Abstract
The Mitotic Exit Network (MEN), a budding yeast Ras-like signal transduction cascade, translates nuclear position into a signal to exit from mitosis. Here we describe how scaffolding the MEN onto spindle pole bodies (SPB—centrosome equivalent) allows the MEN to couple the final stages of mitosis to spindle position. Through the quantitative analysis of the localization of MEN components, we determined the relative importance of MEN signaling from the SPB that is delivered into the daughter cell (dSPB) during anaphase and the SPB that remains in the mother cell. Movement of half of the nucleus into the bud during anaphase causes the active form of the MEN GTPase Tem1 to accumulate at the dSPB. In response to Tem1’s activity at the dSPB, the MEN kinase cascade, which functions downstream of Tem1, accumulates at both SPBs. This localization to both SPBs serves an important role in promoting efficient exit from mitosis. Cells that harbor only one SPB delay exit from mitosis. We propose that MEN signaling is initiated by Tem1 at the dSPB and that association of the downstream MEN kinases with both SPBs serves to amplify MEN signaling, enabling the timely exit from mitosis.
INTRODUCTION
At the end of mitosis, inactivation of cyclin-dependent kinases (CDKs) causes the progression from mitosis into G1, a transition known as mitotic exit. During this transition, the mitotic spindle disassembles, chromosomes decondense, and cells split into two. In the budding yeast Saccharomyces cerevisiae, mitotic CDK inhibition occurs in two steps (reviewed in Sullivan and Morgan, 2007). During the metaphase to anaphase transition, a ubiquitin ligase known as the Anaphase Promoting Complex or Cyclosome (APC/C) targets S phase and mitotic cyclins for degradation, decreasing CDK activity (Irniger et al., 1995; King et al., 1995; Sudakin et al., 1995; Tugendreich et al., 1995). A pool of mitotic cyclins is, however, protected from degradation by an unknown mechanism. This residual pool of mitotic CDK activity sustains anaphase events such as spindle elongation. Once anaphase has been completed, the remaining mitotic CDK pool is inactivated. This inactivation requires the essential protein-phosphatase Cdc14. Cdc14 promotes the synthesis of the CDK inhibitor Sic1 and triggers mitotic cyclin degradation by dephosphorylating the APC/C activator Cdh1(Taylor et al., 1997; Jaspersen et al., 1998; Visintin et al., 1998; Zachariae et al., 1998). Dephosphorylation of CDK substrates by Cdc14 further contributes to exit from mitosis (Bouchoux and Uhlmann, 2011). The second, Cdc14-mediated, wave of mitotic CDK inactivation, is essential. In the absence of Cdc14 activity, cells fail to exit from mitosis and arrest in anaphase.
To maintain genome integrity, mitotic exit is coordinated with nuclear position. Cells only exit from mitosis when a genome complement has been delivered through the bud neck into the bud. This coordination mechanism impinges on Cdc14 activity. When the nucleus fails to translocate into the bud during anaphase, Cdc14 remains inactive and cells arrest in anaphase without exiting from mitosis (Yeh et al., 1995; Bardin et al., 2000; Pereira et al., 2000). The Mitotic Exit Network (MEN), a Ras-like GTPase signal transduction cascade, controls Cdc14 activity in response to nuclear position (Bardin et al., 2000; Pereira et al., 2000). MEN components localize to the cytoplasmic face of spindle pole bodies (SPBs), the yeast equivalent of centrosomes. As described in detail below, this localization allows the MEN to sense nuclear position.
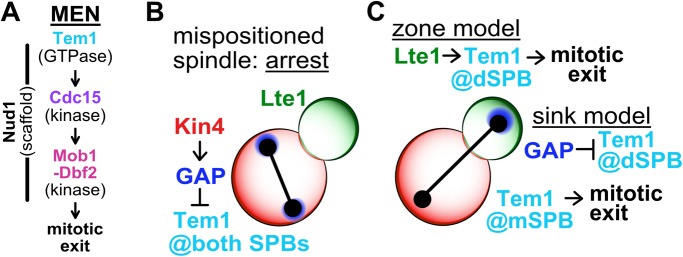
The MEN is composed of the GTPase Tem1, the protein kinase Cdc15, the protein kinase-complex Dbf2-Mob1, and the scaffold protein Nud1 (Figure 1A). During mitosis, Tem1 accumulates at SPBs (Bardin et al., 2000; Pereira et al., 2000; Molk et al., 2004). This appears critical for MEN function as constitutively anchoring Tem1 to SPBs hyperactivates the pathway, making it unresponsive to spindle position (Valerio-Santiago and Monje-Casas, 2011). Tem1 together with the polo-kinase Cdc5 recruits the protein kinase Cdc15 to SPBs (Rock and Amon, 2011). Once active, Cdc15 phosphorylates the MEN scaffold Nud1, creating a SPB docking site for Dbf2-Mob1 (Rock et al., 2013). On recruitment to SPBs, Cdc15 activates Dbf2-Mob1, which in turn activates Cdc14 (Mah et al., 2001; Mohl et al., 2009; Manzoni et al., 2010). Cdc14 is controlled by an inhibitor called Cfi1/Net1 (Shou et al., 1999; Straight et al., 1999; Visintin et al., 1999). From G1 until anaphase this inhibitor sequesters Cdc14 in the nucleolus. During anaphase, activated Dbf2-Mob1 promotes the release of Cdc14 from its inhibitor. As a result, Cdc14 spreads throughout the nucleus and cytoplasm to promote mitotic exit.
FIGURE 1:
The MEN and models describing MEN regulation by spindle position. (A) The MEN. The GTPase Tem1 recruits the protein kinase Cdc15 to SPBs. Activated Cdc15 phosphorylates the SPB-associated scaffold Nud1, recruiting the protein kinase-complex Dbf2-Mob1 to SPBs. Cdc15 activates Dbf2-Mob1 at the SPBs and Dbf2-Mob1 then triggers exit from mitosis. (B, C) The MEN is regulated by spindle position. (B) When anaphase spindle elongation fails to occur along the mother–daughter axis, leaving the spindle and both SPBs in the mother cell (mispositioned spindle), the MEN does not become active and cells arrest in anaphase. Asymmetric localization of the MEN inhibitor Kin4 to the mother cell prevents Tem1 activity at mSPBs by modifying the MEN GAP complex, Bub2-Bfa1. (C) Anaphase spindle elongation along the mother–daughter axis delivers a SPB into the daughter cell, activating the MEN and causing cells to exit from mitosis. The zone model describes a MEN-activating zone in the daughter cell and a MEN-inhibitory zone in the mother cell governed by Lte1 and Kin4, respectively. Tem1 is activated at the dSPB by Lte1, causing mitotic exit. The sink model posits that the dSPB acts as a sink, sequestering the MEN GAP complex away from the mSPB and inhibiting Tem1 at the dSPB. Sequestration leaves Tem1 uninhibited at the mSPB, activating the MEN and causing exit from mitosis.
Spindle position regulates MEN activity. Two models have been proposed to explain this regulation—the “zone model” and the “sink model.” Both models are based on the localization pattern of the MEN GTPase Tem1 and its regulators. Tem1 localizes weakly to both SPBs in metaphase and subsequently becomes enriched on the SPB that migrates into the daughter cell, henceforth dSPB, during anaphase (Bardin et al., 2000; Pereira et al., 2000; Molk et al., 2004). This localization is partially dependent on Tem1’s GTPase activating protein complex Bub2-Bfa1(Pereira et al., 2000; Valerio-Santiago and Monje-Casas, 2011; Caydasi et al., 2012). The MEN GAP complex localizes to both SPBs during metaphase, preferentially accumulating at the SPB that will move into the daughter cell (Lengefeld et al., 2017). During anaphase, Bub2-Bfa1 becomes increasingly asymmetric, localizing exclusively to the dSPB (Pereira et al., 2000). The localization of proteins regulating Tem1 function is also asymmetric. The Tem1 inhibitor Kin4 localizes to the mother cell where it promotes dissociation of the Tem1 anchor Bub2-Bfa1 from SPBs, thereby inhibiting Tem1 (D’Aquino et al., 2005; Pereira and Schiebel, 2005; Fraschini et al., 2006; Maekawa et al., 2007; Caydasi and Pereira, 2009; Scarfone et al., 2015). The Tem1 activator Lte1 localizes to the bud where it inhibits Kin4 from interacting with the dSPB, thereby activating Tem1 (Bertazzi et al., 2011; Falk et al., 2011). Lte1 likely also activates Tem1 in additional ways by promoting Tem1’s GDP exchange activity (Falk et al., 2016a).
Based on these localization patterns, the zone model proposes that the cell is divided into two zones: a Tem1 inhibitory-zone in the mother cell and a Tem1 activating-zone in the daughter cell (Chan and Amon, 2010; Falk et al., 2016b; Figure 1, B and C). Tem1 is activated at the SPB when it traverses the bud neck and moves into the bud. On the dSPB Tem1, together with Cdc5, activates Cdc15. This model, however, leaves the localization and presumably activity of Tem1 and Cdc15 on the mother cell SPB, henceforth mSPB, unexplained. The sink model aims to explain this very shortcoming of the zone model. In the sink model (Figure 1C), the dSPB serves as a sink for the Tem1 GAP, allowing Tem1 to activate Cdc15 at the mSPB (Fraschini et al., 2006; Hotz and Barral, 2014). This model, however, fails to explain the observation that Kin4 inhibits MEN activity in the mother cell (D’Aquino et al., 2005; Pereira and Schiebel, 2005; Fraschini et al., 2006; Maekawa et al., 2007; Scarfone et al., 2015).
In this paper, we aim to distinguish between the zone and sink models by clarifying from which SPB MEN signaling emanates. We find that movement of a SPB into the daughter cell during early anaphase causes the MEN GTPase Tem1 to accumulate at the dSPB. Tem1’s activity at the dSPB then causes the MEN kinases to associate with both the mSPB and the dSPB where they are active. We propose that MEN signaling is initiated at the dSPB, but amplification of the signal at the mSPB allows cells to rapidly exit from mitosis.
RESULTS
Tem1 binds to mother- and bud-localized SPBs via different mechanisms
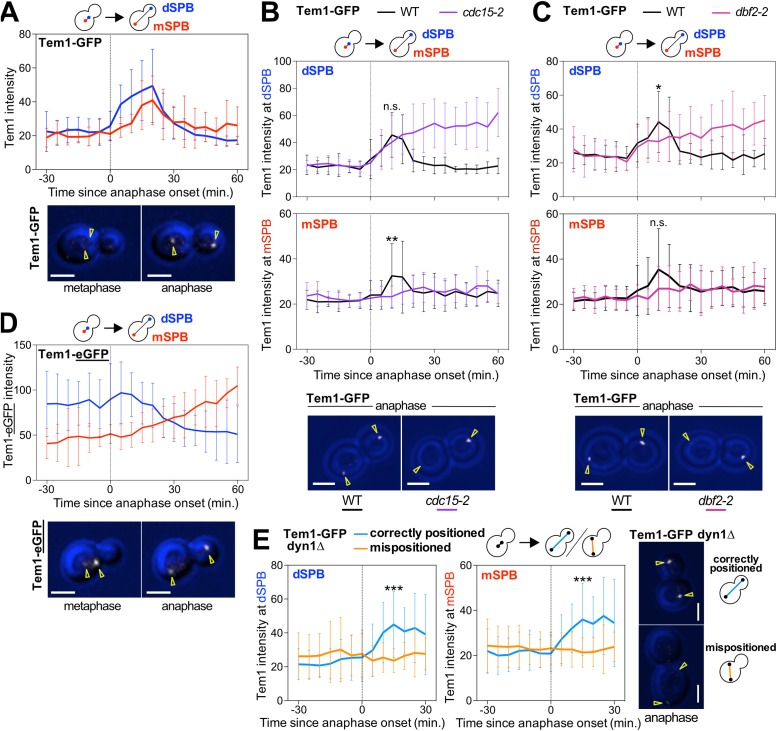
To determine how the MEN senses spindle position, it was important to quantitatively describe the association of Tem1 with SPBs. We analyzed the dynamics of Tem1-GFP binding to SPBs by time-lapse microscopy and quantified the Tem1-GFP signal at SPBs using maximum projections. We chose this Tem1 fusion because it is not known to increase Tem1’s activity (Supplemental Figure S1). Translocation of a SPB into the bud during anaphase caused Tem1-GFP to become enriched at SPBs (Figure 2A). Tem1-GFP first accumulated at the dSPB and subsequently accumulated at the mSPB (Figure 2A). Tem1-GFP disassociated from both SPBs at the time cells exited mitosis and returned to G1. We note that it has previously been reported that Tem1 localizes to SPBs in metaphase (Bardin et al., 2000; Pereira et al., 2000) and subsequently increases in intensity as a SPB crosses the bud neck during anaphase (Molk et al., 2004; Caydasi et al., 2012). In contrast, we did not detect Tem1-GFP on SPBs in metaphase. This is likely due to differences in image acquisition. To minimize light-induced toxicity, prevent metaphase arrest, and prolong image acquisition, we imaged cells with lower intensity. However, importantly, both the study by Molk et al. (2004) and the study by us found that the Tem1-GFP signal at SPBs increases during anaphase.
FIGURE 2:
Tem1 accumulates at the SPB that moves into the daughter cell during anaphase. (A–E) Quantification and representative images of Tem1 localization at the SPBs. Cells were imaged by time-lapse microscopy every 5 min for 3–6 h to determine the intensity (max(SPB)-mean(slide)) of Tem1 at either the mother cell-localized SPB (mSPB) or the daughter cell-localized SPB (dSPB). In metaphase, the SPB closer to the bud neck was designated as the dSPB. Curves show mean and SD for 20 cells per condition. Data were centered at the first frame of anaphase (spindle length >3 μm). Student’s t test was used to assess statistical significance at time points when association of Tem1 with SPBs was maximal in wild-type cells. Representative sample images of a cell in metaphase or anaphase with the indicated fluorescently tagged protein in white. Yellow arrowheads indicate location of the SPBs determined from Tub1 localization. Scale bars are 3 μm. (A, D) Cells containing mCherry-TUB1 and TEM1-GFP (A, Ay22556) or TEM1-yEGFP (D, Ay21613) were imaged at 25°C. Cells were chosen for analysis in which a SPB entering the bud coincided with anaphase onset. (B, C) Wild-type (Ay22556), cdc15-2 (Ay40721), or dbf2-2 (Ay40876) cells containing TEM1-GFP and mCherry-TUB1 were shifted from room-temperature to 32°C and imaged. Cells were chosen for analysis where a SPB entering the bud coincided with anaphase onset. (E) The dyn1Δ cells containing TEM1-GFP and mCherry-TUB1 (Ay22666) were imaged at 25°C. Cells where a SPB entering the bud coincided with anaphase onset (correctly positioned) were compared with cells where a SPB did not enter the bud until at least 30 min after the onset of anaphase (mispositioned).
Tem1 binding to SPBs is partially dependent on BUB2 and BFA1 (Pereira et al., 2000; Valerio-Santiago and Monje-Casas, 2011; Caydasi et al., 2012). The GAP complex, however, does not localize to the mSPB during anaphase (Pereira et al., 2000). How then does Tem1 bind to the mSPB in late anaphase? We hypothesized that Tem1’s effector kinases, Cdc15 and Dbf2-Mob1, which localize to both SPBs during anaphase, recruit Tem1 to the mSPB. Consistent with previous studies (Bardin et al., 2000), enrichment of Tem1 at dSPBs occurred independently of the downstream kinases (Figure 2, B and C). Contrarily, Tem1 accumulation at mSPBs in late anaphase required both CDC15 and DBF2 (Figure 2, B and C). Note that in these experiments, the difference between Tem1 association with the dSPB and mSPB was less obvious in wild-type cells than in the experiment shown in Figure 2A. This is because here we conducted the experiment at 32°C rather than 25°C. At this higher temperature, background fluorescence is increased, making subtle differences in timing of recruitment of Tem1 to the dSPB and mSPB less obvious.
Our analysis of Tem1-GFP localization in MEN kinase mutants leads to two important conclusions. First, Tem1 binding to the mSPB and dSPB is differentially regulated. Tem1 binding to the dSPB is partially dependent on the Bub2-Bfa1 GAP complex. Binding to the mSPB, but not dSPB, requires the activity of the downstream MEN kinases. Second, Tem1 accumulates at the dSPB during early anaphase and only much later at the mSPB. Provided that Tem1 accumulation at SPBs is an indicator of Tem1 activity, it follows that MEN signaling initiates at the dSPB. Its accumulation at the mSPB is a consequence of MEN activation. Next, we determined whether this assumption is correct.
Active Tem1 binds SPBs
MEN activation during early anaphase coincides with Tem1 accumulation at dSPBs. To test whether this correlation reflects Tem1 activity, we examined the localization of a hypermorphic TEM1 allele, the TEM1-eGFP fusion. TEM1-eGFP produces approximately twofold more protein than wild-type TEM1 and has phenotypes consistent with hyperactive Tem1 (Chan and Amon, 2009; Supplemental Figure S1). In contrast to Tem1-GFP, which becomes enriched at the SPBs as cells enter anaphase (Figure 2A), Tem1-eGFP associated with the SPBs prematurely, accumulating at the SPB closest to the bud neck already in metaphase and not becoming further enriched as the SPB moved into the bud (Figure 2D). Thus, in the TEM1-eGFP fusion, increased activity correlates with premature enrichment at SPBs. Two previously published observations further support a causal link between Tem1 binding to SPBs and its activity: 1) Tem1 localization to SPBs is necessary and sufficient to activate the MEN: inhibiting binding of Tem1 to SPBs prevents mitotic exit (Gruneberg et al., 2000; Valerio-Santiago and Monje-Casas, 2011), while artificially tethering Tem1 to SPBs causes hyperactivation of the MEN (Valerio-Santiago and Monje-Casas, 2011). 2) Constitutively activating Tem1 by preventing GTP hydrolysis causes premature and increased Tem1 binding to SPBs (Scarfone et al., 2015). We conclude that Tem1 binds to SPBs when active. We further note that the Tem1-eGFP fusion is not an appropriate tool to assess Tem1 localization or activity.
Tem1 SPB binding responds to movement of the dSPB into the bud
Accumulation of Tem1 at the dSPB could be caused by anaphase onset or translocation of a SPB into the bud. To distinguish between these possibilities, we imaged Tem1-GFP in cells that failed to translocate a SPB into the daughter cell during anaphase, that is, in cells with mispositioned anaphase spindles. In such cells the MEN is inactive and cells arrest in anaphase (Yeh et al., 1995). To increase the rate of spindle mispositioning, we inactivated a component of the spindle position machinery; we deleted the gene encoding cytoplasmic dynein (DYN1). While Tem1-GFP accumulated at the dSPB in dyn1Δ cells with correctly positioned anaphase spindles, it failed to do so in cells with mispositioned anaphase spindles (Figure 2E; D’Aquino et al., 2005). We conclude that a SPB translocating into the bud causes Tem1 to accumulate at the dSPB. This accumulation coincides with MEN activation.
The MEN kinases localize to both SPBs in response to Tem1 activity at the dSPB
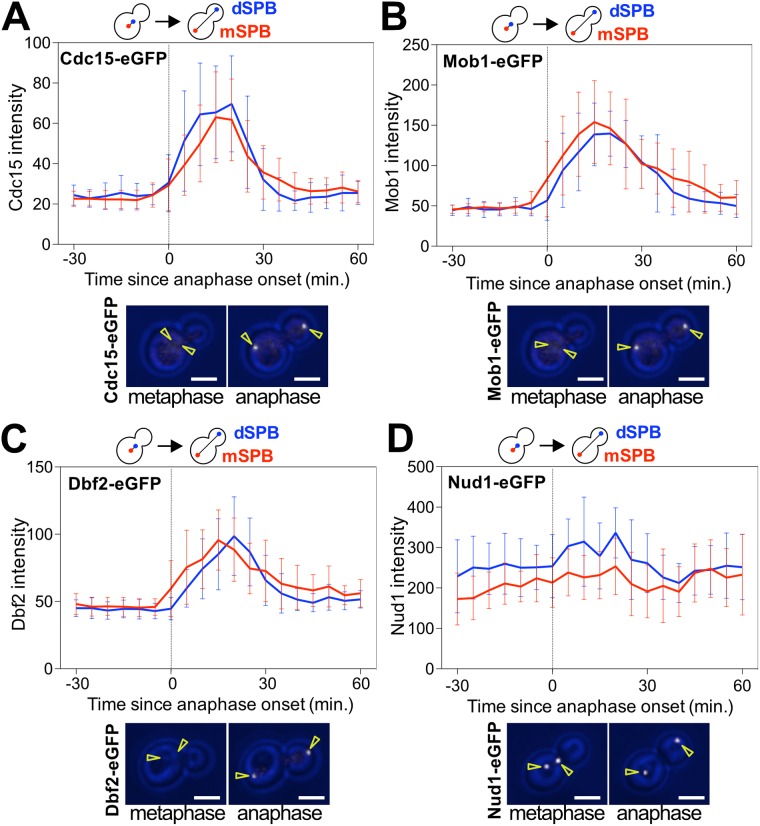
The only essential function of Tem1 is to recruit the MEN kinase Cdc15 to SPBs (Rock and Amon, 2011). Based on this observation, one would predict that Cdc15 binding to SPBs mirrors Tem1 localization. Curiously, this does not appear to be the case. We imaged Cdc15-eGFP by time-lapse microscopy and found that Cdc15-eGFP accumulated at both SPBs early in anaphase. The protein kinase first became enriched at the dSPB and shortly thereafter the mSPB (Figure 3A).
FIGURE 3:
The MEN kinases accumulate at both SPBs at the onset of anaphase. (A–D) Localization of MEN proteins at SPBs in an unperturbed cell cycle. Cells were imaged every 5 min for 3–6 h to determine the intensity (max(SPB)-mean(slide)) of the indicated fluorescently tagged proteins at either the mSPB or the dSPB. Curves show mean and SD for 20 cells per condition. Data were centered at the first frame of anaphase (spindle length > 3 μm). Representative sample images with the indicated fluorescently tagged protein in white. Yellow arrowheads indicate location of the SPBs determined from Tub1 or Spc42 localization. Scale bars are 3 μm. Cells were imaged at 25°C. Cells were chosen for analysis where a SPB entering the bud coincided with anaphase onset. (A) CDC15-eGFP, mCherry-TUB1 (Ay26481). (B) MOB1-eGFP, SPC42-mCherry, CDC14-tdTomato (Ay39323). (C) DBF2-eGFP, SPC42-mCherry, CDC14-tdTomato (Ay39273). (D) NUD1-yEGFP, SPC42-mCherry (Ay40869).
Cdc15 recruits Dbf2-Mob1 to SPBs by creating a docking site for the kinase complex on Nud1 (Rock et al., 2013). As previously reported (Visintin and Amon, 2001; Yoshida and Toh-e, 2001), Dbf2-eGFP and Mob1-eGFP accumulated at both SPBs during anaphase. Our quantitative analysis further revealed that Dbf2-Mob1 became enriched at the mSPB slightly earlier than to the dSPB (Figure 3, B and C). In contrast to the dynamic localization of the MEN kinases, the MEN scaffold Nud1 localized to SPBs throughout the cell cycle (Figure 3D; Adams and Kilmartin, 1999; Elliott et al., 1999; Lengefeld et al., 2017).
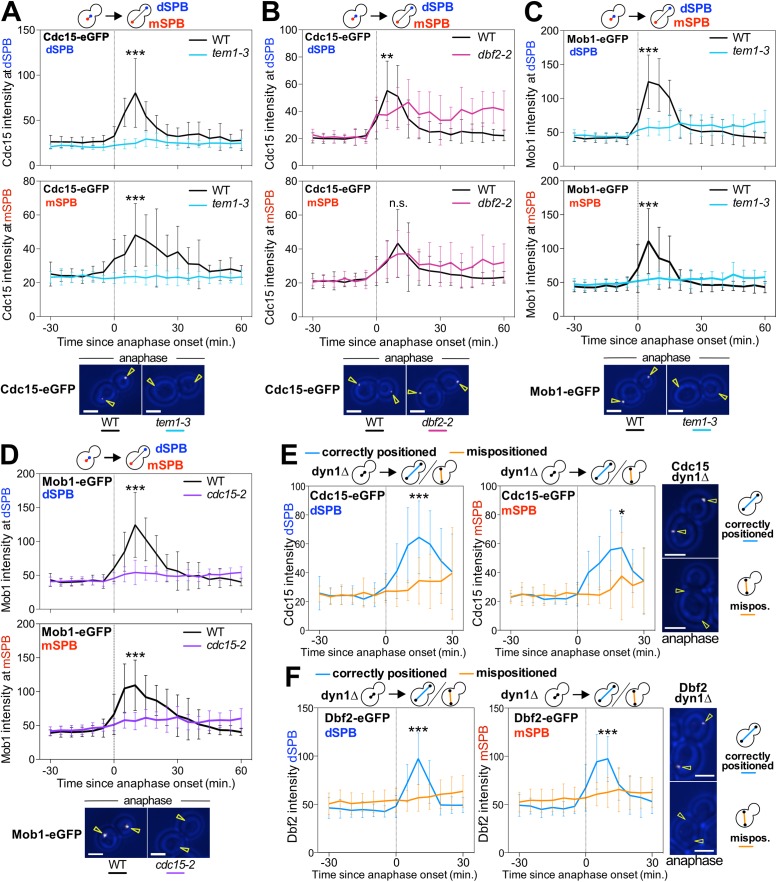
We further confirmed previously established dependencies for MEN component localization to SPBs (Visintin and Amon, 2001). Cdc15’s localization to both SPBs was dependent on TEM1, but not on DBF2 or MOB1. Dbf2-Mob1’s localization to both SPBs required TEM1 and CDC15 (Figure 4, A–D). These data together with previous findings support the model that active Tem1 accumulates at the dSPB. This in turn leads to the recruitment of Cdc15 to both SPBs, which is followed by recruitment of Dbf2-Mob1 to both SPBs (Figure 1A). Importantly, accumulation of the MEN kinases, like accumulation of Tem1 at SPBs, correlates with MEN activity. Cdc15-eGFP and Dbf2-eGFP did not become enriched at SPBs in cells known to harbor an inactive MEN, in cells with mispositioned anaphase spindles (Figure 4, E and F). We conclude that accumulation of MEN components at SPBs reflects their active state and that accumulation of Tem1 at dSPBs induces the downstream kinases to bind to both spindle poles.
FIGURE 4:
Localization dependencies of the MEN kinases. (A–F) Localization of MEN proteins at SPBs in various MEN mutants. Cells were imaged every 5 min for 3–6 h to determine the localization (max(SPB)-mean(slide)) of the indicated fluorescently tagged proteins at either the mSPB or the dSPB. In metaphase, the SPB closer to the bud neck was designated as the dSPB. Curves show mean and SD for 20 cells per condition. Data were centered at the first frame of anaphase (spindle length > 3 μm). Student’s t test was used to assess statistical significance at time points when association of MEN proteins with SPBs was maximal in wild-type cells. Representative sample images with the indicated fluorescently tagged protein are shown in white. Yellow arrowheads indicate location of the SPBs determined from Tub1 or Spc42 localization. Scale bars are 3 μm. (A–D) Cells were imaged after shifting from room-temperature to 34°C. Cells were chosen for analysis where a SPB entering the bud coincided with anaphase onset. (A, B) Wild-type (Ay26481), tem1-3 (Ay30518), or dbf2-2 (Ay40922) cells containing CDC15-eGFP and mCherry-TUB1. (C) Wild-type (Ay33715) or tem1-3 (Ay40390) cells containing MOB1-eGFP and SPC42-mCherry. (D) Wild-type (Ay39323) or cdc15-2 (Ay39353) cells containing MOB1-eGFP, SPC42-mCherry, and CDC14-tdTomato. (E, F) Cells where a SPB entering the bud coincided with anaphase onset (correctly positioned) were compared with cells where a SPB did not enter the bud until at least 30 min after the onset of anaphase (mispositioned). (E) The dyn1Δ cells containing mCherry-TUB1 and CDC15-eGFP (Ay33942) were imaged at 25°C. (F) The dyn1Δ cells containing mCherry-TUB1 and DBF2-eGFP (Ay36721) were imaged at 32°C.
Mitotic exit does not require a SPB in the mother cell
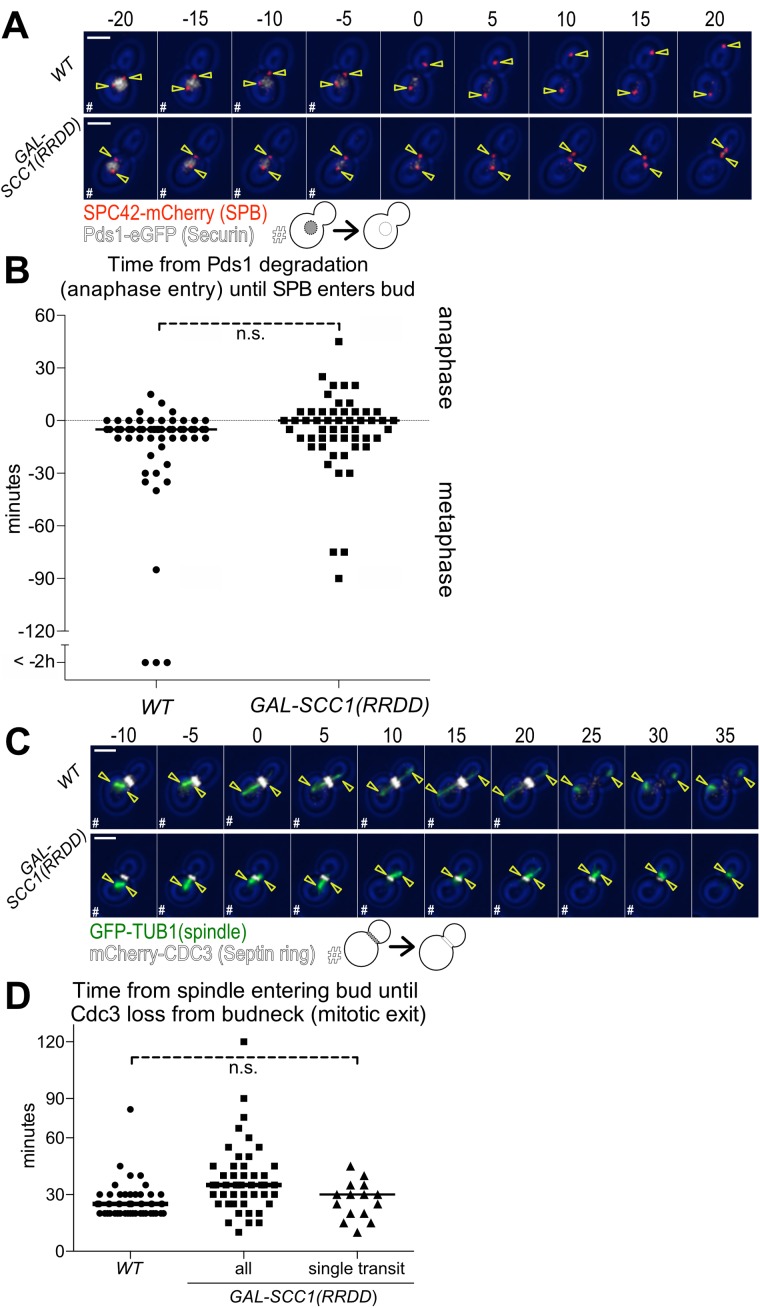
Our results show that the MEN signaling module is assembled in reverse at mSPBs, with Cdc15 and Dbf2-Mob1 recruiting Tem1. A key question arising from these findings is whether accumulation of the MEN kinases at mSPBs is required for MEN signaling. To address this question, we took advantage of a mutation that causes both SPBs to migrate into the bud during anaphase. We overexpressed a noncleavable version of cohesin (Scc1-RRDD; Uhlmann et al., 1999).
Cohesins hold the duplicated sister chromatids together until the protein is cleaved at the onset of anaphase. Cohesin cleavage facilitates anaphase chromosome movement. Preventing anaphase spindle elongation by expressing SCC1-RRDD leads to both SPBs moving into the bud during the time wild-type cells undergo anaphase (Supplemental Figure S2, A and B). We measured when the SPBs move into the bud with respect to the metaphase–anaphase transition as defined by the degradation of the anaphase inhibitor Pds1-eGFP (Cohen-Fix et al., 1996). We found that a SPB first entered the bud within 10 min of anaphase onset in the majority of wild-type cells and cells overexpressing Scc1-RRDD (Figure 5, A and B). Similar results were obtained when we measured the time from the onset of expression of the mitotic-cyclin Clb2, which initiates during late S phase, until a SPB entered the daughter cell (Supplemental Figure S2, A and B). While preventing spindle elongation did not interfere with the ability of SPBs to move into the daughter cell, it did affect how far SPBs moved into the bud. Spindle elongation normally moves one SPB to the tip of the daughter cell. In Scc1–RRDD-expressing cells that failed to elongate their spindle, SPBs moved only halfway into the bud (Figure 5A; Supplemental Figure S2A). Furthermore, preventing spindle elongation resulted in the spindle moving back and forth between the mother cell and the bud in 71% of cells (Figure 5A; Supplemental Figure S2A).
FIGURE 5:
Both SPBs moving into the daughter cell causes exit from mitosis. (A, B) Time from anaphase entry (degradation of Pds1-eGFP) until a SPB enters the bud. Wild-type (Ay40836) or GAL-SCC1(RRDD) (Ay40862) cells containing PDS1-eGFP and SPC42-mCherry were shifted to galactose containing medium at 25°C and imaged every 5 min for 8 h. GAL-SCC1(RRDD) cells were excluded if the SPBs separated (>3 μm). (A) Representative 5 min time-lapse images of a cell in metaphase degrading Pds1-eGFP, transitioning to anaphase, and translocating a SPB in the daughter cell. SPB (Spc42-mCherry) in red, Securin (Pds1-eGFP) in white, and yellow arrowheads indicate location of the SPBs determined from Spc42 localization; scale bar is 3 μm. Metaphase cells containing Pds1-eGFP are marked as “#.” (B) Time from the completion of Pds1-eGFP degradation (anaphase entry) until the first frame a SPB translocates the bud neck (n > 50 cells per condition, median displayed as a solid line, no significant difference according to Student’s t test). (C, D) Time from the spindle entering the bud until mitotic exit (mCherry-Cdc3 loss from the bud neck). Wild-type (Ay39477) or GAL-Scc1(RRDD) (Ay40483) cells containing mCherry-CDC3 and GFP-TUB1 were shifted to galactose containing medium at 25°C and imaged every 5 min for 6 h. GAL-SCC1(RRDD) cells were excluded if the SPBs separated (>3 μm). (C) Representative 5 min time-lapse images of spindle movement into the daughter cell followed by mitotic exit (mCherry-Cdc3 loss from the bud neck). Spindle (GFP-Tub1) in green, Septin ring (mCherry-Cdc3) in white, and yellow arrowheads indicate location of the SPBs determined from Tub1 localization; scale bar is 3 μm. Time points where the cell had not exited mitosis as judged by loss of Cdc3 from the bud neck are marked with “#.” (D) Time from the first frame when part of the spindle translocates the bud neck until loss of Cdc3 ring from the bud neck. The category “all” includes all cells where spindle elongation does not occur and a SPB moves into the bud at some point during imaging. The category “single transit” only encompasses cells in which the spindle transited the bud neck once and remained in the bud (WT n = 52 cells, all n = 51 cells, single transit n = 15 cells, median displayed as solid line, no significant difference according to Student’s t test between control and “single transit” cells.).
We next measured the kinetics of mitotic exit in cells overexpressing Scc1-RRDD by monitoring Cdc3 localization. Cdc3 is a component of the septin ring that localizes to the bud neck from the time of bud formation until CDK inactivation at the end of mitosis (Kim et al., 1991). We found that all cells overexpressing Scc1-RRDD that moved both SPBs into the daughter cell exited from mitosis (Figure 5C), demonstrating that MEN signaling from bud localized SPBs is sufficient for cells to exit from mitosis. We noted that Scc1-RRDD cells exhibited a subtle, yet significant (p = 0.002; Student’s t test) delay in exit from mitosis of 10 min (Figure 5D). We observed a similar 10-min mitotic exit delay when we used Clb2 degradation as a measure of CDK inactivation at the end of mitosis (Supplemental Figure S2, A and C). We hypothesized that the spindle shuttling between mother cell and bud multiple times could be responsible for this delay in exit from mitosis in Scc1–RRDD-expressing cells. Indeed, Scc1–RRDD-expressing cells that underwent a single spindle translocation event, from the mother cell into the bud, only exhibited a 5-min delay that was not statistically significant (Figure 5D). We conclude that the persistent presence of a SPB in the mother cell during anaphase is not required for the MEN to promote exit from mitosis. Thus, our results are inconsistent with the sink model, which posits that MEN signaling emanates from the mSPB.
Two SPBs are necessary for efficient MEN signaling
Having established that the presence of a SPB in the mother cell during anaphase is not required for MEN signaling, we next asked whether the mSPB has a function in MEN signaling not by virtue of where it is localized in the cell but rather because it provides an additional signaling platform. We tested this possibility by analyzing MEN activity in cells that only contained one SPB.
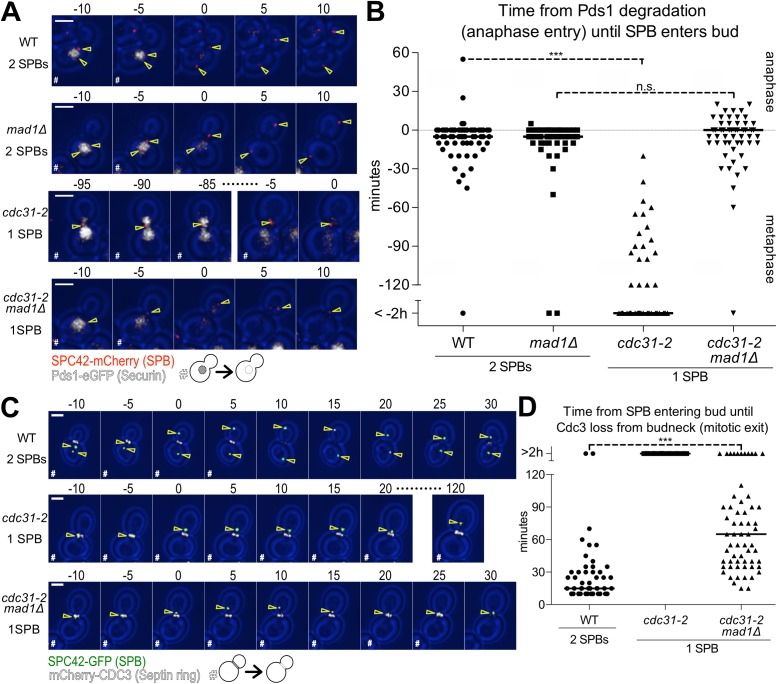
Cells mutant for the SPB half-bridge component CDC31 fail to duplicate their SPBs (Byers, 1981a). EM studies of the SPB in cdc31 mutants indicate that the singular SPB is enlarged but able to nucleate nuclear and cytoplasmic microtubules (Byers, 1981b). As such, assembly of MEN components on this SPB is not expected to be affected. Cells harboring only one SPB, however, fail to form a mitotic spindle and hence activate the spindle assembly checkpoint (SAC), which arrests cells in metaphase (Weiss and Winey, 1996). To prevent the SAC from interfering with mitotic progression, we deleted the SAC gene MAD1. We then determined whether cells with a monopolar spindle progress into anaphase by monitoring the time from Pds1-eGFP degradation (anaphase entry) until a SPB moves into the bud. In wild-type and mad1Δ strains, SPB duplication was followed by a SPB traversing the bud neck 5 min prior to Pds1 degradation (median; Figure 6, A and B). In contrast, cdc31-2 mutants arrested in metaphase and eventually moved their single SPB into the daughter cell without degrading Pds1 (Figure 6, A and B). The cdc31-2, mad1Δ double mutants were able to enter anaphase as judged by Pds1 degradation. Movement of the single SPB across the bud neck coincided with anaphase entry (median; Figure 6, A and B). Only 9% of cdc31-2, mad1Δ double mutants left their single SPB in the mother cell for more than 10 min after Pds1 degradation. It is, however, worth noting that the single SPB of cdc31-2, mad1Δ mutants did not move into the bud as deeply as did wild-type SPBs and occasionally traversed the bud neck multiple times.
FIGURE 6:
Mitotic exit is delayed in cells with a single SPB. (A, B) Time from anaphase entry (degradation of Pds1-eGFP) until a SPB enters the bud. Wild-type (Ay40836), mad1Δ (Ay40891), cdc31-2 (Ay40890), or cdc31-2 mad1Δ (Ay40888) cells containing PDS1-eGFP and SPC42-mCherry were shifted to 34°C and imaged every 5 min for 8 h. The cdc31-2 and cdc31-2 mad1Δ cells with 2 SPBs were excluded from analysis. (A) Representative 5 min time-lapse images of a cell in metaphase degrading Pds1-eGFP, transitioning to anaphase, and translocating a SPB into the daughter cell. SPB (Spc42-mCherry) in red, Securin (Pds1-eGFP) in white, and yellow arrowheads indicate location of the SPBs determined from Spc42 localization; scale bar is 3 μm. Metaphase cells containing Pds1-eGFP are marked with “#.” (B) Time from the completion of Pds1-eGFP degradation (anaphase entry) until the first frame a SPB translocates the bud neck (n > 50 cells, median displayed as solid line, asterisks show significant difference according to Student’s t test). (C, D) Time from a SPB entering the bud until mitotic exit (mCherry-Cdc3 loss from bud neck). Wild-type (Ay40538), cdc31-2 (Ay40536), or cdc31-2 mad1Δ (Ay40534) cells containing mCherry-CDC3 and SPC42-GFP were shifted to 34°C and imaged every 5 min for 8 h. The cdc31-2 and cdc31-2 mad1Δ cells with 2 SPBs were excluded. (C) Representative 5 min time-lapse images of SPB movement into the daughter cell followed by mitotic exit (mCherry-Cdc3 loss from bud neck), except cdc31-2, which arrests in metaphase and therefore fails to exit mitosis. SPBs (Spc42-GFP) in green, Septin ring (mCherry-Cdc3) in white, and yellow arrowheads indicate location of the SPBs determined from Spc42 localization, scale bar is 3 μm. Cells that have not exited mitosis and degraded Cdc3 are marked as “#.” (D) Time from the first frame that a SPB translocates the bud neck until degradation of Cdc3 (n > 50 cells, median displayed as a solid line; asterisks show significant difference according to Student’s t test).
Having established that preventing SPB duplication does not prevent entry of the singular SPB into the bud, we next determined whether cells were able to exit from mitosis with only a single SPB providing a scaffold for MEN signaling. Eighty-two percent of cdc31-2, mad1Δ double mutants were able to exit mitosis within 2 h of the SPB first entering the bud as judged by Cdc3 dissociation from the bud neck (Figure 6, C and D). While a single SPB was sufficient to trigger mitotic exit, the cell cycle transition was delayed in cdc31-2, mad1Δ mutants. Wild-type cells exited mitosis 15 min after a SPB entered the bud compared with 65 min in cdc31-2, mad1Δ mutants with a single SPB (median, Figure 6D).
The cdc31-2 cells spontaneously diploidized (Schild et al., 1981). Although ploidy is not known to affect anaphase duration and the kinetics of exit from mitosis, we wished to examine another mutant defective in SPB duplication that does not undergo spontaneous diploidization, sfi1-3 (Kilmartin, 2003). Like cdc31-2 mad1Δ mutants, sfi1-3 mad1Δ mutants were also delayed in exit from mitosis (Supplemental Figure S3). We conclude that a single SPB in the daughter cell provides sufficient MEN signal to trigger mitotic exit. The fact that this exit occurs with a significant delay suggests that a single SPB is less efficient in promoting this cell cycle transition than two. We propose that MEN kinases assembling at mSPBs serves to amplify MEN signaling.
DISCUSSION
SPBs - MEN signaling hubs
A large body of evidence indicates that MEN components must assemble onto SPBs to promote Cdc14 release from the nucleolus and thereby promote exit from mitosis. First, a near perfect correlation exists between accumulation of MEN components at SPBs and their activity (Bardin et al., 2000; Pereira et al., 2000; Visintin and Amon, 2001; Yoshida and Toh-e, 2001; König et al., 2010; Rock and Amon, 2011; Rock et al., 2013; Campbell et al., 2019; this study). A notable exception to this correlation is the lack of Cdc15 enrichment at SPBs in cdc14-3 mutant cells (I.W.C., unpublished observations), which may indicate that Cdc15 can act at SPBs without accumulating. Second, binding of MEN components to SPBs is necessary for MEN activation. Interfering with localization of Tem1 to SPBs by targeting the protein to the plasma membrane prevents MEN signaling (Valerio-Santiago and Monje-Casas, 2011). Finally, constitutively targeting the MEN components Tem1 or Cdc15 to SPBs is sufficient to activate the pathway (Rock and Amon, 2011; Valerio-Santiago and Monje-Casas, 2011; Scarfone et al., 2015). Together, these data indicate that when a MEN component accumulates at SPBs, the pathway is active.
Given the importance of assembly at SPBs for MEN signaling, it is critical to understand how their binding to the organelle is regulated. We found that the manner by which MEN components associated with SPBs differs between the mother and daughter cell compartment. At the dSPB, Tem1 binding is in part mediated by Bub2-Bfa1. Tem1 in turn recruits Cdc15 which then, by phosphorylating the MEN scaffold Nud1, creates a binding interface for Dbf2-Mob1 at SPBs. Assembly of MEN signaling modules at the mSPB is different. Tem1 binding requires Cdc15 and Dbf2-Mob1. Given that binding of Cdc15 and Dbf2-Mob1 to the mSPB depends on Tem1, this observation indicates that assembly of the MEN at mSPBs only happens after Tem1 activates the MEN at dSPBs. These findings further indicate that once activated, Cdc15 can bind to SPBs independently of upstream components of the pathway. Whether or not Tem1 serves a MEN-activating function at the mSPB once recruited to this spindle pole remains to be determined. Accumulation of the GTPase at the mSPB occurs very late in anaphase—10 min after the dSPB first enters the bud, suggesting that Tem1 signaling from the mSPB is not relevant for exit from mitosis.
Which SPB does the MEN signal from?
A key question regarding MEN regulation is which SPB signals. As described in the Introduction, the zone model posits that MEN activity at the dSPB promotes exit from mitosis, whereas the sink model predicts that the mSPB does (Figure 1, A and B). If, as a large body of evidence suggests, the MEN can only signal from SPBs, the localization pattern of MEN components suggests the following sequence of events: the MEN signal is initiated at the dSPB and then transmitted through the mSPB and dSPB. Tem1 accumulates at the dSPB first and only much later at the mSPB. Cdc15 arrives at the dSPB slightly earlier than on the mSPB. Curiously, Dbf2-Mob1 exhibits the reverse localization pattern. The ultimate MEN component accumulates at the mSPB slightly earlier than at the dSPB. This switch in order of SPB association may be due to differential CDK or Cdc14 activity at the dSPB and mSPB. CDKs phosphorylate Cdc15 and Dbf2-Mob1, thereby inhibiting their activity from S phase until anaphase onset (Jaspersen and Morgan, 2000; König et al., 2010; Campbell et al., 2019). Mitotic CDK activity might be lower at mSPBs, allowing Dbf2-Mob1 to accumulate at the mSPB first.
Based on these localization patterns of MEN components, we propose that only the MEN GTPase Tem1 is regulated by the position of the SPBs (Figure 7). The mother cell-localized MEN-inhibitor Kin4 prevents Tem1 from becoming active at mSPBs, while the daughter cell-localized MEN-activator Lte1 promotes Tem1’s activation at dSPBs (D’Aquino et al., 2005; Pereira and Schiebel, 2005; Fraschini et al., 2006; Maekawa et al., 2007; Chan and Amon, 2010; Bertazzi et al., 2011; Falk et al., 2011, 2016a). Therefore, in early anaphase, Tem1 only accumulates at SPBs that move into the daughter cell (Figure 7, Step 1). The GTPase, together with the Polo kinase Cdc5, then recruits and activates Cdc15 at dSPBs (Figure 7, Step 2). Active Cdc15 binds both SPBs creating a binding site for Dbf2-Mob1 by phosphorylating Nud1 (Figure 7, Step 3). Dbf2-Mob1 binds to the mSPB first, possibly due to lower CDK activity at this SPB (Figure 7, Step 4). Dbf2-Mob1 activated at mSPBs then initiates the translocation of Cdc14 into the cytoplasm, where it antagonizes mitotic CDKs at the dSPB to allow Dbf2-Mob1 to bind the dSPB, further amplifying the MEN signal (Figure 7, Step 5). Signaling from both SPBs allows for the efficient activation of Cdc14 and the prompt exit from mitosis.
FIGURE 7:
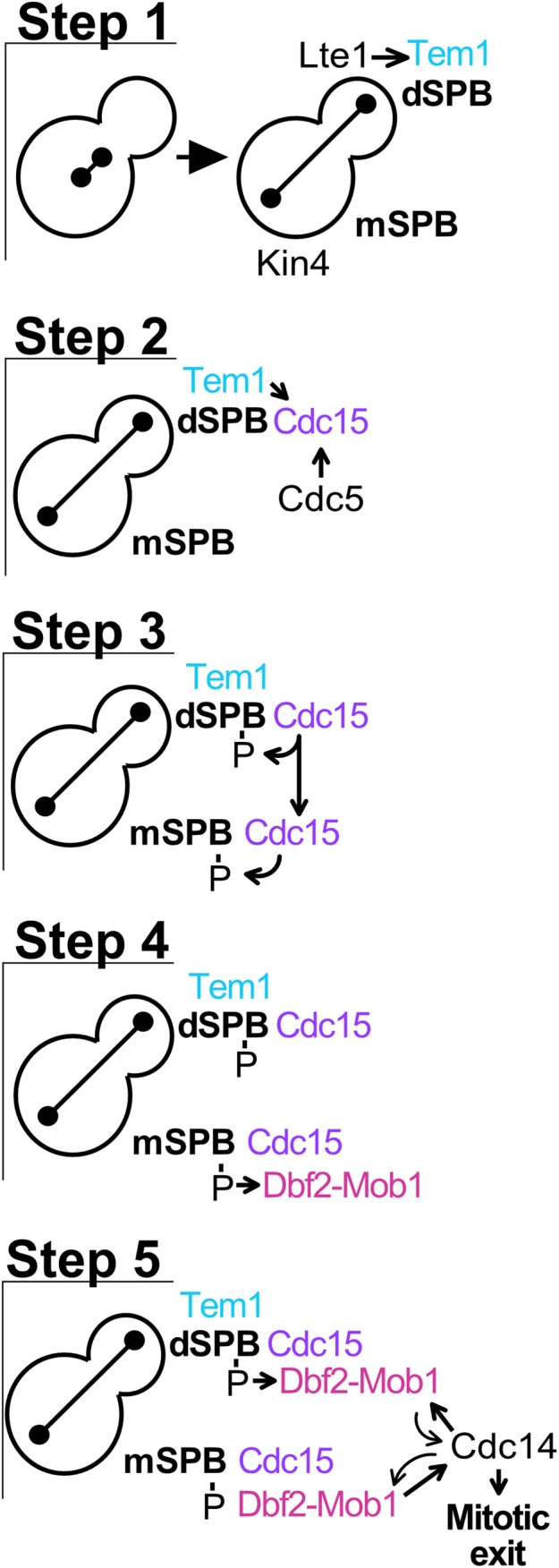
MEN signaling is amplified by localization of the kinase cascade to both SPBs. (Step 1) The mother cell-localized MEN-inhibitor Kin4 prevents Tem1 activation on mSPBs. Anaphase spindle elongation along the mother–daughter axis brings a SPB into contact with the daughter cell-localized MEN-activator Lte1, promoting Tem1 activation and association with the dSPB. (Step 2) Tem1 and Cdc5 recruit Cdc15 to the dSPB. (Step 3) Active Cdc15 binds to both SPBs, creating a binding site for Dbf2-Mob1 by phosphorylating Nud1. (Step 4) Dbf2-Mob1 binds first to the mSPB, becoming active. (Step 5) Dbf2-Mob1 initiates translocation of Cdc14 into the cytoplasm where it promotes Dbf2-Mob1 association with both SPBs by antagonizing CDK activity. Amplification of Dbf2-Mob1 activity originating from both SPBs increases Cdc14 activity, thereby triggering exit from mitosis.
In summary, our localization studies lead to the surprising conclusion that the MEN is not active at one SPB or the other, but rather that different components of the pathway are active at different poles at different times. Tem1 is active at the dSPB early in anaphase, Cdc15 is first active at the dSPB and then the mSPB, whereas Dbf2-Mob1 is active first at the mSPB and then at the dSPB. Having Dbf2-Mob1 active at both SPBs is important. The presence of two SPBs, regardless of whether they are both in the daughter cell or split between mother and daughter cells, is required to create a rapid and controlled mitotic exit signal.
How do the dSPB and mSPB communicate?
An important question raised by our work is how Tem1 at the dSPB causes binding of Cdc15 to the mSPB. It is possible that a small, unobserved population of Tem1 at the mSPB recruits Cdc15. We do not favor this hypothesis because it is inconsistent with the finding that Tem1’s anchor, Bub2-Bfa1, is actively removed from mSPBs by Kin4 (D’Aquino et al., 2005; Pereira and Schiebel, 2005; Fraschini et al., 2006; Maekawa et al., 2007; Caydasi and Pereira, 2009; Scarfone et al., 2015). It is also possible that a pool of cytoplasmic Tem1 activates Cdc15, allowing Cdc15 to associate with mSPBs (Caydasi et al., 2012). Indeed, during meiosis, the MEN does not require a SPB scaffold for signaling (Attner and Amon, 2012). However, we do not favor this hypothesis either, because a large body of evidence indicates that during mitosis Tem1 is only active at the SPBs: 1) inhibiting Tem1 from binding to SPBs prevents mitotic exit (Gruneberg et al., 2000; Valerio-Santiago and Monje-Casas, 2011), 2) tethering Tem1 to SPBs causes hyperactivation of the MEN (Valerio-Santiago and Monje-Casas, 2011), and 3) constitutively active Tem1- Q79L accumulates prematurely at SPBs (Scarfone et al., 2015).
We favor the idea that activation of Cdc15 at the dSPB by Tem1 causes Cdc15 to gain the ability to bind SPBs independently of Tem1 and hence is able to bind the mSPB. Cdc15 is homologous to PAK kinases. These kinases harbor regulatory domains that inhibit kinase activity in trans (Kumar et al., 2017). Binding of Tem1 to Cdc15 and, we speculate, phosphorylation by Cdc5, alleviates this Cdc15 autoinhibition and exposes a SPB-binding interface in the protein kinase. Understanding the molecular mechanisms whereby Tem1 and Cdc5 facilitate expansion of MEN signaling to two SPBs will be critical not only to understanding how the MEN promotes rapid exit from mitosis but also to expanding our knowledge as to how GTPases can control effector kinases.
MATERIALS AND METHODS
Yeast strains and growth conditions
All S. cerevisiae strains in this study are derivatives of W303 (A2587). Strain genotypes are listed in Supplemental Table S1. Growth conditions are described in the figure legends.
Time-lapse microscopy
Growth conditions for time-lapse imaging are described in the figure legends. Cells were imaged on agarose pads (2% agarose, synthetic complete medium containing 2% glucose, unless otherwise noted) affixed to a glass slide. In our analyses, we found that limit of detection for the proteins examined in this study was affected by growth temperature. Growth at higher temperatures (32°C and higher) resulted in a lower signal-to-background ratio. Therefore, we only compared cells grown at the same temperature. Comparison should not be made between graphs, especially when cells were grown at different temperatures.
The microscope platform was a DeltaVision Elite (GE Healthcare Bio-Sciences, Pittsburgh, PA) consisting of an InsightSSI solid state light source, an UltimateFocus hardware autofocus system, and a model IX-71, Olympus microscope controlled by SoftWoRx software. A 60× Plan APO 1.42NA objective and CoolSNAP HQ2 camera were used for image acquisition.
Image processing was performed using the Volocity (PerkinElmer) software package. Analysis was performed on maximum projections. The number of cells, length of analysis, and cell selection criteria are described in the figure legends. Protein intensity was quantified by manually segmenting the SPBs, as identified by Spc42 or Tub1 localization, and measuring maximum intensity within the segmented region (max[SPB]). Slide background was subtracted from the maximum intensity at the SPB (max[SPB]-mean[slide]). Data were centered at the first frame of anaphase (spindle length >3 μm) as defined by Spc42 or Tub1 localization. Pds1-eGFP degradation was defined as the time frame when Pds1-eGFP intensity was reduced below three-fourths of the maximum. A SPB entering the bud was defined as the first time frame a SPB, as identified by Spc42 or Tub1 localization, translocated the bud neck and was observed in the daughter cell. Cdc3 loss from the bud neck was defined by the first time frame a complete ring was no longer observed at the bud neck. Clb2 expression and degradation were manually defined by the presence and absence (respectively) of nuclear Clb2-eGFP. Anaphase durations in Supplemental Figure S1 was measured by monitoring the distance between two SPBs marked by Spc42. Custom MATLAB scripts (MATLAB_R2018b) were used to automatically segment and track cells and SPBs. Anaphase duration was defined as the time between anaphase onset (spindle length/distance between two SPBs >3 μm) and spindle disassembly/relaxation (the time frame after maximum spindle length).
Supplementary Material
Acknowledgments
We thank S. Jaspersen (Stowers, USA), S. Lacefield (Indiana Univeristy, USA), D. Pellman (Harvard, USA), E. Unal (Berkeley, USA), R. Deshaies (Caltech, USA), and E. Schiebel (ZMBH, Germany) for strains and reagents; and members of our laboratory for their critical reading of the manuscript. This work was supported by a grant from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (HD085866). X.Z. was supported by a Helen Hay Whitney postdoctoral fellowship. A.A. is also investigator of the Howard Hughes Medical Institute and the Paul F. Glenn Center for Biology of Aging Research at MIT.
Abbreviations used:
- APC/C
Anaphase Promoting Complex or Cyclosome
- CDK
cyclin-dependent kinase
- SPB
spindle pole body
- dSPB
daughter-cell-localized SPB
- MEN
Mitotic Exit Network
- mSPB
mother-cell-localized SPB
- SAC
spindle assembly checkpoint.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-10-0584) on February 19, 2020.
REFERENCES
- Adams IR, Kilmartin JV. (1999). Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae . J Cell Biol , 809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attner MA, Amon A. (2012). Control of the mitotic exit network during meiosis. Mol Biol Cell , 3122–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin AJ, Visintin R, Amon A. (2000). A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell , 21–31. [DOI] [PubMed] [Google Scholar]
- Bertazzi DT, Kurtulmus B, Pereira G. (2011). The cortical protein Lte1 promotes mitotic exit by inhibiting the spindle position checkpoint kinase Kin4. J Cell Biol , 1033–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchoux C, Uhlmann F. (2011). A quantitative model for ordered Cdk substrate dephosphorylation during mitotic exit. Cell , 803–814. [DOI] [PubMed] [Google Scholar]
- Byers B. (1981a). Cytology of the yeast life cycle. In: The Molecular Miology of the Yeast Saccharomyces Life Cycle and Inheritance, New York: Cold Spring Harbor Laboratory Press, 59–96. [Google Scholar]
- Byers B. (1981b). Multiple roles of the spindle pole bodies in the life cycle of Saccharomyces cerevisiae. In: Molecular Genetics in Yeast: Alfred Benzon Symposia 16, ed. von Wettstein D, Friis J, Kielland-Brandt M, Stenderuped A, Munksgaard, Copenhagen, 119–131. [Google Scholar]
- Campbell IW, Zhou X, Amon A. (2019). The Mitotic Exit Network integrates temporal and spatial signals by distributing regulation across multiple components. Elife , e41139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caydasi AK, Lohel M, Grünert G, Dittrich P, Pereira G, Ibrahim B. (2012). A dynamical model of the spindle position checkpoint. Mol Syst Biol , 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caydasi AK, Pereira G. (2009). Spindle alignment regulates the dynamic association of checkpoint proteins with yeast spindle pole bodies. Dev Cell , 146–156. [DOI] [PubMed] [Google Scholar]
- Chan LY, Amon A. (2009). The protein phosphatase 2A functions in the spindle position checkpoint by regulating the checkpoint kinase Kin4. Genes Dev , 1639–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan LY, Amon A. (2010). Spindle position is coordinated with cell-cycle progression through establishment of mitotic exit-activating and -inhibitory zones. Mol Cell , 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. (1996). Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev , 3081–3093. [DOI] [PubMed] [Google Scholar]
- D’Aquino KE, Monje-Casas F, Paulson J, Reiser V, Charles GM, Lai L, Shokat KM, Amon A. (2005). The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol Cell , 223–234. [DOI] [PubMed] [Google Scholar]
- Elliott S, Knop M, Schlenstedt G, Schiebel E. (1999). Spc29p is a component of the Spc110p subcomplex and is essential for spindle pole body duplication. Proc Natl Acad Sci , 6205–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk JE, Campbell IW, Joyce K, Whalen J, Seshan A, Amon A. (2016a). LTE1 promotes exit from mitosis by multiple mechanisms. Mol Biol Cell , 3991–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk JE, Chan LY, Amon A. (2011). Lte1 promotes mitotic exit by controlling the localization of the spindle position checkpoint kinase Kin4. Proc Natl Acad Sci , 12584–12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk JE, Tsuchiya D, Verdaasdonk J, Lacefield S, Bloom K, Amon A. (2016b). Spatial signals link exit from mitosis to spindle position. Elife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R, D’Ambrosio C, Venturetti M, Lucchini G, Piatti S. (2006). Disappearance of the budding yeast Bub2-Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit. J Cell Biol , 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U, Campbell K, Simpson C, Grindlay J, Schiebel E. (2000). Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J , 6475–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotz M, Barral Y. (2014). The Mitotic Exit Network: new turns on old pathways. Trends Cell Biol , 145–152. [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. (1995). Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell , 269–278. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Tinker-Kulberg RL, Morgan DO. (1998). A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae . Mol Biol Cell , 2803–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Morgan DO. (2000). Cdc14 activates cdc15 to promote mitotic exit in budding yeast. Curr Biol , 615–618. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV. (2003). Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J Cell Biol , 1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HB, Haarer BK, Pringle JR. (1991). Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J Cell Biol , 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Peters J-M, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. (1995). A 20s complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell , 279–288. [DOI] [PubMed] [Google Scholar]
- König C, Maekawa H, Schiebel E. (2010). Mutual regulation of cyclin-dependent kinase and the mitotic exit network. J Cell Biol , 351–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Sanawar R, Li X, Li F. (2017). Structure, biochemistry, and biology of PAK kinases. Gene , 20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengefeld J, Hotz M, Rollins M, Baetz K, Barral Y. (2017). Budding yeast Wee1 distinguishes spindle pole bodies to guide their pattern of age-dependent segregation. Nat Cell Biol , 941–951. [DOI] [PubMed] [Google Scholar]
- Maekawa H, Priest C, Lechner J, Pereira G, Schiebel E. (2007). The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal. J Cell Biol , 423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah AS, Jang J, Deshaies RJ. (2001). Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc Natl Acad Sci USA , 7325–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoni R, Montani F, Visintin C, Caudron F, Ciliberto A, Visintin R. (2010). Oscillations in Cdc14 release and sequestration reveal a circuit underlying mitotic exit. J Cell Biol , 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohl DA, Huddleston MJ, Collingwood TS, Annan RS, Deshaies RJ. (2009). Dbf2–Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis. J Cell Biol , 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molk JN, Schuyler SC, Liu JY, Evans JG, Salmon ED, Pellman D, Bloom K. (2004). The differential roles of budding yeast Tem1p, Cdc15p, and Bub2p protein dynamics in mitotic exit. Mol Biol Cell , 1519–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Höfken T, Grindlay J, Manson C, Schiebel E. (2000). The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell , 1–10. [PubMed] [Google Scholar]
- Pereira G, Schiebel E. (2005). Kin4 kinase delays mitotic exit in response to spindle alignment defects. Mol Cell , 209–221. [DOI] [PubMed] [Google Scholar]
- Rock JM, Lim D, Stach L, Ogrodowicz RW, Keck JM, Jones MH, Wong CC, Yates JR 3rd, Winey M, Smerdon SJ, et al. (2013). Activation of the yeast Hippo pathway by phosphorylation-dependent assembly of signaling complexes. Science , 871–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JM, Amon A. (2011). Cdc15 integrates Tem1 GTPase-mediated spatial signals with Polo kinase-mediated temporal cues to activate mitotic exit. Genes Dev , 1943–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarfone I, Venturetti M, Hotz M, Lengefeld J, Barral Y, Piatti S. (2015). Asymmetry of the budding yeast Tem1 GTPase at spindle poles is required for spindle positioning but not for mitotic exit. PLOS Genet , e1004938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D, Ananthaswamy HN, Mortimer RK. (1981). An endomitotic effect of a cell cycle mutation of Saccharomyces cerevisiae. Genetics , 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ. (1999). Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell , 233–244. [DOI] [PubMed] [Google Scholar]
- Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. (1999). Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell , 245–256. [DOI] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A. (1995). The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell , 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M, Morgan DO. (2007). Finishing mitosis, one step at a time. Nat Rev Mol Cell Biol , 894–903. [DOI] [PubMed] [Google Scholar]
- Taylor GS, Liu Y, Baskerville C, Charbonneau H. (1997). The activity of Cdc14p, an oligomeric dual specificity protein phosphatase from Saccharomyces cerevisiae, is required for cell cycle progression. J Biol Chem , 24054–24063. [DOI] [PubMed] [Google Scholar]
- Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. (1995). CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell , 261–268. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. (1999). Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature , 37–42. [DOI] [PubMed] [Google Scholar]
- Valerio-Santiago M, Monje-Casas F. (2011). Tem1 localization to the spindle pole bodies is essential for mitotic exit and impairs spindle checkpoint function. J Cell Biol , 599–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Amon A. (2001). Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Mol Biol Cell , 2961–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. (1998). The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell , 709–718. [DOI] [PubMed] [Google Scholar]
- Visintin R, Hwang ES, Amon A. (1999). Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature , 818–823. [DOI] [PubMed] [Google Scholar]
- Weiss E, Winey M. (1996). The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol , 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. (1995). Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol , 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Toh-e A. (2001). Regulation of the localization of Dbf2 and mob1 during cell division of saccharomyces cerevisiae. Genes Genet Syst , 141–147. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W. (1998). Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science , 1721–1724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.