Abstract
Breast cancer is the second leading cause of death among women in the US. Targeted therapies exist, however resistance is common and patients resort to chemotherapy. Chemotherapy is also a main treatment for triple negative breast cancer (TNBC) patients; while radiation is delivered to patients with advanced disease to counteract metastasis. Yet, resistance to both chemo- and radiotherapy is still frequent, highlighting a need to provide novel sensitizers. We discovered that MT1-MMP modulates DNA damage responses (DDR) in breast cancer. MT1-MMP expression inversely correlates to chemotherapy response of breast cancer patients. Inhibition of MT1-MMP sensitizes TNBC cells to IR and doxorubicin in vitro, and in vivo in an orthotopic breast cancer model. Specifically, depletion of MT1-MMP causes stalling of replication forks and Double Strand Breaks (DBSs), leading to increased sensitivity to additional genotoxic stresses. These effects are mediated by integrinβ1, as a constitutive active integrinβ1 reverts replication defects and protects cells depleted of MT1-MMP from IR and chemotherapy. These data highlight a novel DNA damage response triggered by MT1-MMP-integrinβ1 and provide a new point of therapeutic targeting that may improve breast cancer patient outcomes.
Keywords: MT1-MMP, breast cancer, radiotherapy, chemotherapy, DDR
Introduction
Breast cancer is the second leading cause of cancer death in women, with an estimated 40,920 women in the United States expected to die from the disease in 2018[1]. Although often curable when localized to the breast and local lymph nodes, metastatic disease is usually not curable. Breast cancer is a heterogeneous disease comprising several molecular subtypes, which are commonly extrapolated into clinical subtypes based on receptor status. In standard clinical practice the specific receptors that are assessed are the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor 2-neu (HER2) receptor. These receptors are both prognostic and also predictive of response to targeted therapy. Indeed, considerable advances have been made in the treatment of certain subtypes of breast cancer, for example, HER2-positive disease. In this subtype, targeted therapies against HER2 have changed the clinical outcome for patients with metastatic disease by providing several effective therapies that can extend survival by many years.
The ER(+) and PR(+) subtypes also have several targeted therapies available using endocrine therapies. However, when the disease becomes metastatic, all patients eventually develop endocrine resistance and eventually require cytotoxic chemotherapy. On the other hand, patients with ER(−), PR(−), and HER2(−) tumors (triple-negative breast cancers (TNBCs)), tend to display an aggressive phenotype and unfortunately, do not have available targeted therapy options as standard of care, relying exclusively on chemotherapy[2].
In contrast to targeted therapies, radiation therapy is a critical component in the management of invasive breast cancer[3]. Radiation therapy is a well-established adjuvant treatment modality in patients that undergo breast conserving surgery; and following mastectomy in those patients at high risk of recurrence. Radiation therapy improves local disease control; reduces morbidity and distress of local recurrence; and improves survival, likely by preventing seeding of distant metastases from persistent reservoirs of local disease[3]. However, resistance to both chemo- and radiotherapy is still a major obstacle to acquiring durable responses. Hence, improving the efficacy of chemo- and radiotherapy will be necessary for blocking metastatic progression and providing sustained cures.
MT1-MMP (aka MMP14) is a zinc-dependent Matrix Metalloproteinase tethered to the plasma-membrane. In humans, 19 out of the 25 known MMPs are soluble and are released in the extracellular milieu whereas 6 family members, including MT1-MMP, are membrane-anchored. All MMPs are produced as zymogens (proenzymes) and require proteolytic cleavage of an inhibitory propeptide for activation. ProMT1-MMP (~64 kDa) is converted into a catalytically active enzyme (~55 kDa) following cleavage by furin in the trans-Golgi network prior to its arrival at the plasma membrane. Due to its location, MT1-MMP orchestrates localized proteolysis of the basement membrane and extracellular matrix (ECM) allowing motile cells to move out of the primary tumor in a directional manner. Additionally, MT1-MMP can cleave and activate a number of substrates such as other MMPs (e.g. MMP2, MMP13) and growth factors (e.g. EGF, CD44, Notch1)[4–11]. Consequently, MT1-MMP is now regarded as a critical player in tumor growth and metastasis for a number of cancers including breast cancer[12–14].
Here we show that MT1-MMP regulates an outside-in DNA Damage Response (DDR) through the modulation of integrinβ1. Inhibition of MT1-MMP sensitizes TNBC cells to ionizing radiation and doxorubicin both in vivo and in vitro through the induction of fork stalling and collapse. This phenomenon is mediated by the reduction of integrinβ1 signaling as restoration of active integrinβ1 protects cells from DNA damage following MT1-MMP inhibition. The targeting of MT1-MMP represent a new point of therapeutic intervention that can not only provide anti-metastatic effects, but can at the same time increase sensitivity of breast cancer, particularly triple negative breast cancer, to radio- and chemotherapy thus enhancing the therapeutic responses of patients.
Materials and Methods
Cell lines, plasmids and Reagents.
MDA-MB-231, MDA-MB-468, BT474, T47D, SKBR3 and MCF7 cells were grown in RPMI-1640 supplemented with 10% FBS and maintained at 37°C with 5% CO2. MDA-MB-231 cells were authenticated in 2013 by STR profiling (BDC Molecular Biology Core Facility, University of Colorado). Doxorubicin was dissolved in DMSO (Sigma Aldrich). The catalytically active MT1-MMP was a gift of Dr. Motoharu Seiki (University of Tokyo, Japan) and was inserted into the pLM-CMV-Ha-puro-PL3 lentiviral plasmid as previously described[11, 15]. Integrinβ1 self-clustering mutant V37N was a gift of Dr. Valerie Weaver (University of California at San Francisco). mRNA silencing of MT1-MMP was performed using shRNA TRCN0000050855 (Sigma Aldrich), previously characterized and validated[11, 15, 16].
Immunohistochemistry:
The Tissue MicroArray (TMA) composed of normal and disease spots (Fig. 1B) was purchased from Novus Biolabs. The TMA comprising breast cancer subclasses was purchased from US Biomax. The slides were incubated O/N at 4°C with an anti-MT1-MMP antibody (Millipore, LEM-2/15.8), followed by incubation for 30 min at RT with the ImmPRESS anti-mouse polymer (Vector laboratories, Burlingame, CA, USA). The H-score was calculated for each core by multiplying the staining score (1+−3+) by the % of positive cells with that score, according to the formula: 1×(%cells 1+)+2×((%cells 2+)+3×(%cells 3+)[17, 18]. These data were analyzed and quantified by Dr. Abdul-Karim.
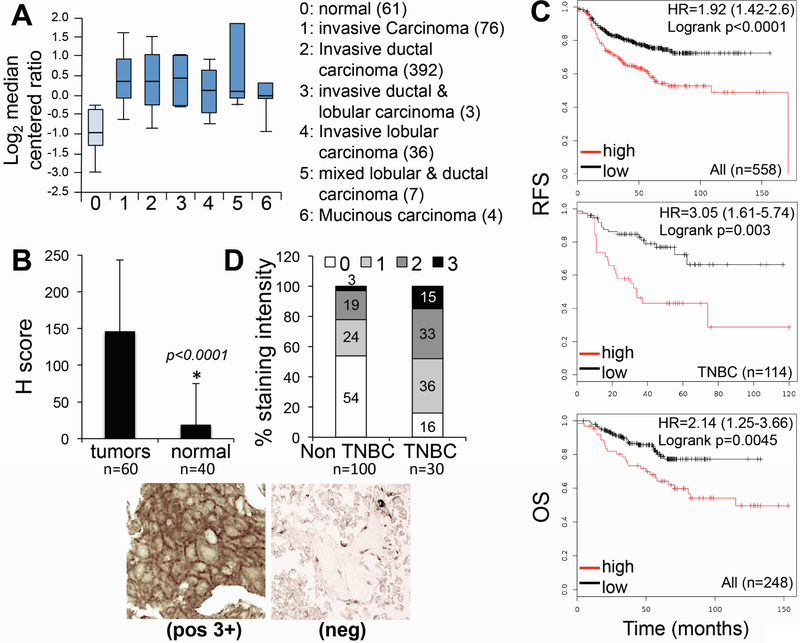
Figure 1: MT1-MMP expression is elevated in breast cancer and inversely correlated to response to chemotherapy.
A) Oncomine data comparing mRNA expression of MMP14 (gene name for MT1-MMP) between normal tissue and different subtypes of invasive breast carcinoma (TCGA breast data set). P values for all types compared to normal are <0.0001. B) Average Hscore between breast cancer and adjacent normal tissue array. Representative sections of a positive tumor (3+ - left panel) and a negative one (0 - right panel) are shown below. C) Kaplan Meier plots of breast cancer showing expression of MT1-MMP relative to patients’ response to chemotherapy. RFS: relapse free survival; OS: Overall Survival. Only KM plots resulting from the “best probe” (Jetset probe25) are included. D) MT1-MMP protein expression TNBCs versus non-TNBCs tumors, assessed as staining intensity for MT1-MMP of a breast cancer TMA comprising several breast cancer subtypes.
Western Blotting:
Cell seeding, collection of protein and Western blot methods were as previously described[19] Membranes were probed with the following antibodies: anti-MT1-MMP (EMD Millipore, MA); anti–integrinβ1 (Cell signaling Technology); anti γH2AX (ser139 JBC301, Millipore); anti p-FAKY397 (Abcam); anti-p-ChK1, anti p-ChK2, anti p-RPA32 (Cell Signaling Technology); β-actin, GAPDH (Santa Cruz Biotechnology).
Colony formation assay:
Radiation was performed with a 137Cs irradiator (Shepherd). A total of 500 to 10,000 cells per plates were stained after 10 days with 0.1% crystal violet. Assays were done ≥3 times with individual samples in triplicate. Colonies with more than 50 cells were counted.
Immunofluorescence:
Cells were fixed in 3% formaldehyde and stained using standard procedures: γH2AX antibody (Abcam, 1:500), secondary Alexa-Fluor 594 anti-mouse (A11032; Invitrogen, 1:500). Slides were mounted in Vectashield with DAPI (Vector Laboratories). Immunofluorescence was observed at ×100, foci were counted using ImageJ from at least 50 nuclei in 5 fields per slide, in triplicate, for each condition.
Comet Assay:
Comet assays were performed as directed (Trevingen), and quantified with ComeScore software. Briefly, 1.5×106 cells were irradiated or left untreated, then cultured for up to 48 hrs. Cells were trypsinized, counted, and number of cells normalized. 5 ul of each cell suspension was mixed with pre-melted low melting agarose, mixed and plated on glass slides provided in the kit and left at 4°C for 30 minutes to allow solidification of the agarose and then immersed in cold lysis buffer for 2 hours to ensure complete lysis. Electrophoresis was carried out at 21 volts for 45 minutes using neutral electrophoresis buffer (1×TBE). Slides were washed twice in milliQ water followed by 70% ethanol for 30 minutes at room temperature. Slides were dried at 37°C overnight and stained using 1:10,000× Syber green DNA stain. Comets were imaged at 10× objective. Comet analysis was done using Comet Score (TriTek). A minimum 50 comets were included per condition.
DNA Fiber Assay:
Cells were pulse-labeled with 50 μmol/L IdU (Sigma-Aldrich) for 40 minutes, followed by treatment with hydroxy Urea to stall replication, then pulsed-labeled 200 μmol/L CldU (Sigma–Aldrich) for 40 minutes. At the end of the CldU pulse, cell suspensions (2.5 μL) were mixed with 7.5 μL of lysis buffer [0.5% SDS, 200 mmol/L Tris-HCl (pH 7.4), 50 mmol/L EDTA] and dropped and spread onto an uncoated glass slide and let dry. DNA spreads were then fixed a 3:1 solution of methanol-acetic acid for 10 min, let dry and then placed in 70% ethanol at 4°C for 1 hour. DNA was denatured with 2.5 mol/L HCl for 30 minutes at 37°C. The slides were blocked in 1% BSA 30 minutes at RT and then incubated with mouse anti-BrdUrd antibody (BD Biosciences) and rat anti-CldU antibody (Abcam). The slides were incubated with secondary fluorescent antibodies (Alexa Fluor 594, or Alexa Fluor 488, Thermo Fisher Scientific). Replication fibers were viewed at 1,000 × magnification on a NIKON 90i fluorescence microscope (photometric cooled mono CCD camera; Nikon). Signals were measured using ImageJ software (NCI/NIH), as previously described[20].
Tumor formation assay:
MDA-MB-231 (2×106) cells expressing shGFP or shMT1-MMP were injected in the inguinal fat pads of Eight-week-old athymic NSG mice. Tumors were allowed to grow to comparable sizes (~200mm3) prior to starting treatment with doxorubicin (2mg/Kg, I.P.) twice weekly. Tumor growth was also monitored twice weekly and quantified using a caliper. 10 mice per group were used for a total of 20 tumors per group.
Results
MT1-MMP is highly expressed in breast cancer and inversely correlated to patients’ response to chemotherapy.
MT1-MMP has been shown to contribute to the aggressiveness of several cancers, including breast cancer[12–14, 21]. We sought to further characterize the importance of MT1-MMP in breast cancer to determine if targeting this protein in this amalgam of diseases might have clinical utility. We interrogated several publicly available data sets and also assessed MT1-MMP protein expression in breast cancer tissue microarrays (TMA). The TCGA data sets from the Oncomine[22] database revealed that MMP14 (gene for MT1-MMP) expression is significantly increased in several breast cancer subtypes with respect to normal breast (Fig. 1A). Likewise, analysis of a breast cancer TMA containing 60 breast carcinomas and 40 adjacent normal tissues revealed significantly elevated expression of the protein in the tumors compared to normal breast epithelium (Fig. 1B, p<0.0001). Furthermore, by using Kaplan Meier Plotter (KMPlot)[23], and by selecting data comprising patients that received chemotherapy, we found that MT1-MMP expression was inversely associated with response to therapy, measured as relapse free survival (RFS) and overall survival (OS), both when all subtypes or just TNBC were considered (Fig. 1C). Notably, TNBC patients only receive chemotherapy as standard of care, supporting a role for MT1-MMP in modulating responses to chemotherapy in these patients. Interestingly, MT1-MMP protein expression is more elevated overall in TNBCs versus non-TNBCs tumors, assessed as staining intensity for MT1-MMP of a breast cancer TMA containing several breast cancer subtypes (Fig. 1D). Together, these data emphasize the need to interrogate the function of MT1-MMP in breast cancer progression and therapeutic response, in particular towards genotoxic therapies.
MT1-MMP sensitizes triple negative breast cancer to DNA damaging agents.
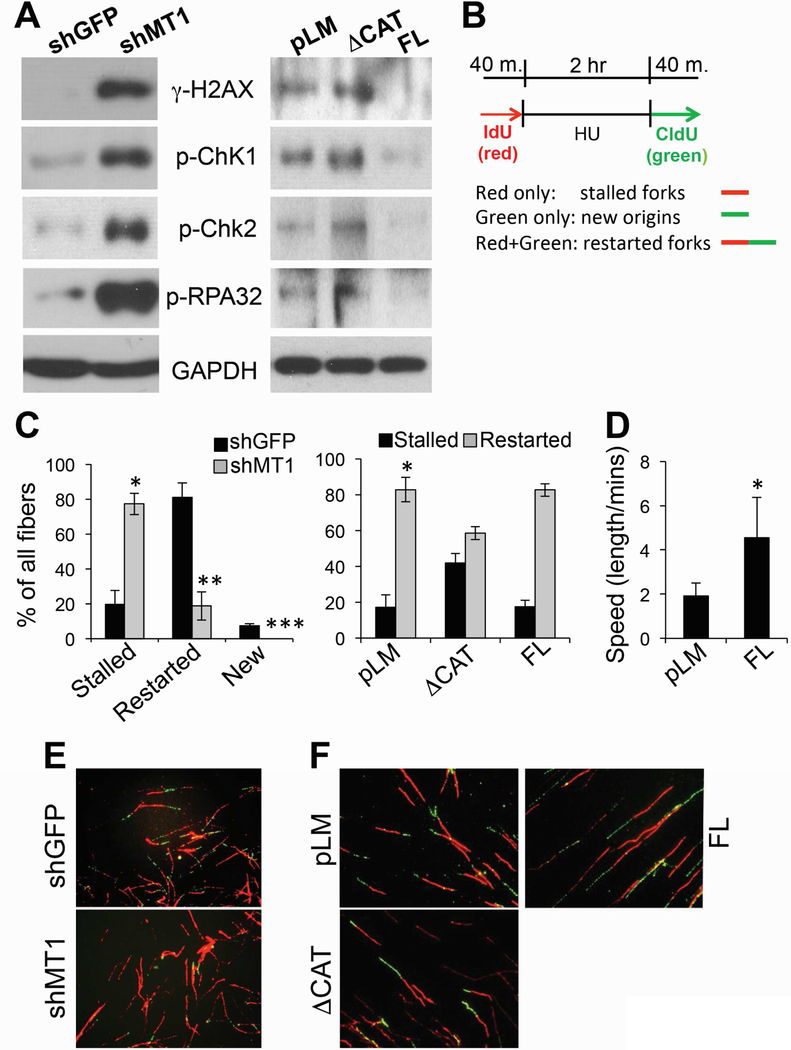
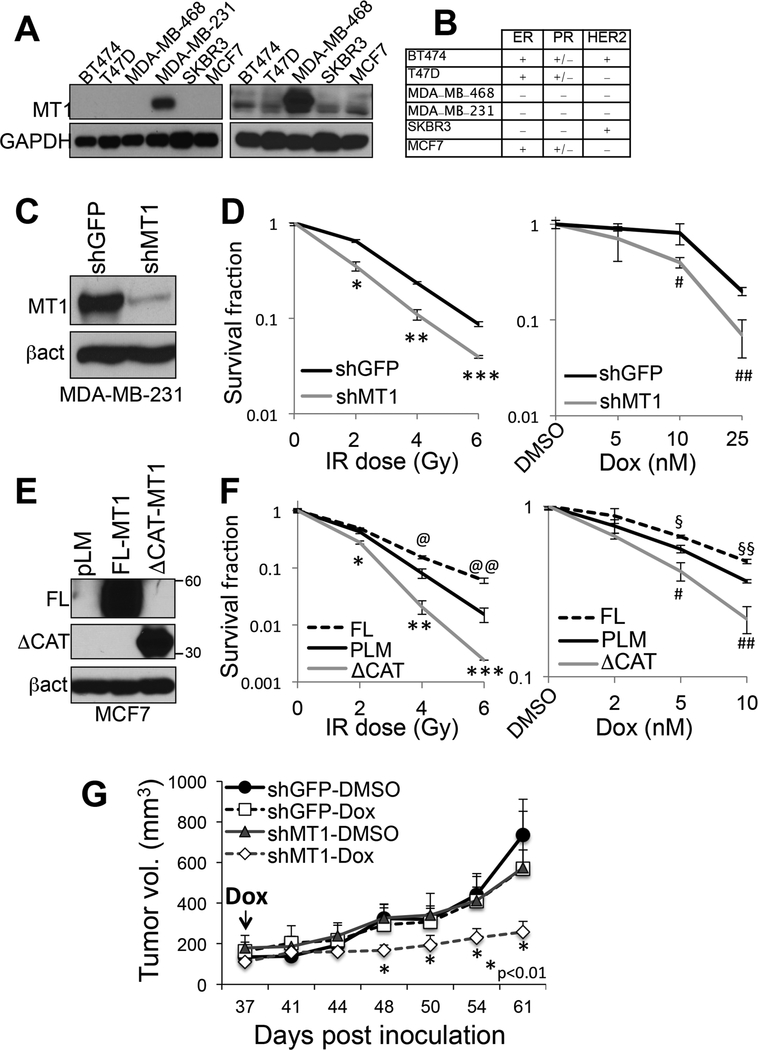
To test the possibility that MT1-MMP may have a role in modulating tumor cell responses to DNA damage, we stably silenced MT1-MMP expression in MDA-MB-231 cells, a triple negative breast cancer cell line that expresses high levels of MT1-MMP with respect to other cell lines (Fig. 2A–C). Cells were then subjected to ionizing radiation (IR) or doxorubicin (DXr), a DNA intercalating agent used clinically for the treatment of breast cancer patients. Survival was then evaluated using clonogenic assays (Fig. 2D). Inhibition of MT1-MMP was sufficient to significantly sensitize cells to both IR and DXR. To further attest this findings, MCF7 cells that express relative lower levels of MT1-MMP with respect to MDA-MB-231 (Fig. 2A), were transduced with a catalytically active and a catalytically dead mutant MT1-MMP (Fig. 2E). The latter lacks the catalytic domain and therefore has a molecular weight of approximately 34 KD versus 55–60 of the full length protein, as previously shown[11]. Interestingly, while the active protein protected cells from IR and DXR, the dead mutant sensitized them similarly to lack of MT1-MMP (Fig. 2F). This suggests MT1-MMP requires its catalytic activity to provide protection from genotoxic stresses and also, the mutant acts as a dominant negative[24], by counteracting the activity of endogenous MT1-MMP. The effects observed were not due to differences in cell growth, survival or senescence, as inhibition of MT1-MMP did not affect any of these features (Suppl. Fig. 1).
Figure 2: Expression of MT1-MMP affects cell responses to genotoxic stresses.
A) Expression of MT1-MMP in MDA-MB-231 (left), and other cell lines after removal of MDA-MB-231 and exposure of the film for a longer time (right). B) ER, PR, HER2 status of the cell lines in A (all cells are BRCA1WT). C) Knock down levels of MT1-MMP in MDA-MB-231. D) Clonogenic assay of the cells in C treated with the indicated IR doses or several doses of doxorubicin for 10 days (the duration of the clonogenic assay). *,**,***p<0.0001; #,##p<0.001, Student’s t test. E) Expression of empty (pLM), active full length (FL) and catalytically truncated (ΔCAT) MT1-MMP in MCF7. F) Clonogenic assay of the cells in E treated with the indicated IR doses or several doses of doxorubicin for 16 days (the duration of the clonogenic assay). *,**,***p<0.0001; @,@@p<0.005; $,$ $p<0.05, Student’s t test. G) MDA-MB-231 orthotopically injected in the fat pad of NSG mice (n=10 per group). DXR (2mg/Kg, i.p.) was administered twice weekly starting at day 37 post-inoculation. Tumor volumes were recorded twice weekly.
Finally, in order to more conclusively demonstrate the relevance of MT1-MMP inhibition as a sensitizer to chemotherapy in breast cancer, we employed an in vivo orthotopic xenograft model of triple negative breast cancer (TNBC). To this end, MDA-MB-231 cells expressing shGFP or shMT1-MMP (Fig. 2C) were injected in the inguinal fat pads of NSG mice. Tumors were allowed to grow to comparable sizes (~200mm3) prior to starting treatment with doxorubicin twice weekly. The data show that inhibition of MT1-MMP significantly enhances the tumor response to doxorubicin (Fig. 2G) indicating inhibition of MT1-MMP can indeed sensitize tumors to this chemiotherapic.
Together, these findings highlight a novel activity of the protease in controlling cancer cell responses to DNA damaging agents.
MT1-MMP affects DNA Double Strand Breaks (DSBs) and replication fork stress.
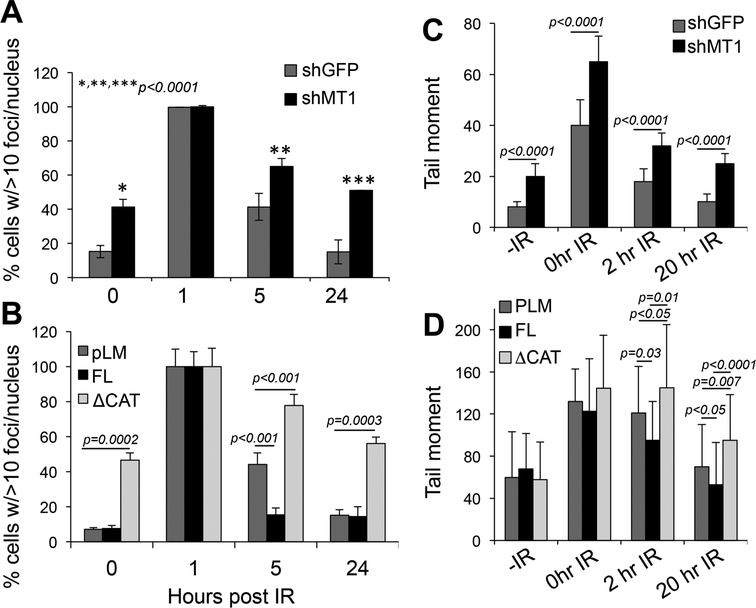
Both IR and DXR are known to cause DNA double strand breaks (DSBs)[25]. Hence to start understanding how MT1-MMP sensitizes cells to DNA damage, we assessed the levels of γH2AX, a marker of DSBs in response to changes in MT1-MMP levels, in MDA-MB-231. This revealed that silencing MT1-MMP alone, without any stressor, was sufficient to induce DNA damage. Figs. 3A and B (and suppl. Fig. 2A) show that cells depleted of MT1-MMP have a 4-fold increase in the number of γH2AX foci than shGFP cells. When treated with 5Gy IR, cells expressed similar levels of γH2AX regardless of the level of MT1-MMP at least at one hour post irradiation. However, the loss of MT1-MMP extended the time cells harbor unrepaired DNA with respect to control cells, which return to normal within 24 hours post irradiation. Conversely, the expression of a catalytically active MT1-MMP in MCF7 cells accelerated the time of recovery after IR, such that γH2AX levels returned to normal after 6 hours vs 24 of control cells (Fig. 3B and suppl. Fig. 2B). On the other hand, the dead mutant behaved similarly to the shMT1-MMP, causing higher levels of γH2AX in untreated cells that is maintained over time.
Figure 3: Inhibition of MT1-MMP increases DNA DSBs.
A) quantification of γH2AX foci in shGFP and shMT1-MMP MDA-MB-231 cells treated with 5Gy IR for the indicated times. B) quantification of γH2AX foci in MCF7 cells expressing an empty vector (pLM), the full length (FL) and the catalytically truncated (ΔCAT) MT1-MMP constructs, treated with 5Gy IR for the indicated times. C-D) Quantification of the tail moment at the indicated times after treatment with 8Gy IR. C - MDA-MB cells expressing shGFP and shMT1-MMP as shown in 2C; D – MCF7 expressing pLM, full length and catalytically truncated MT1-MMP mutant as shown in 2E. P values were calculated by the Student’s t test. Foci were counted in al least 50 cells. Tail moments were measured in at least 50 cells per sample.
To measure damage repair in MT1-MMP proficient and deficient cells, we performed a neutral comet assay (to quantify DSBs) in MDA-MB-231 shGFP and shMT1-MMP stable knockdown cells (Fig. 3C and suppl. Figs. 3A and suppl. Fig. 4). As expected, shMT1-MMP cells began with more DNA damage and accumulated greater damage upon IR. In contrast, control cells displayed more extensive repair within 20 hours post-IR and at 48 hours post 2Gy IR. Again, expression of a catalytically active MT1-MMP in MCF7 cells significantly accelerated the time of repair, whereas the dead mutant resulted in retention of damage (Fig. 3D and suppl. Fig. 3B).
DSBs can result from exogenous stresses, such as ionizing radiation (IR), and chemical compounds (e.g. DXR) or from endogenous insults such as DNA replication stress[26, 27]. The suppression of MT1-MMP alone results in DNA damage, whereas the expression of active MT1-MMP accelerate DNA repair, suggesting that MT1-MMP may modulate endogenous stresses. We therefore tested the possibility that MT1-MMP normally represses replication errors.
A hallmark of replication stress is the formation of excessive single stranded DNA (ssDNA) caused by stalling of replication forks[28], that if unrepaired, collapse into DSBs[29]. Markers of replication stress and DSBs include the activation of ATR/Chk1 and ATM-/Chk2, phosphorylation of H2AX (γH2AX), and of the ssDNA binding protein RPA32. Hence, we assessed the phosphorylation status of these four factors in response to modulation of MT1-MMP. Notably, the phosphorylation of all four factors was increased with MT1-MMP silencing (Fig. 4A), and, conversely, reduced in cells expressing active MT1-MMP (Fig. 4A, B). Again, the dead mutant MT1-MMP behaved similarly to MT1-MMP silencing causing an increase in phosphorylation of these factors. Additionally, MT1-MMP silencing resulted in slight, although consistent, accumulation of cells in G2/M (Suppl. Fig. 5)[30], which is a known response to replication fork stalling and collapse into DNA DSBs[31].
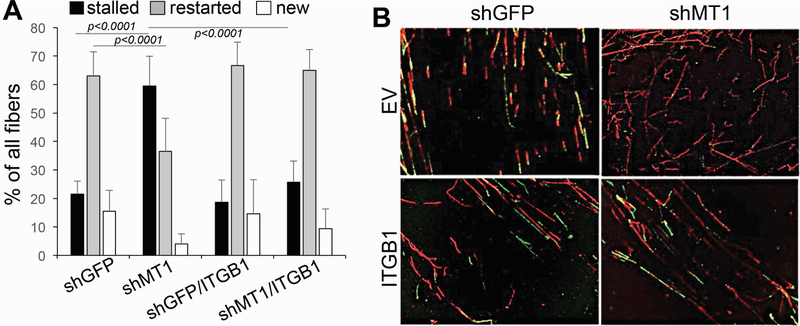
Figure 4: MT1-MMP affects ATM, ATR and replication fork stability.
A) MDA-MB-231 cells expressing shGFP or shMT1-MMP (left), and MCF7 expressing active and catalytically dead MT1-MMP (right), showing phosphorylation levels of H2AX, Chk1, Chk2 and RPA32. B) Scheme of treatment. Red only fibers: stalled forks; Green only fibers: new origin; Red+Green: restarted forks after Hydroxy Urea. C) % of replication forks. A total of 200 DNA fibers were counted in all groups. % of all fibers was calculated by the Student’s t test. *,**,***p<0.0001; D) speed at which forks are produced. *p<0.01. E-F) Representative CldU and IdU stained DNA fibers.
Hence, these data suggest inhibition of MT1-MMP may cause stalling of replication forks. To determine whether MT1-MMP affects DNA replication, we performed a DNA fiber assay using shGFP and shMT1-MMP expressing cells (Fig. 4C, left panel) or cells expressing the catalytically active and dead mutant MT1-MMP (Fig. 4C right panel, D). Cells were incubated with IdU (Iodo-deoxyuridine), followed by the addition of Hydroxy Urea (HU) to deplete the nucleotide pool and stall replication forks. HU was then removed, and the cells were labeled with CldU (chloro-deoxyuridine) to quantify the ability of replication forks to restart. While shGFP (control) cells were able to efficiently restart stalled forks and also fired new origins, shMT1-MMP cells had 4-fold more stalled forks, a 4-fold reduction in restarted forks, and no new origins. On the other hand, the dead mutant MT1-MMP resulted in a 2.4-fold increase in stalled forks and 1.4-fold decrease in restarted forks. Interestingly, while the expression of active MT1-MMP did not overtly affect replication fork dynamics, it did increase the speed at which forks are produced (Fig. 4D). Overall, these data indicate that cells depleted of the membrane bound matrix metalloproteinase MT1-MMP undergo severe replication stress represented by stalled replication forks. This is followed by persistent unrepaired DNA, with accumulation of DSBs[29, 30, 32] that likely increase cellular sensitivity to additional genotoxic stresses such as IR and doxorubicin.
MT1-MMP acts through integrinβ1.
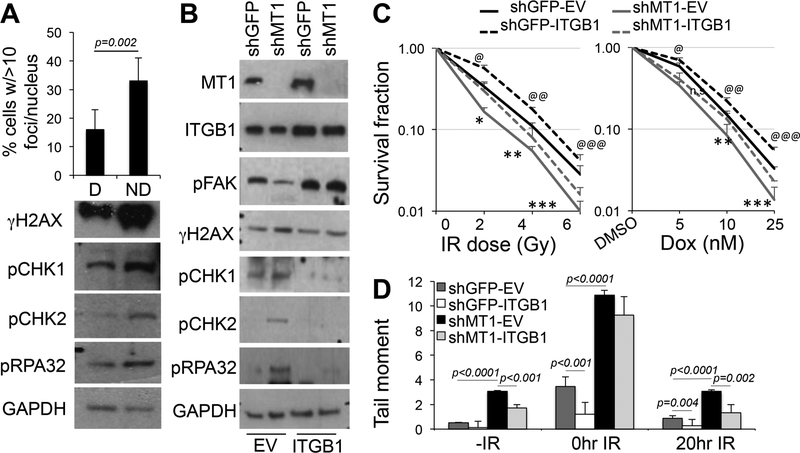
Our data show MT1-MMP requires its catalytic activity to exert its protective effects against IR and chemotherapy, supporting a role of MT1-MMP-derived proteolytic products. MT1-MMP is a major collagenase[33] required by cancer cells to break down collagen in the extracellular matrix (ECM) for invasion[21]; and can process other ECM components including fibronectin and laminins[34]. Hence, we hypothesized that MT1-MMP mediates its protective effects via processing of ECM components. To test this possibility, cells were embedded in synthetic poly(ethylene glycol) hydrogels that incorporate MT1-MMP-sensitive or -insensitive peptide cross-bridges[35, 36]. Interestingly, MT1-MMP proficient cells showed a 2-fold increase in γH2AX foci (Fig. 5A, upper panel) and activation of DDR and replication fork stress markers when seeded in MT1-MMP resistant gels (Fig. 5A, lower panel), indicating MT1-MMP confers its protective effects in part through the processing of ECM components.
Figure 5: MT1-MMP acts through integrinβ1.
A) Quantification of γH2AX foci of MDA-MB-231 cells seeded in MT1-MMP degradable (D) and non-degradable (ND) matrixes for 24 hours. Foci were quantified from at least 50 cells. Data are the mean of two independent experiments. Bottom panel: western blotting to detect γH2AX, pChK1, pChK2, pRPA32 of cells embedded in the matrixes. B) Expression of integrinβ1V37N (ITGB1) in MDA-MB-231 cells expressing shGFP or shMT1-MMP. pFAKY297 and the markers in A are also shown. C) Clonogenic assay of the cells in B treated with the indicated IR doses or several doses of doxorubicin for 15 days (the duration of the clonogenic assay). (Left: *,**,***p<0.001; @,@@,@@@p<0.001; Right: **,***p=0.005; @,@@,@@@p<0.001, Student’s t test). D) Quantification of the tail moment at the indicated times after treatment with 8Gy IR of the cells in B. Tail moments were measured in at least 100 cells per sample. P values were calculated by the Student’s t test.
A major cell surface receptor of ECM collagens is integrinβ1[37]. Intriguingly, integrinβ1 has been implicated in resistance to chemo- and radiotherapy in breast cancer[38]. MT1-MMP has been shown to activate integrinβ1, and drive osteogenic differentiation versus adipogenic through processing of the ECM[36]. Hence, we sought to determine whether MT1-MMP protective effects towards genotoxic stresses were mediated by integrinβ1. MDAMB-231 cells expressing either a control shRNA (shGFP) or shMT1-MMP, were transduced with either an empty vector (EV) or the constitutive active integrinβ1V37N (ITGB1), a mutant that supports integrin self-clustering (Fig. 5B)[36, 39]. Inhibition of MT1-MMP was accompanied by a decrease in integrinβ1 activity as indicated by the reduction of FAK phosphorylation at Y397 (p-FAK), a downstream target of activated integrinβ1[39, 40], as we have previously shown[15]. Reintroduction of activated integrinβ1 restored FAK activation. Interestingly, while inhibition of MT1-MMP induced a DDR resulting in activation of several markers of DNA damage response and fork stress, reconstitution of active integrinβ1 dampened these effects, reducing the phosphorylation of all the tested markers. This implies integrinβ1 should improve/rescue cell viability that is inhibited by reduced MT1-MMP after IR and chemotherapy. Indeed, survival, measured by clonogenic assay, of cells depleted of MT1-MMP and then irradiated or treated with various doses of doxorubicin, was significantly increased after restoration of active integrinβ1 (Fig. 5C). Likewise, the DNA repair capabilities of cells expressing activated integrinβ1 were superior with respect to the corresponding counterparts expressing shGFP or shMT1-MMP, as shown by neutral comet assay (Fig. 5D). The shMT1-MMP cells expressing integrinβ1 showed an almost complete recovery at 20 hours post irradiation similar to control, shGFP expressing cells.
Finally, given that MT1-MMP inhibition causes stalling and collapse of replication forks, we conducted a DNA fiber assay to conclusively determine whether integrinβ1 could protect from replication stress. Indeed, the presence of activated integrinβ1 was sufficient to restore DNA replication as indicated by the increase to the levels of controls of restated forks in cells depleted of MT1-MMP and also promoted the firing of new origins similarly to control cells (Fig. 6).
Figure 6: Integrinβ1 rescues stalling of replication forks caused by MT1-MMP inhibition.
A) % of replication forks. A total of 200 DNA fibers were counted in all groups. % of all fibers was calculated by the Student’s t test. B) Representative CldU and IdU stained DNA fibers. Red: stalled forks; Green: new origin; Red+Green: restarted forks after Hydroxy Urea.
Together, these data demonstrate MT1-MMP controls an outside-in DDR via processing of the ECM and activation of integrinβ1.
Discussion
Despite significant advances in breast cancer treatment, several patients still rely on chemo- and radiotherapy for the management of the disease. However, the efficacy of these treatments is often undermined by the emergence of resistance. Here we demonstrate a novel mechanism by which triple negative breast cancer cells may better withstand genotoxic stresses through the expression of the membrane tethered matrix metalloproteinase MT1-MMP. We show that inhibition of MT1-MMP is sufficient to sensitize cells to IR and doxorubicin by causing stalling and collapse of replication forks. MT1-MMP mediates its protective effects by triggering an outside-in signaling pathway initiated by the processing of the ECM and activation of integrinβ1. Together, these data highlight MT1-MMP as a novel modulator of DNA damage responses and a potential novel radio and chemo-sensitizer.
MT1-MMP is a major collagenase capable of directly degrading fibrillar collagens such as type I collagen, of which the ECM is particularly enriched, and that is normally resistant to other proteases at physiological pH[34]. Due to this capability, MT1-MMP is implicated in the processing and remodeling of several structures during growth[33, 41] and in the processing of the ECM during cancer cell invasion[12–14]. We have previously shown that MT1-MMP is a poor prognostic factor for melanoma patients and it plays an essential role in melanoma metastases[15]. MT1-MMP has also been shown to be involved in breast cancer invasion and poorer patient outcome in association with VEGF[42, 43]. Our data reiterate these previous findings by showing a correlation between MT1-MMP and breast cancer cell progression; but most importantly, show MT1-MMP expression is associated with a poorer outcome in patients that underwent chemotherapy. Hence, MT1-MMP can potentially function as a new predictor of response to these treatments.
Of note, MT1-MMP has also been shown to promote cancer cell invasion through catalytically independent mechanisms. For example, MT1-MMP can induce RAC1 activation via the hemopexin domain [44]. However, our data points at the enzymatic ECM processing role of MT1-MMP in providing protection from genotoxic stresses. In fact, a catalytic dead mutant of MT1-MMP[24] acts as dominant negative likely by counteracting endogenous MT1-MMP, thus inducing DSBs and replication fork stalling, which renders cells more susceptible to radiation and DXR, similarly to inhibition of MT1-MMP. Additionally, our data suggest MT1-MMP is likely to exert its protective effects through the processing of the ECM. Indeed, an MT1-MMP resistant matrix, i.e. an artificial matrix that is uncleavable by MT1-MMP, mimics MT1-MMP inhibition as cells accumulate DNA damage.
The ECM has been shown to contribute to chemotherapy resistance both by functioning as a mechanical barrier to drugs such as doxorubicin, vinblastine and methotrexate which bind to ECM macromolecules making them less capable of penetrating the tumor mass[45]; and by providing anti-apoptotic cues through the interaction of ECM components with cancer cells[46]. This type of resistance, which is also observed in radiotherapy[47], is mostly driven by activation of an integrinβ1 mediated survival signaling pathway. Our work demonstrates that MT1-MMP lays upstream of integrinβ1, at the ECM-integrin interface. Inhibition of MT1-MMP reduces integrinβ1 activity, whereas restoration of active integrinβ1 protects from DNA damage caused by inhibition of MT1-MMP.
The precise mechanism by which MT1-MMP activates integrinβ1 in our system is not known. We have speculated that MT1-MMP does so by cleaving collagen and possibly other components of the ECM, which in turn bind and activate integrinβ1. Studies are underway to address this question and unravel the mechanisms leading from the ECM-MT1-MMP-integrinβ1 to the DNA. Nonetheless, our data suggest that inhibition of MT1-MMP would not only prevent metastatic spread, but it would also sensitize cancer cells to radiation and chemotherapy by disrupting the ECM-cell survival cues.
From a therapeutic stand point, new small molecule inhibitors and neutralizing antibody against specific MMPs are being tested, and now several studies have demonstrated the anti-tumor activity of the specific targeting of MT1-MMP[48–50]. For example, an anti MT1-MMP neutralizing antibody has demonstrated anti-tumor activity in a triple negative breast cancer model, by suppressing several hallmarks of cancer including growth, metastasis, angiogenesis and immunosuppression[51]. Likewise, we have previously shown that a small molecule inhibitor of MT1-MMP catalytic activity exerted anti-melanoma activity by decreasing both growth and metastasis[52]. Hence, given the availability of selective inhibitors and the relevance of MT1-MMP in triple negative breast cancer response to genotoxic therapies, specific targeting of MT1-MMP in combination with standard chemo- and radiotherapy might represent an intriguing new avenue in treating aggressive breast cancer resulting in improved patient survival.
Supplementary Material
Chermotherapy and radiation are still standard of care for many breast cancer patients
Resistance to these therapies is common underscoring a need for novel sensitizers
Depletion of MT1-MMP, via dampening of integrinβ1 signaling, causes DNA replication stress and double strand breaks.
MT1-MMP, a zinc dependenr matrix metalloproteinase bound the the plasma membrane, provides cytoportective effects against chemotherapy and radiation
Tumor cells are sensitized to chemo- and radiotherapy
Acknowledgment:
*This work was supported by NIH grants R01 CA177652 and R21 CA187695 to BB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest
Bibliography
- [1].American Cancer Society. Cancer Facts & Figures, Atlanta: American Cancer Society, (2018). [Google Scholar]
- [2].Santa-Maria CA, Gradishar WJ, Changing Treatment Paradigms in Metastatic Breast Cancer: Lessons Learned, JAMA Oncol, 1 (2015) 528–534; quiz 549. [DOI] [PubMed] [Google Scholar]
- [3].Langlands FE, Horgan K, Dodwell DD, Smith L, Breast cancer subtypes: response to radiotherapy and potential radiosensitisation, Br J Radiol, 86 (2013) 20120601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Poincloux R, Lizarraga F, Chavrier P, Matrix invasion by tumour cells: a focus on MT1-MMP trafficking to invadopodia, J Cell Sci, 122 (2009) 3015–3024. [DOI] [PubMed] [Google Scholar]
- [5].Kajita M, Itoh Y, Chiba T, Mori H, Okada A, Kinoh H, Seiki M, Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration, J Cell Biol, 153 (2001) 893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jin G, Zhang F, Chan KM, Xavier Wong HL, Liu B, Cheah KS, Liu X, Mauch C, Liu D, Zhou Z, MT1-MMP cleaves Dll1 to negatively regulate Notch signalling to maintain normal B-cell development, EMBO J, 30 (2011) 2281–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Koshikawa N, Mizushima H, Minegishi T, Iwamoto R, Mekada E, Seiki M, Membrane type 1-matrix metalloproteinase cleaves off the NH2-terminal portion of heparin-binding epidermal growth factor and converts it into a heparin-independent growth factor, Cancer Res, 70 (2010) 6093–6103. [DOI] [PubMed] [Google Scholar]
- [8].Sabbota AL, Kim HR, Zhe X, Fridman R, Bonfil RD, Cher ML, Shedding of RANKL by tumor-associated MT1-MMP activates Src-dependent prostate cancer cell migration, Cancer Res, 70 (2010) 5558–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Golubkov VS, Aleshin AE, Strongin AY, Potential relation of aberrant proteolysis of human protein tyrosine kinase 7 (PTK7) chuzhoi by membrane type 1 matrix metalloproteinase (MT1-MMP) to congenital defects, J Biol Chem, 286 (2011) 20970–20976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Golubkov VS, Chekanov AV, Cieplak P, Aleshin AE, Chernov AV, Zhu W, Radichev IA, Zhang D, Dong PD, Strongin AY, The Wnt/planar cell polarity protein-tyrosine kinase-7 (PTK7) is a highly efficient proteolytic target of membrane type-1 matrix metalloproteinase: implications in cancer and embryogenesis, J Biol Chem, 285 (2010) 35740–35749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ma J, Tang X, Wong P, Jacobs B, Borden EC, Bedogni B, Noncanonical Activation of Notch1 Protein by Membrane Type 1 Matrix Metalloproteinase (MT1-MMP) Controls Melanoma Cell Proliferation, J Biol Chem, 289 (2014) 8442–8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lodillinsky C, Infante E, Guichard A, Chaligne R, Fuhrmann L, Cyrta J, Irondelle M, Lagoutte E, Vacher S, Bonsang-Kitzis H, Glukhova M, Reyal F, Bieche I, Vincent-Salomon A, Chavrier P, p63/MT1-MMP axis is required for in situ to invasive transition in basal-like breast cancer, Oncogene, 35 (2016) 344–357. [DOI] [PubMed] [Google Scholar]
- [13].Li Y, Cai G, Yuan S, Jun Y, Li N, Wang L, Chen F, Ling R, Yun J, The overexpression membrane type 1 matrix metalloproteinase is associated with the progression and prognosis in breast cancer, Am J Transl Res, 7 (2015) 120–127. [PMC free article] [PubMed] [Google Scholar]
- [14].Perentes JY, Kirkpatrick ND, Nagano S, Smith EY, Shaver CM, Sgroi D, Garkavtsev I, Munn LL, Jain RK, Boucher Y, Cancer cell-associated MT1-MMP promotes blood vessel invasion and distant metastasis in triple-negative mammary tumors, Cancer Res, 71 (2011) 4527–4538. [DOI] [PubMed] [Google Scholar]
- [15].Shaverdashvili K, Wong P, Ma J, Zhang K, Osman I, Bedogni B, MT1-MMP modulates melanoma cell dissemination and metastasis through activation of MMP2 and RAC1, Pigment Cell Melanoma Res, 27 (2014) 287–296. [DOI] [PubMed] [Google Scholar]
- [16].Shaverdashvili K, Zhang K, Osman I, Honda K, Jobava R, Bedogni B, MT1-MMP dependent repression of the tumor suppressor SPRY4 contributes to MT1-MMP driven melanoma cell motility, Oncotarget, 6 (2015) 33512–33522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hirsch FR, Varella-Garcia M, Bunn PA Jr., Di Maria MV, Veve R, Bremmes RM, Baron AE, Zeng C, Franklin WA, Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis, J Clin Oncol, 21 (2003) 3798–3807. [DOI] [PubMed] [Google Scholar]
- [18].John T, Liu G, Tsao MS, Overview of molecular testing in non-small-cell lung cancer: mutational analysis, gene copy number, protein expression and other biomarkers of EGFR for the prediction of response to tyrosine kinase inhibitors, Oncogene, 28 Suppl 1 (2009) S14–23. [DOI] [PubMed] [Google Scholar]
- [19].Zhang K, Wong P, Zhang L, Jacobs B, Borden EC, Aster JC, Bedogni B, A Notch1-neuregulin1 autocrine signaling loop contributes to melanoma growth, Oncogene, 31(43) October 25 (2012) 4609–4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yang X, Pan Y, Qiu Z, Du Z, Zhang Y, Fa P, Gorityala S, Ma S, Li S, Chen C, Wang H, Xu Y, Yan C, Ruth K, Ma Z, Zhang J, RNF126 as a Biomarker of a Poor Prognosis in Invasive Breast Cancer and CHEK1 Inhibitor Efficacy in Breast Cancer Cells, Clin Cancer Res, 24 (2018) 1629–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Marchesin V, Castro-Castro A, Lodillinsky C, Castagnino A, Cyrta J, Bonsang-Kitzis H, Fuhrmann L, Irondelle M, Infante E, Montagnac G, Reyal F, Vincent-Salomon A, Chavrier P, ARF6-JIP¾ regulate endosomal tubules for MT1-MMP exocytosis in cancer invasion, J Cell Biol, 211 (2015) 339–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM, ONCOMINE: a cancer microarray database and integrated data-mining platform, Neoplasia, 6 (2004) 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z, An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients, Breast Cancer Res Treat, 123 (2010) 725–731. [DOI] [PubMed] [Google Scholar]
- [24].Miller MC, Manning HB, Jain A, Troeberg L, Dudhia J, Essex D, Sandison A, Seiki M, Nanchahal J, Nagase H, Itoh Y, Membrane type 1 matrix metalloproteinase is a crucial promoter of synovial invasion in human rheumatoid arthritis, Arthritis Rheum, 60 (2009) 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Banath JP, Olive PL, Expression of phosphorylated histone H2AX as a surrogate of cell killing by drugs that create DNA double-strand breaks, Cancer Res, 63 (2003) 4347–4350. [PubMed] [Google Scholar]
- [26].Bohgaki T, Bohgaki M, Hakem R, DNA double-strand break signaling and human disorders, Genome Integr, 1 (2010) 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Jackson SP, Bartek J, The DNA-damage response in human biology and disease, Nature, 461 (2009) 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mazouzi A, Velimezi G, Loizou JI, DNA replication stress: causes, resolution and disease, Exp Cell Res, 329 (2014) 85–93. [DOI] [PubMed] [Google Scholar]
- [29].Ward IM, Chen J, Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress, J Biol Chem, 276 (2001) 47759–47762. [DOI] [PubMed] [Google Scholar]
- [30].Chanoux RA, Yin B, Urtishak KA, Asare A, Bassing CH, Brown EJ, ATR and H2AX cooperate in maintaining genome stability under replication stress, J Biol Chem, 284 (2009) 5994–6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nam EA, Cortez D, ATR signalling: more than meeting at the fork, Biochem J, 436 (2011) 527–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Downey M, Durocher D, gammaH2AX as a checkpoint maintenance signal, Cell Cycle, 5 (2006) 1376–1381. [DOI] [PubMed] [Google Scholar]
- [33].Holmbeck K, Bianco P, Yamada S, Birkedal-Hansen H, MT1-MMP: a tethered collagenase, J Cell Physiol, 200 (2004) 11–19. [DOI] [PubMed] [Google Scholar]
- [34].Itoh Y, Membrane-type matrix metalloproteinases: Their functions and regulations, Matrix Biol, 44–46 (2015) 207–223. [DOI] [PubMed] [Google Scholar]
- [35].Ehrbar M, Sala A, Lienemann P, Ranga A, Mosiewicz K, Bittermann A, Rizzi SC, Weber FE, Lutolf MP, Elucidating the role of matrix stiffness in 3D cell migration and remodeling, Biophys J, 100 (2011) 284–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tang Y, Rowe RG, Botvinick EL, Kurup A, Putnam AJ, Seiki M, Weaver VM, Keller ET, Goldstein S, Dai J, Begun D, Saunders T, Weiss SJ, MT1-MMP-dependent control of skeletal stem cell commitment via a beta1-integrin/YAP/TAZ signaling axis, Dev Cell, 25 (2013) 402–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yeh YC, Lin HH, Tang MJ, A tale of two collagen receptors, integrin beta1 and discoidin domain receptor 1, in epithelial cell differentiation, Am J Physiol Cell Physiol, 303 (2012) C1207–1217. [DOI] [PubMed] [Google Scholar]
- [38].Park CC, Zhang H, Pallavicini M, Gray JW, Baehner F, Park CJ, Bissell MJ, Beta1 integrin inhibitory antibody induces apoptosis of breast cancer cells, inhibits growth, and distinguishes malignant from normal phenotype in three dimensional cultures and in vivo, Cancer Res, 66 (2006) 1526–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM, Tensional homeostasis and the malignant phenotype, Cancer Cell, 8 (2005) 241–254. [DOI] [PubMed] [Google Scholar]
- [40].Eyckmans J, Boudou T, Yu X, Chen CS, A hitchhiker’s guide to mechanobiology, Dev Cell, 21 (2011) 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Holmbeck K, Bianco P, Caterina J, Yamada S, Kromer M, Kuznetsov SA, Mankani M, Robey PG, Poole AR, Pidoux I, Ward JM, Birkedal-Hansen H, MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover, Cell, 99 (1999) 81–92. [DOI] [PubMed] [Google Scholar]
- [42].Jiang WG, Davies G, Martin TA, Parr C, Watkins G, Mason MD, Mansel RE, Expression of membrane type-1 matrix metalloproteinase, MT1-MMP in human breast cancer and its impact on invasiveness of breast cancer cells, Int J Mol Med, 17 (2006) 583–590. [PubMed] [Google Scholar]
- [43].Yao G, He P, Chen L, Hu X, Gu F, Ye C, MT1-MMP in breast cancer: induction of VEGF-C correlates with metastasis and poor prognosis, Cancer Cell Int, 13 (2013) 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Cao J, Kozarekar P, Pavlaki M, Chiarelli C, Bahou WF, Zucker S, Distinct roles for the catalytic and hemopexin domains of membrane type 1-matrix metalloproteinase in substrate degradation and cell migration, J Biol Chem, 279 (2004) 14129–14139. [DOI] [PubMed] [Google Scholar]
- [45].Di Paolo A, Bocci G, Drug distribution in tumors: mechanisms, role in drug resistance, and methods for modification, Curr Oncol Rep, 9 (2007) 109–114. [DOI] [PubMed] [Google Scholar]
- [46].Hodkinson PS, Mackinnon AC, Sethi T, Extracellular matrix regulation of drug resistance in small-cell lung cancer, Int J Radiat Biol, 83 (2007) 733–741. [DOI] [PubMed] [Google Scholar]
- [47].Nam JM, Chung Y, Hsu HC, Park CC, beta1 integrin targeting to enhance radiation therapy, Int J Radiat Biol, 85 (2009) 923–928. [DOI] [PubMed] [Google Scholar]
- [48].Remacle AG, Cieplak P, Nam DH, Shiryaev SA, Ge X, Strongin AY, Selective function-blocking monoclonal human antibody highlights the important role of membrane type-1 matrix metalloproteinase (MT1-MMP) in metastasis, Oncotarget, 8 (2017) 2781–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Udi Y, Grossman M, Solomonov I, Dym O, Rozenberg H, Moreno V, Cuniasse P, Dive V, Arroyo AG, Sagi I, Inhibition mechanism of membrane metalloprotease by an exosite-swiveling conformational antibody, Structure, 23 (2015) 104–115. [DOI] [PubMed] [Google Scholar]
- [50].Devy L, Huang L, Naa L, Yanamandra N, Pieters H, Frans N, Chang E, Tao Q, Vanhove M, Lejeune A, van Gool R, Sexton DJ, Kuang G, Rank D, Hogan S, Pazmany C, Ma YL, Schoonbroodt S, Nixon AE, Ladner RC, Hoet R, Henderikx P, Tenhoor C, Rabbani SA, Valentino ML, Wood CR, Dransfield DT, Selective inhibition of matrix metalloproteinase-14 blocks tumor growth, invasion, and angiogenesis, Cancer Res, 69 (2009) 1517–1526. [DOI] [PubMed] [Google Scholar]
- [51].Ling B, Watt K, Banerjee S, Newsted D, Truesdell P, Adams J, Sidhu SS, Craig AWB, A novel immunotherapy targeting MMP-14 limits hypoxia, immune suppression and metastasis in triple-negative breast cancer models, Oncotarget, 8 (2017) 58372–58385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Marusak C, Bayles I, Ma J, Gooyit M, Gao M, Chang M, Bedogni B, The thiirane-based selective MT1-MMP/MMP2 inhibitor ND-322 reduces melanoma tumor growth and delays metastatic dissemination, Pharmacol Res, 113 (2016) 515–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.