Abstract
Cytochrome P450cam is an archetypal example of the vast family of heme monooxygenases and serves as a model for an enzyme that is highly specific for both its substrate and reductase. During catalysis, it undergoes significant conformational changes of the F and G helices upon binding its substrate and redox partner, putidaredoxin (Pdx). Recent studies have shown that Pdx binding to the closed camphor-bound form of ferric P450cam results in its conversion to a fully open state. However, during catalytic turnover, it remains unclear whether this same conformational change also occurs or whether it is coupled to the formation of the critical compound I intermediate. Here, we have examined P450cam bound simultaneously by camphor, CN−, and Pdx as a mimic of the catalytically competent ferrous oxy-P450cam-Pdx state. The combined use of double electron–electron resonance and molecular dynamics showed direct observation of intermediate conformational states of the enzyme upon CN− and subsequent Pdx binding. This state is coupled to the movement of the I helix and residues at the active site, including Arg-186, Asp-251, and Thr-252. These movements enable occupation of a water molecule that has been implicated in proton delivery and peroxy bond cleavage to give compound I. These findings provide a detailed understanding of how the Pdx-induced conformational change may sequentially promote compound I formation followed by product release, while retaining stereoselective hydroxylation of the substrate of this highly specific monooxygenase.
Graphical Abstract
The cytochromes P450 comprise an enormous family of heme-containing monooxygenases that catalyze the oxidation of a range of substrates involved in drug metabolism, steroid biosynthesis, and catabolism of xenobiotic compounds as energy sources.1,2 The substrate is oxidized during each catalytic cycle, and O2 is required along with a redox partner, usually a ferredoxin, to provide two electrons to the P450 heme. Using an ordered sequential mechanism, the substrate first binds to the low-spin state of the ferric enzyme to generate a high-spin enzyme–substrate complex.3,4 One-electron reduction is followed by O2 binding and then a second one-electron reduction to give the ferric hydroperoxo species.5,6 This intermediate is short-lived, and with the delivery of a critical proton, the peroxy bond is cleaved to give compound I, the key intermediate capable of substrate hydroxylation via a radical rebound mechanism.7
The camphor-specific P450 from Pseudomonas putida, CYP101A1 (P450cam), has been extensively investigated as a model for structure–function relationships in other P450 variants.8 It shares the general protein fold and catalytic cycle with all known forms of P450.8,9 However, P450cam is unusually specific for its substrate camphor, converting from an open to a closed conformation in response to substrate binding.10,11 It is also highly specific for its redox partner by accepting putidaredoxin (Pdx) and no other electron donor.12,13 X-ray crystallography,14 double electron–electron resonance (DEER),15 and NMR measurements16 all show that Pdx binds to the proximal side of the enzyme, distant from the substrate binding pocket. When oxidized putidaredoxin (Pdxox) binds to camphor/P450cam, it causes conversion of the enzyme from the closed to the fully open state,14,17,18 suggesting that this may account for the well-known effector role of Pdx on P450cam function.19–23 When either Pdxox or reduced putidaredoxin (Pdxred) binds to Fe2+–CO/camphor, the enzyme remains in the closed form, suggesting that the Pdx-induced conformational shift may not occur in the reduced forms.18 However, the Fe2+–CO complex is not a perfect model for the Fe2+–O2 form, so the inability of Pdxred to induce a conformational shift in this state may result from subtle effects of the distal heme coordination environment.24
These results have left important questions unanswered about how Pdx, and no other electron donor, allows the second electron transfer and its subsequent advancement through the catalytic cycle. Central in this debate is whether the enzyme exists in the closed, open, or other uncharacterized intermediate states during the electron transfer from Pdxred to Fe2+–O2/camphor and also during the conversion to the compound I intermediate. Although Pdxox binding results in the conversion of camphor/P450cam to the open state, there are arguments that the fully open state is not ideally suited for catalysis.18,25–27 Camphor binds to the open form of P450cam in multiple orientations, which is not consistent with the observed high regio- and stereoselectivity of its reaction.18 If the enzyme transitioned from the closed to the open state immediately upon Pdxred binding and prior to subsequent catalytic steps, one would expect an increase in the probability of releasing unreacted substrate or reactive oxygen species, and this is also not fully consistent with the highly coupled turnover of this enzyme. These arguments have led to the suggestion that an intermediate state, in between the closed and open forms, may be involved in the critical electron transfer and substrate oxidation steps.18,27 Indeed, recent molecular dynamics calculations support the role of Pdx in favoring conformations that are intermediate between open and closed.25,27 However, only indirect physical evidence for such an intermediate state has been obtained to date. A library of tethered substrates bound to P450cam have been structurally characterized and some of them populate the fully open state, while others induce a conformation that is intermediate between the open and closed form.28 These large substrate analogues are too large to allow closure of the substrate access channel, so it is not clear if the intermediate state observed here is representative of any such state that may occur in the catalytic cycle with native substrates. To obtain a better understanding of the Pdx-induced activation of P450cam during the catalytic cycle, we have performed a series of DEER and molecular dynamics (MD) experiments using CN−/camphor as a mimic of the Fe2+–O2/camphor state. The results clearly show the presence of at least two intermediate conformations of the enzyme that have significant implications on the critical steps of this important reaction.
METHODS
Mutant Construction, Protein Expression, and Purification.
The plasmid pETCAM-C334A was used for P450cam expression in Escherichia coli. In order for site-directed spin-labeling (SDSL), four exposed cysteines, C58, C85, C136, and C285, were mutated to serine, and three mutants, S48C, Y179C, and D251N, were introduced by using overlap extension PCR. Protein expression and purification for P450cam and Pdx were performed as previously described.10 P450cam used in this study had a ratio of A391/A280 > 1.45, and purified Pdx had a ratio of A412/A280 > 0.48. Before spin-labeling, all buffers contained 1 mM dithiothreitol (DTT) to prevent the formation of intermolecular disulfide bonds. Rates of NADH oxidation were measured in 50 mM potassium phosphate buffer, 1 mM camphor, 200 μM NADH, 0.5 μM Pdr, 5 μM Pdx, and 0.5 μM P450cam at 25 °C and pH 7.5.
The NADH concentration was measured by monitoring the absorbance change at 340 nm on an Agilent 8453 UV/visible spectrophotometer, and an extinction coefficient of 6.22 mM−1 cm−1 was used.29,30
EPR Sample Preparation.
P450cam samples were labeled with 4-maleimido-2,2,6,6-tetramethyl-1-piperidinyloxy (4MT). Before spin-labeling, DTT was removed via two sequential PD-10 columns (GE Healthcare) with 100 mM Tris (pH 7.5), and a 10-fold molar excess of 4MT was added for 10 min at room temperature followed by 6× buffer exchange with an Amicon centrifugal filter with 50 mM Tris, 1 mM camphor, 300 mM KCl, and 60 mM KCN to remove the excess of 4MT. The final EPR samples were adjusted to 100 μM P450cam containing 30% glycerol-d8 (Cambridge Isotopes Laboratory), and either 50 mM Tris (for substrate-free) or 50 mM Tris, 300 mM KCl, 1 mM camphor (for substrate-bound), and 60 mM KCN (for the CN− complex), and 200 μM Pdx was added for the P450cam/Pdx complex. The pD of the buffer was 7.6 to prevent maleimides from reacting with additional deprotonated primary amines. For cw-EPR samples, a 100 μL sample volume was loaded into 4 mm OD quartz tubes (Wilmad 706-PQ-9.50), and 15 μL was loaded into a 1.6 mm OD quartz tube (Vitrocom Inc.) for pulsed EPR experiments, and all samples were flash frozen and stored in liquid nitrogen.
EPR Data Collection and Analysis.
X-band continuous wave (CW)-EPR measurements were performed on a Bruker ESC106 spectrometer at 9.5 GHz/0.34 T with a Bruker SHQE resonator. Low-spin EPR spectra were obtained with a modulation frequency of 100 kHz at 50 K, 0.2 mW, and high-spin EPR signals were collected at 15 K, 2 mW. DEER spectra were collected on a Bruker ELEXSYS E580 spectrometer at 34 GHz using a Q-Band EN 5107D2 resonator at 20 K. The four pulsed DEER sequence was
The π/2 and π pulse lengths were 16 and 32 ns for the probe pulses, respectively, 24 ns was used for the pump pulse, and the frequency difference between probe and pump pulses was 80 MHz. DEER time traces and resultant distance distributions were analyzed by DeerAnalysis 2015.31
Molecular Dynamics Simulations.
The starting structures were prepared from crystal structures in Protein Data Bank entries (PDB IDs 3l63, 4jx1, 1o76, and 2a1n). Closed P450cam was from 3l63, camphor/P450cam/Pdx was from 4jx1, CN−/camphor was constructed by replacing the heme iron moiety of 4jx1 with the cyanide-bound heme moiety of 1o76, and the D251N mutant was from 2a1n with the oxygen removed. The cross-linker was removed, and mutants were reverted to the wild type. The parameters for the iron sulfur cluster of Pdx and the heme iron moiety for ferric camphor-bound, camphor-free, and cyanide-bound heme were parametrized using the MCPB.py model in AmberTools 16.32 To parametrize the 4MT spin label, Gaussian 09 was used to optimize the molecular structure and to determine the molecular electrostatic potential (MEP) at the HF/6–31G* level of theory,33 and then the restrained electrostatic potential (RESP) method was used for charge-fitting in AMBER 16.34 Besides the nonstandard parameters, the ff14SB force field and general Amber force field (GAFF) parameters were used to describe the protein and nonprotein molecules.35,36 All crystal structure waters were retained, and each complex was neutralized with potassium ions and solvated in a truncated octahedral box of TIP3P water molecules with at least a 12 Å cushion from the protein atoms. All MD simulations were carried out using the Amber 16 Molecular Dynamics package,37 starting with a total of 10 000 steps of minimization with a Cartesian restraint on all atoms followed by another two cycles with restraint on all heavy atoms and finally with all restraints removed. To anneal the system, the temperature was slowly increased from 0 to 300 K. An 8 ns equilibrium stage was performed using gradually weaker restraints on all heavy atoms, followed by a 200 ns production run. An average pressure was maintained at 1 atm, and a Langevin dynamics approach was used for temperature control with a collision frequency of 1 ps−1, and the SHAKE algorithm was used to constrain bonds involving hydrogen.38 Nonbonding interactions were calculated with a cutoff of 12 A by the particle mesh Ewald (PME) method.39 All simulation trajectories were analyzed using the cpptraj in the AmberTools 16 package and VMD.40,41 For cluster analysis, all structures were aligned on residues 295–405 because these residues are the most structurally invariant,42 and the agglomerative hierarchical clustering algorithms were used with an average linkage.43 F- and G-helix movements compared to the closed state were calculated based on the movement of Lys178 and Lys197 at their Cα atoms, respectively. Figures were made with Matlab and PyMOL.
RESULTS
A Specific Probe for the F Helix Movement.
For this study, it was important to develop a spin-label probe to detect movements of the F helix separately from those of the G helix. Previous DEER studies have firmly established conversion between the open and closed states in response to camphor and Pdx binding.11,15,17,18 Our original strategy placed one of the pairs of spin labels at a fixed position (residue 48) and the other at a position (residue 190) on the tip of the loop between the F and G helices.11,17,18 Crystallographic observations of an intermediate conformation with tethered substrates showed that the movements relative to the closed state were more localized on the F helix than on the G helix.28 Thus, we reasoned that sensitive detection of an intermediate conformation may require a probe that is specific to movements of the F helix. We have previously reported a strategy for placing bifunctional spin labels to separately detect movements of the F and G helices.18 This was successful in detecting G helix movements during conformational changes. However, the F helix probe used in that study did not respond in the expected way, suggesting an undesirable structural effect of placing a bifunctional spin label within the F helix. Here, we introduce a new pair of monofunctional spin-labeling sites to monitor F-helix-specific movements. As shown in Figure 1, one site was placed at a reference position (S48C) and the other within the F helix at Y179C. For these experiments, 4-maleimido-TEMPO (4MT) spin labels were attached via an S–C linkage to prevent cleavage of the label by CN−. The measured rate of NADH oxidation by the spin-labeled F-helix mutant was 85% of the rate observed for the WT enzyme containing no surface accessible cysteines. In contrast, the NADH oxidation rate for the spin-labeled D251N mutant retained only 0.2% of the WT enzyme. As expected, the DEER data of Figure 2a,b and Table 1 show that the binding of camphor to ferric P450cam containing the new probe detects a 5 Å shift of the F helix from the open to the closed conformation, matching the 5 Å movement between the CA carbons of these residues in the crystal structures. In addition, this new F helix probe shows that Pdx binding to closed camphor/P450cam results in conversion to the fully open conformation. These results are fully consistent with previous DEER studies on P450cam labeled on the tip of the F/G loop and with the bifunctional G helix probe, indicating that our new F-helix-specific probe is functioning as expected.
Figure 1.
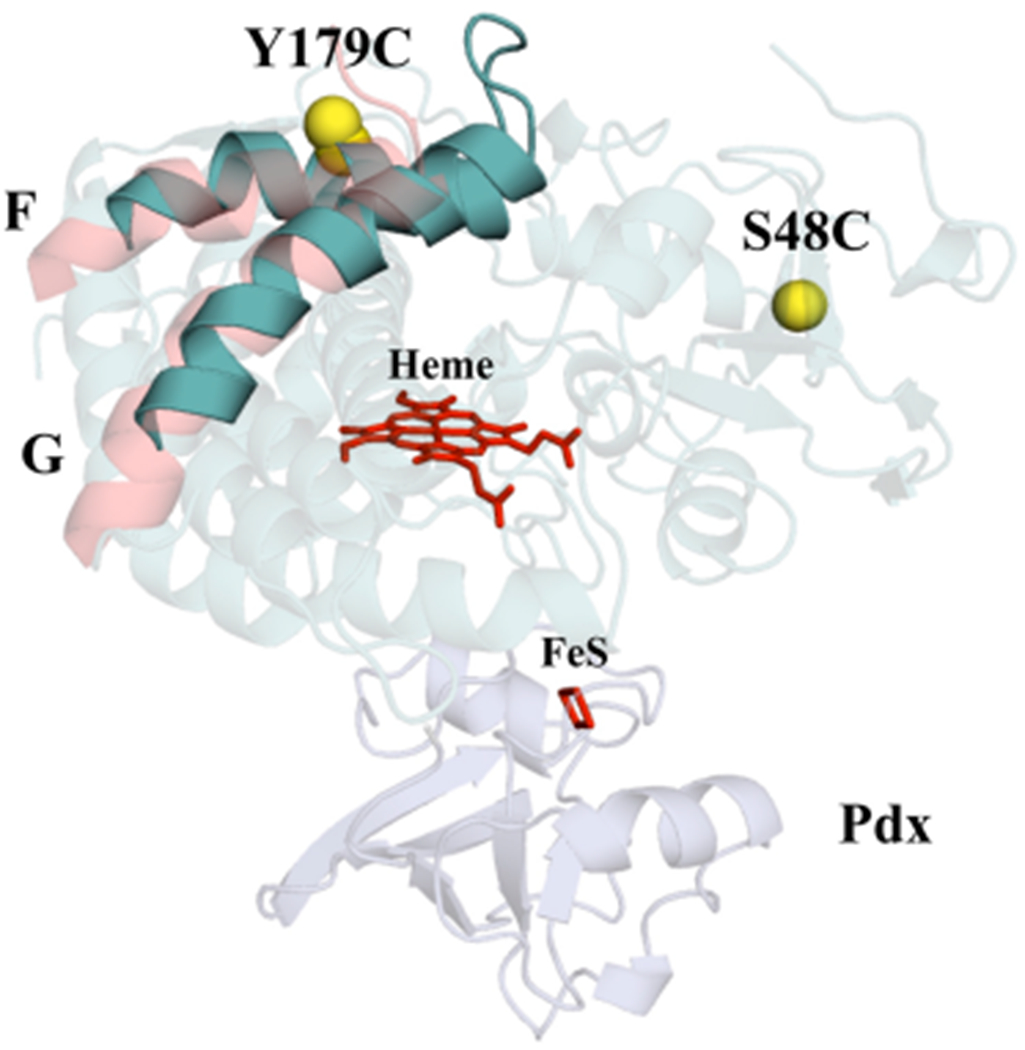
Position of labeling sites on the P450cam structure (PDB ID 3l63) to probe F-helix movements. One 4MT label is attached to a reference position at S48C, and the other 4MT label is attached to the outwardly pointing Y179C in the middle of the F helix. The CA and CB atoms of S48C and Y179C are shown as yellow spheres, and the heme and ion-sulfur cluster are shown as red sticks. Closed P450cam is shown in cyan, the F/G helices in the open form are shown in pink, and Pdx is in blue.
Figure 2.
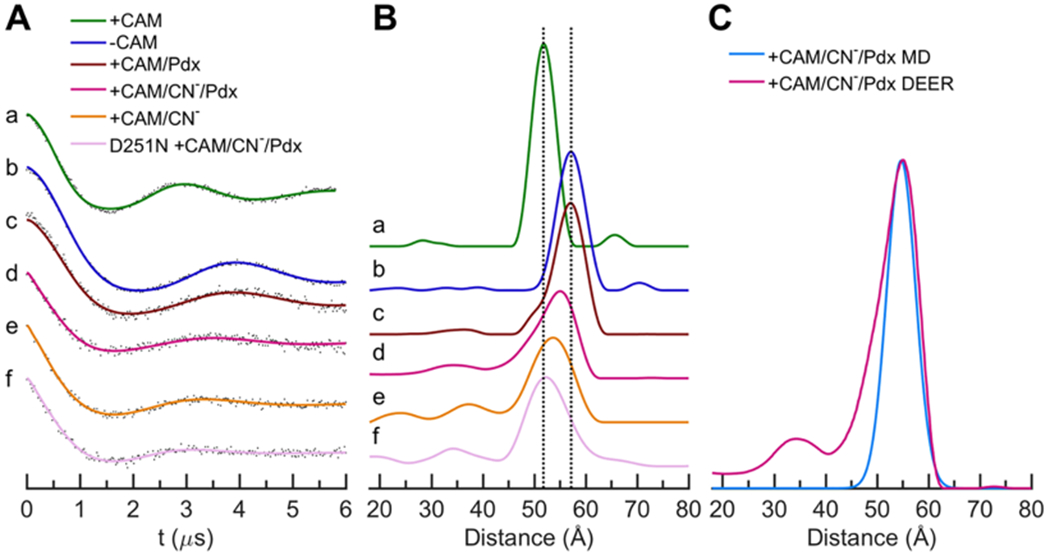
(A) Time-domain DEER data of ferric P450cam labeled with 4MT for different states. Points are the raw DEER echo amplitudes after background subtraction, and the solid traces are the fitted curves. Shown are traces for camphor/P450cam (a, green), P450cam (b, blue), camphor/P450cam/Pdx (c, brown), CN−/camphor/P450cam/Pdx (d, magenta), CN−/camphor/P450cam (e, orange), and CN−/camphor/P450cam(D251N)/Pdx (f, pink). (B) Distance distributions derived from the DEER data of panel A. (C) Comparison of MD (cyan) and DEER (magenta) distance distributions for CN−/camphor/P450cam/Pdx.
Table 1.
Distance Measurements of P450cam in Different States between Residues 48 and 179
| complex | crystal CA–CA (Å) | DEER (Å) (this study) | MD (Å) (this study) |
|---|---|---|---|
| CAM/P450cam | 38.2 (3l63) | 52 (closed) | 51.8 |
| P450cam | 43.5 (3l62) | 57 (open) | |
| CAM/P450cam/Pdx | 43.7 (4jx1) | 57 (open) | 57.3 |
| CN−/CAM/P450cam/Pdx | 55 (intermediate) | 54.8 | |
| CN−/CAM/P450cam | 38.3 (1o76) | 54 (intermediate) | |
| CN−/CAM/P450cam(D251N)/Pdx | 52 (closed) |
An Intermediate Conformation of P450cam Induced by Pdx.
DEER measurements were used to detect an intermediate state in a mimic of the Fe2+–O2/camphor/P450cam complex. The preparation of DEER samples of Pdx bound to Fe2+–O2/camphor/P450cam has to date been unsuccessful due to its rapid rate of auto-oxidation, making it necessary to study structural mimics of the Fe2+–O2 state. Although a number of studies have shown that the Raman, IR, and NMR44–47 properties of the Fe2+–CO complex are sensitive to Pdx binding, our previous DEER studies using the stable Fe2+–CO/camphor clearly showed that Pdxox or Pdxred binding to this closed form did not induce any opening of the enzyme.17,18 However, crystal structures show that the CN− complex is a more accurate structural analogue of the Fe2+–O2 complex than is the Fe2+–CO form. Both O2 and CN− are bound end-on to the heme iron including a bent Fe–C–N bond as is observed for Fe2+–O2 P450cam.6,48 In addition, CN− binding results in a small broadening of the I helix (Table 2), in which the Asp251 peptide carbonyl flips and is accompanied by a minor reorientation of Thr252 as is also seen for Fe2+–O2/P450cam.5,6,48 We thus investigated the effects of camphor binding using the 4MT-labeled F helix probe for the CN−/P450cam complex. Complete CN− binding to DEER samples was verified by EPR (Figures S1 and S2) and UV/vis spectroscopy (Figure S3). The DEER results of Figure 2B traces a and e and Table 1 show that the F helix in the CN−/camphor complex undergoes a small 2 Å shift toward the open position relative to the closed camphor/P450cam form. This shift in the F helix was not observed previously in the crystal structure of CN−/camphor.48,49 Most importantly, Figure 2B trace d and Table 1 show that binding of Pdxox to CN−/camphor induces an additional F-helix movement to a position that is 3 A away from the closed position toward the open state and is nearly midway between the two states. This finding unambiguously shows that, in solution, distinct intermediate conformational states are populated in the CN−/camphor state and in its complex with Pdxox.
Table 2.
Distance Measurements between the Carbonyl Carbon of Gly248 and the β-carbon of Thr252 in Different P450cam Crystal Structures
| crystal structure | I helix groove width (G248C-T252CB) (Å) |
|---|---|
| WT + CAM/Pdx (4jx1) | 5.3 |
| WT + CAM (3l63) | 4.5 |
| D251N + CAM/O2 (2a1n) | 4.7 |
| WT + CAM/O2 (2a1m) | 5.6 |
| WT + CAM/CN (1o76) | 5.8 |
Link between the F Helix Movement and the Catalytic Environment.
An important interaction that may connect the Pdx-induced movement of the F helix with changes in the distal catalytic environment involves a salt bridge between Arg186 on the F helix and Asp251 at the bulge in the I helix, directly above the heme iron. The D251N mutant is catalytically impaired, presumably due to the role of Asp251 in proton delivery and to its effects via the I helix distortion on the positioning of Thr252 and two active site waters that are believed to provide protons during peroxy bond cleavage.6,29,50,51 We thus investigated the effects of the D251N mutant on the ability of Pdx to induce the F helix movement. The DEER data (Figure 2f and Table 1) shows that Pdxox binding to CN−/camphor/P450cam(D251N) leaves the F helix probe at precisely the distance observed for the closed conformation of camphor/P450cam. It is noted that, while the F helix is clearly in the closed state, the width of the distance distribution is somewhat larger for the mutant. In addition, the precise alignment of the D251N mutant distance (52 Å) with that of the camphor-bound closed state along with the reproducibility of the two known open distances in Table 1 (57 Å) provides additional confidence that the reported distance changes of 2–3 Å in the CN−-bound states are significant. Thus, the ability of Pdx to induce the intermediate opening of the F helix in CN−/camphor/P450cam is dependent on the presence of the R186-D251 ion pair.
Molecular Dynamics.
To help interpret our observed DEER measurements in terms of absolute and relative protein structural movements, molecular dynamics simulations were performed. MD trajectories were run for three states of spin-labeled P450cam: the closed state of camphor/P450cam, the open state of camphor/P450cam/Pdx, and the intermediate state of CN−/camphor/P450cam/Pdx. For the latter state, the calculations started with the coordinates of the open camphor/P450cam/Pdx crystal structure (PDB ID 4jx1), to which CN− was added as a ligand to the iron. For each simulation, an initial annealing/equilibration period of 8 ns was followed by a 200 ns production run without restraints on inter-residue distances or any other degrees of freedom. Distance distributions were calculated between the nitroxide nitrogen atoms for the final 100 ns of the MD trajectories of the spin-labeled forms and compared with the observed DEER distance distributions in Figure 2C and Figure S4. These plots and Table 1 show that our MD simulations give excellent predictions (within 0.3 Å) of the absolute F helix position for all three states, and importantly, they predict the observed intermediate position of the F helix for the CN−/camphor complex to within 0.2 Å over a total distance of 55 Å.
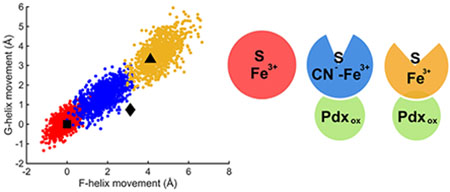
Given the success of the above MD simulations, we investigated the properties of the simulated closed, intermediate, and open states in more detail. Additional MD calculations were performed on the unlabeled forms of the above complexes to remove any possible effects of the spin label on protein conformation. As with the spin-labeled form, the unlabeled CN−/camphor simulations began with the coordinates of the open form of the camphor/P450cam/Pdx crystal structure (PDB ID 4jxl). The total RMSD for P450cam atoms during these runs (Figure S5) shows that each state is stable with an overall fluctuation that is the lowest for the closed camphor/P450cam and highest for the open camphor/P450cam/Pdx, with the CN−/camphor state intermediate between these extremes. As shown in Figure S6 for CN−/camphor, the distance between CA atoms on probe residues 48 and 179 begins at the fully open value of 45 A but settles after about 50 ns to a value about halfway to the closed distance of 38 A. As shown in Figure 3, a scatter plot of the relative movements of the F and G helices during the simulations for each of the three states falls on a linear continuum, implying a coordinated movement of both the F and G helices by about 5 A between the open and closed states. The intermediate conformations predicted for CN−/camphor span the space between the open and closed conformations. These results are similar to recent MD simulations of Batabyal.27 The centers of these helix shift distributions are completely consistent with the previously observed closed and open state structures as shown by the black symbols of Figure 3. However, the intermediate states previously observed with large tethered substrate analogues lie somewhat off this linear MD-predicted distribution and are characterized by a larger shift in the F helix than for the G helix (black diamond of Figure 3). Notably the center of the cluster of MD-predicted intermediate states occurs with a 2–3 Å shift in the F helix, exactly as observed by our DEER result.
Figure 3.
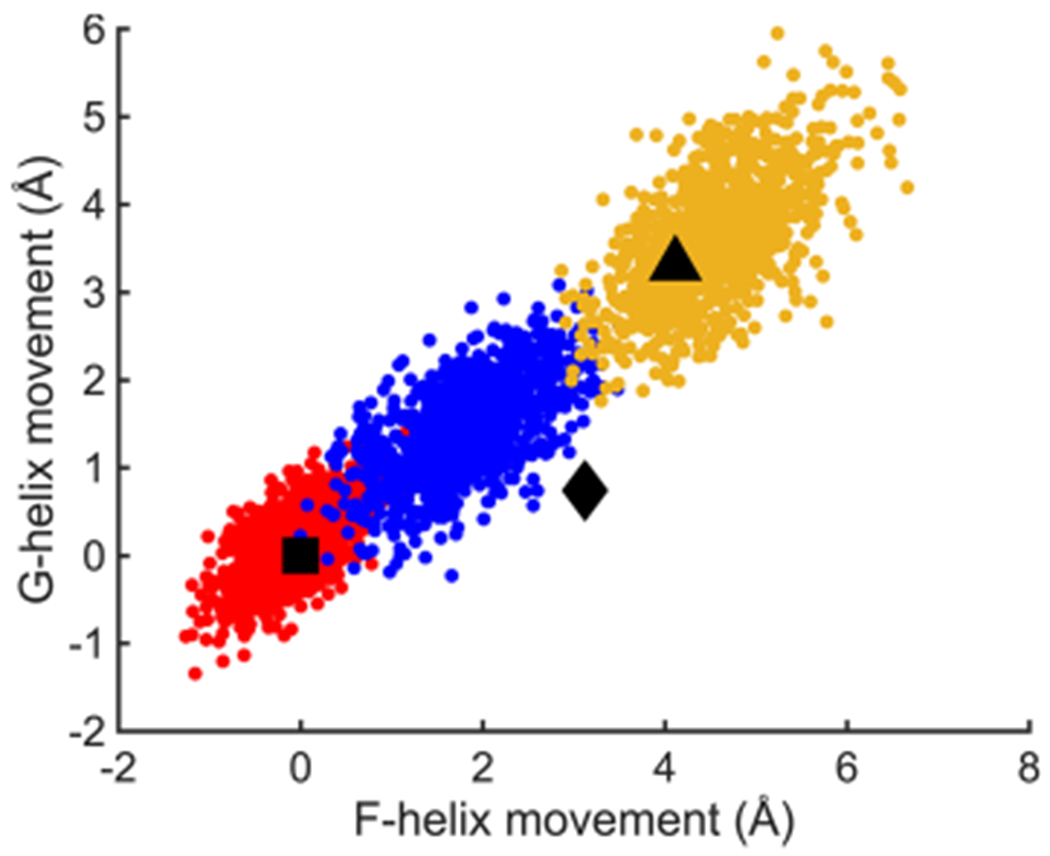
Movements of F and G helices derived from MD trajectory snapshots in three ferric P450cam complexes, camphor/P450cam (red), CN−/camphor/P450cam/Pdx (blue), and camphor/P450cam/Pdx (yellow) relative to the camphor-bound state (PDB ID 3l63). Square, diamond, and triangle symbols represent the movements of F and G helices observed in crystal structures of P450cam in the closed camphor-bound (PDB ID 3l63), intermediate tethered adamantane (camphor analogue)-bound (PDB ID 1re9), and open camphor/Pdx-bound (PDB ID 4jx1) P450cam complexes.
Additional structural insights obtained from these MD simulations are shown in Figures S7–S11 and summarized here. The salt bridge between Arg186 on the F helix and Asp251 on the I helix remains well-formed for the closed camphor/P450cam state and is completely broken in the open camphor/P450cam/Pdx (Figure S7). For the CN−/camphor form, once the enzyme achieves the intermediate state at 50 ns, the salt bridge reforms and remains stable. In the closed state, camphor is mostly oriented correctly for specific hydroxylation and only occasionally flips (Figure S8). The camphor orientation is highly disordered in the open camphor/P450cam/Pdx complex. The intermediate state, once formed, shows camphor predominantly oriented for the correct hydroxylation similar to that of the closed state. The hydrogen bond between Tyr96 and the camphor carbonyl is predominantly formed in the intermediate state but is often broken in the open form (Figure S10). Finally, Figure S11 shows that the aromatic packing of Phe87 and Phe193 in the loop between the F and G helices is largely interrupted in the intermediate state, as it is with the open state.
Structural Prediction of the Pdx-Induced Intermediate State.
The MD-derived structures for the most highly represented cluster for each of the closed camphor/P450cam, intermediate CN−/camphor/P450cam/Pdx, and open camphor/P450cam/Pdx conformations are shown in Figure 4. The active site features of both the closed and open forms (Figure 4A,C) are essentially the same as have been previously reported for these states.8,10,18 This includes the highly disordered occupation of camphor in the open form. For the intermediate state induced by Pdx binding to the CN−/camphor complex (Figure 4B), a number of potentially important features are observed. First, camphor remains highly ordered in its productive orientation and the salt bridge between Arg186 and Asp251 is stretched but intact. Notably, changes include a flip of the peptide bond at Gly248 and the resulting reorientation of Thr252 modulating the width of the I helix bulge. These changes enable population of a water molecule, which is positioned to hydrogen bond to CN−, Thr252, and Val247 in the intermediate state, which is also observed as the “catalytic” water in the ferrous–O2 complex crystal structure.6,48
Figure 4.
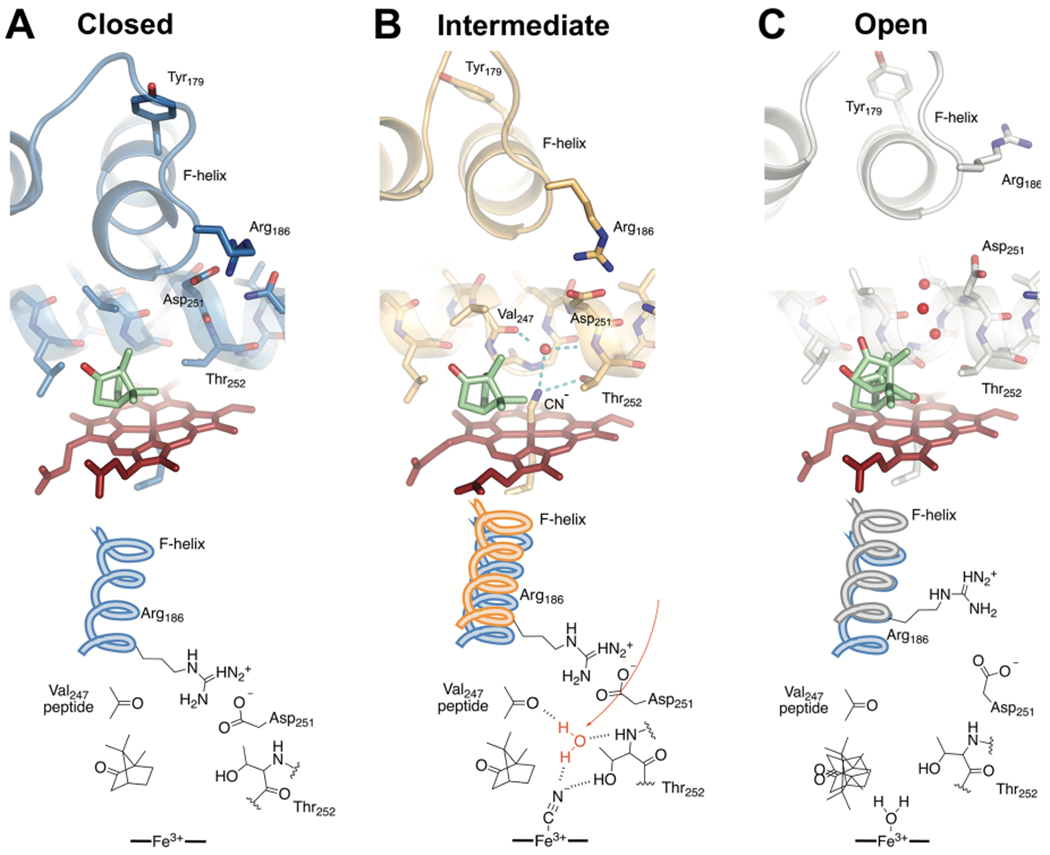
Comparisons of the active site structures predicted from cluster analysis of MD trajectories. Shown are the closed camphor/P450cam (A), intermediate CN−/camphor/P450cam/Pdx (B), and open camphor/P450cam/Pdx (C) complexes representing the most highly populated cluster in each MD simulation. Camphor and heme are shown as green and red sticks, respectively. Water oxygen atoms are shown as red spheres. The bottom panel shows important differences in the interactions at the active site. Both the closed and intermediate structures have an intact ion pair interaction between Arg186 and Asp251. The partially retracted F helix in the intermediate structure (B) provides new interactions with the I helix and the ion pair to favor reorientation of Thr252 and population of the catalytic water (red). We propose this represents the conformation competent for hydroperoxy bond cleavage in the formation of compound I.
DISCUSSION
Functional Implications of the Pdx-Induced Intermediate Open State.
The proposal that Pdx may induce a catalytically active conformation of Fe2+–O2/camphor/P450cam that is in between the closed and fully open conformations has been one of the important recent insights into the function of this important enzyme.18,25 Such an intermediate conformation could explain the long-known effector role of Pdx on enzyme function;12,19–21 it may enable product release as the enzyme proceeds to the fully open state and might prevent loss of specificity and release of reactive oxygen intermediates. Although Pdxox binding has been shown to induce conversion to the fully open state of camphor/P450cam,14,17,18 its effect on Fe2+–O2/camphor has been difficult to test directly. Neither Pdxox nor Pdxred binding caused a change in the closed conformation of the Fe2+–CO/camphor state, but the Fe2+–CO complex is not a perfect mimic of Fe2+–O2. Here, we show by DEER and MD simulations that CN−/camphor itself and progressively its complex with Pdx cause two new intermediate conformations that have significant implications for enzyme function. The crystal structure of CN−/camphor has been reported, in which the bent Fe–C–N bond is a good structural mimic for the Fe–O–O bond of the Fe2+–O2 complex and was observed in the closed conformation.48 However, the DEER data of this study show that, in solution, the CN−/camphor complex results in a partial opening of the F helix, implying that there may be some differences between the crystal and solution forms of this complex. Importantly, Pdx binding causes a further opening of the F helix to a position that is midway between the closed and open states. Notably, this Pdx-induced F helix movement is not observed in the D251N mutant, showing that both the state of heme coordination and distal interactions between the F and I helices must be delicately balanced for the effect to be observed. In our MD simulations of the D251N mutant (based on PDB entry 2a1n) in the CN−/camphor complex, we observe only the closed conformation (Figures S12 and S13), in which Asn251 forms a hydrogen bond with the side chain of Arg186 on the F helix and an additional hydrogen bond with the Val247 carbonyl group on the I-helix, so this alternative state might be keeping I helix closed and preventing the effector function of Pdx. We propose that F helix retraction across the I helix results from the effects of Pdx binding to the proximal binding site and communicates its effects through the C helix to the F/G helices. Our MD simulations of the Pdx-induced intermediate state are fully consistent with our DEER results and allow structural comparison of the closed, intermediate, and open forms. Previous P450cam crystal structures in the presence of large tethered substrates showed sequential movement of the F/G helices, but this study suggests F/G helices move together as a result of Pdx binding. In addition, we propose that contacts between Thr181 on the F helix, which makes contact with Val247/Leu250 in the I helix, in combination with the Arg186-Asp251 salt bridge, result in a linkage between movements of the F and I helices. Thus, retraction of the F helix causes changes in the opening of the I helix bulge, reorientation of Thr-252, and population of the catalytic water that are both implicated in proton transfer during compound I formation.
The results of this work also provide a very relevant point of comparison and contrast to very recent results reported by X-ray crystallography. Follmer et al. have reported the structure of camphor/P450cam/Pdx after soaking crystals in CN−.52 The results agree with this study in some respects, but mainly, in order to make room for CN− binding, movements in the I, F, and G helices are necessary. Significant differences are also observed in that the reported crystal structure indicates that the F and G helices remain in a full open state, and the B’ helix has become disordered to cause exposure of the proposed “channel 2”.53 In addition, the Tyr-96 side chain, which makes a hydrogen bond to camphor in the closed conformation, has flipped out of its normally observed position. Our MD simulations, which agree accurately with the DEER results, are in contrast with the reported structure in two significant ways. First, they indicate that the F helix in the CN−/camphor/P450cam/Pdx complex is in an intermediate and not the fully open conformation. Second, this allows the Arg186-Asp251 salt bridge to remain intact in the intermediate conformation. Our results do show however (Figure S14) that the intermediate state exhibits a distance between Ser83 and Ser102 that fluctuates between values that Follmer et al.53 suggest represents the closed (5 Å) and the open (>7 Å) state of “channel 2” due to disordering of the B’ helix. It should also be noted that the structure of the CN− complex was determined from crystals of the open state of the camphor/P450cam/Pdx complex, which were then soaked in CN−. We have shown that crystals of the open form of P450cam do not convert to the closed state upon soaking in camphor, demonstrating that crystal contacts can interfere with conformational changes that would be seen in solution.18 As acknowledged by Follmer et al.,52 the changes seen upon soaking in CN− may also be influenced by the effects of crystal contacts. The results of this work report the state of the CN−/camphor/P450cam/Pdx complex in solution and may thus more accurately represent the solution state complex of O2/camphor/P450cam/Pdx that is on the catalytic pathway. In this regard, our results suggest that Pdx binding to the presumably closed form of O2/camphor/P450cam causes a conversion toward the open state, but this transition is partially (in the case of CN−) or completely (in the case of CO)18 blocked by critical effects of the distal ligand. This study thus provides an important new view of the long-sought link between the effects of Pdx binding and the structure of the catalytic center of this important enzyme.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Shu-Hao Liou, Dr. Xiaoxiao Shi, Christopher Niedek, and Prof. Thomas Poulos for helpful discussions.
Funding
This work was supported by the National Institutes of Health (GM41049 to D.B.G.; GM126961 to R.D.B; AI130684-02 to L.-P.W.).
ABBREVIATIONS
- DEER
double electron–electron resonance
- P450cam
substrate-free ferric cytochrome P450 from Pseudomonas putida containing the mutations C334A, C58S, C85S, C136S, and C285S
- Pdx
putidaredoxin from Pseudomonas putida
- Pdxox
oxidized Pdx
- Pdxred
reduced Pdx
- camphor/P450cam
the camphor-bound complex of P450cam
- camphor/P450cam/Pdx
the complex of P450cam with both camphor and Pdx
- CN−/camphor/P450cam/Pdx
both camphor and Pdx bound to P450cam, and CN− coordinated to the ferric heme
- CN−/camphor/P450cam(D251N)/Pdx
the indicated quaternary complex containing the Asp251 to Asn251 mutation
- MD
molecular dynamics
- 4MT
4-maleimido-2,2,6,6-tetramethyl-1-piperidinyloxy
- DTT
dithiothreitol
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.9b00192.
CW-EPR spectra, UV/vis spectra, details of molecular dynamics simulations, and NADH oxidation rate measurements (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Sono M, Roach MP, Coulter ED, and Dawson JH (1996) Heme-Containing Oxygenases. Chem. Rev 96, 2841–2888. [DOI] [PubMed] [Google Scholar]
- (2).Ortiz de Montellano PR (2005) Cytochrome P450: Structure, Mechanism, and Biochemistry, Springer Science & Business Media. [Google Scholar]
- (3).Griffin BW, and Peterson JA (1972) Camphor binding of Pseudomonas putida cytochrome P-450. Kinetics and thermodynamics of the reaction. Biochemistry 11, 4740–4746. [DOI] [PubMed] [Google Scholar]
- (4).Lipscomb JD (1980) Electron paramagnetic resonance detectable states of cytochrome P-450cam. Biochemistry 19, 3590–3599. [DOI] [PubMed] [Google Scholar]
- (5).Schlichting I, Berendzen J, Chu K, Stock AM, Maves SA, Benson DE, Sweet RM, Ringe D, Petsko GA, and Sligar SG (2000) The Catalytic Pathway of Cytochrome P450cam at Atomic Resolution. Science 287, 1615–1622. [DOI] [PubMed] [Google Scholar]
- (6).Nagano S, and Poulos TL (2005) Crystallographic Study on the Dioxygen Complex of Wild-type and Mutant Cytochrome P450cam: IMPLICATIONS FOR THE DIOXYGEN ACTIVATION MECHANISM. J. Biol. Chem 280, 31659–31663. [DOI] [PubMed] [Google Scholar]
- (7).Rittle J, and Green MT (2010) Cytochrome P450 Compound I: Capture, Characterization, and C-H Bond Activation Kinetics. Science 330, 933–937. [DOI] [PubMed] [Google Scholar]
- (8).Poulos TL, Finzel BC, and Howard AJ (1987) High-resolution crystal structure of cytochrome P450cam. J. Mol. Biol 195, 687–700. [DOI] [PubMed] [Google Scholar]
- (9).Poulos TL, and Johnson EF (2015) In Cytochrome P450: Structure, Mechanism, and Biochemistry (Ortiz de Montellano PR, Ed.) pp 3–32, Springer International Publishing, Cham. [Google Scholar]
- (10).Lee Y-T, Wilson RF, Rupniewski I, and Goodin DB (2010) P450cam Visits an Open Conformation in the Absence of Substrate. Biochemistry 49, 3412–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Stoll S, Lee Y-T, Zhang M, Wilson RF, Britt RD, and Goodin DB (2012) Double electron–electron resonance shows cytochrome P450cam undergoes a conformational change in solution upon binding substrate. Proc. Natl. Acad. Sci. U. S. A 109, 12888–12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Lipscomb JD, Sligar SG, Namtvedt MJ, and Gunsalus IC (1976) Autooxidation and hydroxylation reactions of oxygenated cytochrome P-450cam. J. Biol. Chem 251, 1116–1124. [PubMed] [Google Scholar]
- (13).Poulos TL (2014) Heme Enzyme Structure and Function. Chem. Rev 114, 3919–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Tripathi S, Li H, and Poulos TL (2013) Structural Basis for Effector Control and Redox Partner Recognition in Cytochrome P450. Science 340, 1227–1230. [DOI] [PubMed] [Google Scholar]
- (15).Liou S-H, Myers WK, Oswald JD, Britt RD, and Goodin DB (2017) Putidaredoxin Binds to the Same Site on Cytochrome P450cam in the Open and Closed Conformation. Biochemistry 56, 4371–4378. [DOI] [PubMed] [Google Scholar]
- (16).Hiruma Y, Hass MAS, Kikui Y, Liu W-M, Ölmez B, Skinner SP, Blok A, Kloosterman A, Koteishi H, Lohr F, Schwalbe H, Nojiri M, and Ubbink M (2013) The Structure of the Cytochrome P450cam–Putidaredoxin Complex Determined by Paramagnetic NMR Spectroscopy and Crystallography. J. Mol. Biol 425, 4353–4365. [DOI] [PubMed] [Google Scholar]
- (17).Myers WK, Lee Y-T, Britt RD, and Goodin DB (2013) The Conformation of P450cam in Complex with Putidaredoxin Is Dependent on Oxidation State. J. Am. Chem. Soc 135, 11732–11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Liou S-H, Mahomed M, Lee Y-T, and Goodin DB (2016) Effector Roles of Putidaredoxin on Cytochrome P450cam Conformational States. J. Am. Chem. Soc 138, 10163–10172. [DOI] [PubMed] [Google Scholar]
- (19).Gunsalus IC, Lipscomb JD, and Meeks JR (1973) CYTOCHROME P-450cam SUBSTRATE AND EFFECTOR INTERACTIONS. Ann. N. Y. Acad. Sci 212, 107–121. [DOI] [PubMed] [Google Scholar]
- (20).Sligar SG, Debrunner PG, Lipscomb JD, Namtvedt MJ, and Gunsalus IC (1974) A Role of the Putidaredoxin COOH-terminus in P-450cam (Cytochrome m) Hydroxylations. Proc. Natl. Acad. Sci. U. S. A 71, 3906–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Glascock MC, Ballou DP, and Dawson JH (2005) Direct Observation of a Novel Perturbed Oxyferrous Catalytic Intermediate during Reduced Putidaredoxin-initiated Turnover of Cytochrome P-450-CAM PROBING THE EFFECTOR ROLE OF PUTIDAREDOXIN IN CATALYSIS. J. Biol. Chem 280, 42134–42141. [DOI] [PubMed] [Google Scholar]
- (22).Pochapsky SS, Pochapsky TC, and Wei JW (2003) A Model for Effector Activity in a Highly Specific Biological Electron Transfer Complex: The Cytochrome P450cam-Putidaredoxin Couple. Biochemistry 42, 5649–5656. [DOI] [PubMed] [Google Scholar]
- (23).Purdy MM, Koo LS, Ortiz de Montellano PR, and Klinman JP (2004) Steady-State Kinetic Investigation of Cytochrome P450cam: Interaction with Redox Partners and Reaction with Molecular Oxygen. Biochemistry 43, 271–281. [DOI] [PubMed] [Google Scholar]
- (24).Raag R, and Poulos TL (1989) Crystal structure of the carbon monoxide-substrate-cytochrome P-450CAM ternary complex. Biochemistry 28, 7586–7592. [DOI] [PubMed] [Google Scholar]
- (25).Hollingsworth SA, Batabyal D, Nguyen BD, and Poulos TL (2016) Conformational selectivity in cytochrome P450 redox partner interactions. Proc. Natl. Acad. Sci. U. S. A 113, 8723–8728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Skinner SP, Liu W-M, Hiruma Y, Timmer M, Blok A, Hass MAS, and Ubbink M (2015) Delicate conformational balance of the redox enzyme cytochrome P450cam. Proc. Natl. Acad. Sci. U. S. A 112, 9022–9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Batabyal D, Richards LS, and Poulos TL (2017) Effect of Redox Partner Binding on Cytochrome P450 Conformational Dynamics. J. Am. Chem. Soc 139, 13193–13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lee Y-T, Glazer EC, Wilson RF, Stout CD, and Goodin DB (2011) Three Clusters of Conformational States in P450cam Reveal a Multistep Pathway for Closing of the Substrate Access Channel. Biochemistry 50, 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Gerber NC, and Sligar SG (1994) A Role for Asp-251 in Cytochrome P-450cam Oxygen Activation. J. Biol. Chem 269, 4260–4266. [PubMed] [Google Scholar]
- (30).De Voss JJ, and Ortiz de Montellano PR (1995) Computer-Assisted, Structure-Based Prediction of Substrates for Cytochrome P450cam. J. Am. Chem. Soc 117, 4185–4186. [Google Scholar]
- (31).Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, and Jung H (2006) DeerAnalysis2006–-a comprehensive software package for analyzing pulsed ELDOR data. Appl Magn. Reson 30, 473–498. [Google Scholar]
- (32).Li P, and Merz KM (2016) MCPB.py: A Python Based Metal Center Parameter Builder. J. Chem. Inf. Model 56, 599–604. [DOI] [PubMed] [Google Scholar]
- (33).Frisch MJ et al. (2016) Gaussian 16, Revision B.01 [Google Scholar]
- (34).Bayly CI, Cieplak P, Cornell W, and Kollman PA (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem 97, 10269–10280. [Google Scholar]
- (35).Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, and Simmerling C (2015) ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput 11, 3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Wang J, Wolf RM, Caldwell JW, Kollman PA, and Case DA (2004) Development and testing of a general amber force field. J. Comput. Chem 25, 1157–1174. [DOI] [PubMed] [Google Scholar]
- (37).Case DA et al. (2017) AMBER16, University of California, San Francisco. [Google Scholar]
- (38).Ryckaert J-P, Ciccotti G, and Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys 23, 327–341. [Google Scholar]
- (39).Darden T, York D, and Pedersen L (1993) Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys 98, 10089–10092. [Google Scholar]
- (40).Roe DR, and Cheatham TE (2013) PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput 9, 3084–3095. [DOI] [PubMed] [Google Scholar]
- (41).Humphrey W, Dalke A, and Schulten K (1996) VMD: Visual molecular dynamics. J. Mol. Graphics 14, 33–38. [DOI] [PubMed] [Google Scholar]
- (42).Markwick PRL, Pierce LCT, Goodin DB, and McCammon JA (2011) Adaptive Accelerated Molecular Dynamics (Ad-AMD) Revealing the Molecular Plasticity of P450cam. J. Phys. Chem. Lett 2, 158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Shao J, Tanner SW, Thompson N, and Cheatham TE (2007) Clustering Molecular Dynamics Trajectories: 1. Characterizing the Performance of Different Clustering Algorithms. J. Chem. Theory Comput 3, 2312–2334. [DOI] [PubMed] [Google Scholar]
- (44).Unno M, Christian JF, Benson DE, Gerber NC, Sligar SG, and Champion PM (1997) Resonance Raman Investigations of Cytochrome P450cam Complexed with Putidaredoxin. J. Am. Chem. Soc 119, 6614–6620. [Google Scholar]
- (45).Unno M, Christian JF, Sjodin T, Benson DE, Macdonald IDG, Sligar SG, and Champion PM (2002) Complex Formation of Cytochrome P450cam with Putidaredoxin: EVIDENCE FOR PROTEIN-SPECIFIC INTERACTIONS IN-VOLVING THE PROXIMAL THIOLATE LIGAND. J. Biol. Chem 277, 2547–2553. [DOI] [PubMed] [Google Scholar]
- (46).Nagano S, Shimada H, Tarumi A, Hishiki T, Kimata-Ariga Y, Egawa T, Suematsu M, Park S-Y, Adachi S. i., Shiro Y, and Ishimura Y (2003) Infrared Spectroscopic and Mutational Studies on Putidaredoxin-Induced Conformational Changes in Ferrous CO-P450cam. Biochemistry 42, 14507–14514. [DOI] [PubMed] [Google Scholar]
- (47).Tosha T, Yoshioka S, Takahashi S, Ishimori K, Shimada H, and Morishima I (2003) NMR Study on the Structural Changes of Cytochrome P450cam upon the Complex Formation with Putidaredoxin: FUNCTIONAL SIGNIFICANCE OF THE PUTI-DAREDOXIN-INDUCED STRUCTURAL CHANGES. J. Biol. Chem 278, 39809–39821. [DOI] [PubMed] [Google Scholar]
- (48).Fedorov R, Ghosh DK, and Schlichting I (2003) Crystal structures of cyanide complexes of P450cam and the oxygenase domain of inducible nitric oxide synthase–structural models of the short-lived oxygen complexes. Arch. Biochem. Biophys 409, 25–31. [DOI] [PubMed] [Google Scholar]
- (49).Batabyal D, Li H, and Poulos TL (2013) Synergistic Effects of Mutations in Cytochrome P450cam Designed To Mimic CYP101D1. Biochemistry 52, 5396–5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Vidakovic M, Sligar SG, Li H, and Poulos TL (1998) Understanding the Role of the Essential Asp251 in Cytochrome P450cam Using Site-Directed Mutagenesis, Crystallography, and Kinetic Solvent Isotope Effect. Biochemistry 37, 9211–9219. [DOI] [PubMed] [Google Scholar]
- (51).Benson DE, Suslick KS, and Sligar SG (1997) Reduced Oxy Intermediate Observed in D251N Cytochrome P450cam. Biochemistry 36, 5104–5107. [DOI] [PubMed] [Google Scholar]
- (52).Follmer AH, Tripathi S, and Poulos TL (2019) Ligand and Redox Partner Binding Generates a New Conformational State in Cytochrome P450cam (CYP101A1). J. Am. Chem. Soc 141, 2678–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Follmer AH, Mahomed M, Goodin DB, and Poulos TL (2018) Substrate-Dependent Allosteric Regulation in Cytochrome P450cam (CYP101A1). J. Am. Chem. Soc 140, 16222–16228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


