Abstract
The majority of research topics declared that most of the recombinant proteins have been expressed by Escherichia coli in basic investigations. But the majority of high expressed proteins formed as inactive recombinant proteins that are called inclusion body. To overcome this problem, several methods have been used including suitable promoter, environmental factors, ladder tag to secretion of proteins into the periplasm, gene protein optimization, chemical chaperones and molecular chaperones sets. Co-expression of the interest protein with molecular chaperones is one of the common methods The chaperones are a group of proteins, which are involved in making correct folding of recombinant proteins. Chaperones are divided two groups including; cytoplasmic and periplasmic chaperones. Moreover, periplasmic chaperones and proteases can be manipulated to increase the yields of secreted proteins. In this article, we attempted to review cytoplasmic chaperones such as Hsp families and periplasmic chaperones including; generic chaperones, specialized chaperones, PPIases, and proteins involved in disulfide bond formation.
Keywords: Molecular chaperone, Inclusion body, Heat shock protein, Peptidyl-prolyl isomerase, Disulfide bond formation
1. Introduction
Stanley Cohen and Herbert Boyer were applied cloning and expression methods to produce recombinant proteins in different organisms [1]. At the beginning of the recombinant protein expression systems, Escherichia coli (E. coli) is a remarkable hosts due to its rapid growth rate, requirement for inexpensive carbon sources, low costs and well-characterized genetic structure [2], [3]. However, E. coli has some defects such as inability for posttranslational modifications, ineffective cleavage of the amino terminal methionine, inability to produce proteins containing complex disulphide bonds, and expression of proteins as insoluble inclusion bodies (IBs) (IBs) [4], [5].
IBs, are inactive form of proteins, which are formed in both prokaryotic and eukaryotic cells that have been miss folded and are not in proper intra-molecular interaction [6]. The natural IBs in some bacteria is like β–lactamase which is a secretion protein [7]. The reasons of IBs formation in protein expression are amount and level of their expression that high level expression of recombinant protein led to IBs formation [8]. The microscopic analysis showed that the size and shape of IBs were different in cytoplasmic and periplasmic spaces [8]. IBs have been cylindrical or ovoid forms [9] and diameter of the shapes changed from 0.5 to 1.3 μm [10]. Moreover, IBs are highly hydrated and have spongy structures that can be simply separated by high speed centrifugation [11], [12]. The studies showed that IBs have dynamic, reversible, and kinetic structure that is consequent of unbalance between soluble and insoluble proteins [9], [13]. Also, shifting of IB proteins in to active forms in in vitro is really hard and need to several approaches containing isolation, solubilisation, and refolding of IBs, which has low efficiency [12], [14]. The solubilisation of IBs are mostly dependent on denaturant agent such as urea, HCL, and guanidine hydrochloride which stimulate disruption of intramolecular interactions [13]. In production of recombinant proteins, several procedures have been used for soluble expression and preventing of IB formation such as using molecular chaperones, low temperature, suitable promoter, ladder tag to secretion of proteins into the periplasm, gene protein optimization and chemical chaperones [5], [15]. Cells normally have molecular chaperones and other factors to correct folding of proteins [16]. Numerous studies demonstrate positive effects of molecular chaperons on correct folding formation of recombinant protein and prevention of IBs formation in the cytoplasm and periplasm [17], [18]. Also molecular chaperons have clinical and experimental application such as immunomodulatory [19], treating neurodegenerative diseases [20], and morbidity of chronic illnesses [20], [21]. In this review we focused on cytoplasmic and periplasmic chaperones category and their applications in recombinant protein expression with correct folding.
2. Molecular chaperones
Molecular chaperones are proteins existing in bacteria and eukaryotic cells which have an important roles to assist cellular homeostasis under normal and detrimental growth situation and cell defences to inhibit aggregation of mis-folded proteins [22], [23]. Various genetic sources have molecular chaperones because these agents have critical roles in cell such as prevention of accumulation and mis-folding protein, and also, help to preparing correct folding of protein [24], [25]. Chaperones do not have steric information of target protein structure to determine exact folding. So, they prevent improper interactions within and between other polypeptides by enhancing the yield of correct folded proteins. However, they don’t increase the amount of folding reactions. Unfolded proteins have hydrophobic residues, which are abnormally exposed to the cell solution that own to form stable inactive accumulations of these proteins. Molecular chaperones are able to bind to hydrophobic residues of unfolded proteins and assist to create the correct folding of protein [22], [26]. Molecular chaperones exist in the cytoplasm and organelles of eukaryotic cells like nucleus, mitochondria, endoplasmic reticulum, chloroplast, and periplasmic space of prokaryotic cells [27]. Molecular chaperones are divided in cytoplasmic and periplasmic categories that have been reviewed (Table 1 ).
Table 1.
Components of molecular chaperones in both prokaryotic and eukaryotic cells.
| Type of chaperone | Classification | Protein | Organism | Subcellar localization | Functions | Cooperating factors | energy consumption | reference |
|---|---|---|---|---|---|---|---|---|
| cytoplasmic | HSP40 | DnaJ | prokaryotic (Escherichia coli) | Cytosol, ER | Co-chaperone | – | [153], [154] | |
| Hsp60 | GroEL | prokaryotic (Escherichia coli) | Prokaryotic cytosol | (Holding and Folding chaperone) Promotes protein folding and preventing aggregation |
GroES, Hspl0, Cpnl0 | ATP (+) | [54], [155] | |
| Hsp58 | Mammals | Mitochondria | ||||||
| Cpn60 | Plants | Chloroplasts | ||||||
| HSP70 | DnaK | prokaryotic (Escherichia coli) | Prokaryotic cytosol | (Folding chaperone) Assisting refolding, Preventing aggregation, increase lysosomal degradation of cytosolic proteins, Iron-sulfur cluster protein assembly Sigma regulation, protein translocation into ER and protein translocation into mitochondria |
DnaJ, GrpE | ATP (+) | [156], [157] | |
| SSA1-4 | prokaryotic (Saccharomyces cerevisiae) | Eukaryotic cytosol | ||||||
| SSB1, 2 | prokaryotic (Saccharomyces cerevisiae) | |||||||
| Hsc73 | Mammals | |||||||
| HscA | prokaryotic (Escherichia coli) | Prokaryotic cytosol | ||||||
| HscC | ||||||||
| KAR2 | prokaryotic (Saccharomyces cerevisiae) | ER | ||||||
| BiP/Grp78 | Mammals | |||||||
| SSC1 | prokaryotic (Saccharomyces cerevisiae) | Mitochondria | ||||||
| ctHsp70 | Plants | Chloroplasts | ||||||
| HSP90 | HtpG | prokaryotic (Escherichia coli) | Prokaryotic cytosol | (Holding chaperone) Assist to folding properly, make stable condition for proteins against heat stress, Regulation of receptors, Protein translocation, Genetic buffering and protein degradation |
HSP9, Grp94 | ATP (+) | [158], [159] | |
| HSP90A | Mammals | Cytosol | ||||||
| HSP90B | Mammals | ER | ||||||
| TRAP | Mammals | Mitochondria | ||||||
| HSP100 | Hsp104 | prokaryotic (Saccharomyces cerevisiae) | Mitochondria | spread of yeast prions and clearance of aggregates | Hsp70, DnaK, ClpP, HsIV | ATP (+) | [41], [160] | |
| ClpB, C, A, X, | prokaryotic (Escherichia coli) | Cytosol | (Disaggregated chaperones) Fold properly and Disaggregate |
|||||
| Hsp78 | prokaryotic (Saccharomyces cerevisiae) | Mitochondria | ||||||
| HsIU | Plants | Chloroplast | ||||||
| sHSP | GroES | prokaryotic (Escherichia coli) | Cytosol | (Holding chaperone) Preventing aggregation during stress, Co-chaperone, provide thermotolerance in vivo and prohibition of apoptosis |
GroEL | ATP (+) | [76], [161] | |
| GrpE | DnaJ, DnaK | |||||||
| Hsp10 | Humans | Cytosol, mitochondria, Chloroplast |
– | |||||
| Hsp27 | Humans | Cytosol, ER, Nucleus | (−) | |||||
| HspB6 | Humans | Cytosol, Nucleus | ||||||
| HspB1 | Humans | Cytosol, Nucleus | ||||||
| periplasmic | Generic | Skp (OmpH) | prokaryotic (Escherichia coli) | Outer membrane | folding and assembling of outer membrane proteins and folding-assisting |
– | (−) | [93] |
| FkpA | periplasm space | |||||||
| PPIases | SurA | prokaryotic (Escherichia coli) | Outer membrane | correct folding, assembly of outer membrane proteins, folding of many newly translocated proteins | – | (−) | [102], [124], [128] | |
| PpiD | ||||||||
| FkpA | ||||||||
| PpiA (RotA) | Humans | Nucleus | ||||||
| disulfide bond formation | DsbA | prokaryotic | periplasm space | Folding of protein into the functional structure | – | (−) | [151], [162] | |
| DsbB | Inner membrane | Cooperates with ubiquinone to generate a disulfide bond | ||||||
| DsbC | periplasm space | As a disulfide bond isomerasation and facilitate folding of the protein | ||||||
| DsbD | Inner membrane | As reductive pathway | ||||||
| DsbE | periplasm space | Facilitate folding of the protein | ||||||
| DsbG | ||||||||
| Specialized | SurA | prokaryotic (Escherichia coli) |
Outer membrane | facilitates correct folding of outer membrane proteins, | – | (−) | [99] | |
| LolA (B,C,D,E) | periplasm space, Inner membrane and Outer membrane | rapid transfer of associated lipoproteins | ATP (+) | [108] | ||||
| PapD and its family | periplasm space and Outer membrane | biogenesis of pilus | (−) | [163] | ||||
| FimC | periplasm space | interacts with each pilus subunit |
(−) | [164] | ||||
2.1. Cytoplasmic chaperones
Most of cytoplasmic chaperones are mainly expressed under stress conditions such as high temperature [28]. Significantly, the chaperones bind to misfolded proteins to unfold them and in turn substrates are released from the chaperones to provide a new chance for unfolded protein to gain correct folding [16]. These chaperones can be categorized into five families based on their molecular weight including heat shock protein100 (Hsp100), Hsp90, Hsp70, Hsp60, and small Hsps (sHsps) that have molecular weight between 12 and 43 KD [19]. Some type of cytoplasmic chaperones have an ATP-dependant mode of substrates interactions (Hsp100, Hsp90, Hsp70, Hsp60) to have functional system, but in contrast, majority of sHsps don’t need ATP consumption [29]. Among them, Hsp90 and Hsp70 are the main chaperones responsible for protein holding to stop association of target protein with another protein [30]. Cytoplasmic chaperones based on their function and mechanisms are divided to three sub-classes including disaggregate chaperones that enhanced the solubilisation of stress-induced accumulated proteins [31], holding chaperones that keep and hold the folded proteins on their surface [32], [33], and folding chaperones that interfere the refolding of protein and enhance the proteolysis of mis-folded proteins [34], [35]. In this section, we focused on the cytoplasmic chaperones based on their molecular weights.
2.1.1. Hsp100
Hsp100 or (Clp) family are Hsps with molecular weight of 100–104 KDa, which give the organisms tolerate excessive stress and have variety of proteolysis functions [36]. Also, Hsp100 is responsible for protein disaggregation and degradation that lead to omitting non-functional and harmful polypeptides that is vital for the maintenance of cellular homeostasis [37]. Unlike the other Hsps, Hsp100 family solubilize accumulated proteins, thermally [38]. At first the members of this family were recognized as components of the two subunit bacterial Clp protease system, which contains regulatory ATPase/chaperones such as (ClpA and ClpX) and proteolytic (ClpP) subunits [39]. Then this family separated to two classes and eight individual subfamilies within this classes [40]. Furthermore, Hsp100 family cooperate with Hsp70, which is another ATP-dependant chaperone system for saving proteins from aggregation. The Hsp70 system gets the solubilized proteins from Hsp100 and refolds them [41]. The Hsp 100 is a member of AAA+ superfamily (ATPases associated with various cellular activities) proteins, which get their energy from hydrolysis of ATP to stimulate creation of correct folding in target proteins [42]. All AAA+ superfamily members have AAA domain which have two motifs for nucleotide binding and hydrolysis. Moreover, the AAA domain is responsible for protein oligomerization that lead to made of hexameric structures with a central pore [43]. Hsp100 chaperones are differ from each other based on the number of AAA domains which are one or two in each protomer and the existence of extra domains [43].
2.1.2. Hsp90
The Hsp90 family are highly conserved and proteins, which exist in all organisms from bacteria to humans and their expressions rises in response to the stress conditions in prokaryotic and eukaryotic cells. This family is often found with other chaperones that they have various functions depend on ATP [44]. In human, two homologous isoforms of Hsp90 are exist in equal amounts known as α and β isoforms [45]. Hsp90 is a constitutive homodimer, which has main inter subunit in association with the COOH-terminal 190 residues [46], the highly conserved 25 kDa NH2-terminal domain that is the ATP binding site [47] and middle domain, which is binding site of proteins and co-chaperones [48], [49]. Most of the Hsp90 substrates are signal transduction proteins such as steroid hormone receptors and signalling kinases [50]. Forasmuch as, cancer cells are so sensitive to Hsp90 inhibition, the Hsp90 active form in these cells are in high level [51], [52]. Therefore, Hsp90 molecule is an attractive target for cancer targeted therapy [53]. The most important function of Hsp90 is to manage protein folding and inhibit protein aggregation [54]. Furthermore, this chaperone has roles in signal-transduction networks, cell-cycle control, protein degradation and protein trafficking [55]. Despite Hsp90, which are the most numerous chaperones in the cell but its functions in in-vitro is poorly understood and it has been proved that have functions with contribution of several cofactors [44].
2.1.3. Hsp70
The Hsp70 family are highly protected class of the heat shock proteins and the most of the organisms contain several members of them. This family have some function in the cells including; binding strongly to partially synthesized peptide sequences, keeping translocation competent structures of ER and mitochondrial precursors in the cytosol, and simplify translocation from inside the target compartment [56]. Actually, the particular roles of Hsp70 proteins are confirmed by their location in diverse subcellular sections [54], the differential expression of Hsp70 at dissimilar phases of growth [57] and their communication with specific groups of Hsp70-associated proteins [58]. Hsp70 family consist of two domains; the 25KDa COOH-terminal domain, which is needed for polypeptide binding and the 44KDa N-terminal highly protected ATPase domain, which is binds to ADP and ATP in the existence of K+ and Mg2+ that lead to hydrolyse ATP. Correct folding of protein is taken by cooperation of both domains [59], [60]. The ATPase domain had similar structure to actin and hexokinase which contain four sub-domains [61]. Hsp70 obtain two conformational conditions based on the binding to ATP or ADP [62]. The ATP-bound form in comparison to ADP-bound form showed high association and dissociation rate constants for substrates. Attachment of substrate to Hsp70-ATP is hastening ATP hydrolysis by Hsp70 which lead to change in folding of substrate, After that ADP replaced with ATP result in releasing of substrate from Hsp70 [63]. Hsp70 known as DnaK that expressed in prokaryotes which participate with DnaJ and GrpE proteins in reaction cycle of DnaK [64]. In addition GrpE assists in the releasing of the bound nucleotide from Hsp70 and DnaJ motivates ATP hydrolysis by Hsp70. Furthermore, DnaJ itself acts as a molecular chaperone by binding to unfolded proteins and probably control affinity of Hsp70 for substrate proteins [65].
2.1.4. Hsp60
This family has several names such as GroEL family or TCP-1. Most of these family members are known as chaperonins. X-ray crystallographic studies showed that GroEL (E. coli chaperone) has 14 identical subunits in two stacked heptameric rings, each containing central hole which bind to unfolded proteins [66]. GroEL simplifies protein folding by cooperation ATP and its co-chaperonin known as GroEs [67]. GroES has a heptameric ring structure that was made by identical 10 kDa subunits that attached to one or both ends of GroEL cylinder [68]. GroEL chaperone have three different functions: first, GroEL form complexes with non-native polypeptides and inhibits their aggregations, so it will reduce the concentration of misfolding protein [69]. Second function is folding substrate that bind to central cavity of GroEL complex, a protected environment without intramolecular interactions [70]. Finally, GroEL seems to be capable to unfold trapped misfolding intermediates, so they are able to have a new chance for correct folding [71].
Members of the Hsp60 family are involved in construction of multiprotein complexes such as Rubisco [72]. Furthermore, Sonoda et al. reported that co-expression of GroEl/GroEs with scFv lead to improve soluble expression that the resultant scFv demonstrated a 4.6 fold rise in antigen-binding activity [73]. Also Moeng et al. expressed five set of molecular chaperones with anti-BNP scFv. They reported that co-expression of GroEL/GroEs with anti-BNP scFv showed more effective than other set of chaperones in soluble expression of the scFv [74].
2.1.5. Small Hsps
In prokaryotic and eukaryotic cells, majority of the small Hsps (sHsps) are constructed in stress conditions and some small Hsps are expressed throughout certain developmental phases [27]. In stress conditions the small Hsps bind to the denatured proteins that finally led to prevent its accumulation in an ATP-independent manner [75], [76]. Small Hsps are in relation with nucleus, cytoskeleton, and membranes. Also, this family consist of 12–43 kDa chaperons that they bind to large multimeric structures [77]. The sHsps endure dynamic assembly into mono- and poly-disperse oligomers that the rate of disassembly influence chaperoning [78]. The structure of the sHsps include a conserved α-crystallin domain, which is enriched in β strands formed in β sheet that is in charge of dimer formation [79]. Most of the sHsps oligomerization and chaperoning are affected by the amino-terminus because the amino terminal domain is sensitive to phosphorylation and has slack structure [80], [81]. Moreover, sHsps have an important role in refolding by cooperation of ATP-dependant chaperones such as DnaK system [82].
2.2. Periplasmic chaperones
The space between cytoplasmic and outer membrane of the gram-negative bacteria is called periplasmic space, which has total volume of 20–40% of the whole cell. It is an aqueous space that has numerous molecules such as membrane derived oligosaccharides, amino acids, peptides, and etc. [83]. The proteins in the periplasm are classified to three classes based on their function: enzymes, chaperones, and high affinity binding proteins for vitally important substrates which are essential for the transport across the cytoplasmic membrane [84]. The periplasm is an oxidizing region of bacterial cells that disulfide bonds formed in protein folding process. Thus, many secreted proteins containing disulfide bonds are in the periplasmic space [85]. Protein folding in the periplasmic space can be catalysed by two groups of overexpressing enzymes including protein disulfide isomerases (PDI), which accelerated the oxidation of disulfide bonds and peptidyl-prolyl-cis-trans isomerase (PPI) that are member of periplasmic chaperones [86]. Periplasmic chaperones are divided to four classes including generic chaperones, specialized chaperones, PPIases, and proteins involved in disulfide bond formation.
2.2.1. Generic chaperones
The members of generic chaperones are homodimers or homotrimer, which hold unfolded proteins and help to its correct folding formation [87]. In result, they have application to periplasmic expression and phage display of recombinant protein [88]. These chaperones can be active in presence and absent of ATP condition in periplasmic space, and have the most generic folding activity [89], [90]. Generic chaperones are including; Skp and FkpA. Skp is an 18 KDa periplasmic protein that has significant role in folding and gathering of outer membrane proteins (OMP) of E. coli, which it means that Skp binds to denatured OMPs [91]. Skp simplifies proper folding of expressed proteins that increased the solubility and affinity of proteins [92]. For instance, Ow et al. expressed SKp and scFv to achieve high soluble expression. In result, they observed high scFv soluble expression [93]. Moreover, Ute Schafer et al. demonstrated that Skp is involved in making and keeping the solubility of early folding intermediates of outer membrane proteins in the periplasmic space of gram negative bacteria [87]. FkpA is V-shaped homodimeric chaperone with two domains which exhibits like PPlase activity [16]. The N-terminal domain, which has α-helixes structure and responsible for dimerization and activity of FkpA [94]. However, the C-terminal domain is capable to catalyse slow refolding cis-trans isomerization reactions in proline containing proteins [95]. In addition, Zhang et al. evaluated the co-expression of FKpA with scFv, which the results demonstrated that co-expression of FKp was suitable for soluble expression of recombinant antibodies in E. coli [96]. Furthermore, Bothmann and Plückthun showed that interacting FKpA with early folding intermediates inhibit their aggregation. Also FKpA binds to partially unfolded proteins and reactivate them [95]. Ow et al. demonstrated that the chaperone activity of FkpA, rather than its PPlase activity. This activity is more critical in overcoming metabolic stresses from misfolded proteins [93].
2.2.2. Specialized chaperones
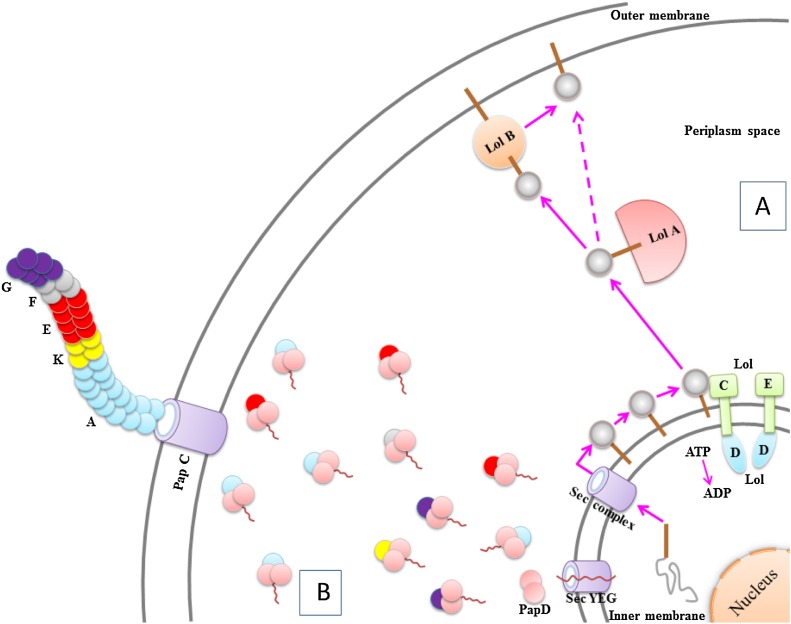
The members of specialized chaperones have two PPIase domains, which have chaperone activity to prepare the maturation of three outer membrane proteins including OmpA, OmpF, LamB [97]. Also, they stimulated folding of unstable proteins and pre-aggregated proteins. Specialized chaperones include SurA, LolA, PapD and FimC. SurA is one of the vital proteins that is required to cell survival during stationary phase [98]. The SurA has four domains consist of N-terminal domain that has approximately 150 amino acid, two parvulin-like PPlase domains that have approximately 100 residues and a C-terminal domain that has approximately 40 amino acid [99]. Three of these domains formed core shape of the complex protein and the second PPlase domain has exhibit chaperone function [100]. SurA is one of the first chaperones for OMP folding that detects Ar-X-Ar motif in unfolded OMPs [101]. Also, Hennecke et al. reported two characteristics of SurA that are essential for peptide binding such as particular pattern of aromatic residues and the direction of their side chains, which are founded mostly in integral outer membrane proteins. In addition, SurA-peptide binding needs neither an active PPlase domain nor the existence of proline, in result this data demonstrate the SurA-peptide binding depends on substrate specificity [102]. Bitto and McKay realized that SurA detects a peptide motif which is features of integral outer membrane proteins [103]. Lol chaperon family consist of five members that by cooperation of ATP-binding transporter LolCDE complex, LolA attached to the bacterial lipoproteins, then the LolA- lipoproteins complex released from inner membrane to transfer of lipoproteins to LolB in the outer membrane [104], [105]. The stage of attaching LolA to lipoproteins just needs ATP [106]. LolA and LolB have a similar structures and have function in monomeric form despite different sequences (Fig. 1 A) [106]. Tajime et al. indicated disruption of the chromosomal LolA gene was mortal for the strains without lipoproteins which LolA is generally vital for the outer membrane localization of lipoproteins [107]. Okuda and Tokuda revealed that shifting lipoproteins via lol proteins happens in a mouth to mouth manner, which means their hydrophobic holes were found to bind lipoproteins inside [108]. PapD is a 28.5 KDa periplasmic protein from a family with 30 members, which has two domains with an immunoglobulin (Ig) like structure [109]. Three important performance of PapD in bacterial cell are attaching and covering pilus subunits to avoid non-specific interactions [110], [111], simplifying transfer of these subunits in the periplasm [112] and assistance to the folding of subunit [111]. As it is represented in the legend to (Fig. 1B) Periplasmic chaperones help in folding pilus subunits and attacking them to the OM usher. At first, transferring fiber subunits to the periplasmic space done by sec system (YEG). Then the chaperones in the periplasm has function with stop aggregation and degradation of pilus subunits by binding their interfacing regions and facilitating correct protein folding before bringing them to the usher/fimbriae complex. Jacob-Dubuisson et al. show that PapD is responsible to made of native-like conformation in pilus subunits [113]. FimC is one of the periplasmic proteins has similar structure, function and the mechanism of action to PapD. Pilus chaperones like FimC identify specific carboxyl terminal motifs on pilus subunits and creating chaperone-subunit complexes [114]. Also, Jones et al. indicated that FimC has a high degree of homology to PapD and it is In accordance with sequence of periplasmic chaperones [115].
Fig. 1.
(A) A Schematic representation of the outer membrane localization of lipoproteins mediated by the Lol system. Binding lipoproteins to LolCDE complex begins the lipoprotein release in the inner membrane and the affinity of the Lol complex for the ATP increases, then LolA gets the lipoprotein from the complex in an ATP-dependent manner and transfer it to LolB in the outer membrane. (B) A schematic overview of the pap chaperone-usher system showing all subunits and their organization.
2.2.3. PPIases
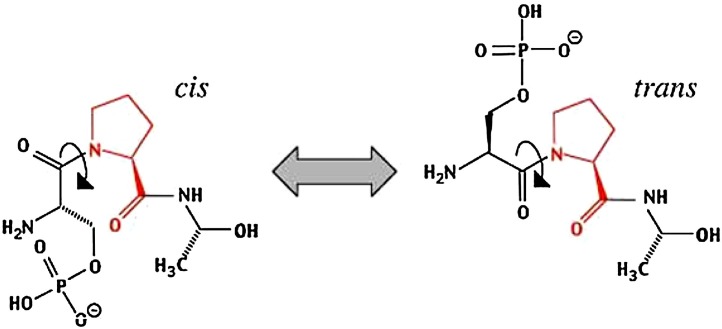
Peptidyl-prolyl isomerases (ppls) are housekeeping proteins and highly conserved in prokaryotic and eukaryotic cells. They exist in the periplasm of E. coli and in eukaryotic cell organelles such as cytosol, mitochondria, endoplasmic reticulum [116], [117]. The PPlases are enzymes that facilitate the slow cis-trans isomerization of peptidyl-prolyl bonds in folding of target proteins (Fig. 2 ) [118]. The first discoveries showed that immunosuppressant drugs attach to PPls and prevent the PPl activity, so PPls are necessary for immune system function [119]. In immune system they act by separating calcineurin, which is a calcium/calmodulin-dependent protein phosphatase. PPlases are divided to three families based on drug specificity and primary sequence homology: 1) the cyclosporine A (CsA)-binding proteins and cyclophillins, 2) rapamycin binding proteins, binding proteins FK506 and FKBPs 3) parvulins, which do not attach immunosuppressant drugs [120], [121]. The SurA, PpiD, FkpA, and PpiA are the members of PPlase family. For example, Juctice et al. show that the four known periplasmic cis-trans prolyl isomerases simplify suitable protein folding by raising the rate of transferring of proline residues between cis-trans state. Also, Pplases are not necessary for cell growth but they have great roles in survival in environmental and pathogenic positions [122]. SurA, as it is mentioned above is specialized for the biogenesis of OMPs and also it has a peptidyl-prolyl cis-trans isomerise activity [99], [100]. PpiD have a single parvulin domain and attached to the inner membrane by an N-terminal transmembrane section. The study showed that N-terminal section of PpiD and SurA have similar structure and have overlapping role in OMP biogenesis [123], [124], [125]. Loss of PpiD in the cells own to modified outer membrane configuration which has negative effects on protein folding process [84], [123]. Also, findings demonstrated that PpiD has chapronic function to help in the early periplasmic folding of proteins [124]. Dartigalongue and Raina showed that a null mutation in PpiD own to completely decreasing in the level and folding of OMPs and to the stimulation of the periplasmic stress response. Null mutation in PpiD and SurA led to death of cells [123]. PpiA has two functions including peptidyl-prolyl cis-trans isomerase function and chaperonic function. PpiA is vital in viral infections like HIV, influenza A virus, SARS coronavirus, vaccinia virus which it might have a role in life cycle of viruses [126], [127]. Tremillon et al. indicated that PpiA gene exists in Lactococcus lactis which expressed under normal and stress conditions. In normal conditions, PpiA protein expressed and released from cell with added protease then exposed at the cell surface. A recombinant soluble form of PpiA was produced and secreted from L.lactis that showed both pplase and chaperone activities [128].
Fig. 2.
A Schematic of Cis/trans isomerization of peptide bond.
2.2.4. Chaperones involved in disulfide bond formation
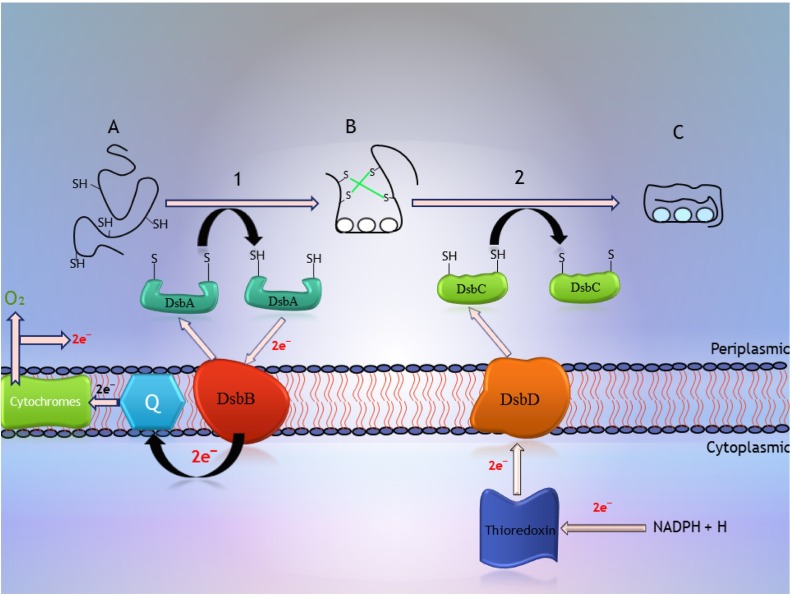
These chaperones known as disulphide bonds proteins (Dsb proteins) have highly conserved motifs which are necessary for disulphide oxido-reductase activity. In numerous secreted proteins, disulphide bonds formation is essential for their folding and stability because disulphide bonds generally stabilized the tertiary structure of the protein. The Dsb proteins reorganized primary disulphide bonds and made new disulphide bonds in target protein. In E. coli the disulphide bonds are made by the Dsb proteins in the periplasm space. In periplasmic spaces Dsb proteins increase the solubility of recombinant proteins and the secretion efficiency [129], [130]. Dsb proteins are including DsbA, DsbB, DsbC, DsbD, DsbG, DsbE, CcmH. (Fig. 3 ).
Fig. 3.
The mechanisms of disulfide bond formation in the periplasm. One of the disulfide bond donors in the periplasm is DsbA, which is oxidized by DsbB and this oxidizing reaction needs respiratory quinone component. Whenever there is more than two cysteine, the incorrect disulfide bond may occur. For preventing that, the disulfide bond isomerase DsbC comes to the rescue and it is oxidized by DsbD.
DsbA(thioredoxin-like thiol-disulphide oxido-reductase) and DsbB(cytoplasmic membrane protein) are two enzymes that help to disulphide bonds formation in periplasmic proteins [131], [132]. The DsbA is a small 21 KDa polypeptide, which its active site contains the motif known as Cys30-Pro31-His32-Cys33 has high oxidizing potential activity. In the oxidative process, disulphide bond formed between two cysteines by DsbA which DsbA need re-oxidized by DsbB [133], [134]. Kishigami et al. recently suggested that synthetized DsbA is quickly oxidized by pre-existing DsbA, whereas oxidation of functional DsbA needs DsbB, which has opposite action with DsbA-reducing enzymes [135]. Moreover, Zhuo et al. were co-expressed DsbA/DsbC proteins and recombinant reteplase in E coli. As result, the co-expression system improved soluble expression of recombinant reteplase, and high yield of soluble reteplase achieved especially with the DsbC co-expression system [136]. The DsbB is a 20 KD periplasmic protein that has four transmembrane (TM) fragments and two periplasmic loops that each loop contains a pair of cysteines [137]. Kishigami and Ito suggested that DsbB stimulated recycling DsbA by disulfide interaction between Cys30 of DsbA and Cys104 of DsbB [137]. DsbC(periplasmic oxido reductase) and DsbD(cytoplasmic membrane protein) have roles in isomerization of non-native disulphide bonds [131], [132]. DsbC is 23 KDa homodimeric soluble protein containing four cysteines residue which these residue act as a catalyst for reorganization of disulfide bonds in protein with multiple disulfide bonds. DsbC deficiency can lead to misfolding of proteins with numerous disulphide bonds [138]. The active site cysteines of DsbC are found in dithiol form in isomerization activity which DsbC reduced by DsbD in this activity [139]. Also DsbC has a reductase activity to proteins with false disulfide bonds and then re-oxidize by DsbA. Studies showed that the periplasmic proteins like RNaseI and MepA needs to DsbC for their stability [140]. In addition, Missiakas et al. indicated that overexpression of DsbC can functionally substitute for DsbA loss [141]. For example purified DsbC protein was capable to catalysed insulin oxidation in a dithiothreitol dependant manner [142]. Heo et al. showed that production of anti-c-met scFv could be more increased by co-expressing molecular chaperones such as GroEl and DsbC with scFv. In E. coli trxB/gor mutant, co-expression of DsbC enhanced the efficiency of functional anti-c-met scFv about 2.5 fold [143]. DsbD has three domains including amino terminal periplasmic domain, hydrophobic core domain containing eight transmembrane fragments and carboxy terminal periplasmic domain [144]. Each of these domains has a pair of cysteines that have necessary role for electron transfer from the cytoplasm into the periplasm space [145]. Kurokawa and his colleagues represented that overexpression of DsbD proteins are responsible for formation and isomerization of disulfide bonds that can significantly increase periplasmic production of human nerve growth factor B (NGF) [129]. DsbD is required for the reduction of DsbG. Also it is needed for the biosynthesis of c-type cytochromes [146]. DsbG has negative charged surface, which is appropriate to interact with folded proteins [147], [148]. It can act as a molecular chaperone and its activity is not based on its active site cysteines [149]. Also, Andersen et al. showed that DsbG acts mainly as an oxidant agent during protein disulfide bond formation. In addition, the substrate range of DsbG could be slighter than the other periplasmic oxidative enzymes such as DsbA and DsbC [150]. DsbE is known as CcmG because implicated in maturation of cytochrome c [151]. CcmG selectively gives electron to apocytochrome c cysteines via CcmH, which is another periplasmic cytochrome c maturation factor with a redox activity Cys-X-X-Cys motif [152]. This proposes that CcmG is capable to identify and selectively cooperate with DsbD and CcmH [151]. Also DsbD is responsible for keeping DsbE in reduced form in the periplasm [145].
3. Conclusion
The eventual objective of amplify production of desired recombinant protein in an E. coli host cell depends on some factors that lead to outstanding out come in production of recombinant protein. As usual, recombinant proteins were expressed as inclusion body in the E. coli. For understanding of some problem we described the details to find suitable ways for enhancing soluble expression of recombinant proteins with correct folding that several approaches are used including; chemical chaperones, gene protein optimization, signal peptide sequences, molecular chaperone, and etc. Molecular chaperones either cytoplasmic or periplasmic are more efficient in production of recombinant proteins with appropriate folding. Cytoplasmic chaperones Hsp100 (Clp), Hsp90, Hsp70 (DnaK/DnaJ/GrpE), Hsp60 (GroEL/GroES) and small Hsps bind to aggregated and misfolded recombinant proteins to unfold them and finally the recombinant proteins acquire their correct folding. The HSP60, HSP70 and their co chaperones used ATP to solubilized and refolded aggregated recombinant protein. Periplasmic chaperones, located in periplasm space, were fold the recombinant protein correctly via formation of disulfide bonds, which finally produce soluble form of recombinant proteins. Efficiency of folding process mediated by periplasmic chaperones depends on type of recombinant protein. Some factors including; enhance the level of some periplasmic chaperones efficiency such as FkpA, and SurA, which each of them facilitate the slow cis-trans isomerization of peptidyl-prolyl bonds in folding of target proteins and DsbA and DsbC which help to form disulphide bridge. These activities have more important roles in overcoming to production of misfolded proteins.
Conflict of interest
The authors declare that they have no competing interests.
Acknowledgments
The authors hope the best for all contributors in Tabriz University of medical science and College of Rab-Rashidi.
References
- 1.Itakura K., Hirose T., Crea R., Riggs A.D., Heyneker H.L., Bolivar F., Boyer H.W. Science. 1977;198:1056–1063. doi: 10.1126/science.412251. [DOI] [PubMed] [Google Scholar]
- 2.Baneyx F. Garland Sci. 2004. Protein expression technologies: current status and future trends; pp. 532–560. Chapter 2. [Google Scholar]
- 3.Veisi K., Farajnia S., Zarghami N., Khoram Khorshid H.R., Samadi N., Khosroshahi S.A., Jaliani H.Z. Adv. Pharm. Bull. 2015;5:621–627. doi: 10.15171/apb.2015.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sørensen H.P., Mortensen K.K. Microbial Cell Fact. 2005;4:1–8. doi: 10.1186/1475-2859-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Marco A. Nat. Protoc. 2007;2:2632–2639. doi: 10.1038/nprot.2007.400. [DOI] [PubMed] [Google Scholar]
- 6.Kopito R.R. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 7.Bowden G.A., Paredes A.M., Georgiou G. Nat. Biotechnol. 1991;9:725–730. doi: 10.1038/nbt0891-725. [DOI] [PubMed] [Google Scholar]
- 8.Mukhopadhyay A. Biotreatment Downstream Processing and Modelling. Springer; 1997. Inclusion bodies and purification of proteins in biologically active forms; pp. 61–109. [DOI] [PubMed] [Google Scholar]
- 9.Carrio M., Villaverde A. J. Biotechnol. 2002;96:3–12. doi: 10.1016/s0168-1656(02)00032-9. [DOI] [PubMed] [Google Scholar]
- 10.Taylor G., Hoare M., Gray D., Marston F. Nat. Biotechnol. 1986;4:553–557. [Google Scholar]
- 11.Carrió M.M., Cubarsi R., Villaverde A. FEBS Lett. 2000;471:7–11. doi: 10.1016/s0014-5793(00)01357-0. [DOI] [PubMed] [Google Scholar]
- 12.Singh S.M., Panda A.K. J. Biosci. Bioeng. 2005;99:303–310. doi: 10.1263/jbb.99.303. [DOI] [PubMed] [Google Scholar]
- 13.Villaverde A., Carrió M.M. Biotechnol. Lett. 2003;25:1385–1395. doi: 10.1023/a:1025024104862. [DOI] [PubMed] [Google Scholar]
- 14.Vallejo L.F., Rinas U. Microb. Cell Fact. 2004;3:11. doi: 10.1186/1475-2859-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schumann W., Ferreira L.C.S. Genet. Mol. Biol. 2004;27:442–453. [Google Scholar]
- 16.Baneyx F., Mujacic M. Nat. Biotechnol. 2004;22:1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- 17.Schwarz E., Lilie H., Rudolph R. Biol. Chem. Hoppe Seyler. 1996;377:411–416. [PubMed] [Google Scholar]
- 18.Ben-Zvi A.P., Goloubinoff P. J. Struct. Biol. 2001;135:84–93. doi: 10.1006/jsbi.2001.4352. [DOI] [PubMed] [Google Scholar]
- 19.Almeida M.B., do Nascimento J.L.M., Herculano A.M., Crespo-López M.E. Biomed. Pharmacother. 2011;65:239–243. doi: 10.1016/j.biopha.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 20.Sõti C., Nagy E., Giricz Z., Vígh L., Csermely P., Ferdinandy P. Br. J. Pharmacol. 2005;146:769–780. doi: 10.1038/sj.bjp.0706396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shigapova N., Török Z., Balogh G., Goloubinoff P., Vígh L., Horváth I. Biochem. Biophys. Res. Commun. 2005;328:1216–1223. doi: 10.1016/j.bbrc.2005.01.081. [DOI] [PubMed] [Google Scholar]
- 22.Hinault M.-P., Ben-Zvi A., Goloubinoff P. J. Mol. Neurosci. 2006;30:249–265. doi: 10.1385/JMN:30:3:249. [DOI] [PubMed] [Google Scholar]
- 23.Priya S., Sharma S.K., Goloubinoff P. FEBS Lett. 2013;587:1981–1987. doi: 10.1016/j.febslet.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 24.Laudanski K., Wyczechowska D. Archivum immunologiae et therapiae experimentalis. 2006;54:103–111. doi: 10.1007/s00005-006-0013-3. [DOI] [PubMed] [Google Scholar]
- 25.Ruddon R.W., Bedows E. J. Biol. Chem. 1997;272:3125–3128. doi: 10.1074/jbc.272.6.3125. [DOI] [PubMed] [Google Scholar]
- 26.Natalello A., Mattoo R.U., Priya S., Sharma S.K., Goloubinoff P., Doglia S.M. J. Mol. Biol. 2013;425:1158–1171. doi: 10.1016/j.jmb.2012.12.025. [DOI] [PubMed] [Google Scholar]
- 27.Waters E.R., Lee G.J., Vierling E. J. Exp. Bot. 1996;47:325–338. [Google Scholar]
- 28.Georgopoulos C.M., Tissières R.I. 1994. The Biology of Heat Shock Proteins and Molecular Chaperones. [Google Scholar]
- 29.Bukau B., Horwich A.L. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 30.Powers M.V., Jones K., Barillari C., Westwood I., Montfort R.L.v., Workman P. Cell Cycle. 2010;9:1542–1550. doi: 10.4161/cc.9.8.11204. [DOI] [PubMed] [Google Scholar]
- 31.Thomas J.G., Baneyx F. Mol. Microbiol. 2000;36:1360–1370. doi: 10.1046/j.1365-2958.2000.01951.x. [DOI] [PubMed] [Google Scholar]
- 32.Kitagawa M., Miyakawa M., Matsumura Y., Tsuchido T. Eur. J. Biochem. 2002;269:2907–2917. doi: 10.1046/j.1432-1033.2002.02958.x. [DOI] [PubMed] [Google Scholar]
- 33.Kuczynska-Wisnik D., Kçdzierska S., Matuszewska E., Lund P., Taylor A., Lipinska B., Laskowska E. Microbiology. 2002;148:1757–1765. doi: 10.1099/00221287-148-6-1757. [DOI] [PubMed] [Google Scholar]
- 34.Deuerling E., Patzelt H., Vorderwülbecke S., Rauch T., Kramer G., Schaffitzel E., Mogk A., Schulze-Specking A., Langen H., Bukau B. Mol. Microbiol. 2003;47:1317–1328. doi: 10.1046/j.1365-2958.2003.03370.x. [DOI] [PubMed] [Google Scholar]
- 35.Xu Y., Hsieh M.Y., Narayanan N., Anderson W.A., Scharer J.M., Moo-Young M., Chou C.P. Biotechnol. Progr. 2005;21:1357–1365. doi: 10.1021/bp0501120. [DOI] [PubMed] [Google Scholar]
- 36.Park C.-J., Seo Y.-S. Plant Pathol. J. 2015;31:323. doi: 10.5423/PPJ.RW.08.2015.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agarwal M., Katiyar-Agarwal S., Sahi C., Gallie D.R., Grover A. Cell Stress Chaperones. 2001;6:219–224. doi: 10.1379/1466-1268(2001)006<0219:athpka>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsell D.A., Kowal A.S., Singer M.A., Lindquist S. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 39.Gottesman S., Squires C., Pichersky E., Carrington M., Hobbs M., Mattick J.S., Dalrymple B., Kuramitsu H., Shiroza T., Foster T. Proc. Natl. Acad. Sci. 1990;87:3513–3517. doi: 10.1073/pnas.87.9.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schirmer E.C., Glover J.R., Singer M.A., Lindquist S. Trends Biochem. Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 41.Glover J.R., Lindquist S. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 42.Tucker P.A., Sallai L. Curr. Opin. Struct. Biol. 2007;17:641–652. doi: 10.1016/j.sbi.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 43.Mogk A., Kummer E., Bukau B. Front. Mol. Biosci. 2015;2:22. doi: 10.3389/fmolb.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bose S., Weikl T., Bugl H., Buchner J. Science. 1996;274:1715–1717. doi: 10.1126/science.274.5293.1715. [DOI] [PubMed] [Google Scholar]
- 45.Jakob U., Buchner J. Trends Biochem. Sci. 1994;19:205–211. doi: 10.1016/0968-0004(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 46.Nemoto T., Ohara-Nemoto Y., Ota M., Takagi T., Yokoyama K. Eur. J. Biochem. 1995;233:1–8. doi: 10.1111/j.1432-1033.1995.001_1.x. [DOI] [PubMed] [Google Scholar]
- 47.Whitesell L., Mimnaugh E.G., De Costa B., Myers C.E., Neckers L.M. Proc. Natl. Acad. Sci. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ratzke C., Mickler M., Hellenkamp B., Buchner J., Hugel T. Proc. Natl. Acad. Sci. 2010;107:16101–16106. doi: 10.1073/pnas.1000916107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prodromou C., Pearl L.H. Curr. Cancer Drug Targets. 2003;3:301–323. doi: 10.2174/1568009033481877. [DOI] [PubMed] [Google Scholar]
- 50.Young J.C., Moarefi I., Hartl F.U. J. Cell Biol. 2001;154:267–274. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Workman P. Cancer Lett. 2004;206:149–157. doi: 10.1016/j.canlet.2003.08.032. [DOI] [PubMed] [Google Scholar]
- 52.Solit D.B., Chiosis G. Drug Discov. Today. 2008;13:38–43. doi: 10.1016/j.drudis.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 53.Li Y., Zhang T., Schwartz S.J., Sun D. Drug Resistance Updates. 2009;12:17–27. doi: 10.1016/j.drup.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frydman J. Annu. Rev. Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 55.Pratt W.B., Krishna P., Olsen L.J. Trends Plant Sci. 2001;6:54–58. doi: 10.1016/s1360-1385(00)01843-4. [DOI] [PubMed] [Google Scholar]
- 56.Craig E.A., Weissman J.S., Hotwicht A.L. Cell. 1994;78:365–372. doi: 10.1016/0092-8674(94)90416-2. [DOI] [PubMed] [Google Scholar]
- 57.Sung D.Y., Kaplan F., Guy C.L. Physiol. Plant. 2001;113:443–451. [Google Scholar]
- 58.May T., Soll J. Plant Cell. 2000;12:53–63. doi: 10.1105/tpc.12.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fink A., Goto U. Marcel Dekker; New York: 1998. Molecular Chaperones in the Life Cycle of Proteins; pp. 123–150. [Google Scholar]
- 60.Hightower L., Leung S. Cell. 1998;12:256–275. [Google Scholar]
- 61.Fink A.L. Physiol. Rev. 1999;79:425–449. doi: 10.1152/physrev.1999.79.2.425. [DOI] [PubMed] [Google Scholar]
- 62.Farr C.D., Witt S.N. Cell Stress Chaperones. 1999;4:77. doi: 10.1379/1466-1268(1999)004<0077:altaeb>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kubo Y., Tsunehiro T., Nishikawa S.-i., Nakai M., Ikeda E., Toh-e A., Morishima N., Shibata T., Endo T. J. Mol. Biol. 1999;286:447–464. doi: 10.1006/jmbi.1998.2465. [DOI] [PubMed] [Google Scholar]
- 64.Liberek K., Marszalek J., Ang D., Georgopoulos C., Zylicz M. Proc. Natl. Acad. Sci. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schröder H., Langer T., Hartl F., Bukau B. EMBO J. 1993;12:4137. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boisvert D.C., Wang J., Otwinowski Z., Norwich A.L., Sigler P.B. Nat. Struct. Mol. Biol. 1996;3:170–177. doi: 10.1038/nsb0296-170. [DOI] [PubMed] [Google Scholar]
- 67.Zahn R., Perrett S., Fersht A.R. J. Mol. Biol. 1996;261:43–61. doi: 10.1006/jmbi.1996.0440. [DOI] [PubMed] [Google Scholar]
- 68.Hunt J.F., Weaver A.J., Landry S.J., Gierasch L., Deisenhofer J. Nature. 1996;379:37–45. doi: 10.1038/379037a0. [DOI] [PubMed] [Google Scholar]
- 69.Buchner J., Schmidt M., Fuchs M., Jaenicke R., Rudolph R., Schmid F.X., Kiefhaber T. Biochemistry. 1991;30:1586–1591. doi: 10.1021/bi00220a020. [DOI] [PubMed] [Google Scholar]
- 70.Mayhew M., Da Silva A., Martin J., Erdjument-Bromage H., Tempst P., Hartl F.U. Nature. 1996;379:420–426. doi: 10.1038/379420a0. [DOI] [PubMed] [Google Scholar]
- 71.Sparrer H., Buchner J. J. Biol. Chem. 1997;272:14080–14086. doi: 10.1074/jbc.272.22.14080. [DOI] [PubMed] [Google Scholar]
- 72.Viitanen P.V., Lubben T.H., Reed J., Goloubinoff P., O'Keefe D.P., Lorimer G.H. Biochemistry. 1990;29:5665–5671. doi: 10.1021/bi00476a003. [DOI] [PubMed] [Google Scholar]
- 73.Sonoda H., Kumada Y., Katsuda T., Yamaji H. Biochem. Eng. J. 2011;53:253–259. [Google Scholar]
- 74.Maeng B.H., Nam D.H., Kim Y.H. World J. Microbiol. Biotechnol. 2011;27:1391–1398. doi: 10.1007/s11274-010-0590-5. [DOI] [PubMed] [Google Scholar]
- 75.Reddy G.B., Das K.P., Petrash J.M., Surewicz W.K. J. Biol. Chem. 2000;275:4565–4570. doi: 10.1074/jbc.275.7.4565. [DOI] [PubMed] [Google Scholar]
- 76.Lee G.J., Vierling E. Plant Physiol. 2000;122:189–198. doi: 10.1104/pp.122.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Buchner J. FASEB J. 1996;10:10–19. [PubMed] [Google Scholar]
- 78.Sun Y., MacRae T.H. Cell. Mol. Life Sci. CMLS. 2005;62:2460–2476. doi: 10.1007/s00018-005-5190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim K.K., Kim R., Kim S.-H. Nature. 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- 80.Jiao W., Qian M., Li P., Zhao L., Chang Z. J. Mol. Biol. 2005;347:871–884. doi: 10.1016/j.jmb.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 81.Peterson J.J., Young M.M., Takemoto L.J. Mol. Vis. 2004;10:857–866. [PubMed] [Google Scholar]
- 82.Lee G.J., Roseman A.M., Saibil H.R., Vierling E. EMBO J. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Duong F., Eichler J., Price A., Leonard M.R., Wickner W. Cell. 1997;91:567–573. doi: 10.1016/s0092-8674(00)80444-4. [DOI] [PubMed] [Google Scholar]
- 84.Rizzitello A.E., Harper J.R., Silhavy T.J. J. Bacteriol. 2001;183:6794–6800. doi: 10.1128/JB.183.23.6794-6800.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Swartz J.R. Curr. Opin. Biotechnol. 2001;12:195–201. doi: 10.1016/s0958-1669(00)00199-3. [DOI] [PubMed] [Google Scholar]
- 86.Choi J., Lee S. Appl. Microbiol. Biotechnol. 2004;64:625–635. doi: 10.1007/s00253-004-1559-9. [DOI] [PubMed] [Google Scholar]
- 87.Schäfer U., Beck K., Müller M. J. Biol. Chem. 1999;274:24567–24574. doi: 10.1074/jbc.274.35.24567. [DOI] [PubMed] [Google Scholar]
- 88.Bothmann H., Plückthun A. Nat. Biotechnol. 1998;16:376–380. doi: 10.1038/nbt0498-376. [DOI] [PubMed] [Google Scholar]
- 89.Walton T.A., Sousa M.C. Mol. Cell. 2004;15:367–374. doi: 10.1016/j.molcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 90.Saul F., Arie J.-P., Vulliez-le Normand B., Kahn R., Betton J.-M., Bentley G. J. Mol. Biol. 2004;335:595–608. doi: 10.1016/j.jmb.2003.10.056. [DOI] [PubMed] [Google Scholar]
- 91.Hayhurst A., Harris W.J. Protein Expr. Purif. 1999;15:336–343. doi: 10.1006/prep.1999.1035. [DOI] [PubMed] [Google Scholar]
- 92.Wang R., Xiang S., Feng Y., Srinivas S., Zhang Y., Lin M., Wang S. Front. Cell. Infect. Microbiol. 2013;3:72. doi: 10.3389/fcimb.2013.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ow D.S.-W., Lim D.Y.-X., Nissom P.M., Camattari A., Wong V.V.-T. Microb. Cell Fact. 2010;9:1. doi: 10.1186/1475-2859-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arié J.P., Sassoon N., Betton J.M. Mol. Microbiol. 2001;39:199–210. doi: 10.1046/j.1365-2958.2001.02250.x. [DOI] [PubMed] [Google Scholar]
- 95.Bothmann H., Plückthun A. J. Biol. Chem. 2000;275:17100–17105. doi: 10.1074/jbc.M910233199. [DOI] [PubMed] [Google Scholar]
- 96.Zhang Z., Song L.-p., Fang M., Wang F., He D., Zhao R., Liu J., Zhou Z.-y., Yin C.-c., Lin Q. Biotechniques. 2003;35:1032–1043. doi: 10.2144/03355rr03. [DOI] [PubMed] [Google Scholar]
- 97.Behrens S., Maier R., de Cock H., Schmid F.X., Gross C.A. EMBO J. 2001;20:285–294. doi: 10.1093/emboj/20.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tormo A., Almiron M., Kolter R. J. Bacteriol. 1990;172:4339–4347. doi: 10.1128/jb.172.8.4339-4347.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bitto E., McKay D.B. Structure. 2002;10:1489–1498. doi: 10.1016/s0969-2126(02)00877-8. [DOI] [PubMed] [Google Scholar]
- 100.Goemans C., Denoncin K., Collet J.-F. Biochimica et Biophysica Acta (BBA)–Mol. Cell Res. 2014;1843:1517–1528. doi: 10.1016/j.bbamcr.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 101.Gallo W.T., Bradley E.H., Falba T.A., Dubin J.A., Cramer L.D., Bogardus S.T., Kasl S.V. Am. J. Ind. Med. 2004;45:408–416. doi: 10.1002/ajim.20004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hennecke G., Nolte J., Volkmer-Engert R., Schneider-Mergener J., Behrens S. J. Biol. Chem. 2005;280:23540–23548. doi: 10.1074/jbc.M413742200. [DOI] [PubMed] [Google Scholar]
- 103.Bitto E., McKay D.B. J. Biol. Chem. 2003;278:49316–49322. doi: 10.1074/jbc.M308853200. [DOI] [PubMed] [Google Scholar]
- 104.Yakushi T., Masuda K., Narita S.-i., Matsuyama S.-i., Tokuda H. Nat. Cell Biol. 2000;2:212–218. doi: 10.1038/35008635. [DOI] [PubMed] [Google Scholar]
- 105.Matsuyama S.i., Yokota N., Tokuda H. EMBO J. 1997;16:6947–6955. doi: 10.1093/emboj/16.23.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Matsuyama S., Tajima T., Tokuda H. EMBO J. 1995;14:3365. doi: 10.1002/j.1460-2075.1995.tb07342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tajima T., Yokota N., Matsuyama S.-i., Tokuda H. FEBS Lett. 1998;439:51–54. doi: 10.1016/s0014-5793(98)01334-9. [DOI] [PubMed] [Google Scholar]
- 108.Okuda S., Tokuda H. Proc. Natl. Acad. Sci. 2009;106:5877–5882. doi: 10.1073/pnas.0900896106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lindberg F., Tennent J., Hultgren S., Lund B., Normark S. J. Bacteriol. 1989;171:6052–6058. doi: 10.1128/jb.171.11.6052-6058.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sauer F.G., Fütterer K., Pinkner J.S., Dodson K.W., Hultgren S.J., Waksman G. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- 111.Soto G.E., Dodson K.W., Ogg D., Liu C., Heuser J., Knight S., Kihlberg J., Jones C.H., Hultgren S.J. EMBO J. 1998;17:6155–6167. doi: 10.1093/emboj/17.21.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jones C.H., Danese P.N., Pinkner J.S., Silhavy T.J., Hultgren S.J. EMBO J. 1997;16:6394–6406. doi: 10.1093/emboj/16.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jacob-Dubuisson F., Pinkner J., Xu Z., Striker R., Padmanhaban A., Hultgren S.J. Proc. Natl. Acad. Sci. 1994;91:11552–11556. doi: 10.1073/pnas.91.24.11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kuehn M.J., Ogg D.J., Kihlberg J., Slonim L.N., Flemmer K., Bergfors T., Hultgren S.J. Science. 1993;262:1234–1241. doi: 10.1126/science.7901913. [DOI] [PubMed] [Google Scholar]
- 115.Jones C.H., Pinkner J.S., Nicholes A.V., Slonim L.N., Abraham S.N., Hultgren S.J. Proc. Natl. Acad. Sci. 1993;90:8397–8401. doi: 10.1073/pnas.90.18.8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.DANIELSON P.E., FORSS-PETTER S., BROW M.A., CALAVETTA L., DOUGLASS J., MILNER R.J., Sutcliffe J.G. DNA. 1988;1(7):261–267. doi: 10.1089/dna.1988.7.261. [DOI] [PubMed] [Google Scholar]
- 117.Davis J.M., Boswell B., Bächinger H. J. Biol. Chem. 1989;264:8956–8962. [PubMed] [Google Scholar]
- 118.Lin L.-N., Hasumi H., Brandts J.F. Biochimica et Biophysica Acta (BBA)–Prot. Struct. Mol. Enzymol. 1988;956:256–266. doi: 10.1016/0167-4838(88)90142-2. [DOI] [PubMed] [Google Scholar]
- 119.Schreiber S.L. Science. 1991;251:283–287. doi: 10.1126/science.1702904. [DOI] [PubMed] [Google Scholar]
- 120.Fischer G., Wittmann-Liebold B., Lang K., Kiefhaber T., Schmid F.X. Nature. 1989;337:476–478. doi: 10.1038/337476a0. [DOI] [PubMed] [Google Scholar]
- 121.Ou W.B., Luo W., Park Y.D., Zhou H.M. Protein Sci. 2001;10:2346–2353. doi: 10.1110/ps.23301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Justice S.S., Hunstad D.A., Harper J.R., Duguay A.R., Pinkner J.S., Bann J., Frieden C., Silhavy T.J., Hultgren S.J. J. Bacteriol. 2005;187:7680–7686. doi: 10.1128/JB.187.22.7680-7686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dartigalongue C., Raina S. EMBO J. 1998;17:3968–3980. doi: 10.1093/emboj/17.14.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Matern Y., Barion B., Behrens-Kneip S. BMC Microbiol. 2010;10:1. doi: 10.1186/1471-2180-10-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Weininger U., Jakob R.P., Kovermann M., Balbach J., Schmid F.X. Protein Sci. 2010;19:6–18. doi: 10.1002/pro.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Watashi K., Shimotohno K. Drug Target Insights. 2007;2:9. [PMC free article] [PubMed] [Google Scholar]
- 127.Nigro P., Pompilio G., Capogrossi M. Cell Death Dis. 2013;4:e888. doi: 10.1038/cddis.2013.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Trémillon N., Morello E., Llull D., Mazmouz R., Gratadoux J.-J., Guillot A., Chapot-Chartier M.-P., Monlezun L., Solé V., Ginisty H. PLoS One. 2012;7:e33516. doi: 10.1371/journal.pone.0033516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kurokawa Y., Yanagi H., Yura T. J. Biol. Chem. 2001;276:14393–14399. doi: 10.1074/jbc.M100132200. [DOI] [PubMed] [Google Scholar]
- 130.Qiu J., Swartz J.R., Georgiou G. Appl. Environ. Microbiol. 1998;64:4891–4896. doi: 10.1128/aem.64.12.4891-4896.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Missiakas D., Raina S. J. Bacteriol. 1997;179:2465. doi: 10.1128/jb.179.8.2465-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Debarbieux L., Beckwith J. Cell. 1999;99:117–119. doi: 10.1016/s0092-8674(00)81642-6. [DOI] [PubMed] [Google Scholar]
- 133.Bader M., Muse W., Ballou D.P., Gassner C., Bardwell J.C. Cell. 1999;98:217–227. doi: 10.1016/s0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- 134.Kishigami S., Kanaya E., Kikuchi M., Ito K. J. Biol. Chem. 1995;270:17072–17074. doi: 10.1074/jbc.270.29.17072. [DOI] [PubMed] [Google Scholar]
- 135.Kishigami S., Akiyama Y., Ito K. FEBS Lett. 1995;364:55–58. doi: 10.1016/0014-5793(95)00354-c. [DOI] [PubMed] [Google Scholar]
- 136.Zhuo X.-F., Zhang Y.-Y., Guan Y.-X., Yao S.-J. J. Biotechnol. 2014;192:197–203. doi: 10.1016/j.jbiotec.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 137.Kishigami S., Ito K. Genes Cells. 1996;1:201–208. doi: 10.1046/j.1365-2443.1996.d01-233.x. [DOI] [PubMed] [Google Scholar]
- 138.Rietsch A., Belin D., Martin N., Beckwith J. Proc. Natl. Acad. Sci. 1996;93:13048–13053. doi: 10.1073/pnas.93.23.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rietsch A., Bessette P., Georgiou G., Beckwith J. J. Bacteriol. 1997;179:6602–6608. doi: 10.1128/jb.179.21.6602-6608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hiniker A., Bardwell J.C. J. Biol. Chem. 2004;279:12967–12973. doi: 10.1074/jbc.M311391200. [DOI] [PubMed] [Google Scholar]
- 141.Missiakas D., Georgopoulos C., Raina S. EMBO J. 1994;13:2013. doi: 10.1002/j.1460-2075.1994.tb06471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shevchik V.E., Condemine G., Robert-Baudouy J. EMBO J. 1994;13:2007. doi: 10.1002/j.1460-2075.1994.tb06470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Heo M.-A., Kim S.-H., Kim S.-Y., Kim Y.-J., Chung J., Oh M.-K., Lee S.-G. Protein Expr. Purif. 2006;47:203–209. doi: 10.1016/j.pep.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 144.Stirnimann C.U., Rozhkova A., Grauschopf U., Grütter M.G., Glockshuber R., Capitani G. Structure. 2005;13:985–993. doi: 10.1016/j.str.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 145.Stewart E.J., Katzen F., Beckwith J. EMBO J. 1999;18:5963–5971. doi: 10.1093/emboj/18.21.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sambongi Y., Ferguson S.J. FEBS Lett. 1994;353:235–238. doi: 10.1016/0014-5793(94)01053-6. [DOI] [PubMed] [Google Scholar]
- 147.Depuydt M., Leonard S.E., Vertommen D., Denoncin K., Morsomme P., Wahni K., Messens J., Carroll K.S., Collet J.-F. Science. 2009;326:1109–1111. doi: 10.1126/science.1179557. [DOI] [PubMed] [Google Scholar]
- 148.Yoon J.Y., Kim J., Lee S.J., Kim H.S., Im H.N., Yoon H.-J., Kim K.H., Kim S.-J., Han B.W., Suh S.W. FEBS Lett. 2011;585:3862–3867. doi: 10.1016/j.febslet.2011.10.042. [DOI] [PubMed] [Google Scholar]
- 149.Shao F., Bader M.W., Jakob U., Bardwell J.C. J. Biol. Chem. 2000;275:13349–13352. doi: 10.1074/jbc.275.18.13349. [DOI] [PubMed] [Google Scholar]
- 150.Andersen C.L., Matthey-Dupraz A., Missiakas D., Raina S. Mol. Microbiol. 1997;26:121–132. doi: 10.1046/j.1365-2958.1997.5581925.x. [DOI] [PubMed] [Google Scholar]
- 151.Edeling M.A., Guddat L.W., Fabianek R.A., Thöny-Meyer L., Martin J.L. Structure. 2002;10:973–979. doi: 10.1016/s0969-2126(02)00794-3. [DOI] [PubMed] [Google Scholar]
- 152.Fabianek R.A., Hofer T., Thöny-Meyer L. Arch. Microbiol. 1999;171:92–100. doi: 10.1007/s002030050683. [DOI] [PubMed] [Google Scholar]
- 153.Guo F., Snapp E.L. J. Cell Sci. 2013;126:1429–1439. doi: 10.1242/jcs.118182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ohta M., Takaiwa F. Plant Signal. Behav. 2014;9:e28194. doi: 10.4161/psb.28194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hartl F.U. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 156.Kampinga H.H., Craig E.A. Nat. Rev. Mol. Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Boston R.S., Viitanen P.V., Vierling E. Post-transcriptional Control of Gene Expression in Plants. Springer; 1996. Molecular chaperones and protein folding in plants; pp. 191–222. [DOI] [PubMed] [Google Scholar]
- 158.Krishna P., Gloor G. Cell Stress Chaperones. 2001;6:238–246. doi: 10.1379/1466-1268(2001)006<0238:thfopi>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rutherford S.L., Lindquist S. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 160.Goloubinoff P., Mogk A., Zvi A.P.B., Tomoyasu T., Bukau B. Proc. Natl. Acad. Sci. 1999;96:13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ehrnsperger M., Gräber S., Gaestel M., Buchner J. EMBO J. 1997;16:221–229. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Inaba K., Ito K. Biochimica et Biophysica Acta (BBA)–Mol. Cell Res. 2008;1783:520–529. doi: 10.1016/j.bbamcr.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 163.Sauer F.G., Barnhart M., Choudhury D., Knight S.D., Waksman G., Hultgren S.J. Curr. Opin. Struct. Biol. 2000;10:548–556. doi: 10.1016/s0959-440x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 164.Remaut H., Tang C., Henderson N.S., Pinkner J.S., Wang T., Hultgren S.J., Thanassi D.G., Waksman G., Li H. Cell. 2008;133:640–652. doi: 10.1016/j.cell.2008.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]