Abstract
Intermediate filament (IF) cytoskeletal networks simultaneously support mechanical integrity and influence signal transduction pathways. Marked remodeling of the keratin IF network accompanies collective cellular morphogenetic movements that occur during early embryonic development in the frog Xenopus laevis. While this reorganization of keratin is initiated by force transduction on cell–cell contacts mediated by C-cadherin, the mechanism by which keratin filament reorganization occurs remains poorly understood. In this work, we demonstrate that 14-3-3 proteins regulate keratin reorganization dynamics in embryonic mesendoderm cells from Xenopus gastrula. 14-3-3 colocalizes with keratin filaments near cell–cell junctions in migrating mesendoderm. Coimmunoprecipitation, mass spectrometry, and bioinformatic analyses indicate 14-3-3 is associated with Keratin 19 (K19) in the whole embryo and, more specifically, mesendoderm tissue. Inhibition of 14-3-3 results in both the decreased exchange of keratin subunits into filaments and blocks keratin filament recruitment toward cell–cell contacts. Synthetically coupling 14-3-3 to K19 through a unique fusion construct conversely induces the localization of this keratin population to the region of cell–cell contacts. Taken together, these findings indicate that 14-3-3 acts on keratin IFs and is involved in their reorganization to sites of cell adhesion.
INTRODUCTION
Intermediate filaments (IFs) comprise a diverse group of structurally conserved proteins that assemble into long fibrils that function as a “cellular skeleton.” Found across many of the tissues of chordate eukaryotes, these cordlike macromolecules form intricate networks in both the cytoplasmic and the nuclear compartments of cells. Keratins, a subtype of IF proteins, are expressed in epithelia as well as a few additional cell types, including cells of the early vertebrate embryo.
IFs have a well-established role in providing structural integrity within cells through binding to protein scaffolds that experience transmission of forces (Sanghvi-Shah and Weber, 2017). These interfaces include hemidesmosomes at cell–matrix contacts as well as desmosomes and classical cadherins at cell–cell junctions. IFs, including keratins, are essential at these locations to confer mechanical resistance and withstand strains encountered by cells (Acehan et al., 2008; Weber et al., 2012; Conway et al., 2013).
In vitro assembly assays have demonstrated autonomous formation of IF networks that are dense, notoriously insoluble, and subject to little change (Sanghvi-Shah and Weber, 2017). Despite these observations, emerging evidence in the cellular environment has shown that these networks are, in fact, subject to ongoing modification (Vikstrom et al., 1992; Windoffer et al., 2004; Kolsch et al., 2010). In conjunction with descriptions of ongoing filament network maintenance, IF networks have been shown to undergo dramatic and large-scale reorganization in response to onset of mechanical stresses (Conway et al., 2013). For instance, application of shear stress to cells results in both remodeling of cytoplasmic keratin IFs and an increase in dynamic exchange rate within these filaments (Sivaramakrishnan et al., 2009). In our own previous work, we have shown that collectively migrating mesendoderm cells from the frog embryo demonstrate ongoing reorganization of keratin IFs to cell–cell contacts under tension (Weber et al., 2012). Application of force to these cells through the cell–cell adhesion receptor C-cadherin induces a rapid shift in the cellular keratin network from widespread cytoplasmic to proximal to the cadherin junction experiencing tension. This distinct keratin recruitment presumably fortifies the mechanically sensitive junction and provides an instructive cue for directional cellular migration away from the site bearing mechanical load.
While several studies address the manner in which IFs are continually being formed (Windoffer et al., 2004; Kolsch et al., 2010), few to date have interrogated the processes that govern the disassembly and reorganization of IF networks. The phosphorylation status of IF proteins is a critical factor that determines features such as filament protein solubility, assembly, and disassembly behaviors and even aggregation of filament proteins (Ridge et al., 2005; Snider and Omary, 2014). These modifications have cellular implications that include changes in signaling, cellular growth, and disease manifestation, among others. Several kinases and phosphatases have been shown to act on IF proteins (Ridge et al., 2005; Omary et al., 2006; Sivaramakrishnan et al., 2009; Ju et al., 2015). However, how phosphorylation and dephosphorylation alters IF conformation and protein-binding partners is largely unknown. Moreover, a substantial gap also exists in identification of IF-binding proteins that direct IF disassembly and/or recruitment of IFs to distinct cellular compartments.
The 14-3-3 proteins are a family of seven isoforms that have been demonstrated to bind to sequence motifs containing phosphorylated serine or threonine residues (Muslin et al., 1996; Yaffe et al., 1997). 14-3-3 homo- and heterodimers have been shown to exert control over target substrates by alternatively limiting or enhancing affinity of these molecules for other binding partners (Tzivion et al., 2000; Margolis et al., 2006; Zhou et al., 2010; Obsil and Obsilova, 2011). In this manner, 14-3-3 proteins are recognized to modulate the activity of a variety of IFs (Li et al., 2006; Miao et al., 2013), as well as other cytoskeleton associated proteins such as motors and scaffold molecules (Jin et al., 2004; Roberts et al., 2013; Sehgal et al., 2014; Vishal et al., 2018).
We have previously demonstrated a mechanosensitive reorganization of the keratin network to cadherin-mediated cell–cell contacts (Weber et al., 2012). It is abundantly clear that the keratin network dynamics must be regulated to determine where and when such rearrangement events occur. We propose that 14-3-3 regulates the force-triggered reorganization of the widespread keratin network toward the cell–cell junction. In this study, we investigate a fundamental role for 14-3-3 interaction with Keratin 19 (K19) in modulating IF dynamic exchange and network reorganization toward cell–cell contacts. This interplay between the keratin network and the 14-3-3 proteins may underlie signaling events that occur during collective cell migration, which in turn regulate the migratory behavior itself.
RESULTS
14-3-3 protein expression is ubiquitous throughout early embryonic stages and tissues
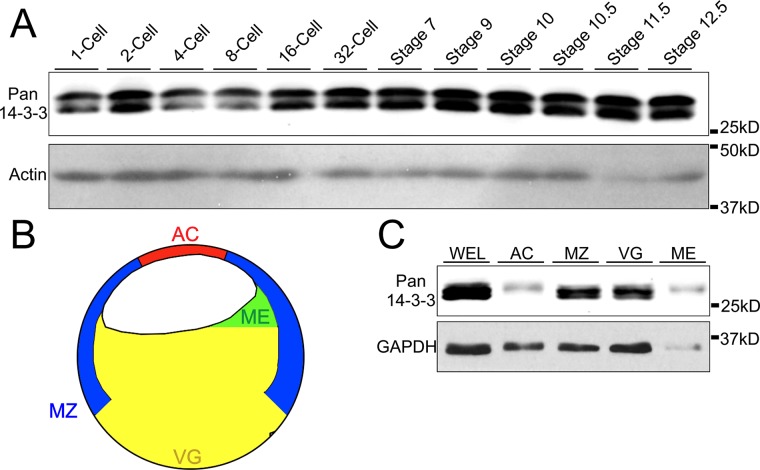
Our previous work demonstrated reorganization of the keratin network in mesendoderm of the Xenopus frog embryo (Weber et al., 2012). If 14-3-3 is to be a potential regulator of keratin IFs in the collectively migrating mesendoderm, 14-3-3 must be present in this tissue during the particular period of development in which this tissue differentiates and migrates, namely gastrulation. Previous work by Lau, Wu, and Muslin examined mRNA expression levels and patterns of six different 14-3-3 isoforms during stages 2–38 of Xenopus embryonic development (Lau et al., 2006). While mRNA expression was found to be abundant for 14-3-3 β, ε, and τ during stages 2–14, mRNA is not necessarily indicative of protein expression in the embryo, which could be translationally regulated and/or maternally derived. We first confirmed whether 14-3-3 protein expression in developmental stages leading up to late gastrulation paralleled previously published mRNA expression data. Lysates were collected at time points beginning at one-cell stage through late gastrula (NF Stage 12.5), and immunoblot was performed using an antibody reactive with all isoforms of 14-3-3 (Figure 1A). Expression of 14-3-3 was detected throughout these early embryonic stages. Although 14-3-3 protein levels did increase slightly following midblastula transition (stages 7–9) when zygotic mRNA is transcriptionally up-regulated, we saw relatively high and persistent levels of 14-3-3 proteins present throughout early development. To further examine the extent to which 14-3-3 was present in various embryonic tissues, gastrulating embryos were dissected into several regions (Figure 1B), and protein lysates were prepared. We examined the expression of 14-3-3 proteins in the animal cap, marginal zone, vegetal hemisphere, and mesendoderm tissues compared with that of whole embryos (Figure 1C). 14-3-3 was found to be present in all tissues of Xenopus gastrula with varying expression levels. 14-3-3 expression was neither exclusive to nor absent from any one particular region, suggesting broadly ubiquitous functions for 14-3-3 across different tissue types. Expression levels of 14-3-3 were greater relative to the housekeeping protein GAPDH in some tissues, including mesendoderm (Figure 1C).
FIGURE 1:
14-3-3 protein expression is ubiquitous across early embryonic stages and tissues. (A) Whole embryo lysates (1% Triton X-100) were immunoblotted for 14-3-3 using a pan antibody that detects multiple isoforms. Each lane represents approximately 50 μg. (B) Colored schematic of a bisected Xenopus embryo at gastrula depicting major tissue divisions. The tissues include the animal cap (AC), mesendoderm (ME), marginal zone (MZ), vegetal hemisphere (VG), and whole embryo lysate (WEL). (C) Embryos were dissected into separate tissues and corresponding lysates (1% Triton X-100) were immunoblotted using pan 14-3-3 antibody to examine expression across the gastrulating embryo. Each lane represents a portion of protein equivalent to approximately 1 embryo.
K19 associates with 14-3-3 in whole embryos and collectively migrating tissues
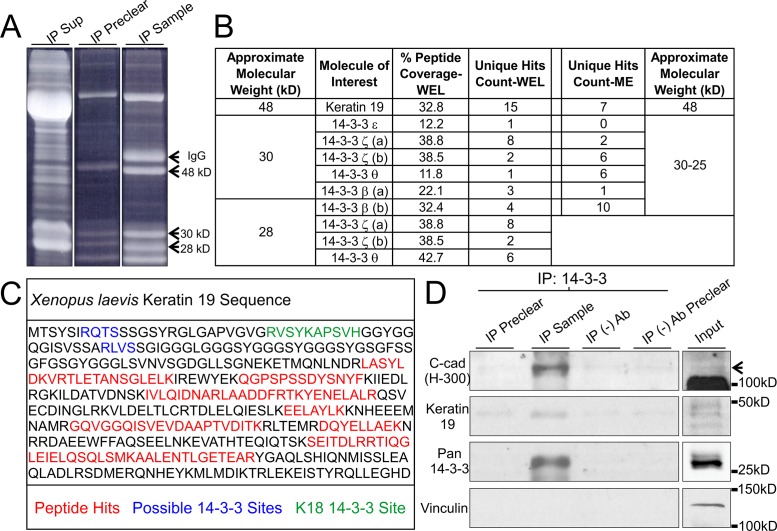
We next sought to identify specifically which 14-3-3 protein isoforms were present in gastrula and examine whether association with keratin IF proteins could be detected. Rather than rely on antibody specificity for 14-3-3 isoforms in Xenopus, we took a proteomics approach to determine which 14-3-3 isoforms were expressed in the frog embryo and with which proteins 14-3-3 were associated. Coimmunoprecipitation was performed using pan-14-3-3 antibody with NP-40 solubilized protein lysates from whole embryos or from mesendoderm tissue only. Samples were then separated by gel electrophoresis and stained to visualize protein band pattern (Figure 2A). Several distinct protein bands were visible in 14-3-3 immunoprecipitated lysates at the expected molecular weight for 14-3-3 proteins, between 28 and 30 kDa. This molecular weight range for 14-3-3 is due to both the difference in amino acid sequences and posttranslational modifications. In addition to the isolated 14-3-3 isoforms, an especially prominent band was detected at approximately 48 kDa. These bands were selected for proteomic screening using liquid chromatography, tandem mass spectrometry (LC/MS-MS).
FIGURE 2:
Xenopus K19 associates with 14-3-3 proteins and C-cadherin. (A) Pan 14-3-3 immunoprecipitates (1% Tergitol type NP-40) from whole embryo lysates prior to band extraction and processing using LC/MS-MS. Prominent bands at 48, 30, and 28 kDa were processed. Heavy chain IgG from the antibody used for IP was not excised. (B) Table summary of relevant proteins detected in gel extracts processed using LC/MS-MS. Experiments were conducted using 14-3-3 immunoprecipitates from whole embryo lysates (WEL) as well as lysates from mesendoderm (ME) tissue only. Analysis was performed using Scaffold 4.7.3. (C) Summary schematic of K19 peptides (red) detected in the 48 kDa sample. Peptides are depicted within the context of the K19 primary structure and alongside described (green) and predicted (blue) possible 14-3-3 interaction sites. (D) 14-3-3 proteins were immunoprecipitated (1% Tergitol type NP-40) from whole embryo lysates and immunoblotted for C-cadherin, K19, and Vinculin. C-cadherin band is denoted by an arrow. The bottom band is yolk protein from sample.
Analysis of peptides identified by mass spectrometry confirmed the presence of multiple 14-3-3 protein isoforms (Figure 2B). Expanded datasets from the LC/MS-MS analyses can be found in Supplemental Figure S1. Several unique peptides as well as substantial total protein sequence coverage were found for 14-3-3 isoforms ζ, β, and θ in both whole embryo lysates and mesendoderm tissue alone (Figure 2B). Isoform 14-3-3ε was detected with lower unique peptide and coverage spectra than other isoforms and was not detected in mesendoderm. Interestingly, in the analysis of the 48 kDa band, K19 was detected with many unique peptides and substantial coverage in 14-3-3 coimmunoprecipitates from both Xenopus whole embryo lysates and mesendoderm (Figure 2C). Only a single peptide was isolated that has identical conserved sequence with human K19, and the majority of peptide sequences correspond specifically to protein product of the Xenopus laevis krt19.L gene, not krt19.S. To our knowledge, this is the first time K19 has been shown to associate with 14-3-3.
14-3-3 is known to bind protein substrates containing particular primary amino acid sequences. Notably, the motif sequence RXXS/TP is a preferred target, but other variations do exist (Muslin et al., 1996; Yaffe et al., 1997; Johnson et al., 2010). Several keratins, including Keratin 18 (K18), have been identified as phosphoserine-modified to become 14-3-3 substrates (Ku et al., 1998). Using three different bioinformatics analyses, we examined whether K19 might also contain similar binding phosphorylated sites and motifs that could provide 14-3-3 docking sites (Supplemental Figure S2). Xenopus K19 contains three putative phosphorylated sites, S10, S33 and S52, which are highly conserved across phyla (Figure 2C and Supplemental Figure S2). S33 is of particular interest because it has previously been shown to be the primary phosphorylation site of K19 in several signaling and IF remodeling events (Zhou et al., 1999; Ju et al., 2015). We confirmed the association between 14-3-3 and K19 by immunoprecipitating 14-3-3 from whole embryo lysates and immunoblotting for K19 (Figure 2D). Indeed, K19 was present in 14-3-3-immunoprecipitated samples by immunoblot analysis as well.
The keratin IF network has been shown to be reorganized proximal to cell–cell adhesions in mesendoderm cells (Weber et al., 2012). This reorganization of the keratin network occurs as a consequence of tugging forces on cell–cell interactions (Weber et al., 2012). In Xenopus gastrula, the classical cadherin protein C-cadherin provides the primary means of cell–cell adhesion (Heasman et al., 1994). Since keratin IFs are recruited to C-cadherin in mesendoderm cells as a function of applied force on the adhesions, we next investigated whether 14-3-3 was also associated with C-cadherin. 14-3-3 coimmunoprecipitated lysates were positive when immunoblotted for C-cadherin (Figure 2D). Interestingly, although vinculin has been implicated in mechanosensation in both cadherin cell–cell adhesions and focal adhesions by several reports (Riveline et al., 2001; le Duc et al., 2010), we did not find vinculin to be associated with 14-3-3 (Figure 2D).
Keratin filaments colocalize with 14-3-3 at cell boundaries
Because 14-3-3 was found to biochemically associate with K19, we performed immunofluorescence to assess the relationship between 14-3-3 and keratin filament networks in situ. To determine the localization of 14-3-3 proteins in mesendoderm, immunofluorescence was performed on sagittal sections of gastrulating Xenopus embryos. We found that several tissues showed compartmentalization of 14-3-3 rather than diffuse cytoplasmic distribution. In mesendoderm, 14-3-3 proteins localized clearly and distinctly to cell boundaries at lower magnifications (Supplemental Figure S3). At higher magnifications, we were able to more precisely assess the subcellular distribution of 14-3-3, especially in relation to keratin filaments.
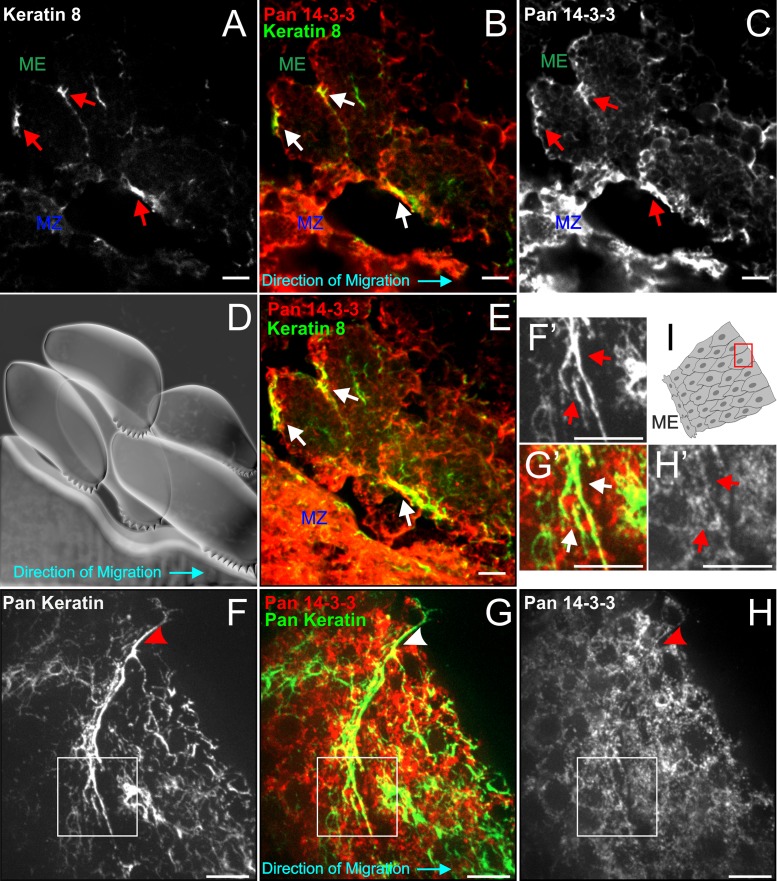
Keratin filaments in the early Xenopus gastrula are comprised of obligate heterodimers of Keratin 8 (K8) with either K18 and/or K19 (Franz et al., 1983; Suzuki et al., 2017; Briggs et al., 2018). As seen previously (Weber et al., 2012), keratin filaments, as visualized using antibody to K8, were found localized to the posterior of migratory mesendoderm cells, where they link to cell–cell contacts (Figure 3A). 14-3-3 shows strong colocalization with these densities of keratins present at points of cell–cell adhesion (Figure 3, B–E). Similar colocalization was also seen in coimmunofluorescence labeling of mesendoderm explants with a pan-keratin antibody and pan-14-3-3 (Figure 3, F–I). Regions of signal overlap are filamentous in shape in both sagittal whole embryo sections (visualizing cells in X-Z) and explanted tissue (Figure 3, E and F’–H’). Rather than extending across the entire filament density, the colabeling occurs specifically at the area proximal to the cell–cell adhesion. 14-3-3 also appeared to exhibit concentrated intensity in lamellipodia of mesendoderm cells, where keratin filaments are notably absent. Interestingly, 14-3-3 labeling in lamellipodia lacked the filamentous pattern observed in the posterior of cells.
FIGURE 3:
Filaments recruited to cell–cell adhesions associate with 14-3-3. (A–C) Single plane confocal images showing a sagittal perspective of a cryosectioned gastrulating embryo labeled immunocytochemically for 14-3-3 proteins (red) and K8 (green). Areas where filamentous colocalization was detected in B are illustrated by arrows in all three panels. (D) Cartoon schematic depicting orientation of cells in A–C. The blue arrow indicates direction of tissue migration. (E) Maximum intensity projection of confocal z-stack of cells shown in A–D to show more comprehensive filament distribution. (F–H) Leading edge of a mesendoderm explant demonstrating association between 14-3-3 and keratins (pan-keratin antibody labeling) at a cell–cell interface (arrowheads). Closer inspection of the keratin morphology at this area (F’–H’) reveals filamentous 14-3-3 labeling. (I) Explant schematic depicting the cell pair (F–H) relative to the rest of the tissue. Images are z-stacks (maximum intensity projection). Scale bars are 10 μm.
14-3-3 proteins are distributed proximal to cell–cell adhesions
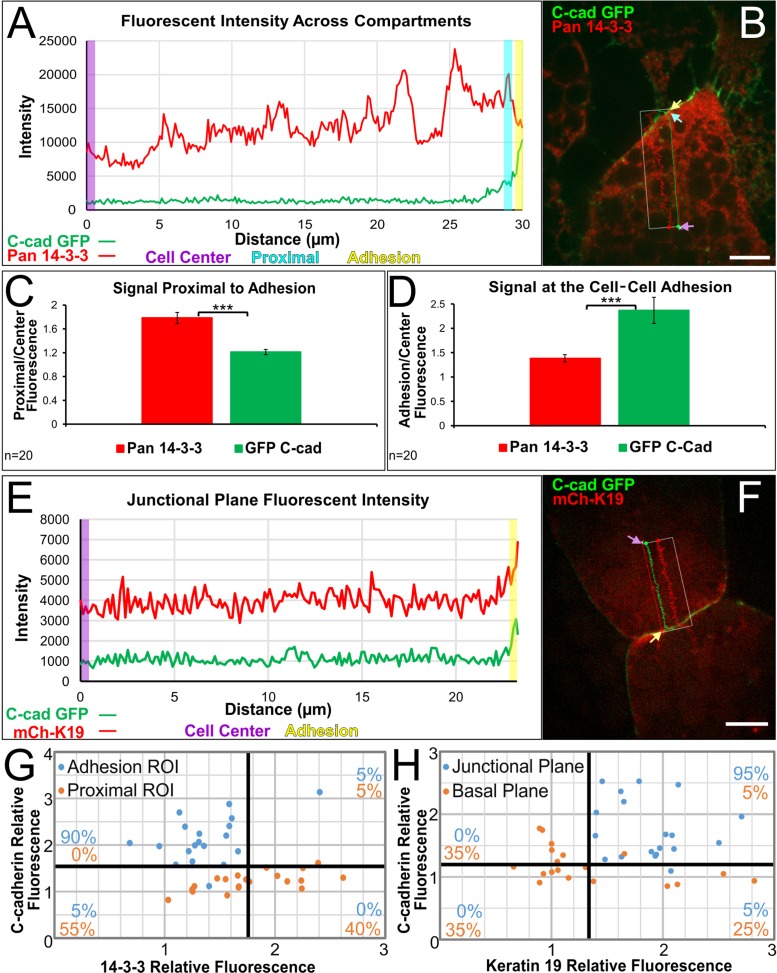
C-cadherin colocalizes with keratin filaments at points of cell–cell contact in mesendoderm and coimmunoprecipitates with keratin in lysates from Xenopus gastrula (Weber et al., 2012). Given that coimmunoprecipitation of 14-3-3 also shows association with C-cadherin, we examined how the subcellular localization of each relate to one another. Curiously, although 14-3-3 appears at the cell periphery at low magnification and resolution (Supplemental Figure S3), an appreciable separation between 14-3-3 and C-cadherin is observed at higher resolution (Figure 4, A and B; Supplemental Figure S4). 14-3-3 is increasingly concentrated nearer to cell–cell contacts (Figure 4C) as compared with more centrally located regions of the cytoplasm. However, at the more precise point of cell–cell contact, indicated by C-cadherin-eGFP labeling, we noted a decrease in 14-3-3 signal intensity (Figure 4D). Average mean fluorescence intensities for C-cadherin were high in adhesion ROIs, but this did not correspond with high 14-3-3 intensities (Figure 4G). In contrast, K19 colocalized with C-cadherin at the cell–cell contact (Figure 4, E and F) and coordinately increased in intensity at the cell–cell contact (Supplemental Figure S4). Adhesion ROIs analyzed in the junctional plane exhibited high fluorescence intensity of both C-cadherin and K19 (Figure 4H). This positioning of K19 mirrors the colocalization previously observed among C-cadherin, plakoglobin, and K8 in these cells (Weber et al., 2012). Taken together with evidence of biochemical association, these data suggest that the association between 14-3-3 and C-cadherin is likely indirect. 14-3-3 appears to localize with keratin IFs and concentrate near cell–cell contacts, but not exactly at the cell–cell contact itself.
FIGURE 4:
14-3-3 proteins are distributed proximally to cell–cell adhesions. (A) Representative linescan analysis of fluorescence across cellular compartments indicated in B. Lines extended from the approximate middle of the cell to the area just prior to C-cadherin signal and to the onset of C-cadherin signal. Measurements were taken from the cell center (purple rectangle), proximal to the adhesion (teal rectangle), and at the adhesion (yellow rectangle). Each rectangle represents 0.5 μm in length. (B) Immunofluorescence image of a leading edge mesendoderm cell labeled for 14-3-3 and expressing C-cadherin (X. laevis origin, eGFP label). Colored arrows indicate regions represented by rectangles in A. (C, D) Comparison of the mean increase in 14-3-3 signal and C-cadherin signal from the cell center to the area proximal to the cell–cell adhesion (in C) and at the cell–cell adhesion (in D). Analysis was performed using paired sample t tests, ***p < 0.001. Error bars are ± SEM. (E) Representative linescan analysis of fluorescence across cellular compartments indicated in F. Lines extended from the approximate middle of the cell to the onset of C-cadherin signal. Measurements were taken from the cell center (purple rectangle) and at the adhesion (yellow rectangle). Each rectangle represents 0.5 μm in length. (F) Immunofluorescence image of mesendoderm cells expressing C-cadherin (eGFP) and mCherry-K19 (X. laevis origin). Colored arrows indicate regions represented by rectangles in E. Scale bars are 10 μm. (G) Scatterplot of the mean relative fluorescence intensities of C-cadherin and 14-3-3 in 20 cells near and at the cell–cell contact zone. (H) Scatterplot of the mean relative fluorescence intensities of C-cadherin and K19 in 20 cells imaged by confocal in the basal plane and a higher junctional plane. Percentages indicate the proportion of corresponding mean intensities in each quadrant.
IF dynamic exchange is disrupted by 14-3-3 inhibition
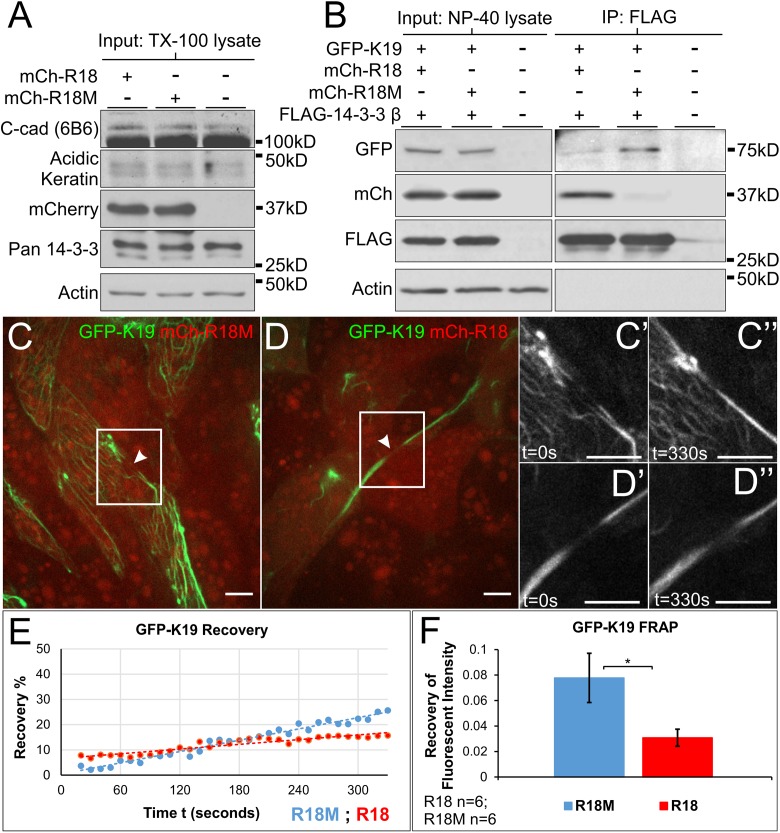
The observation that 14-3-3 associates with keratins and colocalizes with filaments implies a shared functional relationship. Given that 14-3-3 proteins have been shown to have a role in disassembly of IFs (Li et al., 2006; Miao et al., 2013) and to associate with keratin filaments induced to remodel by okadaic acid treatment (Strnad et al., 2002), one possible function of this association is to facilitate modification of pre-existing filament networks. Modification of polymerized filaments involves addition or subtraction of IF proteins along the length of a filament polymer, known as lateral exchange or dynamic exchange (Vikstrom et al., 1992; Colakog˘lu and Brown, 2009; Nöding et al., 2014). Keratin networks show filament remodeling and 14-3-3-dependent increases in dynamic exchange rate after exposure to shear stress (Sivaramakrishnan et al., 2009). Since changes in mechanical stress are continually transduced across junctions during collective cell migration and result in keratin reorganization (Weber et al., 2012), we examined whether 14-3-3 might have a role in the keratin filament organization in mesendoderm explants. To functionally inhibit 14-3-3, we expressed a short peptide sequence (R18) previously shown to bind 14-3-3 with high affinity and block its binding to endogenous substrates (Petosa et al., 1998; Wang et al., 1999). A negative control peptide (R18M) containing two point mutations was used for comparison. We created mCherry-tagged R18 and R18M constructs so that we could readily identify living cells in which R18 or R18M was expressed. No change in overall protein expression of C-cadherin, acidic keratins, 14-3-3, or actin resulted from R18 expression (Figure 5A). The ability of mCherry-R18, but not mCherry-R18M, to bind 14-3-3 was confirmed by coimmunoprecipitation analyses (Figure 5B). As mCherry-R18 bound to 14-3-3, a corresponding decrease in the association of 14-3-3 with K19 was observed (Figure 5B). Thus, R18 functions as a competitive inhibitor of 14-3-3 association with K19.
FIGURE 5:
Mesendoderm keratin filament dynamic exchange is decreased by 14-3-3 inhibition. (A) Immunoblot analyses of protein extracts (1% Triton X-100) of stage 10.5 Xenopus embryos expressing mCh-R18 or mCh-R18M. (B) Coimmunoprecipitation (1% Tergitol type NP-40) performed using stage 10.5 lysates expressing human FLAG-14-3-3 β with either mCherry-R18 or mCherry-R18M. (C, D) Still images from photobleach and recovery time lapse movies (Supplemental Movie S1). Mesendoderm explants expressing either 14-3-3 inhibitor peptide mCherry-R18 or control peptide mCherry-R18M (red) with eGFP-K19 (green) were exposed to GFP photobleaching and fluorescence recovery at the site was measured. (C’–D’’) Enlarged view of the region of filament bleaching (white boxes) during recovery measurements. The time annotations in seconds refer to start of capture (t = 0 s) and end of capture (t = 330 s). (E) Representative analysis plotting fluorescence recovery against time of image capture. (F) Comparison of mean eGFP-K19 recovery rate in explants expressing either mCherry-R18 or mCherry-R18M. Analysis was performed using a one-tailed t test, *p < 0.05. Error bars are ± SEM. Scale bars are 10 μm.
Movie 1A.
FRAP movies. Recovery of eGFP-K19 fluorescence was measured for mesendoderm explants expressing mCherry-R18M (Movie A) and mCherry-R18 (Movie B). Photobleaching was performed using the 488 laser at 100% power in the area indicated by the ROI box for 120 seconds. Images were taken every 10 seconds after photobleaching (330 seconds total). Scale bars are 10 μm.
Movie 1B.
FRAP movies. Recovery of eGFP-K19 fluorescence was measured for mesendoderm explants expressing mCherry-R18M (Movie A) and mCherry-R18 (Movie B). Photobleaching was performed using the 488 laser at 100% power in the area indicated by the ROI box for 120 seconds. Images were taken every 10 seconds after photobleaching (330 seconds total). Scale bars are 10 μm.
To examine the role of 14-3-3, we coexpressed mCherry-R18 or R18M with eGFP-K19. R18 expression did not initially result in any apparent changes to the keratin network detectable by fluorescence microscopy. Through early gastrulation and explant preparation, no gross morphological changes were observed in the overall keratin network or subcellular localization of filaments. We suspect that this may be because of a delayed onset of R18 expression until after midblastula transition. Nonetheless, subtler effects on the IF dynamics were observed in native mesendoderm explants. Fluorescence recovery after photobleaching (FRAP) experiments of eGFP-K19 revealed that 14-3-3 inhibition results in attenuation of dynamic exchange rate when compared with that of explants expressing control peptide R18M (Figure 5, C–F). Despite inherent differences in the width and subcellular location of filaments bleached across trials, the lower recovery rate in 14-3-3 inhibited samples persisted (Figure 5, E–F). Though additional reports provide evidence of different times of lateral exchange onset that range from within an hour to 8 h (Colakog˘lu and Brown, 2009; Nöding et al., 2014), we detect more rapid differences in recovery after 14-3-3 inhibition that occurs within minutes (Figure 5). This finding demonstrates that the interaction between 14-3-3 and K19 is required for modification of filaments that occurs on an appreciably fine timescale in motile tissues.
14-3-3 is required for targeting of keratin to cell–cell adhesions
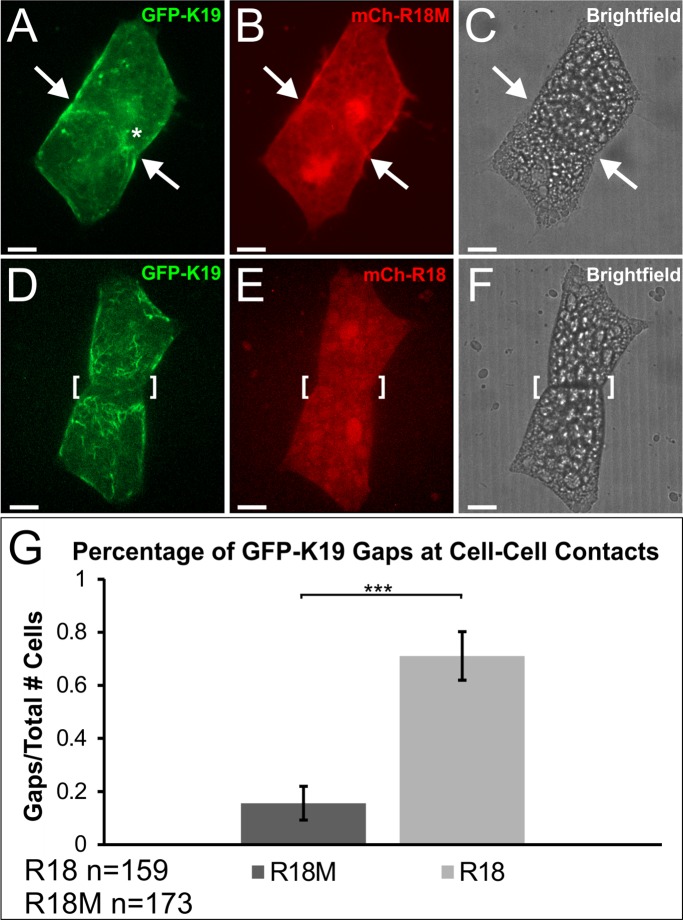
Force transmitted through mesendoderm cell–cell contacts induces both changes in the keratin network throughout the cell and recruits filaments to sites of transduced tension (Weber et al., 2012). We were surprised to find that expression of R18 did not dramatically alter the distribution of keratin IFs in the mesendoderm given its association with keratin and role in dynamic exchange. We reasoned that this could be because expression of R18 did not occur until after relatively stable cell–cell adhesions were formed. To determine whether 14-3-3 is required for localization of filaments to cell junctions, mesendoderm tissue expressing mCherry-R18 or R18M and eGFP-K19 was dissociated into single cells, plated on fibronectin substrate, and imaged as cells collided and formed cell pairs. Our work and others’ have shown that many migratory cells polarize in opposite directions on collision, resulting in substantial force at the cell–cell junction (Nelson and Chen, 2003; Maruthamuthu et al., 2011; Weber et al., 2012; Mertz et al., 2013). We additionally showed that keratin IFs are recruited to cell–cell adhesions between mesendoderm cells as a function of force on C-cadherin (Weber et al., 2012). This was also the case in mesendoderm cells expressing the control construct mCherry-R18M (Figure 6, A–C). When 14-3-3 was inhibited by expression of mCherry-R18, a distinct zone lacking fluorescent K19 filaments was observed at points of de novo cell–cell adhesion (Figure 6, D–F). This K19-deplete zone was observed at a significantly higher frequency in 14-3-3-inhibited cells after establishment of cell–cell contact relative to controls (Figure 6). These regions lacking K19 IFs had a size range of 2 to 17 μm with an average of 7 μm when measured from the cell–cell contact to the onset of visible filament fluorescence. The zone was surprisingly uniform along the width of the cell–cell contact in these cells and very few R18-positive cells were found to have K19 targeted to cell–cell contacts (Figure 6G).
FIGURE 6:
14-3-3 is necessary for targeting of keratin to cell–cell contacts. (A–C) Mesendoderm cell pair establishing de novo cell–cell contact after collision. Cells are expressing mCherry-R18M and eGFP-K19. The arrowhead depicts the cell–cell adhesion where keratin densities (asterisk) have localized. Image stack is 35 slices (10.54 μm). (D–F) Postcollision mesendoderm cell pair expressing mCherry-R18 and eGFP-K19. The bracket depicts the cell–cell adhesion that demonstrates a gap where keratin filaments have failed to reorganize. Image stack is 40 slices (10.92 μm). (G) Comparison of the number of keratin gaps at cell–cell adhesions in postcollision cell pairs expressing either mCherry-R18M or mCherry-R18. Analysis was performed using a z-test for proportions from two samples, ***p < 0.001. Error bars represent the 99.9% CI for each sample proportion. Fluorescent images are z-stacks (maximum intensity projection). Brightfield images are single planes. Scale bars are 20 μm.
14-3-3 association with K19 is sufficient for cell–cell adhesion targeting
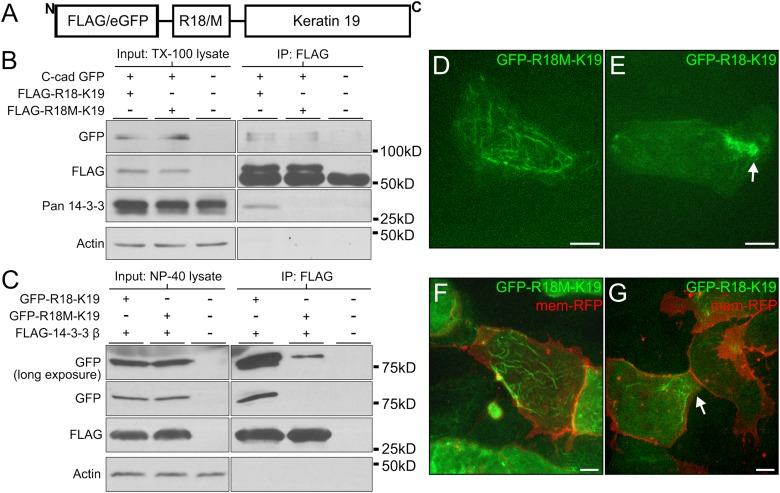
Since inhibition of 14-3-3 prevented keratin localization to cell–cell contacts, we wanted to see whether forced association of K19 with 14-3-3 would be sufficient to drive targeting to cell junctions. 14-3-3 endogenously binds to phosphorylated serine and threonine residues contained in motifs within certain substrates (Yaffe et al., 1997; Johnson et al., 2010). Mutation of serine residues to nonphosphorylatable alanines (e.g., S33A of hK18) eliminates binding between keratins and 14-3-3; however, mutation of serine to phosphomimetic amino acids (e.g., S33D of hK18) is similarly dysfunctional at promoting association between 14-3-3 and keratins, presumably because of differences in the side chain compared with phosphate groups (Ku et al., 1998). To promote the association of K19 with 14-3-3, we created an eGFP-R18-K19 fusion protein (Figure 7A). This construct consists of eGFP for visualization, R18 to promote association with 14-3-3, and full-length K19 from X. laevis such that all native regulatory elements in the head domain of K19 remain in place. Immunoprecipitation analyses showed robust association of R18-K19 fusion proteins with both endogenous 14-3-3 (Figure 7B) and exogenous FLAG-14-3-3 (Figure 7C). Note that FLAG-14-3-3 also associated with eGFP-R18M-K19 (Figure 7C), albeit at a much reduced level, likely via the normal intrinsic phosphorylation sites within K19. Since R18 is a high affinity peptide for 14-3-3, this fusion protein results in a traceable K19 protein with a constitutive signaling site for 14-3-3 binding that is phosphorylation independent.
FIGURE 7:
14-3-3 proteins target keratins to cell–cell adhesions. (A) Schematic of fusion peptides created by the insertion of R18 or R18M (R18/M) into FLAG/eGFP-K19. This construct was generated using full-length K19.L from X. laevis. (B) Protein lysates from stage 10.5 Xenopus embryos expressing C-cadherin-eGFP and either FLAG-R18-K19 or FLAG-R18M-K19 were prepared in 1% Triton X-100. FLAG constructs were immunoprecipitated and analyzed by immunoblot for associated proteins. (C) Protein lysates from stage 10.5 Xenopus embryos expressing human FLAG-14-3-3β and either eGFP-R18-K19 or eGFP-R18M-K19 were prepared in 1% Tergitol-type NP-40. FLAG constructs were immunoprecipitated and analyzed by immunoblot for associated proteins. (D, E) Explanted mesendoderm cells mosaically expressing eGFP-R18M-K19 (in D) or eGFP-R18-K19 (in E). (F, G) Explanted mesendoderm coexpressing mem-RFP and eGFP-R18M-K19 (in F) or eGFP-R18-K19 (in G). Arrows indicate areas where filament densities have localized. Images are z-stacks (maximum intensity projection). Scale bars are 10 μm.
Mosaic expression of these fusion constructs was established in mesendoderm tissue by injecting DNA, and the IF distribution in live mesendoderm cells, ∼4 rows back from the leading edge, was examined by confocal microscopy. Expressed eGFP-R18M-K19 in mesendoderm explants forms filaments spread throughout the cell body but absent from cell protrusions (Figure 7, D and F), typical of keratin networks described previously in unpolarized mesendoderm (Weber et al., 2012). In mesendoderm expressing eGFP-R18-K19, the fluorescence appeared as densities that preferentially localize to areas of cell–cell adhesion and lack widespread expression throughout the cytosol (Figure 7, E and G). In the most robust instances, eGFP-R18-K19 localized as an intense aggregate exclusively at the cell–cell boundary (Figure 7E). In other cases, distinct filaments could be seen and the population was limited to near the cell–cell adhesion (Figure 7G). The forced association of K19 with 14-3-3 through this fusion construct created a distribution that in many ways was the exact opposite of when 14-3-3 was inhibited (compare Figure 7E with Figure 6D). To examine the extent to which this change in filament compartmentalization was related to association with cell–cell contacts, C-cadherin-eGFP association with 3xFLAG-R18-K19 was compared with that of 3xFLAG-R18M-K19 (Figure 7B). Interestingly, the association of R18-K19 with C-cadherin-eGFP appeared to be roughly equivalent to that of R18M-K19. These data indicate that K19 association with 14-3-3 is important for targeting K19 populations to the cell–cell junctional region, but does not induce extended coupling of K19 with cadherins themselves.
DISCUSSION
Our findings in the current study lead us to propose that 14-3-3 proteins are responsible for targeting keratin IFs to sites of cell–cell adhesion that are transmitting tension. Expression of a fluorescent keratin fused to a high affinity 14-3-3-binding domain causes filaments assembled from these keratins to mobilize to cell–cell junctions. In reciprocal support, inhibition of 14-3-3 through expression of a peptide inhibitor results in the failed targeting of keratin filament to sites of de novo cell–cell adhesion. These observations provide evidence for the first time that 14-3-3 functions as a critical spatial determinant of the IF network in response to mechanical loading at cell contacts.
Cytoskeletal structures must be able to efficiently sense tension and undergo remodeling in order to fortify cells against changes in the direction, magnitude, and nature of tension transmitted. Rapid changes in force application require concomitant responses by cytoskeletal proteins, necessitating mechanistic filament assembly strategies that are effective within a short time scale. Cellular mechanisms that induce changes within a shorter time scale than that required for biosynthesis include pathways that make posttranslational modifications to pre-existing proteins. IF proteins, which have no known intrinsic catalytic activity, are modified by other molecules that respond to changes in cellular tension, such as kinases (Loschke et al., 2015). It follows that accessory proteins recognize and bind these posttranslational modifications and accordingly alter the filament dynamics and spatial localization of IF proteins. These interactions can reorganize filament populations to particular locales within the cell and change polymerization–depolymerization characteristics. Our data support a functional role for 14-3-3 in mediating keratin IF dynamic exchange and reorganization in cells that are actively migrating and encountering changes in the direction and magnitude of loading forces.
We show here an interaction between 14-3-3 and keratin proteins. Indeed, 14-3-3 proteins have been demonstrated to interact with a variety of IFs (Tzivion et al., 2000; Li et al., 2006; Miao et al., 2013), including some keratin types (Ku et al., 1998; Kim et al., 2006; Margolis et al., 2006; Sivaramakrishnan et al., 2009; Boudreau et al., 2013). We show for the first time an interaction between K19 and 14-3-3. We do not yet know the specific 14-3-3 isoform that binds endogenously. Our finding of particularly high expression of 14-3-3 β and ζ is in agreement with recent independent data from Peshkin et al. (2019). Previous work has shown that K18 directly binds to 14-3-3 when phosphorylated on serine 33, but an association between K19 and 14-3-3 was not observed (Ku et al., 1998). Nonetheless, this serine residue is conserved between K18 and K19, conserved across species, and is a putative phosphorylation target in K19 (Ku et al., 1998; Zhou et al., 1999; Ju et al., 2015). 14-3-3 associates with a number of Type I acidic keratins aside from K18 (Kim et al., 2006; Boudreau et al., 2013). Serine residues 10, 33 (S35 in human), and 52 (S54 in human) of K19 are predicted to be phosphorylated by several kinases including protein kinase C and to be 14-3-3-binding sites by our bioinformatics analyses (Supplemental Figure S2). Regardless of which specific phosphorylation site provides for interaction with 14-3-3, an association is evident by our experimental observations and supported by numerous predictive computational analyses. While one might contend that K19 could be associating with 14-3-3 indirectly through other keratins, K19 was uniquely abundant in our 14-3-3 immunoprecipitation samples.
What is the interaction of 14-3-3 with K19 actually doing to the IFs? At a whole-cell level, association of keratins with 14-3-3 is necessary for keratin network disassembly during mitosis (Liao and Omary, 1996). Knockdown of 14-3-3 has been shown to result in abnormal increases in keratin filament abundance and decreases in soluble filament precursors (Boudreau et al., 2013). Fluid flow shear stress of alveolar cells results in phosphorylation of known 14-3-3 sites in K18 along with increases in dynamic exchange that are 14-3-3 dependent (Sivaramakrishnan et al., 2009). Similarly, we see decreased FRAP recovery when 14-3-3 is inhibited in eGFP-K19 expressing mesendoderm tissue, which is known to have intrinsic mechanical stresses through the cell–cell contacts (Weber et al., 2012; Sonavane et al., 2017). We expect that expression of R18 peptide results in inhibition of 14-3-3 in multiple contexts throughout the cell rather than perturbation of the subset of 14-3-3 molecules that associate with the keratin–cadherin complex. Nonetheless, we find it remarkable that 14-3-3 inhibition still resulted in a distinct change in keratin subcellular distribution as a consequence of cell–cell adhesion signals.
We find that 14-3-3 provides for targeting of K19 toward cadherin-mediated cell–cell contacts. While 14-3-3 inhibition resulted in the establishment of a keratin deplete zone proximal to the contact, forced association of K19 with 14-3-3 conversely localized the filament population to the cell–cell contact compartment.14-3-3 is observed decorating the filamentous keratin population proximal to these adhesions. Despite finding biochemical association between 14-3-3 and C-cadherin, we note that at high resolution these molecules do not precisely colocalize. This suggests that there are additional components that may facilitate interaction between 14-3-3/keratin–protein complexes and other molecules of the cell–cell junction and which may play a role in the reorganization event. 14-3-3 has been demonstrated to associate with a number of molecules important for establishment of cytoskeletal networks at the cell–cell adhesion (Acehan et al., 2008) including kinesin (Sehgal et al., 2014), plakoglobin (Sehgal et al., 2014; Vishal et al., 2018), and plakophilin (Jin et al., 2004; Roberts et al., 2013). The association and function of 14-3-3 in relation to keratin IFs may provide an interface between cytoskeletal networks and a host of scaffolding proteins and signal transduction pathways (Margolis et al., 2006; Loschke et al., 2016; Sonavane et al., 2017). In turn, the targeting of keratin to sites of tension due to local signaling/phosphorylation events could promote a positive feedback loop for keratin filament assembly dynamics (Ridge et al., 2005; Woll et al., 2007; Ju et al., 2015; Sonavane et al., 2017). The enriched presence of 14-3-3 near both the cell–cell adhesion and in the lamellipodia suggests either non–IF-related functions in the latter region or a possible function in IF transport or formation (Windoffer et al., 2004, 2011). Further elucidation of these macromolecular complexes in polarized migratory cells will be the subject of ongoing studies.
The significance of K19 association with 14-3-3 to the development of the early embryo remains uncertain. Previous work has shown important roles for 14-3-3 in Xenopus development through the targeted knockdown of different isoforms by morpholino oligonucleotides (Lau et al., 2006). Knockdown of certain 14-3-3 isoforms caused severe gastrulation defects including exogastrulation and failed mesodermal patterning (Lau et al., 2006). Knockdown or inhibition of K8, the lone Type II keratin in early embryogenesis, similarly induces exogastrulation (Klymkowsky et al., 1992; Torpey et al., 1992). The specific functions of K18 and K19 in early embryonic development have yet to be determined, but it is likely that there exists some functional redundancy as evidenced by murine knockouts (Magin et al., 1998). Our finding of K19 in the mesendoderm is in agreement with two recent –omics reports broadly examining mRNA and protein expression patterns in the Xenopus embryo (Briggs et al., 2018; Peshkin et al., 2019). It is notable that Xenopus K19 has only a single amino acid in the tail domain and human K19 has only 13 amino acids, making this keratin structurally deviant from other IF proteins (Herrmann et al., 1996; Kirmse et al., 2007; Lee et al., 2012). This has been speculated to result in keratin networks with greater dynamic potential (Hofmann and Franke, 1997; Fradette et al., 1998) and are thus suited to a dynamic tissue such as collectively migrating mesendoderm.
Molecular control of reorganization of filaments is a process that is as critical to cellular migration as it is to cellular and junctional integrity. The results of the current study lend insights into the mechanisms of active tissues ranging from cellular sheets in migration to three-dimensional tissues that balance changes in loading (Winklbauer et al., 1992; Weber et al., 2012). These observations can also inform cancer models that detect roles for 14-3-3 proteins and keratins in promoting invasiveness of cells (Boudreau et al., 2013; Cheung et al., 2013; Deng et al., 2013; Ju et al., 2015). Decrease in the ability of IFs to form the networks that fortify sealed junctions across cells, enable cells to form resistant tissues, and coordinate the activity of tissues has implications for essential processes including barrier function, embryonic tissue patterning, wound healing, and cancer progression.
MATERIALS AND METHODS
Xenopus embryo in vitro fertilization and staging
Embryos were obtained and cultured using standard methods and staged according to Nieuwkoop and Faber (1994). Embryos were dejellied in 2% cysteine and cultured at 15°C in 0.1× Modified Barth’s saline (MBS; 1× MBS: 88 mM NaCl, 1 mM KCl, 2.5 mM NaHCO3, 0.35 mM CaCl2, 0.5 mM MgSO4, 5 mM HEPES, pH 7.8).
Xenopus embryo protein lysate preparation and Western blot
Xenopus whole embryos were solubilized in lysis buffer (50 mM Tris-HCl pH 7.5, 1 mM phenylmethylsulfonofluoride, 1% mammalian protease inhibitor cocktail [Sigma-Aldrich, P2714], 0.2 mM H2O2, 3 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 10 mM sodium fluoride, sodium β-glycerophosphate [10 mg/ml], 1% Triton X-100 [Sigma-Aldrich, T-9284], or 1% Tergitol type NP-40 [Spectrum Biosciences, T1279]). Protein samples were prepared in 2× Laemmli buffer supplemented with 5% β-mercaptoethanol, incubated on a 95°C heat block for 5 min, and loaded onto 12% SDS–PAGE gels. Proteins were transferred onto nitrocellulose membrane prior to incubation with antibodies.
Antibodies
Several antibodies were used throughout this study: pan 14-3-3 pAb (Santa Cruz, K19 sc-629), K8 mAb (DSHB, 1h5), C-Cadherin mAb (DSHB, 6B6), pan Cadherin pAb (Santa Cruz, H-300 sc-10733), Vinculin mAb (Millipore, MAB3574), K19 mAb (Progen, 61010), pan keratin mAb (Sigma Aldrich, C2562), Actin-HRP (Sigma-Aldrich,A3854), GAPDH mAb (Abcam, mAbcam 9484), GFP mAb (Invitrogen, A-11120), GFP mAb (Santa Cruz, B-2 sc-9996), mCherry pAb (BioVision, 5993-100), and FLAG mAb (Sigma-Aldrich, F1804). The 1h5 anti-XCK1(8) monoclonal antibody developed by Michael Klymkowsky and 6B6 anti-C-cadherin monoclonal antibody developed by Barry Gumbiner were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biology, University of Iowa (Iowa City, IA 52242).
Plasmids
The following plasmids were used in this study: pCS2-mCherry-R18, pCS2-mCherry-R18M, pCS2-eGFP-K19 (derived from X. laevis), pcDNA3-FLAG-HA-14-3-3β (Addgene plasmid #8999, human derived), pCS2-FLAG-HA-14-3-3β (from human), pCS2-C-cadherin-eGFP (from X. laevis, created by B. Gumbiner, University of Washington), pCS2-eGFP-R18-K19 (from X. laevis), pCS2-eGFP-R18M-K19 (from X. laevis), pCS2-3xFLAG-R18-K19 (from X. laevis), pCS2-3xFLAG-R18M-K19 (from X. laevis), and pCS2-mem-RFP. Oligonucleotides that translate to the described R18 and R18M primary sequences (Wang et al. 1999) were commercially synthesized (Genewiz) and cloned into pCS2 vector backbone by the author. All constructs containing K19 were created using full-length X. laevis krt19.L (NP_001084992.1) obtained as cDNA from GE Dharmacon (MXL1736-202774753) and subcloned into pCS2 vector backbone by the author. pCS2-FLAG-HA-14-3-3β was subcloned using 1478 pcDNA3 flag HA-14-3-3β (human derived), a gift from William Sellers (Addgene plasmid #8999), as the origination source for the fusion construct. The mem-RFP construct was a kind gift from Megason and Fraser (Megason and Fraser, 2003).
RNA constructs and microinjection
RNA was prepared via in vitro transcription (Promega, P1420). DNA or RNA was diluted to microinject concentrations of 200–500 pg in 5-nl pulses for the following constructs: pCS2-eGFP-K19, pCS2-mCherry-R18, pCS2-mCherry-R18M, pCS2-FLAG-HA-14-3-3β, pCS2-C-cadherin-eGFP, pCS2-mem-RFP, pCS2-3xFLAG/eGFP-R18-K19, and pCS2-3xFLAG/eGFP-R18M-K19. Embryos utilized for microscopy were injected dorsally into both blastomeres at the two-cell stage to target mesendoderm. Embryos utilized for immunoprecipitation were twice injected at the one-cell stage in the animal cap on both sides peripheral to the germinal vesicle.
Immunoprecipitation
Whole embryos or dissected mesendoderm for immunoprecipitation were processed in aforementioned lysis buffer with 1% Tergitol type NP-40 or 1% Triton X-100 as indicated in figure legends. Endogenous IP lysates (equivalent to 40 embryos) were initially incubated with 50 μl Protein-G agarose bead slurry (Roche Diagnostics, 11243233001) for an hour. Beads were pelleted by centrifugation at 3000 rpm for 5 min (4°C). The IP lysate was then removed and incubated with primary antibody (5 μg pan 14-3-3, Santa Cruz) for overnight immunoprecipitation. The Protein-G beads were washed three times in lysis buffer for 10 min. Bead samples were stored in 2× Laemmli buffer with 5% β-mercaptoethanol at –80°C for subsequent gel electrophoresis. After overnight primary incubation, endogenous IP lysates were incubated with 50 μl Protein-G agarose bead slurry overnight. FLAG IP lysates (equivalent to 40 embryos) were incubated with 40 μl agarose slurry covalently linked to anti-FLAG M2 mAb (Sigma-Aldrich, A2220) for overnight immunoprecipitation. IP samples were pelleted by centrifugation at 3000 rpm for 5 min (4°C), and lysates were removed for storage as supernatant samples. Beads were washed with three exchanges of lysis buffer (1% Tergitol type NP-40 or 1% Triton X-100 depending on IP detergent) for 10 min. IP bead samples were dissociated in 25 μl of 2× Laemmli buffer with 5% β-mercaptoethanol, incubated on a 95°C heat block for 5 min, and loaded onto 12% SDS–PAGE polyacrylamide gels for electrophoresis. Gels were either stained using Sypro Red or transferred onto nitrocellulose membrane prior to incubation with antibodies. All incubation and wash steps were performed at 4°C using a vertical rotator. All centrifugation steps were performed at 4°C.
LC/MS-MS proteomic analysis
All reagents used for proteomics work were filtered and meticulous precautions were taken to avoid dust and other potential sources of keratin contamination. Prior to staining with Sypro Red dye (Invitrogen, S12000), SDS–PAGE gels were incubated twice in fixative solution consisting of 50% methanol and 7% glacial acetic acid for 30 min per incubation. After decanting the second fixative solution, the gel was placed in a fresh dish and incubated in 60 ml of Sypro Red dye overnight. The staining solution was decanted and the gel was incubated in a wash solution of 10% methanol and 7% glacial acetic acid for 30 min. Afterward, the gel was washed three times in 100 ml of commercial ultrapure water before gel imaging and further preparation for LC/MS-MS. All incubations and washes were performed at room temperature on a flat rotator.
LC/MS-MS was performed by the Center for Advanced Proteomics Research (CAPR) at the Rutgers New Jersey Medical School. Sypro Red-labeled SDS–PAGE gel sections were excised at the facility and in-gel trypsin digestion was performed. The resulting peptides were C18 desalted and analyzed by LC/MS-MS on the Q Exactive instrument. The MS/MS spectra were searched against the NCBI X. laevis database using MASCOT (v.2.3) search engines on the Proteome Discoverer (V1.4) platform. The protein false discovery rate is less than 1%. The mass spectrometry data were obtained from an Orbitrap instrument funded in part by National Institutes of Health (NIH) grant NS046593 for the support of the UMDNJ Neuroproteomics Core Facility. Information in tables was derived utilizing Scaffold 4.7.3 with a protein and peptide false discovery rate of 1%.
Mesendoderm dissection and dissociation
Stage 10.5 embryos in 0.5× MBS were dissected at the animal cap surface to reveal the mesendodermal mantle. The dorsal side of the embryo was identified and the mesendoderm leading edge was excised from the rest of the tissue. These cells were pipette transferred into a solution of Ca2+/Mg2+ free 1× MBS on 1% Ca2+/Mg2+ free agarose and allowed to dissociate for 30 min at room temperature. Dissociated cells were transferred to fibronectin (Sigma-Aldrich, F4759) matrix (200 μl of 1:7.5 in water; incubated on MatTek glass bottom microwell dishes, P35G-1.5-14-C) and imaged. Fibronectin incubation was performed overnight at 4°C.
Dorsal marginal zone explant preparation
DMZ explants were excised and plated as described in (Davidson et al., 2004). Stage 10.5 embryos in 0.5x MBS were carefully dissected at the animal cap surface to reveal the mesendodermal mantle. The embryo was then bisected and the dorsal side of the embryo was used. The remaining animal cap cells and endodermal cells were trimmed away, leaving the mesoderm, mesendoderm, and bottle cells of the embryo. The explant was flattened on fibronectin (Sigma-Aldrich, F4759) matrix (100 μl of 1:7.5 in water; incubated on MatTek glass bottom microwell dishes, P35G-1.5-14-C) and silicone grease was used to mount a coverslip. Fibronectin incubation was performed overnight at 4°C. Explants were slightly compressed and allowed to attach and migrate for 2 h (live imaging experiments) or 5 h (samples to be fixed). To fix explants, the coverslip was removed just prior to incubation in 100% methanol.
Immunofluorescence
Embryos and dorsal marginal zone explants were fixed in ice-cold 100% methanol or Dent’s fixative (80% methanol, 20% DMSO) and incubated at –20°C overnight. Embryos were rehydrated in partial changes of 0.1× MBS. Embryos were transferred into vinyl molds with dimensions of 15 mm × 15 mm × 5 mm (Electron Microscopy Sciences, 62534-15) containing tissue freezing medium (Electron Microscopy Sciences, 72592) and briefly immersed in liquid nitrogen to flash freeze. Frozen embryo blocks were stored at –80°C until sectioned (Leica 819, 14035838925; 40 μm per slice) at –20°C using a temperature-controlled cryostat and mounted on slides (VWR, 48311-703). Prior to blocking, embryo sections and explants were rehydrated in 1× Tris-buffered saline (TBS). Samples were permeablized in 1x TBS with 0.25% Triton X-100 for 10 min before blocking in 5% goat serum for 1 h at room temperature. Incubation in primary antibody (30 min at 37°C) was followed by three buffer exchanges with 1× TBS and incubation in Alexa Fluor 488– or 555–conjugated goat anti-mouse (Invitrogen, A11029; A21424) and/or goat anti-rabbit (Invitrogen, A11034; A21429) IgG (30 min at 37°C). Explants were imaged after three buffer exchanges with 1× TBS. Embryonic sections were serially dehydrated using 1× TBS with increasing percentages of methanol and cleared using benzyl benzoate/benzyl alcohol. Slides were coverslipped (Corning, 2980-245), sealed with multiple coats of nail polish, and left for drying overnight (4°C) before imaging.
Linescan analysis and quantification
Linescan measurements of fluorescence were conducted using the linescan tool in the profile tab of the Zen 2.3 lite software application. Two types of measurements were conducted to compare fluorescence of C-cadherin-eGFP and 14-3-3 across cellular compartments. The compartments measured included the approximate center of the cell (called “center”), the area nearby but not at the cell–cell adhesion (called “proximal”), and the cell–cell adhesion proper (called “adhesion”). To measure change in fluorescence at the proximal and adhesion compartments, lines were drawn from the center and extended to the area just prior to C-cadherin-eGFP signal (proximal compartment) or to the C-cadherin GFP signal (adhesion compartment).
Ratios were produced to analyze relative changes in fluorescence. These ratios were calculated using the average of the last five proximal or adhesion measurements versus the average of the first five center measurements. Five measurements in a linescan represented approximately 0.5 μm. This produced four ratios for each cell: proximal/center and adhesion/center for C-cadherin GFP, and proximal/center and adhesion/center for 14-3-3.
To compare the ratios of change in fluorescence across compartments, two statistical comparisons were conducted. These included a comparison of the proximal/center ratios of C-cadherin GFP and 14-3-3, as well as a comparison of the adhesion/center ratios of C-cadherin and 14-3-3. Ratios paired as described were compared statistically using a paired sample t test (one-way) to determine whether statistically significant differences were present. Scatterplots were generated of each cell’s relative average C-cadherin intensity and corresponding 14-3-3 intensity at regions proximal to the cortex and at the adhesion. The bold horizontal line indicates the median relative C-cadherin intensity across both proximal and adhesion ROIs. The bold vertical line indicates the mean relative 14-3-3 intensity in ROIs proximal to the cortex.
This method was also utilized to compare change in fluorescence between C-cadherin GFP and mCherry-K19 across compartments in different cellular planes. The compartments measured included the center and adhesion zones. To measure fluorescence in compartments in different cellular planes, lines were drawn from the center and extended to the adhesion on an image corresponding to the basal plane of the cell (called “basal plane”) and on an image corresponding to the plane that includes the cell–cell adhesion junction (called “junctional plane”). Using the aforementioned method, line measurements were utilized to produce ratios. Four ratios of adhesion/center intensities were derived: basal and junctional planes for both C-cadherin GFP and mCh-K19.
To compare the ratios of change in fluorescence across compartments in different cellular planes, two statistical comparisons were conducted. These included a comparison of the C-cadherin GFP adhesion/center ratios from the basal plane and the junction plane and a comparison of the mCh-K19 adhesion/center ratios from the basal plane and the junction plane. Ratios paired as described were compared statistically using a paired sample t test (one-way) to determine whether statistically significant differences were present. Scatterplots were generated of each cell’s relative average C-cadherin intensity and corresponding mCh-K19 intensity at the adhesion in the imaged basal plane and the higher junctional plane. The bold horizontal line indicates the mean relative C-cadherin intensity in the basal plane. The bold vertical line indicates the mean relative mCh-K19 intensity in the basal plane.
FRAP experiments
Filaments chosen for photobleaching were imaged every 15 s over the course of 1 min to capture intensity of fluorescence prior to bleaching. The filament was then bleached using 100% power on the 488 channel for a duration of 2 min. The area of photobleaching was determined by a capture of the photobleaching mask acquired at the end of the 2-min bleach, along with the capture of the filament segment with reduced fluorescence at the start of the recovery period. Images of recovery of fluorescence within the bleach zone were captured in 10-s intervals beginning after the conclusion of the bleach period.
Recovery of fluorescence was measured using the Zen 2.3 lite application. A region of interest (ROI) was aligned to the bleached segment of the filament in each of the postbleach time points to determine the mean value of fluorescence intensity (mFI). This ROI was also used to measure the intensity of the bleach zone before photobleaching. An ROI was similarly used to measure a nonbleached control area of the filament proximal to the bleach zone. Using the method described by Zheng et al. (2011) with minor alterations, the rate of photobleaching (rate r) was first established by determining the ratio of the control ROI mFI in each postbleach time point to the average mFI across prebleach control ROIs [r = mFIc ÷ mFIc0]. The normalized fluorescence intensity (NFI) was established by determining the ratio of the difference between the mFI of the initial experimental postbleach time point and each subsequent experimental postbleach time point to the rate established for that time point [NFI = (mFIb# – mFIb1] ÷ r]. The recovery percentage (rec) of each time point was established by determining the ratio of the NFI in each experimental postbleach time point to the average mFI across experimental prebleach ROIs and multiplying that ratio by 100 [rec# = NFI# ÷ (mFIb0) × 100]. Time point series to be measured for recovery of fluorescence in this way were first scrutinized for clarity of measurement. Exclusion criteria for recovery of fluorescence experiments included unclear bleach zones, measurement zones that were obstructed during the series by crossing or merging filaments, gross reorganization of filaments that resulted in loss of the measurement zone, and readily apparent movement of the measurement zone and proximal regions such that they did not remain in the plane of focus.
Analysis of fluorescence recovery was conducted by plotting the percentage of recovery for each postbleach time point for a given series against time increments (seconds) and fitting a trendline to the data. Variation in time was controlled for by using the same time duration for each analysis (20 s to 330 s). The slope of the trendline was determined for each analysis and the mean slope was compared across R18 and R18M groups using a one-way t test in which the sample variances were shown to be equal via an F test.
Gap quantification
Z-stacks of eGFP-K19 expressing mesendoderm cells after collision were observed for the presence of a filament-depleted zone proximal to a cell–cell contact. These gaps were counted and a proportion of total gaps to total cells was calculated for cells expressing mCh-R18 and cells expressing mCh-R18M. To detect whether a statistical difference in these ratios was present, a z-test for proportions from two samples was utilized. The 99.9% confidence interval for each sample proportion was calculated assuming a normal distribution. The length of gaps from the adhesion to the first visible filaments was measured using the linescan tool in the profile tab of the Zen 2.3 lite software application.
Image acquisition
Images for live and fixed samples were taken using a Zeiss Observer spinning disk confocal microscope using a 40× 1.3 NA or 63× 1.4 NA Apochromat objective, unless specified otherwise.
Supplementary Material
Acknowledgments
We thank the members of the Weber Laboratory and the Rutgers Department of Biological Sciences for their useful feedback during the course of this work. Special thanks to D. Brian Weber for the illustration presented in Figure 3. We also thank the many investigators who provided reagents for these studies. We thank Tong Liu and Hong Li of the New Jersey Medical School CAPR for their assistance in processing and obtaining the mass spectrometry data. Mass spectrometry data were obtained from an Orbitrap instrument funded in part by NIH grant NS046593, for the support of the UMDNJ Neuroproteomics Core Facility. This research was supported by USPHS grant R15-HD084254 to G.F.W.
Abbreviations used:
- FRAP
fluorescence recovery after photobleaching
- IF
intermediate filament
- K8
Keratin 8
- K18
Keratin 18
- K19
Keratin 19
- LC/MS-MS
liquid chromatography, tandem mass spectrometry
- MBS
modified Barth’s saline
- mFI
mean fluorescence intensity
- NFI
normalized fluorescence intensity
- ROI
region of interest.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-06-0373) on February 19, 2020.
REFERENCES
- Acehan D, Petzold C, Gumper I, Sabatini DD, Mueller EJ, Cowin P, Stokes DL. (2008). Plakoglobin is required for effective intermediate filament anchorage to desmosomes. J Invest Dermatol , 2665–2675. [DOI] [PubMed] [Google Scholar]
- Boudreau A, Tanner K, Wang D, Geyer FC, Reis-Filho JS, Bissell MJ. (2013). 14-3-3sigma stabilizes a complex of soluble actin and intermediate filament to enable breast tumor invasion. Proc Natl Acad Sci USA , E3937–E3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JA, Weinreb C, Wagner DE, Megason S, Peshkin L, Kirschner MW, Klein AM. (2018). The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science , eaar5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. (2013). Collective invasion in breast cancer requires a conserved basal epithelial program. Cell , 1639–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colakog˘lu G, Brown A. (2009). Intermediate filaments exchange subunits along their length and elongate by end-to-end annealing. J Cell Biol , 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway DE, Breckenridge MT, Hinde E, Gratton E, Chen CS, Schwartz MA. (2013). Fluid shear stress on endothelial cells modulates mechanical tension across VE-cadherin and PECAM-1. Curr Biol , 1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson LA, Keller R, DeSimone D. (2004). Patterning and tissue movements in a novel explant preparation of the marginal zone of Xenopus laevis. Gene Expr Patterns , 457–466. [DOI] [PubMed] [Google Scholar]
- Deng M, Zhang WTang HYe QLiao QZhou YWu MXiong WZheng YGuo X, et al. (2013). Lactotransferrin acts as a tumor suppressor in nasopharyngeal carcinoma by repressing AKT through multiple mechanisms. Oncogene , 4273–4283. [DOI] [PubMed] [Google Scholar]
- le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D, de Rooij J. (2010). Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J Cell Biol , 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradette J, Germain L, Seshaiah P, Coulombe PA. (1998). The type I keratin 19 possesses distinct and context-dependent assembly properties. J Biol Chem , 35176–35184. [DOI] [PubMed] [Google Scholar]
- Franz JK, Gall L, Williams MA, Picheral B, Franke WW. (1983). Intermediate-size filaments in a germ cell: Expression of cytokeratins in oocytes and eggs of the frog Xenopus. Proc Natl Acad Sci USA , 6254–6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J, Ginsberg D, Geiger B, Goldstone K, Pratt T, Yoshida-Noro C, Wylie C. (1994). A functional test for maternally inherited cadherin in Xenopus shows its importance in cell adhesion at the blastula stage. Development , 49–57. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Häner M, Brettel M, Müller SA, Goldie KN, Fedtke B, Lustig A, Franke WW, Aebi U. (1996). Structure and assembly properties of the intermediate filament protein vimentin: the role of its head, rod and tail domains. J Mol Biol , 933–953. [DOI] [PubMed] [Google Scholar]
- Hofmann I, Franke WW. (1997). Heterotypic interactions and filament assembly of type I and type II cytokeratins in vitro: viscometry and determinations of relative affinities. Eur J Cell Biol , 122–132. [PubMed] [Google Scholar]
- Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, Metalnikov P, O’Donnell P, Taylor P, Taylor L, et al. (2004). Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol , 1436–1450. [DOI] [PubMed] [Google Scholar]
- Johnson C, Crowther S, Stafford MJ, Campbell DG, Toth R, MacKintosh C. (2010). Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem J , 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JH, Oh SLee KMYang WNam KSMoon HGNoh DYKim CGPark GPark JB, et al. (2015). Cytokeratin19 induced by HER2ERK binds and stabilizes HER2 on cell membranes. Cell Death Differ , 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Wong P, Coulombe PA. (2006). A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature , 362–365. [DOI] [PubMed] [Google Scholar]
- Kirmse R, Portet S, Mücke N, Aebi U, Herrmann H, Langowski J. (2007). A quantitative kinetic model for the in vitro assembly of intermediate filaments from tetrameric vimentin. J Biol Chem , 18563–18572. [DOI] [PubMed] [Google Scholar]
- Klymkowsky MW, Shook DR, Maynell LA. (1992). Evidence that the deep keratin filament systems of the Xenopus embryo act to ensure normal gastrulation. Proc Natl Acad Sci USA , 8736–8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolsch A, Windoffer R, Wurflinger T, Aach T, Leube RE. (2010). The keratin-filament cycle of assembly and disassembly. J Cell Sci , 2266–2272. [DOI] [PubMed] [Google Scholar]
- Ku NO, Liao J, Omary MB. (1998). Phosphorylation of human keratin 18 serine 33 regulates binding to 14-3-3 proteins. EMBO J , 1892–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau JMC, Wu C, Muslin AJ. (2006). Differential role of 14-3-3 family members in Xenopus development. Dev Dyn , 1761–1776. [DOI] [PubMed] [Google Scholar]
- Lee C-H, Kim M-S, Chung BM, Leahy DJ, Coulombe PA. (2012). Structural basis for heteromeric assembly and perinuclear organization of keratin filaments. Nat Struct Mol Biol , 707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Guo Y, Teng JL, Ding MX, Yu ACH, Chen JG. (2006). 14-3-3 gamma affects dynamics and integrity of glial filaments by binding to phosphorylated GFAP. J Cell Sci , 4452–4461. [DOI] [PubMed] [Google Scholar]
- Liao J, Omary MB. (1996). 14-3-3 proteins associate with phosphorylated simple epithelial keratins during cell cycle progression and act as a solubility cofactor. J Cell Biol , 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschke F, Homberg M, Magin TM. (2016). Keratin isotypes control desmosome stability and dynamics through PKCa; J Invest Dermatol , 202–213. [DOI] [PubMed] [Google Scholar]
- Loschke F, Seltmann K, Bouameur JE, Magin TM. (2015). Regulation of keratin network organization. Curr Opin Cell Biol , 56–64. [DOI] [PubMed] [Google Scholar]
- Magin TM, Schröder R, Leitgeb S, Wanninger F, Zatloukal K, Grund C, Melton DW. (1998). Lessons from keratin 18 knockout mice: Formation of novel keratin filaments, secondary loss of keratin 7 and accumulation of liver-specific keratin 8-positive aggregates. J Cell Biol , 1441–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis SS, Perry JA, Forester CM, Nutt LK, Guo Y, Jardim MJ, Thomenius MJ, Freel CD, Darbandi R, Ahn JH, et al. (2006). Role for the PP2A/B56δ phosphatase in regulating 14-3-3 release from Cdc25 to control mitosis. Cell , 759–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruthamuthu V, Sabass B, Schwarz US, Gardel ML. (2011). Cell-ECM traction force modulates endogenous tension at cell-cell contacts. Proc Natl Acad Sci USA , 4708–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megason SG, Fraser SE. (2003). Digitizing life at the level of the cell: high-performance laser-scanning microscopy and image analysis for in toto imaging of development. Mech Dev , 1407–1420. [DOI] [PubMed] [Google Scholar]
- Mertz AF, Che Y, Banerjee S, Goldstein JM, Rosowski KA, Revilla SF, Niessen CM, Marchetti MC, Dufresne ER, Horsley V. (2013). Cadherin-based intercellular adhesions organize epithelial cell-matrix traction forces. Proc Natl Acad Sci USA , 842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao LQ, Teng JL, Lin JQ, Liao XZ, Chen JG. (2013). 14-3-3 proteins interact with neurofilament protein-L and regulate dynamic assembly of neurofilaments. J Cell Sci , 427–436. [DOI] [PubMed] [Google Scholar]
- Muslin AJ, Tanner JW, Allen PM, Shaw AS. (1996). Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell , 889–897. [DOI] [PubMed] [Google Scholar]
- Nelson CM, Chen CS. (2003). VE-cadherin simultaneously stimulates and inhibits cell proliferation by altering cytoskeletal structure and tension. J Cell Sci , 3571–3581. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. (1994). Normal table of Xenopus laevis (Daudin): a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. New York: Garland Pub, 252. [Google Scholar]
- Nöding B, Herrmann H, Köster S. (2014). Direct observation of subunit exchange along mature vimentin intermediate filaments. Biophys J , 2923–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obsil T, Obsilova V. (2011). Structural basis of 14-3-3 protein functions. Semin Cell Dev Biol , 663–672. [DOI] [PubMed] [Google Scholar]
- Omary MB, Ku NO, Tao GZ, Toivola DM, Liao J. (2006). “Heads and tails” of intermediate filament phosphorylation: multiple sites and functional insights. Trends Biochem Sci , 383–394. [DOI] [PubMed] [Google Scholar]
- Peshkin L, Lukyanov A, Kalocsay M, Gage RM, Wang D, Pells TJ, Karimi K, Vize PD, Wühr M, Kirschner MW. (2019). The protein repertoire in early vertebrate embryogenesis. BioRxiv 571174. [Google Scholar]
- Petosa C, Masters SC, Bankston LA, Pohl J, Wang BC, Fu HI, Liddington RC. (1998). 14-3-3 zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J Biol Chem , 16305–16310. [DOI] [PubMed] [Google Scholar]
- Ridge KM, Linz L, Flitney FW, Kuczmarski ER, Chou YH, Omary MB, Sznajder JI, Goldman RD. (2005). Keratin 8 phosphorylation by protein kinase C delta regulates shear stress-mediated disassembly of keratin intermediate filaments in alveolar epithelial cells. J Biol Chem , 30400–30405. [DOI] [PubMed] [Google Scholar]
- Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, Kam Z, Geiger B, Bershadsky AD. (2001). Focal contacts as mechanosensors: Externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol , 1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts BJ, Reddy R, Wahl JK. (2013). Stratifin (14-3-3 sigma) limits plakophilin-3 exchange with the desmosomal plaque. PLoS One , 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghvi-Shah R, Weber GF. (2017). Intermediate filaments at the junction of mechanotransduction, migration, and development. Front Cell Dev Biol , 10.3389/fcell.2017.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal L, Mukhopadhyay ARajan AKhapare NSawant MVishal SSBhatt KAmbatipudi SAntao NAlam H, et al. (2014). 14-3-3 gamma-mediated transport of plakoglobin to the cell border is required for the initiation of desmosome assembly in vitro and in vivo. J Cell Sci , 2174–2188. [DOI] [PubMed] [Google Scholar]
- Sivaramakrishnan S, Schneider JL, Sitikov A, Goldman RD, Ridge KM. (2009). Shear stress induced reorganization of the keratin intermediate filament network requires phosphorylation by protein kinase C zeta. Mol Biol Cell , 2755–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider NT, Omary MB. (2014). Post-translational modifications of intermediate filament proteins: mechanisms and functions. Nat Rev Mol Cell Biol , 163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonavane PR, Wang C, Dzamba B, Weber GF, Periasamy A, DeSimone DW. (2017). Mechanical and signaling roles for keratin intermediate filaments in the assembly and morphogenesis of Xenopus mesendoderm tissue at gastrulation. Development , 4363–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Windoffer R, Leube RE. (2002). Induction of rapid and reversible cytokeratin filament network remodeling by inhibition of tyrosine phosphatases. J Cell Sci , 4133–4148. [DOI] [PubMed] [Google Scholar]
- Suzuki KT, Suzuki M, Shigeta M, Fortriede JD, Takahashi S, Mawaribuchi S, Yamamoto T, Taira M, Fukui A. (2017). Clustered Xenopus keratin genes: A genomic, transcriptomic, and proteomic analysis. Dev Biol , 384–392. [DOI] [PubMed] [Google Scholar]
- Torpey N, Wylie CC, Heasman J. (1992). Function of maternal cytokeratin in Xenopus development. Nature , 413–415. [DOI] [PubMed] [Google Scholar]
- Tzivion G, Luo ZJ, Avruch J. (2000). Calyculin A-induced vimentin phosphorylation sequesters 14-3-3 and displaces other 14-3-3 partners in vivo. J Biol Chem , 29772–29778. [DOI] [PubMed] [Google Scholar]
- Vikstrom KL, Lim SS, Goldman RD, Borisy GG. (1992). Steady-state dynamics of intermediate filament networks. J Cell Biol , 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishal SS, Tilwani S, Dalal SN. (2018). Plakoglobin localization to the cell border restores desmosome function in cells lacking 14-3-3γ. Biochem Biophys Res Commun , 1998–2003. [DOI] [PubMed] [Google Scholar]
- Wang B, Yang H, Liu YC, Jelinek T, Zhang L, Ruoslahti E, Fu H. (1999). Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry , 12499–12504. [DOI] [PubMed] [Google Scholar]
- Weber GF, Bjerke MA, DeSimone DW. (2012). A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration. Dev Cell , 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windoffer R, Beil M, Magin TM, Leube RE. (2011). Cytoskeleton in motion: the dynamics of keratin intermediate filaments in epithelia. J Cell Biol , 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windoffer R, Woll S, Strnad P, Leube RE. (2004). Identification of novel principles of keratin filament network turnover in living cells. Mol Biol Cell , 2436–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winklbauer R, Selchow A, Nagel M, Angres B. (1992). Cell interaction and its role in mesoderm cell migration during Xenopus gastrulation. Dev Dyn , 290–302. [DOI] [PubMed] [Google Scholar]
- Woll S, Windoffer R, Leube RE. (2007). p38 MAPK-dependent shaping of the keratin cytoskeleton in cultured cells. J Cell Biol , 795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. (1997). The structural basis for 14-3-3 : phosphopeptide binding specificity. Cell , 961–971. [DOI] [PubMed] [Google Scholar]
- Zheng C-Y, Petralia RS, Wang Y-X, Kachar B. (2011). Fluorescence recovery after photobleaching (FRAP) of fluorescence tagged proteins in dendritic spines of cultured hippocampal neurons. J Vis Exp , 2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QQ, Kee YSPoirier CCJelinek COsborne JDivi SSurcel AWill MEEggert USMüller-Taubenberger A, et al. (2010). 14-3-3 Coordinates microtubules, Rac, and Myosin II to control cell mechanics and cytokinesis. Curr Biol , 1881–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Liao J, Hu L, Feng L, Omary MB. (1999). Characterization of the major physiologic phosphorylation site of human keratin 19 and its role in filament organization. J Biol Chem , 12861–12866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.