- 42.Frey A.D. Expression of rat β(1,4)-N-acetylglucosaminyltransferase III in Nicotiana tabacum remodels the plant-specific N-glycosylation. Plant Biotechnol. J. 2009;7:33–48. doi: 10.1111/j.1467-7652.2008.00370.x. [DOI] [PubMed] [Google Scholar]
- 43.Pasupuleti M. End-tagging of ultra-short antimicrobial peptides by W/F stretches to facilitate bacterial killing. PLoS ONE. 2009;4:e5285. doi: 10.1371/journal.pone.0005285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yazici H. Engineered chimeric peptides as antimicrobial surface coating agents toward infection-free implants. ACS Appl. Mater. Interfaces. 2016;8:5070–5081. doi: 10.1021/acsami.5b03697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de la Fuente-Núñez C. d-Enantiomeric peptides that eradicate wild-type and multi-drug resistant biofilms and protect against lethal Pseudomonas aeruginosa infections. Chem. Biol. 2015;22:196–205. doi: 10.1016/j.chembiol.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bechinger B. Membrane insertion and orientation of polyalanine peptides: a (15)N solid-state NMR spectroscopy investigation. Biophys. J. 2001;81:2251–2256. doi: 10.1016/S0006-3495(01)75872-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Migliolo L. Structural and functional characterization of a multifunctional alanine-rich peptide analogue from Pleuronectes americanus. PLoS ONE. 2012;7:e47047. doi: 10.1371/journal.pone.0047047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts M.J. Chemistry for peptide and protein PEGylation. Adv. Drug Deliv. Rev. 2012;64(Suppl.):116–127. doi: 10.1016/s0169-409x(02)00022-4. [DOI] [PubMed] [Google Scholar]
- 49.Hamley I.W. PEG–peptide conjugates. Biomacromolecules. 2014;15:1543–1559. doi: 10.1021/bm500246w. [DOI] [PubMed] [Google Scholar]
- 50.Harris K.S. Efficient backbone cyclization of linear peptides by a recombinant asparaginyl endopeptidase. Nat. Commun. 2015;6:10199. doi: 10.1038/ncomms10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thakkar A. Global analysis of peptide cyclization efficiency. ACS Comb. Sci. 2013;15:120–129. doi: 10.1021/co300136j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.da Silva A.V.R. The effects of the C-terminal amidation of mastoparans on their biological actions and interactions with membrane-mimetic systems. Biochim. Biophys. Acta Biomembr. 2014;1838:2357–2368. doi: 10.1016/j.bbamem.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 53.Aoki W., Ueda M. Characterization of antimicrobial peptides toward the development of novel antibiotics. Pharmaceuticals. 2013;6:1055–1081. doi: 10.3390/ph6081055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vriens K. Antifungal plant defensins: mechanisms of action and production. Molecules. 2014;19:12280. doi: 10.3390/molecules190812280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ongey E.L., Neubauer P. Lanthipeptides: chemical synthesis versus in vivo biosynthesis as tools for pharmaceutical production. Microb. Cell Factor. 2016;15:97. doi: 10.1186/s12934-016-0502-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nilsson B.L. Chemical synthesis of proteins. Annu. Rev. Biophys. Biomol. Struct. 2005;34:91–118. doi: 10.1146/annurev.biophys.34.040204.144700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vlieghe P. Synthetic therapeutic peptides: science and market. Drug Discov. Today. 2010;15:40–56. doi: 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 58.Becker Y. Dengue fever virus and Japanese encephalitis virus synthetic peptides, with motifs to fit HLA class I haplotypes prevalent in human populations in endemic regions, can be used for application to skin Langerhans cells to prime antiviral CD8+ cytotoxic T cells (CTLs): a novel approach to the protection of humans. Virus Genes. 1994;9:33–45. doi: 10.1007/BF01703433. [DOI] [PubMed] [Google Scholar]
- 59.Parachin N.S. Expression systems for heterologous production of antimicrobial peptides. Peptides. 2012;38:446–456. doi: 10.1016/j.peptides.2012.09.020. [DOI] [PubMed] [Google Scholar]
- 60.Mulder K. Current scenario of peptide-based drugs: the key roles of cationic antitumor and antiviral peptides. Front. Microbiol. 2013;4:321. doi: 10.3389/fmicb.2013.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gordon Y.J. A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs. Curr. Eye Res. 2005;30:505–515. doi: 10.1080/02713680590968637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nawrot R. Plant antimicrobial peptides. Folia Microbiol. 2013;59:181–196. doi: 10.1007/s12223-013-0280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gleba Y. Engineering viral expression vectors for plants: the ‘full virus’ and the ‘deconstructed virus’ strategies. Curr. Opin. Plant Biol. 2004;7:182–188. doi: 10.1016/j.pbi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 64.Gleba Y.Y. Plant viral vectors for delivery by Agrobacterium. In: Palmer K., Gleba Y., editors. Plant Viral Vectors. Springer; 2014. pp. 155–192. [DOI] [PubMed] [Google Scholar]
- 65.Gleba Y. Viral vectors for the expression of proteins in plants. Curr. Opin. Biotechnol. 2007;18:134–141. doi: 10.1016/j.copbio.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 66.Gupta R.M., Musunuru K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR–Cas9. J. Clin. Investig. 2014;124:4154–4161. doi: 10.1172/JCI72992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar V., Jain M. The CRISPR–Cas system for plant genome editing: advances and opportunities. J. Exp. Bot. 2015;66:47–57. doi: 10.1093/jxb/eru429. [DOI] [PubMed] [Google Scholar]
- 68.Bortesi L., Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015;33:41–52. doi: 10.1016/j.biotechadv.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 69.Hsu P.D. Development and applications of CRISPR–Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ran F.A. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mali P. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.O’Neill J. HM Government and the Welcome Trust; 2014. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. [Google Scholar]
- 73.Nagel T.E. The Developing world urgently needs phages to combat pathogenic bacteria. Front. Microbiol. 2016;7:882. doi: 10.3389/fmicb.2016.00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eeckhout D. Isolation and characterization of recombinant antibody fragments against CDC2a from Arabidopsis thaliana. Eur. J. Biochem. 2000;267:6775–6783. doi: 10.1046/j.1432-1033.2000.01770.x. [DOI] [PubMed] [Google Scholar]
- 75.Kaur R., Vrati S. Development of a recombinant vaccine against Japanese encephalitis. J. Neurovirol. 2003;9:421–431. doi: 10.1080/13550280390218454. [DOI] [PubMed] [Google Scholar]
- 76.Mulder K.C. Production of a modified peptide clavanin in Pichia pastoris: cloning, expression, purification and in vitro activities. AMB Express. 2015;5:1–8. doi: 10.1186/s13568-015-0129-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.López-Abarrategui C. Cm-p5: an antifungal hydrophilic peptide derived from the coastal mollusk Cenchritis muricatus (Gastropoda: Littorinidae) FASEB J. 2015;29:3315–3325. doi: 10.1096/fj.14-269860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saravanan R. Design of short membrane selective antimicrobial peptides containing tryptophan and arginine residues for improved activity, salt-resistance, and biocompatibility. Biotechnol. Bioeng. 2014;111:37–49. doi: 10.1002/bit.25003. [DOI] [PubMed] [Google Scholar]
- 79.Streatfield S.J. Corn as a production system for human and animal vaccines. Vaccine. 2003;21:812–815. doi: 10.1016/s0264-410x(02)00605-9. [DOI] [PubMed] [Google Scholar]
- 80.Sojikul P. A plant signal peptide-hepatitis B surface antigen fusion protein with enhanced stability and immunogenicity expressed in plant cells. Proc. Natl. Acad. Sci. U. S. A. 2003;100:2209–2214. doi: 10.1073/pnas.0438037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vaquero C. Transient expression of a tumor-specific single-chain fragment and a chimeric antibody in tobacco leaves. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11128–11133. doi: 10.1073/pnas.96.20.11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takagi H. Biochemical safety evaluation of transgenic rice seeds expressing T cell epitopes of Japanese cedar pollen allergens. J. Agric. Food Chem. 2006;54:9901–9905. doi: 10.1021/jf061848v. [DOI] [PubMed] [Google Scholar]
- 83.Deslouches B. De novo generation of cationic antimicrobial peptides: influence of length and tryptophan substitution on antimicrobial activity. Antimicrob. Agents Chemother. 2005;49:316–322. doi: 10.1128/AAC.49.1.316-322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sedor J. Cathepsin-G interferes with clearance of Pseudomonas aeruginosa from mouse lungs. Pediatr. Res. 2007;61:26–31. doi: 10.1203/01.pdr.0000250043.90468.c2. [DOI] [PubMed] [Google Scholar]
- 85.Yazici H. Biological response on a titanium implant-grade surface functionalized with modular peptides. Acta Biomater. 2013;9:5341–5352. doi: 10.1016/j.actbio.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paranjape S.M. Modulation of proinflammatory activity by the engineered cationic antimicrobial peptide WLBU-2. F1000 Res. 2013;2:36. doi: 10.12688/f1000research.2-36.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Giles F.J. A phase III, randomized, double-blind, placebo-controlled, study of iseganan for the reduction of stomatitis in patients receiving stomatotoxic chemotherapy. Leuk. Res. 2004;28:559–565. doi: 10.1016/j.leukres.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 88.Tan H.H. Topical antibacterial treatments for acne vulgaris: comparative review and guide to selection. Am. J. Clin. Dermatol. 2004;5:79–84. doi: 10.2165/00128071-200405020-00002. [DOI] [PubMed] [Google Scholar]
- 89.Sader H.S. Omiganan pentahydrochloride (MBI 226), a topical 12-amino-acid cationic peptide: spectrum of antimicrobial activity and measurements of bactericidal activity. Antimicrob. Agents Chemother. 2004;48:3112–3118. doi: 10.1128/AAC.48.8.3112-3118.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kavanagh K., Dowd S. Histatins: antimicrobial peptides with therapeutic potential. J. Pharm. Pharmacol. 2004;56:285–289. doi: 10.1211/0022357022971. [DOI] [PubMed] [Google Scholar]
- 91.Moore A. The big and small of drug discovery. EMBO Rep. 2003;4:114–117. doi: 10.1038/sj.embor.embor748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Finlay B.B., Hancock R.E.W. Can innate immunity be enhanced to treat microbial infections? Nat. Rev. Microbiol. 2004;2:497–504. doi: 10.1038/nrmicro908. [DOI] [PubMed] [Google Scholar]
- 93.Lienard D. Suspension-cultured BY-2 tobacco cells produce and mature immunologically active house dust mite allergens. Plant Biotechnol. J. 2007;5:93–108. doi: 10.1111/j.1467-7652.2006.00221.x. [DOI] [PubMed] [Google Scholar]
- 94.Del Prete G. Plant-derived recombinant F1, V, and F1-V fusion antigens of Yersinia pestis activate human cells of the innate and adaptive immune system. Int. J. Immunopathol. Pharmacol. 2009;22:133–143. doi: 10.1177/039463200902200115. [DOI] [PubMed] [Google Scholar]
- 95.Thanavala Y. Immunogenicity of transgenic plant-derived hepatitis B surface antigen. Proc. Natl. Acad. Sci. U. S. A. 1995;92:3358–3361. doi: 10.1073/pnas.92.8.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang G.G. Production of HIV-1 p24 protein in transgenic tobacco plants. Mol. Biotechnol. 2002;20:131–136. doi: 10.1385/MB:20:2:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gu Q. Cloning of Helicobacter pylori urease subunit B gene and its expression in tobacco (Nicotiana tabacum L.) Plant Cell Rep. 2005;24:532–539. doi: 10.1007/s00299-005-0962-8. [DOI] [PubMed] [Google Scholar]
- 98.Lenzi P. Translational fusion of chloroplast-expressed human papillomavirus type 16 L1 capsid protein enhances antigen accumulation in transplastomic tobacco. Transgenic Res. 2008;17:1091–1102. doi: 10.1007/s11248-008-9186-3. [DOI] [PubMed] [Google Scholar]
- 99.Koya V. Plant-based vaccine: mice immunized with chloroplast-derived anthrax protective antigen survive anthrax lethal toxin challenge. Infect. Immun. 2005;73:8266–8274. doi: 10.1128/IAI.73.12.8266-8274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pogrebnyak N. Severe acute respiratory syndrome (SARS) S protein production in plants: development of recombinant vaccine. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9062–9067. doi: 10.1073/pnas.0503760102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tregoning J.S. Protection against tetanus toxin using a plant-based vaccine. Eur. J. Immunol. 2005;35:1320–1326. doi: 10.1002/eji.200425453. [DOI] [PubMed] [Google Scholar]
- 102.Ma S. Induction of oral tolerance to prevent diabetes with transgenic plants requires glutamic acid decarboxylase (GAD) and IL-4. Proc. Natl. Acad. Sci. U. S. A. 2004;101:5680–5685. doi: 10.1073/pnas.0307420101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee M.Y.T. Expression of viral capsid protein antigen against Epstein–Barr virus in plastids of Nicotiana tabacum cv. SR1. Biotechnol. Bioeng. 2006;94:1129–1137. doi: 10.1002/bit.20948. [DOI] [PubMed] [Google Scholar]
- 104.Wu L. Expression of foot-and-mouth disease virus epitopes in tobacco by a tobacco mosaic virus-based vector. Vaccine. 2003;21:4390–4398. doi: 10.1016/s0264-410x(03)00428-6. [DOI] [PubMed] [Google Scholar]