Abstract
In many organisms, positive and negative signals cooperate to position the division site for cytokinesis. In the rod-shaped fission yeast Schizosaccharomyces pombe, symmetric division is achieved through anillin/Mid1-dependent positive cues released from the central nucleus and negative signals from the DYRK-family polarity kinase Pom1 at cell tips. Here we establish that Pom1’s kinase activity prevents septation at cell tips even if Mid1 is absent or mislocalized. We also find that Pom1 phosphorylation of F-BAR protein Cdc15, a major scaffold of the division apparatus, disrupts Cdc15’s ability to bind membranes and paxillin, Pxl1, thereby inhibiting Cdc15’s function in cytokinesis. A Cdc15 mutant carrying phosphomimetic versions of Pom1 sites or deletion of Cdc15 binding partners suppresses division at cell tips in cells lacking both Mid1 and Pom1 signals. Thus, inhibition of Cdc15-scaffolded septum formation at cell poles is a key Pom1 mechanism that ensures medial division.
INTRODUCTION
Accurate positioning of cell division is crucial for both cellular function and integrity. Studies in prokaryotic and eukaryotic cells revealed two major positioning systems: local positive signals and distal inhibitory signals (Oliferenko et al., 2009). In the rod-shaped fission yeast Schizosaccharomyces pombe, anillin-like Mid1 released from the centrally positioned nucleus is the positive cue for assembly of the contractile ring (CR) for cytokinesis (Rincon and Paoletti, 2016). Mid1 recruits CR proteins such as myosin II Myo2, formin Cdc12, and F-BAR protein Cdc15 into equatorial cortical nodes (Wu et al., 2006; Laporte et al., 2011). These nodes coalesce into a CR, which constricts and serves as the landmark for septum formation (Willet et al., 2015b; Rincon and Paoletti, 2016). In the absence of mid1, CR placement is aberrant, leading to oblique, frequently off-center septa (Chang et al., 1996; Sohrmann et al., 1996). However, in mid1∆ cells, aberrant division remains restricted to the central region of the cell due to inhibitory signals generated by cell tip-localized DYRK-family kinase Pom1 (Huang et al., 2007). In the absence of both Mid1 and tip-localized Pom1, CRs form and persist at cell tips leading to tip septation and subsequent cell death; compromising Cdc15 function but not the function of several other CR proteins suppresses tip septation (Huang et al., 2007).
Cdc15 is an essential component of the CR, necessary for cytokinesis and septation (Fankhauser et al., 1995). It acts as a key anchor for the CR and is a platform for the assembly of additional CR components (Wachtler et al., 2006; Vjestica et al., 2008; Roberts-Galbraith et al., 2009, 2010; Laporte et al., 2011; Arasada and Pollard, 2014; McDonald et al., 2015). Cdc15 is also one of the first components detected at the incipient CR (Wu et al., 2006) and one of the most abundant CR proteins (Wu and Pollard, 2005).
Here we establish that Pom1 prevents septation at cell tips even if the positive cue, Mid1, is moved proximal to cell ends. We find that Pom1 phosphorylation of Cdc15 antagonizes Cdc15’s ability to bind membranes and paxillin, Pxl1, thereby inhibiting Cdc15’s scaffolding function. A Cdc15 mutant carrying phosphomimetic versions of Pom1 sites or deletion of Cdc15 binding partners suppresses division at cell tips in cells lacking both Mid1 and Pom1 signals. Thus, inhibition of Cdc15-scaffolded septum formation at cell poles is a key Pom1 mechanism that contributes to medial division.
RESULTS
Pom1 kinase activity prevents division at cell tips
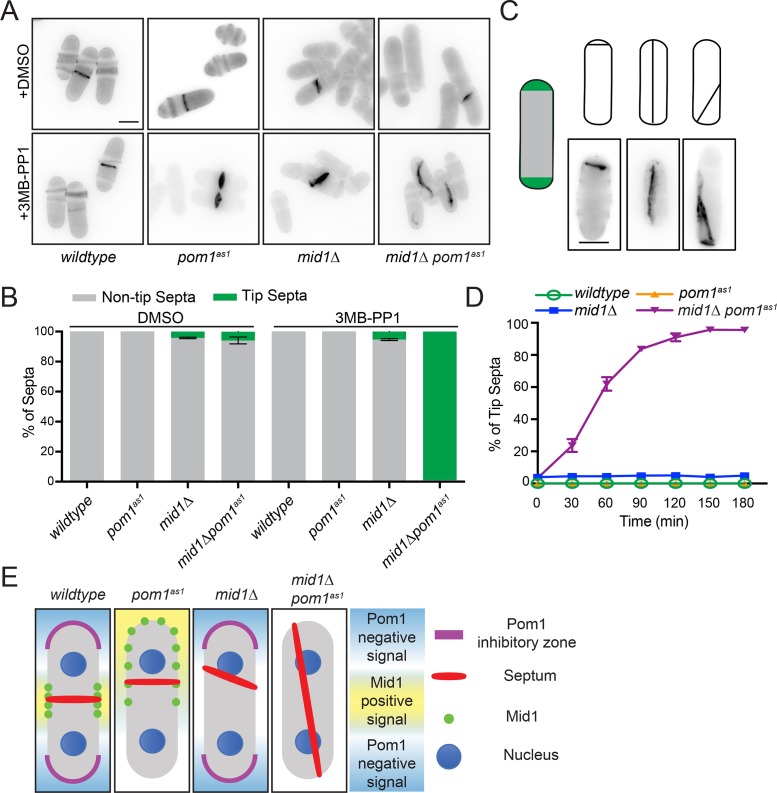
Pom1 is localized to cell tips by the microtubule-deposited Tea1/Tea4 complex (Hachet et al., 2011). Deletion of pom1, tea1, or tea4 is synthetically lethal with mid1∆, and double mutants form septa at cell tips before dying (Huang et al., 2007). To validate that the signals blocking septum formation at the cell tip, known as “septum tip occlusion,” are generated by Pom1 kinase activity, we used pom1(T778G) (pom1as1) (Padte et al., 2006) to specifically inhibit kinase activity with the ATP analogue 3MB-PP1 in mid1Δ cells. After 2 h of 3MB-PP1 treatment, we observed tip septation in almost all septated mid1Δ pom1as1 cells (Figure 1, A–C). Septa were scored as tip septa if they met one of three criteria (Figure 1C): 1) one or two septa within the hemispheric end(s) of the cell; 2) septa anchored at both cell ends and spanning the long cell axis; 3) oblique septa anchored at one cell end. The incidence of tip septa in mid1Δ pom1as1 increased over time (Figure 1D), with >95% of septated cells showing tip septa after a 2 h incubation with 1 μM 3MB-PP1. These phenotypes were not observed in either single mutant or in the double mutant treated with vehicle (Figure 1, A–D). These results suggest that the specific signal preventing formation of division septa at cell tips arises from Pom1 kinase activity (see Figure 1E).
FIGURE 1:
Pom1 kinase activity is required to inhibit tip septa. (A) Representative single z-section images of the indicated strains treated for 2 h with vehicle (DMSO) or ATP analog (3MB-PP1). Cells were fixed and stained with calcofluor. (B) Quantification of tip septa from images acquired as in A from three biological replicates, n > 300 septated cells. Graph shows mean and SEM. (C) Schematics and example images of the three types of tip septa scored. (D) Time course of tip septa appearance. 3MB-PP1 was added at time 0. Samples were prepared as in A. Measurements are from three biological replicates, n > 300 septated cells. (E) Model of the effects of Pom1 kinase activity and Mid1 in division site placement in the context of previous work (Huang et al., 2007; Rincon and Paoletti, 2016).
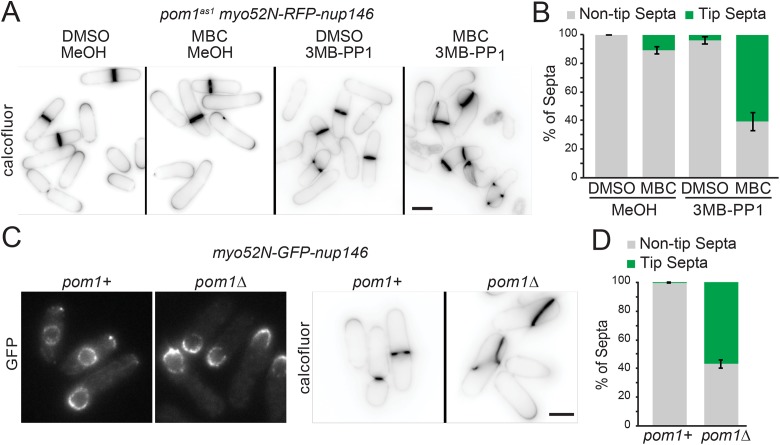
Mid1-dependent signaling can also be disrupted by displacing the nucleus (Daga and Chang, 2005; Almonacid et al., 2009). We expressed a chimeric protein of myosin-V Myo52 motor domain fused to a nuclear pore complex protein Nup146 (Myo52N-RFP-Nup146), which displaces the nucleus toward cell ends by transporting it along actin cables (Lo Presti et al., 2012). A penetrant nuclear displacement phenotype also requires microtubule depolymerization with methyl benzimidazole-2-gamma-carbamate (MBC) (Lo Presti et al., 2012). Simultaneous nuclear displacement and Pom1 inhibition increased tip septation (Figure 2, A and B). Nuclear displacement in pom1∆ led to tip septation even without the addition of MBC (Figure 2, C and D), likely due to an imbalance of actin forces in monopolar pom1∆ causing off-center nuclei (Bahler and Pringle, 1998). Thus, even in the presence of Mid1, Pom1 is required for tip occlusion.
FIGURE 2:
Pom1 kinase inhibits division site placement even if the positive Mid1 cue is proximal to the cell tip. (A) Representative images of pom1as1nmt81::myo52N-RFP-nup146 cells grown for 24 h in medium lacking thiamine to induce expression of Myo52-Nup146 and treated for 4 h with vehicle (MeOH) or 1 μM 3MB-PP1, and vehicle (DMSO) or 25 μg/ml MBC to displace the nucleus, and stained with calcofluor. (B) Quantification of tip septa as in A from four biological replicates, n > 490 septated cells. Graphs show mean and SEM. (C) Representative images of Myo52N-GFP-Nup146 in pom1+ and pom1Δ cells (left) and cells stained with calcofluor (right). (D) Quantification of tip septa as in C from three biological replicates, n > 326 septated cells. Graphs show mean and SEM. Scale bar, 5 µm.
Pom1 phosphorylates Cdc15 for septa tip occlusion
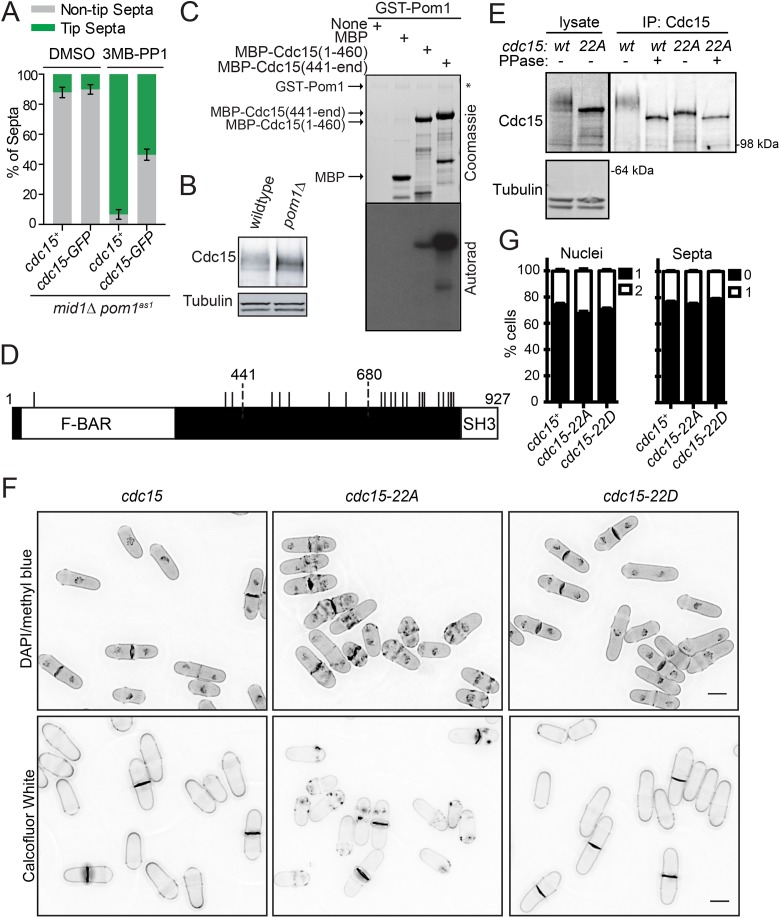
C-terminal tagging of Cdc15 with GFP blocked tip septa formation in mid1-18 tea1Δ cells, suggesting that cdc15-GFP is hypomorphic and that Cdc15 inhibition is a mechanism of tip occlusion (Huang et al., 2007). cdc15-GFP also inhibited tip septation in mid1Δ pom1as1 (Figure 3A). Given that Cdc15 is highly phosphorylated during interphase (Fankhauser et al., 1995; Roberts-Galbraith et al., 2010) and that multiple Cdc15 phosphorylation events are reduced on Pom1 inhibition (Kettenbach et al., 2015), we and others (Huang et al., 2007; Lee et al., 2018) hypothesized that Cdc15 is a Pom1 substrate. Indeed, when examined by SDS–PAGE, the slow-migrating, phosphorylated forms of Cdc15 were reduced in pom1Δ cells (Figure 3B), and recombinant Pom1 efficiently phosphorylated recombinant N-terminal (Cdc15N; amino acids [aa]1–460) and C-terminal (Cdc15C; aa441–end) fragments of Cdc15 in vitro (Figure 3C) (Lee et al., 2018).
FIGURE 3:
Pom1 can phosphorylate Cdc15 on 22 sites. (A) Quantification of tip septa as in Figure 1, A and B. (B) Denatured protein lysates separated by SDS–PAGE and immunoblotted for the indicated proteins. (C) In vitro kinase reactions were separated by SDS–PAGE. Coomassie-stained gel of inputs and autoradiographs detecting 32P incorporation are shown. (D) Schematic of Cdc15 to scale with Pom1 phosphorylation sites indicated by black lines. (E) Denatured lysates and IP-phosphatase assays were separated by SDS–PAGE and immunoblotted for the indicated proteins. (F) Representative images of DAPI and methyl blue (top panels) or calcofluor (bottom panels) stained cells of the indicated cdc15 genotype. Single z-sections of 0.5 µm are shown in the top panels and maximum projections are shown in the bottom panels. Scale bar, 5 μm. (G) Quantification of nuclei and septation indices from images acquired as in E for the DAPI/methyl blue-stained cells. Graph shows mean and SEM from three biological replicates, n ≥ 940 cells.
To test if Pom1-mediated Cdc15 phosphorylation is important to prevent septum formation at cell tips, we first identified all of the sites in Cdc15 that can be phosphorylated by Pom1. Phosphoamino acid analysis revealed that Pom1 phosphorylates Cdc15C predominantly on serines and phosphorylates Cdc15N on both serines and threonines (Supplemental Figure S1A). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of Cdc15C phosphorylated by Pom1 identified 12 of 17 sites closely matching the consensus sequence for DYRK kinases (RPX(S/T)P) (Himpel et al., 2000) (Supplemental Figure S2). All 12 sites were previously identified as phosphorylated in vivo (Wilson-Grady et al., 2008; Roberts-Galbraith et al., 2010; Kettenbach et al., 2015; Swaffer et al., 2018). The remaining four sites matching the DYRK consensus sequence (S813, S821, S831, S836) were not identified in our MS/MS analysis, but S813 and S836 were identified in other phosphoproteomic datasets (Kettenbach et al., 2015; Swaffer et al., 2018). A Cdc15C mutant with these 16 consensus sites mutated to alanine was still phosphorylated by Pom1 (data not shown). Thus, we mutated a remaining RXXS site (S785) and two additional residues that we identified through MS/MS as highly phosphorylated by Pom1 in vitro (T492, S732). The resulting Cdc15C-19A mutant abolished phosphorylation by Pom1 in vitro (Supplemental Figure S1B). We identified three sites (S43, S405, T419) in Cdc15N that both match the DYRK consensus sequence and are phosphorylated in vivo (Roberts-Galbraith et al., 2010; Kettenbach et al., 2015). Mutating these sites to alanine abolished phosphorylation of Cdc15N by Pom1 (Supplemental Figure S1C). Thus, Cdc15 can be phosphorylated by Pom1 on 22 sites (Figure 3D; Supplemental Figure S1, C–E).
To evaluate the role of Pom1-mediated Cdc15 phosphorylation, we mutated these 22 residues to Ala (to abolish phosphorylation) or Asp (to partially mimic phosphorylation) and integrated the cdc15-22A and cdc15-22D alleles at the endogenous cdc15 locus. SDS–PAGE of Cdc15-22A showed that most of the phosphorylation-induced gel retardation of Cdc15 had been eliminated (Figure 3E), validating the successful identification of in vivo phosphosites. Cdc15-22A was still phosphorylated, consistent with the ≥35 phosphorylation sites previously identified on Cdc15 (Roberts-Galbraith et al., 2010) and with Kin1 kinase phosphorylating Cdc15 on nonoverlapping sites (Lee et al., 2018). Both the cdc15-22A and cdc15-22D phosphomutants were viable with normal mitotic and septation indices; no off-center or tip septa were observed in these mutants, but abnormal cell wall deposits were observed in the medial regions of cdc15-22A cells (Figure 3, F and G).
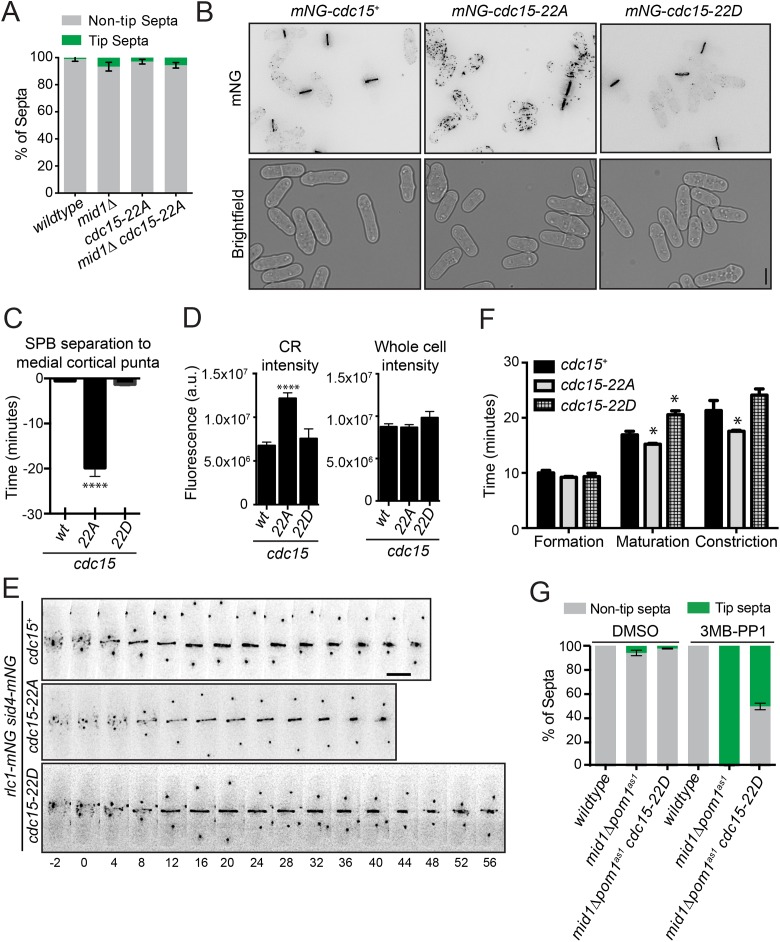
When combined with mid1∆, mid1∆ cdc15-22A did not form tip septa (Figure 4A), which might have been expected if Cdc15 was the sole Pom1 substrate involved in Pom1-mediated tip occlusion. However, we found that the Pom1 phosphosites are important for Cdc15 function. First, by visualizing Cdc15-22A tagged at the N-terminus with mNeonGreen (mNG), we found that mNG-Cdc15-22A localized abnormally to cortical puncta throughout the cell cycle (Figure 4B), as previously observed for other phosphoablating Cdc15 mutants (Roberts-Galbraith et al., 2010). Co-imaging with spindle pole body (SPB) marker Sid4-mNG showed that mNG-Cdc15-22A arrived at the division site 19.8 ± 6.6 min before mitotic onset (i.e., SPB separation) compared with 0.5 ± 1 and 1.3 ± 1.3 min for mNG-Cdc15 and mNG-Cdc15-22D, respectively (Figure 4C). mNG-Cdc15-22A abundance in the fully formed, preconstriction CR was also increased, although the whole cell fluorescence intensities of mNG-Cdc15, mNG-Cdc15-22A, and mNG-Cdc15-22D were comparable (Figure 4D). By time-lapse imaging with Rlc1-mNG and Sid4-mNG as markers of the CR and SPB, respectively, we found that cdc15-22A and cdc15-22D cells had altered cytokinesis dynamics (Figure 4, E and F). Though the length of CR formation (node appearance to complete ring) was similar in wild type, cdc15-22A, and cdc15-22D, the periods of maturation (interval between CR formation and constriction initiation) and constriction (start to end of CR diameter decrease) were shorter in cdc15-22A and longer in cdc15-22D (Figure 4F). Altogether, these results indicate that Pom1 phosphorylation inhibits Cdc15 function in cytokinesis.
FIGURE 4:
A Cdc15 phosphomimetic suppresses tip septa formation. (A) Quantification of tip septa as described in Figure 1, A–C. (B) Representative live-cell images of the indicated strains. Fluorescent images were deconvolved and maximum projected. (C) Average arrival of Cdc15 at the division site relative to SPB separation (time 0). Measurements are from three biological replicates, n ≥ 12 cells. (D) Quantification of the fluorescence intensity of Cdc15 in the CR (left) or whole cell (right) of the indicated strains from three biological replicates, n ≥ 33 cells. All images used for quantification were not deconvolved and were sum projected. (E) Representative montages from live-cell, time-lapse imaging of indicated strains. Images were acquired every 2 min and every 4 min are shown. Numbers indicate minutes from SPB separation. (F) Length of cytokinesis stage based on imaging as in E. Measurements are from three biological replicates, n ≥ 18 cells. (G) Quantification of tip septa phenotype as described in Figure 1, A–C, except n > 250 cells. Graphs show mean and SEM; wt, wildtype. *p < 0.05, one-way ANOVA with Tukey’s post-hoc analysis. ****p < 0.0001, Brown–Forsythe and Welch ANOVA tests with Dunnett T3 correction for multiple comparisons to wt. Scale bar, 5 µm.
Given Pom1’s inhibitory effect on Cdc15 function, we predicted that Pom1-mediated Cdc15 phosphorylation prevents tip septation in mid1Δ pom1as1 cells. Indeed, the percentage of tip septa was significantly reduced in mid1∆ pom1as1 cdc15-22D cells (Figure 4G). We conclude that Cdc15 is a key substrate in the Pom1-mediated tip occlusion pathway.
Pom1 phosphorylation differentially inhibits Cdc15 binding interactions
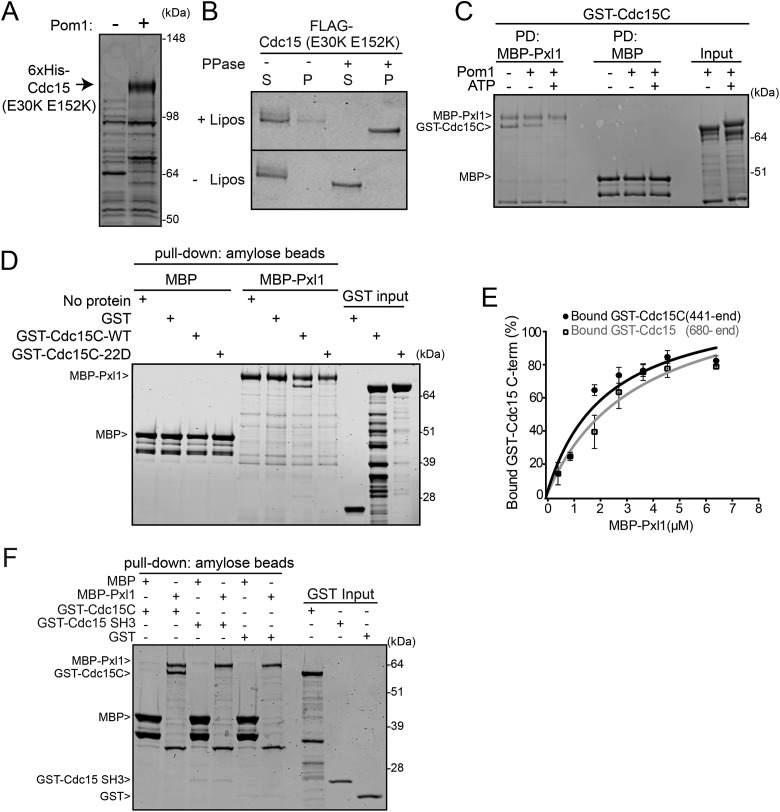
Cdc15 has an N-terminal F-BAR domain that oligomerizes and binds to both membranes and the formin Cdc12 (Carnahan and Gould, 2003; McDonald et al., 2015; Willet et al., 2015a), followed by a predicted intrinsically disordered region (IDR) (Mangione et al., 2019). The C-terminal SH3 domain of Cdc15 binds several CR components (Roberts-Galbraith et al., 2009; Ren et al., 2015) including paxillin Pxl1, which promotes CR integrity (Ge and Balasubramanian, 2008; Pinar et al., 2008; Martin-Garcia et al., 2018), and Fic1, which promotes CR disassembly (Bohnert and Gould, 2012) and septum formation based on its orthologue in Saccharomyces cerevisiae (Nishihama et al., 2009; Devrekanli et al., 2012). Cdc15 dephosphorylation on mitotic onset correlates with its redistribution from the cytosol to the CR and its binding to at least some protein partners (Roberts-Galbraith et al., 2010). Consistent with Cdc15 phosphostate driving these changes, multiple Cdc15 mutants containing different subsets of phosphorylation sites mutated to Ala accumulate on the cell cortex and precociously interact with protein partners (Roberts-Galbraith et al., 2010). In vitro, complete Cdc15 dephosphorylation also leads to its oligomerization, which increases its avidity for membranes (McDonald et al., 2015). These results have led to a model in which phosphorylation induces a closed, dimeric (rather than oligomeric), and inactive Cdc15 conformation. However, whether Cdc15 phosphorylation directly alters its membrane binding and/or SH3-mediated protein binding is unknown; therefore, we tested if these properties were inhibited by Pom1 phosphorylation.
To test membrane binding, full-length Cdc15 that cannot oligomerize (Cdc15-E30K E152K) (McDonald et al., 2015) was coexpressed in bacteria with Pom1. Coexpression with Pom1 resulted in Cdc15 hyperphosphorylation and allowed full-length Cdc15-E30K E152K to be purified (Figure 5A). Oligomerization-competent Cdc15 could also be coproduced with Pom1, but could not be purified without the use of high salt and detergent (data not shown). After purification, Pom1-phosphorylated Cdc15-E30K E152K was treated with lambda phosphatase or vehicle control and then incubated with liposomes rich in phosphoinositides (Folch fraction). While hyperphosphorylated Cdc15-E30K E152K did not copellet with liposomes, dephosphorylated Cdc15-E30K E152K was found exclusively in the liposome pellet (Figure 5B). Therefore, Pom1-mediated phosphorylation blocks Cdc15’s ability to bind membranes.
FIGURE 5:
Pom1 inhibits Cdc15 binding to membranes and Pxl1. (A) Coomassie-stained SDS–PAGE of proteins purified from bacteria expressing Cdc15-E30K E152K in the presence or absence of Pom1. (B) Cdc15-E30K E152K coexpressed with Pom1 was purified from bacteria and then treated with lambda phosphatase (PPase) or control before adding to Folch fraction liposomes (lipos). Coomassie staining of supernatant (S) and pellet (P) after pelleting and SDS–PAGE. (C) GST-Cdc15C was phosphorylated in vitro with GST-Pom1 and incubated with MBP or MBP-tagged Pxl1 bound to amylose resin. After pelleting, proteins bound to the amylose resin were separated by SDS–PAGE and stained with Coomassie. PD, pull down. (D) Coomassie-stained SDS–PAGE of binding assays between the indicated MBP-tagged and GST-tagged proteins. (E) Binding curves with varying concentrations of MBP-Pxl1 on MBP nanobody resin and GST-Cdc15 C-terminal variants in solution. Quantification was done from three biological replicates as detailed in the methods. Error bars show SEM. (F) Coomassie-stained SDS–PAGE of binding assays between the indicated MBP-tagged and GST-tagged proteins.
We next tested if Pom1-mediated phosphorylation of Cdc15C (aa441–end) affects binding to SH3 interactors Fic1 and Pxl1. We found that Fic1 still bound phosphorylated Cdc15C (Supplemental Figure S3A) and full-length Cdc15-E30K E152K phosphorylated in bacteria by Pom1 (Supplemental Figure S3B). Cdc15 immunoprecipitates contained Fic1 at all cell cycle stages (Supplemental Figure S3, C and D), further indicating that this interaction is not governed by Cdc15 phosphostate.
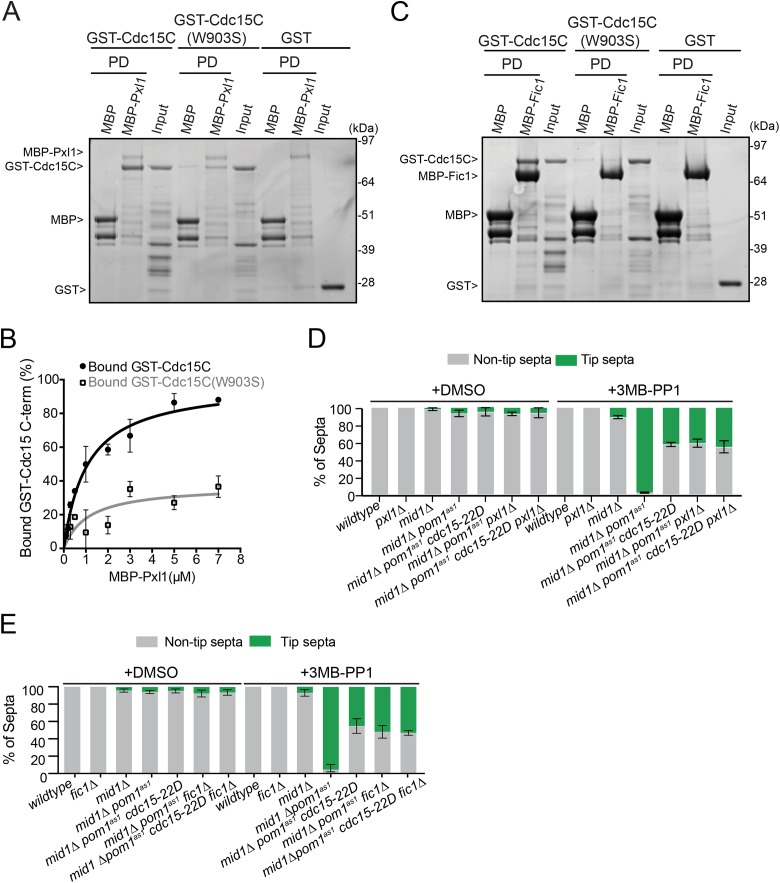
In contrast to Fic1, Pom1-mediated phosphorylation of Cdc15C blocked its binding to Pxl1 (Figure 5C). Similarly, mutating the Pom1 phosphorylation sites in Cdc15C to Asp prevented Pxl1 binding (Figure 5D). That Pxl1 and Fic1 binding to Cdc15 was differentially regulated by phosphorylation was unexpected because both were previously found to require the Cdc15 SH3 domain for binding (Roberts-Galbraith et al., 2009) and because Pom1 phosphorylation of the Cdc15 C-terminus occurs in the IDR and not in the SH3 domain (Figure 3D). We reasoned that Pxl1 might bind a more extensive region of Cdc15, comprising both the SH3 domain and a part of the IDR. To test this, we performed binding experiments with Pxl1 and various Cdc15 segments. While Cdc15 residues 680–end bound to Pxl1 comparably to Cdc15C (441-end) (Figure 5E), the Cdc15 SH3 domain alone did not bind Pxl1 (Figure 5F), explaining the failure to identify Pxl1 in Cdc15 SH3 pull downs that successfully identified Fic1 (Ren et al., 2015). Mutating the SH3 domain (W903S) within Cdc15C to abolish binding to PXXP motifs (Saksela and Permi, 2012) substantially decreased Pxl1 binding (Figure 6, A and B) and eliminated Fic1 binding (Figure 6C). Thus, Pxl1 binds Cdc15 through both the SH3 domain and IDR, and Pom1 phosphorylation of the IDR disrupts this interaction. This novel function of the Cdc15 IDR may contribute to its requirement for viability and successful cytokinesis (Mangione et al., 2019).
FIGURE 6:
Pxl1 interacts with the central region and SH3 domain of Cdc15. (A, C) Coomassie-stained SDS–PAGE of binding assays between the indicated MBP-tagged and GST-tagged proteins. Amylose resin was used to pull down (PD) proteins. (B) Binding curves with varying concentrations of MBP-Pxl1 on MBP nanobody resin and GST-Cdc15C and GST-Cdc15C (W903S) in solution. Quantification was done from three biological replicates. Error bars show SEM. (D, E) Quantification of tip septa as in Figure 1, A–C.
We hypothesized that a key mechanism of Pom1-mediated septa tip occlusion is inhibition of Cdc15 scaffolding. Indeed, deleting components of the CR scaffolded by Cdc15 (e.g., pxl1 or fic1) suppressed tip septation in mid1∆ pom1as1cells to a similar degree as cdc15-22D (Figure 6, D and E). Additionally, combining cdc15-22D with pxl1∆ or fic1∆ suppressed tip septation to the same extent as each mutant individually (Figure 6, D and E), indicating that Fic1 and Pxl1 promote cell division via their interaction with Cdc15.
DISCUSSION
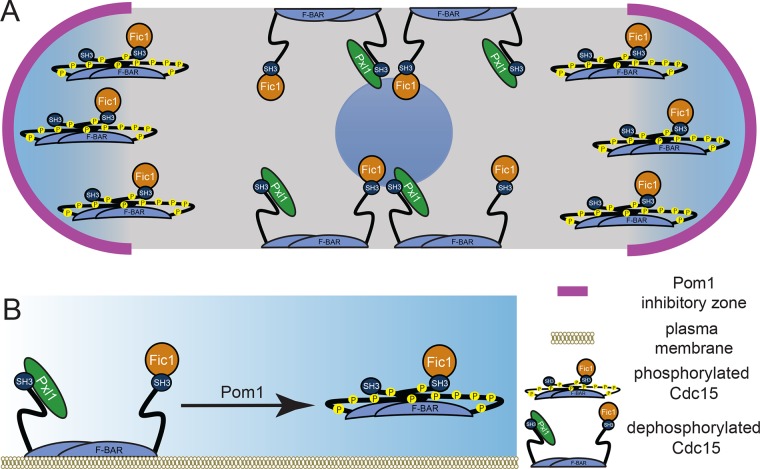
The DYRK-family kinase Pom1 is critical for division site placement in S. pombe, providing both Mid1-dependent and Mid1-independent signals that prevent division site establishment at cell tips (Huang et al., 2007). By demonstrating that Pom1 phosphorylation of the F-BAR protein Cdc15 is a major part of the Mid1-independent mechanism, our findings expand Pom1’s known roles of phosphorylating tropomyosin Cdc8 to partly destabilize actin cables (Palani et al., 2019) and phosphorylating the SAD-like kinase Cdr2 to control division timing and fine-tune the Mid1-dependent signal (Celton-Morizur et al., 2006; Padte et al., 2006; Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009; Bhatia et al., 2014; Rincon et al., 2014; Kettenbach et al., 2015). By phosphorylating Cdc15, Pom1 inhibits a major membrane-binding scaffold of the CR, thereby destabilizing the CR at cell tips. In either wild-type or mid1∆ cells, we propose that Cdc15 can bind membrane and stabilize the CR only where Pom1 is not active (Figure 7A). In Schizosaccharomyces japonicus, division site placement is Mid1 independent, relying predominantly on Pom1 that is also proposed to inhibit S. japonicus Cdc15 function at cell tips (Gu et al., 2015). Thus, the molecular mechanisms we report here by which Pom1 phosphorylation inhibits F-BAR Cdc15 at cell tips to ensure medial division are likely conserved.
FIGURE 7:
Models of Pom1 inhibition of Cdc15 function. (A) Model of Pom1 inhibition of Cdc15 membrane binding at cell tips in a single interphase cell with the nucleus in the middle. (B) Molecular model of Pom1 inhibition of Cdc15 membrane binding.
We previously proposed a molecular model in which an electrostatic threshold triggers a conformational switch in Cdc15 that inhibits all of its functions (Roberts-Galbraith et al., 2010). However, our analysis of Pom1 phosphorylation shows that it affects two Cdc15 protein interactions differently and surprisingly, that one protein binds not only the SH3 domain but also the IDR (Figure 7B). Furthermore, the effects of cdc15-22A and cdc15-22D on cytokinesis dynamics are not identical to those of cdc15-27A and cdc15-27D, respectively, which share only 14 phosphosites (Roberts-Galbraith et al., 2010). Therefore, it remains possible that phosphorylation events affect Cdc15 function or conformation differently and could fine-tune Cdc15 to allow it to participate in distinct processes. Though we have focused on Cdc15’s cytokinesis function, Cdc15 also functions in endocytosis (Arasada and Pollard, 2011; MacQuarrie et al., 2019). It remains to be determined whether phosphorylation affects Cdc15 interaction with other SH3 domain-binding partners (Ren et al., 2015). This is an intriguing question given that Cdc15 is never completely dephosphorylated (Roberts-Galbraith et al., 2010) and thus a complex hierarchy of phosphorylation events could be regulating its various scaffolding activities throughout the cell cycle.
The localization of Pom1 at poles suggests that Cdc15 phosphorylation is spatially regulated, with membrane binding specifically inhibited at cell poles. However, several mutations that globally reduce Cdc15 function but do not carry positional information, such as hypomorphic cdc15-GFP, phosphomimetic cdc15-22D, or deletion of Cdc15 binding partners, partly alleviate the tip septation phenotype of cells lacking Pom1 and Mid1. Similarly, previous work showed that Pom1’s function in division site placement is less sensitive to Pom1 distribution than its function in controlling division timing (Bhatia et al., 2014; Gerganova et al., 2019). In addition, in S. japonicus hyphae, Pom1 does not form marked concentration gradients from cell poles, though it still controls division site placement (Kinnaer et al., 2019), suggesting that spatial control of Pom1 function is not absolutely essential to its function in controlling division placement. These observations suggest that Pom1 signal combines with other anisotropies to make the cell center more favorable for Cdc15 function and subsequent CR and septa formation.
A shared feature of all cdc15 phosphoablating mutants characterized thus far is enhanced cortical localization throughout the cell cycle. This is consistent with our findings that Pom1 phosphorylation of Cdc15 blocks its membrane binding in vitro, although it remains to be determined whether lipid binding is prevented by a conformational change that obscures the binding interface and/or if the net negative charge repels negatively charged lipids. Interestingly, in no case has enhanced Cdc15 membrane association been sufficient to drive CR formation, and tip septation does not occur in mid1∆ cdc15-22A cells. While a second Pom1 substrate in tip occlusion has been identified as Cdc8 tropomyosin, still additional Pom1 substrates must require dephosphorylation for septation in S. pombe because the penetrance of tip septa formation in the cdc8-S125A phosphomutant is low (Palani et al., 2019). The transglutaminase-like protein Cyk3, which promotes septum synthesis (Pollard et al., 2012), is a confirmed Pom1 substrate (Kettenbach et al., 2015; Lee et al., 2018) and the phosphorylation of two other CR proteins, Rga7 and Imp2, is also modulated by Pom1 (Kettenbach et al., 2015). The consequences of these phosphorylation events have not yet been determined, and it will be interesting to explore whether Pom1 phosphorylates this entire CR network to prevent septation at cell tips.
MATERIALS AND METHODS
Experimental model and subject details
Supplemental Table S1 lists S. pombe strains used in this study. Cells were cultured in rich medium YES (5 g/l yeast extract, 30 g/l glucose, 225 mg/l adenine) with supplements or Edinburgh Minimal Media media (EMM; 3 g/l potassium hydrogen phthalate, 20 g/l glucose, 2.2 g/l disodium phosphate) plus selective supplements.
Fusion proteins were produced in Escherichia coli Rosetta2(DE3) pLysS cells. Bacteria were grown in Terrific Broth media (12 g/l tryptone, 24 g/l yeast extract, and 4 ml/l glycerol) with antibiotics. For MBP-Pxl1, 150 µM ZnCl2 was added to the culture media.
Strain construction
Transformation of yeast with plasmid or linear DNA was accomplished using electroporation or lithium acetate methods, respectively. cdc15 was tagged at the 3′ end of its ORF with GFP:kanR using a pFA6 cassette as previously described (Bahler et al., 1998). Integration of tag was verified using whole cell PCR and/or microscopy.
For integration of cdc15 alleles at the endogenous locus, cdc15+/cdc15::ura4+ was transformed with pIRT2 vector with cdc15 5′ and 3′ noncoding regions, the desired cdc15 allele, and where indicated, sequences encoding mNG (Roberts-Galbraith et al., 2009; Willet et al., 2015a). Spores were germinated on selective medium to select for cdc15::ura4+ haploids that have the vector. Haploid integrants were recovered based on resistance to 5-fluoroorotic acid, and integration plus loss of vector was verified by growth on selective media, PCR and/or microscopy, and finally sequencing.
Introduction of tagged loci or cdc15 mutants into other genetic backgrounds was accomplished using standard S. pombe mating, sporulation, and tetrad dissection techniques.
Molecular biology methods
GST-Pom1 was described previously (Hachet et al., 2011). All other expression and integration vectors were constructed using standard molecular biology techniques. Mutations were made with a QuikChange Multi-lightning mutagenesis kit (Agilent Technologies). All constructs were sequenced for verification.
Tip septa quantification
Except for Figure 2, A–D, to inhibit Pom1as1 in vivo, cells were grown in YES at 32°C to midlog phase and then treated with 4-amino-1-tert-butyl-3-(3-methylbenzyl)pyrazolo[3,4-3]pyrimidine (3MB-PP1) (Toronto Research Chemical; A602960 or Cayman Chemical; 17860) at a final concentration of 1 µg/ml for 2 h unless otherwise indicated. Vehicle-treated cells were grown in equal volume of dimethyl sulfoxide (DMSO) (Sigma; D2650). Cells were fixed and stained with calcofluor and imaged (see Microscopy and image analysis).
Nuclear displacement assays and their microscopy analysis
To displace nuclei, strains containing integrated Myo52-Nup146 fusion construct (Lo Presti et al., 2012) under control of the nmt81 promoter were grown in EMM-AL medium without thiamine for 24 h to induce expression of the fusion protein. pom1as1 strains were subsequently treated with 1 µM 3MB-PP1 in methanol and/or 25 µl MBC in DMSO (or vehicle) for 4 h at 30°C to inactivate Pom1as1 and depolymerize microtubules, respectively. Cells were stained with calcofluor white and imaged on a DeltaVision platform (Applied Precision) composed of a customized inverted microscope (IX-71; Olympus), a 60× /1.42 NA oil objective, a camera (PrimeBSI CMOS; Photometrics), and a color-combined unit illuminator (Insight SSI 7; Social Science Insights). Images were acquired using softWoRx v4.1.2 software (Applied Precision).
Denatured lysis and immunoprecipitation-phosphatase assays for detecting Cdc15 phosphorylation status
For Figure 3E, pellets were snap frozen in dry ice–ethanol baths and then lysed by bead disruption in NP-40 lysis buffer (Gould et al., 1991) with the addition of 0.5 mM diisopropyl fluorophosphate (Sigma-Aldrich). Lysates were denatured by boiling at 95°C for 1 min. Cdc15 was immunoprecipitated from lysates using anti-Cdc15(1-405) polyclonal antibody (VU326; Cocalico Biologicals; Roberts-Galbraith et al., 2009) and protein A sepharose (GE Healthcare; 17-5280-04). The sepharose was washed three times with NP-40 lysis buffer and two times with 1× phosphatase buffer (50 mM HEPES, pH 7.4, 150 mM NaCl). Samples were treated with lambda protein phosphatase (New England Biolabs; P0753) according to manufacturer’s protocol and reactions were stopped by the addition of sample buffer. Immunoblot analysis was performed as previously described (Roberts-Galbraith et al., 2009). Briefly, to best visualize different Cdc15 phospho-species, proteins were resolved by SDS–PAGE using freshly poured (within 24 h) 8% Tris-Glycine gels at 150 V for 2.25 h. Then, protein samples were transferred to PVDF membrane (Immobilon P; EMD Millipore). Anti-Cdc15(1-405) polyclonal antibody (VU326) or anti-α-tubulin monoclonal antibody (B512) (Sigma-Aldrich) was used as primary antibodies in immunoblotting. Secondary antibodies were conjugated to Alexa Fluor 680 (Invitrogen) or IRDye800 (LI-COR Biosciences) and visualized using an Odyssey machine (LI-COR Biosciences).
Block and release
For the block-and-release experiment using the cdc25-22 background shown in Supplemental Figure S3D, cells were arrested in G2 by shifting to the restrictive temperature (36°C) for 3.5 h and then released back into the cell cycle by shifting to the permissive temperature (25°C). Cell pellets were collected at the time of release (0 min) and at subsequent time points as indicated. Additionally, cells were fixed in ice-cold 70% ethanol for staining with 4′,6-diamidino-2-phenylindole (DAPI) and methyl blue to visualize nuclei and septa, respectively (see Microscopy and image analysis section).
Native lysis and coimmunoprecipitation of Cdc15 and Fic1
For Supplemental Figure S3, C and D, 30 OD of cells were lysed with FAST-PREP and proteins were extracted with 2 × 0.6 ml NP40 buffer. Cell lysate was incubated with 4 µl anti-Cdc15 serum (VU326) for 1 h followed by the addition of 80 µl 50% slurry of protein A sepharose and nutation for an additional 1 h. Beads were then washed 4× in 0.6 ml NP40 buffer and were resuspended in 40 µl 2× SDS sample loading buffer. Samples (20 µl) were resolved by 3–8% Tris-Acetate gel at 125 V for 1 h, 20 min for Supplemental Figure S3C and 120 V for 2 h, 30 min for Supplemental Figure S3D. Resolved protein was transferred to immobilon–FL PVDF membrane (12 V for 18 h) and immunoblotted with anti-Cdc15 serum (VU326) (1:2,000), anti-FLAG antibody (1:1,000) (Sigma-Aldrich; P2983), and anti-PSTAIRE (1:10,000) (Sigma-Aldrich; P7962).
In vitro kinase assays
For Figure 3C and Supplemental Figure S1, B and C, radioactive in vitro kinase assays were performed in 30 mM Tris, pH 8, 100 mM NaCl, 10 mM MgCl2, 1 mM EGTA, 10% glycerol, 1 mM DTT, 10 µM ATP, and 2 µCi [32P]-ATP (PerkinElmer BLU002250UC) with 0.5 µg GST-Pom1 and 2 µg substrate in a 25 µl reaction. After 30 min at 30°C, reaction was stopped by boiling in sample buffer and analyzed by SDS–PAGE. Inputs were detected by Coomassie Blue (Sigma-Aldrich; B0770) and 32P incorporation was detected by autoradiography.
For Supplemental Figure S2, cold kinase assays for the identification of phosphorylation sites were performed the same way except without hot ATP, with 40 mM ATP, and in 10 µl final volume. A second dose of GST-Pom1 was added after 45 min and the reaction was stopped after 90 min.
For Figure 5C and Supplemental Figure S3A, before the addition to binding assays, cold kinase assays were performed in 30 mM Tris, pH 8, 100 mM NaCl, 10 mM MgCl2, 1 mM EGTA, 10% glycerol, 1 mM DTT with 2 mM ATP, 0.5 µg GST-Pom1, and 1 µg GST-Cdc15 (aa 441-end) as substrate in a 25 µl reaction. After 30 min at 30°C, reaction was stopped by boiling in sample buffer and analyzed by SDS–PAGE or added directly to beads for binding assay.
Phosphoaminoacid analysis
For Supplemental Figure S1A, radioactive in vitro kinase assays were performed as described above, except that after SDS–PAGE proteins were transferred to PVDF membranes (Immobilon P; EMD Millipore). Proteins were visualized by staining membrane with REVERT Total Protein stain (LI-COR Biosciences). Then, bands corresponding to the substrate were cut from the membrane. Membrane-bound proteins were subjected to partial acid hydrolysis using boiling 6 M HCl for 1 h. Hydrolyzed amino acids were separated by two-dimensional thin-layer electrophoresis (Boyle et al., 1991).
Phosphorylation site identification by mass spectrometry
Trichloroacetic acid-precipitated proteins were digested and analyzed by two-dimensional LC-MS/MS as described previously (Chen et al., 2013), except that the following modifications were made. Proteins were digested by trypsin, chymotrypsin, and elastase. The number of salt elution steps was reduced to 6 (i.e., 0, 25, 50, 100, 600, 1000, and 5000 mM ammonium acetate). Peptide identifications were filtered and assembled using Scaffold (version 4.8.4; Proteome Software) and phosphorylation sites were analyzed using Scaffold PTM (version 3.1.0) using the following filters: minimum of 99% protein identification probability; minimum of five unique peptides; minimum of 95% peptide identification probability.
Microscopy and image analysis
Microscope.
All images except for Figure 2, A–D, were acquired using a Personal DeltaVision microscope system (GE Healthcare, Issaquah, WA), which includes an Olympus IX71 microscope, 60×/NA 1.42 PlanApo and 100×/NA 1.40 UPlanSApo objectives, fixed- and live-cell filter wheels, a Photometrics CoolSnap HQ2 camera, and softWoRx imaging software.
Sample preparation.
For live-cell fluorescence imaging (Figure 4, B–F), strains were grown overnight to log phase in YES media at 25°C in a shaking water bath. Cells were imaged in YES media at 23–29°C.
Yeast cells were fixed by adding ice-cold 70% ethanol while vortexing and then incubating at 4°C for at least 15 min. Cells were fixed at a ratio of 0.5 OD cells per 1 ml 70% ethanol.
For visualizing tip septa, approximately 0.5 OD of fixed cells were washed once with phosphate-buffered saline (PBS), pH 7.5, and then resuspended in 20 µl of 50 µg/ml Calcofluor White (Sigma; 18909) and incubated at room temperature for 5 min. Then, cells were washed once with PBS and imaged immediately.
For visualizing nuclei and cell wall, approximately 0.5 OD of fixed cells were washed once with PBS, pH 7.5, and then resuspended in 50 µl of 1 mg/ml methyl blue (Sigma; M6900) and incubated at room temperature for 30 min. Then, cells were pelleted and mixed 1:1 with 5 µg/ml DAPI (Sigma; D9542) before imaging immediately.
Time-lapse imaging.
For Figure 4, C, E, and F, time-lapse imaging was performed using an ONIX microfluidics perfusion system (CellASIC ONIX, EMD Millipore). Fifty microliters of a 40 × 106 cells/ml YES suspension were loaded into Y04C plates for 5 s at 8 psi. YES medium flowed into the chamber at 5 psi throughout imaging. Time-lapse images were obtained at 2 min intervals. Ten Z-series optical sections were taken at 0.5 µm spacing. Stage temperature was maintained at 29°C. Images were deconvolved with 10 iterations using softWoRx and maximum intensity Z-projections were generated using ImageJ (Schindelin et al., 2012). Cytokinesis stages were defined as follows: formation is the time from Rlc1 appearance in nodes to coalescence into a ring; maturation is the period between formation and constriction; constriction is the time from first decrease in Rlc1 ring diameter to a single point.
Quantifying fluorescence intensity.
For all intensity measurements, the background was subtracted by creating a region of interest (ROI) in the same image where there were no cells. The raw intensity of the background was divided by the area of the background, which was multiplied by the area of the ROI. This number was subtracted from the raw integrated intensity of that ROI. For CR intensity quantification, an ROI was drawn around the CR and measured for raw integrated density. Three biological replicates were performed. All images for intensity analyses of the CR and whole cell were not deconvolved and the z-slices were sum projected.
Recombinant protein purification
Bacteria were grown in Terrific Broth media with antibiotics to log-phase (OD595 1-1.5) at 36°C. For MBP-Pxl1, 150 µM ZnCl2 was added during the growth of the culture to log-phase. Then, induction was initiated by incubating for 15 min on ice before adding 0.4 mM IPTG (Fisher Scientific; BP1755). Protein was produced for 16–18 h at 18°C.
For MBP and GST fusion proteins, frozen cell pellets were lysed in either MBP buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM DTT) or GST buffer (4.3 mM NaHPO4, 137 mM NaCl, 2.7 mM KCl, 1 mM DTT) with the addition of 200 µg/ml lysozyme (Sigma-Aldrich; L6876), cOmplete EDTA-free protease inhibitor cocktail (Roche), and 0.1% NP-40 (US Biologicals; N3500). Buffers were modified for MBP-Pxl1 to exclude EDTA. Continuous agitation on ice for 20 min was used to suspend the cell pellet. Then, lysates were sonicated three times for 30 s, with at least a 30 s pause between sonications (Sonic Dismembrator Model F60, Fisher Scientific; power 15 W). Lysates were cleared for 15–30 min at 10–13K rpm. Cleared lysate was then used in a batch purification protocol by adding either amylose (New England Biolabs; E8021L) or GST-bind (EMD Millipore; 70541) resin for 2 h at 4°C. Then, resin was washed three times for 5 min at 4°C with the appropriate buffer. To elute proteins from beads, dry beads were resuspended in an equal volume of appropriate elution buffer and nutated for 30 min at 4°C. MBP fusion proteins were eluted in MBP buffer supplemented with 10 mM maltose (Fisher Scientific; M75-100) and GST proteins were eluted in GST elution buffer (50 mM Tris-HCl, pH 8, 10 mM glutathione; Sigma-Aldrich; G4251). The supernatant was separated from the resin to a fresh tube; 100 mM NaCl was added to GST fusion proteins. Eluted fusion proteins were then aliquoted, snap frozen, and stored at –80°C.
To purify Flag-Cdc15(E30KE152K) co-expressed with Pom1, frozen cell pellets were lysed in 50 mM Tris, pH 7.4, 150 mM NaCl with the addition of 200 µg/ml lysozyme (Sigma-Aldrich; L6876), cOmplete EDTA-free protease inhibitor cocktail (Roche), and 0.1% NP-40 (US Biologicals; N3500). Lysates were sonicated four times for 30 s, with at least a 30s pause between sonications (Sonic Dismembrator Model F60, Fisher Scientific; power 15 W). Lysates were cleared for 15 min at 13K rpm. Cleared lysate was incubated with Anti-FLAG(R) M2 Magnetic Beads, affinity-isolated antibody (Millipore Sigma; M8823) for 2 h at 4°C. Then, beads were washed 4 times for 5 min at 4°C with 50 mM Tris, pH 7.4, 150 mM NaCl buffer. FLAG fusion protein was eluted in 1 ml 50 mM Tris, pH7.4,150 mM NaCl, 200µg/ml 3xFlag peptide (Sigma-Aldrich; F4799) for 20 min in room temperature. The eluate was concentrated to 100 µl and was cleared for 15 min at 13K rpm in 4°C.
To purify His-Cdc15(E30KE152K) plus or minus co-expression with Pom1, frozen pellets were lysed in 50 mM Na2PO4, pH 8.0, 300 mM NaCl,1%NP-40 (US Biologicals; N3500), 5 mM imidazole (Fisher, 03196-500), 5 mM β-mercaptoethanol (Sigma; M3148), phenylmethylsulfonyl fluoride (Sigma; P7626), benzamidine (B-6506), and cOmplete EDTA-free protease inhibitor cocktail (Roche). Continuous agitation on ice for 20 min was used to suspend the cell pellet. Then, lysates were sonicated four times for 30 s, with at least a 30-s pause between sonications (Sonic Dismembrator Model F60, Fisher Scientific; power 15 W). Lysates were cleared for 15 min at 13K rpm. Cleared lysate was incubated with cOMPLETE His-tag purification resin (Roche) for 1.5 h at 4°C. The beads were washed with the lysis buffer for three times. Finally, the washed beads were resuspended in lysis buffer to make 1:1 slurry.
Protein concentration was calculated from Coomassie Brilliant Blue G (Sigma-Aldrich; B0770) staining of SDS–PAGE-separated purified proteins and bovine serum albumin standards (Sigma-Aldrich).
Lipid binding assay
For Figure 5A, lipids were obtained from Sigma-Aldrich (B1502, Brain extract from bovine brain, Type I, Folch Fraction). In vitro liposome preparation was performed as previously described (McDonald and Gould, 2016). For liposome co-pelleting assay, 10 µg of Flag-Cdc15(E30KE152K) was incubated for 30 min at 30°C with lambda phosphatase (New England Biolabs, P0753) and 1 mM MnCl2. Then the volume was made up to 100 µl by liposome buffer (50 mM Tris, pH 7.4, 150 mM NaCl) and a clearing spin was done at 13k rpm 4°C for 15 min. Protein (100 µl) was added to 100 µl of 1 mg/ml liposome samples without disturbing any pellets that may form and incubated in room temperature for 15 min. Reactions were spun at 150,000 × g for 15 min at 25°C. Supernatants (unbound fraction) were collected from the reaction. The pellets (bound fraction) were resuspended in 200 µl liposome buffer (50 mM Tris, pH 7.4, 150 mM NaCl). Sample buffer was added to both bound and unbound fractions and 20 µl samples from all experimental conditions were run on SDS–PAGE for Coomassie staining analysis.
In vitro binding assays
For Figure 5, C, D, and F, and Supplemental Figure S3, A and B, 1 µM of recombinant fusion proteins immobilized with the indicated resin was incubated with 1 µM binding partners or 25 µl kinase assay samples in a 50 µl binding reaction. Binding buffer was 20 mM Tris, pH 7.0, 150 mM NaCl, 0.1% NP-40 for MBP-Pxl1. This buffer plus 2 mM EDTA was used for MBP-Fic1. After nutating for 2 h at 4°C, beads were washed with binding buffer three times for 5 min and then boiled with 30 µl sample buffer. Proteins were resolved in SDS–PAGE for Coomassie staining analysis.
For Figures 5E and 6B, 50 µg of recombinant MBP-Pxl1 was immobilized with 100 µl MBP-Trap agarose beads (Chromotek, mbta-20) in binding buffer (20 mM Tris, pH 7.4, 150 mM NaCl) overnight. The beads were then washed three times with 500 µl of binding buffer. Finally, a 1:1 slurry of the MBP Trap-bound proteins was used for the assay. Recombinant GST-Cdc15C(441-end), GST-Cdc15C(441–end, W903S), or GST-Cdc15(680–end) (1 µM) was added to different concentrations of MBPTrap-bound Pxl1 (0.1 to 5 µM) in a total reaction volume of 50 µl. Binding reaction was incubated for 2 h at 4°C. After 2 h, supernatants were collected and boiled with sample buffer. Proteins were resolved in SDS–PAGE and then transferred to PVDF membrane (Immobilon P; EMD Millipore). Anti-GST (Rabbit polyclonal) antibody was used to quantitate the percentage of unbound GST-Cdc15 fragments in the supernatants.
For Supplemental Figure S3B, 1 µM of recombinant Flag-Cdc15(E30K E152K) that had been coproduced with Pom1 was incubated for 30 min at 30°C with vehicle or 0.5 µl lambda phosphatase (New England Biolabs, P0753) and 1 mM MnCl2 in a final volume of 30 µl with binding buffer (50 mM Tris, pH 7.4, 150 mM NaCl). This reaction was then immobilized with 1 µM of MBP-Fic1 in a total of 50 µl binding reaction for 1 h at 4°C. The beads were washed with the binding buffer (50 mM Tris, pH 7.4, 150 mM NaCl) three times for 5 min and were boiled with 30 µl sample buffer. Proteins were resolved in SDS–PAGE for Coomassie staining analysis.
Quantification and statistical analysis
Calculations of mean, SEM, and statistical significances were performed with Prism 6.0 or Prism 8.0 (GraphPad Software). Sample size (n), replicates (N), and statistical tests are included in the figure graph or legends. For all image analyses, no raw data were excluded with the exception of cells that were not in focus or if a cell moved during imaging.
Supplementary Material
Acknowledgments
We are grateful to Liping Ren for exceptional technical assistance and Sierra Cullati and Alaina Willet for constructive comments on the manuscript. We thank Caitlin Anderson and David Kovar for their advice on producing recombinant Pxl1. M.C.M. was supported by NIH grants F31GM119252 and T32GM007347, C.E.S. was supported by NIH T32GM00855421 and American Heart Association 17PRE33410245, and N.A.M. was supported by American Heart Association 15PRE21780003. This work was supported by Swiss National Science Foundation grants 31003A_155944 and 310030B_176396 to S.G.M. and NIH grants R01GM101035 and R35GM131799 to K.L.G.
Abbreviations used:
- aa
amino acid
- CR
contractile ring
- DAPI
4′,6-diamidino-2-phenylindole
- DMSO
dimethyl sulfoxide
- EMM
Edinburgh Minimal Media
- IDR
intrinsically disordered region
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- MBC
methyl benzimidazole-2-gamma-carbamate
- mNG
mNeonGreen
- PBS
phosphate-buffered saline
- ROI
region of interest
- SPB
spindle pole body
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-01-0026) on February 26, 2020.
REFERENCES
- Almonacid M, Moseley JB, Janvore J, Mayeux A, Fraisier V, Nurse P, Paoletti A. (2009). Spatial control of cytokinesis by Cdr2 kinase and Mid1/anillin nuclear export. Curr Biol , 961–966. [DOI] [PubMed] [Google Scholar]
- Arasada R, Pollard TD. (2011). Distinct roles for F-BAR proteins Cdc15p and Bzz1p in actin polymerization at sites of endocytosis in fission yeast. Curr Biol , 1450–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasada R, Pollard TD. (2014). Contractile ring stability in S. pombe depends on F-BAR protein Cdc15p and Bgs1p transport from the Golgi complex. Cell Rep , 1533–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Pringle JR. (1998). Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev , 1356–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe . Yeast , 943–951. [DOI] [PubMed] [Google Scholar]
- Bhatia P, Hachet O, Hersch M, Rincon SA, Berthelot-Grosjean M, Dalessi S, Basterra L, Bergmann S, Paoletti A, Martin SG. (2014). Distinct levels in Pom1 gradients limit Cdr2 activity and localization to time and position division. Cell Cycle , 538–552. [DOI] [PubMed] [Google Scholar]
- Bohnert KA, Gould KL. (2012). Cytokinesis-based constraints on polarized cell growth in fission yeast. PLoS Genet , e1003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle WJ, van der Geer P, Hunter T. (1991). Phosphopeptide mapping and phosphoamino acid analysis by two- dimensional separation on thin-layer cellulose plates. Methods Enzymol , 110–149. [DOI] [PubMed] [Google Scholar]
- Carnahan RH, Gould KL. (2003). The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. J Cell Biol , 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celton-Morizur S, Racine V, Sibarita JB, Paoletti A. (2006). Pom1 kinase links division plane position to cell polarity by regulating Mid1p cortical distribution. J Cell Sci , 4710–4718. [DOI] [PubMed] [Google Scholar]
- Chang F, Woollard A, Nurse P. (1996). Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J Cell Sci , 131–142. [DOI] [PubMed] [Google Scholar]
- Chen JS, Broadus MR, McLean JR, Feoktistova A, Ren L, Gould KL. (2013). Comprehensive proteomics analysis reveals new substrates and regulators of the fission yeast clp1/cdc14 phosphatase. Mol Cell Proteomics , 1074–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga RR, Chang F. (2005). Dynamic positioning of the fission yeast cell division plane. Proc Natl Acad Sci USA , 8228–8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devrekanli A, Foltman M, Roncero C, Sanchez-Diaz A, Labib K. (2012). Inn1 and Cyk3 regulate chitin synthase during cytokinesis in budding yeasts. J Cell Sci , 5453–5466. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V. (1995). The S. pombe cdc15 gene is a key element in the reorganization of F- actin at mitosis [published erratum appears in Cell 1997 Jun 27;89(7):1185]. Cell , 435–444. [DOI] [PubMed] [Google Scholar]
- Ge W, Balasubramanian MK. (2008). Pxl1p, a paxillin-related protein, stabilizes the actomyosin ring during cytokinesis in fission yeast. Mol Biol Cell , 1680–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerganova V, Floderer C, Archetti A, Michon L, Carlini L, Reichler T, Manley S, Martin SG. (2019). Multi-phosphorylation reaction and clustering tune Pom1 gradient mid-cell levels according to cell size. Elife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KL, Moreno S, Owen DJ, Sazer S, Nurse P. (1991). Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J , 3297–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Yam C, Oliferenko S. (2015). Rewiring of cellular division site selection in evolution of fission yeasts. Curr Biol , 1187–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O, Berthelot-Grosjean M, Kokkoris K, Vincenzetti V, Moosbrugger J, Martin SG. (2011). A phosphorylation cycle shapes gradients of the DYRK family kinase Pom1 at the plasma membrane. Cell , 1116–1128. [DOI] [PubMed] [Google Scholar]
- Himpel S, Tegge W, Frank R, Leder S, Joost HG, Becker W. (2000). Specificity determinants of substrate recognition by the protein kinase DYRK1A. J Biol Chem , 2431–2438. [DOI] [PubMed] [Google Scholar]
- Huang Y, Chew TG, Ge W, Balasubramanian MK. (2007). Polarity determinants Tea1p, Tea4p, and Pom1p inhibit division-septum assembly at cell ends in fission yeast. Dev Cell , 987–996. [DOI] [PubMed] [Google Scholar]
- Kettenbach AN, Deng L, Wu Y, Baldissard S, Adamo ME, Gerber SA, Moseley JB. (2015). Quantitative phosphoproteomics reveals pathways for coordination of cell growth and division by the conserved fission yeast kinase pom1. Mol Cell Proteomics , 1275–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnaer C, Dudin O, Martin SG. (2019). Yeast-to-hypha transition of Schizosaccharomyces japonicus in response to environmental stimuli. Mol Biol Cell , 975–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Coffman VC, Lee IJ, Wu JQ. (2011). Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol , 1005–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ME, Rusin SF, Jenkins N, Kettenbach AN, Moseley JB. (2018). Mechanisms connecting the conserved protein kinases Ssp1, Kin1, and Pom1 in fission yeast cell polarity and division. Curr Biol , 84–92.e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti L, Chang F, Martin SG. (2012). Myosin Vs organize actin cables in fission yeast. Mol Biol Cell , 4579–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQuarrie CD, Mangione MC, Carroll R, James M, Gould KL, Sirotkin V. (2019). The S. pombe adaptor protein Bbc1 regulates localization of Wsp1 and Vrp1 during endocytic actin patch assembly. J Cell Sci , 10.1242/jcs.233502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangione MC, Snider CE, Gould KL. (2019). The intrinsically disordered region of the cytokinetic F-BAR protein Cdc15 performs a unique essential function in maintenance of cytokinetic ring integrity. Mol Biol Cell , 2790–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Berthelot-Grosjean M. (2009). Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature , 852–856. [DOI] [PubMed] [Google Scholar]
- Martin-Garcia R, Arribas V, Coll PM, Pinar M, Viana RA, Rincon SA, Correa-Bordes J, Ribas JC, Perez P. (2018). Paxillin-mediated recruitment of calcineurin to the contractile ring is required for the correct progression of cytokinesis in fission Yeast. Cell Rep , 772–783.e774. [DOI] [PubMed] [Google Scholar]
- McDonald NA, Gould KL. (2016). Characterization of cytokinetic F-BARs and other membrane-binding proteins. Methods Mol Biol , 181–189. [DOI] [PubMed] [Google Scholar]
- McDonald NA, Vander Kooi CW, Ohi MD, Gould KL. (2015). Oligomerization but not membrane bending underlies the function of certain F-BAR proteins in cell motility and cytokinesis. Dev Cell , 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Mayeux A, Paoletti A, Nurse P. (2009). A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature , 857–860. [DOI] [PubMed] [Google Scholar]
- Nishihama R, Schreiter JH, Onishi M, Vallen EA, Hanna J, Moravcevic K, Lippincott MF, Han H, Lemmon MA, Pringle JR, Bi E. (2009). Role of Inn1 and its interactions with Hof1 and Cyk3 in promoting cleavage furrow and septum formation in S. cerevisiae. J Cell Biol , 995–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliferenko S, Chew TG, Balasubramanian MK. (2009). Positioning cytokinesis. Genes Dev , 660–674. [DOI] [PubMed] [Google Scholar]
- Padte NN, Martin SG, Howard M, Chang F. (2006). The cell-end factor pom1p inhibits mid1p in specification of the cell division plane in fission yeast. Curr Biol , 2480–2487. [DOI] [PubMed] [Google Scholar]
- Palani S, Koster DV, Hatano T, Kamnev A, Kanamaru T, Brooker HR, Hernandez-Fernaud JR, Jones AME, Millar JBA, Mulvihill DP, Balasubramanian MK. (2019). Phosphoregulation of tropomyosin is crucial for actin cable turnover and division site placement. J Cell Biol , 3548–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinar M, Coll PM, Rincon SA, Perez P. (2008). Schizosaccharomyces pombe Pxl1 is a paxillin homologue that modulates Rho1 activity and participates in cytokinesis. Mol Biol Cell , 1727–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard LW, Onishi M, Pringle JR, Lord M. (2012). Fission yeast Cyk3p is a transglutaminase-like protein that participates in cytokinesis and cell morphogenesis. Mol Biol Cell , 2433–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Willet AH, Roberts-Galbraith RH, McDonald NA, Feoktistova A, Chen JS, Huang H, Guillen R, Boone C, Sidhu SS, et al. (2015). The Cdc15 and Imp2 SH3 domains cooperatively scaffold a network of proteins that redundantly ensure efficient cell division in fission yeast. Mol Biol Cell , 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon SA, Bhatia P, Bicho C, Guzman-Vendrell M, Fraisier V, Borek WE, Alves Fde L, Dingli F, Loew D, Rappsilber J, et al. (2014). Pom1 regulates the assembly of Cdr2-Mid1 cortical nodes for robust spatial control of cytokinesis. J Cell Biol , 61–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon SA, Paoletti A. (2016). Molecular control of fission yeast cytokinesis. Semin Cell Dev Biol , 28–38. [DOI] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Chen JS, Wang J, Gould KL. (2009). The SH3 domains of two PCH family members cooperate in assembly of the Schizosaccharomyces pombe contractile ring. J Cell Biol , 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Ohi MD, Ballif BA, Chen JS, McLeod I, McDonald WH, Gygi SP, Yates JR, 3rd, Gould KL. (2010). Dephosphorylation of F-BAR protein Cdc15 modulates its conformation and stimulates its scaffolding activity at the cell division site. Mol Cell , 86–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksela K, Permi P. (2012). SH3 domain ligand binding: what’s the consensus and where’s the specificity? FEBS Lett , 2609–2614. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann M, Fankhauser C, Brodbeck C, Simanis V. (1996). The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev , 2707–2719. [DOI] [PubMed] [Google Scholar]
- Swaffer MP, Jones AW, Flynn HR, Snijders AP, Nurse P. (2018). Quantitative phosphoproteomics reveals the signaling dynamics of cell-cycle kinases in the fission yeast Schizosaccharomyces pombe. Cell Rep , 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vjestica A, Tang XZ, Oliferenko S. (2008). The actomyosin ring recruits early secretory compartments to the division site in fission yeast. Mol Biol Cell , 1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtler V, Huang Y, Karagiannis J, Balasubramanian MK. (2006). Cell cycle-dependent roles for the FCH-domain protein Cdc15p in formation of the actomyosin ring in Schizosaccharomyces pombe. Mol Biol Cell , 3254–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willet AH, McDonald NA, Bohnert KA, Baird MA, Allen JR, Davidson MW, Gould KL. (2015a). The F-BAR Cdc15 promotes contractile ring formation through the direct recruitment of the formin Cdc12. J Cell Biol , 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willet AH, McDonald NA, Gould KL. (2015b). Regulation of contractile ring formation and septation in Schizosaccharomyces pombe. Curr Opin Microbiol , 46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Grady JT, Villen J, Gygi SP. (2008). Phosphoproteome analysis of fission yeast. J Proteome Res , 1088–1097. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Pollard TD. (2005). Counting cytokinesis proteins globally and locally in fission yeast. Science , 310–314. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. (2006). Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol , 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.