Abstract
The large GTPase Dynamin 2 (Dyn2) is known to increase the invasiveness of pancreatic cancer tumor cells, but the mechanisms by which Dyn2 regulates changes in the actin cytoskeleton to drive cell migration are still unclear. Here we report that a direct interaction between Dyn2 and the actin-bundling protein alpha-actinin (α-actinin) 4 is critical for tumor cell migration and remodeling of the extracellular matrix in pancreatic ductal adenocarcinoma (PDAC) cells. The direct interaction is mediated through the C-terminal tails of both Dyn2 and α-actinin 4, and these proteins interact at invasive structures at the plasma membrane. While Dyn2 binds directly to both α-actinin 1 and α-actinin 4, only the interaction with α-actinin 4 is required to promote tumor cell invasion. Specific disruption of the Dyn2–α-actinin 4 interaction blocks the ability of PDAC cells to migrate in either two dimensions or invade through extracellular matrix as a result of impaired invadopodia stability. Analysis of human PDAC tumor tissue additionally reveals that elevated α-actinin 4 or Dyn2 expression are predictive of poor survival. Overall, these data demonstrate that Dyn2 regulates cytoskeletal dynamics, in part, by interacting with the actin-binding protein α-actinin 4 during tumor cell invasion.
INTRODUCTION
Metastasis is the process by which tumor cells invade from the site of the primary tumor to colonize within secondary tissues (Steeg, 2016). This invasive dissemination process, rather than the primary tumor, is the actual cause of most cancer-related deaths (Valastyan and Weinberg, 2011; Lambert et al., 2017). Defining the protein-centric machinery that drives tumor cell invasion will lead to a deeper understanding of the mechanisms by which tumor cells gain this abnormal ability to move through the body, and generate new strategies about how cancer therapies should be designed to limit the metastatic potential of tumor cells.
The large GTPase Dynamin 2 (Dyn2) is overexpressed in tumor cells and enhances their invasiveness (McNiven et al., 2000; Kruchten and McNiven, 2006; Eppinga et al., 2012; Razidlo et al., 2013; Xu et al., 2014). Dyn2 is classically known as a mechanoenzyme that catalyzes the scission of clathrin-coated vesicles from the plasma membrane, and Dyn2 also participates in other forms of endocytosis and intracellular trafficking (Hinshaw, 2000; Ferguson and De Camilli, 2012). Dyn2 is also required for multiple processes during cell migration, such as formation of lamellipodia, which allow for directional cell migration (Wang et al., 2011; Menon et al., 2014; Yamada et al., 2016; Wong et al., 2019). Dyn2 participates in cell migration by interacting with the actin cytoskeleton in multiple ways. First, it is able to interact with actin filaments directly and can bundle actin filaments to change the physical properties of the actin cytoskeleton to allow for changes in cell shape and the formation of invasive protrusions (Gu et al., 2010; Chuang et al., 2019). In addition, Dyn2 regulates cytoskeletal remodeling by binding to and recruiting cytoskeletal proteins, including cortactin and Arp 2/3, to the leading edge of migrating tumor cells (Krueger et al., 2003; Orth and McNiven, 2003). This suggests that Dyn2 is at the center of a multiprotein complex that is able to enhance tumor cell migration, but there is still a limited understanding of the cytoskeletal proteins Dyn2 interacts with, and how Dyn2 is able to regulate the activity of these proteins to drive changes in actin organization.
In the growth cone of neurons, which represents a nonmalignant model of cell migration, we have previously identified that Dyn2 is in a complex with the actin-bundling protein α-actinin 1 and enhances its recruitment to the base of the cell (Kurklinsky et al., 2011). Similarly, α-actinin was found to coprecipitate with Dyn2 in prostate cancer cells (Hara et al., 2007), and it was demonstrated in U2OS osteosarcoma cells that Dyn2 can alter the localization of α-actinin 1 and the general organization of actomyosin networks (Mooren et al., 2009). There are four α-actinin isoforms: α-actinin 2 and 3 are expressed in muscle cells, while α-actinin 1 and 4 are the nonmuscle isoforms and are expressed in most cell types (Sjoblom et al., 2008; Murphy and Young, 2015). The α-actinin proteins form homo- or heterodimers and cross-link actin filaments to stabilize actin networks at structures such as focal adhesions (Knudsen et al., 1995; Edlund et al., 2001). Owing to this ability to regulate actin networks, the α-actinin proteins have also been implicated in tumor cell migration; most specifically α-actinin 4, which is amplified in multiple types of cancers (Fukumoto et al., 2015; Honda, 2015). Overexpression of α-actinin 4 enhances the migration of multiple types of cancer cells and drives the formation of invasive protrusions at the leading edge, and there is also evidence for a role for α-actinin 4 in transcriptional regulation by binding to transcription factors to change gene expression patterns in tumor cells (Khurana et al., 2011; Aksenova et al., 2013; Shao et al., 2014). While it has been demonstrated that α-actinin 4 can enhance tumor cell metastasis, the mechanism by which α-actinin 4 alters the actin cytoskeleton to enhance cell motility remains poorly defined. In addition, the relative contribution of the different α-actinin isoforms to tumor cell migration has not been analyzed in depth.
Both Dyn2 and α-actinin 4 are overexpressed in pancreatic ductal adenocarcinomas (PDACs), which is one of the most aggressive forms of cancers and is associated with high rates of metastasis (Kikuchi et al., 2008; Welsch et al., 2009; Eppinga et al., 2012). As previous data suggested that Dyn2 and α-actinin can functionally interact in neurons (Kurklinsky et al., 2011), we hypothesized that the interaction between Dyn2 and α-actinin is necessary for establishing the cytoskeletal networks required for PDAC cell migration. In this study, we first demonstrate that Dyn2 can bind directly to both nonmuscle isoforms of α-actinin, and map the binding regions in these proteins that mediate this interaction. Using deletion mutant forms of both Dyn2 and α-actinin 1 and 4, which block the binding between these proteins, we demonstrate the interaction between Dyn2 and α-actinin 4 is required for PDAC migration in 2D, as well as remodeling of the extracellular matrix (ECM) to facilitate invasion through the tumor microenvironment. Finally, we demonstrate the Dyn2–α-actinin 4 interaction is required for the stability of invadopodia, which are invasive protrusions utilized by PDAC cells to degrade extracellular matrix proteins by secreting matrix metalloproteases (MMPs) at the leading edge. These findings provide new information about how Dyn2 modulates the actin cytoskeleton by binding to actin-binding proteins and establishes a multiprotein complex that promotes tumor cell migration.
RESULTS
Dynamin 2 interacts with α-actinin 1 and 4 at the leading edge of tumor cells
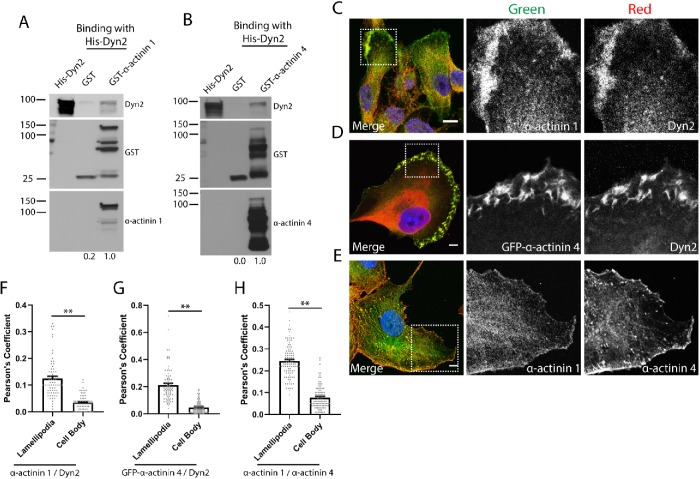
We previously identified that Dyn2 is in a complex with the actin-bundling protein α-actinin 1 in neurons, and overexpression of Dyn2 could drive the recruitment of α-actinin 1 to the base of the cell (Kurklinsky et al., 2011). This observation suggests that Dyn2 may be able to regulate changes in organization of the actin cytoskeleton by binding to actin regulatory proteins and recruiting them to discrete regions of the cell. To determine whether Dyn2 can interact directly with α-actinin 1, pull down of glutathione S-transferase (GST)-tagged α-actinin 1 was used to test direct binding to purified His-Dyn2. We found that Dyn2 can bind directly to α-actinin 1 (Figure 1A). Interestingly, Dyn2 can also interact directly with the closely related isoform α-actinin 4 (Figure 1B). We predicted that Dyn2 may regulate cytoskeletal dynamics via direct binding to both of these α-actinin isoforms.
FIGURE 1:
Dyn2 binds to α-actinin 1 and 4 and colocalizes in lamellipodia of migrating tumor cells. (A, B) GST pull down of full-length GST–α-actinin 1 (A) or GST–α-actinin 4 (B) was performed to test direct binding to full-length His-Dyn2. GST proteins were blotted with α-actinin 1 and α-actinin 4 antibodies to validate their identity. n = 3 independent experiments, densitometry was performed to measure binding, and the relative average binding values are listed below each lane. (C–E) Immunofluorescence of α-actinin 1/4 and Dyn2 in PANC-1 cells reveals these proteins colocalize in lamellipodia in PDAC cells. The region highlighted in the Merge image is shown in the individual channel insets. Scale bars: 10 μm. (F–H) Pearson’s coefficients were measured to quantify where α-actinin 1/4 and Dyn2 colocalize in tumor cells. For each cell analyzed, the colocalization between indicated proteins was quantified in the lamellipodia and in the cell body. Graphed data represent the mean ± SEM, and data points represent individual cells. Between 70 and 101 cells were quantified across three independent experiments. Scale bars: 10 μm. Student’s t test was used to measure statistical significance. ** indicates p < 0.01.
We next determined where Dyn2 and α-actinin colocalize in pancreatic tumor cells, which may suggest particular processes regulated by the interaction between these proteins. The most striking colocalization between α-actinin 1/4 and Dyn2 occurred on lamellipodia or other plasma membrane protrusions that form the leading edge of migratory tumor cells (Figure 1, C and D). Both α-actinin and Dyn2 are enriched on the leading edge, suggesting that cell migration may be regulated by interaction between α-actinin and Dyn2. Additionally, immunofluorescence was used to compare the localization of α-actinin 1 and α-actinin 4 in tumor cells. Both proteins localized to the lamellipodia and focal adhesions in tumor cells, but there were also distinct localizations of the two proteins, which suggested they may have nonoverlapping functions (Figure 1E). The colocalization of these proteins was quantified using Pearson’s coefficients in regions corresponding to the lamellipodia and the cell body, and we observed the colocalization between Dyn2 and α-actinin 1/4 is enhanced in the lamellipodia of PDAC cells (Figure 1, F–H), indicating the functional role of this protein–protein interaction may involve tumor cell migration. To verify these structures are lamellipodia, we additionally performed immunofluorescence to measure the colocalization between Dyn2 and α-actinin 1/4 with cortactin, a known lamellipodia protein (Bryce et al., 2005). Cortactin is enriched in the lamellipodia of PANC-1 cells, and we observed enhanced colocalization between cortactin and each of Dyn2 and α-actinin 1/4 in the lamellipodia as opposed to the cell body (Supplemental Figure 1, A–C). As Dyn2 and α-actinin could be recruited to the lamellipodia of tumor cells, GFP–α-actinin 4 and mCherry-Dyn2 were expressed in PANC-1 cells that were then viewed by confocal microscopy to visualize the dynamics of the two proteins during cell migration. We found that Dyn2 and α-actinin 4 are heavily recruited to the leading edge and membrane ruffles during migration of multiple PDAC tumor cell lines, as well as on intracellular puncta that appear to be invadopodia, another actin-rich protrusive structure used by PDAC cells to remodel the extracellular matrix (Supplemental Movies S1 and S2). These observations support the concept that the interaction between Dyn2 and α-actinin 1/4 is involved in multiple processes that support tumor cell migration.
Movie S1.
Live cell imaging of DanG cells co-expressing GFP-α-actinin 1 WT and mCherry-Dyn2 WT shows co-localization of both proteins on the lamellipodia at the leading edge and on invadopodia in the center of the cell during migration. The image was acquired at a rate of 1 frame per 30 seconds.
Movie S2.
Live cell imaging of DanG cells co-expressing GFP-α-actinin 4 WT and dsRed-Dyn2 WT shows co-localization of the two proteins on lamellipodia and membrane ruffles of PDAC tumor cells, as well as on invadopodia which disassemble during the movie. The image was acquired at a rate of 1 frame per 30 seconds.
Dyn2 binds directly to α-actinin 1 and 4
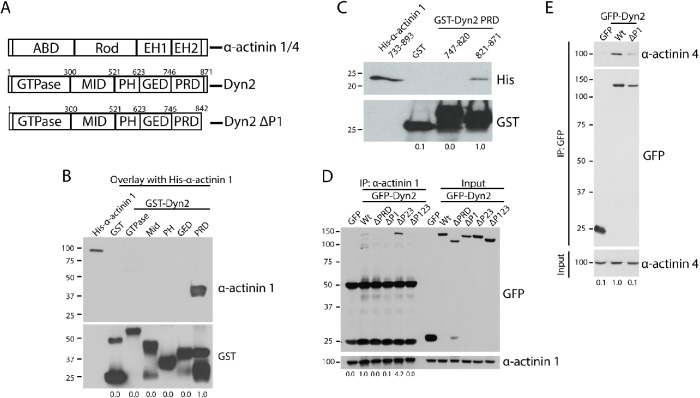
The observation that Dyn2 binds to and colocalizes with the α-actinin isoforms at the leading edge of tumor cells indicates that this protein interaction may regulate tumor cell invasion. To test the function of the Dyn2–α-actinin interaction in tumor cell migration and invasion, the binding regions of these proteins were mapped to generate deletion mutants that cannot interact. We first determined which region in Dyn2 is required for binding to α-actinin 1. The Dyn2 protein consists of a GTPase domain that allows Dyn2 to act as a mechanoenzyme, a middle domain that mediates Dyn2 oligomerization, a pleckstrin-homology (PH) domain that allows Dyn2 to bind to lipid membranes, and a GTPase effector domain (GED) and proline-rich domain (PRD) domain that allow Dyn2 to bind to effector proteins (Figure 2A). Using GST-tagged forms of the individual Dyn2 domains, we tested which domain could interact with α-actinin 1 (Figure 2A). Using an overlay assay with full-length His–α-actinin 1, it was determined that the Dyn2 PRD, which is responsible for mediating many interactions between Dyn2 and other proteins, is able to bind to α-actinin 1 (Figure 2B). Deleting the entire PRD from Dyn2 would disrupt multiple protein interactions, so the binding region for α-actinin 1 to the PRD was refined further. To this end, GST-tagged N-terminal and C-terminal halves of the Dyn2 PRD were generated, and using GST pull down the C-terminal half of the Dyn2 PRD was identified as required for binding the C-terminal tail of α-actinin 1 (residues 821–871; Figure 2C). The C-terminal tail of Dyn2 has previously been divided into five regions based on the presence of proline-rich repeats named P1 through P5 (McNiven et al., 2000), so we tested whether deletion of these smaller regions in the PRD could disrupt binding to α-actinin 1. GFP-tagged forms of Dyn2 were constructed and expressed in HeLa cells, and then immunoprecipitation of α-actinin 1 was used to detect changes in the Dyn2–α-actinin 1 interaction. Full-length Dyn2 was able to be coimmunoprecipitated with α-actinin 1, and this was prevented by deletion of the entire PRD. Deletion of the C-terminal P1 region blocked the interaction between α-actinin 1 and Dyn2, whereas deleting the P2-3 region in the Dyn2 PRD still allowed for an interaction between α-actinin 1 and Dyn2. These data demonstrate that the Dyn2 P1 region is critical for the interaction (Figure 2D). As a direct interaction between Dyn2 and α-actinin 4 was also identified, this experiment was repeated for α-actinin 4, and we found that the same P1 region of the Dyn2 PRD is responsible for mediating the interaction between both α-actinin isoforms (Figure 2E). In summary, deletion of the P1 (rat Dyn2: Δ843–871, human Dyn2: Δ846–869) in Dyn2 can block the binding interaction with α-actinin 1 and 4.
FIGURE 2:
The P1 region in the Dyn2 PRD binds to α-actinin 1 and 4. (A) Schematic diagrams showing the domain structure of Dyn2 and both α-actinin 1 and 4. The α-actinin proteins contain an actin-binding domain (ABD), a rod domain, and two calmodulin-like EF-hand domains (EH1/2). Dyn2 contains a GTPase domain, a middle domain (MID), a pleckstrin-homology domain (PH), a GTPase effector domain (GED), and a proline-rich domain (PRD). Also depicted is the Dyn2 ΔP1-binding mutant that does not bind to α-actinin 1/4 (rat Dyn2: Δ843-871; human Dyn2: Δ846-869). (B) An overlay binding assay was used to test which domain of Dyn2 binds to full-length His–α-actinin 1. GST was used as a control to show no binding. n = 3 independent experiments. (C) GST pull down of the N-terminal half of the Dyn2 PRD (amino acids 747–820) and the C-terminal half (amino acids 821–870) was performed to test direct binding with His–α-actinin 1 EH1/2 domains. n = 3 independent experiments. (D, E) Immunoprecipitation of α-actinin 1 (D) or α-actinin 4 (E) from cells expressing different GFP-Dyn2 deletion mutants was performed to determine the α-actinin binding region in the Dyn2 PRD. The Dyn2 deletion mutants tested were deletion of the entire PRD (amino acids 747–871), deletion of the P1 region (amino acids 843–871), deletion of the P2-3 region (amino acids 820–844), and deletion of the P1-P3 region (amino acids 820–871). n = 4 independent experiments (n = 2 independent experiments for GFP-Dyn2 ΔP123). (E) Binding with α-actinin 4 was tested with GFP-Dyn2 WT and GFP-Dyn2 ΔP1 using a GFP-Trap pull down. n = 4 independent experiments. For B–E, relative average binding values are listed below each condition.
As we have previously shown that Dyn2 can interact with the actin-binding protein cortactin, and this interaction also occurs in the C-terminal region of the Dyn2 PRD (McNiven et al., 2000), it was important to confirm that the Dyn2 ΔP1 deletion mutant was specific for the α-actinin proteins, and did not disrupt other functions of Dyn2. Immunoprecipitation of cortactin followed by Western blot was used to test binding to different GFP-tagged Dyn2 constructs. As shown previously, Dyn2 coimmunoprecipitated with cortactin, and deletion of the entire PRD of Dyn2 blocked binding this interaction. Importantly, cortactin was still able to interact with Dyn2 ΔP1, which does not interact with α-actinin 1 or 4 (Supplemental Figure 1D). This supports the premise that a Dyn2 ΔP1 protein cannot bind the α-actinin proteins, and functional effects are likely not caused by disrupting the interaction between Dyn2 and cortactin.
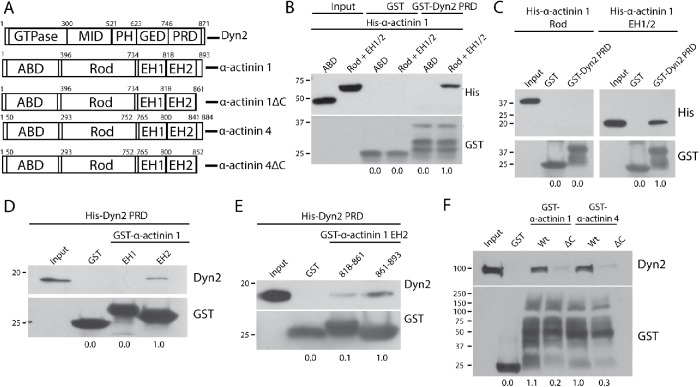
We next mapped the region in the α-actinin proteins that binds to Dyn2. The α-actinin proteins contain an N-terminal actin-binding domain (ABD), a rod domain that mediates dimerization, and C-terminal EF-hand domains that bind calcium and regulate protein activity (Figure 3A). We first generated a His-tagged α-actinin 1 ABD and the rod + EH domains. GST pull down of the Dyn2 PRD revealed that the C-terminal half of α-actinin 1 binds directly to Dyn2 (rod + EH1/2; Figure 3B). Subsequently, it was tested whether the rod or EH domains were responsible for binding Dyn2. Pull-down experiments using purified proteins revealed that the Dyn2 PRD binds directly to the C-terminal region containing the EH domains (Figure 3C), and more specifically, the C-terminal EH2 domain (Figure 3D). This region included the EH2 domain, as well as a short C-terminal tail of α-actinin 1 adjacent to the EH2 domain. We then tested whether the EH2 domain itself or the C-terminal tail of α-actinin 1 bound to Dyn2. GST pull down of His-Dyn2 PRD revealed that the C-terminal fragment of α-actinin 1 mediates the Dyn2–α-actinin interaction (residues 861–893; Figure 3E). Importantly, this region is not part of the conserved EH2 domain, so deletion of the region should not affect the ability of α-actinin to bind calcium, or disrupt other protein–protein interactions that occur in the EH domains of α-actinin. We next generated deletions of this C-terminal tail within full-length α-actinin 1 to test whether this region is required for binding to Dyn2. Based on sequence similarity between α-actinin 1 and α-actinin 4, we predicted the same C-terminal region in both proteins would be required for binding to Dyn2 in cell lysates. GST-tagged α-actinin 1 or 4, wild type (WT) or with a deletion of the C-terminal tail, was used for a GST pull down from DanG cell lysates. Indeed, deletion of amino acids 861–892 in α-actinin 1, or amino acids 853–884 in α-actinin 4, disrupts the interaction between α-actinin and Dyn2 (ΔC; Figure 3F), confirming that the C-terminal tails of the α-actinin proteins mediate binding to Dyn2.
FIGURE 3:
The C-terminal tails of α-actinin 1 and 4 are responsible for binding to Dyn2. (A) Schematic diagrams showing the domain structure of Dyn2 and both α-actinin isoforms. Also depicted are the α-actinin 1ΔC (rat α-actinin 1: Δ862–893; human α-actinin 1: Δ861–892) and α-actinin 4ΔC (human α-actinin 4: Δ853–884) forms that cannot bind Dyn2. (B) GST pull down of GST-Dyn2 PRD was used to test binding to different regions in α-actinin 1. His–α-actinin 1 proteins representing the ABD (amino acids 1–395) and the rod + EH 1/2 domains (amino acids 396–893) were generated. GST was used as a control and showed no binding interaction. n = 3 independent experiments. (C) GST pull down of GST-Dyn2 PRD was used to test whether the Rod domain (amino acids 396–733) or the EH1/2 domain (amino acids 734–893) of α-actinin 1 is responsible for binding to Dyn2. n = 3 independent experiments. (D) Proteins representing the EH1 domain (amino acids 734–817) and EH2 domain (amino acids 818–893) were used to test binding to the Dyn2 PR. n = 2 independent experiments. (E) The α-actinin 1 EH2 domain was further separated into an N-terminal half (amino acids 818–861) and a C-terminal half (862–893) to further characterize the Dyn2-binding region. n = 3 independent experiments. (F) GST–α-actinin proteins were generated with deletions of the identified Dyn2-binding region in human α-actinin 1 (amino acids 861–892), and the predicted Dyn2-binding region in human α-actinin 4 (amino acids 853–884) based on sequence similarity to α-actinin 1. The GST fusion proteins were used in pull-down assays with lysates from DanG cells. n = 3 independent experiments. For B–F, relative average binding values are listed below each condition.
The interaction between Dyn2 and α-actinin 4 is required for efficient PDAC cell migration and invasion
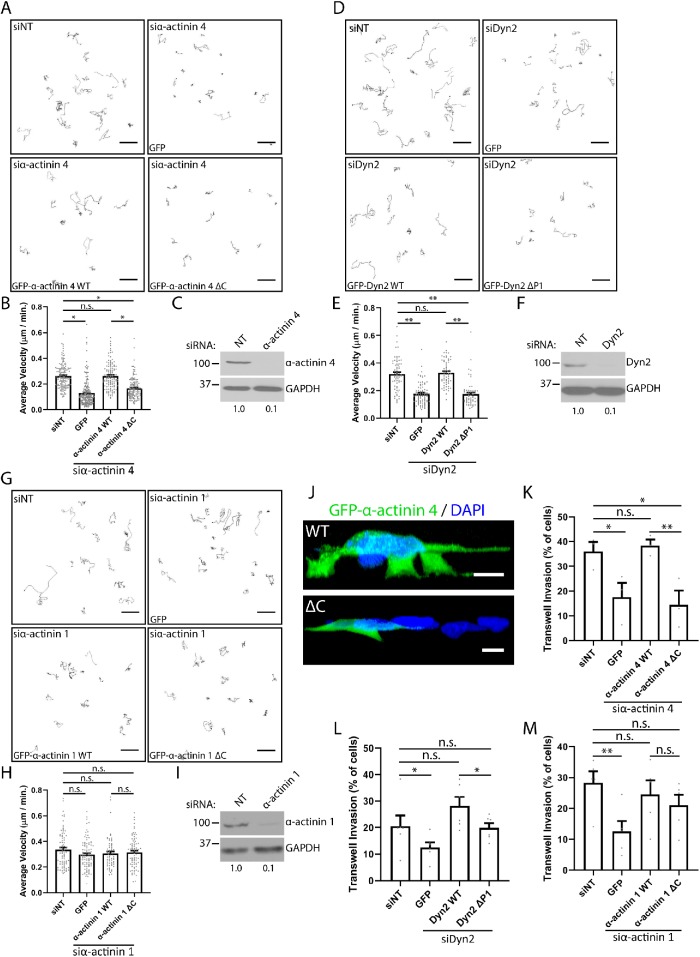
As Dyn2 colocalizes with α-actinin at the lamellipodia of pancreatic tumor cells (Figure 1, C and D), we tested whether disrupting the binding between these proteins could impair tumor cell migration using the binding mutants identified in Figures 2 and 3. To test this, the velocity of PANC-1 cells during unstimulated 2D cell migration was measured by plating the cells in glass dishes and imaging overnight using confocal microscopy, and analyzing the images using the Manual Tracking plug-in on ImageJ to calculate cell velocity. PANC-1 cells were treated with small interfering RNA (siRNA) to knock down α-actinin 1, α-actinin 4, or Dyn2, and then either the GFP-tagged WT or binding mutant form of the protein was reexpressed into the cells. As predicted, knockdown of α-actinin 4 reduced the velocity of PANC-1 cells during cell migration. This defect was rescued by reexpression of GFP–α-actinin 4 WT, but not GFP-α-actinin 4 ΔC, which disrupts binding to Dyn2 (Figure 4, A–C). Similarly, knockdown of Dyn2 reduced migration velocity. This was rescued by GFP-Dyn2 WT, but not by GFP-Dyn2 ΔP1, which disrupts the interaction with α-actinin 1 and 4 (Figure 4, D–F). This suggests that not only are α-actinin 4 and Dyn2 each required for PDAC cell migration, but that the ability of these proteins to interact is also crucial for this process. Interestingly, unlike α-actinin 4, knockdown of α-actinin 1 did not impact cell migration velocity (Figure 4, G–I). Further, reexpressing GFP–α-actinin 1 WT or GFP–α-actinin 1 ΔC, which disrupts binding to Dyn2, also had no effect on migration velocity. These data suggest that the interaction between α-actinin 1 and Dyn2 is not required for PDAC cell migration, but that it is specific for α-actinin 4.
FIGURE 4:
The interaction between Dyn2 and α-actinin 4 is required for 2D and invasive migration of PDAC cells. (A–I) The velocity of PANC-1 cells during 2D migration was quantified after knockdown of α-actinin 4 (A, B), Dyn2 (D, E), or α-actinin 1 (G, H) and reexpression of the WT or binding mutant protein. Representative cell tracks are shown for each condition, with each path representing a single cell. Graphed data represent the mean ± SEM, and data points represent individual cells. n = 3 independent experiments, with between 62 and 192 cells quantified across all experiments. Western blots showing the efficiency of siRNA knockdown for α-actinin 4 (C), Dyn2 (F), and α-actinin 1 (I) are shown, and values below blots represent quantification from densitometry. (J–M) Transwell migration assays were performed to measure the ability of PANC-1 cells to undergo invasive migration after manipulating α-actinin– Dyn2 binding. Reconstructions of z-stack images of PANC-1 cells expressing either GFP–α-actinin 4 WT or GFP–α-actinin 4 ΔC invading through transwell filters are shown (J). Transwell invasion efficiency was quantified after knockdown of α-actinin 4 (K), Dyn2 (L), or α-actinin 1 (M) and reexpression of the WT or binding mutant protein. Graphed data represent the mean ± SEM, and data points represent average values for each experiment. α-Actinin 4: n = 3 biological replicates, between 334 and 549 cells per condition in each experiment. α-Actinin 1: n = 5 biological replicates, between 132 and 538 cells per condition in each experiment. Dyn2: n = 6 biological replicates, between 167 and 581 cells per condition in each experiment. Scale bars: 50 μm (A, D, G), 10 μm (J). Student’s t test was used to measure statistical significance. * indicates p < 0.05; ** indicates p < 0.01.
The importance of the α-actinin–Dyn2 interaction in tumor cell migration was tested further using chemotactic transwell migration assays. PANC-1 cells were treated with siRNA, as in the 2D migration experiments, but for this assay the tumor cells were plated on top of a gelatin-coated filter to provide a barrier to cell migration. The percentage of cells that migrated across the filter was measured by scoring the Hoechst-stained nuclei on the top and bottom of the filter following incubation. Using this assay, knockdown of either α-actinin 4 or Dyn2 reduced transwell migration (Figure 4, K and L). Reexpression of GFP–α-actinin 4 WT was able to rescue transwell invasion; however, impaired invasion persisted in cells reexpressing GFP–α-actinin 4 ΔC (Figure 4K). Following Dyn2 knockdown, reexpression of either GFP-Dyn2 WT or GFP-Dyn2 ΔP1 rescued cell invasion; however, the WT protein significantly enhanced invasion beyond the effect of the deletion mutant (Figure 4L). This may suggest that there are other functions of Dyn2 in supporting cell invasion in addition to its binding to α-actinin 4, but this interaction is required for Dyn2 to exert its maximal effect on tumor cell invasion. These data are consistent with the findings in Figure 4, A–E, providing further evidence that the interaction between α-actinin 4 and Dyn2 is required for PDAC cell migration. We also generated 3D reconstructions of PANC-1 cells expressing either GFP–α-actinin 4 WT or ΔC, and found the cells expressing GFP–α-actinin 4 WT formed multiple protrusions to drive invasion, in contrast to the cells expressing GFP–α-actinin 4 ΔC (Figure 4J). Interestingly, while knockdown of α-actinin 1 reduced transwell migration for PANC-1 cells, both GFP–α-actinin 1 WT and GFP–α-actinin 1 ΔC, which disrupts binding to Dyn2, were able to restore transwell migration to a similar extent (Figure 4M). Thus, in contrast to α-actinin 4, the interaction between Dyn2 and α-actinin 1 does not regulate tumor cell migration.
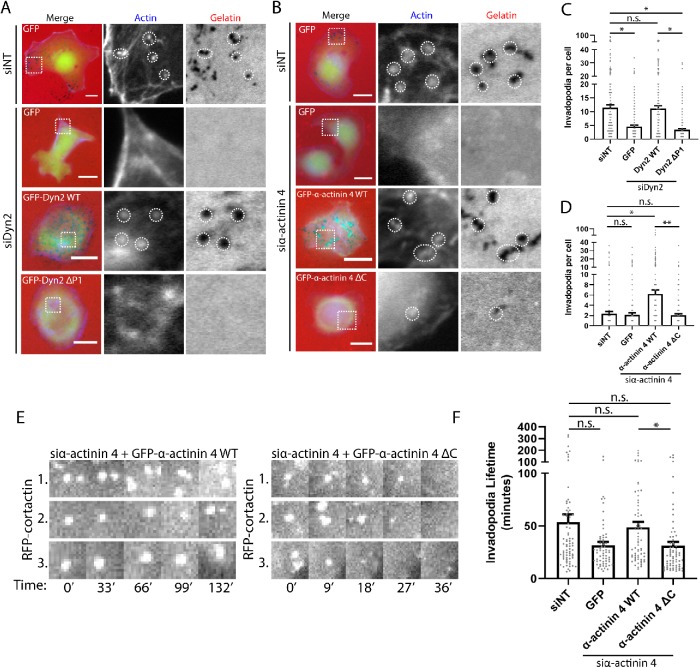
The observation that disrupting the α-actinin 4–Dyn2 interaction impaired cell invasive migration through a gelatin substrate led us to hypothesize that these two proteins are required for the formation of matrix-degrading structures called invadopodia. PDAC cells and other tumor cells utilize invadopodia at the leading edge to concentrate matrix metalloproteinases to enhance ECM degradation to facilitate cell invasion through confined environments. Both α-actinin and Dyn2 have been reported to localize to invadopodia previously (Baldassarre et al., 2003; Yamada et al., 2016; Yamaguchi et al., 2017), but we sought to determine whether they colocalize at invadopodia, and whether the interaction between these proteins is required for ECM remodeling. To test this, we first performed immunofluorescence imaging of DanG cells, a PDAC cell line that forms multiple prominent invadopodia. Staining of GFP–α-actinin 1 and Dyn2 revealed colocalization on functional invadopodia, which were localized above regions of gelatin degradation (Figure 5A). Similarly, α-actinin 4 colocalized at invadopodia puncta with GFP-Dyn2 (Figure 5B). Additional immunofluorescence analysis of Dyn2 and α-actinin 1/4 with Tks5, a commonly used marker for invadopodia (Lock et al., 1998), was performed and showed a striking colocalization of Dyn2 and α-actinin 1/4 with Tks5 at sites of gelatin degradation, confirming these structures as functional invadopodia in PDAC cells (Supplemental Figure 2, A–C).
FIGURE 5:
Dyn2 colocalizes with α-actinin at invadopodia, and the interaction between Dyn2 and α-actinin 4 is required for ECM remodeling. (A, B) Immunofluorescence staining of Dyn2 and α-actinin 1/4 on fluorescent gelatin-coated coverslips shows colocalization of these proteins at invadopodia actively degrading matrix. The region highlighted in the Merge image is shown in the individual channel insets. Images were collected from n = 3 experiments. (C–E) Gelatin degradation assays were used to measure invadopodia activity in DanG cells expressing the WT or deletion mutant forms of each protein after knockdown of the endogenous protein. DanG cells were plated on Cy3-fluorescent gelatin-coated coverslips for 7 h to allow for gelatin degradation, and then fixed. (F–H) The area of matrix degradation was quantified by dividing the total area of degraded gelatin by the cell area. Graphed data represent the mean ± SEM, and data points represent average values for independent experiments relative to the nontargeting control. α-Actinin 4: n = 5 biological replicates, 9–42 cells per condition in each experiment. α-Actinin 1: n = 3 biological replicates, 20–34 cells per condition in each experiment. Dyn2: n = 3 biological replicates, 26–37 cells per condition in each experiment. Scale bars: 5 μm (A, B) and 10 μm (C–E). Student’s t test was used to measure statistical significance. * indicates p < 0.05; ** indicates p < 0.01.
To functionally test the role of the α-actinin–Dyn2 interaction in degradation of the extracellular matrix, we utilized a gelatin degradation assay (Artym et al., 2006). DanG cells were plated on a fluorescent gelatin substrate, and invadopodia-mediated matrix degradation was visualized by a loss of fluorescence. Knockdown-rescue experiments were again used in cells expressing either the WT or binding mutant of α-actinin 1, α-actinin 4, and Dyn2 to test whether there was a difference in gelatin degradation. Consistent with the results from the migration assays, knockdown of either α-actinin 4 or Dyn2 reduced the area of matrix degradation. This defect was rescued by reexpression of WT α-actinin 4 or Dyn2, but not by GFP–α-actinin 4 ΔC or GFP-Dyn2 ΔP1, suggesting that the α-actinin 4–Dyn2 interaction is required for invasive matrix degradation (Figure 5, C, D, F, and G). In contrast, knockdown of α-actinin 1 did not impact matrix degradation, and there was no difference in the degree of matrix degradation between cells reexpressing GFP–α-actinin 1 WT and GFP–α-actinin 1 ΔC (Figure 5, E and H), supporting the specific role of the α-actinin 4-Dyn2 interaction in matrix remodeling.
As invadopodia are a central mechanism by which PDAC cells perform matrix remodeling, we next sought to address whether disrupting the α-actinin 4–Dyn2 interaction might impair the formation or stability of invadopodia. First the number of invadopodia in fixed cells was measured after knockdown and reexpression of either the α-actinin 4 or Dyn2 WT protein or binding mutant. This was achieved by counting the regions of degraded fluorescent gelatin that colocalized with actin puncta, which suggests functional invadopodia. There was a marked reduction in the number of invadopodia in cells reexpressing the binding mutant form of either Dyn2 or α-actinin 4 compared with the respective WT proteins, confirming that the Dyn2–α-actinin 4 interaction is required for the presence of functional invadopodia (Figure 6, A–D). To address whether invadopodia formation and/or turnover were attenuated by disrupting α-actinin 4–Dyn2 binding, we used live-cell imaging of RFP-cortactin, a known invadopodia protein, and measured lifetimes of individual invadopodia in cells expressing WT or binding mutant GFP–α-actinin 4. Expression of GFP–α-actinin 4 ΔC caused a significant reduction in invadopodial lifetime, indicating that invadopodia turnover occurs faster than in cells expressing GFP–α-actinin 4 WT (Figure 6, E and F). These data suggest that the interaction between α-actinin 4 and Dyn2 is not necessarily required for the formation of invadopodia, but rather this interaction is required for stabilizing invadopodia to allow for ECM remodeling to occur during tumor cell invasion.
FIGURE 6:
Disrupting the Dyn2–α-actinin 4 interaction decreases invadopodia number and stability in PDAC cells. (A–D) Invadopodia number in DanG cells was quantified after plating cells on Cy3- fluorescent gelatin for 7 h to allow for invadopodia-based matrix degradation to occur. Knockdown of α-actinin 4 (A) or Dyn2 (B) was performed, and then either the WT or binding mutant protein was reexpressed in the cells. Cells were stained with phalloidin to mark actin, and invadopodia number was measured by counting the number of actin puncta that overlapped a spot of degraded gelatin per cell (C, D). Between 24 and 41 cells were scored per condition in each of three to four independent experiments for α-actinin 4, or 29–51 cells per condition in each of five independent experiments for Dyn2. Graphed data represent the mean ± SEM, and data points represent values for individual cells. (E, F) Live-cell microscopy of RFP-cortactin, a known invadopodia protein, was used to measure the lifetime of invadopodia after knockdown of α-actinin 4 and reexpression of GFP–α-actinin 4 WT or GFP–α-actinin ΔC in DanG cells. Cells were imaged once every 3 min for 6 h, and invadopodia present halfway through imaging were selected for quantification. Invadopodia lifetime was quantified using the Invadopodia Tracker ImageJ plug-in. Images show a representative cell expressing GFP–α-actinin 4 WT with multiple RFP-cortactin–positive invadopodia that persist, and a cell expressing GFP–α-actinin 4 ΔC with RFP-cortactin–positive invadopodia that turn over within minutes. At least 10 cells were scored per experiment over five independent experiments. Graphed data represent the mean ± SEM, and data points represent average invadopodia lifetime per cell. Scale bars: 10 μm. Student’s t test was used to measure statistical significance. * indicates p < 0.05; ** indicates p < 0.01.
Taken together, these data indicate that Dyn2 and α-actinin 4 regulate invadopodia formation and PDAC cell invasion. As these multidomain proteins have additional cellular roles, this does not exclude the possibility that other functions of Dyn2 or α-actinin 4 also promote tumor cell invasion. For example, Dyn2’s GTPase activity has previously been implicated in tumor cell invasion (Baldassarre et al., 2003). Indeed, following Dyn2 knockdown, reexpression of Dyn2 K44A, a dominant negative GTPase-deficient mutation, is not able to rescue 2D migration velocity (Supplemental Figure 3, A–C), transwell invasion (Supplemental Figure 3D), or invadopodia number (Supplemental Figure 3, E and F). Thus, Dyn2 may promote tumor cell invasion via multiple mechanisms, including an interaction with α-actinin 4.
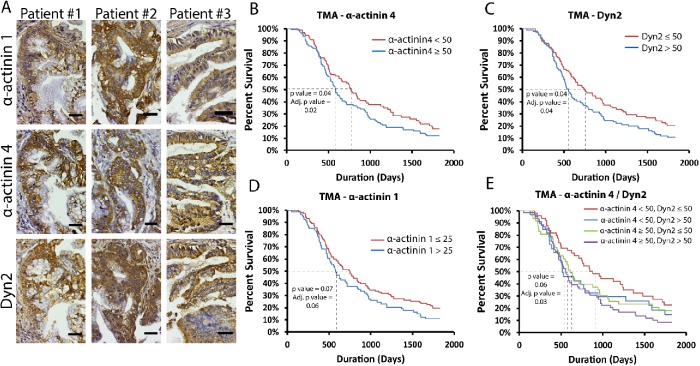
As the findings described above indicate that Dyn2 and α-actinin 4 interact directly to promote tumor cell invasion and migration, we next used histological analysis of PDAC patient tissue samples to evaluate expression of α-actinin 1/4 and Dyn2 in human tumors. A pancreatic tumor tissue microarray including tissue samples from 205 unique primary PDAC tumors were stained with antibodies against α-actinin 1, α-actinin 4, or Dyn2 (Figure 7A). Each sample was scored based on the intensity and extent of the staining, and patients were stratified based on low or high expression of each protein based on the median staining intensity. Patients with the highest levels of α-actinin 4 or Dyn2 had lower mean survival times compared with patients with lower expression (Figure 7, B and C). In contrast, α-actinin 1 expression did not correlate with a statistically significant difference in patient survival (Figure 7D). Additionally, patients were grouped into independent categories and that enabled a correlative comparison between protein expression and survival. From this analysis it appeared that patients with low α-actinin 4 and low Dyn2 expression had significantly longer survival than patients with overexpression of one or both proteins (Figure 7E). Overall, these data support the concept that the overexpression of Dyn2 or α-actinin 4 is enhancing the aggressiveness of PDAC, and the interaction between these proteins may be a critical mediator of tumor cell invasion.
FIGURE 7:
Dyn2 and α-actinin 4 overexpression in PDAC patients is correlated with worse prognosis. (A) Immunohistochemistry of tissue microarray (TMA) slides shows the localization of α-actinin 1 and 4, and Dyn2 in human PDAC tissue. Serial sections of primary PDAC tumors from three unique patients are shown. Both α-actinin isoforms and Dyn2 are expressed in the neoplastic ducts. (B–D) Kaplan Meier survival curves were generated based on the expression of α-actinin 4 (B), Dyn2 (C), and α-actinin 1 (D) in 205 PDAC patient tissue cores represented in the TMA slides. H-scores were calculated based on intensity and extent of the staining, and patients were stratified into groups based on the median H-score value, which are listed in the graph legends. (E) Patients were further grouped into four categories based on the expression of both α-actinin 4 and Dyn2, stratified by the H-scores listed in the graph legend. Scale bars: 25 μm. Likelihood ratio test was used to measure statistical significance; adjusted p values take into account patient demographics (age at diagnosis, sex, obesity [BMI ≥ 30], and patient-reported diabetes).
DISCUSSION
In addition to a well-defined role as a mechanoenzyme in membrane remodeling events, Dyn2 is also known to regulate cytoskeletal architecture. In this study we present new data that Dyn2 can bind directly to α-actinin, and this interaction is required for regulating cytoskeletal dynamics during tumor cell migration. The colocalization of Dyn2 and α-actinin 4 on invadopodia (Figure 5, A and B) and the loss of invadopodia stability by disrupting the Dyn2–α-actinin 4 interaction (Figure 6, C, D, and F) suggests that this protein–protein interaction controls the maintenance of invasive membrane protrusions, as well as the membrane–cytoskeletal remodeling required for migration (Figure 4). Thus, the interaction between Dyn2 and α-actinin 4 during tumor cell invasion allows Dyn2 to coordinate changes in actin organization to migrate through diverse environments during metastasis.
Dyn2 interacts with multiple actin-binding proteins to control cytoskeletal remodeling and invadopodial dynamics. The nucleation of actin filaments to form invadopodia is driven in part by proteins like Arp 2/3 and cortactin, which are both known Dyn2-associated proteins, and through these interactions Dyn2 is able to regulate the initiation of invadopodia formation (Krueger et al., 2003; Destaing et al., 2013; Zhang et al., 2016). The observations described here suggest the interaction between Dyn2 and α-actinin 4 is required for the stabilization of invadopodia after they are formed. Live-cell analysis of invadopodia lifetime revealed invadopodia formation after expressing GFP–α-actinin 4 ΔC, which does not bind Dyn2, but decreased invadopodial lifetime (Figure 6F). It has previously been shown that Dyn2 can influence the localization of α-actinin in the cell (Mooren et al., 2009; Kurklinsky et al., 2011), so it is possible that Dyn2 binds to α-actinin 4 to recruit it to the growing invadopodia to stabilize these structures during invasion. Previous investigations into the role of the α-actinin isoforms in invadopodia formation suggest that the appearance of the α-actinin proteins at invadopodia is concurrent with the appearance of F-actin, so Dyn2 may recruit α-actinin 4 to stabilize the newly formed F-actin networks at these sites (Yamaguchi et al., 2017).
As the interaction between Dyn2 and α-actinin 4 appears to optimize tumor cell migration (Figure 4, A–E), it is also possible that this interaction is important for regulating the organization or stability of actin filaments at the leading edge of tumor cells during migration. In addition to the role in invadopodia formation, 2D migration experiments (Figure 4) indicate a role for the Dyn2–α-actinin 4 interaction in cell migration independent of interactions with an ECM. This suggests the interaction between these two proteins may be required for processes such as lamellipodia formation or organization of the actin cytoskeleton to polarize cells for migration. Both Dyn2 and α-actinin are known to bind directly to actin filaments and act as actin-bundling and cross-linking proteins, which affect the mechanical properties of actin filaments (Gu et al., 2010; Foley and Young, 2014). It was recently identified that Tks5, a scaffold protein that is required for invadopodia formation, binds to Dyn2 and enhances the stiffness of Dyn2-bundled actin filaments during the process of myoblast fusion, which involves an invadopodia-like structure that drives cell–cell fusion (Chuang et al., 2019). The C-terminal EF-hand domains of α-actinin are important for regulating the ability of α-actinin to bind to actin filaments (Prebil et al., 2016), so the binding of Dyn2 to the C-terminus of α-actinin 1/4 could regulate its actin-bundling functions. The GTPase function of Dyn2 is also required for invadopodia formation (Baldassarre et al., 2003), but it is unknown whether α-actinin regulates Dyn2 GTPase activity.
The observation that the interaction between Dyn2 and α-actinin 1 was not required for tumor cell migration and invasion (Figure 4, G, H, and M) suggests functional differences between α-actinin 1 and 4. The α-actinin isoforms are very similar in terms of amino acid sequence, and they share the same domain structure; however, to date there has not been much investigation into the unique roles of the α-actinin isoforms in different cellular processes. Both α-actinin 1 and 4 have been shown previously to be involved in invadopodia formation; however, overexpression of only α-actinin 4 was able to enhance invadopodia formation (Yamaguchi et al., 2017). In addition, there is evidence that α-actinin 4, and not α-actinin 1, decreases focal adhesion formation in colorectal adenocarcinoma cells (Fukumoto et al., 2015), but most studies on the nonmuscle α-actinin protein have only addressed the function of one specific isoform. Our data support that both α-actinin isoforms are involved in ECM remodeling and invadopodia formation (Figures 4, K and M, and 5, A and B), as both isoforms localize to invadopodia, and knockdown of either reduced transwell invasion of PDAC cells through gelatin-coated filters. However, blocking the interaction between Dyn2 and α-actinin 1 had no effect on these processes. This could be caused by α-actinin 4 compensating for the function of α-actinin 1, or it may suggest that the recruitment of α-actinin 4 to invadopodia by Dyn2 is uniquely required to recruit additional proteins or initiate certain events to drive invadopodia formation. This system is made even more complex by the fact that the α-actinin isoforms can form heterodimers with one another (Foley and Young, 2013), and it is still unknown what the purpose of the different forms of α-actinin dimers are in regulating cellular processes.
In addition to unique functions observed between the α-actinin isoforms in regulating tumor cell migration, there is also evidence that the expression of α-actinin 1 and 4 is regulated differently in cancer. While α-actinin 1 expression is not observed to be altered in human cancers, α-actinin 4 expression is increased in multiple types of cancers, including PDAC, normally as a result of a gene amplification event (Kikuchi et al., 2008; Honda, 2015). Increased expression of α-actinin 4 is typically correlated with worse patient survival, which is what we observed in our analysis of human PDAC patient tissue samples (Figure 7B). It has also been suggested that α-actinin 4 amplification could be a predictive biomarker for what type of therapy to use to treat PDAC patients (Watanabe et al., 2015). We did not observe a worse prognosis for patients showing enhanced expression of both α-actinin 4 and Dyn2 compared with the overexpression of either protein individually (Figure 7E). As binding between α-actinin 4 and Dyn2 enhances PDAC cell invasion this might suggest that overexpression of either protein is able to enhance the function of the α-actinin 4 and Dyn2-binding interaction in promoting tumor cell migration. As Dyn2 and α-actinin 4 are part of a larger cytoskeletal and membrane remodeling complex in PDAC cells, further investigation would be required to determine whether overexpression of other binding partners of Dyn2 or α-actinin 4 similarly worsen disease progression and cooperate with Dyn2 and α-actinin 4 in regulating cell invasion.
In summary, the findings presented here indicate a new mechanism through which Dyn2 is able to participate in the process of tumor cell invasion, namely, by binding to the actin-binding protein α-actinin 4 to provide a link to regulation of the actin cytoskeleton. This provides new understanding of the molecular machinery that drives tumor cell invasion and could give insight into better methods for treating tumors to target these mechanisms that drive metastasis and cancer progression.
MATERIALS AND METHODS
Cell culture, antibodies, and reagents
PANC-1 and BxPC-3 human pancreatic cancer cell lines and HeLa cervical cancer cell line were obtained from the American Type Culture Collection. DanG human pancreatic cancer cells were provided by Daniel Billadeau (Mayo Clinic). PANC-1, DanG, and HeLa cells were cultured in DMEM with 10% fetal bovine serum (FBS) and penicillin/streptomycin, and BxPC-3 cells were cultured in RPMI media with 10% FBS and penicillin/streptomycin. Cell lines were screened for mycoplasma contamination by DAPI staining and PCR.
Antibodies used in this article were as follows: α-actinin 1 (Santa Cruz; sc-1782, and Abcam; ab68194), α-actinin 4 (Abcam; ab108198), Dyn2 (purification described previously; Henley et al., 1998), GAPDH (Cell Signaling; D16H11), GST (Santa Cruz; sc-138), GFP (Roche), His epitope tag (Cell Signaling; 27E8), and cortactin (Cao et al., 2003). Actin was stained using phalloidin-tetramethylrhodamine or phalloidin–Alexa Fluor 647 (Sigma).
Transfection of siRNA and expression constructs
RNAi-mediated protein knockdown was performed using the lipofectamine RNAiMax reagent (Invitrogen) according to the manufacturer’s protocol in Opti-MEM media. Cells were treated with siRNAs at 50 nM for 48–72 h before experiments were performed, and knockdown was confirmed by Western blotting. The siRNA sequences used in the article were as follows: nontargeting (On-Target Plus #D-001810; Dharmacon), human α-actinin 1 (sense: CACAGAUCGAGAACAUCGAAGUU; Dharmacon), human α-actinin 4 (sense: CCACAUCAGCUGGAAGGAUGGUCUU; Dharmacon), human Dyn2 (sense: GACAUGAUCCUGCAGUUCA; Dharmacon).
Expression of fluorescent constructs was performed using the lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocol. The fluorescent marker is tagged to the C-terminus of the protein for constructs used in the article. pEGFP-N1 α-actinin 1 was provided by Carol Otey at the University of North Carolina–Chapel Hill (Edlund et al., 2001; Addgene plasmid #11908), and human pEGFP-N1 α-actinin 4 was provided by Alan Wells at the University of Pittsburgh. Human Dyn2 was provided by Daniel Billadeau (Mayo Clinic) and cloned into pEGFP-N1. Rat Dyn2 in pEGFP-N1 was described previously (Cao et al., 1998). Dyn2 K44A was generated by site-directed mutagenesis as described previously (Cao et al., 2000). Rat α-actinin 1 constructs used in binding assays were constructed from rat brain cDNA. Deletions were introduced using QuikChange site-directed mutagenesis with modifications. Primers used for key constructs were as follows: human GFP– α-actinin 1 ΔC: sense: 5′-CCCGACCAGGCTGAGTACTGCGTACCGCGGGCCCGGGATCCA-3′; human GFP–α-actinin 4 ΔC: sense: 5′-CCCGACCAGGCCGAGTACTGCGTACCGCGGGCCCGGGATCCA-3′; human GFP-Dyn2 ΔP1: sense: 5′-CGGATCCCCCCAGGGATTCCCCGAATTCTGCAGTCGACGGTACCG-3′. RNAi-resistant versions of α-actinin 4 were generated by site-directed mutagenesis using the primers (sense: 5′-AACTTCCACATCAGCTGGAAAGACGGACTTGCCTTCAATGCCC-3′). DanG cell lines stably overexpressing GFP, siRNA-resistant GFP–α-actinin 4 WT or GFP–α-actinin 4 ΔC were generated by transfection using lipofectamine 2000 and selection with G418 (800 µg/ml selection, 400 µg/ml maintenance), and enriched using fluorescence-activated cell sorting. The stable cell lines were used for live-cell imaging of invadopodial lifetime.
Protein purification and binding experiments
His-tagged proteins were purified by transforming BL-21 pLysS competent cells, which were then induced with 0.3 mM isopropyl b-d-1-thiogalactopyranoside (IPTG). Cells were lysed with 10 ml of lysis buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5 mM MgCl2, 10 µM 2-mercaptoethanol, 10% glycerol, 5 mM imidazole, 0.1% Triton X-100, protease inhibitor), sonicated, and centrifuged to collect the supernatant. ProBond resin (Invitrogen) was prepared according to the manufacturer’s instructions, and then incubated with the supernatant for 1–2 h at 4°C. The solution was then added to a purification column and washed with 10 ml of wash buffer 1 (same as lysis buffer except with 20 mM imidizole) and then 10 ml of wash buffer 2 (20 mM Tris-HCl, pH 8.0, 100 mM KCl, 5 mM MgCl2, 20% glycerol, 30 mM imidizole, 0.1% Triton X-100, proteinase inhibitor). Elutions were collected in elution buffer (same as wash buffer 2 except with 300 mM imidizole), and then the protein was dialyzed in cold phosphate-buffered saline (PBS) buffer with 0.04% azide for 24–48 h.
GST-tagged proteins were purified by transforming and inducing BL-21 competent cells as described above. After centrifugation the pellet was resuspended in 10 ml of PBS + protease inhibitor + 5 mM dithiothreitol (DTT) and sonicated. Triton X-100 was added to a final concentration of 1%, and mixed for 30 min at 4°C while rotating, and then centrifuged. Glutathione sepharose 4B beads (Sigma; #17-0755-05) were added and incubated for 2 h at 4°C with rotation, then spun down at 500 × g for 5 min. The supernatant was removed and the beads were washed with 1 ml of PBS + protease inhibitor three to four times. After the last wash, 300 μl of PBS was added to the beads and protein purity was assessed by running the protein on a gel and staining with Coomassie blue.
GST pull-down experiments were performed by first washing the GST–protein beads four times with 1 ml of TEN100 buffer (20 mM Tris, pH 7.4, 0.1 mM EDTA, 100 mM NaCl). The GST–protein fusion was incubated with the indicated whole-cell extract or purified His proteins at 4°C while rotating for 1 h. The beads were then washed with 1.0 ml of NETN buffer (0.5% NP40, 0.1 mM EDTA, 20 mM Tris, pH 7.4, 300 mM NaCl). The bound proteins were eluted by boiling in 1× sample buffer and analyzed by Western blotting.
For overlay binding assays, the protein sample was run on a SDS–PAGE gel and transferred to a polyvinylidene fluoride (PVDF) membrane and then blocked in 5% milk + basic buffer (20 mM HEPES, pH 7.5, 50 mM KCl, 10 mM MgCl2, 1 mM DTT, 0.1% NP40) overnight at 4°C. To test overlay binding, the second protein sample was incubated with the membrane at a concentration of 2–3 μg/ml in 1–2% milk + basic buffer overnight at 4°C. The membrane was washed with PBS-T and then probed with primary and secondary antibodies. The membrane was incubated with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher) and exposed to HyBlot CL Autoradiography film (Thomas Scientific).
For co-IP of Dyn2 with α-actinin from cell lysates, cells were lysed in 500 μl of NP-40 lysis buffer + protease inhibitors (20 mM Tris-HCl, 137 mM NaCl, 10% glycerol, 1% NP-40, 2 mM EDTA, pH 8.0), sonicated on ice, and centrifuged to collect the supernatant. Protein A or protein G beads (Sigma) were prepared according to the manufacturer’s instructions and added to 1 mg of protein lysate, and the solution was incubated at 4°C with rocking for 20–30 min to preclear and then centrifuged. Primary antibody (3–5 μg) was added to the supernatant and incubated with rocking at 4°C for 3 h, and then 100 μl of beads was added to each sample and incubated for an additional 1.5 h at 4°C. Samples were washed three times with lysis buffer followed by centrifugation, and after the last wash 15 μl of 4× sample buffer was added to each sample, boiled for 5 min, and then analyzed by Western blotting.
Protein samples for Western blotting were collected in NP40 lysis buffer + protease inhibitors. SDS–PAGE sample buffer (4×, 8% SDS, 30% glycerol, 1 M Tris-HCl, 0.4% bromophenol blue, 20% beta-mercaptoethanol, pH 6.8) was added to a final concentration of 1×, samples were boiled for 5 min, and run on a SDS–PAGE gel. PVDF membranes (Immobilon) were used for transfer, and the membranes were blocked in 5% milk (BioRad) or 5% bovine serum albumin (BSA) in PBS for 1 h. Membranes were washed three times for 5 min each with PBS, then incubated with primary antibody in blocking buffer (1× PBS, 0.05% Tween, 1% BSA) at 4°C overnight. Membranes were washed three times with PBS-T, and then incubated with secondary HRP antibody for 45 min at room temperature. Blots were imaged using enhanced chemiluminescence (Thermo Fisher), exposed to film, and developed using an X-OMAT processor. Densitometry was performed using ImageJ.
Immunofluorescence and live-cell microscopy
Cultured PDAC cells were prepared for immunofluorescence as described previously (Wang et al., 2011). Coverslips were incubated with primary antibodies in blocking buffer (5% goat serum, 5% glycerol, 0.04% sodium azide in D-PBS) overnight at 4°C, and incubated with secondary antibodies in blocking buffer for 1 h at room temperature. Alexa Fluor (Thermo Fisher) secondary antibodies were used at a concentration of 1:500. Coverslips were mounted onto glass slides using Prolong Gold (Thermo Fisher). Images were acquired using a Zeiss Axio Observer epifluorescence microscope and Zeiss LSM 780 confocal microscope (Carl Zeiss, Oberkochen, Germany) controlled by Zeiss Zen software (2012 SP1 black edition). Images processed uniformly using Adobe Photoshop software. Protein colocalization analysis was performed using the Coloc2 plug-in for ImageJ.
Live-cell imaging of GFP–α-actinin 4 and GFP-Dyn2 was performed on a Zeiss LSM 780 confocal microscope. Cells were plated into 35 mm glass-bottom imaging dishes (MatTek Corporation) before imaging. Cells were maintained in a 37°C, 5% CO2 environment over the course of imaging.
2D and transwell migration assays
For 2D migration assays, PANC-1 cells were plated after knockdown and reexpression in eight-well glass-bottom slides (Ibidi; #80827). Cells were imaged overnight on a Zeiss LSM 780 confocal microscope at 10× magnification, with one image acquired every 15 min. Cell velocity was calculated for individual cells using the Manual Tracking plug-in for ImageJ (Schneider et al., 2012). The track images represent the migration tracks for individual cells during the experiment.
For transwell migration assays, following knockdown and reexpression of the indicated proteins, PANC-1 cells were seeded in blind well chambers on polycarbonate track-etch (PCTE) filters with 12-µm pores that were coated with 0.1% gelatin (Neuroprobe BW200L; PFA12) and allowed to invade for 4 h (Dyn2 manipulations) or 16 h (α-actinin 1 or 4 manipulations). Cells were seeded in serum-free medium and migrated in a chemotactic gradient toward medium containing 10% FBS before fixation and Hoechst staining (Invitrogen; #H3570). The number of cells on the top and bottom of each filter was scored using fluorescence microscopy, and the percentage of cells that invaded across the filter was graphed for each condition.
Gelatin degradation assays
Fluorescent gelatin degradation assays were performed by coating coverslips with 0.2% gelatin and either Oregon Green–488 gelatin (Invitrogen; #G13186) or Cy3-gelatin (EMD Millipore; 8:1 ratio) as described previously (Artym et al., 2006). Following knockdown and reexpression of the indicated proteins, DanG cells were seeded onto the gelatin-coated coverslips for 7 h before fixation and staining with phalloidin-647 (Sigma). The area of matrix degradation was measured using ImageJ and normalized to the total cell area. The number of invadopodia per cell was measured by counting the number of actin puncta that colocalized with gelatin degradation spots.
Invadopodia lifetime analysis was performed using DanG cells stably expressing GFP, siRNA-resistant GFP–α-actinin 4 WT, or siRNA-resistant GFP–α-actinin 4 ΔC. Cells were treated with α-actinin 4 siRNA as described previously, and transfected with RFP-cortactin as an invadopodia marker. The cells were imaged using an Axio Observer.Z1/7 microscope (Carl Zeiss MicroImaging) with a 63× water lens (NA 1.2) with stage top incubation set at 37°C and 5% CO2. Images were acquired every 3 min for 121 frames (6 h elapsed time) with a Zeiss Axiocam 702 monochrome camera and illuminated by a Colibri 7 LED light source. The microscope was controlled by, and the images acquired with, Zen 2.3 Pro (blue edition) software (Carl Zeiss Microscopy GmbH, 2011). At least 10 cells for each condition were imaged over five independent experiments. All invadopodia present in the cell at frame 50 were chosen for analysis, and the ImageJ plug-in Invadopodia Tracker (Sharma et al., 2013) was used to calculate invadopodia lifetimes for each cell.
Immunohistochemistry
Tissue microarray (TMA) slides of paraffin embedded human PDAC tumors were provided by the Mayo Clinic SPORE in Pancreatic Cancer. All patients provided written informed consent, and the study was approved by the Mayo Clinic IRB. The TMA was constructed in 2012, and tissue was collected from patients at the time of surgical resection. All patients in this TMA had at least one treatment of gemcitabine following surgery. Immunohistochemistry was performed using the Cell and Tissue Staining Kit (R&D Systems) according to the manufacturer’s protocols. For each antibody, 1 mM EDTA (pH 8.1) was used as the antigen retrieval buffer. Tissue samples were counterstained with hematoxylin (Richard-Allan Scientific; #7221) as a counterstain. Verification of PDAC and scoring of immunohistochemical staining was performed by a pathologist. Kaplan Meier survival curves were generated based on the expression of α-actinin 1, α-actinin 4, or Dyn2 in 205 PDAC patient tissue cores represented in the TMA slides. Patients were grouped into high or low expression based on the median H-score values, which are based on the intensity of staining (0–3 scale) multiplied by the extent of the tissue that is stained (0–100% scale). The H-scores from multiple cores per patient on the TMAs were averaged for each subject. p values were based on median survival values and were calculated using a log rank statistical test. Adjusted p values were calculated using a likelihood ratio statistical test and take into account patient demographics (age at diagnosis, sex, obesity [BMI ≥ 30], and patient-reported diabetes). To address hypotheses focusing on the prognostic value of the protein expression, subject level data was used with Cox Proportional Hazards Regression models to fit the relationship with overall survival. Likelihood ratio tests (LRTs) were used to assess statistical significance for any differences observed between groups, with critical values determined based upon a chi-square distribution (0.95) with 1 degree of freedom.
Statistical analysis
Statistical analysis of data was performed using Microsoft Excel. Student’s t test was used to identify statistical significance for all experiments, unless noted otherwise, and graphed data represent the mean ± SEM. * indicates a p value below 0.05, and ** indicates a p value below 0.01. In very rare cases, statistical outliers were identified and excluded using the interquartile range rule.
Supplementary Material
Acknowledgments
This research was supported by R01 CA104125 from the National Cancer Institute (M.A.M., G.L.R.), the Mayo Clinic SPORE in Pancreatic Cancer (P50 CA102701), the Mayo Clinic Center for Cell Signaling in Gastroenterology (P30 DK084567), and the Mayo Clinic Graduate School in Biomedical Sciences (K.M.B.). Special thanks to Micah Schott and Cody Rozeveld for technical assistance.
Abbreviations used:
- Dyn2
Dynamin 2
- ECM
extracellular matrix
- MMPs
matrix metalloproteinases
- PDAC
pancreatic ductal adenocarcinoma
- PRD
proline-rich domain.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-07-0395) on January 22, 2020.
REFERENCES
- Aksenova V, Turoverova L, Khotin M, Magnusson KE, Tulchinsky E, Melino G, Pinaev GP, Barlev N, Tentler D. (2013). Actin-binding protein alpha-actinin 4 (ACTN4) is a transcriptional co-activator of RelA/p65 sub-unit of NF-kB. Oncotarget , 362–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artym VV, Zhang Y, Seillier-Moiseiwitsch F, Yamada KM, Mueller SC. (2006). Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res , 3034–3043. [DOI] [PubMed] [Google Scholar]
- Baldassarre M, Pompeo A, Beznoussenko G, Castaldi C, Cortellino S, McNiven MA, Luini A, Buccione R. (2003). Dynamin participates in focal extracellular matrix degradation by invasive cells. Mol Biol Cell , 1074–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce NS, Clark ES, Leysath JL, Currie JD, Webb DJ, Weaver AM. (2005). Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr Biol , 1276–1285. [DOI] [PubMed] [Google Scholar]
- Cao H, Garcia F, McNiven MA. (1998). Differential distribution of dynamin isoforms in mammalian cells. Mol Biol Cell , 2595–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Orth JD, Chen J, Weller SG, Heuser JE, McNiven MA. (2003). Cortactin is a component of clathrin-coated pits and participates in receptor-mediated endocytosis. Mol Cell Biol , 2162–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Thompson HM, Krueger EW, McNiven MA. (2000). Disruption of Golgi structure and function in mammalian cells expressing a mutant dynamin. J Cell Sci (Pt 11), 1993–2002. [DOI] [PubMed] [Google Scholar]
- Chuang MC, Lin SS, Ohniwa RL, Lee GH, Su YA, Chang YC, Tang MJ, Liu YW. (2019). Tks5 and Dynamin-2 enhance actin bundle rigidity in invadosomes to promote myoblast fusion. J Cell Biol , 1670–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destaing O, Ferguson SM, Grichine A, Oddou C, De Camilli P, Albiges-Rizo C, Baron R. (2013). Essential function of dynamin in the invasive properties and actin architecture of v-Src induced podosomes/invadosomes. PLoS One , e77956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund M, Lotano MA, Otey CA. (2001). Dynamics of alpha-actinin in focal adhesions and stress fibers visualized with alpha-actinin-green fluorescent protein. Cell Motil Cytoskeleton , 190–200. [DOI] [PubMed] [Google Scholar]
- Eppinga RD, Krueger EW, Weller SG, Zhang L, Cao H, McNiven MA. (2012). Increased expression of the large GTPase dynamin 2 potentiates metastatic migration and invasion of pancreatic ductal carcinoma. Oncogene , 1228–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, De Camilli P. (2012). Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol , 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley KS, Young PW. (2013). An analysis of splicing, actin-binding properties, heterodimerization and molecular interactions of the non-muscle alpha-actinins. Biochem J , 477–488. [DOI] [PubMed] [Google Scholar]
- Foley KS, Young PW. (2014). The non-muscle functions of actinins: an update. Biochem J , 1–13. [DOI] [PubMed] [Google Scholar]
- Fukumoto M, Kurisu S, Yamada T, Takenawa T. (2015). α-Actinin-4 enhances colorectal cancer cell invasion by suppressing focal adhesion maturation. PLoS One , e0120616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Yaddanapudi S, Weins A, Osborn T, Reiser J, Pollak M, Hartwig J, Sever S. (2010). Direct dynamin-actin interactions regulate the actin cytoskeleton. EMBO J , 3593–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Honda K, Shitashige M, Ono M, Matsuyama H, Naito K, Hirohashi S, Yamada T. (2007). Mass spectrometry analysis of the native protein complex containing actinin-4 in prostate cancer cells. Mol Cell Proteomics , 479–491. [DOI] [PubMed] [Google Scholar]
- Henley JR, Krueger EW, Oswald BJ, McNiven MA.(1998). Dynamin-mediated internalization of caveolae. J Cell Biol , 85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE. (2000). Dynamin and its role in membrane fission. Annu Rev Cell Dev Biol , 483–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K. (2015). The biological role of actinin-4 (ACTN4) in malignant phenotypes of cancer. Cell Biosci , 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S, Chakraborty S, Cheng X, Su YT, Kao HY. (2011). The actin-binding protein, actinin alpha 4 (ACTN4), is a nuclear receptor coactivator that promotes proliferation of MCF-7 breast cancer cells. J Biol Chem , 1850–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Honda K, Tsuda H, Hiraoka N, Imoto I, Kosuge T, Umaki T, Onozato K, Shitashige M, Yamaguchi U, et al. (2008). Expression and gene amplification of actinin-4 in invasive ductal carcinoma of the pancreas. Clin Cancer Res , 5348–5356. [DOI] [PubMed] [Google Scholar]
- Knudsen KA, Soler AP, Johnson KR, Wheelock MJ. (1995). Interaction of alpha-actinin with the cadherin/catenin cell-cell adhesion complex via alpha-catenin. J Cell Biol , 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruchten AE, McNiven MA. (2006). Dynamin as a mover and pincher during cell migration and invasion. J Cell Sci , 1683–1690. [DOI] [PubMed] [Google Scholar]
- Krueger EW, Orth JD, Cao H, McNiven MA. (2003). A dynamin-cortactin-Arp2/3 complex mediates actin reorganization in growth factor-stimulated cells. Mol Biol Cell , 1085–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurklinsky S, Chen J, McNiven MA. (2011). Growth cone morphology and spreading are regulated by a dynamin-cortactin complex at point contacts in hippocampal neurons. J Neurochem , 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AW, Pattabiraman DR, Weinberg RA. (2017). Emerging biological principles of metastasis. Cell , 670–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock P, Abram CL, Gibson T, Courtneidge SA. (1998). A new method for isolating tyrosine kinase substrates used to identify fish, an SH3 and PX domain-containing protein, and Src substrate. EMBO J , 4346–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNiven MA, Kim L, Krueger EW, Orth JD, Cao H, Wong TW. (2000). Regulated interactions between dynamin and the actin-binding protein cortactin modulate cell shape. J Cell Biol , 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon M, Askinazi OL, Schafer DA. (2014). Dynamin2 organizes lamellipodial actin networks to orchestrate lamellar actomyosin. PLoS One , e94330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooren OL, Kotova TI, Moore AJ, Schafer DA. (2009). Dynamin2 GTPase and cortactin remodel actin filaments. J Biol Chem , 23995–24005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy ACH, Young PW. (2015). The actinin family of actin cross-linking proteins—a genetic perspective. Cell Biosci , 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, McNiven MA. (2003). Dynamin at the actin-membrane interface. Curr Opin Cell Biol , 31–39. [DOI] [PubMed] [Google Scholar]
- Prebil SD, Slapsak U, Pavsic M, Ilc G, Puz V, Ribeiro EDA, Anrather D, Hartl M, Backman L, Plavec J, et al. (2016). Structure and calcium-binding studies of calmodulin-like domain of human non-muscle α-actinin-1. Sci Rep-Uk , 27383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razidlo GL, Wang Y, Chen J, Krueger EW, Billadeau DD, McNiven MA. (2013). Dynamin 2 potentiates invasive migration of pancreatic tumor cells through stabilization of the Rac1 GEF Vav1. Dev Cell , 573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods , 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Li S, Watkins SC, Wells A. (2014). α-Actinin-4 is required for amoeboid-type invasiveness of melanoma cells. J Biol Chem , 32717–32728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VP, Entenberg D, Condeelis J. (2013). High-resolution live-cell imaging and time-lapse microscopy of invadopodium dynamics and tracking analysis. Methods Mol Biol , 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom B, Salmazo A, Djinovic-Carugo K. (2008). Alpha-actinin structure and regulation. Cell Mol Life Sci , 2688–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeg PS. (2016). Targeting metastasis. Nat Rev Cancer , 201–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastyan S, Weinberg RA. (2011). Tumor metastasis: molecular insights and evolving paradigms. Cell , 275–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cao H, Chen J, McNiven MA. (2011). A direct interaction between the large GTPase dynamin-2 and FAK regulates focal adhesion dynamics in response to active Src. Mol Biol Cell , 1529–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Ueno H, Watabe Y, Hiraoka N, Morizane C, Itami J, Okusaka T, Miura N, Kakizaki T, Kakuya T, et al. (2015). ACTN4 copy number increase as a predictive biomarker for chemoradiotherapy of locally advanced pancreatic cancer. Br J Cancer , 704–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch T, Keleg S, Bergmann F, Bauer S, Hinz U, Schmidt J. (2009). Actinin-4 expression in primary and metastasized pancreatic ductal adenocarcinoma. Pancreas , 968–976. [DOI] [PubMed] [Google Scholar]
- Wong BS, Shea DJ, Mistriotis P, Tuntithavornwat S, Law RA, Bieber JM, Zheng L, Konstantopoulos K. (2019). A direct podocalyxin–dynamin-2 interaction regulates cytoskeletal dynamics to promote migration and metastasis in pancreatic cancer cells. Cancer Res , 2878–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Teng LH, Silva SD, Bijian K, Al Bashir S, Jie S, Dolph M, Alaoui-Jamali MA, Bismar TA. (2014). The significance of dynamin 2 expression for prostate cancer progression, prognostication, and therapeutic targeting. Cancer Med , 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Takeda T, Michiue H, Abe T, Takei K. (2016). Actin bundling by dynamin 2 and cortactin is implicated in cell migration by stabilizing filopodia in human non-small cell lung carcinoma cells. Int J Oncol , 877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Ito Y, Miura N, Nagamura Y, Nakabo A, Fukami K, Honda K, Sakai R. (2017). Actinin-1 and actinin-4 play essential but distinct roles in invadopodia formation by carcinoma cells. Eur J Cell Biol , 685–694. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Nolan M, Yamada H, Watanabe M, Nasu Y, Takei K, Takeda T. (2016). Dynamin2 GTPase contributes to invadopodia formation in invasive bladder cancer cells. Biochem Biophys Res Commun , 409–414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.