Abstract
The infectious bronchitis virus (IBV) is continuously evolving through point mutation and recombination of their genome, subsequently the emergence of IBV variants complicates disease control. The objective of this study was to investigate genetic characterization of new IBV variants isolated from commercial chicken flocks in Korea collected between 2005 and 2010. Phylogenetic analysis revealed that all new IBV isolates belonged to Korean group II (K-II), which included the nephropathogenic IBV strains. However, the isolates formed a new gene cluster that was distinguished from the two distinct K-II subgroups (KM91-like and QX-like). Recombination events were identified in the S1 gene, with their putative parental strains being the KM91-like or QX-like subgroup. In addition, two crossover sites were observed in the S1 gene of IBV isolates. These results suggest that natural genetic recombination between heterologous strains classified into different genetic groups has occurred and may have caused the emergence of new IBV strains. This finding provides important information on IBV evolution and is essential for the effective control of IB in Korea.
Keywords: Infectious bronchitis virus, Spike glycoprotein, Sequence analysis, Recombination analysis
1. Introduction
Infectious bronchitis virus (IBV) causes an acute and highly contagious viral disease of chickens that is characterized by respiratory signs, nephritis, and reduced egg reproduction (Cavanagh and Naqi, 2003). Among the IBV structural proteins, the spike glycoprotein S1 subunit is responsible for attachment to the host cellular membrane, induction of neutralization, and hemagglutination of inhibiting antibodies (Casais et al., 2003, Cavanagh, 2007). Because of the high rates of S1 gene mutation, a number of IBV serotypes or antigenic variant strains have been reported in many countries (Cavanagh, 2007). Moreover, another evolutionary process involving recombination between co-circulating heterotypic IBV strains has increased the genetic diversity of IBV viruses (Lee and Jackwood, 2000). However, not all recombinant strains of IBV cause epidemics. While most recombinant strains have disappeared due to the unfitness for the host environment, some strains, such as GA98 strain derived from DE072 (Lee et al., 2001), have continuously persisted in the field long enough to maintain an epidemic (Lee and Jackwood, 2001).
In Korea, IB was first described in 1986 (Rhee et al., 1986), and Massachusetts (Mass) type live IB attenuated vaccine as well as inactivated killed oil-emulsion vaccine have been applied to prevent and control the incidence of the disease. Despite extensive vaccination, IBV infection has remained a problem in the Korean poultry industry (Lee et al., 2004). Since the early 1990s, Korean nephropathogenic IBV, represented by the KM91 strain, has exacted economic losses in Korea (Kim et al., 2004). New nephropathogenic IBVs related to the QXIBV strain originating from China have been introduced and become wide-spread since 2000 (Lee et al., 2008). More recently, we isolated new IBV strains that formed a new gene cluster in the phylogenetic tree of the IBV S1 gene. These new strains have been continuously isolated in Korea over time and outbreaks have been increasing annually. Nevertheless, the genetic features of these IBV strains are still unknown.
The present study conducted a phylogenetic analysis and recombinant identification on the S1 gene to investigate the genetic characteristics of new emerging Korean IBV isolates. The results reveal that these isolates are genetically close to nephropathogenic strains. It is further demonstrated that the newly emerged IBVs have been generated by natural recombination between circulating KM91-like and QX-like strains.
2. Materials and methods
2.1. Viruses
Ten IBV field isolates from commercial chickens in Korea between 2005 and 2010 were used (Table 1 ). All isolates were propagated in 10-day-old specific-pathogen free embryonated chicken eggs (Hy-Vac.com) at 37 °C for 48 h. Allantoic fluid from eggs infected with each isolate was harvested and frozen at −70 °C until use.
Table 1.
Field isolates of IBV used in this study.
| IBV isolate | Type of birds | Age | Province | Year of isolation | Lesionsa | Tissuesb |
|---|---|---|---|---|---|---|
| K716 | Broiler | 26 days | Gyeongi | 2005 | RS | Tra, Kid, C.T |
| K1585 | Broiler breeder | 280 days | Gyeongi | 2007 | RS, ED | C.T |
| K183 | Korean native chicken | 230 days | Gyeongbuk | 2007 | RS, ED | C.T |
| K426 | Broiler | 42 days | Gyeongbuk | 2008 | NS | Tra |
| K33 | Broiler | 21 days | Gyeongi | 2009 | RS, NS | Kid |
| K40 | Broiler | 25 days | Chungbuk | 2009 | RS, NS | Tra, Kid, C.T |
| K74 | Broiler | 21 days | Gyeongi | 2009 | RS, NS | Kid |
| K344 | Broiler | 27 days | Chungbuk | 2009 | RS | Tra, C.T |
| K88 | Broiler | 15 days | Chungnam | 2010 | RS, NS | Tra, Kid, C.T |
| K147 | Broiler | 28 days | Chonnam | 2010 | RS, NS | Tra, Kid, C.T |
Lesions: RS = respiratory signs, ED = egg drop, NS = nephritis.
Tissues used for IBV isolation: Tra = trachea, Kid = kidney, C.T = caecal tonsil.
2.2. Viral RNA extraction, reverse transcription-polymercase chain reaction (RT-PCR) and DNA sequencing
Viral RNA was extracted from virus-infected allantoic fluid using an RNeasy minikit (Qiagen) following the manufacturer's instructions. The primer sets (forward: 5′-TAG TGA CCC TTT TGT GTG CAC TAT-3′ and reverse: 5′-GTT TGT ATG TAC TCA TCT GTA AC-3′) were designed for the amplification of the IBV S1 gene. RT was done using Moloney murine leukemia virus reverse transcriptase (Invitrogen) and the reverse primer. For PCR, EX-Taq polymerase (Takara Bio) and forward primer were added to the RT reaction. PCR was performed by 40 cycles of denaturation (94 °C, 10 s), annealing (53 °C, 90 s), and polymerization (72 °C, 90 s). The pre-denaturation step was at 94 °C for 20 s and post-polymerization step was at 72 °C for 5 min. The amplified sequences were analyzed by 1.5% agarose gel electrophoresis.
The amplified DNA products were purified using a gel extraction kit (Elpis Biotech) and the purified products were ligated into a pGEM-T Vector (Promega). Nucleotide sequencing was performed with BigDye Terminator v3.1 Cycle Sequencing Kit and products were analyzed on the ABI PRISM 3730xl genetic analyzer (Applied Biosystems). The nucleotide sequences for the S1 genes have been submitted to GenBank under accession numbers HM486953–HM486962.
2.3. Phylogenetic and sequence analysis of S1 gene
The nucleotide sequences of the S1 gene of the IBV isolates were compiled and the amino acid sequences were deduced using Bioedit version 7.0.9.0 (Hall, 1999). The Korean IBV reference strains, which are in three genetic groups (Korean group I, Korean group II and Mass group), were retrieved from the GenBank database (Table 2 ). Sequence analysis of partial S1 gene nucleotide positions 25–1611 (numbering is in reference to the Mass41 strain) were performed with the Clustal W multiple alignment method. The phylogenetic tree was constructed with the neighbor-joining method with 1000 bootstrap replicates using MEGA version 4 (Tamura et al., 2007).
Table 2.
IBV reference strains included in this study.
| Strain | Country | Accession numbers |
|---|---|---|
| K283 | Korea | FJ807923 |
| K463/04 | Korea | FJ807924 |
| K1019/03 | Korea | FJ807927 |
| K1255/03 | Korea | FJ807928 |
| K1583/04 | Korea | FJ807931 |
| LS2 | China | AY278246 |
| QXIBV | China | AF193423 |
| LX4 | China | AY189157 |
| K961/04 | Korea | FJ807926 |
| K507-01 | Korea | AY257064 |
| KM91 | Korea | FJ807946 |
| K774-01 | Korea | AY257065 |
| K748-01 | Korea | AY790358 |
| K151/98 | Korea | FJ807937 |
| K069-01 | Korea | AY257061 |
| K446-01 | Korea | AY257063 |
| M41 | USA | X04722 |
| K110/06 | Korea | FJ807921 |
| H120 | Netherlands | EU822341 |
| K203-02 | Korea | AY257067 |
| K210-02 | Korea | AY257068 |
| B4 | Korea | FJ807932 |
| K348/99 | Korea | FJ807940 |
| K571/99 | Korea | FJ807942 |
| K620/02 | Korea | FJ807944 |
| RB86 | Korea | FJ807947 |
| KC90 | Korea | FJ807945 |
| K576/99 | Korea | FJ807943 |
| K451/99 | Korea | FJ807941 |
| K242/99 | Korea | FJ807939 |
| K152/98 | Korea | FJ807938 |
| K1277/03 | Korea | FJ807930 |
| K1257/03 | Korea | FJ807929 |
| K154/05 | Korea | FJ807922 |
| K035/04 | Korea | FJ807920 |
2.4. Analysis of recombination
Putative recombinant sequences were identified with the soft package of SimPlot (Lole et al., 1999). The consecutive IBV nucleotide sequences from the partial S1 gene based on the multiple alignment results were introduced into similarity plots with SimPlot version 3.5.1. The nucleotide identity was calculated using the Kimura 2-parameter method with a transition–transversion ratio of 2 in each window of 200 bp. The window was successively furthered along the alignment using a 20-bp increment. Furthermore, the phylogenetic trees based on the different fragments of S1 gene were constructed as an evidence for recombination (Worobey et al., 2002).
3. Results
3.1. Phylogenetic analysis
A phylogenetic tree was generated to describe the relationship between nucleotide sequences of the S1 gene of IBV isolates and other field strains in Korea. The Korean IBV strains were divided into two distinct genetic groups: Korean group I (K-I) and Korean group II (K-II). K-I included respiratory strains that split into a unique genetic group, while all nephropathogenic strains were included in K-II. In addition, K-II was divided into two separate subgroups: KM91-like and QX-like. A phylogenetic tree constructed from the nucleotide sequences of the S1 gene showed that all the new IBV strains isolated in this study belonged to K-II (Fig. 1 ). However, the IBV isolates formed a new gene cluster that was distinguished from two distinct subgroups of K-II (KM91-like and QX-like).
Fig. 1.
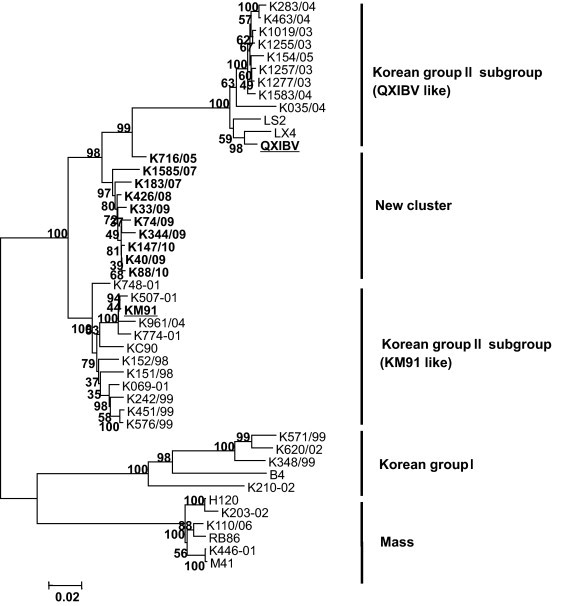
IBV phylogenetic tree of S1 genes. The tree is based on the sequence of the S1 genes from the 10 IBV isolates and the 35 reference strains, and ClustalW alignment method for S1 nucleotide positions 25–1611 corresponding to those of strain Mass41 (GenBank accession number X04722). The IBV isolates are denoted in bold and two reference strains (KM91 and QXIBV) are underlined. The provisional designations, including genogroups and sub-genogroups, are indicated on the right.
3.2. Sequence comparisons
Sequencing results showed that partial S1 genes of the IBV isolates contained mutations, insertions and/or deletions, resulting in different lengths of nucleotides and, consequently, different numbers of amino acids being encoded. The S1 gene of nine IBV isolates (K1585/07, K183/07, K426/08, K33/09, K40/09, K74/09, K344/09, K88/10 and K147/10) had 532 amino acids and one isolate (K716/05) had 530 amino acids, which were encoded by 1596 and 1590 nucleotides, respectively. In addition, the analysis and comparison of nucleotide and deduced amino acid sequences of the 10 IBV isolates with the KM91 and QXIBV strain were performed. The nucleotide and amino acid identities of S1 gene sequences among the 10 IBV isolates ranged from 93.7% to 99.4%. They shared 90.2–91.7% amino acid similarity to the S1 gene of the KM91 strain and 88.7–90.7% similarity to that of the QXIBV strain.
In addition, the possible recombination regions of the S1 genes were identified with the Simplot program (version 3.5.1.) and the individual amino acid sequences were compared (Fig. 2 ). One potential crossover region (amino acids 288–306) and two different amino acid characteristic regions similar to QXIBV and KM91 were detected on the S1 gene of nine IBV isolates (K1585/07, K183/07, K426/08, K33/09, K40/09, K74/09, K344/09, K88/10 and K147/10). The former part of S1 gene sequence showed high similarity with QX-IBV (95.4–97.5%) and the later part of that with KM91 (93.8–95.5%). The K716/05 also had one potential crossover regions (amino acids 346–359) and two different amino acid characteristic regions similar with QXIBV and KM91. The former part of S1 gene sequence showed high similarity with QX-IBV (96.8%) and the later part of that with KM91 (94.1%).
Fig. 2.
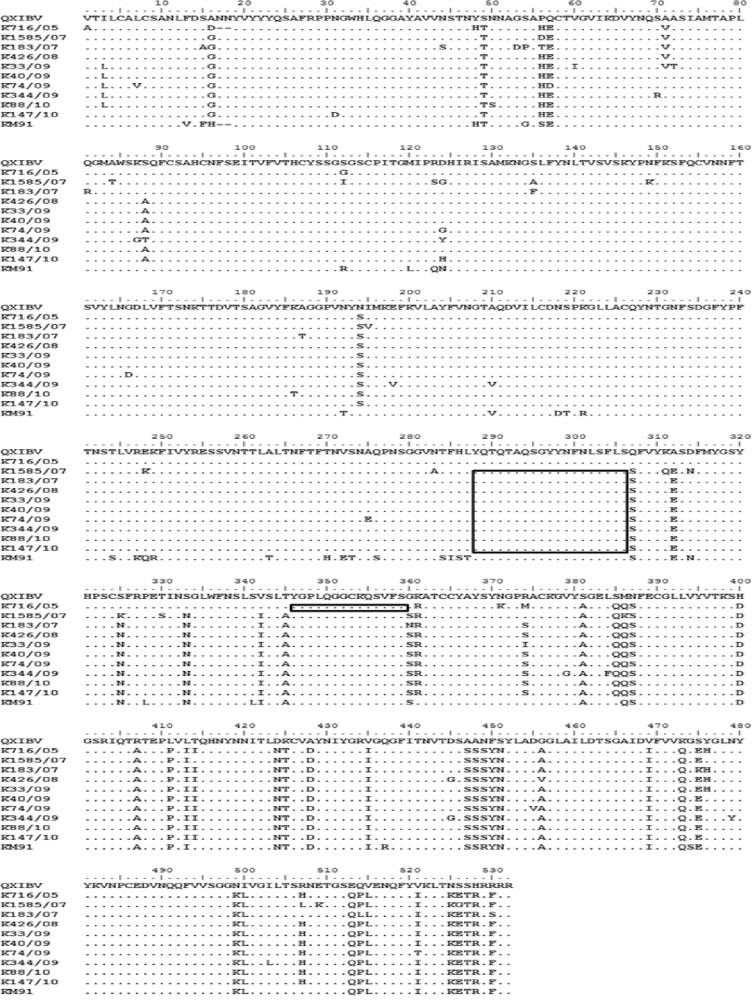
Amino acid alignment of the S1 genes of KM91, QXIBV and 10 IBV isolates. The amino acid sequence of the S1 gene of QXIBV is shown at the top of the panel and that of KM91 is at the bottom. Gaps (dashes) were introduced in order to align the sequences and the dots indicate the residues identical to those of QXIBV. Potential two crossover regions are boxed.
3.3. Identification of putative recombinant sequence
The KM91 and QXIBV strains were used as putative parental strains and the 10 IBV isolates were queried in the Simplot analysis. The strains were considered as recombinants if any crossover event took place between two putative parental strains. The recombination breakpoints were determined by the maximization of χ 2 analysis (Lole et al., 1999). The parental strain KM91 was replaced with QXIBV when the 10 IBV isolates were queried. The S1 gene sequences of nine IBV isolates (K1585/07, K183/07, K426/08, K33/09, K40/09, K74/09, K344/09, K88/10 and K147/10) exhibited greater affinity with one putative parent QXIBV strain before the break point (861 bp), while the S1 gene shared higher sequence similarity with the KM91 strain after position 861 bp (Fig. 3A). The S1 gene sequence of the K716/05 strain exhibited greater affinity with one putative parent QXIBV strain in the region from 1 to 1118, while the S1 gene shared higher sequence similarity with the KM91 strain after position 1118 bp (Fig. 4A). Furthermore, when phylogenetic trees inferred for nucleotide sequences of each recombination region in the S1 gene of IBV isolates were constructed, a significant discrepancy was found between these phylogenetic trees (Fig. 3, Fig. 4). These findings also provide important evidence that IBV isolates descended from two putative parents, the KM91-like and QXIBV-like subgroups.
Fig. 3.
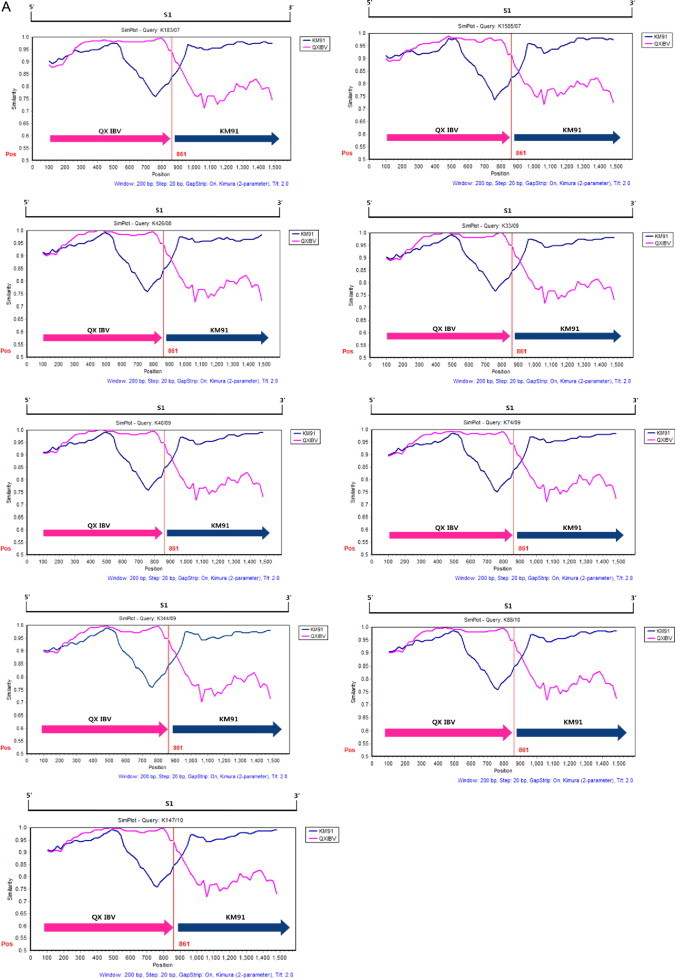
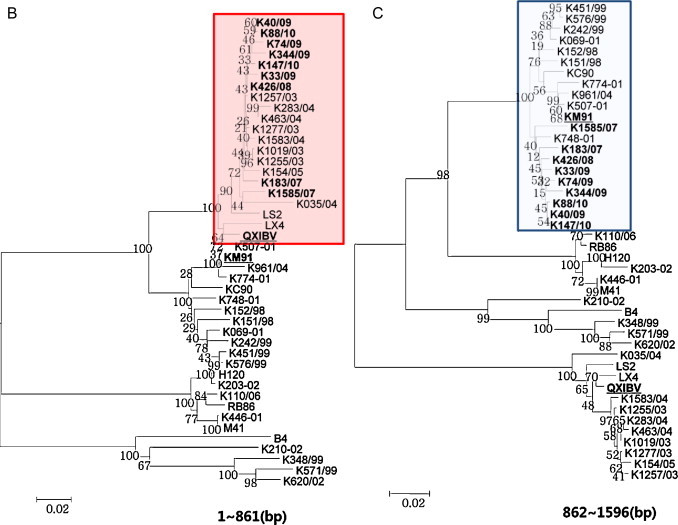
Evidence for recombination in the S1 gene of nine IBV isolates. (A) The S1 gene similarity from Simplot analysis of nine IBV isolates (K1585/07, K183/07, K426/08, K33/09, K40/09, K74/09, K344/09, K88/10 and K147/10) and its putative parents (QXIBV (Pink) and KM91 (Blue)). The query sequence is indicated on the upper part of the figure. The y-axis gives the percentage of identity within a sliding window 200 bp wide centered on the position plotted, with a step size between plots of 20 bp. The S1 gene scale was given at the top of the plot. The red vertical line shows the recombination point at position 861. (B) The maximum-likelihood trees of the S1 gene segment in regions 1–861. The nine IBV isolates were included in the QXIBV cluster (Pink). (C) The maximum-likelihood trees of the S1 gene segment in regions 861–1594. The eight IBV isolates belong to the KM91 cluster (Blue). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Fig. 4.
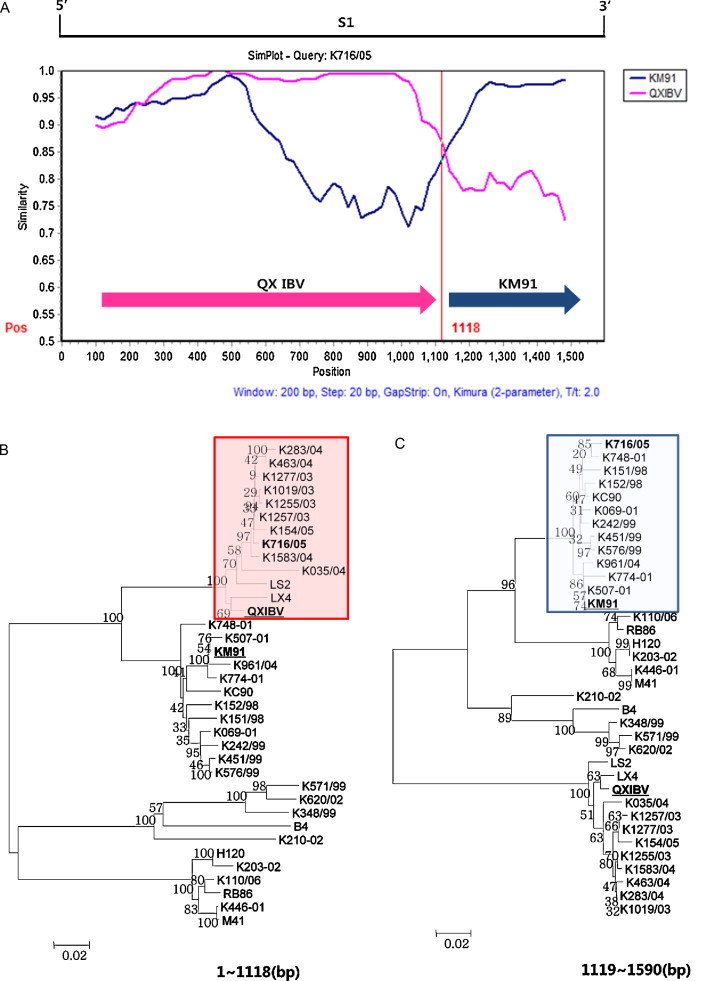
Evidence for recombination in the S1 gene of the K716/05 strain. (A) Results from Simplot analysis of K716/05 strain are shown. The analysis was carried out using K716/05 strain as a query. QXIBV (Pink) and KM91 (Blue) were used as putative parental strains. The y-axis gives the percentage of identity and the S1 gene scale was given at the top of the plot. The red vertical lines show the recombination point at position 1118. (B) The phylogenetic tree of the region from position 1–1118. The IBV isolate K716/05 was included in the QXIBV cluster (Pink). (C) The phylogenetic trees of the region from position 1118–1590. The K716/05 strain belonged to the KM91 cluster (Blue). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
4. Discussion
Recombination is one of the important IBV evolution mechanisms in the generation of new variant viruses in the field (Lee and Jackwood, 2000). In particular, the use of live vaccines could produce new variant viruses by recombination with field strains resulting from the spread of vaccine strains (Farsang et al., 2002). In this study, the result of recombination analysis provided the evidence that homologous recombination had occurred in the IBV isolates (Fig. 3, Fig. 4). However, the Mass type strain has only been used in Korea as a live vaccine before the use of a nephropathogenic strain of live-attenuated IBV vaccine was begun in 2010 (Lee et al., 2010). Furthermore, the IBV isolates in this study were obtained prior to the use of nephropathogenic strain of live-attenuated IBV vaccine, and they were almost always isolated from unvaccinated broiler chickens. Therefore, the new IBV isolates might have been generated naturally by recombination resulting from co-circulation of KM-91 like and QX-like field strains. To our knowledge, this is the first report of natural recombination among nephropathogenic IBV strains in Korea.
Previous studies of IBV have demonstrated that putative crossover sites are detected at a relatively conserved sequence near the hypervariable region of the S1 gene (Song and Lee, 2000, Wang et al., 1994). However, presently, the two crossover sites were located at a conserved sequence within the hypervariable region. Especially, most of the IBV isolates experienced crossover events at nucleotide 861 of the S1 gene (Fig. 3), indicating that this site may serve as a possible region for template switching in viral RNA synthesis and is a breakpoint candidate for genetic recombination within the S1 gene of Korean nephropathogenic IBV strains. In addition, considering the fact that genetic recombination of IBV can occur in multiple genes (Chen et al., 2009, Kuo et al., 2010, Mase et al., 2009), further studies involving recombination events in several genes might provide more information about the genetic characterization of IBV isolates.
Although the generation of many recombinant strains can occur naturally (Jia et al., 1995, Wang et al., 1993), each recombinant strain displays different levels of fitness for a host environment (Domingo et al., 1985). For instance, the EJ95 recombinant strain disappeared from Korea due to unfitness for the host environment (Song and Lee, 2000), while the GA98 strain has continuously persisted in the field long enough to maintain an epidemic in North America (Lee, 2002). Unpublished results from an epidemiological investigation have shown that the IBV isolates used in this study were isolated continuously in different regions of Korea from 2005 to 2010, and that outbreaks of these isolates have been increasing annually. These observations support the possibility that IBV recombinant strains have the potential to spread widely and cause epidemics in chickens. In addition, since the S protein of coronavirus is related to host range and cell tropism (Casais et al., 2003, Kuo et al., 2000), alterations in the pathogenic and antigenic characteristic in IBV isolates could be expected. How recombinant strains from heterologous strains demonstrate tissue tropism or pathogenicity in a host is not clear, but it has been reported that vaccination with commercial vaccines does not ensure complete protection against challenge with recombinant strains (Liu et al., 2006, Liu et al., 2009). Therefore, further studies should investigate the cross-protective efficacy of commercial IBV vaccines against these isolates for effective control of IB in Korea.
In conclusion, the present study indicates that new emerging IBV isolates might have originated from natural recombination between co-circulating Korean nephropathogenic IBVs. Moreover, the observations support the possibility that these isolates might increase the genetic diversity of IBVs in the field and could hamper disease control in Korea. Therefore, more intensive investigations on biological and antigenic characteristics of the recombinant IBV isolates and detailed assessments of the efficacy of the current vaccines against these isolates are needed.
Acknowledgments
This work was supported by grant no. 610001-03-1-SU000 from Technology Development Program for Agriculture and Forestry, Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea. We thank Hyo-Sun Joo and Byoung-Yoon Kim for their technical assistance, Sang-Won Lee for excellent technical support.
References
- Casais R., Dove B., Cavanagh D., Britton P. Recombinant avian infectious bronchitis virus expressing a heterologous spike gene demonstrates that the spike protein is a determinant of cell tropism. J. Virol. 2003;77:9084–9089. doi: 10.1128/JVI.77.16.9084-9089.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Naqi S. Diseases of Poultry. 11th ed. Iowa State University Press; Ames, IA: 2003. Infectious bronchitis; pp. 101–109. [Google Scholar]
- Chen H.W., Huang Y.P., Wang C.H. Identification of Taiwan and China-like recombinant avian infectious bronchitis viruses in Taiwan. Virus Res. 2009;140:121–129. doi: 10.1016/j.virusres.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E., Martinez-Salas E., Sobrino F., de la Torre J.C., Portela A., Ortin J., Lopez-Galindez C., Perez-Brena P., Villanueva N., Najera R., Vandepol S., Steinhauer D., Depolo N., Holland J.J. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance – a review. Gene. 1985;40:1–8. doi: 10.1016/0378-1119(85)90017-4. [DOI] [PubMed] [Google Scholar]
- Farsang A., Ros C., Renstrom L.H., Baule C., Soos T., Belak S. Molecular epizootiology of infectious bronchitis virus in Sweden indicating the involvement of a vaccine strain. Avian Pathol. 2002;31:229–236. doi: 10.1080/03079450220136530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Comput. Methods Programs Biomed. 1999;41:95–98. [Google Scholar]
- Jia W., Karaca K., Parrish C.R., Naqi S.A. A novel variant of avian infectious bronchitis virus resulting from recombination among three different strains. Arch. Virol. 1995;140:259–271. doi: 10.1007/BF01309861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Song C.S., Mo I.P., Kim S.H., Sung H.W., Yoon H.S. An outbreak of nephropathogenic infectious bronchitis in commercial pullets. Res. Rep. Rural Dev. Admin. 2004;34:28–31. [Google Scholar]
- Kuo L., Godeke G.J., Raamsman M.J., Masters P.S., Rottier P.J. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 2000;74:1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo S.M., Wang C.H., Hou M.H., Huang Y.P., Kao H.W., Su H.L. Evolution of infectious bronchitis virus in Taiwan: characterisation of RNA recombination in the nucleocapsid gene. Vet. Microbiol. 2010;144:293–302. doi: 10.1016/j.vetmic.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.W. Evolution of avian infectious bronchitis virus: genetic drift and recombination. Korean J. Vet. Serv. 2002;25:97–103. [Google Scholar]
- Lee C.W., Hilt D.A., Jackwood M.W. Identification and analysis of the Georgia 98 serotype, a new serotype of infectious bronchitis virus. Avian Dis. 2001;45:164–172. [PubMed] [Google Scholar]
- Lee C.W., Jackwood M.W. Evidence of genetic diversity generated by recombination among avian coronavirus IBV. Arch. Virol. 2000;145:2135–2148. doi: 10.1007/s007050070044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.W., Jackwood M.W. Origin and evolution of Georgia 98 (GA98), a new serotype of avian infectious bronchitis virus. Virus Res. 2001;80:33–39. doi: 10.1016/s0168-1702(01)00345-8. [DOI] [PubMed] [Google Scholar]
- Lee E.K., Jeon W.J., Lee Y.J., Jeong O.M., Choi J.G., Kwon J.H., Choi K.S. Genetic diversity of avian infectious bronchitis virus isolates in Korea between 2003 and 2006. Avian Dis. 2008;52:332–337. doi: 10.1637/8117-092707-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Lee H.J., Youn H.N., Kwon J.S., Lee Y.J., Kim J.H., Lee J.B., Park S.Y., Choi I.S., Song C.S. Characterization of a novel live attenuated infectious bronchitis virus vaccine candidate derived from a Korean nephropathogenic strain. Vaccine. 2010;28:2887–2894. doi: 10.1016/j.vaccine.2010.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.K., Sung H.W., Kwon H.M. S1 glycoprotein gene analysis of infectious bronchitis viruses isolated in Korea. Arch. Virol. 2004;149:481–494. doi: 10.1007/s00705-003-0225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Chen J., Han Z., Zhang Q., Shao Y., Kong X., Tong G. Infectious bronchitis virus: S1 gene characteristics of vaccines used in China and efficacy of vaccination against heterologous strains from China. Avian Pathol. 2006;35:394–399. doi: 10.1080/03079450600920984. [DOI] [PubMed] [Google Scholar]
- Liu S., Zhang X., Wang Y., Li C., Liu Q., Han Z., Zhang Q., Kong X., Tong G. Evaluation of the protection conferred by commercial vaccines and attenuated heterologous isolates in China against the CK/CH/LDL/97I strain of infectious bronchitis coronavirus. Vet. J. 2009;179:130–136. doi: 10.1016/j.tvjl.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mase M., Inoue T., Yamaguchi S., Imada T. Genetic diversity of avian infectious bronchitis viruses in Japan based on analysis of s2 glycoprotein gene. J. Vet. Med. Sci. 2009;71:287–291. doi: 10.1292/jvms.71.287. [DOI] [PubMed] [Google Scholar]
- Rhee Y.O., Kim J.H., Mo I.P., Yoon H.S., Choi S.H., Namgoong S. Outbreaks of infectious bronchitis in Korea. J. Perinat. Med. 1986;26:277–282. [Google Scholar]
- Song C.S., Lee Y.J. Molecular and epidemiological characteristics of infectious bronchitis virus isolated in Korea. Korean J. Poultry Sci. 2000;2:91–98. [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Wang L., Junker D., Collison E.W. A novel variant of avian infectious bronchitis virus resulting from recombination among three different strains. Virology. 1993;192:710–716. [Google Scholar]
- Wang L., Junker D., Hock L., Ebiary E., Collisson E.W. Evolutionary implications of genetic variations in the S1 gene of infectious bronchitis virus. Virus Res. 1994;34:327–338. doi: 10.1016/0168-1702(94)90132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M., Rambaut A., Pybus O.G., Robertson D.L. Questioning the evidence for genetic recombination in the 1918 “Spanish flu” virus. Science. 2002 doi: 10.1126/science.296.5566.211a. (New York, NY 296, 211 discussion 211) [DOI] [PubMed] [Google Scholar]


