Abstract
Collective cell migration plays crucial roles in tissue remodeling, wound healing, and cancer cell invasion. However, its underlying mechanism remains unknown. Previously, we showed that the RhoA-targeting guanine nucleotide exchange factor Solo (ARHGEF40) is required for tensile force–induced RhoA activation and proper organization of keratin-8/keratin-18 (K8/K18) networks. Here, we demonstrate that Solo knockdown significantly increases the rate at which Madin-Darby canine kidney cells collectively migrate on collagen gels. However, it has no apparent effect on the migratory speed of solitary cultured cells. Therefore, Solo decelerates collective cell migration. Moreover, Solo localized to the anteroposterior regions of cell–cell contact sites in collectively migrating cells and was required for the local accumulation of K8/K18 filaments in the forward areas of the cells. Partial Rho-associated protein kinase (ROCK) inhibition or K18 or plakoglobin knockdown also increased collective cell migration velocity. These results suggest that Solo acts as a brake for collective cell migration by generating pullback force at cell–cell contact sites via the RhoA-ROCK pathway. It may also promote the formation of desmosomal cell–cell junctions related to K8/K18 filaments and plakoglobin.
INTRODUCTION
Collective cell migration plays important roles in various pathophysiological processes such as developmental tissue morphogenesis, epithelial wound healing, and cancer cell invasion (Friedl and Gilmour, 2009; Friedl et al., 2012; Rorth, 2009; Haeger et al., 2015). When epithelial cells assemble as a sheet, one of them on the edge is transformed into a leader cell and migrates to the exterior along with the cells inside that follow it. The protrusion of collectively migrating cells forms a finger-like structure and moves in a particular direction at a nearly constant speed (Yamaguchi et al., 2015; Mayor and Etienne-Manneville, 2016b). The movement of cells in collective cell migration differs from that of solitary migrating cells. Unlike the latter, the former is regulated by cell–cell contacts including adherens junctions and desmosomes. Synchronization of the direction and speed of collectively migrating cells appear to be regulated by mechanical forces generated at these points of contact (Rorth, 2009; Tambe et al., 2011; Li et al., 2012; Weber et al., 2012; Mayor and Etienne-Manneville, 2016b). Thus, the coordinated nature of collective migration relies heavily on the activity of cell–cell adhesion sites. Nevertheless, its underlying mechanism remains unknown.
Actin cytoskeletal reorganization is vital for cell motility, migration, and contractile force formation. It regulates forces at cell–cell contact sites and coordinates the cell population in collective cell migration (Jaffe and Hall, 2005; Charras and Sahai, 2014; Zegers and Friedl, 2014; van Helvert et al., 2018). The leader cell at the head of the finger-like structure spreads lamellipodia, migrates to the free edge, and pulls the follower cells. The cell–cell contacts linked to the actin cytoskeleton are subjected to the pulling forces of the leader cells. These forces reorganize the actin cytoskeletal structures and coordinate migration polarity and speed in collective cell migration (Weber et al., 2012; Haeger et al., 2015; Hayer et al., 2016; Mayor and Etienne-Manneville, 2016b; van Helvert et al., 2018). Reorganization of the actin cytoskeleton is regulated by Rho family small GTPases (Jaffe and Hall, 2005), which are encoded by ∼20 genes in the human genome. They form specific actin structures with the help of their respective downstream effectors (Heasman and Ridley, 2008). To construct appropriate high-order actin structures in response to various stimuli, Rho GTPases must be strictly regulated by upstream signaling molecules. About 80 Rho-guanine nucleotide exchange factors (Rho-GEFs) are encoded in the human genome. They may regulate Rho GTPases with spatiotemporal specificity in response to various external stimuli (Cook et al., 2014). We previously showed that Solo (ARHGEF40), a Dbl-related Rho-GEF, participates in cyclic stretch–induced endothelial cell orientation and tensile force–induced RhoA activation via cell–cell adhesions (Abiko et al., 2015). We also demonstrated that Solo binds to keratin-8/keratin-18 (K8/K18) filaments. This interaction is required for force-induced RhoA activation and stress fiber formation (Fujiwara et al., 2016, 2019; Ohashi et al., 2017). Thus, it is predicted that Solo is involved in cell responses to mechanical forces through cell–cell adhesions (Abiko et al., 2015).
In addition to actin filaments, intermediate filaments (IFs) form another major cytoskeleton with many subtypes in vertebrates. IFs function in cellular resistance to external forces. They are anchored to the desmosomes at cell–cell adhesion sites and to hemidesmosomes at the basal areas of cells where they stabilize epidermal and epithelial tissues (Coulombe and Wong, 2004; Herrmann et al., 2007). Defects in desmosomal proteins and keratin filaments impair tissue integrity and underlie various skin and heart diseases (Herrmann et al., 2007; Eriksson et al., 2009; Broussard et al., 2015; Toivola et al., 2015). Weber and colleagues reported that K8/K18 and plakoglobin accumulate at the cell–cell adhesion sites subjected to the pulling force and participate in the migration of Xenopus early embryo mesendoderm cells in a direction opposite to that of the pulling force (Weber et al., 2012). Therefore, IFs and desmosomal proteins seem to regulate the mechanical force generation and harmonized motility of cells during collective migration by tight coordination with the regulation of actin cytoskeletal remodeling (Sanghvi-Shah and Weber, 2017).
In the present study, we show that Solo knockdown accelerates collective cell migration but does not affect the migration velocity of solitary cultured cells. We provide evidence that Solo maintains RhoA activity and the proper organization of K8/K18 filaments in collectively migrating epithelial cells. We also demonstrate that partial ROCK inhibition or K18 or plakoglobin knockdown accelerates collective cell migration. Our results suggest that Solo serves as a brake for collective cell migration by regulating the RhoA-ROCK signaling pathway and desmosomal cell–cell adhesions.
RESULTS
Solo knockdown accelerates collective migration
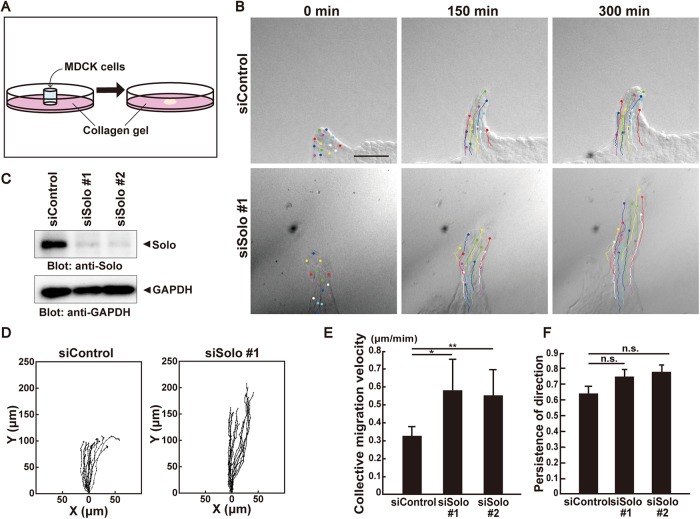
When Madin-Darby canine kidney (MDCK) cells were cultured on soft collagen gel and formed a sheet, one of the cells at the edge was converted to a leader cell and migrated to the exterior where it was followed by some of the interior cells (Figure 1, A and B, and Supplemental Movie 1). The cell population protruding from the sheet formed a finger-like shape. These cells migrated in a particular direction at nearly constant speed (Haga et al., 2005; Yamaguchi et al., 2015). To investigate the role of Solo in collective cell migration, we performed time-lapse imaging of the finger-like protrusion of MDCK cells transfected with either control or Solo siRNAs. Recordings were made every 10 min for 5 h (Figure 1B and Supplemental Movie 1). Immunoblot analysis revealed that endogenous Solo was markedly down-regulated in MDCK cells transfected with two independent Solo-targeting small interfering RNA (siRNAs; Figure 1C). Both control and Solo-knockdown leader cells led followers in the same manner (Supplemental Movie 1). To analyze the migration velocity and the persistence of direction of individual cells, we traced the trajectories of the nuclei of the follower cells in the finger-like protrusions (Figure 1D). Collective cell migration speed was significantly increased by treatment with two independent Solo-targeting siRNAs. The average migration velocities of the control and Solo-knockdown cells were 0.32 and 0.57 μm min−1, respectively (Figure 1E). On the other hand, Solo knockdown had no significant effect on the persistence of direction of cell migration measured by the ratio of the net translocation distance to the cumulative length of the migration path (Figure 1, D and F). These results suggest that Solo retards the collective migration of MDCK cells.
FIGURE 1:
Solo knockdown accelerates collective cell migration. (A) Schematic representation of the collective cell migration assay on collagen gel. For further details, see Materials and Methods. (B) Differential interference contrast (DIC) images of finger-like protrusions comprised of control or Solo siRNA-transfected MDCK cells at 0, 150, and 300 min after the start of observation. Trajectories of individual cells were overlaid. Scale bar = 100 μm. (C) Effect of Solo knockdown on endogenous Solo expression in MDCK cells. Cells were transfected with control or Solo-targeting siRNAs and incubated for 48 h. Cells were harvested, lysed, and analyzed by immunoblotting with anti-Solo antibody. (D) Trajectories of individual cells in the protrusions in B. Cells were tracked every 10 min for 5 h. Intersection of the x- and y-axes was taken as the position of the cell nucleus at the starting point. (E) Velocities of the collective migration of the control and Solo-knockdown cells are shown as average velocities of the tracked cells in each condition. (F) Persistence of direction of individual migrating cells in the control and Solo-knockdown protrusions is represented as the distance between the starting and ending points divided by the length of trajectory. A value close to 1 indicates that the cells maintain their direction and positional relation to the rest of the cell population during migration. Cell tracks are representative of four independent experiments. In E and F, data are means ± SD of three independent experiments (≥10 cells/experiment). *, P < 0.05; **, P < 0.01 (one-way ANOVA followed by Dunnett’s test).
Movie S1.
Time-lapse observation of finger-like protrusions comprising control or Solo siRNA-transfected MDCK cells on collagen gel. Images were obtained every 10 min. Trajectories of individual cells were overlaid. This movie corresponds to Fig. 1B. showing finger-like protrusions comprising control- or Solo siRNA-transfected MDCK cells.
Solo knockdown has no effect on solitary cultured cell migration
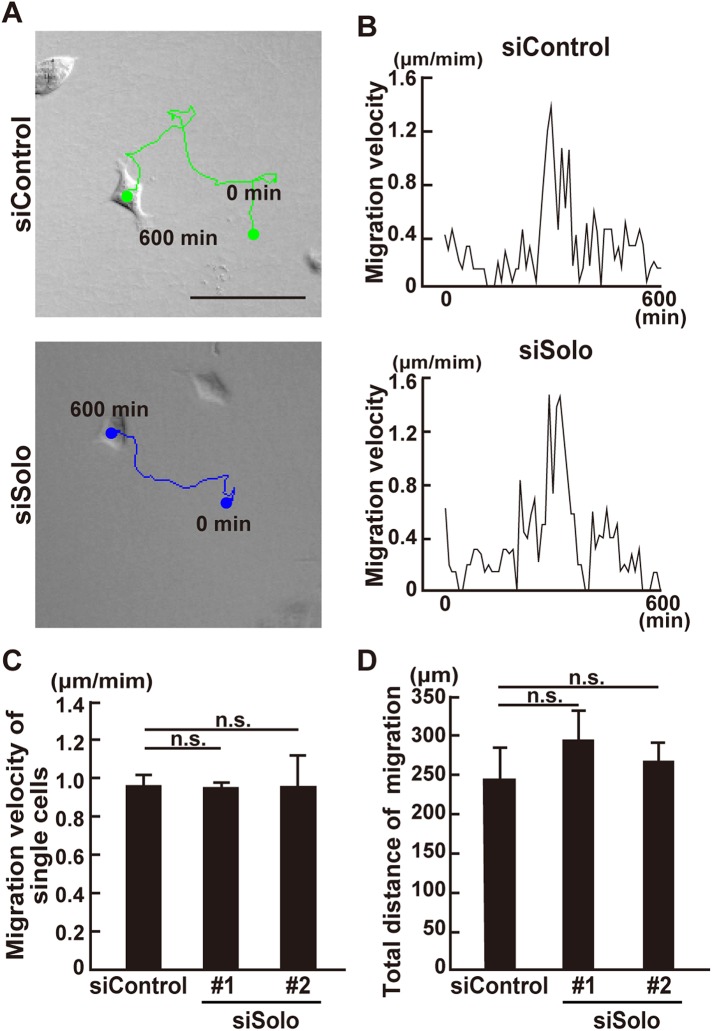
The increase in the velocity of collective cell migration may be the result of 1) the increase in the migration speed of the individual cells, 2) the proper alignment of the polarity of the individual cells in the protrusions, and 3) the reduction of the opposing forces at the cell–cell contact sites. To determine the reason for the increase in the velocity of the collective migration of Solo-knockdown cells, we analyzed the effect of Solo knockdown on the migration speed of solitary cultured MDCK cells. Cells were treated either with control or Solo siRNAs and cultured on collagen gel at low cell density to minimize cell–cell contact. Cell locomotion was then analyzed by time-lapse imaging (Figure 2A and Supplemental Movie 2). Migration velocity and frequency of the solitary cultured cells were measured by tracing individual cell trajectories (Figure 2, A and B). Solitary cultured cell migration was discontinuous. Both control and Solo-knockdown cells started and stopped at random time intervals (Figure 2B). Thus, we selected cells that continuously migrated at >0.5 μm min−1 for >30 min and analyzed their migration velocity (Figure 2C). We also measured their total migration distance for 10 h to estimate cell locomotion frequency. The migration velocity and total distance of the solitary cultured Solo-knockdown cells did not significantly differ from those of the control cells (Figure 2, C and D). Therefore, the Solo knockdown–induced increase in collective cell migration velocity was not likely the result of the increase in individual cell migration velocity. The average migration velocity of the solitary cultured control cells (0.95 μm min−1) was approximately three times greater than that of the collectively migrating control cells (0.32 μm min−1; Figures 1E and 2C). Thus, the collective cell migration speed was decreased.
FIGURE 2:
Effect of Solo knockdown on solitary MDCK cell motility. (A) DIC images of control and Solo-knockdown migrating solitary MDCK cells on collagen gel at the last frame. Cells were transfected with control or Solo-targeting siRNAs, incubated for 24 h, and seeded at low cell density on collagen gel. After overnight incubation, the cells were observed every 10 min for 10 h. Positions of the nuclei were tracked and their trajectories were overlaid. Scale bar = 100 μm. (B) Lengths of the cell migration paths during each 10 min period. (C) Migration velocities of control and Solo-knockdown solitary cells. Values were calculated by averaging the velocities when the cells migrate at >0.5 μm min−1 for >30 min. (D) Total migratory distance of the control and Solo-knockdown solitary cells for 10 h. Data are means ± SD of three independent experiments (8–26 cells/experiment). n. s., not significant (one-way ANOVA followed by Dunnett’s test).
Movie S2.
Time-lapse observation of control or Solo siRNA-transfected solitary cultured cells on collagen gel. Images were obtained every 10 min. Trajectories of the cells were overlaid. This movie corresponds to Fig. 2A.
Solo knockdown has no effect on migration polarity or leader cell characteristics during collective cell migration
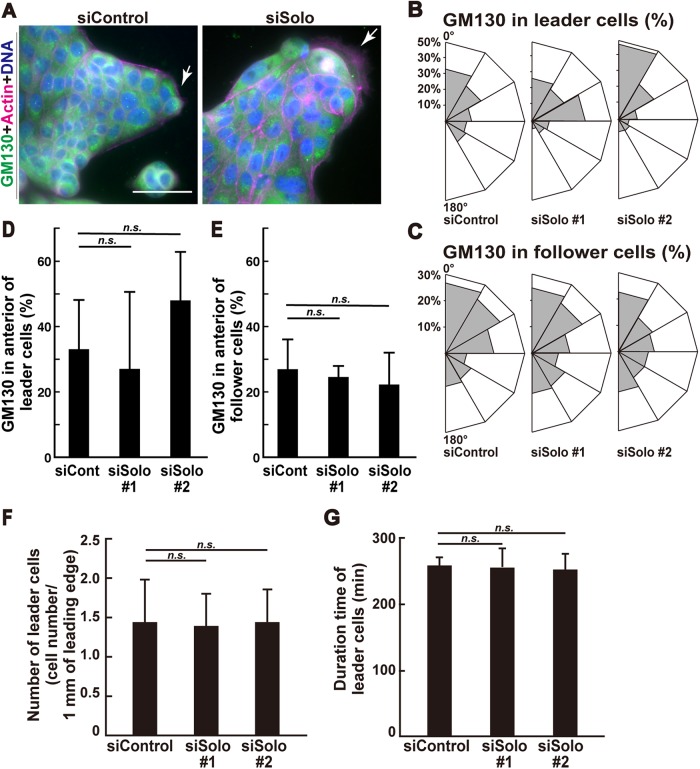
Alignment of migrating cell polarity is important for the determination of collective cell migration velocity (Chiapparo et al., 2016; Plutoni et al., 2016). To identify migration polarity direction in individual cells, we stained the Golgi apparatus marker GM130, which is usually located in front of the nuclei of migrating cells (Bershadsky and Futerman, 1994). The migration polarities of control and Solo-knockdown leader cells (arrows in Figure 3A) and follower cells in the protrusions were analyzed. As previously reported (Reffay et al., 2011), GM130 in the control leader and follower cells was polarized to the anterior direction (Figure 3, B and C) but the polarization of the follower cells was comparatively weak. Similar results were obtained for the Solo-knockdown cells. Solo knockdown had no significant effect on leader or follower cell migration polarity in collective migration (Figure 3, D and E).
FIGURE 3:
Effects of Solo knockdown on cell polarity and leader cell characteristics in the protrusions. (A) Immunofluorescence images of F-actin, Golgi apparatus, and nuclei in control and Solo-knockdown cells in finger-like protrusions. Collectively migrating cells on collagen gel were fixed and stained with anti-GM130 antibody for Golgi (green), Alexa Fluor 568–phalloidin for F-actin (red), and DAPI for nucleus (blue). Arrows indicate leader cells. Scale bar = 50 μm. (B, C) Angular distributions showing percentage of control or Solo-knockdown leader cells (B) or follower cells (C) whose Golgi apparatus were located within six equal sections (indicated as the total percentage of the left and right sides). Migration direction is indicated as 0°. (D, E) Percentage of leader cells (D) or follower cells (E) whose Golgi apparatus were located within 30° of the front sides in B and C. (F) Number of leader cells within 1 mm length of the edge around the control or Solo-knockdown protrusions. (G) Duration of leader cells in the control or Solo-knockdown protrusions. Life spans of leader cells were measured by time-lapse observation. Data are means ± SD of three independent experiments (48–78 cells/experiment). n. s., not significant (one-way ANOVA followed by Dunnett’s test).
The generation frequency and life span of leader cells may also influence collective cell migration velocity. Therefore, we evaluated the frequency of the conversion of peripheral cells to leader cells in the protrusion over 5 h as well as the life span of the leader cells. There were no significant differences between the control and Solo-knockdown cells in terms of the conversion frequency and life span of the leader cells (Figure 3, F and G). Therefore, the increase in collective cell migration velocity caused by Solo knockdown is not the result of aligning the migrating polarity or increasing the conversion frequency or life span of the leader cells.
Polarized Solo localization at the cell–cell contact sites in collectively migrating cells
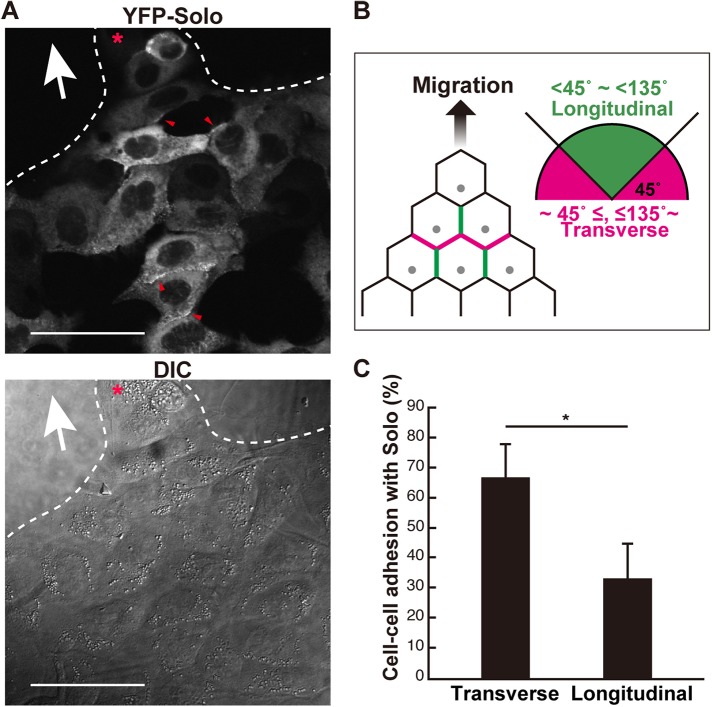
We previously showed that Solo is unevenly distributed in the cell–cell contact sites of endothelial cells (Abiko et al., 2015). Therefore, we investigated whether Solo localization in the cell–cell contact regions is associated with the migration polarity of collectively migrating cells. To this end, we used MDCK cells constitutively expressing yellow fluorescence protein (YFP)–Solo. To identify the position of Solo relative to the migration direction, we categorized the localization of Solo in the cell–cell contact sites (Figure 4A, red arrowheads) into positions transverse and longitudinal to the direction of cell migration (Figure 4B). Solo was significantly located at the anterior and posterior cell–cell contact regions but not at those at the lateral ends (Figure 4C). Thus, Solo localizes primarily at the cell–cell contact regions that lie transverse to the direction of cell migration. In these regions, Solo may be involved in the generation of intercellular contractile force. The pulling force derived from the backward-facing cell may be reversely oriented against the traction force of the forward cells. In this way, the collective migration speed may be reduced.
FIGURE 4:
Polarized Solo localization at the cell–cell contact sites during collective cell migration. (A) DIC and confocal microscopic fluorescence images of YFP-Solo–expressing MDCK cells. Collectively migrating YFP-Solo–expressing MDCK cells were fixed and the localization of YFP-Solo at the cell–cell contact sites was visualized in a single plane of cross-sectional images. Red arrowheads indicate Solo accumulation at the cell–cell contact sites. The red asterisk indicates the leader cell and the white arrow indicates the putative migration direction. Scale bars = 50 μm. (B) The longitudinal contact site was defined as its angle >45° and <135° relative to the migration direction (green lines). The transverse contact site is indicated by magenta lines. (C) The graph shows the rates of the transverse and longitudinal contact sites wherein YFP-Solo accumulated. Data are means ± SD of three independent experiments (25–47 cells/experiment in B). *, P < 0.05 (one-way ANOVA followed by Dunnett’s test).
We also investigated the localization of a Ser-18 and Ser-19 dual-phosphorylated myosin light chain (ppMLC), which is a maker of tensile force–subjected actomyosin, and RhoA at the cell–cell contact sites in collectively migrating cells (Supplemental Figure S1). RhoA was located almost equally at all cell–cell contact sites and partially colocalized with Solo; however, it was not accumulated in the cell–cell contact sites where Solo was accumulated (Supplemental Figure S1A). Alternatively, ppMLC was located as puncta at the cell–cell contact sites where Solo was accumulated (Supplemental Figure S1B).
Partial ROCK inhibition increases collective cell migration velocity
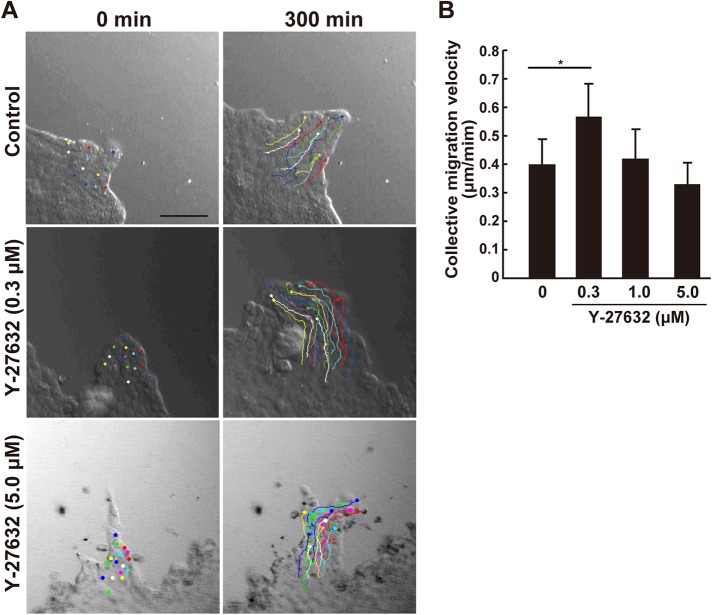
We hypothesized that the increase in collective migration velocity by Solo knockdown is caused by a reduction in the pullback forces from the rearward cells. Therefore, we examined whether a decrease in the myosin-dependent contractile forces accelerates collective MDCK cell migration. Accordingly, MDCK cells were exposed to various concentrations of Y-27632, an inhibitor of Rho-associated protein kinase (ROCK). The collective migratory speed was determined to be slightly lower following treatment with 5 μM Y-27632; while it was significantly accelerated by 0.3 μM Y-27632 (Figure 5, A and B, and Supplemental Movie 3). It seems likely that the low dose of Y-27632 accelerated collective cell migration by partially inhibiting ROCK activity and reducing the ROCK-induced pullback force at the cell–cell contact sites, whereas the high dose of Y-27632 decelerated collective migration by more severely inhibiting ROCK and ROCK-induced cellular activities associated with cell migration. Thus, myosin-dependent contractile force likely acts as a brake for collective cell migration. Accordingly, the increase in migration speed due to partial inhibition of myosin activity may represent a decrease in pullback force. Furthermore, as Solo targets RhoA, our data suggest that Solo reduces collective cell migration velocity by generating a pullback force via RhoA activation.
FIGURE 5:
Partial ROCK inhibition accelerates collective cell migration. Dose-dependent effect of Y-27632 on collective MDCK cell migration velocity. (A) DIC images of migrating cell protrusion in the presence of the indicated Y-27632 concentration at the start (0 min) and end (300 min) of the time-lapse observation. Trajectories of the individual cells were overlaid. Cells were tracked every 5 min for 5 h. Scale bar = 100 μm. (B) Velocities of the collective cell migration are shown as average velocities of the tracked cells in each condition. Data are means ± SD of three independent experiments (29–32 cells/experiment). *, P < 0.05 (one-way ANOVA followed by Dunnett’s test).
Movie S3.
Time-lapse observation of finger-like protrusions comprising MDCK cells treated with the indicated dose of Y-27632. Images were obtained every 5 min. Trajectories of the cells were overlaid. This movie corresponds to Fig. 5A.
Effects of Solo knockdown on RhoA and Rac1 activity
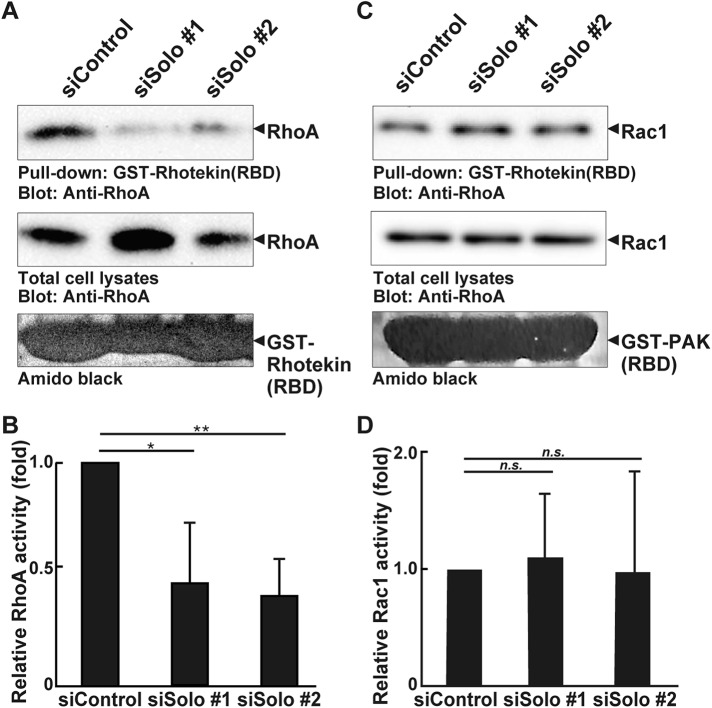
We investigated the contribution of Solo to cellular RhoA activity by active RhoA pulldown assays. We used the glutathione S-transferase (GST)-fused RhoA-binding domain of rhotekin (Figure 6, A and B). The active form of RhoA was decreased in the Solo siRNA-transfected MDCK cells relative to that of the control siRNA-transfected cells. RhoA down-regulation may increase Rac activity (Sander et al., 1999; Raftopoulou and Hall, 2004). To determine whether Solo knockdown activates Rac1, we also evaluated the latter by GST pull-down assays using the GST-fused Rac-binding domain of PAK. Solo knockdown had no significant effect on Rac1 activity in MDCK cells (Figure 6, C and D). These results suggest that Solo knockdown accelerates collective cell migration by reducing the activity of RhoA but not that of Rac1.
FIGURE 6:
Effects of Solo knockdown on RhoA and Rac1 activity in MDCK cells. (A, C) Control and Solo siRNA-transfected MDCK cells were incubated for 48 h until they formed a cell sheet. The cells were then lysed. Active RhoA and Rac1 were analyzed by GST pull-down assays using GST-rhotekin (RBD) and GST-PAK3 (PBD), respectively. (B, D) Relative RhoA and Rac1 activities are shown. The value of the control cells is taken as 1.0. Each value is the mean ± SD of four independent experiments. *, P < 0.05; **, P < 0.01; n.s., not significant (one-way ANOVA followed by Dunnett’s test).
Solo is required for the polarized localization of keratin filaments during collective cell migration
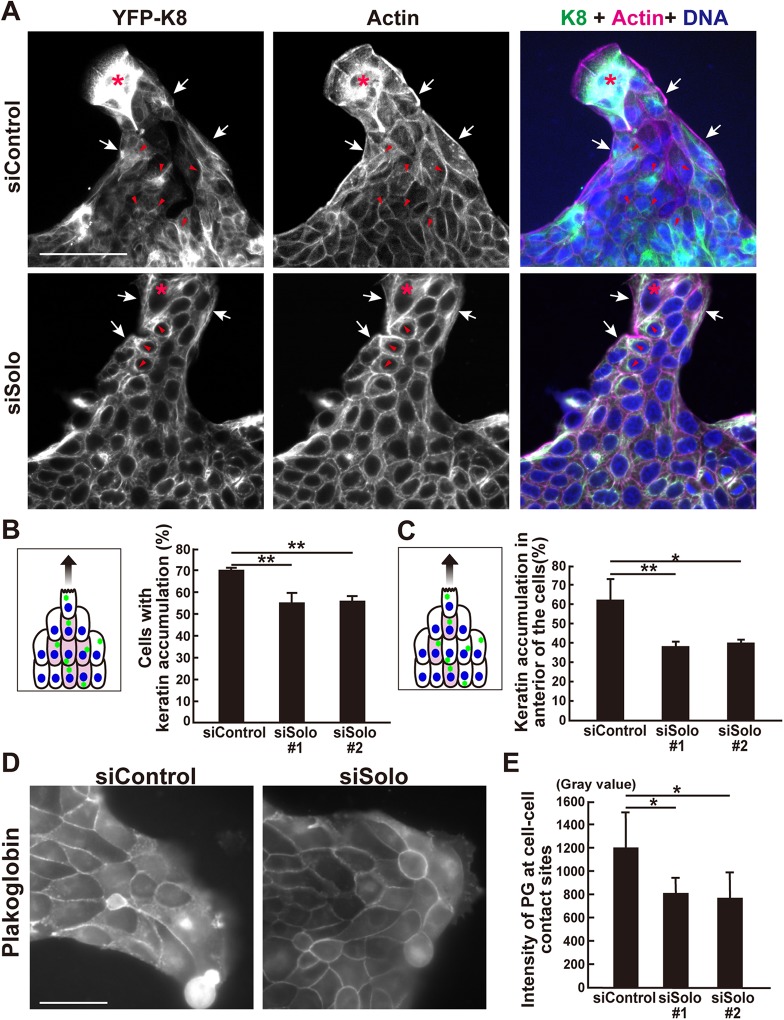
Our results suggest that the acceleration of collective cell migration by Solo knockdown is not the result of changes in the motility or polarity of individual cells. We considered that Solo knockdown accelerates collective cell migration by destabilizing cell–cell contact structures. We previously showed that Solo is required for the proper organization of K8/K18 networks and localization of plakoglobin (PG) in MDCK cells (Fujiwara et al., 2016). PG is a component of adherens junctions and desmosomes. Therefore, Solo knockdown may accelerate collective cell migration by perturbing K8/K18 networks and desmosomes. To investigate the role of Solo in collective migration, we analyzed the effects of its knockdown on K8/K18 organization in MDCK cells constitutively expressing YFP-K8. We previously showed that Solo knockdown caused the loss of K8/K18 filaments at the cell edge region of spreading MDCK cells cultured at low density (Fujiwara et al., 2016). As the cells were cultured at high density in the present study, we observed no similar effect of Solo knockdown on edge region K8/K18 networks of collectively migrating cells (Figure 7A). Unlike in spreading MDCK cells, K8/K18 filaments of collectively migrating cells showed a distinct polarity. The K8/K18 filaments of cells along the migrating edge accumulated on the periphery of the cell nuclei, which appeared independent of Solo (Figure 7A, white arrows). In contrast, K8/K18 filaments of cells within the migrating body accumulated in front of the nuclei (Figure 7A, red arrowheads). Furthermore, the ability to accumulate and polarize the location of keratin filaments appeared Solo dependent. Indeed, Solo knockdown significantly decreased the number of interior cells accumulating keratin filaments (Figure 7B). Solo knockdown also significantly reduced the number of interior cells with anterior keratin filament localization (Figure 7C).
FIGURE 7:
Effects of Solo knockdown on K8/K18 filaments and plakoglobin localization during collective cell migration. (A) Immunofluorescence images of YFP-K8, F-actin, and nuclei in control and Solo-knockdown YFP-K8–expressing MDCK cells in the finger-like protrusions. The collectively migrating YFP-K8–expressing MDCK cells were fixed and the K8/K18 networks (green), F-actin (magenta), and nuclei (blue) were visualized. Accumulation of K8/K18 filaments in the peripheral and the interior cells are indicated by white arrows and red arrowheads, respectively. Asterisks indicate leader cells. Scale bar = 100 μm. (B) Schematic representation of K8/K18 accumulation (green dots) in the cells in the finger-like protrusion. Colored cells are the interior cells of the protrusion with K8/K18 accumulation. These were counted and the percentage of interior cells in the control- and Solo-knockdown finger-like protrusions are shown in the graph. (C) An interior cell was divided into four parts in a radial manner at 90° based on the migration direction axis and the cell center. The front section was defined as the front side. The model indicates that the colored cells in the interior area accumulated K8/K18 on the front side of the nucleus. The percentage of these interior cells in the control and Solo-knockdown finger-like protrusions are shown in the graph. (D) Immunofluorescence images of PG at the cell–cell contact sites in the control and Solo-knockdown finger-like protrusions. The cells were fixed and stained with anti-PG antibody. Scale bar = 50 μm. (E) PG intensities at the cell–cell contact sites of the control and Solo-knockdown cells. Data are mean ± SD of three independent experiments (60–140 cells/experiment in B and C; 100 cells in E). *, P < 0.05; **, P < 0.01 (one-way ANOVA followed by Dunnett’s test).
We also assessed the effect of Solo knockdown on PG localization by measuring PG immunofluorescence intensity in the cell–cell contact sites. Solo knockdown significantly decreased PG localization in the cell–cell contact sites of the collectively migrating cell population (Figure 7, D and E). Therefore, Solo is involved in polarized K8/K18 filament accumulation at the anterior region of the cells and PG localization at the cell–cell junctions.
Next, we examined the effects of Y-27632 on the organization of K8 networks and PG localization at the cell–cell contact sites in collectively migrating cells. Accordingly, MDCK cells or YFP-K8 expressing MDCK cells were exposed to 0.3 or 5 μM Y-27632. Results show that the number of cells with accumulated K8 filaments decreased following Y-27632 treatment in a dose-dependent manner (Supplemental Figure S2, A and B), indicating that ROCK activity is required for the accumulation of K8 filaments in collectively migrating cells. Alternatively, Y-27632 treatment had no significant effect on the intensity of plakoglobin at the cell–cell contact sites (Supplemental Figure S2, C and D).
Further, to investigate the correlation in localization of Solo and K8 filaments at cell–cell contact sites, we expressed CFP-Solo in YFP-K8–expressing MDCK cells and analyzed the localization of K8 filaments surrounding the cell–cell contact sites where Solo was accumulated (Supplemental Figure S1C). We found that thick K8 bundles were located along the cell–cell contacts where Solo was accumulated and thin K8 bundles perpendicular to the cell–cell contact sites were anchored to the positions of Solo-positive dots (Supplemental Figure S1C, blue arrowheads). We previously reported that Solo exhibits punctate localization along K8 fibers on the ventral side of the cell and partially colocalizes with K8 fibers (Fujiwara et al., 2016). Hence, the localization of Solo and K8 filaments at the cell–cell contact sites was similar to that observed at the basal membrane.
Keratin-18 or plakoglobin knockdown accelerates collective cell migration
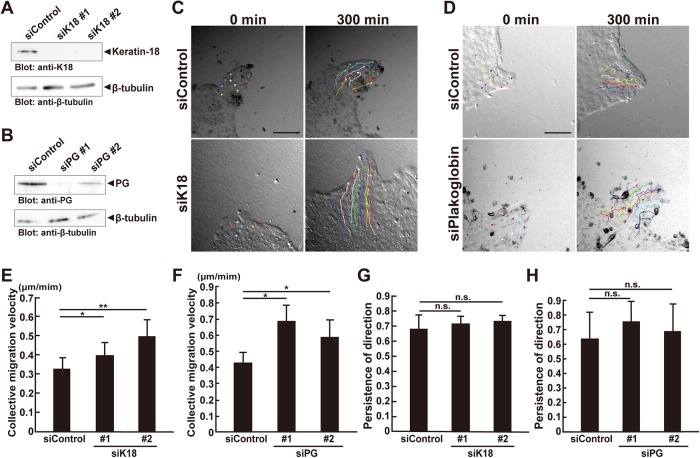
Previous wound-healing assays showed that the knockdown of keratins or desmosomal components accelerated collective cell migration (Morley et al., 2003; Wong and Coulombe, 2003; Long et al., 2006; Sechler et al., 2015). We evaluated the effects of K18 or PG knockdown on collective MDCK cell migration. Endogenous K18 and PG were down-regulated by treating the MDCK cells with K18- and PG-targeting siRNAs, respectively (Figure 8, A and B). K18 and PG knockdown significantly accelerated collective cell migration as did Solo knockdown (Figure 8, C–F, and Supplemental Movie 4). However, they had no apparent effect on the persistence of migrating direction (Figure 8, G and H). These results indicate that K8/K18 filaments and PG function as brakes for collective cell migration. As Solo knockdown perturbed proper K8/K18 network organization and PG localization, Solo may decelerate collective cell migration by regulating K8/K18 networks and desmosomal structure.
FIGURE 8:
K18 or plakoglobin knockdown accelerates collective cell migration. (A, B) Knockdown effects of K18 or PG on K18 or PG expression in MDCK cells. Cells were transfected with control, K18-, or PG-targeting siRNAs and incubated for 48 h. Cells were harvested, lysed, and analyzed by immunoblotting with anti-K18 or anti-PG antibody, respectively. (C, D) DIC images of the migration of the control, K18-, or PG-siRNA–transfected finger-like protrusions at the start (0 min) and end (300 min) of the time-lapse observation. Trajectories of individual cells were overlaid. Scale bars = 100 μm. (E–H) Velocities of the collective migration and persistence of the direction of collectively migrating control, K18-, or PG-knockdown cells were analyzed as indicated in Figure 1. Data are means ± SD of five independent experiments in E and G (25–37 cells/experiment), and three independent experiments in F and H (15–30 cells/experiment). *, P < 0.05; **, P < 0.01; n.s., not significant (one-way ANOVA followed by Dunnett’s test).
Movie S4.
Time-lapse observation of finger-like protrusions comprising the control, keratin-18- or plakoglobin-siRNA-transfected MDCK cells on collagen gel. Images were obtained every 5 min. Trajectories of individual cells were overlaid. This movie corresponds to Figs. 8C and 8D.
We also examined the effects of K18 or PG knockdown on RhoA activity in MDCK cells and found that the active form of RhoA was significantly decreased in K18 or PG siRNA-transfected cells, compared with that in control siRNA-transfected cells (Supplemental Figure S3, A and B), indicating that K18 and PG are involved in RhoA activation.
DISCUSSION
Rho-GEFs are involved in collective migration
In the present study, we illustrated the role of Solo as a brake for collective cell migration. Previous studies have described similar roles for other Rho-GEFs, namely, p115RhoGEF and LARG, in collective cell migration (Medlin et al., 2010; Kher et al., 2014). Specifically, knockdown of p115RhoGEF served to down-regulate junctional E-cadherin in epithelial cells and accelerate collective mammary epithelial cell migration in wound-healing assays (Kher et al., 2014). Additionally, p115RhoGEF has been reported as to become activated following application of tensile force through JAM, a tight junction component (Scott et al., 2016). These results suggest that p115RhoGEF suppresses collective cell migration by regulating the tensile force–induced adherens junction and tight junction reorganization occurring at intercellular adhesions. Further, LARG overexpression was found to inhibit collective breast and colorectal cancer cell migration (Ong et al., 2009), while its knockdown accelerates collective cell migration of smooth muscle cells in wound-healing assays (Medlin et al., 2010). LARG is also involved in RhoA activation in response to application of mechanical force to integrins at cell–substrate adhesions (Guilluy et al., 2011). These results suggest that LARG decelerates collective cell migration by regulating FA dynamics. Similarly, we previously showed that Solo is involved in tensile force–induced RhoA activation and stress fiber formation, while its knockdown serves to decrease keratin bundle formation and plakoglobin at the site of cell–cell adhesions, as well as actin stress fibers (Fujiwara et al., 2016). Hence, it is likely that Solo acts to decelerate collective cell migration by generating a pullback force and promoting desmosomal junctions at cell–cell contact sites. Thus, the Rho-GEFs, p115RhoGEF, LARG, and Solo appear to exert individual yet cooperative roles in collective cell migration by differentially regulating the force-induced cytoskeletal organization and dynamics at cell–cell and cell–substrate adhesion sites.
Solo decelerates collective cell migration by regulating the mechanical force responses at the cell–cell contacts
We showed that the velocity of collectively migrating cells is >1/3 lower than that of solitary cultured cells on collagen gels. Whereas solitary cells started and stopped at random time intervals, collectively migrating cells continuously moved at nearly the same speed and maintained the direction of migration. To sustain collective cell migration velocity and direction, the individual cells in the population must maintain cell–cell contacts, sense tensile forces from neighboring cells, and generate pullback forces to balance the external forces. Pullback force in the opposite direction of migration attenuates the traction force of the forward cells and functions as a brake for collective cell migration. The pullback force at the rearward cell–cell contact region may also contribute to persistent cell migration (Tambe et al., 2011; Trepat and Fredberg, 2011; Weber et al., 2012; Mayor and Etienne-Manneville, 2016a). Balance of the forces in the cell population might be a trade-off between the deceleration of migration and the cooperative and continuously aligned migration of the cell population (Mayor and Etienne-Manneville, 2016b). In the present study, we showed that Solo knockdown accelerates collectively migrating cells but not solitary cells. Thus, Solo is required to generate pullback to balance the tensile force at the cell–cell contact regions. The migration direction and persistence were not perturbed by Solo knockdown. Therefore, Solo is involved in generating a force opposing the tensile forces from the frontal neighboring cells. But, it does not affect the direction of migration. We also showed that partial ROCK inhibition with a low concentration of Y-27632 accelerates collective migration. The collective invasion of cancer cells into 3D Matrigel was also promoted by Y-27632 (Omelchenko et al., 2003). We previously reported that Solo activates RhoA in response to a cadherin- or integrin-mediated pulling force (Abiko et al., 2015; Fujiwara et al., 2016). These findings suggest that Solo acts as a brake for collective cell migration by generating contractile forces at the cell–cell contact sites and activating the Rho-ROCK pathway in response to tensile forces at cell–cell contact sites.
ROCK exhibits multifaceted roles in cell migration by spatially and temporally regulating various cellular activities, including actin and keratin filament reorganization, cell–cell and cell–substrate adhesions, cell polarity formation, and traction force generation. The low Y-27632 dose may have partially suppressed ROCK activity, thereby limiting its cellular effects without complete interference. Alternatively, a high dose of Y-27632 would significantly suppress ROCK-mediated cellular activities, resulting in inhibition of cell migration. In this study, 0.3 μM Y-27632 accelerated collective cell migration, likely by preferentially reducing the pullback force induced by the Rho-ROCK signal at the cell–cell contact sites; while 5 μM of Y-27632 caused a slight deceleration in collective migration, possibly due to inhibitory effects at this dose of Y-27632 on various ROCK-mediated cellular activities required for cell migration.
Solo regulates collective cell migration through keratin filaments and desmosomal proteins
Previous studies showed that defects in keratin filaments or desmosomal proteins accelerate collective cell migration. Mutations of the keratin-14 and keratin-5 genes are responsible for the symptoms of epidermolysis bullosa simplex and accelerated collective epidermal cell migration in wound-healing assays (Morley et al., 2003). The outgrowth of keratinocytes from skin explants was accelerated in keratin-6 knockout mice (Wong and Coulombe, 2003). Defects in the K8/K18 or desmosomal proteins (periplakin or desmoplakin) disrupted the integrity of epithelial cell sheets and accelerated collective cell migration (Long et al., 2006; Wang et al., 2018). We previously showed that Solo binds to K8/K18 filaments via multiple K8/K18-binding domains and that Solo is required for the proper organization of K8/K18 networks in MDCK and MCF10A epithelial cells (Fujiwara et al., 2016, 2018). Solo knockdown reduced plakoglobin localization at the cell–cell adhesions (Fujiwara et al., 2016). Therefore, Solo knockdown may accelerate collective migration by disorganizing the K8/K18 networks and desmosome structures. A major function of keratin filaments is to strengthen tissues by stiffening cells and cell–cell contacts. Perturbations of the keratin filament networks and desmosomes weaken the cell–cell contact structure and decrease pullback toward the forward cells, which, in turn, probably accelerate collective cell migration (Matsuzawa et al., 2018).
Keratin-6 (K6) knockdown in epidermal cell sheets accelerates collective cell migration by increasing focal adhesion (FA) turnover and destabilizing FAs through the unbinding of K6 filaments to myosin II in FAs and the decrease in desmoplakin at the cell–cell contact sites (Wang et al., 2018). Solo knockdown down-regulated RhoA and attenuated stress fibers in MDCK cells (Fujiwara et al., 2016). Consequently, Solo knockdown increases FA turnover and should accelerate both collective and solitary migrating cells. In the present study, however, we demonstrated that the migration speed of solitary cells did not increase. This discrepancy could be explained by the relative differences in experimental conditions including substrate stiffness. In the present study, we seeded MDCK cells on soft collagen gel in which the FAs were already destabilized. Therefore, Solo knockdown likely had negligible influence on FA turnover.
Physiological functions of Solo in epithelial cell populations
Solo stimulates RhoA activity, binds to keratin filaments, and accumulates in regions where contractile forces are applied. For these reasons, Solo may be activated by pulling forces from neighboring cells and responds by pulling back on them. When cells are subjected to pulling forces, Solo may accumulate at the cell–cell contact sites and increase contractile forces there until a proper balance with the external force is achieved. Mechanical force perception and balance at cell–cell contacts in epithelial tissues play important roles in tissue homeostasis (Mammoto and Ingber, 2010; Guillot and Lecuit, 2013; Rubsam et al., 2018). Solo knockdown may accelerate collective cell migration by inducing disequilibrium between the pulling and pullback forces at the cell–cell contact sites in epithelial cell sheets. However, the roles of these forces at the cell–cell and cell–substrate contact sites as well as those of the leader and follower cells in collective cell migration are not yet fully understood. Further studies are needed to elucidate the roles of the forces and their regulatory mechanisms in collective cell migration.
MATERIALS AND METHODS
Reagents and antibodies
Y-27632 was purchased from Wako Pure Chemical Industries (Osaka, Japan). Alexa Fluor 633–conjugated phalloidin was purchased from Thermo Fisher Scientific (Waltham, MA). A rabbit antiserum recognizing human and dog Solo was raised against the C-terminal peptide (LSRQSHARALSDPTTPL) of human Solo (Abiko et al., 2015). The following antibodies were purchased: anti-RhoA (sc-418; Santa Cruz Biotechnology, Dallas, TX), anti-GAPDH (ab8245; Abcam, Cambridge, UK), anti-GM130 (CSB-PA600856ESR1HU; CUSABIO, Baltimore, MD), anti-K18 (Ks 18.04; Progen, Heidelberg, Germany), anti-plakoglobin (Clone 15; BD Biosciences, Franklin Lakes, NJ), anti-phosphomyosin light chain (Thr18/Ser19; 3674S; Cell Signaling Technology, Danvers, MA), Alexa Fluor 568–conjugated anti-rabbit immunoglobulin G (IgG) and Alexa Fluor 488–conjugated anti-mouse IgG (Thermo Fisher Scientific), and horseradish peroxidase–conjugated anti-mouse IgG and anti-rabbit IgG (GE Healthcare, Little Chalfont, UK).
Plasmid construction and siRNAs
The expression plasmids encoding mCherry- and ECFP-Solo were constructed by inserting Solo cDNA into the pmCherry-C1 and pECFP-C1 vectors, respectively (Clontech, Mountain View, CA). The siRNAs targeting dog Solo, K18, and plakoglobin were designed using the siDirect website (http://sidirect2.rnai.jp) and purchased from Sigma-Aldrich, St. Louis, MO. The siRNA sequences are as follows: 5′-GAGCUGAAAGAGGAACUCAAACC-3′ (Solo siRNA #1), 5′-CAGCCUUUACGCCCAGUACGUGA-3′ (Solo siRNA #2), 5′-GAGUUGGAUGCCCCCAAAUCUCA-3′ (K18 siRNA #1), 5′-AGGCGUUGAGGCAGCAAAACAGG-3′ (K18 siRNA #2), 5′-GUCUCUGGACCUUGCGCAA-3′ (plakoglobin siRNA #1), 5′-CGAAGACAGCUCGCUGCUA-3′ (plakoglobin siRNA #2). MISSION siRNA Universal Negative Control was used as a negative control siRNA and was purchased from Sigma-Aldrich.
Cell culture and transfection
MDCK cells were cultured in DMEM supplemented with 10% fetal bovine serum, 100 U ml−l penicillin, and 0.1 mg streptomycin. MDCK cells stably expressing YFP-Lifeact, YFP-K8, and YFP-Solo were established on selection medium containing G418 (Fujiwara et al., 2016). Cells were transfected with siRNAs using RNAiMAX (Thermo Fisher Scientific, Waltham, MA). The siRNAs were used at a final concentration of 50 nM. Two days after transfection, the cells were used in the assays.
Observation of collective and solitary cell migration
To observe collective cell migration, 1 ml of 1.6 mg ml−l collagen (Cellmatrix Type I-P; Nitta Gelatin, Osaka, Japan) was gelatinized in a 35-mm dish. Then 2.5 × 104 MDCK cells were seeded in a 5-mm glass ring set on the center of the collagen gel sheet in the dish. After the cells were incubated overnight, the glass ring was removed. After culture for 4 h, the cells were subjected to time-lapse imaging under the LSM 710 confocal microscope (Carl Zeiss AG, Jena, Germany) fitted with an EC Plan N 10× objective lens (NA 0.3). A microscope stage incubator was used to keep the cells at 37°C and under 5% CO2 and high relative humidity. Images were obtained every 5 or 10 min for >5 h using the definite-focus function to maintain the focal plane on the cells during time-lapse imaging. To observe solitary cell migration, 1 × 104 MDCK cells were seeded on gelatinized collagen in a 35-mm dish. After incubation overnight, the cells were subjected to time-lapse imaging in the same manner as that for collective cell migration.
Immunofluorescence staining and imaging
Cells were fixed and permeabilized with 4% (wt/vol) paraformaldehyde and 0.5% (wt/vol) Triton X-100 in phosphate-buffered saline (PBS) at room temperature for 20 min. After being washed with PBS, the cells were stained with anti-GM130, anti-plakoglobin, or anti-RhoA antibody diluted in Can-Get-Signal immunostaining solution (Toyobo, Osaka, Japan). The secondary antibodies, Alexa Fluor 633–phalloidin and 4′,6-diamidino-2-phenylindole (DAPI) were diluted with 2% fetal calf serum in PBS. Fluorescence images were obtained under an inverted fluorescence microscope (DMI 6000B; Leica Microsystems, Wetzlar, Germany) fitted with a PL Apo 63× oil-immersion objective lens (NA 1.3). The images were analyzed with ImageJ (National Institutes of Health, Bethesda, MD).
Image analysis
Image analysis was performed in ImageJ. Collective cell migration velocities were measured by tracking the cell migration trajectories with the manual tracking plugin of ImageJ. The collective cell migration velocity was calculated by dividing the lengths of the trajectories of the cells in the tip of a finger-like protrusion by the observation time. Persistence of the direction of migration was calculated by dividing the lengths of the cell trajectories by the distances between the starting and ending points. More than 10 cells per tip of each finger-like protrusion were tracked. Solitary cell velocity was calculated by dividing the length of the cell trajectories by the time during which the cells continuously moved at >0.5 μm min−1 for >30 min. Plakoglobin fluorescence signal intensities at the cell–cell contact sites in collective migrating cells in the absence or presence of 0.3 or 5 μM of Y-27632 were measured with ImageJ. The migration polarity of the collectively migrating cells was determined from the angles between the migration direction and the vectors from the centers of the nuclei to the centers of the GM130 signals (Golgi apparatus) in the leader or follower cells. The positions of Solo and K8/K18 bundle accumulations relative to the migration direction were also determined. The migration direction of the cells population was established using the direction from a cell of interest to the leader cell of the finger-like protrusion. To identify the polarized Solo localization, a line of cell–cell contact sites where Solo accumulated was categorized as transverse or longitudinal depending on its angle in relation to the migration direction. To assess K8/K18 accumulation and its position in a cell, the cells were divided into anterior, posterior, and lateral quadrants, counted, and categorized according to the K8/K18 accumulations in each area.
Immunoblotting
Cells were lysed with ice-cold lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% wt/vol NP-40, 10% wt/vol glycerol, 10 mM MgCl2, 0.2 mM Na3VO4, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 10 μg ml−l leupeptin, and 2 μg ml−l pepstatin A). Cell lysates were subjected to immunoblot analysis as previously described (Ohashi et al., 2000).
Pull-down assay
The activities of RhoA and Rac1 were evaluated by pull down of their GTP-bound forms using the GST-tagged RhoA-binding domain (RBD) and the Rac1-binding domain (PBD) of PAK1 as previously described (Nishita et al., 2002). MDCK cells were transfected with control, Solo, keratin-18, or plakoglobin siRNAs, incubated for 48 h, lysed with lysis buffer, and incubated with GST-rhotekin (RBD) or GST-PAK1 (PBD) bound to glutathione-Sepharose at 4°C for 30 min. Sepharoses were washed off with lysis buffer and analyzed by immunoblotting with anti-RhoA or anti-Rac1 antibodies. RhoA or Rac1 activity was normalized to the total amount of RhoA or Rac1 in each condition.
Statistical analysis
Data are expressed as means ± SD of more than three independent experiments. Data processing was conducted in Prism v. 4 (GraphPad Software, La Jolla, CA). P values were calculated by one-way analysis of variance (ANOVA) followed by Dunnett’s test for multiple dataset comparisons. In all cases, P < 0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Naoki Honzawa for technical assistance. This work was supported by the Japan Agency for Medical Research and Development (Grant no. JP18gm5810015h0003 to K.O.), https://www.amed.go.jp/en/index.html; the Japan Society for the Promotion of Science (JSPS) KAKENHI (Grants no. JP23112005 and no. JP16K07335 to K.O., Grants no. JP15K14469, no. JP16H00749, and no. JP18K19280 to K.M., and Grant no. JPT17K151180 to S.F.), http://www.jsps.go.jp/j-grantsinaid/; and JSPS (Fellowship no. JPA16J041330 to S.F.), http://www.jsps.go.jp/j-pd/.
Abbreviations used:
- DAPI
4′,6-diamidino-2-phenylindole
- DTT
dithiothreitol
- FA
focal adhesion
- GEF
guanine nucleotide exchange factor
- GST
glutathione S-transferase
- IFs
intermediate filaments
- K8
keratin-8
- K18
keratin-18
- MDCK
Madin-Darby canine kidney
- PBS
phosphate-buffered saline
- PG
plakoglobin
- ROCK
Rho-associated protein kinase
- siRNA
small interfering RNA
- YFP
yellow fluorescence protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-07-0357) on February 12, 2020.
REFERENCES
- Abiko H, Fujiwara S, Ohashi K, Hiatari R, Mashiko T, Sakamoto N, Sato M, Mizuno K. (2015). Rho guanine nucleotide exchange factors involved in cyclic-stretch-induced reorientation of vascular endothelial cells. J Cell Sci , 1683–1695. [DOI] [PubMed] [Google Scholar]
- Bershadsky AD, Futerman AH. (1994). Disruption of the Golgi apparatus by brefeldin A blocks cell polarization and inhibits directed cell migration. Proc Natl Acad Sci USA , 5686–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broussard JA, Getsios S, Green KJ. (2015). Desmosome regulation and signaling in disease. Cell Tissue Res , 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras G, Sahai E. (2014). Physical influences of the extracellular environment on cell migration. Nat Rev Mol Cell Biol , 813–824. [DOI] [PubMed] [Google Scholar]
- Chiapparo G, Lin XH, Lescroart F, Chabab S, Paulissen C, Pitisci L, Bondue A, Blanpain C. (2016). Mesp1 controls the speed, polarity, and directionality of cardiovascular progenitor migration. J Cell Biol , 463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DR, Rossman KL, Der CJ. (2014). Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene , 4021–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe PA, Wong P. (2004). Cytoplasmic intermediate filaments revealed as dynamic and multipurpose scaffolds. Nat Cell Biol , 699–706. [DOI] [PubMed] [Google Scholar]
- Eriksson JE, Dechat T, Grin B, Helfand B, Mendez M, Pallari HM, Goldman RD. (2009). Introducing intermediate filaments: from discovery to disease. J Clin Invest , 1763–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Gilmour D. (2009). Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol , 445–457. [DOI] [PubMed] [Google Scholar]
- Friedl P, Locker J, Sahai E, Segall JE. (2012). Classifying collective cancer cell invasion. Nat Cell Biol , 777–783. [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Matsui TS, Ohashi K, Deguchi S, Mizuno K. (2018). Solo, a RhoA-targeting guanine nucleotide exchange factor, is critical for hemidesmosome formation and acinar development in epithelial cells. PLoS One , e0195124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Matsui TS, Ohashi K, Mizuno K, Deguchi S. (2019). Keratin-binding ability of the N-terminal Solo domain of Solo is critical for its function in cellular mechanotransduction. Genes Cells , 390–402. [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Ohashi K, Mashiko T, Kondo H, Mizuno K. (2016). Interplay between Solo and keratin filaments is crucial for mechanical force-induced stress fiber reinforcement. Mol Biol Cell , 954–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot C, Lecuit T. (2013). Mechanics of epithelial tissue homeostasis and morphogenesis. Science , 1185–1189. [DOI] [PubMed] [Google Scholar]
- Guilluy C, Swaminathan V, Garcia-Mata R, O’Brien ET, Superfine R, Burridge K. (2011). The Rho GEFs LARG and GEF-H1 regulate the mechanical response to force on integrins. Nat Cell Biol , 722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeger A, Wolf K, Zegers MM, Friedl P. (2015). Collective cell migration: guidance principles and hierarchies. Trends Cell Biol , 556–566. [DOI] [PubMed] [Google Scholar]
- Haga H, Irahara C, Kobayashi R, Nakagaki T, Kawabata K. (2005). Collective movement of epithelial cells on a collagen gel substrate. Biophys J , 2250–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayer A, Shao L, Chung M, Joubert LM, Yang HW, Tsai FC, Bisaria A, Betzig E, Meyer T. (2016). Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells. Nat Cell Biol , 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman SJ, Ridley AJ. (2008). Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol , 690–701. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Bar H, Kreplak L, Strelkov SV, Aebi U. (2007). Intermediate filaments: from cell architecture to nanomechanics. Nat Rev Mol Cell Biol , 562–573. [DOI] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. (2005). Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol , 247–269. [DOI] [PubMed] [Google Scholar]
- Kher SS, Struckhoff AP, Alberts AS, Worthylake RA. (2014). A novel role for p115RhoGEF in regulation of epithelial plasticity. PLoS One , e85409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Hartley R, Reiss B, Sun Y, Pu J, Wu D, Lin F, Hoang T, Yamada S, Jiang J, et al (2012). E-cadherin plays an essential role in collective directional migration of large epithelial sheets. Cell Mol Life Sci , 2779–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HA, Boczonadi V, McInroy L, Goldberg M, Maatta A. (2006). Periplakin-dependent re-organisation of keratin cytoskeleton and loss of collective migration in keratin-8-downregulated epithelial sheets. J Cell Sci , 5147–5159. [DOI] [PubMed] [Google Scholar]
- Mammoto T, Ingber DE. (2010). Mechanical control of tissue and organ development. Development , 1407–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa K, Himoto T, Mochizuki Y, Ikenouchi J. (2018). α-Catenin controls the anisotropy of force distribution at cell-cell junctions during collective cell migration. Cell Rep , 3447–3456. [DOI] [PubMed] [Google Scholar]
- Mayor R, Etienne-Manneville S. (2016a). The front and rear of collective cell migration. Nat Rev Mol Cell Biol , 97–109. [DOI] [PubMed] [Google Scholar]
- Mayor R, Etienne-Manneville S. (2016b). The front and rear of collective cell migration. Nat Rev Mol Cell Biol , 97–109. [DOI] [PubMed] [Google Scholar]
- Medlin MD, Staus DP, Dubash AD, Taylor JM, Mack CP. (2010). Sphingosine 1-phosphate receptor 2 signals through leukemia-associated RhoGEF (LARG), to promote smooth muscle cell differentiation. Arterioscler Thromb Vasc Biol , 1779–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley SM, D’Alessandro M, Sexton C, Rugg EL, Navsaria H, Shemanko CS, Huber M, Hohl D, Heagerty AI, Leigh IM, et al (2003). Generation and characterization of epidermolysis bullosa simplex cell lines: scratch assays show faster migration with disruptive keratin mutations. Br J Dermatol , 46–58. [DOI] [PubMed] [Google Scholar]
- Nishita M, Aizawa H, Mizuno K. (2002). Stromal cell-derived factor 1α activates LIM kinase 1 and induces cofilin phosphorylation for T-cell chemotaxis. Mol Cell Biol , 774–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K, Fujiwara S, Mizuno K. (2017). Roles of the cytoskeleton, cell adhesion and rho signalling in mechanosensing and mechanotransduction. J Biochem , 245–254. [DOI] [PubMed] [Google Scholar]
- Ohashi K, Nagata K, Maekawa M, Ishizaki T, Narumiya S, Mizuno K. (2000). Rho-associated kinase ROCK activates LIM-kinase 1 by phosphorylation at threonine 508 within the activation loop. J Biol Chem , 3577–3582. [DOI] [PubMed] [Google Scholar]
- Omelchenko T, Vasiliev JM, Gelfand IM, Feder HH, Bonder EM. (2003). Rho-dependent formation of epithelial “leader” cells during wound healing. Proc Natl Acad Sci USA , 10788–10793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong DC, Ho YM, Rudduck C, Chin K, Kuo WL, Lie DK, Chua CL, Tan PH, Eu KW, Seow-Choen F, et al (2009). LARG at chromosome 11q23 has functional characteristics of a tumor suppressor in human breast and colorectal cancer. Oncogene , 4189–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plutoni C, Bazellieres E, Le Borgne-Rochet M, Comunale F, Brugues A, Seveno M, Planchon D, Thuault S, Morin N, Bodin S, et al (2016). P-cadherin promotes collective cell migration via a Cdc42-mediated increase in mechanical forces. J Cell Biol , 199–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftopoulou M, Hall A. (2004). Cell migration: Rho GTPases lead the way. Dev Biol , 23–32. [DOI] [PubMed] [Google Scholar]
- Reffay M, Petitjean L, Coscoy S, Grasland-Mongrain E, Amblard F, Buguin A, Silberzan P. (2011). Orientation and polarity in collectively migrating cell structures: statics and dynamics. Biophys J , 2566–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P. (2009). Collective cell migration. Annu Rev Cell Dev Biol , 407–429. [DOI] [PubMed] [Google Scholar]
- Rubsam M, Broussard JA, Wickstrom SA, Nekrasova O, Green KJ, Niessen CM. (2018). Adherens junctions and desmosomes coordinate mechanics and signaling to orchestrate tissue morphogenesis and function: an evolutionary perspective. Cold Spring Harb Perspect Biol , a029207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander EE, ten Klooster JP, van Delft S, van der Kammen RA, Collard JG. (1999). Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J Cell Biol , 1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghvi-Shah R, Weber GF. (2017). Intermediate filaments at the junction of mechanotransduction, migration, and development. Front Cell Dev Biol , 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DW, Tolbert CE, Burridge K. (2016). Tension on JAM-A activates RhoA via GEF-H1 and p115 RhoGEF. Mol Biol Cell , 1420–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechler M, Borowicz S, Van Scoyk M, Avasarala S, Zerayesus S, Edwards MG, Kumar Karuppusamy Rathinam M, Zhao X, Wu PY, Tang K, et al (2015). Novel role for γ-catenin in the regulation of cancer cell migration via the induction of hepatocyte growth factor activator inhibitor type 1 (HAI-1). J Biol Chem , 15610–15620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambe DT, Hardin CC, Angelini TE, Rajendran K, Park CY, Serra-Picamal X, Zhou EHH, Zaman MH, Butler JP, Weitz DA, et al (2011). Collective cell guidance by cooperative intercellular forces. Nat Mater , 469–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivola DM, Boor P, Alam C, Strnad P. (2015). Keratins in health and disease. Curr Opin Cell Biol , 73–81. [DOI] [PubMed] [Google Scholar]
- Trepat X, Fredberg JJ. (2011). Plithotaxis and emergent dynamics in collective cellular migration. Trends Cell Biol , 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helvert S, Storm C, Friedl P. (2018). Mechanoreciprocity in cell migration. Nat Cell Biol , 8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Chen S, Liu HB, Parent CA, Coulombe PA. (2018). Keratin 6 regulates collective keratinocyte migration by altering cell-cell and cell-matrix adhesion. J Cell Biol , 4314–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber GF, Bjerke MA, DeSimone DW. (2012). A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration. Dev Cell , 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P, Coulombe PA. (2003). Loss of keratin 6 (K6) proteins reveals a function for intermediate filaments during wound repair. J Cell Biol , 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Mizutani T, Kawabata K, Haga H. (2015). Leader cells regulate collective cell migration via Rac activation in the downstream signaling of integrin β1 and PI3K. Sci Rep , 7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegers MM, Friedl P. (2014). Rho GTPases in collective cell migration. Small GTPases , e28997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.