Abstract
Hepatitis C virus (HCV) mortality and morbidity is a world health misery with an approximate 130–150 million chronically HCV tainted and suffering individuals and it initiate critical liver malfunction like cirrhosis, hepatocellular carcinoma or liver HCV cancer. HCV NS5B protein one of the best studied therapeutic target for the identification of new drug candidates to be added to the combination or multiple combination medication recently approved. During the past few years, NS5B has thus been an important object of attractive medicinal chemistry endeavors, which induced to the surfacing of betrothal preclinical drug molecules. In this scenario, the current review set limit to discuss research published on NS5B and few other therapeutic functional inhibitors concentrating on hit investigation, hit to lead optimization, ADME parameters evaluation, and the SAR data which was out for each compound type and similarity taken into consideration. The discussion outlined in this specific review will surly helpful and vital tool for those medicinal chemists investigators working with HCV research programs mainly pointing on NS5B and set broad spectrum identification of creative anti HCV compounds. This mini review also tells each and every individual compound ability related how much they are active against NS5B and few other targets.
Keywords: NS5B, HCV, Viral genomes, Inhibitors, Interferon, Liver cirrhosis
Graphical abstract
The prime objective of this work is to provide insights on HCV medicinal chemistry. The various chemical classes of HCV inhibitors targeting non-structural polyprotein were reviewed.
Highlights
-
•
Hepatitis C infection causes severe liver cirrhosis and carcinoma.
-
•
The new acute HCV infections are raising every year and mortality rate become serious concern.
-
•
The plausible list of anti-HCV drugs and clinical agents were listed in this review.
-
•
The divergent medicinal scaffolds as anti-HCV agents were presented as per their targets.
1. Introduction
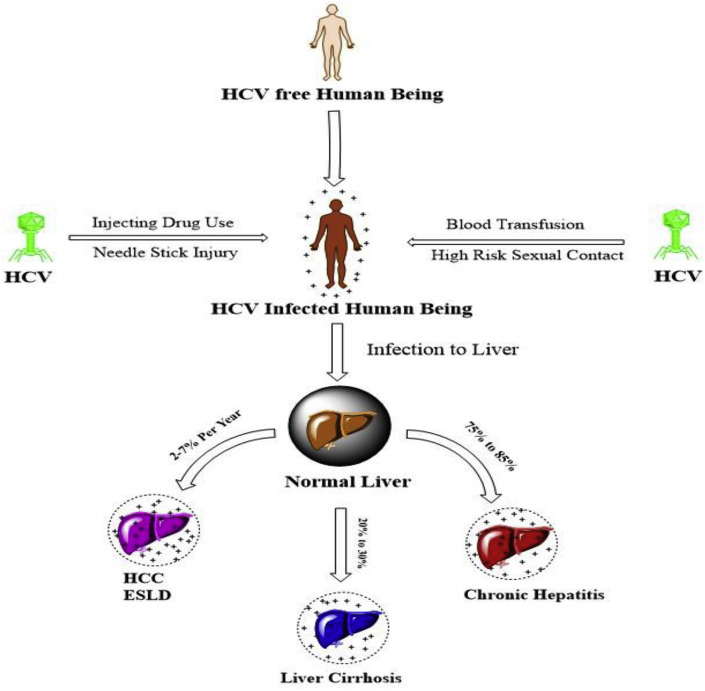
The HCV is an infectious disease caused by Hepatitis C virus, which primarily affects the liver to develop liver Cirrhosis and carcinoma. The HCV causing virus is mainly transmitted to humans by transfusion of human body fluids similar to HIV [1]. According to the world health organization (WHO), 3–5 lakh death cases among 130–150 million hepatitis C virus infected individuals are being reporting each year across the world [2]. The existence of hepatitis C was suggested in the 1970s and is discovered in 1989 [3]. Hepatitis C infects only humans, chimpanzees according to few studies it is also found the in blood samples of horses in the form of non primate hepacivirus (NPHV) [4]. About 85% viral persistence in the liver of HCV infected individuals is reported. The infection is petite, if the diagnosis become delayed it leads into chronic infection (in 70–80% of cases) and development of liver cirrhosis increases the death aspects of the infected individuals (Fig. 1 ).
Fig. 1.
Routes of transmission and severity of infection of HCV.
In few cases, those with cirrhosis will go on to develop liver failure and life threatening cancer [5]. The symptoms of HCV is not provoked immediately and thus majority of infected individuals may not be physically ill [2]. HCV infections can be quickly detected through serological assays by confirming the presence of antibodies related to HCV virus. The other virological assays include ELISA, RT-PCR and immunoblot assays can be employed for the respective detection. The vaccine therapy for prototypes of Hepatitis A and B is available and vaccination program to combat Hepatitis C is still under development. Though, the treatment has been progressed rapidly, but morbidity and mortality rates are still predicted to rise, efficacious and tolerable therapies are urgently needed [6]. The standard drug therapy for the treatment of HCV infection is combination of peginterferon (PEG-Inf) and ribavirin, with either boceprevir or telaprevir cured the 65% of HCV positive people [7]. The drug ribavirin is also a best choice for other viral diseases like respiratory syndrome caused by coronavirus [8].
2. Epidemiology
The WHO declared that, 71 million individuals among the total population are being suffering with chronic hepatitis C infections across the globe; European (1.5%), Eastern Mediterranean (2.3%) and other (0–1%) countries are under major risk for this disease (Fig. 2 a) [9]. In 2013–14, the toll of 17.5% chronic cases among the total diagnosed HCV infections in European Union has stunned the health agencies to initiate preventive strategies and treatment regimen on priority. The prime concerns in the spreading the acute and chronic HCV infections is through the contact of blood products by intravenous mode of Drug Use (IDU), unsafe surgical procedures and untested body fluid transfusion. The others mode includes sharing of injectible syringes, inadequate sterilization of medical equipments and sexual communication (Fig. 2b) [10,11]. According to Centre for Disease Control and Prevention (CDC), the number new acute infections are being increasing the United States from last few years. As per surveillance data from the CDC, the number of estimated acute infections from 1982 to 2016 and number of death cases from 2010 to 2016 has been graphed and presented Fig. 2c and d [12]. The significant number of death cases due chronic HCV infections are raising among the affected individuals is noticed. The mortality rate in Canada from 2003 to 2017 is 27.67% and the chronic situation turns into hepatic carcinoma in 36.66% HCV positive hospitalized individuals [[13], [14], [15]]. Similarly, the adult age to old age (20–60yrs) group of the male population are prone to be victims of the HCV infections than females [11]. The antiviral therapy to combat HCV infections is available and the dosage regimen criterion is varied with respect to health agencies. In recent, the difficulties in prescribing direct acting antiviral drugs (DAA) and other instructions has been directed by the European Association for the Study of the Liver (EASL) [16].
Fig. 2.
a) Countries with chronic hepatitis C infections; b) Mode of transmission of acute and chronic HCV infections, c) Estimated cases reported in USA from 1982 to 2016 and d) The number death cases reported in USA from 2010 to 2016.
3. Genome of the hepatitis C virus
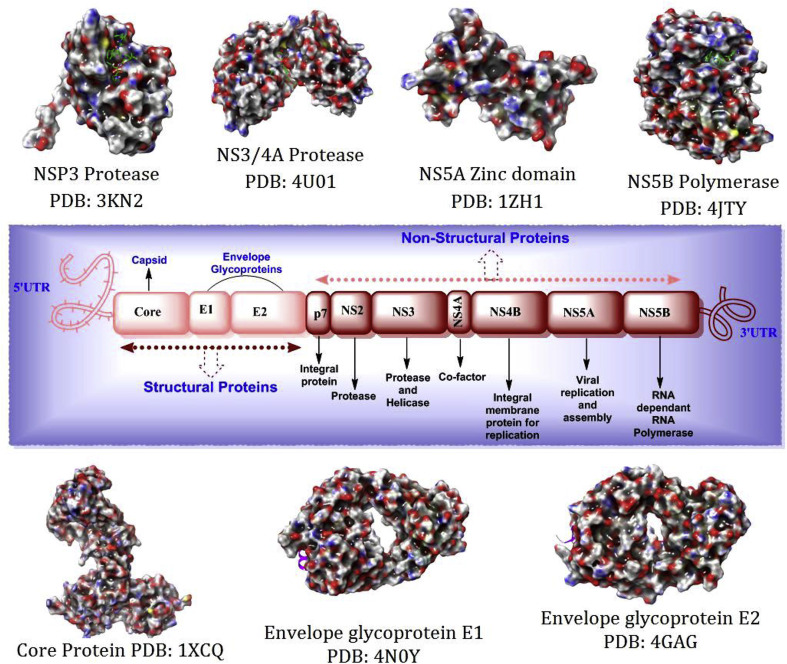
The single stranded positive sence enveloped RNA virus (HCV) is the member of hepacivirus genus and belongs to the Flaviviridae family [17,18]. Hepatitis C virus demonstrate seven types of genotypes (G1, G1a, G1b, G2, G3, G4, G5, G6 and mixed or others) in the nature [5]. Among them Genotype-1 is the most common in South America and Europe, 70% of reported cases are caused by genotype-1 in USA only, while, 20% of the population infected by genotype-2 and nearly 1% each of the other genotypes [19]. The open reading frame (ORF) of HCV genome encodes approximately 3000 amino acids; structural proteins located near to N-terminal regions and non-structural polyproteins present on left over part of the ORF. The HCV viral genome is comprised of structural (Core Protein (Capsid), E1, E2 and Frameshift protein) and nonstructural polyproteins (P7, NS2, NS3S, NS4A/NS4B & NS5A/NS5B) (Fig. 3 ). The structural polyproteins are reported to have the functions at viral entry period into the cell. Among them, core protein plays foremost task in formation of nucleocapsid-like particles (NLPs) to interact with vital cellular proteins [20,21]. The envelope glycoproteins (E1 and E2) are the predominant components of the structural proteins helpful in the viral entry and fusion into the host cell. The E2 glycoprotein networks with critical receptor proteins present on the host surface via charged interactions and is also beneficial in host recognition [22]. The existence of F frameshift protein is reported in chronic infections, role is still imprecise and under exploration.
Fig. 3.
Complete genome of the typical HCV virus and its structural and non-structural polyproteins [[23], [24], [25], [26], [27], [28], [29]].
The non-structural polyproteins are required for viral reproduction within the congregation unit. The P7 is a tiny integral membrane protein important for linking the α-helical transmembrane domains and the HCV infectivity depends on the mutations of this protein [30]. The short lived NS2 protein is accountable for associating of virion with endoplasmic reticulum (ER). It consists of zinc binding site when unite with NS3 and liable for cleavage [31]. The multifunctional role of NS3-NS4A protein by which it affects the host cell pathways made it as druggable target; it is due to occurrence of serine protease domain at N-terminal and helicase/NTPase domain at C-terminal ends. It also acts as protease to catalyze the NS3/4A, NS4A/4B, NS4B/5A and NS5A/5B intersections [32]. The NS3-4A serine protease complex involves in the cleavage activity and protective role to bypass the innate immunity of the host cell. The above specified activity of NS3 is only commenced when it is associated with NS4A as a co-factor. The NS3 helicase/NTPase has numerous functions such as stimulating RNA, RNA binding, unwinding of RNA and altering protein conformation followed by NTP hydrolysis [31]. The integral membrane protein (NS4B) anchors the transmembrane and replication complexes present on the ER [33]. The N-terminal of metalloprotein NS5A is vital for viral assembly and for interacting with RNA; the C-terminal is critical for development of lipid raft composites which is beneficial for viral replication. It also has several other functions depending on its interaction regimen with various cellular proteins [34]. The NS5B is also termed as tail-anchored membrane protein which has the RNA-dependant-RNA polymerase (RdRp) activity, significant for post-translational amendments of α-helical transmembrane situated in C-terminal. Whereas, the N-terminal of the NS5B (RdRp) has three typical binding sites termed as 'fingers', 'palm' and 'thumb' regions. The HCV viral RNA synthesis is depends on the interactions among subdomain of the thumb and finger region results in altering the catalytic site [35]. Apart from the specified roles, the NS5B could interact with numerous cellular proteins to regulate the viral replication and these affirmative facts made it as precise target for anti-HCV drug discovery programs. The in depth anti-HCV agents which are discovered or designed by targeting the non-structural polyproteins were discussed in later sections.
4. The viral life cycle
The HCV viral entry into the host cell is mediated by attaching on a few HCV receptors present on the surface. The CD81 factor, scavenger receptor B type I (SR-BI), dendritic cell-specific intercellular adhesion molecules (DC-SIGN & L-SIGN), low-density lipoprotein receptor (LDL-R), asialoglycoprotein receptor and glycosaminoglycans were experimentally proved to be interacting with HCV particles during early stages to enable the viral entry into the host cell. The fusion between viral and cellular membrane leads to release the viral nucelocapsid into the host cytoplasm by receptor-mediated endocytosis and sometimes this process is pH dependant. The breakdown of nucleocapsid releases the free positively charged RNA particles into the cytoplasm. The RNA synthesis via internal ribosome entry site (IRES) occurs by interacting with other cellular proteins. The post-translational modifications take place to execute the bulky precursor non-structural polyproteins by targeting ER membrane. The maturity of core proteins, conformational changes and cleavage into its essential components acquires in this stage. The formation of HCV replication complex is another key step in viral reproduction initiated by NS4B protein by recruiting the cellular proteins to provoke vesicle formation. The replication complex is also formed by protein-protein interactions. The NS5B RdRp plays the significant deal in aggregation of positively charged single-strand RNA particles by various mechanisms. The maturity of the virions and viral assembly commenced in the ER membrane and Golgi apparatus [31,36].
5. Some potent drugs used for treat hepatits C disease
The current anti-HCV therapy includes with the combination of pegylated interferon (Peg-IFN), ribavirin (RBV) and two recently approved antiviral agents, boceprevir or telaprevir (targeting the viral NS3/4A protease) [37]. Sustained virologic response (SVR) –which is tantamount to cure is achieved only in a subset of treated patients, depending on a combination of viral and host-cell genetic factors [37]. For example, a human polymorphism at the IL28B gene is associated with poor interferon response [37]. Almost all patients undergoing interferon-based treatment also experience detrimental adverse effects, including flu-like symptoms, anemia, and depression [37]. Thus, current anti-HCV therapies are often not tolerated and becoming ineffective for most of the patients, and novel direct-acting antiviral drugs are required for safer, more efficacious treatment [37]. Direct-acting antiviral agents (DAA) have the potential to improve SVR rates and minimize treatment duration. As HCV NS3/4A protease cleaves the four known sites along the virally encoded polyprotein [38] and also hydrolyzes two human proteins, TRIF and MAVS, which are part of the innate immune system, there by confounding the innate immune response to viral infection [37]. Inhibition of the HCV NS3/4A protease (chymotrypsin-like serine protease) is a prime strategy for many of structure-based therapeutic agents.
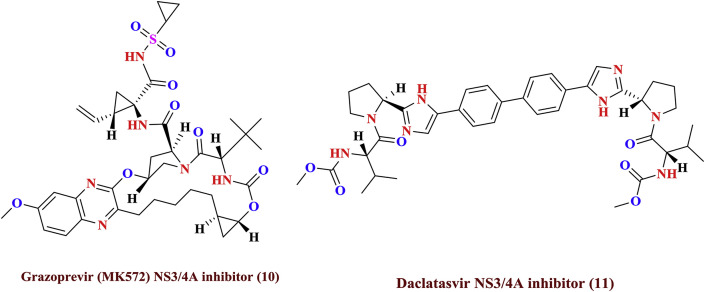
In general, treatment is recommended for those with proven HCV infection and signs of liver inflammation [39]. As of 2010, treatments consist of a combination of pegylated interferon α and the antiviral drug ribavirin for a period of 24 or 48 weeks, depending on HCV genotype 1 b. The cure rates by resulting treatment are reported as 70, 80% for genotype 2, 3, and 45–70% for other genotypes respectively [40]. The amalgamation of ribavirin and pegylated interferon-α-2a evidenced as superior procedure for treating HCV infections than grouping with pegylated interferon-α-2b [41]. Combining either boceprevir or telaprevir with ribavirin and pegylated interferon-α improves antiviral response for hepatitis C genotype-1 [42]. Adverse effects with treatment are common, with half of people getting flu like symptoms and a third experiencing emotional problem. Treatment during the first six months is more effective than once hepatitis C has become chronic [43]. If someone develops a new infection and it has not cleared after eight to twelve weeks, 24 weeks of pegylated interferon therapy is recommended [43]. In people with thalassemia, ribavirin appears to be useful but increases the need for transfusions [44]. Some of the targeted anti-HCV agents (1 to 11) indicated for treatment are shown in Fig. 4 and Fig. 5 .
Fig. 4.
Presently available potent drugs for HCV treatment.
Fig. 5.
Presently available potent drugs for HCV treatment.
Sofosbuvir with ribavirin and interferon appears to be around 90% effective in those with genotype 1, 4, 5, or 6 disease [45]. Sofosbuvir with just ribavirin appears to be 70–95% effective in type 2 and 3 disease [40,45].
This benefit is somewhat offset by a greater rate of adverse effects [6]. Treatment that contains ledipasvir and sofosbuvir for genotype 1 has success rates of around 93–99% but is very expensive [45]. In genotype 6 infection, pegylated interferon and ribavirin is effective in 60–90% of cases [46]. There is some tentative data for simeprevir use in type 6 disease as well [46].
6. Nucleoside, NON-NUCLEOSIDE and protease inhibitors as anti- hepatitis C viral agents in clinical trials
The goal of treatment for HCV infections is by eradicating the viral infections, which can be done by using a combination of drugs. The current standard-of-care involves taking ribavirin plus one of two newly approved therapies, Sovaldi (sofosbuvir) or Olysio (simeprevir), and in many cases pegylated interferon as well. The length of treatment, which can range from 12 to 48 weeks, depends on the person's HCV genotype (genetic structure of the virus), whether the person is eligible to take interferon and whether he or she is waiting for a liver transplant. Many other hepatitis C drugs are in the late stages of development. These medications are called “direct-acting antivirals,” or DAAs, because they are designed specifically to attack the viral proteins. As more DAAs become available, many people may be able to combine these highly effective medications without the need for either pegylated interferon or ribavirin, which is hopeful news because those two drugs, especially interferon, can cause serious side effects. Here's more specific information about each type, or class, of approved HCV treatment along with drugs in the late stages of development.
Interferons: Interferons are the protein substances made by the immune system and named by their specific ability to interfere with viral reproduction. In addition, interferon signals the immune system to recognize and respond to microorganisms, including viral and bacterial infections. Infected cells release interferon to trigger the immune response. There are three types of interferon: α, β and γ. Interferon α is used to treat viral hepatitis and some types of cancer. Ex: Interferon α, Interferon β and Interferon γ.
Numbers of pharmaceutical companies have invested sententious effort in developing HCV NS3/4A protease inhibitors. As mentioned below boceprevir and telaprevir are NS3/4A Protease inhibitors recently approved by Food and Drug Administration, highlighting a vital milestone in the treatment of HCV infection and drug development for the driven two decades [37]. Both boceprevir and telaprevir are linear ketoamide compounds that form a reversible, covalent bond with the catalytic serine of NS3/4A protease [37]. Development of resistance is the one of the significant problem seen in the majority of patients undergoing protease inhibitor therapy and is suspected due to mutations in the NS3/4A region [1]. The R155K mutation in genotype 1a causes resistance against nearly all inhibitors and it is rarely observed in genotype 1b patients [37]. The distinct set of mutations stirred in genotype 1b patients depending on the class of protease inhibitor used; A156 mutates in response to treatment with linear ketoamide protease inhibitors while macrocyclic inhibitors more commonly selective for D168A and R155K variants. Mutations at V36, T54, and V36 + A155 are also associated with resistance to ketoamide inhibitors [37].
Several non-covalent protease inhibitors have also advanced into human clinical trials (12, 13); these inhibitors include both linear BI 201335 (14) [37], BMS-650032 (15) [1], and macrocyclic compounds, containing either a P1—P3 danoprevir (16), TMC435(17) or a P2—P4 vaniprevir (18), MK-5172 (19) macrocycle [37]. Proof of concept of antiviral efficacy was first demonstrated in 2002 with the macrocyclic inhibitor BILN-2061 (ciluprevir) (20) [38,47], which was later discontinued due to concerns about its cardiotoxicity [37]. The pan HCV inhibitor (21) was originated from the structural modification of class of peptide based HCV protease inhibitors and exhibited the potent antiviral activity against both genotypes of 1b and 3a (EC50 = 0.62 and 0.26 nM).
The other protease inhibitors include anusaprevir with (15) IC50 = 1 & 4 nM against GT- 1 & 4, ciluprevir (20) EC50 = 9.5 nM examined as good anti-HCV agents. The NS3/4A protease inhibitors rapidly reduce HCV RNA titers when administered as monotherapy and substantially improve SVR rates when given in combination with Peg-IFN and RBV. The mechanism of single-site mutations at residues R155, A156 and D168 confer resistance against most of HCV protease inhibitors has not been elucidated in atomic detail [37]. The macrocyclic molecules like danoprevir (12), MK-5172 (10) and vaniprevir (18) exhibited significant activity against HCV with IC50 = 0.24, 0.34 and 3 nM respectively [37]. Few benzimidazole derivatives were illustrated with high activity against HCV (22 & 23, EC50 = 0.83 nM) [47]. The four chemically representative of protease inhibitors are telaprevir (4), MK-5172 (10), danoprevir (12) and vaniprevir (18) exhibit distinct susceptibilities to the protease variants R155K, A156T and D168A.
Recently discovered thienoimidazole derivatives (24) [48] were found to be active against NS5A of HCV EC50 = 17, 8 nM (GT-1a, GT-1b respectively), hydroxamino-α-pyranone carboxamides (25) also exhibited activity against HCV with EC50 = 0.18 μM [49]. The pyrimidine based sulfonamide (26) analogues were also tested for their anti-HCV efficacy. The analogues of benzimidazole, thienoimidazole, pyranone, benzidine and other potent class of anti-HCV agents (21 to 26) were provided in Fig. 6, Fig. 7, Fig. 8 .
Fig. 6.
NS3/4A Protease Inhibitors, part-I.
Fig. 7.
Benzimidazole and thienoimidazole-based HCV NS5A inhibitors.
Fig. 8.
Recently published anti-HCV molecules.
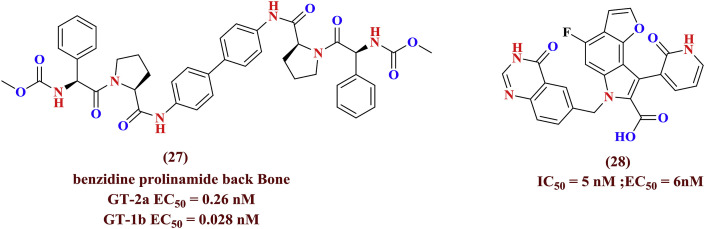
The benzidine prolinamide derivatives exerted anti-HCV properties against genotypes 2a and 1b demonstrated its potency towards genotype 1b (EC50 = 0.028 nM). Other heterocyclic molecules (27, 28) (Fig. 9 ) [50] are showed good result against HCV (EC50 = 0.26 nM, 6 nM and IC50 = 5 nM) infections respectively [51].
Fig. 9.
Recently published anti-HCV molecules.
An effort to find a precise HCV NS5B polymerase inhibitor, the indole derived libraries were screened and a hit was co-crystallized (28–1, IC50 = 53 nM, EC50 = 4800 nM) with enzyme was elaborated the accurate role of pyridone carbonyl group with active site residues. The better pharmacokinetic parameters of another indole pyridone carbonyl compound (28–2, IC50 = 5 nM, EC50 = 90 nM) with irreversible enzyme inhibition was reported. Its potency was increased by 7-fold due to supplementary hydrophobic interactions. The SAR studies and lead optimization of 28–2 made another potent compound (28–3, IC50 = 8 nM and 28–4, EC50 = 1 nM) [[52], [53], [54]]. Instead of carboxylic acid, presence of bulkier sulfonamides at C-2 position of indole ring (28–5, IC50 = 39 nM, EC50 = 11 nM) was found to be significant for increasing the HCV NS5B enzyme inhibition [55]. The incorporation of furone ring on the indole nucleus (28–6, IC50 = 6–15 nM) was reported as positive sign for exerting NS5B polymerase inhibition [56]. The pyridone anti-HCV agents were depicted in Fig. 10 .
Fig. 10.
Pyridone anti-HCV agents as NS5B inhibitors.
The peptide based derivatives (29) (IC50 = 70 nm), heterocyclic compounds like thiaphene dihydroxy pyrimidine derivatives (30, IC50 = 2.6 μM) [57], thiaphene dihydroxy pyrimidine derivatives (31 & 32, IC50 = 0.14 & 0.15 μM) [57], thiazline dihydroxy pyrimidine derivatives (33, IC50 = 0.76 μM), were evaluated as HCV NS5B inhibitors (29 to 33) (Fig. 11 ).
Fig. 11.
Peptide and thiaphene-dihydroxy pyrimidine derivatives as HCV NS5B inhibitors.
At initial oxopyrimidine derivatives (34 & 35, IC50 = 27 & 3.8 μM) were investigated as HCV NS5B inhibitors. An additional hetercyclic agents such as pteridine string of molecules (36 & 37, IC50 = 15 & 0.5 μM) proved as efficient HCV NS5B inhibitors [57]. The other class of N-acylopyrrolidine derivatives (38 & 39, IC50 = 0.3 & 12 μM) are screened as potent anti-HCV agents. The representatives chemical structures of compound 34 to 39 were depicted in Fig. 12 .
Fig. 12.
Oxopyrimidine, Pteridine N-acylpyrrolidines derivatives as HCV NS5B inhibitors.
The benzofuran derivatives (40, EC50 = 0.02–0.5 μM), cinnamides as metal chelators (41) [58], and benzamide derivatives (42) were exhibited fascinating activity against HCV infections. The chemogentic based NS5B RNA polymerase inhibitors were discovered as HCV replication inhibitors but the significant binding analysis was not mentioned (42–1, EC50 = 1.1 μM) [59] (Fig. 13 ). The structure-based docking approach played a major role in identification of mercaptobenzoxazole (42–2, IC50 = 2.01 μM) as polymerase inhibitor [60].
Fig. 13.
HCV NS5B inhibitors part-III.
An allosteric site of NS5B emerged as attractive target for structure-based design and discovery of small antiviral agents. Inhibition of the conformational changes of polymerase allosteric site required in the protein synthesis is become valid strategy to develop new medications. Quinoline derivatives also proven as having an important anti-HCV nature (43, IC50 = 0.008 nM in the allosteric site of NS5B enzyme) [58]. Pyranoindole (44) and benzimidazole derivatives (45 & 46, IC50 = 3.5 μM & < 6 nM) [58] are proved to be interacting with the allosteric site [61] of the NS5B target of HCV to exert anti-HCV activity (Fig. 14 ) [58].
Fig. 14.
Allosteric inhibitors of Polymerase enzyme.
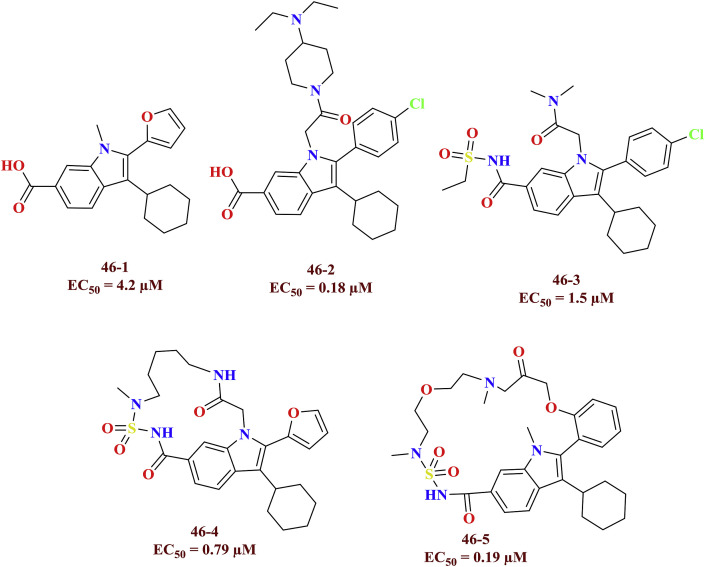
The indole carboxylic acid linked with furon (46–1), aryl rings (46–2) and sulfonamide moieties (46–3) were exhibited vital allosteric inhibition at finger loop region of the NS5B polymerase. The macrocyclic HCV NS5B polymerase inhibitors (46–4 and 46–5) were discovered based on the previously tested anti-HCV molecules as finger loop inhibitors [62]. The HCV NS5B finger loop inhibitors were shown in Fig. 15 .
Fig. 15.
HCV NS5B finger loop inhibitors.
The vital role of internal ribosome entry site (IRES) in protein synthesis via cap mediated conversion of the viral genomic RNA made it as elegant druggable target. The benzimidazole derivatives (47 & 48 EC50 = 1.5 μM) were identified as altering the conformations of IRES domain II and become promising scaffolds which disrupts ribosomal assembly. Similarly, a set of imidazopyrimidines (49, EC50 = 5.4 μM), other heterocyclic molecules i. e, limiquimid (50 EC50 = 1 μM) and sumitomo (51, EC50 = 20 nM) [58], deazapurine based small molecules (52 & 53, EC50 = 100, & 102 nM), tested as positive HCV IRES-IIA Sub domain inhibitors and they are presented in Fig. 16 .
Fig. 16.
HCV IRES-IIA Sub domain inhibitors and agonists.
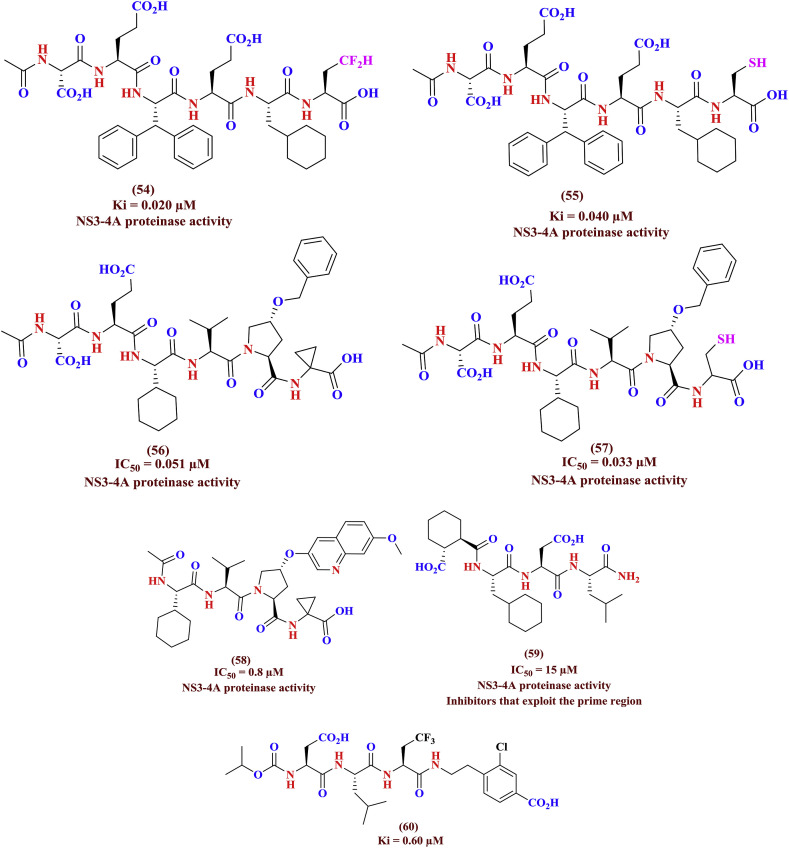
Variations in the patterns of resistant mutations arise from the complex interplay between genotype, replication rates, mutation rates, and the resulting effect of mutations on viral fitness and drug potency. Clearly, despite the benefits of combination therapy in improving SVR rates, the emergence of resistance challenges the long-term efficacy of NS3/4A protease inhibitors is being noticed [37]. The protease inhibitors danoprevir, TMC435, and boceprevir protrude from the substrate envelope in regions that correlate with known sites of resistance mutations. Notably, the large P2 moieties of danoprevir and TMC435 bind in the S2 subsite and extensively interact with residues R155, D168, and A156, which mutate to confer multi-drug resistance. The discovery of peptide based boceprevir as NS3-4A protease inhibitor by structure-based techniques produced new juncture for design few more penta and hexa peptide based anti-HCV agents. The typical α-ketoacid (54) derived from above strategy yielded the anti-HCV activity (Ki = 0.020 μM) and replacement of α-ketoacid with cystiene (Ki = 0.040 μM) (55) also retained the antiviral efficacy and found less when compared with parent scaffold (54). The modification of peptidyl cystiene, other P2 residue with cyclic proline and its allied esters were tested as HCV NS3-4A protease inhibitors (56 to 58). The alteration of phenyl group of P3 residue (56, 57) (IC50 = 0.051, 0.033 μM) with other aryl ring such as quinoline (58, IC50 = 0.8 μM) was reported as not favorable for increasing the anti-HCV efficacy. Further, the tri and tetrapeptidyl agents (59, IC50 = 15 μM) were tested as least active compounds when compared with other peptide inhibitors. The 2-amino butyric acid on tripeptide agents (60, Ki = 0.60 μM) shown to have significant role in inhibition of NS3-4A protease enzyme. The peptide based NS3-4A protease inhibitors described against HCV infections were shown in Fig. 17 .
Fig. 17.
Peptide based NS3/4A Protease Inhibitors.
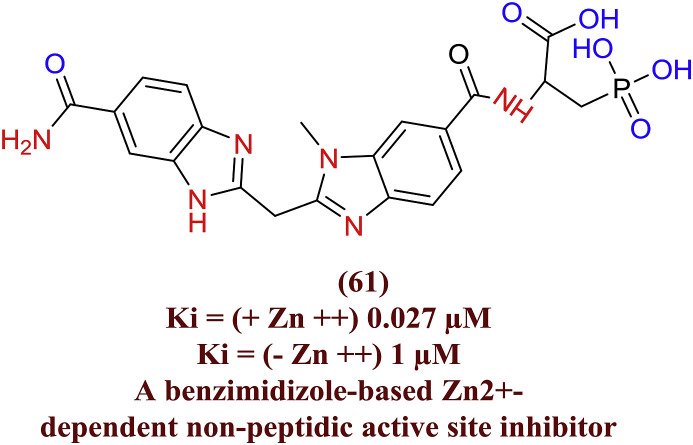
The potent serine protease phosphonoalanine based bisbenzimidazolemethane analogues (61) displayed HCV NS3-4A protease inhibition and it is zinc dependant (Zn2+). The importance of bidentate benzimidazole and their interaction with NS3-4A protease catalytic residues (His57 and Ser195) were experimented as crucial for the inhibition of enzyme. They also investigated the positive role of Zn2+ in presence of peptide based protease inhibitors (Fig. 18 ) [63].
Fig. 18.
Bisbenzimidazolemethane analogues as HCV NS3-4A protease inhibitor.
The benzimidazole-5-carboxylic acid analogue (62) was initially screened as HCV NS5B polymerase inhibitor with IC50 = 1.6 μM. The molecular volume expansion to achieve the desirable interactions essential to induce the conformational changes of polymerase lead its significant carboxamide derivatives (62–1, IC50 = 0.019 μM) as much potent precise NS5B polymerase inhibitor [64]. In another aspect, the α,γ-Diketo acids such as dichloro derivative (63, IC50 = 45 nM) revealed as specific and reversible inhibitor of HCV NS5B polymerase enzyme [65]. The quinazoline carboxaimde (63–1, IC50 = 15.7 μM) was initially tested as antiviral agent targeting HCV NS5B polymerase [66]. Later it was as metal ion chelating agent of HCV NS5B polymerase has been described by it mechanistic studies and revealed that it has multiple functions in the receptor site [67]. The compound 62 to 63–2 were shown in Fig. 19 .
Fig. 19.
Benzimidazole, α,γ-Diketo acids and quinazolines as HCV NS5B polymerase inhibitors.
The benzothiadiazine scaffold is the major role makers in most of the diuretics exists in the market. The antiviral efficiency of benzothiadiazines among its other activities such as anticancer and CNS effects made it a new class of scaffold in the HCV medicinal chemistry. In this context, high throughput screening (HTS) of proprietary library from GlaxoSmithKline (GSK) against HCV polymerase assay yielded the promising class of quinolone based benzothiadiazine analogues (64, IC50 = 200 μM) as promising NS5B inhibitors. The bulkier alkyl substitution on nitrogen of quinolone was investigated as crucial for the identified class of benzothiadiazine analogues. The structural activity relation (SAR) studies and other lead optimization strategy afforded much strong NS5B inhibitors (65 & 66, IC50 = < 0.01 μM). Since, the function of bulkier alkyl chain explained as vital in inhibiting the NS5B polymerase and the its conversion with other parallel molecular size chains has been demonstrated (67 to 69, IC50 = < 0.3 μM) as successful approach. Further, surrogating quinolone with naphthyridine (70), pyridine (71) and pyridazine (72, 73) rings and their NS5B polymerase inhibitory activity were detailed as meaningful to amplify the antiviral properties of benzothiadiazine analogues. The position of imidazopyridazine (74), imidazopyrimidine (75), quinazolinone (76), naphthalene (77 to 79) and bridged quinolone (80) agents were proved to be advantageous anti-HCV agents. The reinstating of core with diazaphosphinin (81) might not be significant for NS5B polymerase inhibition when compared with earlier benzothiadiazine analogues. The effect of pyrrole ring at quinolone's position (82) by keeping constant, swapping benzothiadiazine nucleus with benzisothiazole (83) and diazaphosphinin (84) has been explored for the NS5B inhibitory efficiency. The comprehensive review on NS5B polymerase inhibitors was previously mentioned by D. Das et al. [68] and were depicted in Fig. 20 .
Fig. 20.
Benzothiadiazine class of molecules as HCV NS5B, Palm site inhibitors.
The new set of heterocyclic small molecule non-nucleoside agents (85) against HCV NS5B polymerase were identified by combined high throughput screening (HTS) and structure-based drug design (SBDD) approaches. The lead modifications to improve the hit analogue succeeded to exert potent class of acryalamide derivatives (86) as NS5B polymerase inhibitors were presented in Fig. 21 [69].
Fig. 21.
(2Z)-2-Benzoylamino-3-(4-phenoxyphenyl) acrylic acid palm site NS5B inhibitors.
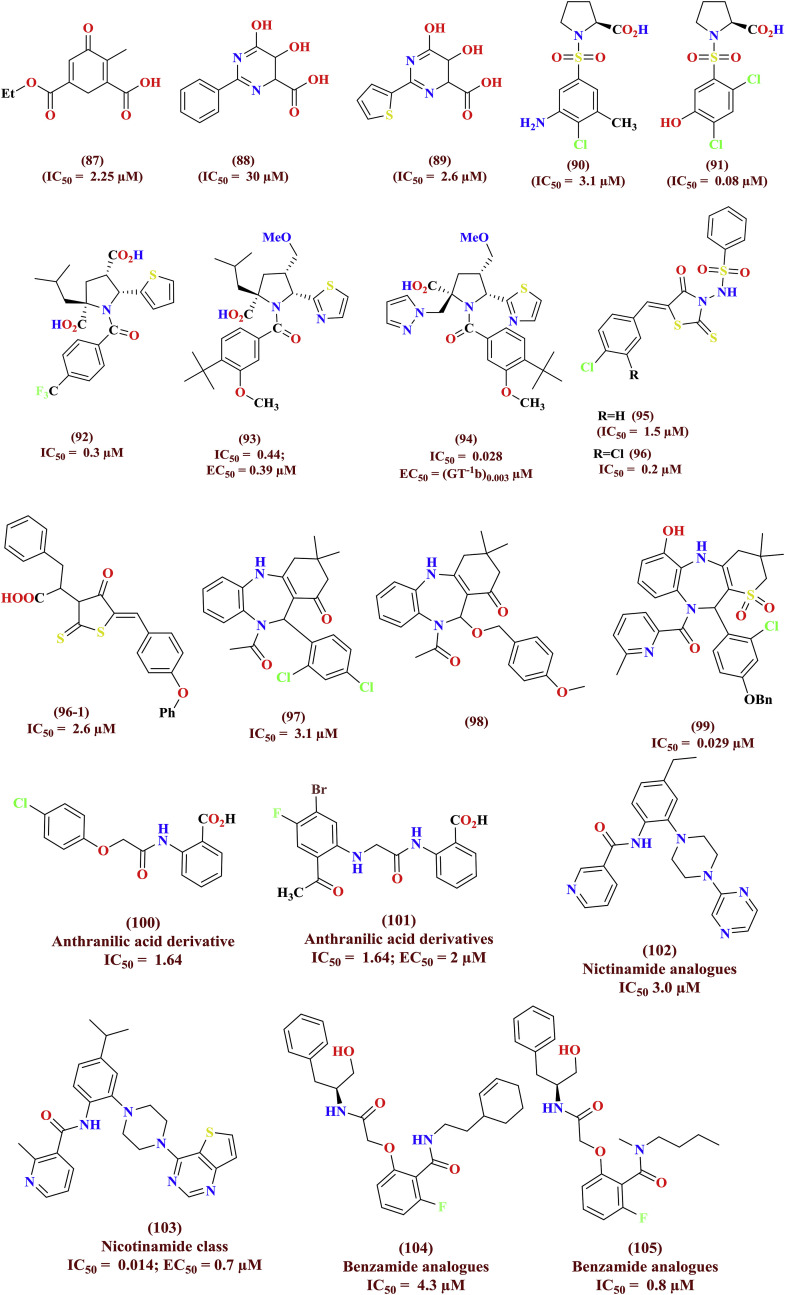
The diketoacid such as mecanoic acid (87) was tested as inhibitor of HCV NS5B polymerase, later it converted with dihydroxypyrimidinecarboxylic acid (88) and structure optimization led thiophene derivative (89) as potent HCV inhibitor. The HTS based assay of a series of libraries against HCV NS5B polymerase offered proline lniked sulfonamide (90, IC50 = 3.1 μM) and found to selective towards HCV when compared with other polymerases. The SAR studies and lead optimization protocol afforded hydroxy derivative (91, IC50 = 80 nM) to potent NS5B polymerase inhibitor over identified HIT [70]. The antiviral N-acylpyrolidine library (92 & 93) afforded clinical candidate (94) against HCV infections were discovered by HTS screening. The HTS assay against HCV NS5B polymerase identified a series of rhodanine analogues (95, 96 and 96–1) as promising antiviral agents and optimized by having chloro substitutions as essential for efficiency [71]. The benzodiazepine antipsychotic scaffold (97 to 99) targeting NS5B polymerase was scrutinized via SBBD platform and experimented as anti-HCV agents. The other palm site amide based NS5B inhibitors include anthranalamides (100 & 101), nicotinamides (102 & 103) and benzamide (104 & 105) were illustrated in Fig. 22 . The comprehensive literature on diverse scaffolds as HCV NS5B polymerase inhibitors were reviewed by Sofia et al. [72].
Fig. 22.
Divergent class of heterocyclic NCEs as palm site NS5B inhibitors.
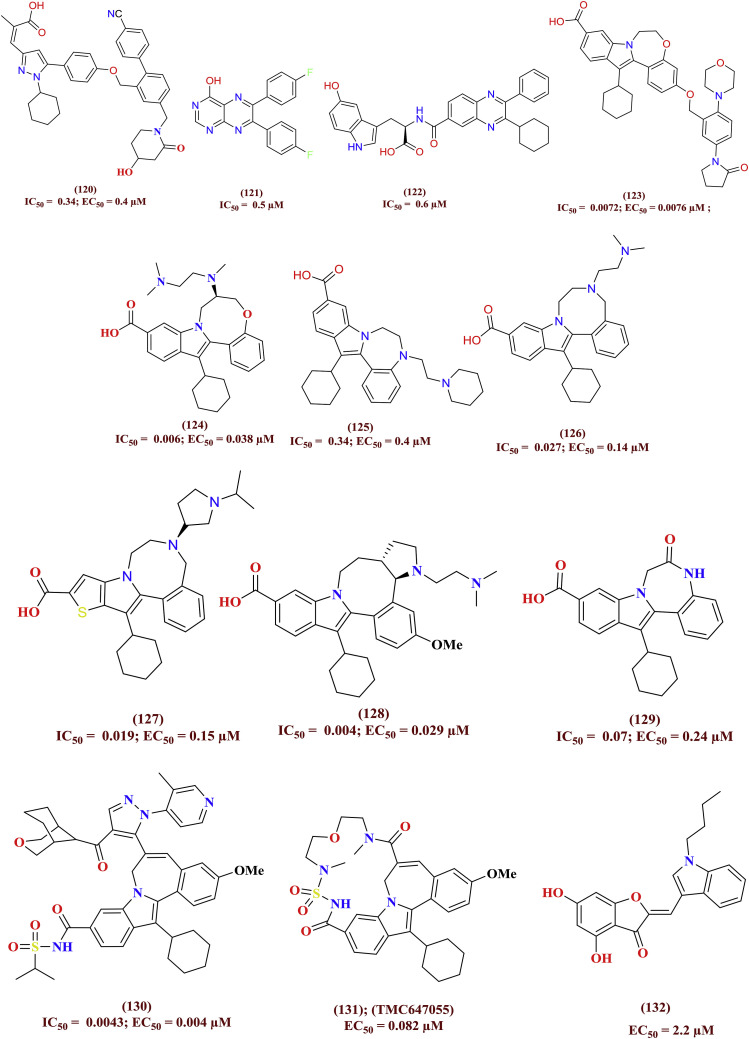
The mechanism behind binding of non-nucleoside inhibitors is to block the GTP activity within the thumb site I of the HCV NS5B polymerase. With respect to these aspects, the varieties of heterocyclic small molecules were tested for their polymerase inhibition capabilities. Among them benzimidazole (106 to 111), indole (112 to 116), thienopyrrole (117 to 119), pyrrazole (120), pteridine (121), quinoxoline (122), benzoxazapinoindole (123, 124), benzodiazepinoindole (125 to 129), indole sulfonamide (130, 131) and aurone (132) derivatives were reported to have significant NS5B polymerase inhibitory activity. A few potent among thumb inhibitors were entered in clinical trials. The molecular levels interactions and their allied biological profile has been presented by Sofia et al. [72]. The precise molecular structures of the HCV NS5B inhibitors (106 to 132) were shown in Fig. 23 .
Fig. 23.
Divergent class of heterocyclic molecules as NS5B thumb site-I inhibitor.
The precise role of thumb II site NS5B inhibitors is under investigation. The small heterocyclic agents include dihydropyrone (133 to 136), benzamide (137), thiophene (138), rhodanine (139 to 143), pyranoindole (144 to 147), carbazole (148), benzoisoquinolone (149, 150), Quinoline (151, 152), imidazolepyridine (153 to 156), thiophene based amides (157, 158) and anthranilic acid derived amides (158–1 to 158–6) [[73], [74], [75], [76]] were investigated as crucial anti-HCV agents. The plausible thumb site II NS5B inhibitors were presented in Fig. 24 .
Fig. 24.
Divergent class of heterocyclic molecules as NS5B thumb site-II inhibitor.
7. Recent patent updates ON HCV inhibitors
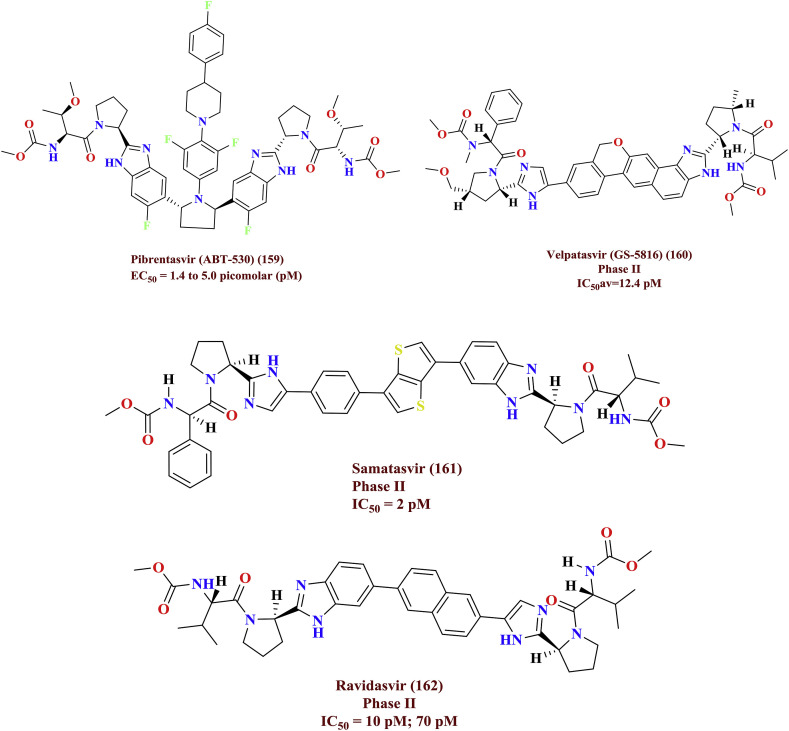
Pibrentasvir (159, ABT-530) is a novel, pan-genotypic hepatitis C virus (HCV) NS5A inhibitor exhibited against all genotypes 1 to 6 with EC50 ranging from 1.4 to 5.0 pM concentrations. Combination of pibrentasvir with other class of HCV inhibitors yielded synergistic inhibitory effect on HCV replication. In summary, pibrentasvir turn out to be a next generation HCV NS5A inhibitor which evidenced an anti-HCV activity against pan-genotypes [77]. Velpatasvir (160) demonstrated the potency against genotype 2 and 3 with IC50 av = 12.4 pM; it also repressed three other mutated forms without altering its inhibitory concentrations IC50av = 12.3 pM [78]. Whereas, the elevated inhibitory levels of samatasvir (161, IC50=≤2 pM) against all genotypes except 4a and 6a has been investigated [79]. Ravidasvir (162, IC50=10 pM) is also a similar analogue to samatasvir is in phase II trials. The phase II trial compounds were depicted in Fig. 25 .
Fig. 25.
Upcoming prominent inhibitors for Hepatitis C viral disease.
The benzofurone based piperidinyl (163, IC50=56.91 & 34.3 nM) and pyridooxazinyl (164, IC50=3.21 & 2.43 nM) derivatives demonstrated HCV NS5B inhibitory activity against genotype 1a and 1b respectively [80,81]. An indole analogue BI 207524 (165, EC50=11 & 29 nM) was discovered as a potent HCV NS5B polymerase thumb pocket 1 inhibitor, the pharmacokinetics profile with respect to clearance of the compound is very low and later its clinical investigation has been halted [82]. The genotoxicity of the clinical candidate BI 207524 further reduced by the structural alterations and this ideology afforded non-genotoxic HCV NS5B thumb site I inhibitor (165, EC50=27 & 14 nM) [83]. The preclinical investigations of potent drug candidate beclabuvir (BMS-791325) (166, EC50=3 & 6 nM) appeared as safe for human use and has been approved for HCV treatment in Japan. The beclabuvir based BMS-961955 (167, EC50=4.3 & 7.9 nM) fluro compound is advanced to clinical trials. The representative structures of 163 to 167 were provided in Fig. 26 .
Fig. 26.
HCV NS5B inhibitors as potent clinical candidates.
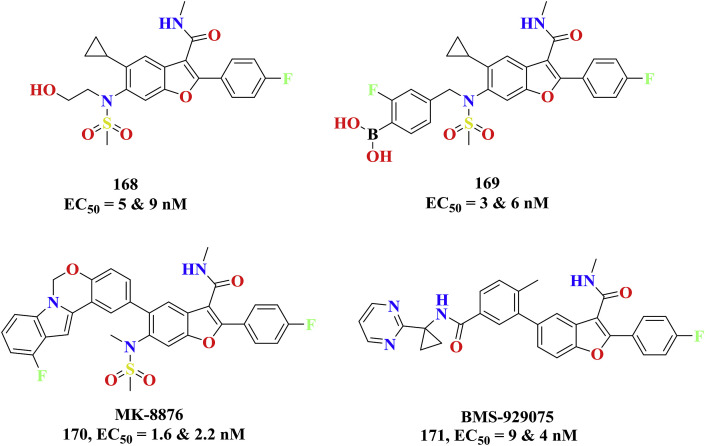
The benzofurone group of substance HCV-796 entered into the clinical trials after displaying the prerequisite HCV NS5B inhibition and pharmacokinetic profile. The combination therapy with other anti-HCV agents, interferon were studied and the due to the liver toxicity concerns it has been abandoned from the trials (168, 1a & 1b, EC50=5 & 9 nM) [84]. The potency of HCV-796 compound was further increased to 5 fold by modifying the aliphatic alcohol with boronic acid (GSK5852) (169, 1a & 1b, EC50=3 & 6 nM) and resulting structure was successful to inhibit the mutated HCV isoforms [85]. The ring extension of cylcopropyl of HCV-796 with bulkier group like oxazinoindole yielded MK-8876 as new potent clinical candidate as HCV NS5B polymerase inhibitors against both genotypes of wild (170, 1a & 1b, EC50=1.6 & 2.2 nM) and mutant type (EC50=2.7 & 1.1 nM) [86]. Additionally, the replacement of oxazinoindole of MK-8876 with linear aryl rings provided BMS-929075 as pan HCV NS5B inhibitor (171, 1a & 1b, EC50=9 & 4 nM) as clinical candidate with improvised oral bioavailability and pharmacokinetics [87]. The benzofurone clinical investigative HCV NS5B candidates were shown in Fig. 27 .
Fig. 27.
HCV NS5B inhibitors as potent benzofurone clinical candidates.
The introduction of hetero atom at C6 position in the above benzofurone (benzofurone ring) category of anti-HCV agents led another series of potent HCV NS5B inhibitors as new series of clinical agents. Among them BMS-986139 (172, EC50=9 & 3 nM) was become ineffective at the stage phase I due to microcrystallization problems in toxicological studies. The second generation of pan HCV NS5B polymerase inhibitors i. e, BMT-052 against both genotypes (173) were developed by modifying with addition of deuterium group in BMS-986139 and observed to be efficient in its metabolic stability and has escalating HCV NS5B enzyme inhibition (EC50 = 4 & 4 nM) [88]. The non-nucleotide NS5B palm site inhibitors dasabuvir (174) has been approved for the treatment of the HCV infections and can be safely administered with other anti-HCV agents. The chemical structures of the 172–174 were provided in Fig. 28 .
Fig. 28.
HCV NS5B inhibitors as potent pyridofurone conjugates.
8. Rationality for selection of current scaffold and designing
2-pyranone [89] is a six-membered cyclic unsaturated ester that shares chemical and physical properties reminiscent of alkene and aromatic compounds. It is highly abundant in bacteria, microbial, plant, insect and animal systems and takes part in many different types of biological processes such as key biosynthetic intermediates, and as metabolites. Derivatives of 2-pyrones are utilized as precursors for the production of biologically significant compounds such as pheromones [90], α-chymotrypsin [91]. Numerous reviews are existing on microbial 2-pyranones, dihydro-2-pyranones and secondary metabolites of fungi [90] as a wide range of anti-fungal, cytotoxic, neurotoxic and phytotoxic agents. Investigations with respect to their anti-tumor and HIV protease inhibiting qualities [91] have enlightened the medicinal importance of 2-pyrones. Therefore, we previously scrutinized this skeleton as design, computer aided structural exploration [92,93] and synthesis of pyranine based new carboxamide analogues for the discovery of antiviral drug candidates. The plausible approach for 2-pyronone scaffold with existing potent anti-HCV agents has been sketched in Fig. 29 .
Fig. 29.
Potential clinical molecules can be used to design derive the hybrid molecules [50,[94], [95], [96], [97]].
9. Conclusion and future perspective
The cases with new acute infections are being raising every year and the mortality rate against chronic HCV infections are distressing the several nations. The mutations in the HCV viral strains, immune response to HCV disease are alleged be a foremost rate limiting issues for ongoing anti-HCV therapy and the precise, inexpensive pan-genotype direct acting anti-virals are required at the earliest. In this context, several direct acting HCV NS5B RNA polymerase inhibitors are employed as essential anti-virals that can be utilized either in blending with PEG-IFN, RBV or in mix with other DAAs in without interferon regimens. Similarly as with the advancement of very dynamic antiretroviral HIV treatment, the anti-HCV therapy would require to combat with the pan-genotype HCV viral burdens. This process delivered the several HCV NS5B polymerase inhibitors including both nucleoside and non-nucleoside based inhibitors. A few new class of nucleoside HCV inhibitors exhibited promising anti-HCV regimen and are in human clinical trials. The development of nucleotide prodrugs to deliver nucleosides into hepatocytes is another powerful approach in nucleoside drug discovery and is useful in treating the liver associated HCV infections. The remarkable nucleoside 5′-monophosphate prodrugs has been entered into the clinical improvement and has indicated an extraordinary adequacy. In overall aspects, the current review might provide the significant information on the existing anti-HCV treatment strategy, development of target based anti-HCV agents and the other clinical agents.
Acknowledgements
NMG,TB Thanks CSIR India. SSJ is thankful for the Science and Engineering Research. Board (SERB) for the funding project (PDF/2017/001556). Tuniki Balaraju greatly thankful IISER-K/DoFA/PDF/DCS/2014/1197 and remembers Dr. Ashoke Sharon, Dr. Chandralata Bal and Dr. Naresh Chandra Bal for their generous advises to write the Paper.
References
- 1.Aceijas C., Rhodes T. Global estimates of prevalence of HCV infection among injecting drug users. Int. J. Drug Pol. 2007;18:352–358. doi: 10.1016/j.drugpo.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization, Hepatitis C, Fact sheet N°164 (http://www.searo.who.int/thailand/factsheets/fs0018/en/). (Accessed on 28th October 2018)doi.org/10.1016/S0140- 6736(14)62401-62406.
- 3.Houghton M. The long and winding road leading to the identification of the hepatitis C virus. J. Hepatol. 2009;51:939–948. doi: 10.1016/j.jhep.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Hayashi S., Tanak T., Moriishi K., Hirayama K., Yamada A., Hotta K. Seroepidemiology of non-primate hepacivirus (NPHV) in Japanese native horses. J. Vet. Med. Sci. 2018;80:186–189. doi: 10.1292/jvms.17-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakano T., Lau G.M.G., Lau G.M.L., Sugiyama M., Mizokami M. An updated analysis of hepatitis C virus genotypes and subtypes based on the complete coding region. Liver Int. 2012;32:339–345. doi: 10.1111/j.1478-3231.2011.02684.x. [DOI] [PubMed] [Google Scholar]
- 6.Nelson K.E., Williams C.M. 2007. Infectious Disease Epidemiology: Theory and Practice; p. 299. [Google Scholar]
- 7.Stone A.E.L., Giugliano S., Schnell G., Cheng L., Leahy K.F., Golden-Mason L., Gale M., Rosen H.R. Hepatitis C virus pathogen associated molecular pattern (PAMP) triggers production of lambda-interferons by human plasmacytoid dendritic cells. PLoS Pathog. 2013;9:1–14. doi: 10.1371/journal.ppat.1003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganta N.M., Jadav S.S., Kumar A., Doy N., Mohanty N.P. The updates on Middle East Respiratory Syndrome coronavirus (MERS-CoV) epidemiology, pathogenesis, viral genome and currently available drugs. J. Pharm. Chem. 2016;3:10–18. doi: 10.14805/jphchem.2016.art47. [DOI] [Google Scholar]
- 9.World Health organization, Hepatitis C http://www.who.int/news-room/fact-sheets/detail/hepatitis-c (Accessed on 28th October 2018).
- 10.Maheshwari A., Thuluvath P.J. Management of acute hepatitis C. Clin. Liver Dis. 2010;14:169–176. doi: 10.1016/j.cld.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 11.European Centre for Disease Prevention and Control . ECDC; 2016. Annual Epidemiological Report 2015. Hepatitis C. Stockholm. [Google Scholar]
- 12.Centers for Disease Control and Prevention, Viral Hepatitis. https://www.cdc.gov/hepatitis/hcv/statisticshcv.htm (Accessed on 28th October 2018).
- 13.Database Study. 2012–2016. Impact of Availability of Direct-acting Antivirals for Hepatitis C on Canadian Hospitalization Rates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolotin S., Fed J.J., Garber G., Wong W.W.L., Guerra F.M., Mazzulli T. Population- based estimate of hepatitis C virus prevalence in Ontario, Canada. PLoS One. 2018;13 doi: 10.1371/journal.pone.0191184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah H., Bilodeau M., Burak K.W., Cooper C., Klein M., Ramji A., Smyth D., Feld J.J. The management of chronic hepatitis C: 2018 guideline update from the Canadian Association for the Study of the Liver. CMAJ (Can. Med. Assoc. J.) 2018;90:E677–E687. doi: 10.1503/cmaj.170453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.European Association for the Study of the Liver EASL recommendations on treatment of hepatitis C. J. Hepatology. 2018;69:461–511. doi: 10.1016/j.jhep.2018.03.02. 2018. [DOI] [PubMed] [Google Scholar]
- 17.Rosen H.R. Clinical practice. Chronic hepatitis C infection. N. Engl. J. Med. 2011;3:2429–2438. doi: 10.1056/NEJMcp1006613. [DOI] [PubMed] [Google Scholar]
- 18.Sofia M.J., Chang W., Furman P.A., Mosley R.T., Ross B.S. J. Med. Chem. 2012;55:2481–2531. doi: 10.1021/jm201384j. [DOI] [PubMed] [Google Scholar]
- 19.Ajantha G.S., Kulkarni R.D., Jeevan S., Shubhada C., Pavithra J. Indian J. Pathol. Microbiol. 2008;51:376–378. doi: 10.4103/0377-4929.42515. [DOI] [PubMed] [Google Scholar]
- 20.Baumert T.F., Ito S., Wong D.T., Liang T.J. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J. Virol. 1998;72:3827–3836. doi: 10.1128/jvi.72.5.3827-3836.1998. PMCID: PMC109606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanchard E., Hourioux C., Brand D., Ait-Goughoulte M., Moreau A., Trassard S., Sizaret P.Y., Dubois F., Roingeard P. Hepatitis C virus-like particle budding: role of the core protein and importance of its Asp111. J. Virol. 2003;77:10131–10138. doi: 10.1128/JVI.77.18.10131-10138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cocquerel L., Wychowski C., Minner F., Penin F., Dubuisson J. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a major role in the processing, subcellular localization, and assembly of these envelope proteins. J. Virol. 2000;74:3623–3633. doi: 10.1128/JVI.74.8.3623-3633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menez R., Housden N.G., Harrison S., Jolivet-Reynaud C., Gore M.G., Stura E.A. Different crystal packing in Fab-protein L semi-disordered peptide complex. Acta Crystallogr. D. 2005;61:744–749. doi: 10.1107/S0907444905006724. 2005. [DOI] [PubMed] [Google Scholar]
- 24.Kong L., Kadam R.U., Giang E., Ruwona T.B., Nieusma T., Culhane J.C., Stanfield R.L., Dawson P.E., Wilson I.A., Law M. Structure of hepatitis C virus envelope glycoprotein E1 antigenic site 314-324 in complex with antibody IGH526. J. Mol. Biol. 2015;427:2617–2628. doi: 10.1016/j.jmb.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potter J.A., Owsianka A.M., Jeffery N., Matthews D.J., Keck Z.Y., Lau P., Foung S.K., Taylor G.L., Patel A.H. Toward a hepatitis C virus vaccine: the structural basis of hepatitis C virus neutralization by AP33, a broadly neutralizing antibody. J. Virol. 2012;86:12923–12932. doi: 10.1128/JVI.02052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tellinghuisen T.L., Marcotrigiano J., Rice C.M. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature. 2005;435:374–379. doi: 10.1038/nature03580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hucke O., Coulombe R., Bonneau P., Bertrand-Laperle M., Brochu C., Gillard J., Joly M.A., Landry S., Lepage O., Llinas-Brunet M., Pesant M., Poirier M., Poirier M., McKercher G., Marquis M., Kukolj G., Beaulieu P.L., Stammers T.A. MolecularDynamics simulations and structure-based rational design lead to allosteric HCV NS5BPolymerase thumb pocket 2 inhibitor with picomolar cellular replicon potency. J. Med. Chem. 2014;57:1932–1943. doi: 10.1021/jm4004522. [DOI] [PubMed] [Google Scholar]
- 28.Parsy C.C., Alexandre F.R., Bidau V., Bonnaterre F., Brandt G., Caillet C., Cappelle S., Chaves D., Convard T., Derock M., Gloux D., Griffon Y., Lallos L.B., Leroy F., Liuzzi M., Loi A.G., Moulat L., Chiara M., Rahali H., Roques V., Rosinovsky E., Savin S., Seifer M., Standring D., Surleraux D. Discovery and structural diversity of the hepatitis C virus NS3/4A serine protease inhibitor series leading to clinical candidate IDX320. Bioorg. Med. Chem. Lett. 2015;25:5427–5436. doi: 10.1016/j.bmcl.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Nair L.G., Sannigrahi M., Bogen S., Pinto P., Chen K.X., Prongay A., Tong X., Cheng K.C., Girijavallabhan V., George Njoroge F. P4 capped amides and lactams as HCV NS3 protease inhibitors with improved potency and DMPK profile. Bioorg. Med. Chem. Lett. 2010;20:567–570. doi: 10.1016/j.bmcl.2009.11.094. [DOI] [PubMed] [Google Scholar]
- 30.Sakai A., Claire M.S., Faulk K., Govindarajan S., Emerson S.U., Purcell R.H., Bukh J. The p7 polypeptide of hepatitis C virus is critical for infectivity and contains functionally important genotype-specific sequences. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11646–11651. doi: 10.1073/pnas.1834545100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chevaliez S., Pawlotsky J.M. In: HCV Genome and Life Cycle, Hepatitis C Viruses: Genomes and Molecular Biology. Tan S.L., editor. Horizon Bioscience; Norfolk (UK): 2006. https://www.ncbi.nlm.nih.gov/books/NBK1630/pdf/Bookshelf_NBK1_630.pdf/ [PubMed] [Google Scholar]
- 32.Pawlotsky J.M. Therapy of hepatitis C: from empiricism to cure. Hepatology. 2006;43(Suppl. 1):S207–S220. doi: 10.1002/hep.21064. [DOI] [PubMed] [Google Scholar]
- 33.Gretton S.N., Taylor A.I., McLauchlan J. Mobility of the hepatitis C virus NS4B protein on the endoplasmic reticulum membrane and membraneassociated foci. J. Gen. Virol. 2005;86:1415–1421. doi: 10.1099/vir.0.80768-0. [DOI] [PubMed] [Google Scholar]
- 34.Kim J.Y., Wang L., Lee J., Ou J.J. Hepatitis C virus induces the localization of lipid rafts to autophagosomes for its RNA replication. J. Virol. 2017;91 doi: 10.1128/JVI.00541-17. e00541-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hepatitis C. Viruses: genomes and molecular biology. In: Tan S.L., editor. Chapter: Biochemical Activities of the HCV NS5B RdRp. Horizon Bioscience; Norfolk (UK): 2006. https://www.ncbi.nlm.nih.gov/books/NBK1629/ [Google Scholar]
- 36.Kim C.W., Chang K.M. Hepatitis C virus: virology and life cycle. Clin. Mol. Hepatol. 2013;19:17–25. doi: 10.3350/cmh.2013.19.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romano K.P., Ali A., Aydin C., Soumana D., Özen A., Deveau L.M., Silver C., Cao H., Newton A., Petropoulos C.J., Huang W., Schiffer C.A. PLoS Pathog. 2012;8:22. doi: 10.1371/journal.ppat.1002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolykhalov A., Mihalik K., Feinstone S., Rice C. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3' nontranslated region are essential for virus replication in vivo. J. Virol. 2000;74:2046–2061. doi: 10.1128/jvi.74.4.2046-2051.2000. PMID: 10644379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilkins T., Malcolm J.K., Raina D., Schade R.R. Hepatitis C: diagnosis and treatment. Am. Fam. Physician. 2010;81:1351–1357. PMID: 20521755. [PubMed] [Google Scholar]
- 40.Liang T.J., Ghany M.G. Current and future therapies for hepatitis C virus infection. N. Engl. J. Med. 2013;368:1907–1917. doi: 10.1056/NEJMra1213651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Awad T., Thorlund K., Hauser G., Stimac D., Mabrouk M., Gluud C. Peginterferon alpha-2a is associated with higher sustained virological response than peginterferon alfa-2b in chronic hepatitis C: systematic review of randomized trials. Hepatology. 2010;51:1176–1184. doi: 10.1002/hep.23504. [DOI] [PubMed] [Google Scholar]
- 42.Lamarre D., Anderson P.C., Bailey M., Beaulieu P., Bolger G., Bonneau P., Bös M., Cameron D.R., Cartier M., Cordingley M.G., Faucher A.M., Goudreau N., Kawai S.H., Kukolj G., Lagacé L., LaPlante S.R., Narjes H., Poupart M.A., Rancourt J., Sentjens R.E., George R. St, Simoneau B., Steinmann G., Thibeault D., Tsantrizos Y.S., Weldon S.M., Yong C.L., Llinàs-Brunet M. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature. 2003;426:186–189. doi: 10.1038/nature02099. [DOI] [PubMed] [Google Scholar]
- 43.Ozaras R., Tahan V. Acute hepatitis C: prevention and treatment. Expert Rev. Anti Infect. Ther. 2009;7:351–361. doi: 10.1586/eri.09.8. [DOI] [PubMed] [Google Scholar]
- 44.Alavian S.M., Tabatabaei S.V. Treatment of chronic hepatitis C in polytransfused thalassaemic patients: a meta-analysis. J. Viral Hepat. 2010;17:236–244. doi: 10.4084/MJHID.2017.003. [DOI] [PubMed] [Google Scholar]
- 45.Hoofnagle J.H., Sherker A.H. Therapy for Hepatitis C-The cost of success. N. Engl. J. Med. 2014;370:1552–1553. doi: 10.1056/NEJMe1401508. [DOI] [PubMed] [Google Scholar]
- 46.Bunchorntavakul C., Chavalitdhamrong D., Tanwandee T. Hepatitis C genotype 6: a concise review and response-guided therapy proposal. World J. Hepatol. 2013;5:496–504. doi: 10.4254/wjh.v5.i9.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinrichsen H., Benhamou Y., Wedemeyer H., Reiser M., Sentjens R.E., Calleja J.L., Forns X., Erhardt A., Crönlein J., Chaves R.L., Yong C.L., Nehmiz G., G.G Gastroenterology. 2004;127:1347–1355. doi: 10.1053/j.gastro.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 48.Wang C., Jia L., Huang H., Qiu D., Valera L., Huang X., Sun J.H., Nower P.T., O'Boyle D.R., Gao M., Fridell R.A. In vitro activity of BMS-790052 on hepatitis C virus genotype 4 NS5A. Antimicrob. Agents Chemother. 2012;56:1588–1590. doi: 10.1128/AAC.06169-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schinazi R.F., Coats S.J., Bassit L.C., Lennerstrand J., Nettles J.H., Hurwitz S.J. Approaches for the development of antiviral compounds: the case of hepatitis C virus. Handb. Exp. Pharmacol. 2009;189:25–51. doi: 10.1007/978-3-540-79086-0_2. [DOI] [PubMed] [Google Scholar]
- 50.Konreddy A.K., Toyama M., Ito W., Bal C., Baba M., Sharon A. Synthesis and anti-HCV activity of 4-hydroxyamino α-pyranone carboxamide analogues. ACS Med. Chem. Lett. 2014;5:259–263. doi: 10.1021/ml400432f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bae I.H., Choi J.K., Chough C., Keum S.J., Kim H., Jang S.K., Kim B.M. Potent hepatitis C virus NS5A inhibitors containing a benzidine core. ACS Med. Chem. Lett. 2014;5:255–258. doi: 10.1021/ml4003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anilkumar G.N., Lesburg C.A., Selyutin O., Rosenblum S.B., Zeng Q., Jiang Y., Chan T.-Y., Pu H., Vaccaro H., Wang L.I. Novel HCV NS5B polymerase inhibitors: discovery of indole 2-carboxylic acids with C3-heterocycles. Bioorg. Med. Chem. Lett. 2011;21:5336–5341. doi: 10.1016/j.bmcl.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 53.Chen K.X., Bancha V., Weiying Y., Mousumi S., Francisco V., Tin-Yau C., Srikanth V., Anilkumar G.N., Zeng Q., Bennet F. Structure–activity relationship (SAR) Development and discovery of potent indole-based inhibitors of the Hepatitis C Virus (HCV) NS5B Polymerase. J. Med. Chem. 2012;55:754–765. doi: 10.1021/jm201258k. [DOI] [PubMed] [Google Scholar]
- 54.Chen K.X., Lesburg C.A., Vibulbhan B., Yang W., Chan T.-Y., Venkatraman S., Velazquez F., Zeng Q., Bennett F., Anilkumar G.N. A novel class of highly potent irreversible hepatitis C virus NS5B polymerase inhibitors. J. Med. Chem. 2012;55:2089–2101. doi: 10.1021/jm201322r. [DOI] [PubMed] [Google Scholar]
- 55.De Francesco R., Tomei L., Altamura S., Summa V., Migliaccio G. Approaching a new era for hepatitis C virus therapy: inhibitors of the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase. Antivir. Res. 2003;58:1–16. doi: 10.1016/s0166-3542(03)00028-7. [DOI] [PubMed] [Google Scholar]
- 56.Velázquez F., Venkatraman S., Lesburg C.A., Duca J., Rosenblum S.B., Kozlowski J.A., Njoroge F.G. Synthesis of new 4, 5-dihydrofuranoindoles and their evaluation as HCV NS5B polymerase inhibitors. Org. Lett. 2012;14:556–559. doi: 10.1021/ol203177g. [DOI] [PubMed] [Google Scholar]
- 57.Chen K.X., Venkatraman S., Anilkumar G.N., Zeng Q., Lesburg C.A., Vibulbhan B., Velazquez F., Chan T.Y., Bennet F., Jiang Y., Pinto P., Huang Y., Selyutin O., Agrawal S., Huang H.C., Li C., Cheng K.C., Shih N.Y., Kozlowski J.A., Rosenblum S.B., Njoroge F.G. Discovery of SCH 900188: a potent hepatitis C virus NS5B polymerase inhibitor prodrug as a development candidate. ACS Med. Chem. Lett. 2014;5:244–248. doi: 10.1021/jm201258k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gopalsamy A., Lim K., Ciszewski G., Park K., Ellingboe J.W., Bloom J., Insaf S., Upeslacis J., Mansour T.S., Krishnamurthy G., Damarla M., Pyatski Y., Ho D., Howe A.Y.M., Orlowski M., Feld B., O'Connell J. Discovery of pyrano[3,4-b]indoles as potent and selective HCV NS5B polymerase inhibitors. J. Med. Chem. 2004;47:6603–6608. doi: 10.1021/jm0401255. [DOI] [PubMed] [Google Scholar]
- 59.Jin G., Lee S., Choi M., Kim G.W., Oh J.W., Lee C., Lee K. Chemical genetics-based discovery of indole derivatives as HCV NS5B polymerase inhibitors. Eur. J. Med. Chem. 2014;75:413–425. doi: 10.1016/j.ejmech.2014.01.062. [DOI] [PubMed] [Google Scholar]
- 60.Yu W., Jinlong L., Jie Q., Mingjie H., Ming W., Fenghua G., Dongmei L., Zhangyong H., Lingbao K., Weiqiang H., Jianping L. Discovery of Novel Hepatitis C Virus NS5B Polymerase Inhibitors by Combining Random Forest, Multiple e- Pharmacophore Modeling and Docking. PLoS One. 2016;11 doi: 10.1371/journal.pone.0148181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wyles D.L., Kaihara K.A., Schooley R.T. Synergy of a hepatitis C virus (HCV) NS4A antagonist in combination with HCV protease and polymerase inhibitors. Antimicrob. Agents Chemother. 2008;52:1862–1864. doi: 10.1128/AAC.01208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.David M.G., Sandrine V., Lin T.-I., Abdellah T., Lili H., Maxwell D.C., Katie A., Jan M.B., Maxime C., Erna C., Pascale D., Stefen L., Els F., Van D.H., Iris Vanden S., Leen V., Marie-Clude R., Gregory F., Origene N., Kristof V.E., Kenneth S., Pierre R. Finger-loop inhibitors of the HCV NS5b polymerase. Part 1: discovery and optimization of novel 1,6- and 2,6-macrocyclic indole series. Bioorg. Med. Chem. Lett. 2012;22:4431–4436. doi: 10.1016/j.bmcl.2012.03.097. [DOI] [PubMed] [Google Scholar]
- 63.David S., Anthny R.G., Joane L., Richard G., Jason M.H., Wang R.W., Emma J.S., Kyle E., James W.J., James M.C., Ken R., Steve W., Kap-Sun Y., Nicholas A.M., Dennis H., Andrew J.S., Brian L.V., Jeffrey R.S. Highly potent non-peptidic inhibitors of the HCV NS3/NS4A serine protease. Bioorg. Med. Chem. Lett. 2002;12:3129–3133. doi: 10.1016/S0960-894X(02)00680-7. [DOI] [PubMed] [Google Scholar]
- 64.Pierre L.B., Michael B., Yves B., Patrick D., Gulrez F., Jean G., James G., Sylvie G., Gintette M., Marc-Andre P., Serge V. Non-nucleoside inhibitors of the hepatitis C virus NS5B polymerase: discovery of benzimidazole 5-carboxylic amide derivatives with low-nanomolar potency. Bioorg. Med. Chem. Lett. 2002;14:967–971. doi: 10.1016/j.bmcl.2003.12.032. K. G. [DOI] [PubMed] [Google Scholar]
- 65.Vincenzo S., Aslessia P., Paola P., Victor G.M., Raffaele D.F., Sergio A., Licia T., Uwe K., Philippe N. Discovery of α,γ-diketo acids as potent selective and reversible inhibitors of hepatitis C virus NS5b RNA-dependent RNA polymerase. J. Med. Chem. 2004;47:14–17. doi: 10.1021/jm0342109. [DOI] [PubMed] [Google Scholar]
- 66.Deore R.R., Chen G.S., Chen C.S., Chang P.T., Chuang M.H., Chern T.R., Wang H.C., Chern J.W. 2-hydroxy-1-oxo-1,2-dihydroisoquinoline-3-carboxylic acid with inbuilt β-N-hydroxy-γ-keto-acid pharmacophore as HCV NS5B polymerase inhibitors. Curr. Med. Chem. 2012;19:613–624. doi: 10.2174/092986712798918833. [DOI] [PubMed] [Google Scholar]
- 67.Deore R.R., Chen G.S., Chang P.T., Chen T.R., Lai S.Y., Chuang M.H., Lin H.H., Kung F.L., Chen C.S., Chiou C.T., Wang ChemMedChem. 2012;7:850–860. doi: 10.1002/cmdc.201200058. 7. [DOI] [PubMed] [Google Scholar]
- 68.Das D., Hong J., Chen S.H., Wang G., Beigelman L., Seiwert S.D., Buckman B.O. Recent advances in drug discovery of benzothiadiazine and related analogs as HCV NS5B polymerase inhibitors. Bioorg. Med. Chem. 2011;19:4690–4703. doi: 10.1016/j.bmc.2011.06.079. [DOI] [PubMed] [Google Scholar]
- 69.Pfefferkorn J.A., Greene M.L., Nugent R.A., Gross R.J., Mitchell M.A., Finzel B.C., Harris M.S., Wells P.A., Shelly J.A., Anstandt R.A., Kilkuske R.E., Kopta L.A., J Schwende F. Inhibitors of HCV NS5B polymerase. Part 1: evaluation of the southern region of (2Z)-2-(benzoylamino)-3-(5-phenyl-2-furyl)acrylic acid. Bioorg. Med. Chem. Lett. 2005;15:2481–2486. doi: 10.1016/j.bmcl.2005.03.066. [DOI] [PubMed] [Google Scholar]
- 70.Ariamala G., Rajiv C., Kitae L., Gregory C., Mengxiao S., Kevin J.C., Steven F.S., Kristine S., Joel B., John W.E., Atul A., Girija K., Anita Y.M.H., Mark O., Boris F., John O., Tarek S.M. Discovery of proline sulfonamides as potent and selective hepatitis C virus NS5b polymerase inhibitors. Evidence for a new NS5b polymerase binding site. J. Med. Chem. 2006;49:3052–3055. doi: 10.1021/jm060168g. [DOI] [PubMed] [Google Scholar]
- 71.Bhargav P., Ramalingam K., Nikhil K., Gurkumar K.R., Amartya B., Payal A., Aaditya B., Maulik R.P., Dibyendu D., Sanjal K., Neerja K.B., Tanaji T.T. Design and synthesis of L- and D-phenylalanine derived rhodanines with novel C5-arylidenes as inhibitors of HCV NS5B polymerase. Bioorg. Med. Chem. 2013;21:3262–3271. doi: 10.1016/j.bmc.2013.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Micheal J.S., Wonsuk C., Phillip A.F., Ralph T.M., Bruce S.R. Nucleoside, nucleotide, and non-nucleoside inhibitors of hepatitis C virus NS5B RNA- dependent RNA-polymerase. J. Med. Chem. 2012;55:2481–2531. doi: 10.1021/jm201384j. [DOI] [PubMed] [Google Scholar]
- 73.Canales E., Carlson J.S., Appleby T., Fenaux M., Lee J., Tian Y., Tirunagari N., Wong M., Watkins W.J. Tri-substituted acylhydrazines as tertiary amide bioisosteres: HCV NS5B polymerase inhibitors. Bioorg. Med. Chem. Lett. 2012;22:4288–4292. doi: 10.1016/j.bmcl.2012.05.025. [DOI] [PubMed] [Google Scholar]
- 74.Stammers T.A., Coulombe R., Duplessis M., Fazal G., Gagnon A., Garneau M., Goulet S., Jakalian A., Laplante S., Rancourt J., Thavonekhem B., Wernic D., Kukolji G., Beaulieu P.L. Anthranilic acid-based Thumb Pocket 2 HCV NS5B polymerase inhibitors with sub-micromolar potency in the cell-based replicon assay. Bioorg. Med. Chem. Lett. 2013;24:6879–6885. doi: 10.1016/j.bmcl.2013.09.102. [DOI] [PubMed] [Google Scholar]
- 75.Stammers T.A., Coulombe R., Rancourt J., Thavonekham B., Fazal G., Goulet S., Jakalian A., Wernic D., Tsantrizos Y., Poupart M.A., Bos M., McKercher G., Thauvette L., Kukoji G., Beaulieu P.L. Bioorg. Med. Chem. Lett. 2013;23:2585–2589. doi: 10.1016/j.bmcl.2013.02.110. [DOI] [PubMed] [Google Scholar]
- 76.Goulb A.G., Gurukumar K.R., Basu A., Bdzhola V.G., Bilokin Y., Yarmoluk S.M., Lee J.C., Talele T.T., Nichols D.B., Kaushik-Basu N. Discovery of new scaffolds for rational design of HCV NS5B polymerase inhibitors. Eur. J. Med. Chem. 2012;58:258–264. doi: 10.1016/j.ejmech.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ng T.I., Krishnan P., Pilot-Matias T., Kati W., Schnell G., Beyer J., Reisch T., Lu L., Dekhtyar T., Irvin M., Tripathi R., Maring C., Randolph J.T., Wagner R., Collins C. In vitro antiviral activity and resistance profile of the next-generation hepatitis C virus NS5A inhibitor pibrentasvir. Antimicrob. Agents Chemother. 2017;61:1–37. doi: 10.1128/AAC.02558-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Foster G.R., Afdhal N., Roberts S.K., Bräu N., Gane E.J., Pianko S., Lawitz E., Thompson A., Shiffman M.L., Cooper C., Towner W.J., Conway B., Ruane P., Bourlière M., Asselah T., Berg T., Zeuzem S., Rosenberg W., Agarwal K., Stedman C.A.M., Mo H., Dvory-Sobol H., Han L., Wang J., McNally J., Osinusi A., Brainard D.M., McHutchison J.G., Mazzotta F., Tran T.T., Gordon S.C., Patel K., Reau N., Mangia A., Sulkowski M. Sofosbuvir and velpatasvir for HCV genotype 2 and 3 infection. N. Engl. J. Med. 2015;373:2608–2617. doi: 10.1056/NEJMoa1512612. [DOI] [PubMed] [Google Scholar]
- 79.Ivanenkov Y.A., Aladinskiy V.A., Bushkov N.A., Ayginin A.A., Majouga A.G., Ivachtchenko A.V. Expert Opin. Ther. Pat. 2017;27:401–414. doi: 10.1080/13543776.2017.1272573. [DOI] [PubMed] [Google Scholar]
- 80.Shwen H., Zhong L., Xing D., Dong X., Clare L., Nicholas Z., Ravi N., Anandan P., Casey C.M., Peng L., Xuanjia P., Richard S. Heterocyclic compounds and methods of use thereof for treatment of hepatitis c. PCT Int. Appl. 2014 WO 2014205592 A1 20141231. [Google Scholar]
- 81.Xing D., Hong L., Anandan P., Shuwen H., Ravi N., Dong X., Nicolas Z., Qun D., Casey C.M., Xuanjia P., Peng L., Richard S. Substituted benzfuron compounds and methods of use thereof for the treament of viral diseases. PCT Int. Appl. 2014 WO 2014209727 A1 20141231. [Google Scholar]
- 82.Pierre L.B., Paul C.A., Richard B., Michael B., Yves B., Christian B., Michael G.C., Gulrez F., Michel G., James R.G., Stephen K., Martin M., McKrecher G., Marc-Andre P., Timothy S., Bounkham T., Dominik W., Jianmin D., George K. J. Med. Chem. 2014;57:10130–10143. doi: 10.1021/jm501532z. [DOI] [PubMed] [Google Scholar]
- 83.Beaulieu P.L., Bolger G., Duplessis M., Gagnon A., Garneau M., Stammers T., Kukoji G., Duan J. Aza follow-ups to BI 207524, a thumb pocket 1 HCV NS5B polymerase inhibitor. Part 1: mitigating the genotoxic liability of an aniline metabolite. Bioorg. Med. Chem. Lett. 2015;25:1135–1139. doi: 10.1016/j.bmcl.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 84.Kneteman N.M., Howe A.Y., Gao T., Lewis J., Pevear D., Lund G., Douglus D., Mercer D.F., Tyrrell D.L., Immermann F., Chaudhary I., Speth J., Villano S.A., O'Connell J., Cellett M. HCV796: a selective nonstructural protein 5B polymerase inhibitor with potent anti-hepatitis C virus activity in vitro, in mice with chimeric human livers, and in humans infected with hepatitis C virus. Hepatology. 2009;49:745–752. doi: 10.1002/hep.22717. [DOI] [PubMed] [Google Scholar]
- 85.Voitenleitner C., Crosby R., Walker J., Remlinger K., Vamathevan J., Wang A., You S., Johnson J., 3rd, Woldu E., Van Horn S., Horton J., Creech K., Shotwell J.B., Hong Z., Hamatake R. In vitro characterization of GSK2485852, a novel hepatitis C virus polymerase inhibitor. Antimicrob. Agents Chemother. 2013;57:5216–5224. doi: 10.1128/AAC.00874-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McComas C.C., Palani A., Chang W., Holloway M.K., Lesburg C.A., Li P., Liverton N., Meinke P.T., Olsen D.B., Peng X., Soll R.M., Ummat A., Wu J., Wu J., Zorn N., Ludmerer S.W. Development of a new structural class of broadly acting HCV non-nucleoside inhibitors leading to the discovery of MK-8876. ChemMedChem. 2017;12:1436–1448. doi: 10.1002/cmdc.201700228. [DOI] [PubMed] [Google Scholar]
- 87.Yeung K.S., Beno B.R., Parcella K., Bender J.A., Grant-Young K.A., Nickel A., Gunaga P., Anjanappa P., Bora R.O., Selvakumar K., Rigat K., Wang Y.K., Liu M., Lemm J., Mosure K., Sheriff S., Wan C., Witmer M., Kish K., Hanumegowda U., Zhuo X., Shu Y.Z., Parker D., Haskell R., Ng A., Gao Q., Colston E., Raybon J., Grasela D.M., Santone K., Gao M., Meanwell N.A., Sinz M., Soars M.G., Knipe J.O., Roberts S.B., Kadow J.F. Discovery of a hepatitis C virus NS5B replicase palm site allosteric inhibitor (BMS-929075) advanced to phase 1 clinical studies. J. Med. Chem. 2017;60:4369–4385. doi: 10.1021/acs.jmedchem.7b00328. [DOI] [PubMed] [Google Scholar]
- 88.Parcella K., Eastman K., Yeung K.S., Grant-Young K.A., Zhu J., Wang T., Zhang Z., Yin Z., Parker D., Mosure K., Fang H., Wang Y.K., Lemm J., Zhuo X., Hanumegowda U., Liu M., Rigat K., Donoso M., Tuttle M., Zvyaga T., Haarhoff Z., Meanwell N.A., Soars M.G., Roberts S.B., Kadow J.F. Improving metabolic stability with deuterium: the discovery of BMT-052, a Pan-genotypic HCV NS5B polymerase inhibitor. ACS Med. Chem. Lett. 2017;8:771–774. doi: 10.1021/acsmedchemlett.7b00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.De Clercq E. Dancing with chemical formulae of antivirals: a panoramic view (Part 2) Biochem. Pharmacol. 2013;86:1397–1410. doi: 10.1016/j.bcp.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 90.Manns M.P., Foster G.R., Rockstroh J.K., Zeuzem S., Zoulim F., Houghton M. Nat. Rev. Drug Discov. 2007;6:991–1000. doi: 10.1038/nrd2411. [DOI] [PubMed] [Google Scholar]
- 91.A. E. L. Stone, S. Giugliano, G. Schnell, L. Cheng, K. F. Leahy, L. Golden-Mason, M. Gale, H. R. Rosen, PLoS Pathog., doi: 10.1371/journal.ppat.1003316. [DOI] [PMC free article] [PubMed]
- 92.Shah U., Jayne C., Chackalamannil S., Velázquez F., Guo Z., Buevich A., a Howe J., Chase R., Soriano A., Agrawal S., Rudd M.T., Mccauley J.A., Liverton N.J., Romano J., Bush, Coleman P.J., Grisé-Bard C., Brochu M.-C., Charron S., Aulakh V., Bachand B., Beaulieu P., Zaghdane H., Bhat S., Han Y., Vacca J.P., Davies I.W., Weber A.E., Venkatraman S., Hcv G., Protease N.S., Shah U., Jayne C., Chackalamannil S., Vela F., Guo Z., Buevich A., a Howe J., Chase R., Soriano A., Agrawal S., Rudd M.T., Mccauley J.A., Liverton N.J., Romano J., Bush K., Coleman P.J., Grise C., Brochu M.-C., Charron S., Aulakh V., Bachand B., Beaulieu P., Zaghdane H., Bhat S., Han Y., Vacca J.P., Davies I.W., Weber A.E., Venkatraman S. ACS Med. Chem. Lett. 2014;5:1–6. doi: 10.1021/ml400466p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Balaraju T., Kumar A., Bal C., Chattopadhyay D., Jena N., Bal N.C., Sharon A. Aromatic interaction profile to understand the molecular basis of raltegravir resistance. Struct. Chem. 2013;24:1499–1512. doi: 10.1007/s11224-012-0181-1. [DOI] [Google Scholar]
- 94.Turner S.R., Strohbach J.W., Tommasi R.A., Aristoff P.A., Johnson P.D., Skulnick H.I., Dolak L.A., Seest E.P., Tomich P.K., Bohanon M.J., Horng M.M., Lynn J.C., Chong K.T., Hinshaw R.R., Watenpaugh K.D., Janakiraman M.N., Thaisrivongs S. Tipranavir (PNU-140690): a potent, orally bioavailable nonpeptidic HIV protease inhibitor of the 5,6-dihydro-4-hydroxy-2-pyrone sulfonamide class. J. Med. Chem. 1998;41:3467–3476. doi: 10.1021/jm9802158. [DOI] [PubMed] [Google Scholar]
- 95.Thiyagarajan A., Salim M.T.A., Balaraju T., Bal C., Baba M., Sharon A. Structure based medicinal chemistry approach to develop 4-methyl-7-deazaadenine carbocyclic nucleosides as anti-HCV agent. Bioorg. Med. Chem. Lett. 2012;22:7742–7747. doi: 10.1016/j.bmcl.2012.09.072. [DOI] [PubMed] [Google Scholar]
- 96.Kasula M., Balaraju T., Toyama M., Thiyagarajan A., Bal C., Baba M., Sharon A. A conformational mimetic approach for the synthesis of carbocyclic nucleosides as anti- HCVleads. ChemMedChem. 2013;8:1673–1680. doi: 10.1002/cmdc.201300277. [DOI] [PubMed] [Google Scholar]
- 97.Balaraju T., Konreddy A.K., Parveen A., Toyama M., Ito W., Karampuri S., Baba M., Sharon A., Bal C. Synthesis and anti-HCV determinant motif identification in pyranone carboxamide scaffold. Bioorg. Med. Chem. Lett. 2015;25:5224–5227. doi: 10.1016/j.bmcl.2015.09.060. [DOI] [PubMed] [Google Scholar]