Abstract
Cellular responsiveness to environment, including changes in extracellular matrix (ECM), is critical for normal processes such as development and wound healing, but can go awry, as in oncogenesis and fibrosis. One type of molecular pathway contributing to this responsiveness is the BMP signaling pathway. Owing to their broad and potent functions, BMPs and their pathways are regulated at multiple levels. In Caenorhabditis elegans, the BMP ligand DBL-1 is a regulator of body size. We previously showed that DBL-1/BMP signaling determines body size through transcriptional regulation of cuticle collagen genes. We now identify feedback regulation of DBL-1/BMP through analysis of four DBL-1–regulated collagen genes. Inactivation of any of these genes reduces DBL-1/BMP signaling, measured by a pathway activity reporter. Furthermore, depletion of these collagens reduces GFP::DBL-1 fluorescence and acts unexpectedly at the level of dbl-1 transcription. We conclude that cuticle, a specialized ECM, impinges on DBL-1/BMP expression and signaling. Interestingly, the feedback regulation of DBL-1/BMP signaling by collagens is likely to be contact independent due to physical separation of the cuticle from DBL-1–expressing cells in the ventral nerve cord. Our results provide an entry point into a novel regulatory mechanism for BMP signaling, with broader implications for mechanical regulation of gene expression.
INTRODUCTION
Cells are exquisitely sensitive to their environments. Cells not only respond to their environments but also influence them; for example, through creation and maintenance of extracellular matrix (ECM). Cells maintain the homeostasis of ECM by regulating the production of secreted collagens, other ECM components, and ECM-modifying enzymes. Loss of this regulation can result in cell death or fibrosis. One group of ECM regulators is the transforming growth factor beta (TGF-β) family. TGF-β signaling regulates collagen synthesis and in turn is regulated by collagens and responsive to changes in matrix stiffness (Chang, 2016; Kim et al., 2018). However, in vitro studies are limited in their ability to recapitulate the complexities of cell–matrix interactions, much less the indirect interactions of cells within organs and ECM, and in vivo systems have thus far been scarce.
We have established Caenorhabditis elegans as an in vivo model to study the interplay between TGF-β signaling and ECM. The C. elegans ECM includes both basement membrane, which lines internal organs, and cuticle, which is secreted apically from hypodermal (epidermal) tissue to protect the organism and is shed at each molt (Kramer, 2005; Page and Johnstone, 2007; Forman-Rubinsky et al., 2017). The cuticle is composed of multiple collagen layers coated by a lipid-rich epicuticle (Chisholm and Hsiao, 2012; Chisholm and Xu, 2012). Cuticle collagens are encoded by a large multigene family with more than 170 members. Some cuticle collagens are expressed in each developmental stage in which a cuticle is synthesized, while others are expressed stage specifically (Jackson et al., 2014). Accordingly, the cuticle at each developmental stage contains different specific collagens and is organized into distinct structures. For example, the lateral ridges known as alae are only present on L1, dauer, and adult cuticles (Page and Johnstone, 2007).
The TGF-β family includes bone morphogenetic proteins (BMPs), highly conserved members that include vertebrate BMPs, Drosophila Dpp, and C. elegans DBL-1. The DBL-1/BMP ligand is not only highly conserved at the sequence level but also signals through a conserved, canonical pathway. This pathway includes the type I and type II receptors and intracellular Smad signal transducers, which are activated by the receptor complex and regulate the transcription of target genes (Gumienny and Savage-Dunn, 2013). The DBL-1 ligand is secreted by ventral cord motor neurons that are nestled in the basal side of the hypodermis (Suzuki et al., 1999). Interestingly, a body size-regulating TGF- β ligand is also produced by motor neurons in Drosophila (Moss-Taylor et al., 2019). Receptors and Smad signal transducers for C. elegans DBL-1 are present in the hypodermis, which receives the DBL-1 signal (Yoshida et al., 2001; Wang et al., 2002). The DBL-1 signaling pathway transcriptionally regulates gene products that function in a number of developmental and homeostatic functions, but the pathway is not essential for viability. There is growing evidence that DBL-1 signaling modulates cuticle structure and function. For example, the DBL-1 pathway plays a major role in regulating body size and morphology, permeability to drugs, and resistance to infection, traits associated with cuticle function (Schultz et al., 2014). Loss of dbl-1 causes physical changes in cuticle morphology (Schultz et al., 2014). Finally, genes encoding cuticle collagens and other ECM constituents and enzymes are known transcriptional targets of the DBL-1 pathway (Liang et al., 2007; Luo et al., 2010; Roberts et al., 2010).
We previously characterized four DBL-1–regulated collagen genes, col-41, rol-6, col-141, and col-142, and demonstrated that they are involved in body size regulation (Madaan et al., 2018). Here we report that these four DBL-1–regulated collagens reciprocally modulate the DBL-1 pathway. Using three different activity reporters, we show that depletion of individual collagen genes impacts the DBL-1 pathway at the levels of Smad activity, DBL-1 protein, and dbl-1 expression. We propose a model in which the presence of these cuticle collagens modifies properties of the cuticle to which DBL-1–expressing cells are sensitive.
RESULTS
Depletion of DBL-1–regulated cuticle collagen gene products impacts DBL-1 pathway activity
To determine whether cuticle collagens affect DBL-1 pathway activity, we used an artificial transcriptional reporter containing multiple Smad-binding sites driving expression of GFP (RAD-SMAD reporter; Tian et al., 2010). We used RNA interference (RNAi) to deplete expression of four cuticle collagen genes, col-41, rol-6, col-141, and col-142, in animals carrying this RAD-SMAD reporter. RNAi was performed with dsRNA targeting the gene-specific amino-termini that do not encode Gly-X-Y repeats. At the L4 stage, depletion of rol-6 caused significant reduction in RAD-SMAD activity in the hypodermis (>40% reduction, p < 0.001; Figure 1A). Compared with control animals fed bacteria carrying the empty vector, the depletion of col-141 or col-142 reduced RAD-SMAD activity in L4 animals (20–25% reduction). The reduction in activity was statistically significant in some but not all trials. In 1-d-old young adults, a more pronounced reduction in fluorescence was seen after rol-6, col-141, and col-142 depletion (50–80% reduction, p < 0.001; Figure 1B). At both L4 and adult stages, RNAi targeting col-41 led to a large variance in fluorescence intensity between individual nuclei, with many nuclei showing reduced expression (Figure 1, A and B). We conclude that DBL-1 Smad signaling in the hypodermis is somehow impacted by the collagen composition of the cuticle.
FIGURE 1:
Loss of cuticle collagen gene function reduces activity of the DBL-1/BMP pathway. (A) L4 animals. (B) Young adults. RNAi was used to deplete collagen gene function. Pathway activity was assayed using a reporter containing Smad-binding sites. AU: arbitrary units. Each data point depicts the fluorescence intensity of an individual nucleus. Bars show mean and SD of the data. Statistical significance compared with control was determined by Brown–Forsythe and Welch analysis of variance (ANOVA) tests. **p < 0.01; ***p < 0.001.
Intensity of GFP::DBL-1 is reduced on loss of DBL-1–regulated collagens
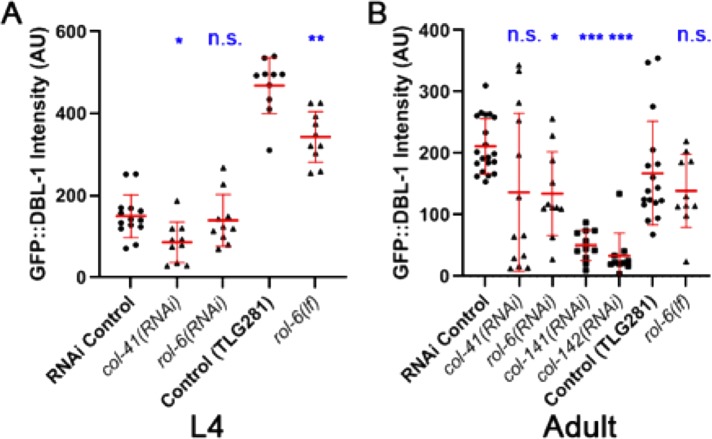
The effect of collagen gene manipulation on the RAD-SMAD transcriptional reporter could occur either at the level of the ligand or on downstream components of the signaling pathway, or possibly both. To distinguish between these possibilities, we first asked whether collagen gene manipulation alters DBL-1 protein distribution. We quantified fluorescence of functional GFP-tagged DBL-1 expressed from an integrated transgene (dbl-1(oe)) in animals fed on bacteria transformed with different RNAi targets (Schultz et al., 2014). We found that RNAi depletion of any of the collagen products of genes col-41, rol-6, col-141, or col-142 reduces the intensity of GFP::DBL-1 fluorescence in adults compared with our control (C06C3.5 pseudogene RNAi) (35–85% reduction depending on the gene; Figure 2B), although the reduction on col-41(RNAi) does not reach statistical significance (p = 0.26). These results support the RAD-SMAD experiment results (Figure 1). We also found that RNAi of col-41, but not of rol-6, caused a significant reduction in GFP intensity at the L4 stage (∼40% reduction for col-41(RNAi); Figure 2A). As in the RAD-SMAD experiment, RNAi of col-41 caused a large variance in fluorescence intensity (Figures 1 and 2). Finally, we verified these RNAi results in a mutant background: rol-6(e187n1270), a loss-of-function mutant (hereafter rol-6(lf); Kramer and Johnson, 1993). We measured the fluorescence intensity of GFP::DBL-1 in the rol-6(lf);dbl-1(oe) strain compared with dbl-1(oe). In rol-6(lf) mutants, GFP::DBL-1 fluorescence was significantly reduced by ∼25% in L4 animals (Figure 2A) and slightly, but not significantly, reduced in adults (Figure 2B). Taken together, these results are consistent with the conclusion that loss of these cuticle collagen isoforms leads to reduction in DBL-1 protein levels (Figure 2). The reduced pathway signaling, as evidenced by reduced RAD-SMAD activity (Figure 1), is therefore likely due, at least in part, to changes in DBL-1 accumulation. In addition to these four DBL-1–regulated collagens, loss of a subset of other cuticle collagens, but not all tested, also reduces GFP::DBL-1 accumulation (Lakdawala et al., 2019).
FIGURE 2:
GFP::DBL-1 accumulation responds to manipulation of cuticle collagen genes. (A) L4 animals. (B) Young adults. A strain carrying texIs100, which expresses functional GFP-tagged DBL-1, was treated with collagen RNAi or crossed with rol-6(lf) as indicated. Fluorescence intensity was measured in L4 (A) or 1-d-old adults (B). AU: arbitrary units. Each data point depicts the intensity in a single animal. Bars show mean and SD of the data. Statistical significance compared with control was determined by Brown–Forsythe and Welch ANOVA tests. *p < 0.05; **p < 0.01; ***p < 0.001.
Manipulation of DBL-1–regulated cuticle collagen genes suppresses long body size phenotype of dbl-1 overexpression
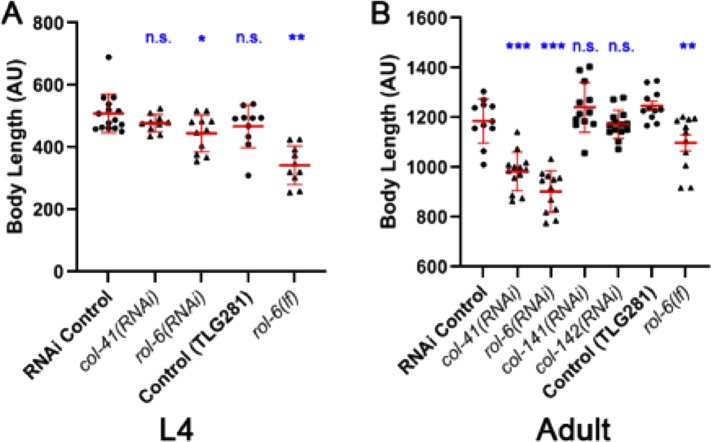
We previously showed that cuticle collagen genes are effectors of DBL-1 body size regulation (Madaan et al., 2018). In the previous study, using targeted RNAi, overexpression strains, and loss-of-function mutants, we found that col-41 is a positive regulator, rol-6 is a dose-dependent regulator, and col-141 is a negative regulator of body size (Madaan et al., 2018). To elucidate further how these cuticle collagens interact with the DBL-1 pathway in body length regulation, we capitalized on the strain described above in which overexpression of functional GFP-tagged DBL-1 results in a long body size. First, we examined the interaction of TLG281 (dbl-1(oe)) with RNAi targeting col-41 or rol-6, which cause small body size phenotypes in the wild type. If these genes act downstream of dbl-1 for body size regulation, we expect their depletion to suppress the long body size phenotype of dbl-1(oe). dbl-1(oe) animals fed on bacteria expressing col-41(RNAi) showed a significant reduction of their long body size in adult animals compared with our control (pseudogene C06C3.5 RNAi) (Figure 3B). We also observed that feeding rol-6(RNAi) caused a significant reduction in the long body size phenotype of dbl-1(oe) in L4 and adult animals (Figure 3, A and B). Finally, we examined rol-6(lf) mutants that result in reduced body length in wild-type animals (Kramer and Johnson, 1993; Madaan et al., 2018). In CS636 (rol-6(lf) expressing GFP::DBL-1), we observed a significant reduction of the long body size phenotype compared with dbl-1(oe) animals for both L4 and adult stages (Figure 3, A and B). Together, these results support our hypothesis that col-41 and rol-6 act downstream of DBL-1 to regulate body size.
FIGURE 3:
Genetic interactions between cuticle collagen genes and DBL-1 overexpression in body size regulation. (A) L4 animals. (B) Young adults. A strain carrying texIs100, a multicopy array that overexpresses GFP::DBL-1 causing a long body size phenotype, was treated with collagen RNAi or crossed with rol-6(lf) as indicated. Body length was measured in L4 (A) or 1-d-old adults (B). AU: arbitrary units. Each data point depicts the length of an individual animal. Bars show mean and SD of the data. Statistical significance compared with control was determined by Brown–Forsythe and Welch ANOVA tests. *p < 0.05; **p < 0.01; ***p < 0.001.
In a previous study, we showed that col-141(RNAi) causes an increase in body size, while overexpression of col-141 and col-142 reduces body size in wild-type animals (Madaan et al., 2018), suggesting that COL-141 and COL-142 negatively impact body length in C. elegans. We tested for genetic interactions between dbl-1(oe) and col-141(RNAi) or col-142(RNAi). If these genes act independently in body size regulation, then we would expect additive effects, with RNAi of col-141 or col-142 further increasing the long body size of dbl-1(oe). When col-141 or col-142 were depleted by RNAi in the dbl-1(oe) background, no further increase in body size was seen (Figure 3B). This result is consistent with these genes acting in the same pathway as DBL-1 to regulate body size. Combined with our previously published data (Madaan et al., 2018) and the results from the reporter constructs (Figures 1 and 2), these new results (Figure 3) provide evidence of bidirectional signaling between the DBL-1 pathway and cuticle collagen genes.
DBL-1–regulated collagens affect expression of a dbl-1 transcriptional reporter
We observed that the genetic manipulation of cuticle collagens caused a reduction in GFP::DBL-1 protein levels (Figures 1 and 2), but these experiments do not distinguish the level at which such regulation occurs. We considered two models: regulation of dbl-1 transcription and regulation of DBL-1 protein distribution or stability. To distinguish between these possibilities, we used a transgenic strain (BW1935) that expresses GFP driven by the dbl-1 promoter (dbl-1p::gfp) (Suzuki et al., 1999). We RNAi depleted each of these four DBL-1–regulated collagen genes in this strain and determined fluorescence intensity of the nuclear-localized GFP in ventral cord motor neurons in the middle 50% of the worm body axis. Remarkably, we found that loss of these cuticle components is associated with a significant reduction of the expression of the dbl-1p::gfp transcriptional reporter at both the L4 and the adult stages for all conditions tested (20–35% at L4 and 40–60% in adults; Figure 4). The large variability in Smad reporter activity and GFP::DBL-1 levels following col-41(RNAi) was not seen for the dbl-1 transcriptional reporter. These results suggest that cuticle collagen genes are capable of regulating DBL-1 at the level of transcription, an unexpected finding because of the physical separation of DBL-1–expressing neurons from the cuticle.
FIGURE 4:
Loss of cuticle collagen gene function reduces expression of a DBL-1 transcriptional reporter. (A) L4 animals. (B) Young adults. A strain carrying ctIs43, a transgene expressing GFP driven by the dbl-1 promoter, was treated with collagen RNAi as indicated. Fluorescence intensity was measured in L4 (A) or 1-d-old adults (B). AU: arbitrary units. Each data point depicts the intensity of an individual nucleus. Bars show mean and SD of the data. Statistical significance compared with control was determined by Brown–Forsythe and Welch ANOVA tests. *p < 0.05; **p < 0.01; ***p < 0.001.
To exclude the possibility that reduction of dbl-1p::gfp expression is due to a general effect on transgene expression under collagen RNAi conditions rather than a specific effect on the dbl-1 promoter, we analyzed the expression of an independent transgene under the same conditions. We analyzed the effects of collagen gene RNAi on an ectopic, hypodermally expressed GFP::DBL-1 construct (strain TLG633). While exposure to GFP-targeted RNAi significantly reduced GFP::DBL-1 fluorescence, exposure to col-41, rol-6, col-141, and col-142 RNAi caused no significant difference (Supplemental Figure S1). We therefore conclude that inhibition of collagen genes by RNAi does not cause a general reduction of transgene expression, nor does it act directly on GFP::DBL-1, but rather likely acts at the level of dbl-1 transcription.
Finally, to validate these RNAi results in altered collagen genetic backgrounds, we crossed the dbl-1 transcriptional reporter into three collagen mutant strains: rol-6(lf), col-141 col-142(oe), and a col-141(lf) deletion allele generated by CRISPR/Cas9 genome editing (Madaan et al., 2018). We quantified the GFP intensity from dbl-1p::gfp in young adults as a measure of dbl-1 transcription levels. Surprisingly, in rol-6(lf) mutants, GFP expression became undetectable (Figure 5B). Thus, loss of rol-6 has a major effect on dbl-1 expression. Similarly, col-141(lf) mutants have slightly reduced dbl-1 expression (Figure 5, C and E). In contrast, col-141 col-142(oe) animals have normal levels of dbl-1 reporter expression (Figure 5, D and E). These results from the rol-6(lf) and col-141(lf) mutant strains corroborate the findings from the respective RNAi experiments. We conclude that cuticle collagens participate in feedback regulation of DBL-1.
FIGURE 5:
DBL-1 transcriptional reporter expression in collagen mutant and overexpression strains. The transgene ctIs43, expressing GFP driven by the dbl-1 promoter, is visualized in A, wild-type; B, rol-6(lf); C, col-141(lf); and D, col-141 col-142(oe) 1-d-old adult hermaphrodites. (A–D) Representative GFP fluorescence images. (A’–D’) The same animals visualized with Nomarski DIC optics. Ventral is downward. (E) Fluorescence intensity was measured in 1-d-old adults. Fluorescence was completely undetectable in rol-6(lf) animals and could not be quantitated. AU: arbitrary units. Each data point depicts the intensity of an individual nucleus. Bars show mean and SD of the data. Statistical significance compared with control was determined by Brown–Forsythe and Welch ANOVA tests. ***p < 0.001.
DISCUSSION
In this work, we investigated the reciprocal interactions between the DBL-1/BMP pathway and the specific cuticle collagens, COL-41, ROL-6, COL-141, and COL-142. In previous work, we showed that col-41, rol-6, col-141, and col-142 are transcriptional targets of DBL-1 signaling (Madaan et al., 2018). They are positive (col-41), negative (col-141), or dose-sensitive (rol-6) regulators of body size (Madaan et al., 2018). Moreover, DBL-1 signaling mutants have altered cuticle morphology and function (Schultz et al., 2014). Here we extended those studies to provide evidence that these cuticle collagen genes act downstream of DBL-1 in body size regulation. We used three different measures of DBL-1 pathway signaling to demonstrate that these cuticle collagens, in turn, feed back on DBL-1 signaling. Interestingly, this postulated feedback regulation is consistent with a previous study that shows the ADAMTS-secreted metalloprotease ADT-2 modulates both cuticle collagen organization and DBL-1 signaling activity (Fernando et al., 2011).
These four DBL-1–regulated collagen genes can be divided into two groups based on temporal expression patterns and loss-of-function phenotypes. One group consists of col-41 and rol-6, which have a peak of expression in the second larval (L2) stage, and loss of either gene product causes a mild small body size phenotype (Kamath et al., 2003; Simmer et al., 2003; Madaan et al., 2018). DBL-1 signaling activates expression of these genes at the L2 stage and represses their expression in adults (Madaan et al., 2018). Loss of col-41 caused reduction in dbl-1 expression, GFP::DBL-1 protein fluorescence, and Smad reporter activity (Figures 1, 2, and 4). Strikingly, col-41(RNAi) consistently resulted in a large variation in Smad activity reporter and GFP::DBL-1 levels, and in some experiments this variation may have contributed to a lack of statistical significance. This variance was not seen in the level of the dbl-1 transcriptional reporter. One possible explanation is that in col-41(RNAi) animals, a compensatory mechanism occurs posttranscriptionally that restores DBL-1 protein levels in a subset of neurons. This intriguing observation invites further elucidation.
Loss of rol-6 had the most profound effects on DBL-1 signaling, causing significant reductions in expression of a Smad activity reporter, GFP::DBL-1 fusion protein, and a dbl-1 transcriptional reporter. C. elegans body size is also highly sensitive to the level of ROL-6. Reduction of the rol-6 transcript by rol-6(RNAi) or by a rol-6(lf) mutation reduces body size and significantly suppresses the long body size of dbl-1(oe) (Madaan et al., 2018; Figure 3). Furthermore, rol-6(oe) is also small, indicating a dose sensitivity for this gene product in body size regulation. These results extend ROL-6′s functions to include impacting DBL-1 signaling.
The second group comprises col-141 and col-142, which have a peak of expression in the adult stage (Jackson et al., 2014). Loss of col-141 causes a mild long body size phenotype. DBL-1 Smads directly regulate these genes, repressing expression at the L2 stage and activating their expression in adults (Madaan et al., 2018). In spite of the opposite functions of col-41 and rol-6 in body size determination, col-141 and col-142 act similarly to those genes in reducing DBL-1 signaling activity, as seen in three independent activity reporters (Figures 1, 2, and 4). Like col-141(RNAi), col-141(lf) reduced dbl-1 reporter transcription (Figures 4 and 5); however, col-141 col-142(oe), which has a small body size phenotype, does not affect the dbl-1 transcriptional reporter. These results dissociate the collagens’ effects on body length from DBL-1 pathway activity, so the effect of collagen loss on DBL-1 signaling likely depends on other properties of the cuticle in these backgrounds.
It is well established in various contexts that members of the TGF-β family, including BMPs, regulate ECM, with important consequences for biological function, which can be called “forward” regulation of the ECM (Zeisberg et al., 2007; Kim et al., 2018). Moreover, the ECM can participate in “reverse” or “feedback” regulation of TGF-β or BMP signaling. For example, type IV collagen controls BMP signaling in Drosophila by directly binding Dpp (Wang et al., 2008; Bunt et al., 2010; Sawala et al., 2012). In addition, the ECM protein fibrillin directly binds TGFβs and BMPs (Sengle et al., 2011). Mutations in FBN1 result in Marfan syndrome, for which some of the pathologies are attributed to increased TGF-β bioavailability (Neptune et al., 2003). Similarly, the interaction between fibrillin1 and BMP4 regulates BMP4 bioavailability (Nistala et al., 2010). Each of these interactions occurs via direct physical contact between ligands and the ECM. Our work, in contrast, supports a novel model of contact-independent regulation of BMP ligand expression and activity by the ECM (Figure 6). The cuticle forms the outermost barrier between the animal and its environment. Its components are synthesized by the underlying hypodermis and secreted to the apical (outer) surface. DBL-1 is secreted from neurons apposed to the basal (internal) side of the hypodermis. DBL-1–expressing cells are thus physically separated from the cuticle, making physical interactions between these cells and cuticle collagens unlikely.
FIGURE 6:
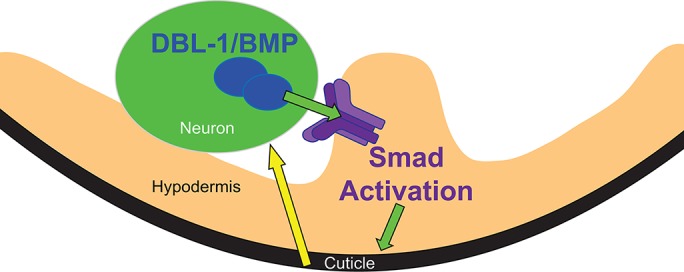
Model for DBL-1/BMP pathway regulation by cuticle collagens. Green arrows represent forward DBL-1 signaling that regulates cuticle collagen expression and body size. DBL-1/BMP is expressed by and secreted from neurons of the ventral cord. It binds to receptors on the hypodermis (purple complex) and activates Smad signaling. Smad activation causes changes in gene expression leading to altered collagen composition of the cuticle. Yellow arrow depicts feedback signaling from the cuticle to a DBL-1/BMP–expressing cell.
How can a contact-independent mechanism operate? Biomechanical properties of a tissue can influence gene expression. For example, mesenchymal stem cells commit to different differentiated lineages depending on matrix stiffness (Engler et al., 2006). In another example, compression of Drosophila embryos (which may mimic forces generated during morphogenetic movements) induces ectopic expression of the transcription factor Twist (Farge, 2003). Alterations of the C. elegans cuticle have also been shown to cause changes in signaling pathways in nonhypodermal tissues. For example, loss of some cuticle collagens alters widely expressed SKN-1/Nrf transcription factor activity and stress responses (Dodd et al., 2018). Interestingly, classical work on the GLP-1/Notch receptor in C. elegans identified mutations in seven cuticle collagen genes that suppress the pharyngeal and germline phenotypes of partial-loss-of-function glp-1 alleles (Maine and Kimble, 1989). They also found the severity of the body size defect associated with loss of collagen function did not correlate with glp-1 suppression. In this work, we show a similar effect on BMP signaling by loss of a different set of cuticle collagens. The reduction of DBL-1/BMP expression does not correlate with the alteration in body size in these collagen knockdowns. Specifically, RNAi inhibition of col-41 or rol-6 results in small animals, while RNAi against col-141 results in long animals, but each of these treatments reduces DBL-1 reporter activity. Therefore, we conclude that it is not the animal’s size but some other feature of the altered cuticle that is responsible for the feedback regulation.
Our model summarizes these observations (Figure 6). The neurons expressing DBL-1 may experience biomechanical forces, such as tension or compression, which modulate gene expression, including dbl-1 expression. Alterations in the collagen composition of the cuticle affect these forces, leading to changes in gene expression. This phenomenon may be a more widespread mechanism of contact-independent regulation of gene expression and cell signaling, for which the C. elegans cuticle may be a useful model. Future studies will identify the biomechanical or other properties of the C. elegans cuticle responsible for this proposed feedback. In summary, the DBL-1/BMP pathway provides a unique in vivo model to study bidirectional interactions between cell signaling and the ECM in the context of the intact organism. We propose that reciprocal interactions permit robust yet environmentally responsive control of body size.
MATERIALS AND METHODS
Strains
C. elegans strains were grown at 20°C using standard methods (Brenner, 1974). All experiments were performed at 20°C. In addition to strains generated for this work, the following strains were used: LW2436 jjIs2436[pCXT51(RAD-SMAD) + LiuFD61(mec-7p::mRFP)] (Tian et al., 2010), TLG281 rrf-3(pk1426); texIs100[dbl-1p::GFP::dbl-1 + ttx-3p::mRFP]; dbl-1(nk3) (Beifuss and Gumienny, 2012), TLG633 texIs121[rol-6p::GFP::dbl-1 + ttx-3p::GFP], BW1935 unc-119(ed3); ctIs43[dbl-1p::GFP + unc-119(+)]; him-5(e1490) (Suzuki et al., 1999), MT2709 rol-6(e187n1270) (a.k.a rol-6(lf)) (Kramer and Johnson, 1993), CS643 qcEx133[rol-6(+) + myo-2p::GFP] (a.k.a. rol-6(oe), CS678 col-141(qc25[2xNLS::GFP]) (a.k.a. col-141(lf)), and CS637 qcEx131[col-141(+) col-142(+) + myo-2p::GFP] (a.k.a. col-141 col-142(oe)) (Madaan et al., 2018).
RNAi
RNAi by feeding was performed as described (Kamath et al., 2003; Beifuss and Gumienny, 2012; Liang et al., 2013). RNAi clones that target the unique (non-Gly-X-Y repeat) regions of collagen genes were generated by cloning PCR fragments encoding these regions into the L4440 vector and transforming bacterial strain HT115 (primer sequences are available on request) (Madaan et al., 2018).
RAD-SMAD reporter, dbl-1 transcriptional reporter, and ectopic GFP::DBL-1 imaging
Fluorescence images were taken with a Zeiss AxioCam MRm camera on a Zeiss Apotome microscope using a 40× objective. For fluorescence quantification, images were taken at 40×. For RAD-SMAD and dbl-1p::gfp reporters, fluorescence intensity was determined by manually selecting each individual nucleus and quantifying intensity using ImageJ software. For the RAD-SMAD reporter, we analyzed hypodermal nuclei. For the dbl-1 transcriptional reporter, we analyzed ventral cord motor neuron nuclei. For the hypodermal GFP::DBL-1 transgene, we selected one or two equivalent regions of interest displaying GFP fluorescence in each animal and quantified fluorescence intensity. In each case, we avoided the head and tail regions. Empty vector (L4440) RNAi was used as a control. Each experiment was performed in two or three trials, and representative trials are shown. In addition to the parent reporter strains, crosses were performed to generate CS657 ctIs43; rol-6(e187n1270), CS675 ctIs43; col-141(qc25), and CS677 ctIs43; qcEx131.
Body size measurements and GFP::DBL-1 imaging
Populations of TLG281 rrf-3(pk1426); dbl-1(nk3); texIs100 animals were staged by a standard bleaching technique and grown overnight at 20°C in M9 to hatch and arrest at the L1 stage (Stiernagle, 2006). Starved hatchlings were plated on bacteria expressing double-stranded RNAi against each gene of interest and grown at 20°C for 5 d, until all animals were young adults. RNAi against the unrelated pseudogene C06C3.5 was used as a control. In addition to the RNAi treatments, the rol-6(e187n1270) allele was crossed with texIs100 to generate CS636 rol-6(e187n1270); texIs100.
Adult animals were immobilized with 0.2 mM levamisole in M9 buffer on a 10% agarose pad before imaging (Fang-Yen et al., 2012). Confocal images were acquired using either a Nikon A1R confocal system or a Nikon Eclipse Ti-E confocal system. We used a 60×/1.40 NA oil CFI Plan Apo VC objective and NIS-Elements Advanced Research software (Nikon Instruments, Melville, NY) to measure and quantify the intensity of the GFP::DBL-1 protein under different conditions. The intensity of the GFP-tagged protein for a selected area was calculated by calculating the mean intensity associated with this area minus the mean intensity of the background. Body length was measured using 10× magnification and NIS AR software.
Supplementary Material
Acknowledgments
We thank Jun (Kelly) Liu for providing strains and helpful conversations. We thank lab members and Alicia Meléndez for helpful feedback on the manuscript. We are grateful to the Chalfie lab for providing comments on the preprint version of the manuscript. This work was supported in part by National Institutes of Health (NIH) R15GM112147 to C.S.D. and R01GM097591 to T.L.G., and Texas Woman’s University (TWU) departmental and TWU Office of Research and Sponsored Programs funds to T.L.G. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40OD010440). This work was carried out in partial fulfillment of the requirements for the Ph.D. degree from The Graduate Center of City University of New York (U.M. and E.J.C.).
Abbreviations used:
- BMP
bone morphogenetic protein
- ECM
extracellular matrix
- RNAi
RNA interference
- TGF-β
transforming growth factor beta.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-07-0390) on February 12, 2020.
REFERENCES
- Beifuss KK, Gumienny TL. (2012). RNAi screening to identify postembryonic phenotypes in C. elegans . J Vis Exp e3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans . Genetics , 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunt S, Hooley C, Hu N, Scahill C, Weavers H, Skaer H. (2010). Hemocyte-secreted type IV collagen enhances BMP signaling to guide renal tubule morphogenesis in Drosophila. Dev Cell , 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. (2016). Agonists and antagonists of TGF-β family ligands. Cold Spring Harb Perspect Biol , a021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm AD, Hsiao TI. (2012). The Caenorhabditis elegans epidermis as a model skin. I: development, patterning, and growth. Wiley Interdiscip Rev Dev Biol , 861–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm AD, Xu S. (2012). The Caenorhabditis elegans epidermis as a model skin. II: differentiation and physiological roles. Wiley Interdiscip Rev Dev Biol , 879–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd W, Tang L, Lone JC, Wimberly K, Wu CW, Consalvo C, Wright JE, Pujol N, Choe KP. (2018). A damage sensor associated with the cuticle coordinates three core environmental stress responses in Caenorhabditis elegans . Genetics , 1467–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. (2006). Matrix elasticity directs stem cell lineage specification. Cell , 677–689. [DOI] [PubMed] [Google Scholar]
- Fang-Yen C, Gabel CV, Samuel AD, Bargmann CI, Avery L. (2012). Laser microsurgery in Caenorhabditis elegans. Methods Cell Biol , 177–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farge E. (2003). Mechanical induction of Twist in the Drosophila foregut/stomodeal primordium. Curr Biol , 1365–1377. [DOI] [PubMed] [Google Scholar]
- Fernando T, Flibotte S, Xiong S, Yin J, Yzeiraj E, Moerman DG, Melendez A, Savage-Dunn C. (2011). C. elegans ADAMTS ADT-2 regulates body size by modulating TGFβ signaling and cuticle collagen organization. Dev Biol , 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman-Rubinsky R, Cohen JD, Sundaram MV. (2017). Lipocalins are required for apical extracellular matrix organization and remodeling in Caenorhabditis elegans . Genetics , 625–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny TL, Savage-Dunn C. (2013). TGF-β signaling in C. elegans. In: WormBook: the Online Review of C elegans Biology, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson BM, Abete-Luzi P, Krause MW, Eisenmann DM. (2014). Use of an activated β-catenin to identify Wnt pathway target genes in Caenorhabditis elegans, including a subset of collagen genes expressed in late larval development. G3 , 733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature , 231–237. [DOI] [PubMed] [Google Scholar]
- Kim KK, Sheppard D, Chapman HA. (2018). TGF-β1 signaling and tissue fibrosis. Cold Spring Harb Perspect Biol , a022293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JM. (2005). Basement membranes. In: WormBook: The Online Review of C elegans Biology, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JM, Johnson JJ. (1993). Analysis of mutations in the sqt-1 and rol-6 collagen genes of Caenorhabditis elegans . Genetics , 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakdawala MF, Madhu B, Faure L, Vora M, Padgett RW, Gumienny TL. (2019). Genetic interactions between the DBL-1/BMP-like pathway and dpy body size-associated genes in Caenorhabditis elegans . Mol Biol Cell , 3151–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Xiong S, Savage-Dunn C. (2013). Using RNA-mediated interference feeding strategy to screen for genes involved in body size regulation in the nematode C. elegans . J Vis Exp , 4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Yu L, Yin J, Savage-Dunn C. (2007). Transcriptional repressor and activator activities of SMA-9 contribute differentially to BMP-related signaling outputs. Dev Biol , 714–725. [DOI] [PubMed] [Google Scholar]
- Luo S, Kleemann GA, Ashraf JM, Shaw WM, Murphy CT. (2010). TGF-β and insulin signaling regulate reproductive aging via oocyte and germline quality maintenance. Cell , 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaan U, Yzeiraj E, Meade M, Clark JF, Rushlow CA, Savage-Dunn C. (2018). BMP signaling determines body size via transcriptional regulation of collagen genes in Caenorhabditis elegans. Genetics , 1355–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine EM, Kimble J. (1989). Identification of genes that interact with glp-1, a gene required for inductive cell interactions in Caenorhabditis elegans . Development , 133–143. [DOI] [PubMed] [Google Scholar]
- Moss-Taylor L, Upadhyay A, Pan X, Kim MJ, O’Connor MB. (2019). Body size and tissue-scaling is regulated by motoneuron-derived activins in Drosophila melanogaster . Genetics , 1447–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neptune ER, Frischmeyer PA, Arking DE, Myers L, Bunton TE, Gayraud B, Ramirez F, Sakai LY, Dietz HC. (2003). Dysregulation of TGF-β activation contributes to pathogenesis in Marfan syndrome. Nat Genet , 407–411. [DOI] [PubMed] [Google Scholar]
- Nistala H, Lee-Arteaga S, Smaldone S, Siciliano G, Carta L, Ono RN, Sengle G, Arteaga-Solis E, Levasseur R, Ducy P, et al (2010). Fibrillin-1 and -2 differentially modulate endogenous TGF-β and BMP bioavailability during bone formation. J Cell Biol , 1107–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AP, Johnstone IL. (2007). The cuticle. In: WormBook: The Online Review of C elegans Biology, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AF, Gumienny TL, Gleason RJ, Wang H, Padgett RW. (2010). Regulation of genes affecting body size and innate immunity by the DBL-1/BMP-like pathway in Caenorhabditis elegans . BMC Dev Biol , 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawala A, Sutcliffe C, Ashe HL. (2012). Multistep molecular mechanism for bone morphogenetic protein extracellular transport in the Drosophila embryo. Proc Natl Acad Sci USA , 11222–11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz RD, Bennett EE, Ellis EA, Gumienny TL. (2014). Regulation of extracellular matrix organization by BMP signaling in Caenorhabditis elegans . PLoS One , e101929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengle G, Ono RN, Sasaki T, Sakai LY. (2011). Prodomains of transforming growth factor beta (TGFβ) superfamily members specify different functions: extracellular matrix interactions and growth factor bioavailability. J Biol Chem , 5087–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH. (2003). Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol , E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T. (2006). Maintenance of C. elegans. In: WormBook: The Online Review of C elegans Biology, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Yandell MD, Roy PJ, Krishna S, Savage-Dunn C, Ross RM, Padgett RW, Wood WB. (1999). A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans . Development , 241–250. [DOI] [PubMed] [Google Scholar]
- Tian C, Sen D, Shi H, Foehr ML, Plavskin Y, Vatamaniuk OK, Liu J. (2010). The RGM protein DRAG-1 positively regulates a BMP-like signaling pathway in Caenorhabditis elegans. Development , 2375–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Harris RE, Bayston LJ, Ashe HL. (2008). Type IV collagens regulate BMP signalling in Drosophila. Nature , 72–77. [DOI] [PubMed] [Google Scholar]
- Wang J, Tokarz R, Savage-Dunn C. (2002). The expression of TGFβ signal transducers in the hypodermis regulates body size in C. elegans . Development , 4989–4998. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Morita K, Mochii M, Ueno N. (2001). Hypodermal expression of Caenorhabditis elegans TGF-β type I receptor SMA-6 is essential for the growth and maintenance of body length. Dev Biol , 32–45. [DOI] [PubMed] [Google Scholar]
- Zeisberg EM, Tarnavski O, Zeisberg M, Dorfman AL, McMullen JR, Gustafsson E, Chandraker A, Yuan X, Pu WT, Roberts AB, et al (2007). Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med , 952–961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.