Abstract
Proteoglycans, a class of carbohydrate-modified proteins, often modulate growth factor signaling on the cell surface. However, the molecular mechanism by which proteoglycans regulate signal transduction is largely unknown. In this study, using a recently developed glycoproteomic method, we found that Windpipe (Wdp) is a novel chondroitin sulfate proteoglycan (CSPG) in Drosophila. Wdp is a single-pass transmembrane protein with leucin-rich repeat (LRR) motifs and bears three CS sugar chain attachment sites in the extracellular domain. Here we show that Wdp modulates the Hedgehog (Hh) pathway. In the wing disc, overexpression of wdp inhibits Hh signaling, which is dependent on its CS chains and the LRR motifs. The wdp null mutant flies show a specific defect (supernumerary scutellar bristles) known to be caused by Hh overexpression. RNA interference knockdown and mutant clone analyses showed that loss of wdp leads to the up-regulation of Hh signaling. Altogether, our study demonstrates a novel role of CSPGs in regulating Hh signaling.
INTRODUCTION
Spatial and temporal regulation of growth factor signaling pathways is essential to proper development and disease prevention. Cell surface signaling events, such as ligand–receptor interactions, are often modulated by proteoglycans (Xu and Esko, 2014). Proteoglycans are carbohydrate-modified proteins that are found on the cell surface and in the extracellular matrix. They are composed of a core protein and one or more glycosaminoglycans (GAGs) covalently attached to specific serine residues on the core protein. GAGs are long, unbranched, and highly sulfated polysaccharide chains consisting of a repeating disaccharide unit. Based on the composition of the disaccharide units, proteoglycans are classified into several types, including heparan sulfate proteoglycans (HSPGs) and chondroitin sulfate proteoglycans (CSPGs).
HSPGs function as coreceptors by interacting with a wide variety of ligands to modulate signaling activities (Lindahl and Li, 2009; Xu and Esko, 2014). Drosophila offers a powerful model system to study the functions of HSPGs in vivo because of its sophisticated molecular genetic tools and minimal genetic redundancy in genes encoding core proteins and HS synthesizing/modifying enzymes (Takemura and Nakato, 2015; Nakato and Li, 2016). In vivo studies using the Drosophila model have shown that HSPGs orchestrate information from multiple ligands in a complex extracellular milieu and sculpt the signal response landscape in a tissue (Nakato and Li, 2016). However, the molecular mechanisms of coreceptor activities of HSPGs still remain a fundamental question. Our previous studies predict that there are unidentified molecules involved in the molecular recognition events on the cell surface (Akiyama et al., 2008).
In addition to HS, Drosophila produces CS, another type of GAG (Toyoda et al., 2000). CSPGs are well known as major structural components of the extracellular matrix. CSPGs have also been shown to modulate signaling pathways, including Hedgehog (Hh), Wnt, and fibroblast growth factor signaling (Cortes et al., 2009; Townley and Bülow, 2018). Given the structural similarities between CS and HS, CSPGs may have modulatory, supportive, and/or complementary functions to HSPGs. However, the mechanisms by which CSPGs function as a coreceptor are unknown. In contrast to a large number of studies on HSPGs, very few CSPGs have been identified and analyzed in Drosophila (Momota et al., 2011). Unlike HSPGs, CSPG core proteins are not well conserved between species (Olson et al., 2006). Therefore, the identification of CSPGs cannot rely on the sequence homology to mammalian counterparts.
Recently, we have developed a glycoproteomic method to identify novel proteoglycans (Noborn et al., 2015, 2016, 2018). Briefly, this method includes trypsinization of protein samples, followed by enrichment of glycopeptides using strong anion exchange chromatography. After enzymatic digestion of HS/CS chains, the glycopeptides bearing a linkage glycan structure common to HS and CS chains are identified using nano-liquid chromatography-tandem mass spectrometry (nLC-MS/MS). This method has successfully identified novel CSPGs in humans (Noborn et al., 2015) and Caenorhabditis elegans (Noborn et al., 2018).
To study the function of CSPGs in signaling, we applied the glycoproteomic method to identify previously unrecognized CSPGs in Drosophila. We found that Windpipe (Wdp) is a novel CSPG and affects Hh signaling. Overexpression of wdp inhibits Hh signaling in the wing disc. This inhibitory effect of Wdp on Hh signaling is dependent on its CS chains and leucin-rich repeat (LRR) motifs. Consistent with the overexpression analysis, loss of wdp increases Hh signaling: wdp null mutant flies show a specific defect (supernumerary scutellar [SC] bristles) known to be caused by Hh overexpression. Our study highlights a novel function of CSPGs in cell signaling.
RESULTS
A glycoproteomic approach identified Wdp as a novel Drosophila CSPG
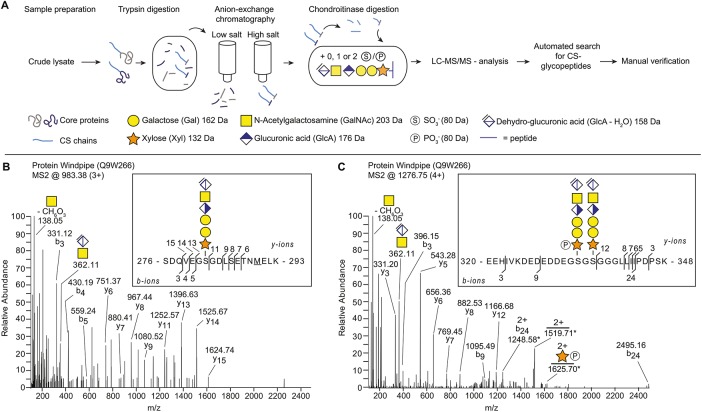
We investigated the potential presence of CSPGs in Drosophila using our recently developed glycoproteomic approach that identifies core proteins and their CS attachment sites. A general workflow for the sample preparation, CS-glycopeptide enrichment, LC-MS/MS analysis, and the subsequent data analysis is shown in Figure 1A. Briefly, Drosophila third-instar larvae were collected from two different genotypes (wild type [WT] [Oregon-R] and a loss-of-function mutant for tout-velu [ttv524]) and the material was homogenized in ice-cold acetone. The ttv encodes a Drosophila HS polymerase, and ttv mutants lack HS chains (The et al., 1999; Toyoda et al., 2000). The samples were incubated with trypsin and then passed over an anion exchange column equilibrated with a low-salt buffer. This procedure enriches for CS-attached glycopeptides as the matrix retains anionic polysaccharides and their attached peptides, whereas neutral or positively charged peptides flow through the column. The bound structures were eluted stepwise with three buffers of increasing sodium chloride concentrations. The resulting fractions were treated with chondroitinase ABC. This procedure reduces the lengths of the CS chains and generates a residual hexasaccharide structure still attached to the core protein. The chondroitinase-treated fractions were analyzed with positive mode nLC-MS/MS and an automated search strategy was used to identify CS-modified peptides in the generated data set (Noborn et al., 2015).
FIGURE 1:
Identification of Wdp as a novel CSPG in Drosophila. (A) Scheme for identifying CSPGs in Drosophila. The workflow includes the enrichment of proteoglycans from fly extract, enzymatic hydrolysis and subsequent analysis, and interpretation of mass spectra. (B, C) MS2 fragment mass spectra of Wdp protein (UniProt: Q9W266) showing two unique CS linkage region-substituted glycopeptides. (B) Peptide (SDQVEGSGDLSETNMELK) identified with one hexasaccharide structure and one methionine oxidation (m/z 983.38; 3+). (C) Peptide (EEHIVKDEDEDDEGSGSGGGLLIIPDPSK) identified with two hexasaccharide structures where one of the xyloses was modified with one phosphate (m/z 1276.75; 4+). This spectrum contains doubly charged fragments at m/z 1248.58; 2+, m/z 1519.71; 2+ and 1625.70; 2+, for which the second isotopic peak represents the largest peak of the isotopic distribution. For these doubly charged fragments, the masses of the second isotopic peaks are annotated and are denoted with an asterisk. All other annotated ions in B and C are from monoisotopic masses.
The analysis revealed the Wdp protein as a novel CSPG, which was modified with three CS-polysaccharides on two unique peptides (Figure 1, B and C). We detected Wdp glycopeptides from both WT and ttv mutant samples, further supporting that Wdp bears CS chains, not HS. One of the identified precursor ions (m/z 983.38; 3+) equated to the mass of a peptide with a SDQVEGSGDLSETNMELK sequence, derived from the middle part of the protein (amino acids 276–293) (Figure 1B). The peptide was modified with one hexasaccharide structure and one methionine oxidation. The measured mass (2947.1186 Da) deviated –3.27 ppm from the theoretical value. The other identified precursor ion (m/z 1276.76; 4+) equated to the mass of a peptide with a EEHIVKDEDEDDEGSGSGGGLLIIPDPSK sequence, located in proximity to the previous peptide (amino acids 320–348) (Figure 1C). The peptide was found to be modified with two hexasaccharide structures and where one of the hexasaccharides was modified with one phosphate modification. The measured mass (5102.9389 Da) deviated +3.05 ppm from the theoretical value. Detailed inspection of the spectra revealed several b- and y-ions as well as the prominent diagnostic oxonium ion at m/z 362.1, corresponding to the disaccharide [GlcAGalNAc-H2O+H]+ (Figure 1, B and C). Furthermore, one of the glycans in Figure 1C was found modified with one phosphate group at a xylose residue (peptide + xylose + phosphate, m/z 1625.70; 2+).
Wdp is a single-pass transmembrane protein containing four LRR motifs in the extracellular domain (Huff et al., 2002). The three CS attachment sites (S282, S334, and S336) revealed by our glycoproteomic analysis are located in the extracellular domain. Interestingly, a recent study reported that Wdp negatively regulates JAK–STAT signaling by promoting internalization and lysosomal degradation of the Domeless (Dome) receptor (Ren et al., 2015). We further investigated the role of Wdp, a novel CSPG, in signal transduction.
Overexpression of wdp inhibits Hh signaling
The growth and patterning of the Drosophila wing are controlled by multiple signaling pathways, including Decapentaplegic (Dpp; a Drosophila BMP), Wingless (Wg; a Drosophila Wnt), and Hh signaling. To determine the role of wdp in these developmental signaling pathways, we first asked whether overexpression of wdp affects adult wing morphology. wdp was overexpressed using BxMS1096-GAL4, which drives high levels of UAS transgene expression broadly in the wing pouch (Capdevila and Guerrero, 1994; Mace and Tugores, 2004; Tripura et al., 2011). We found that wdp overexpression (BxMS1096>wdp) results in reduced wing size compared with control flies (BxMS1096>) (Figure 2, A–D). In addition, the distance between longitudinal wing veins 3 and 4 (L3 and L4) was aberrantly narrower (Figure 2, A–C and E). This decreased distance between L3 and L4 is indicative of reduced Hh signaling during wing development (Mullor et al., 1997b; Strigini and Cohen, 1997).
FIGURE 2:
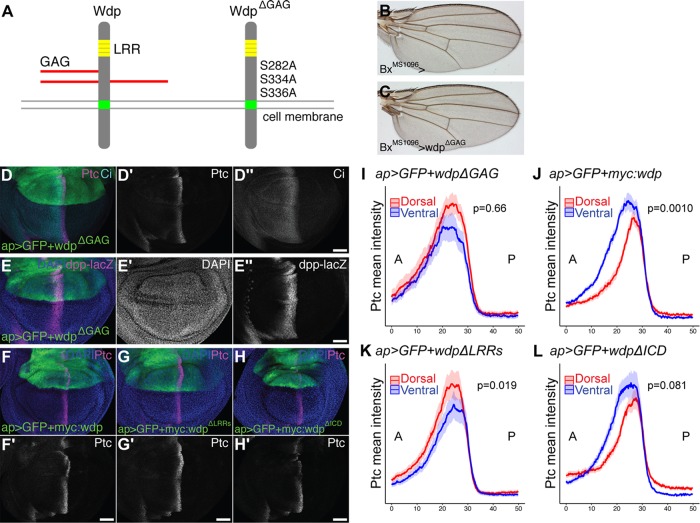
Overexpression of wdp reduces the Hh-signaling-active domain. (A–C) A control adult wing (A) and adult wings expressing UAS-wdp with BxMS1096-GAL4 (B, C). Longitudinal wing veins 3 and 4 are marked (L3 and L4, respectively). (D, E) Quantification of the whole adult wing area (D) and the ratio of the area of the L3–L4 domain relative to the whole wing (E) for BxMS1096-GAL4/+ (control), BxMS1096-GAL4>wdp (wdp), and BxMS1096>wdpΔGAG (wdpΔGAG); n.s., not significant; ***p < 0.001. The Wilcoxon rank sum test was used to test the statistical significance (n = 12 in control; n = 14 in wdp; n = 18 in wdpΔGAG). (F–K) Control wing discs (ap>GFP) (F, H, and J) and wing discs overexpressing wdp with ap-GAL4 (ap>GFP+wdp) (G, I, and K) were immunostained for the expression of Ptc, Ci (F and G), dpp-lacZ (H and I), En, and hh-lacZ (J and K). Nuclei were stained with DAPI. The expression domains of Ptc, Ci, and dpp-lacZ were reduced by wdp overexpression in the dorsal compartment compared with those in the ventral compartment. En expression induced by high-level Hh signaling in the anterior compartment was also diminished by wdp overexpression. Note that the hh-lacZ expression was not affected by wdp overexpression. Anterior to the left; dorsal to the top. Scale bars: 50 µm. (L, M) Signal intensity plots of the Ptc expression in the dorsal (red) and ventral (blue) compartments in ap>GFP (L) and ap>GFP+wdp (M). Solid lines indicate the average intensity of Ptc staining, and shaded areas show the standard error of the mean (n = 21 in L; n = 17 in M). The difference between the distance (x value) of the Ptc mean intensity maximum and the x value of two-thirds of the maximum on the anterior (A) side was measured for each peak. These values were used to compare the shape of the Ptc expression patterns between the dorsal and ventral compartments. P values were calculated using the Wilcoxon rank sum test.
Hh is produced in the posterior compartment of the wing disc and spreads toward the anterior compartment where it induces target gene expression in a concentration-dependent manner (Briscoe and Thérond, 2013; Gradilla and Guerrero, 2013; Hartl and Scott, 2014). Expression of high-threshold target genes, such as Patched (Ptc; the Hh receptor) (Capdevila et al., 1994) and Engrailed (En) (Patel et al., 1989), are induced in anterior cells near the anteroposterior compartment boundary by high levels of Hh signaling (Figure 2, F and J; Jia et al., 2004). Lower levels of Hh signaling induce the expression of dpp and the accumulation of full-length cubitus interruptus (Ci; the transcriptional factor of Hh signaling) in a broader region (more distant away from the anteroposterior boundary) (Figure 2, F and H). To determine whether Hh signaling is indeed affected by wdp, we overexpressed wdp specifically in the dorsal compartment of the wing disc using apterous (ap)-GAL4. We found that wdp overexpression in the dorsal compartment reduced the expression domains of both “high-threshold” targets (Ptc and En) and “low-threshold” targets (dpp-lacZ10638, a reporter for dpp expression, and full-length Ci) compared with those in the ventral compartment (Figure 2, G and I). To quantify the effect of wdp overexpression on Hh signaling, we generated signal intensity plots of the Ptc expression in the dorsal (red) and ventral (blue) compartments. In control wing discs (ap>GFP), the Ptc signal is slightly higher in the dorsal compartment (red) compared with the ventral compartment (blue), but the shape of the Ptc peak is similar in both compartments (Figure 2L). On the other hand, in discs overexpressing wdp in the dorsal compartment (ap>GFP+wdp), the width of Ptc-positive cells became significantly narrower specifically in the dorsal compartment (Figure 2M). Notably, overexpression of wdp did not affect the pattern of a hh transcriptional reporter hh-lacZP30 (Figure 2K). Together, these results suggest that wdp acts as a negative modulator of Hh signaling without affecting hh transcription.
We next asked whether Wdp affects other developmental pathways: Dpp and Wg signaling. When wdp is overexpressed using ap-GAL4 or hh-GAL4 (a posterior compartment-specific GAL4 driver), we did not observe apparent defects in Dpp signaling activity, which was monitored by the expression of phosphorylated Mad (pMad) and Spalt major (Salm) (readouts of Dpp signaling) (Supplemental Figure S1). Similarly, no changes in expression of Senseless (Sens) and Distal-less (Dll) (readouts of Wg signaling) were detected (Supplemental Figure S1). Thus, we did not detect any effect of Wdp on the Dpp and Wg downstream responses. This is consistent with a previous report (Ren et al., 2015).
We also noticed that overexpression of wdp induces massive apoptosis, as detected with anti-cleaved Caspase-3 antibody (Supplemental Figure S2B). This likely contributed to the smaller adult wing phenotype observed in BxMS1096>wdp flies. It was recently reported that Hh signaling is required for cell survival in wing disc cells (Lu et al., 2017). To determine whether reduced Hh signaling is responsible for the observed apoptosis, we first asked if reduced Hh signaling results in apoptosis. We inhibited Hh signaling either by expressing an RNA interference (RNAi) construct (TRiP.HMC03577) targeting smoothened (smo) (Supplemental Figure S2E) or by overexpressing ptc in the dorsal compartment using ap-GAL4. We found that neither treatment increased signals for cleaved Caspase-3 (Supplemental Figure S2, F and G), indicating that reduced Hh signaling is not sufficient to induce massive apoptosis in the wing disc. Furthermore, coexpression of a constitutively active form of Smo with Wdp did not suppress apoptosis in the wing disc (Supplemental Figure S2H). These results suggest that overexpression of wdp induces apoptosis, independent of reduced Hh signaling.
CS chains and LRR motifs are necessary for Wdp to inhibit Hh signaling
Next, we asked whether the CS chains of Wdp are required for its function. In a CSPG core-protein, CS is attached to specific serine residues in the consensus serine-glycine dipeptide surrounded by acidic amino acids (Esko and Zhang, 1996). We generated a UAS-wdpΔGAG construct in which all three GAG-attachment serine residues (S282, S334, and S336) were substituted with alanine residues so that CS cannot be attached to the core protein (Figure 3A). The UAS-wdp and UAS-wdpΔGAG constructs were inserted in the same genomic location (ZH-86Fb) using the phiC31 site-specific integration system in order to ensure the same expression level of the UAS transgenes (Groth et al., 2004; Bischof et al., 2007).
FIGURE 3:
Wdp negatively regulates Hh signaling in a GAG-dependent manner. (A) A schematic drawing of WT Wdp and a mutant form of Wdp (WdpΔGAG). (B, C) A control adult wing (B) and a wing expressing UAS-wdpΔGAG with BxMS1096-GAL4 (C). (D, E) Wing discs expressing UAS-wdpΔGAG with ap-GAL4 were immunostained for Ptc, Ci (D), and dpp-lacZ (E). (F–H) Wing discs expressing UAS-3xMyc:wdp (F), UAS-3xMyc:wdpΔLRRs (G), and UAS-3xMyc:wdpΔICD (H) with ap-Gal4 were immunostained for Ptc. Nuclei were stained with DAPI. Scale bars: 50 µm. (I–L) Signal intensity plots of the Ptc expression in the dorsal (red) and ventral (blue) compartments in wing discs overexpressing UAS-wdpΔGAG (I, n = 12), UAS-myc:wdp (J, n = 18), UAS-myc:wdpΔLRRs (K, n = 25), or UAS-myc:wdpΔICD (L, n = 23) with ap-Gal4. Solid lines indicate the average intensity of Ptc staining and shaded areas show the standard error of the mean. P-values were calculated using the Wilcoxon rank sum test via the method described in Figure 2.
Overexpression of wdpΔGAG by BxMS1096-GAL4 did not decrease the whole wing size (Figures 3, B and C, and 2D). No reduction in the distance between L3 and L4 was observed in BxMS1096>wdpΔGAG adult wings either (Figures 3, B and C, and 2E). Consistent with this, the expression of Ptc, En, Ci, and dpp-lacZ in the wing disc was not affected by wdpΔGAG overexpression in the dorsal compartment of the wing disc (Figure 3, D, E, and I). These results indicate that CS chains are required for Wdp’s activity to down-regulate Hh signaling.
To determine whether the LRR motifs and/or the intracellular domain of Wdp are necessary for inhibiting Hh signaling, we generated several Myc-tagged mutant constructs (Supplemental Figure S3) and examined their activities. Consistent with the earlier result (Figure 2, G and M), expression of a Myc-tagged WT Wdp (Myc:Wdp) led to a narrower Ptc expression domain (Figure 3, F and J). We found that a mutant wdp construct lacking LRR motifs (Myc:WdpΔLRRs) failed to inhibit Hh signaling (Figure 3, G and K). Thus, in addition to CS chains, the LRR motifs of Wdp are required for inhibiting Hh signaling. On the other hand, a truncated construct lacking the intracellular domain (Myc:WdpΔICD) retained the ability to inhibit Hh signaling (Figure 3, H and L). This result suggests that the Wdp intracellular domain may be dispensable for its function as a Hh-signaling regulator.
Wdp expression in the wing disc
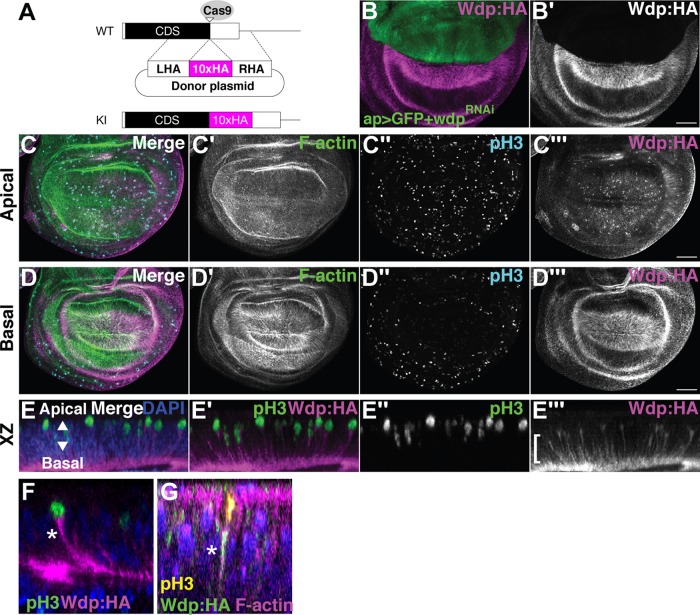
To monitor Wdp expression, we generated transgenic flies (wdpKI.HA and wdpKI.OLLAS) expressing epitope-tagged Wdp protein from its endogenous locus. We inserted a spaghetti monster GFP with 10 copies of HA or OLLAS tags near the C-terminus of Wdp (after Q652; Figure 4A) using CRISPR–Cas9-mediated homology-directed repair (Gratz et al., 2014; Ren et al., 2014). Expression of a wdpRNAi construct (TRiP.HMC06302) using ap-GAL4 in wdpKI.HA/+ flies led to the loss of Wdp:HA staining specifically in the dorsal compartment (Figure 4B). This result validated the specificity of HA staining reflecting Wdp localization as well as the efficacy of RNAi-mediated knockdown of wdp. Expression of Wdp:HA and Wdp:OLLAS was detected in the eye disc, adult midgut, and tracheal system (Supplemental Figure S4), consistent with previous reports (Huff et al., 2002; Ren et al., 2015).
FIGURE 4:
Wdp expression in the wing disc. (A) Schematic of CRISPR–Cas9-mediated gene editing of wdp for generating wdpKI.HA. Ten copies of an HA epitope tag (smGFP-HA) were inserted in frame near the stop codon of the wdp coding sequence (CDS). Only the last exon is shown. CDS, the black box; smGFP-HA, the magenta box; LHA, left homology arm; RHA, right homology arm; Cas9 target site, the open triangle. (B) Anti-HA staining of a wing disc from ap-GAL4 UAS-GFP wdpKI.HA/UAS-wdpRNAi. Wdp:HA is not detectable in the dorsal compartment of a wing disc expressing wdpRNAi (TRiP.HMC06302) with ap-GAL4. (C, D) A wing disc homozygous for wdpKI.HA was stained with Alexa Fluor 568–conjugated phalloidin (F-actin), anti–phospho histone H3 antibody (pH3, mitotic nuclei), and anti-HA antibody. Apical (C) and basal (D) sections of the same disc are shown. Intense staining of Wdp:HA was observed in mitotic cells (C’’ and C’’’) and on the basal side of wing disc epithelium (D’’’). (E–G) Optical cross-sections of 3D-reconstructed images show the accumulation of Wdp:HA in the basal projection (bracket) of apically translocating mitotic cells (E–E’’’). Wdp:HA staining was particularly strong in the basolateral membrane of mitotic cells (asterisk in F) and is colocalized with F-actin (asterisk in G). Nuclei were stained with DAPI (E–G). Scale bars: 50 µm.
In the wing disc, Wdp:HA is expressed in most of the wing cells with enrichment in the basal side, as detected by anti-HA antibody (Figure 4, C and D). This result was confirmed by anti-OLLAS antibody staining of the wdpKI.OLLAS wing discs (Supplemental Figure S4, B and C). During mitosis, the nuclei of wing disc cells translocate to the apical surface to execute cell division, a phenomenon known as interkinetic nuclear migration (Ragkousi and Gibson, 2014). While both the nucleus and the bulk cytoplasm move toward the apical surface, these cells maintain connectivity with the basal side of the epithelium via long, thin, and F–actin-rich basal processes throughout mitosis (Meyer et al., 2011). Interestingly, Wdp:HA staining was particularly strong in the basolateral membrane of mitotic cells (Figure 4, E and F). The Wdp:HA signal is colocalized with F-actin, detected by Phalloidin staining (Figure 4G). Thus, Wdp is enriched in these basal processes during mitosis. At this point, however, biological significance of this localization of Wdp in the basal extensions of mitotic cells is unknown.
Loss of wdp leads to higher levels of Hh signaling
To determine whether loss of wdp affects Hh signaling activity, we examined the effect of wdp RNAi knockdown in the wing disc. The effect of wdp knockdown on Hh signaling was assessed using the Ptc expression level as a readout of the Hh signaling activity. The wdpRNAi expression using ap-GAL4 significantly increased the signal intensity of Ptc staining only in the dorsal compartment (Figure 5A). As stated earlier, the patterns of the Ptc signal in the dorsal and ventral compartments are similar in control wing discs (ap>GFP; Figure 2L). As an additional control, overexpression of flp (ap>flp) did not change the Ptc levels (Figure 5B). On the other hand, Ptc signal intensity plots showed that in discs expressing wdpRNAi with ap-Gal4 (ap>GFP+wdpRNAi), the Ptc peak in the dorsal compartment is significantly higher and broader than that in the ventral compartment (Figure 5C). In addition, we observed that the dpp-lacZ expression domain was expanded anteriorly by wdp knockdown (Figure 5, D and E). The effect of wdp RNAi knockdown on Ci protein levels was less obvious compared with its effect on Ptc and dpp-lacZ (Figure 5A"). It is possible that a high threshold target gene is more sensitive to the change in the level of Wdp than a low threshold target. Thus, wdp RNAi knockdown results in a moderate increase in Hh signaling.
FIGURE 5:
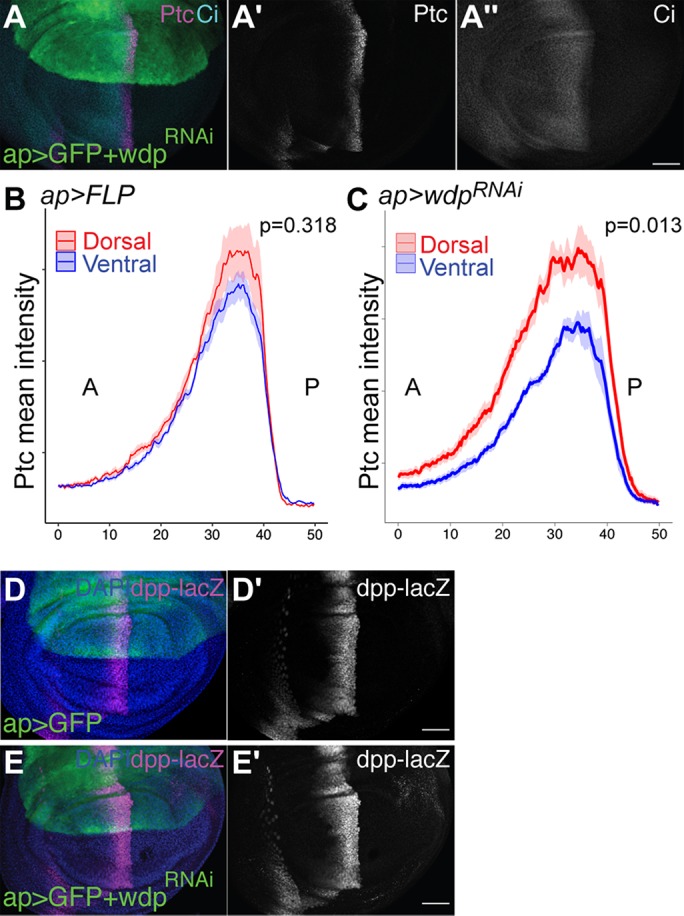
wdp RNAi knockdown leads to increased Hh signaling. (A) A wing disc expressing UAS-wdpRNAi (TRiP.HMC06302) with ap-GAL4 was immunostained for Ptc and Ci. (B, C) Signal intensity plots of the Ptc expression in the dorsal compartment (red) and ventral compartment (blue) in wing discs expressing UAS-Flp (B) and UAS-wdpRNAi (C). Solid lines indicate the average intensity of Ptc staining and shaded areas show the standard error of the mean (n = 10). P values were calculated using the Wilcoxon rank sum test via the method described in Figure 2. (D, E) A control wing disc (D) and a wing disc expressing UAS-wdpRNAi (TRiP.HMC06302) with ap-GAL4 (E) were immunostained for dpp-lacZ. Nuclei were stained with DAPI. Scale bars: 50 µm.
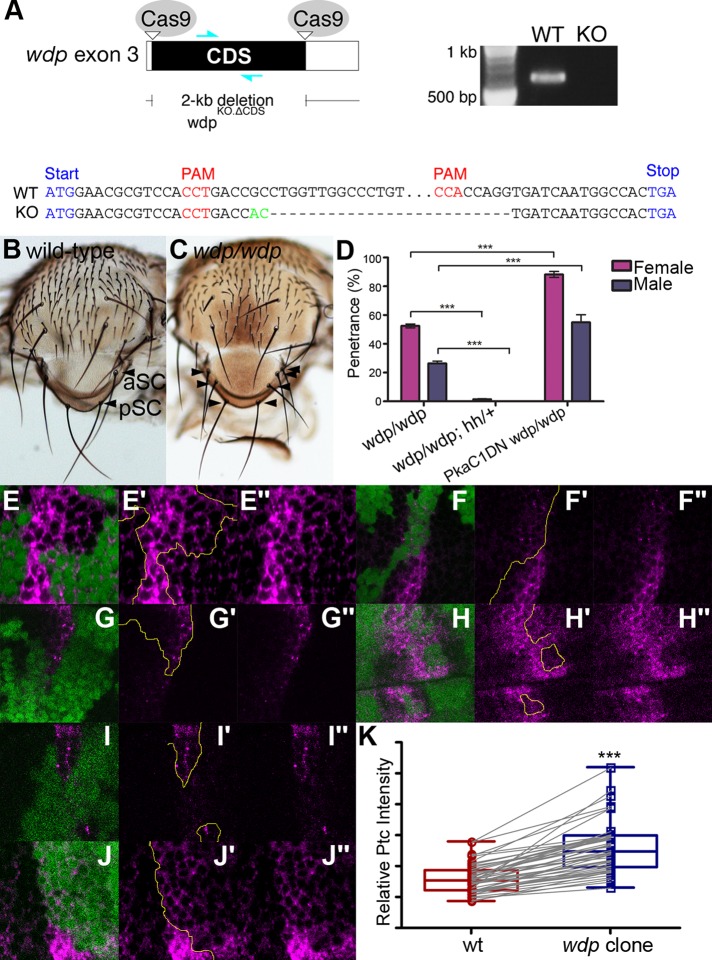
To confirm the wdp knockdown phenotypes, we generated a loss-of-function allele of wdp (wdpKO.ΔCDS), in which most of the wdp coding sequence was removed using CRISPR–Cas9-mediated defined deletion (Figure 6A; Gratz et al., 2013). The wdp homozygous null mutants are viable and fertile. However, adult flies show duplication (or less frequently, triplication) of specific mechanosensory bristles, SC bristles, with a penetrance of 52.5% (females) and 26.4% (males) (Figure 6, B–D). The number and position of these notal mechanosensory bristles, called macrochaetes, are controlled by prepattern genes (Jan and Jan, 1990). For example, Wg and Dpp act as prepattern genes to regulate the specification of presutural (PS) and dorsocentral (DC) bristles, respectively (Whittle and Phillips, 1993; Couso et al., 1994; Tomoyasu et al., 1998). The formation of SC bristles is controlled by Hh signaling (Mullor et al., 1997a). Hh overexpression results in supernumerary SC bristles (Porter et al., 1996). Thus, loss of wdp function led to a known specific phenotype caused by hh overexpression.
FIGURE 6:
The wdp mutant phenotypes. (A) Schematic of the generation of a wdp loss-of-function allele (wdpKO.ΔCDS) lacking most of the wdp CDS using the CRISPR–Cas9 system (top left). A primer set shown as cyan arrows was used for PCR-based genotyping (top right). The results are shown for the WT and wdpKO.ΔCDS allele (KO). Genomic sequencing of the wdp endogenous locus targeted by CRISPR–Cas9 showed a deletion of most of the wdp CDS (bottom). A small insertion is shown in green. (B, C) Control (WT; B) and wdp/wdp (C) SC bristles. Anterior and posterior SC bristles are marked (aSC and pSC, respectively). The wdp homozygous mutants show duplication or triplication of SC bristles (arrowheads). (D) Genetic interactions between wdp and hh or PKa-C1 on the notum phenotype. Penetrance of SC phenotypes is shown. Deleting one copy of hh in wdp homozygotes almost completely suppressed the SC phenotype (wdp/wdp; hhAC/+), while Pka-C1DN significantly enhanced it (Pka-C1DN wdp/wdp). The bristles were scored for at least 200 specimens per genotype. (E–J) Somatic mosaic clones of wdpKO were induced in the wing pouch using Act5C-GAL4 UAS-FLP. Homozygous wdpKO mutant cells are marked by loss of GFP (green) and discs were stained with anti-Ptc antibody (magenta). The clone borders are marked with yellow lines. Increased Ptc expression was observed in wdp mutant clones. Zoom-out images of two wing discs bearing three wdp mutant clones for 6F, G, and H are shown in Supplemental Figure S5 to indicate the relative positions of these clones in the wing disc. (K) Boxplots showing the effect of wdp clones on Ptc staining signal intensity. Ptc staining signal intensity in randomly selected wdp mutant clones was compared with that in immediate neighboring WT cells (n = 49 pairs). ***p < 0.001. The Wilcoxon rank sum test was used to compare two paired samples.
We found that deleting one copy of hh in wdp homozygotes (wdp/wdp; hh/+) almost completely suppressed the SC phenotype (Figure 6D). We also examined the genetic interactions between wdp and cyclic AMP-dependent protein kinase catalytic subunit 1 (Pka-C1). Pka-C1 is involved in Hh signaling (Pan and Rubin, 1995) and a dominant-negative allele of Pka-C1, Pka-C1DN, enhances Hh signaling (Pan and Rubin, 1995). We found that the penetrance of the SC phenotype was significantly increased by Pka-C1DN (Pka-C1DN wdp/wdp; Figure 6D). These observations further support the idea that wdp negatively regulates Hh signaling. wdp does not affect PS or DC bristles, consistent with the specific effect of wdp on the Hh pathway.
wdpKO.ΔCDS homozygous mutant clones were induced in the wing pouch using the FLP–FRT system Actin5C-GAL4 UAS-FLP. The effect of wdp mutant clones on Hh signaling was examined using anti-Ptc antibody. Six examples for such mutant clones are shown in Figure 6, E–J. Consistent with the RNAi knockdown results, we observed a modest increase of Ptc expression in cells mutant for wdp. The change in the level of Ptc is modest but highly consistent: the increased Ptc staining was observed in at least one clone in all wing discs we examined (n = 33). To quantify this effect of wdp mutant clones, Ptc staining signal intensity in wdp null mutant clones (GFP-negative) was compared with that in immediate neighboring WT cells (GFP-positive). This analysis showed a significant increase in the Ptc protein level in wdp mutant clones (Figure 6K). Taken together, our results consistently show that wdp negatively regulates Hh signaling in the Drosophila wing.
DISCUSSION
Our glycoproteomic analysis identified Wdp as a novel CSPG. Apart from Wdp, we did not find any additional novel core proteins in this study. However, some previously established core proteins were also identified, which were found with both CS and/or HS modifications (unpublished data). In a recent glycoproteomic study of C. elegans, we identified 15 novel chondroitin core proteins, in addition to the nine previously established (Noborn et al., 2018). The reason for this discrepancy with regard to the number of identified core proteins in the two model organisms is unclear, but it may suggest that optimization of sample preparation is necessary for identifying additional CSPGs in Drosophila.
Although Wdp was found modified with CS in both WT and ttv backgrounds, general assessment of spectral intensities suggests that Wdp was present in higher abundance in the ttv samples. Earlier studies in zebrafish, mammalian cells, and C. elegans indicated that reduced HS sulfation results in increased CS sulfation (Dierker et al., 2016). Thus, it is not surprising to see a compensatory increase of CS synthesis in a strain lacking HS polymerase (ttv). It should be noted that we did not detect Wdp modified with HS in WT flies, although we explicitly looked for this variant.
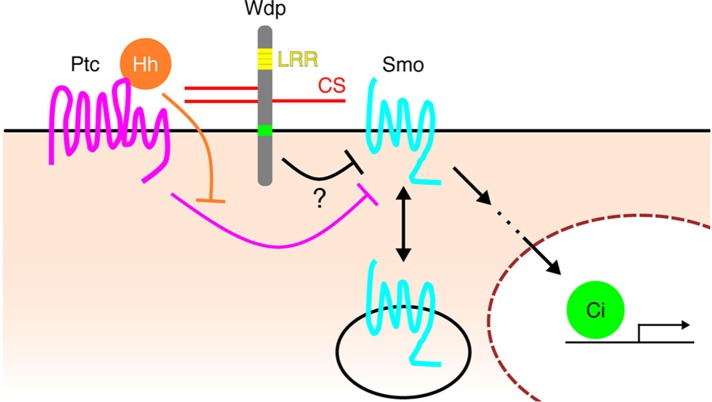
Our genetic analyses of Wdp showed that it acts as a negative modulator of Hh signaling in a CS- and LRR motif-dependent manner. It has also been reported that Wdp negatively regulates JAK–STAT signaling and controls adult midgut homeostasis and regeneration (Ren et al., 2015). The authors showed that Wdp interacts with the Dome receptor and promotes its endocytosis and lysosomal degradation. Although we do not know the mechanism by which Wdp regulates Hh signaling, it is possible that Wdp modulates these two pathways via a similar mechanism: by controlling the stability of cell surface components of the pathways. Hh signaling is controlled by two key membrane proteins—Ptc and Smo. In the absence of Hh, Ptc inhibits the phosphorylation of Smo, which is internalized and degraded (Zhu et al., 2003). In the presence of Hh, restriction of Ptc on Smo is relieved, allowing Smo to accumulate on the cell surface and activate Hh signaling. Our preliminary observation showed that knockdown of wdp increases Smo protein levels (data not shown). Thus, Wdp may down-regulate Hh signaling by affecting Smo levels (e.g., disrupting Smo translocation to the cell membrane or the stability of Smo on the cell surface; Figure 7). However, this does not exclude other possibilities for Wdp action, such as sequestering the ligand, inhibiting Ptc in its Smo phosphorylation/activation, and competing with a HSPG coreceptor. In mice, sulfated CS is necessary for Indian hedgehog (Ihh) signaling in the developing growth plate (Cortes et al., 2009). Although Ihh and Sonic hedgehog (Shh) have been shown to bind to CS (Zhang et al., 2007; Cortes et al., 2009; Whalen et al., 2013), the molecular mechanisms of CSPG function in Hh signaling remain to be elucidated.
FIGURE 7:
Model for the function of Wdp in modulating Hh signaling. The seven-pass transmembrane protein Smo is a key transducer of Hh signaling. In the absence of Hh, Ptc keeps Smo in intracellular vesicles. Hh inhibits the activity of Ptc, allowing Smo to translocate to the plasma membrane and activate downstream signaling. Our preliminary study shows that wdp RNAi knockdown increases the level of Smo protein (data not shown). Given that Wdp negatively regulates JAK–STAT signaling by promoting the degradation of the Dome receptor (Ren et al., 2015), it may down-regulate Hh signaling via a similar mechanism: reducing Smo protein levels. However, the molecular mechanism of Wdp activity in Hh signaling remains elusive.
It is worth noting that both JAK–STAT and Hh signaling, the two pathways negatively controlled by Wdp, are also regulated by HSPGs. Dally-like, a glypican family of HSPGs, positively regulates Hh signaling (Desbordes and Sanson, 2003; Lum et al., 2003; Williams et al., 2010; Yan et al., 2010; Kim et al., 2011). In the developing ovary, Dally up-regulates the JAK–STAT pathway (Hayashi et al., 2012). Given the importance of precise dosage control of oncogenic pathways, such as JAK–STAT and Hh signaling, this dual proteoglycan system could play an important role in fine-tuning the signaling output in order to prevent cancer formation. In vertebrates, HSPGs and CSPGs show opposing effects in neural systems. For example, axon growth is typically promoted by HSPGs but inhibited by CSPGs (Bandtlow and Zimmermann, 2000; Kantor et al., 2004; Silver and Miller, 2004; Van Vactor et al., 2006; Matsumoto et al., 2007; Coles et al., 2011). Our findings suggest that such competing effects of HSPGs and CSPGs may be a general mechanism for the precise control of signaling cascades and pattern formation.
In addition to its functions in signaling, Wdp may play other roles. We found that overexpression of wdp results in massive apoptosis in the wing disc, independent of Hh signaling inhibition (Supplemental Figure S2). Since CSPGs are well known for structural functions, an excess amount of Wdp may affect the epithelial integrity of the wing disc, leading to subsequent apoptosis. Our observation that Wdp is enriched on the basal side of the wing disc and adult midgut cells (Figure 4 and Supplemental Figure S4G) suggests that Wdp may interact with components of the basement membrane, which surrounds these organs.
Previous studies also reported that wdp is associated with aggressive behaviors in Drosophila species. wdp is up-regulated in the head of socially isolated male flies, which exhibit more aggressive behaviors than males raised in groups (Wang et al., 2008). Also, wdp expression is slightly higher in the brain of Drosophila prolongata, which is more aggressive compared with its closely related species (Kudo et al., 2017). Since CSPGs are important in neuronal patterning (Saied-Santiago and Bülow, 2018), it is interesting to define the molecular mechanisms by which Wdp affects Drosophila behavior.
In mammals, there is a class of CSPG molecules with LRR motifs (small leucine-rich proteoglycans, or SLRPs). A number of SLRP members are known as causative genes of human genetic disorders (Bech-Hansen et al., 2000; Pusch et al., 2000; Schaefer and Iozzo, 2008). Although Wdp does not have cysteine-rich regions that are commonly found in mammalian SLRPs, MARRVEL (version 1.1) (Wang et al., 2017) reports that wdp is a potential Drosophila ortholog of the human NYX gene (nyctalopin), a member of SLRPs (DIPOT score 1; Hu et al., 2011). Mutations in NYX cause X-linked congenital stationary night blindness (Bech-Hansen et al., 2000; Pusch et al., 2000). Further studies on Wdp will provide a novel insight into the function of these disease-related human counterparts.
MATERIALS AND METHODS
Preparation of GAG-glycopeptides and LC-MS/MS analysis
GAG-glycopeptide samples were prepared from WT (Oregon-R) and ttv mutant (ttv524) third-instar larvae as previously described (Noborn et al., 2015, 2018). Briefly, 200–400 third-instar larvae (wet weight, 200–400 mg) were lyophilized and homogenized using a motor pestle in 1 ml of ice-cold acetone. After extensive washes with acetone, the insoluble fraction was recovered by centrifugation. After overnight desiccation, the pellet was dissolved in 1.5 ml 1% CHAPS lysis buffer and boiled for 10 min at 96°C. The sample was adjusted to 2 mM MgCl2 and incubated with 3 μl Benzonase (MilliporeSigma, Burlington, MA) at 37°C for 3 h. After heat-inactivation of Benzonase, the sample was centrifuged and the supernatant was collected in a new tube.
An aliquot of the preparation (1 mg of protein) was further used. The sample was reduced and alkylated in 1 ml 50 mM NH4HCO3 and trypsinized at 37°C overnight with 20 μg trypsin (Promega, Madison, WI). The digested samples were applied onto DEAE (GE Healthcare, Chicago, IL) columns (600 μl) at 4°C. The columns were washed with three different low-salt washing solutions at 4°C: 50 mM Tris-HCl, 100 mM NaCl, pH 8.0; 50 mM NaAc, 100 mM NaCl, pH 4.0; and 100 mM NaCl. The glycopeptides that were bound to DEAE were eluted stepwise with four buffers with increasing sodium chloride concentrations at 4°C: 4 ml 250 mM NaCl, 400 mM NaCl, 800 mM NaCl, and 3 ml 1500 mM NaCl. Each fraction was desalted using PD10-columns (GE Healthcare).
All fractions were lyophilized and the salt-free samples were then individually treated with 1 mU of chondroitinase ABC (Sigma-Aldrich, St. Louis, MO) for 3 h at 37°C. Prior to MS analysis, the samples were desalted using a C18 spin column (8 mg resin) according to the manufacturer’s protocol (Thermo Fisher Scientific, Waltham, MA) and lyophilized. LC-MS/MS analysis was performed as previously described (Noborn et al., 2015, 2018). In brief, the samples were analyzed on a Q Exactive mass spectrometer coupled to an Easy-nLC 1000 system (Thermo Fisher Scientific). The glycopeptides (2-μl injection volume) were separated using an analytical column with Reprosil-Pur C18-AQ particles (Dr. Maisch GmbH, Ammerbuch, Germany). The following gradient was run at 300 nl/min; from 7 to 35% B-solvent (acetonitrile in 0.2% formic acid) over 75 min to 100% B-solvent over 5 min, with a final hold at 100% B-solvent for 10 min. The A-solvent was 0.2% formic acid. Spectra were recorded in positive ion mode and MS scans were performed at 70,000 resolution with a mass range of m/z 600–1800. The MS/MS analysis was performed in a data-dependent mode, with the top 10 most abundant precursor ions in each MS scan selected for fragmentation (MS2) by higher-energy collision dissociation (HCD) with a normalized collision energy value of 30%. The MS2 scans were performed at a resolution of 35,000 (at m/z 200). The data analyses were performed as previously described (Noborn et al., 2015) with some small adjustments. In brief, the HCD.raw spectra were converted to Mascot .mgf format using Mascot distiller (version 2.3.2.0, Matrix Science, London, UK). The ions were presented as singly protonated in the output Mascot file. Searches were performed using an in-house Mascot server (version 2.3.02) with the enzyme specificity set to Trypsin, and then to Semitrypsin, allowing for one or two missed cleavages, in subsequent searches on Drosophila sequences of the UniprotKB (42, 507, sequences, 2018-06-18). The peptide tolerance was set to 10 parts per million (ppm) and fragment tolerance was set to 0.01 Da. The searches were allowed to include variable modifications at serine residues of the residual hexasaccharide structure [GlcA(-H2O)GalNAcGlcAGalGalXyl-O-] with 0 (C37H55NO30, 993.2809 Da), 1 (C37H55NO33S, 1073.2377 Da), or 2 (C37H55NO36S2, 1153.1945 Da) sulfate groups attached.
Drosophila strains
The following fly strains were used in this study: Oregon-R, w1118 (Bloomington Drosophila Stock Center [BDSC] #5905), ttv524, ap-GAL4, hh-GAL4, BxMS1096-GAL4 (BDSC #8860), AB1-GAL4 (BDSC #1824), elavC155>mCD8:GFP (BDSC #5146), UAS-GFP (BDSC #1521), UAS-tdTomato (BDSC #36327 and #36328), UAS-FLP (BDSC #4539 and #4540), UAS-ptc (BDSC #44614), nub-GAL4 (BDSC #25754), Act5C-GAL4 (BDSC #3954), FRT42D 2xUbi-GFP, UAS-smo:GFP (BDSC #44624), UAS-FLAG:smoAct (BDSC #44621), UAS-wdpRNAi (TRiP.HMC06302, BDSC #66004), UAS-wdpRNAi (TRiP.HM05118, BDSC #28907), UAS-smoRNAi (TRiP.HMC03577, BDSC #53348), hh-lacZP30 (a gift from Gary Struhl), dpp-lacZ10638 (BDSC #12379), vas-Cas9 (BDSC #55821), and esg-GAL4 (DGRC #113886). The UAS-wdp, UAS-wdpΔGAG, UAS-3xMyc:wdp, UAS-3xMyc:wdpΔGAG, UAS-3xMyc:wdpΔLRRs, UAS-3xMyc:wdpΔICD, wdpKO.ΔCDS, wdpKI.HA, wdpKI.OLLAS flies were generated in this study. A full list of genotypes used in this study can be found in Supplemental Table S1.
For constructing UAS-wdp, wdp CDS (corresponding to wdp-RA–E in FlyBase) was inserted into the XhoI- and XbaI-digested pJFRC7 vector (a gift from Gerald Rubin, Howard Hughes Medical Institute; Addgene # 26220) using NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs [NEB], Ipswich, MA, E2621S). Similarly, wdpΔGAG (S282A, S334A, and S336A), Myc:wdp, Myc:wdpΔLRRs, and Myc:wdpΔICD were inserted into the pJFRC7 vector. The UAS transgenic flies were generated using phiC31 integrase-mediated transgenesis at the ZH-86Fb attP (FBti0076525) integration site (Bischof et al., 2007). Because these UAS transgenes were integrated into an identical genomic location, the phenotypic differences reflect the activity of each construct rather than differential expression levels due to positional effects (Kleinschmit et al., 2010; Dejima et al., 2013; Kleinschmit et al., 2013). Embryonic injection was performed by BestGene (Chino Hills, CA). Primers used in this study will be available on request.
To generate the wdpKO.ΔCDS allele, two sgRNAs (pU6-sgRNA-wdp-1 and pU6-sgRNA-wdp-2) were introduced to delete the wdp CDS. To construct sgRNA plasmids, 5′-CTTCGACAGGGCCAACCAGGCGGTC-3′ and 5′-AAACGACCGCCTGGTTGGCCCTGTC-3′ were annealed (pU6-sgRNA-wdp-1), and 5′-CTTCGAGTGGCCATTGATCACCTGG-3′ and 5′-AAACCCAGGTGATCAATGGCCACTC-3′ (pU6-sgRNA-wdp-2) were annealed and ligated in the BbsI-digested pU6-BbsI-chiRNA plasmid (a gift from Melissa Harrison, University of Wisconsin; Kate O’Connor-Giles, Brown University; and Jill Wildonger, University of Wisconsin; Addgene #45946). A mixture of 50 ng/µl of pU6-sgRNA-wdp-1 and pU6-sgRNA-wdp-2 was injected into the embryos of the vas-Cas9 flies, which express Cas9 under the control of the germline vasa regulatory elements, by BestGene. The wdpKO.ΔCDS allele was screened by PCR and verified by Sanger sequencing.
To generate the wdpKI.HA allele, we constructed a donor plasmid, which contained a Gly-Gly-Ser linker, smGFP-HA, and approximately 1-kb homology arms to wdp flanking the linker and smGFP-HA, for homology-directed repair. The smGFP-HA and the wdp homology sequences on either side of the targeted DSB were PCR-amplified from pJFRC201-10XUAS-FRT>STOP>FRT-myr:smGFP-HA (a gift from Gerald Rubin; Addgene plasmid #63166) and genomic DNA extracted from the vas-Cas9 flies, respectively. These fragments were cloned into the pHD-DsRed-attP backbone (a gift from Melissa Harrison, Kate O’Connor-Giles, and Jill Wildonger; Addgene #51019) using NEBuilder HiFi DNA Assembly Master Mix (NEB, E2621S). Similarly, we generated a donor plasmid with OLLAS tags amplified from pJFRC210-10XUAS-FRT>STOP>FRT-myr:smGFP-OLLAS (a gift from Gerald Rubin; Addgene plasmid #63170). A mixture of 50 ng/µl of pU6-sgRNA-wdp-2 and 125 ng/µl of each donor plasmid was injected into the vas-Cas9 embryos by BestGene. The wdpKI.HA and wdpKI.OLLAS alleles were screened by PCR and verified by Sanger sequencing.
In mosaic analysis, the wdpKO. ΔCDS homozygous clones were generated by FLP/FRT-mediated mitotic recombination (Xu and Rubin, 1993). The FLP expression was induced by Act5C-GAL4 UAS-FLP.
Flies were raised on a standard cornmeal fly medium at 25°C unless otherwise indicated.
Immunohistochemstry
Wing discs were dissected from third-instar wandering larvae in phosphate-buffered saline (PBS, pH 7.4) and subsequently fixed in 3.7% formaldehyde in PBS for 15 min at room temperature. After three 10-min washes with PBST (PBS containing 0.1% [vol/vol] Triton X-100 [Sigma, T8532]), the samples were incubated in primary antibodies overnight at 4°C. After three 10-min washes with PBST, the samples were incubated with Alexa Fluor–conjugated secondary antibodies (1:500; Thermo Fisher Scientific) overnight at 4°C or 2 h at room temperature. After three 10-min washes with PBST, the samples were stained with 1 µg/ml DAPI (4’6-diamidino-2-phenylindole) (Thermo Fisher Scientific, 62248) and subsequently mounted in VECTASHIELD Antifade Mounting Medium (Vector Laboratories, Burlingame, CA, H-1000). F-actin was stained with Alexa Fluor 568 phalloidin (Thermo Fisher Scientific, A12380). Adult midguts were dissected and immunostained as previously described (Takemura and Nakato, 2017). Images were acquired on a LSM710 confocal microscope (Carl Zeiss, Oberkochen, Germany). For quantification of Ptc and Smo staining, images were acquired with the same condition, and fluorescence intensity was measured in a set area with Fiji (Schindelin et al., 2012). No statistical methods were used to predetermine sample size. All n numbers represent biological replicates. Experiments were not randomized or blinded.
The primary antibodies used were as follows: mouse anti–Ptc Apa 1 (1:20, Developmental Studies Hybridoma Bank [DSHB], Iowa City, IA), rat anti–Ci 2A1 (1:20, DSHB), chicken anti–β-galactosidase (1:2000, Abcam), mouse anti–En 4D9 (1:20, DSHB), rabbit anti-pH3 (1:1000, Millipore, 06-570), rat anti–HA 3F10 (1:200, Roche, 11867423001), rabbit anti–HA C29F4 (1:1000, Cell Signaling, 3724), mouse anti–Smo 20C6 (1:50, DSHB), rabbit anti-pSmad3 (1:1000, Epitomics, 1880-1), rabbit anti-Salm (1:30; a gift from Scott Selleck, Penn State University), mouse anti-Dll (1:500; a gift from Dianne Duncan, Washington University), guinea pig anti-Sens (1:1000; a gift from Hugo Bellen, Baylor College of Medicine), rabbit anti-cleaved Caspase-3 (1:200, Cell Signaling, 9661), mouse anti–c-Myc 9E10 (1:1000, Sigma-Aldrich, M5546), rat anti–OLLAS L2 (1:500, Novus Biologicals, NBP1-06713), mouse anti–Arm N2 7A1 (1:50, DSHB), mouse anti–Pros MR1A (1:50, DSHB), and mouse anti–Fas3 7G10 (1:50, DSHB). Alexa 488–, Alexa 546–, Alexa 568– and Alexa 633–conjugated secondary antibodies (Thermo Fisher Scientific) were used at a dilution of 1:500.
Preparation of adult wings and notums
The right wings from female flies were dehydrated in ethanol and subsequently with xylene. Adult cuticles of the notum were boiled in 2.5 N sodium hydroxide, washed in distilled water, and dehydrated in 2-propanol (Fujise et al., 2001). The specimens were mounted in Canada balsam (Benz Microscope, BB0020).
Supplementary Material
Acknowledgments
We thank Melissa Harrison, Kate O’Connor-Giles, Jill Wildonger, Gary Struhl, Scott Selleck, Hugo Bellen, Gerald Rubin, Dianne Duncan, the Bloomington Drosophila Stock Center (National Institutes of Health [NIH] P40OD018537), the Transgenic RNAi Project at Harvard Medical School (NIH/National Institute of General Medical Sciences R01-GM08947), and the Drosophila Genomics Resource Center (NIH 2P40OD010949) for sharing fly strains and plasmids. We also thank the Proteomics Core Facility at the Sahlgrenska Academy, University of Gothenburg, Sweden, for running all the MS analyses. This work was supported by NIH (R35 GM131688) to H.N. and by the Swedish Research Council (8266) to G.L. M.T. held postdoctoral fellowships from the Japan Society for the Promotion of Science and the Uehara Memorial Foundation.
Abbreviations used:
- ap
apterous
- Ci
cubitus interruptus
- CSPG
chondroitin sulfate proteoglycan
- Dll
Distal-less
- Dpp
Decapentaplegic
- DSHB
Developmental Studies Hybridoma Bank
- En
Engrailed
- GAG
glycosaminoglycan
- HCD
higher-energy collision dissociation
- Hh
Hedgehog
- HSPG
heparan sulfate proteoglycan
- Ihh
Indian hedgehog
- LRR
leucin-rich repeat
- NEB
New England Biolabs
- nLC-MS/MS
nano-liquid chromatography-tandem mass spectrometry
- PBS
phosphate-buffered saline
- Pka-C1
protein kinase catalytic subunit 1
- ppm
parts per million
- Ptc
Patched
- Salm
Spalt major
- SC
scutellar
- Sens
Senseless
- SLRP
small leucine-rich proteoglycan
- Smo
smoothened
- Wdp
Windpipe
- Wg
Wingless
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-06-0327) on February 12, 2020.
REFERENCES
- Akiyama T, Kamimura K, Firkus C, Takeo S, Shimmi O, Nakato H. (2008). Dally regulates Dpp morphogen gradient formation by stabilizing Dpp on the cell surface. Dev Biol , 408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandtlow CE, Zimmermann DR. (2000). Proteoglycans in the developing brain: new conceptual insights for old proteins. Physiol Rev , 1267–1290. [DOI] [PubMed] [Google Scholar]
- Bech-Hansen NT, Naylor MJ, Maybaum TA, Sparkes RL, Koop B, Birch DG, Bergen AA, Prinsen CF, Polomeno RC, Gal A, et al (2000). Mutations in NYX, encoding the leucine-rich proteoglycan nyctalopin, cause X-linked complete congenital stationary night blindness. Nat Genet , 319–323. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA , 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Thérond PP. (2013). The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol , 416–429. [DOI] [PubMed] [Google Scholar]
- Capdevila J, Guerrero I. (1994). Targeted expression of the signaling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. EMBO J , 4459–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila J, Pariente F, Sampedro J, Alonso JL, Guerrero I. (1994). Subcellular localization of the segment polarity protein patched suggests an interaction with the wingless reception complex in Drosophila embryos. Development , 987–998. [DOI] [PubMed] [Google Scholar]
- Coles CH, Shen Y, Tenney AP, Siebold C, Sutton GC, Lu W, Gallagher JT, Jones EY, Flanagan JG, Aricescu AR. (2011). Proteoglycan-specific molecular switch for RPTPσ clustering and neuronal extension. Science , 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes M, Baria AT, Schwartz NB. (2009). Sulfation of chondroitin sulfate proteoglycans is necessary for proper Indian hedgehog signaling in the developing growth plate. Development , 1697–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couso JP, Bishop SA, Martinez Arias A. (1994). The wingless signalling pathway and the patterning of the wing margin in Drosophila. Development , 621–636. [DOI] [PubMed] [Google Scholar]
- Dejima K, Takemura M, Nakato E, Peterson J, Hayashi Y, Kinoshita-Toyoda A, Toyoda H, Nakato H. (2013). Analysis of Drosophila glucuronyl C-5 epimerase: implications for developmental roles of heparan sulfate sulfation compensation and 2-O sulfated glucuronic acid. J Biol Chem , 34384–34393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbordes SC, Sanson B. (2003). The glypican Dally-like is required for Hedgehog signalling in the embryonic epidermis of Drosophila. Development , 6245–6255. [DOI] [PubMed] [Google Scholar]
- Dierker T, Shao C, Haitina T, Zaia J, Hinas A, Kjellen L. (2016). Nematodes join the family of chondroitin sulfate-synthesizing organisms: Identification of an active chondroitin sulfotransferase in Caenorhabditis elegans. Sci Rep , 34662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esko JD, Zhang L. (1996). Influence of core protein sequence on glycosaminoglycan assembly. Curr Opin Struct Biol , 663–670. [DOI] [PubMed] [Google Scholar]
- Fujise M, Izumi S, Selleck SB, Nakato H. (2001). Regulation of dally, an integral membrane proteoglycan, and its function during adult sensory organ formation of Drosophila. Dev Biol , 433–448. [DOI] [PubMed] [Google Scholar]
- Gradilla A-C, Guerrero I. (2013). Hedgehog on the move: a precise spatial control of Hedgehog dispersion shapes the gradient. Curr Opin Genet Dev , 363–373. [DOI] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM. (2013). Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics , 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’Connor-Giles KM. (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics , 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. (2004). Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics , 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl TA, Scott MP. (2014). Wing tips: The wing disc as a platform for studying Hedgehog signaling. Methods , 199–206. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Sexton TR, Dejima K, Perry DW, Takemura M, Kobayashi S, Nakato H, Harrison DA. (2012). Glypicans regulate JAK/STAT signaling and distribution of the Unpaired morphogen. Development , 4162–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N, Mohr SE. (2011). An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics , 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff JL, Kingsley KL, Miller JM, Hoshizaki DK. (2002). Drosophila windpipe codes for a leucine-rich repeat protein expressed in the developing trachea. Mech Dev , 173–176. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. (1990). Genes required for specifying cell fates in Drosophila embryonic sensory nervous system. Trends Neurosci , 493–498. [DOI] [PubMed] [Google Scholar]
- Jia J, Tong C, Wang B, Luo L, Jiang J. (2004). Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature , 1045–1050. [DOI] [PubMed] [Google Scholar]
- Kantor DB, Chivatakarn O, Peer KL, Oster SF, Inatani M, Hansen MJ, Flanagan JG, Yamaguchi Y, Sretavan DW, Giger RJ, Kolodkin AL. (2004). Semaphorin 5A is a bifunctional axon guidance cue regulated by heparan and chondroitin sulfate proteoglycans. Neuron , 961–975. [DOI] [PubMed] [Google Scholar]
- Kim M-S, Saunders AM, Hamaoka BY, Beachy PA, Leahy DJ. (2011). Structure of the protein core of the glypican Dally-like and localization of a region important for hedgehog signaling. Proc Natl Acad Sci USA , 13112–13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmit A, Koyama T, Dejima K, Hayashi Y, Kamimura K, Nakato H. (2010). Drosophila heparan sulfate 6-O endosulfatase regulates Wingless morphogen gradient formation. Dev Biol , 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmit A, Takemura M, Dejima K, Choi PY, Nakato H. (2013). Drosophila heparan sulfate 6-O-endosulfatase Sulf1 facilitates wingless (Wg) protein degradation. J Biol Chem , 5081–5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo A, Shigenobu S, Kadota K, Nozawa M, Shibata TF, Ishikawa Y, Matsuo T. (2017). Comparative analysis of the brain transcriptome in a hyper-aggressive fruit fly, Drosophila prolongata. Insect Biochem Mol Biol , 11–20. [DOI] [PubMed] [Google Scholar]
- Lindahl U, Li J-P. (2009). Interactions between heparan sulfate and proteins-design and functional implications. Int Rev Cell Mol Biol , 105–159. [DOI] [PubMed] [Google Scholar]
- Lu J, Wang D, Shen J. (2017). Hedgehog signalling is required for cell survival in Drosophila wing pouch cells. Sci Rep , 11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA. (2003). Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science , 2039–2045. [DOI] [PubMed] [Google Scholar]
- Mace K, Tugores A. (2004). The product of the split ends gene is required for the maintenance of positional information during Drosophila development. BMC Dev Biol , 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Irie F, Inatani M, Tessier-Lavigne M, Yamaguchi Y. (2007). Netrin-1/DCC signaling in commissural axon guidance requires cell-autonomous expression of heparan sulfate. J Neurosci , 4342–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EJ, Ikmi A, Gibson MC. (2011). Interkinetic nuclear migration is a broadly conserved feature of cell division in pseudostratified epithelia. Curr Biol , 485–491. [DOI] [PubMed] [Google Scholar]
- Momota R, Naito I, Ninomiya Y, Ohtsuka A. (2011). Drosophila type XV/XVIII collagen, Mp, is involved in Wingless distribution. Matrix Biol , 258–266. [DOI] [PubMed] [Google Scholar]
- Mullor JL, Calleja M, Capdevila J, Guerrero I. (1997a). Hedgehog activity, independent of decapentaplegic, participates in wing disc patterning. Development , 1227–1237. [DOI] [PubMed] [Google Scholar]
- Mullor JL, Calleja M, Capdevila J, Guerrero I. (1997b). Hedgehog activity, independent of decapentaplegic, participates in wing disc patterning. Development , 1227–1237. [DOI] [PubMed] [Google Scholar]
- Nakato H, Li J-P. (2016). Functions of Heparan Sulfate Proteoglycans in Development: Insights From Drosophila Models. Int Rev Cell Mol Biol , 275–293. [DOI] [PubMed] [Google Scholar]
- Noborn F, Gomez Toledo A, Green A, Nasir W, Sihlbom C, Nilsson J, Larson G. (2016). Site-specific identification of heparan and chondroitin sulfate glycosaminoglycans in hybrid proteoglycans. Sci Rep , 34537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noborn F, Gomez Toledo A, Nasir W, Nilsson J, Dierker T, Kjellén L, Larson G. (2018). Expanding the chondroitin glycoproteome of Caenorhabditis elegans. J Biol Chem , 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noborn F, Gomez Toledo A, Sihlbom C, Lengqvist J, Fries E, Kjellén L, Nilsson J, Larson G. (2015). Identification of chondroitin sulfate linkage region glycopeptides reveals prohormones as a novel class of proteoglycans. Mol Cell Proteomics , 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson SK, Bishop JR, Yates JR, Oegema K, Esko JD. (2006). Identification of novel chondroitin proteoglycans in Caenorhabditis elegans: embryonic cell division depends on CPG-1 and CPG-2. J Cell Biol , 985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Rubin GM. (1995). cAMP-dependent protein kinase and hedgehog act antagonistically in regulating decapentaplegic transcription in Drosophila imaginal discs. Cell , 543–552. [DOI] [PubMed] [Google Scholar]
- Patel NH, Martín-Blanco E, Coleman KG, Poole SJ, Ellis MC, Kornberg TB, Goodman CS. (1989). Expression of engrailed proteins in arthropods, annelids, and chordates. Cell , 955–968. [DOI] [PubMed] [Google Scholar]
- Porter JA, Ekker SC, Park WJ, von Kessler DP, Young KE, Chen CH, Ma Y, Woods AS, Cotter RJ, Koonin EV, Beachy PA. (1996). Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell , 21–34. [DOI] [PubMed] [Google Scholar]
- Pusch CM, Zeitz C, Brandau O, Pesch K, Achatz H, Feil S, Scharfe C, Maurer J, Jacobi FK, Pinckers A, et al (2000). The complete form of X-linked congenital stationary night blindness is caused by mutations in a gene encoding a leucine-rich repeat protein. Nat Genet , 324–327. [DOI] [PubMed] [Google Scholar]
- Ragkousi K, Gibson MC. (2014). Cell division and the maintenance of epithelial order. J Cell Biol , 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W, Zhang Y, Li M, Wu L, Wang G, Baeg G-H, You J, Li Z, Lin X. (2015). Windpipe controls Drosophila intestinal homeostasis by regulating JAK/STAT pathway via promoting receptor endocytosis and lysosomal degradation. PLoS Genet , e1005180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Yang Z, Xu J, Sun J, Mao D, Hu Y, Yang S-J, Qiao H-H, Wang X, Hu Q, et al (2014). Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep , 1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saied-Santiago K, Bülow HE. (2018). Diverse roles for glycosaminoglycans in neural patterning. Dev Dyn , 54–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer L, Iozzo RV. (2008). Biological functions of the small leucine-rich proteoglycans: from genetics to signal transduction. J Biol Chem , 21305–21309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J, Miller JH. (2004). Regeneration beyond the glial scar. Nat Rev Neurosci , 146–156. [DOI] [PubMed] [Google Scholar]
- Strigini M, Cohen SM. (1997). A Hedgehog activity gradient contributes to AP axial patterning of the Drosophila wing. Development , 4697–4705. [DOI] [PubMed] [Google Scholar]
- Takemura M, Nakato H. (2015). Genetic approaches in the study of heparan sulfate functions in Drosophila. Methods Mol Biol , 497–505. [DOI] [PubMed] [Google Scholar]
- Takemura M, Nakato H. (2017). Drosophila Sulf1 is required for the termination of intestinal stem cell division during regeneration. J Cell Sci , 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The I, Bellaiche Y, Perrimon N. (1999). Hedgehog movement is regulated through tout velu-dependent synthesis of a heparan sulfate proteoglycan. Mol Cell , 633–639. [DOI] [PubMed] [Google Scholar]
- Tomoyasu Y, Nakamura M, Ueno N. (1998). Role of dpp signalling in prepattern formation of the dorsocentral mechanosensory organ in Drosophila melanogaster. Development , 4215–4224. [DOI] [PubMed] [Google Scholar]
- Townley RA, Bülow HE. (2018). Deciphering functional glycosaminoglycan motifs in development. Curr Opin Struct Biol , 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda H, Kinoshita-Toyoda A, Selleck SB. (2000). Structural analysis of glycosaminoglycans in Drosophila and Caenorhabditis elegans and demonstration that tout-velu, a Drosophila gene related to EXT tumor suppressors, affects heparan sulfate in vivo. J Biol Chem , 2269–2275. [DOI] [PubMed] [Google Scholar]
- Tripura C, Chandrika NP, Susmitha VN, Noselli S, Shashidhara LS. (2011). Regulation and activity of JNK signaling in the wing disc peripodial membrane during adult morphogenesis in Drosophila. Int J Dev Biol , 583–590. [DOI] [PubMed] [Google Scholar]
- Van Vactor D, Wall DP, Johnson KG. (2006). Heparan sulfate proteoglycans and the emergence of neuronal connectivity. Curr Opin Neurobiol , 40–51. [DOI] [PubMed] [Google Scholar]
- Wang J, Al-Ouran R, Hu Y, Kim S-Y, Wan Y-W, Wangler MF, Yamamoto S, Chao H-T, Comjean A, Mohr SE, et al (2017). MARRVEL: integration of human and model organism genetic resources to facilitate functional annotation of the human genome. Am J Hum Genet , 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Dankert H, Perona P, Anderson DJ. (2008). A common genetic target for environmental and heritable influences on aggressiveness in Drosophila. Proc Natl Acad Sci USA , 5657–5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen DM, Malinauskas T, Gilbert RJC, Siebold C. (2013). Structural insights into proteoglycan-shaped Hedgehog signaling. Proc Natl Acad Sci USA , 16420–16425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle R, Phillips R. (1993). In search of the machinery for spatial patterning of animal epithelia. Bioessays , 757–759. [DOI] [PubMed] [Google Scholar]
- Williams EH, Pappano WN, Saunders AM, Kim M-S, Leahy DJ, Beachy PA. (2010). Dally-like core protein and its mammalian homologues mediate stimulatory and inhibitory effects on Hedgehog signal response. Proc Natl Acad Sci USA , 5869–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Esko JD. (2014). Demystifying heparan sulfate-protein interactions. Annu Rev Biochem , 129–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Rubin GM. (1993). Analysis of genetic mosaics in developing and adult Drosophila tissues. Development , 1223–1237. [DOI] [PubMed] [Google Scholar]
- Yan D, Wu Y, Yang Y, Belenkaya TY, Tang X, Lin X. (2010). The cell-surface proteins Dally-like and Ihog differentially regulate Hedgehog signaling strength and range during development. Development , 2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, McLellan JS, Ayala AM, Leahy DJ, Linhardt RJ. (2007). Kinetic and structural studies on interactions between heparin or heparan sulfate and proteins of the hedgehog signaling pathway. Biochemistry , 3933–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu AJ, Zheng L, Suyama K, Scott MP. (2003). Altered localization of Drosophila Smoothened protein activates Hedgehog signal transduction. Genes Dev , 1240–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.