Abstract
The outbreak of Middle East respiratory syndrome-coronavirus (MERS-CoV) in South Korea in April 2015 led to 186 infections and 37 deaths by the end of October 2015. MERS-CoV was isolated from the imported patient in China. The envelope (E) protein, a small structural protein of MERS-CoV, plays an important role in host recognition and infection. To identify the conserved epitopes of the E protein, sequence analysis was performed by comparing the E proteins from 42 MERS-CoV strains that triggered severe pandemics and infected humans in the past. To predict the potential B cell epitopes of E protein, three most effective online epitope prediction programs, the ABCpred, Bepipred, and Protean programs from the LaserGene software were used. All the nucleotides and amino acids sequences were obtained from the NCBI Database. One potential epitope with a suitable length (amino acids 58–82) was confirmed and predicted to be highly antigenic. This epitope had scores of >0.80 in ABCpred and level 0.35 in Bepipred programs. Due to the lack of X-ray crystal structure of the E protein in the PDB database, the simulated 3D structure of the E protein were also predicted using PHYRE 2 and Pymol programs. In conclusion, using bioinformatics methods, we analyzed the genome sequence of MERS-CoV and identified a potential B-cell epitope of the E protein, which might significantly improve our current MERS vaccine development strategies.
Keywords: MERS-CoV, bioinformatics, linear B-cell epitope
1. Introduction
Middle East respiratory syndrome (MERS, so the early once called it a kind of virus like SARS) is a newly described disease in humans and was first reported after the identification of a novel beta coronavirus (MERS-CoV) from a patient who died of a severe respiratory illness in Saudi Arabia in September 2012 [1]. Coronavirus is a canonical and ancient virus system that can be divided into four categories based on their genome sequence: Alphacoronavirus, Betacoronavirus, Gammacoronavirus , and Deltacoronavirus coronavirus. Since it was discovered coronavirus was considered to be relatively harmless to humans until the outbreaks of SARS and MERS in 2003 and 2012, respectively. MERS-CoV is a new type of coronavirus identified after the discovery of SARS-CoV, belongs to the Beta coronavirus lineage C [2], which causes severe acute respiratory disease with a high fatality rate. Moreover, the virus rapidly spread from the Middle East to many other countries including South Korea in 2015, demonstrating a global epidemic trend. However, no effective antiviral drug or vaccine has been developed to treat MERS-CoV.
Coronavirus is a class of enveloped RNA virus with a 27–3l kb long single-stranded positive-sense genome. The genome includes two large replicase open reading frames, ORF1a and ORF1b, encoding two viral replicase polyproteins. The region downstream of ORF1 contains at least 10 small ORFs, encoding the spike protein (S), small envelope protein (E), membrane protein (M), nucleocapsid protein (N) and the assumed nonstructural proteins [3]. Among these proteins, the E protein is a relatively smaller but massively expressed virus envelope protein and plays an important role in virus membrane packaging [4]. MERS-CoV is a new type coronavirus with a 30.1 kb long genome. Similar to the other coronaviruses, the E protein is a small structural protein in MERS-CoV which was identified as the structural component of the virus. Previous studies suggest that the E proteins are scattered on the surface of the virus envelope and play various important roles in regulating the viral life cycles in some coronaviruses [3], [5], [6], [7]. Understanding the MERS-CoV E protein structure and function could possibly find therapeutic targets to prevent and control the coronaviruses related diseases.
Using bioinformatics methods, we analyzed the MERS CoV E protein's sequence and its secondary and 3D structures. E protein was found to be stable, hydrophobic and highly conserved among different coronavirus stains. Various secondary structures were identified in E protein and its 3D structure was predicted. Finally one potential B-cell linear epitope amino acids position 58-82 was identified at C-terminal of E protein. Thus our study suggests that E protein could possibly be a good candidate for B cell-line epitopes in preparing monoclonal antibodies, vaccines and anti-viral inhibitors against MERS-CoV infection in the future.
2. Methods
2.1. Genome Sequences Comparison Analysis
The sequences of MERS-CoV E protein and other coronavirus E proteins were also compared using the Clustal W multiple sequence alignment program (Bioedit 7). The nucleotide and amino acid sequences of the E proteins from 42 MERS-CoV strains (Fig. 1) were aligned using the Clustal W multiple sequence alignment program (Bioedit 7). All sequences are available at the NCBI database (http://www.ncbi.nlm.nih.gov/nucleotide/ ).
Fig. 1.
Sequence alignment of 42 strains coronavirus E protein.
2.2. Analysis of Protein Primary Structure and Physical Properties of the MERS-CoV E Protein
The complete sequence of the MERS-CoV E protein contains 82 amino acids(GeneBank ID: AGV08384.1) was obtained from the NCBI database. General physical properties of the E protein, including the theoretical pI value, amino acid composition and molecular weight, were analyzed using the online ProtParam tool (http://web.expasy.org/protparam/ ).
2.3. Prediction of Secondary Structure and B Cell Linear Epitope
The secondary structure and linear B-cell epitopes of the E protein were predicted by ABCpred, Bepipred, and Protean programs in LaserGene software. Selected sequences were downloaded from NCBI database and analyzed by each program. ABCpred uses a recurrent method which is based on a neural network algorithm [8] (http://www.imtech.res.in/raghava/abcpred/ABC_submission.html). The lengths of amino acids were set to 12-mer, 14-mer, 16-mer, 18-mer, and 20-mer and the scoring threshold was set to 0.8. Since there is no evidence showing that 20 amino acids is the optimal length for B-cell epitopes, a length of 15–20 amino acids is generally considered. Optimal length is determined by the overall score, higher score indicates a better chance that a sequence is an antigen epitope. Bepipred was developed by Larsen et al. [9] ( http://www.cbs.dtu.dk/services/BepiPred/) and it implements a hidden Markov model and a propensity scale method as described by Parker et al. [10]. A threshold of 0.350 was used. For this threshold the sensitivity is 0.49 and the specificity is 0.75. The Protean program in LaserGene software uses the Garnjer-Robson, Chou-Fasman and Karplus-Schulz methods to predict the protein's secondary structure. The parameters of each residue were determined by Pα, Pβ, PT, and PC values [11]. According to the Kyte-Doolittle standard for hydrophilic amino acids, potential B cell linear epitopes were usually the hydrophilic regions on the protein surface (as described by Emini). Karplus-Schulz method was used to identify the flexible regions of the E protein. Jameson-Wolf method was used to predict the antigenic index. The significance of the parameters has been described previously [12], [13], [14]. All the resulting sequences were collected and aligned using Clustal W multiple sequence alignment programs. Overlapped regions were considered as the potential epitopes.
2.4. Prediction of 3D Structure
PHYRE 2 server was used to model multiple domains of the MERS-CoV E protein with high confidence based on the high scoring template [15]. The predicted 3D structures were delivered through emails and downloaded as PDB files, which were further visualized and modeled using the molecular modeling tool PyMOL (Version 1.7.4 Schrödinger, LLC) [16]. All the final 3D structures were generated as ball-and-stick models with distinct colors using PyMOL as previously described [17].
3. Results
3.1. MERS-CoV E Protein is Highly Conserved Among Different Coronavirus Stains
MERS-CoV strain was compared with the other 41 coronavirus stains including SARS coronavirus (SARS-CoV, NP_828854), human coronavirus (HCoV 229E, NP_073554.1), canine coronavirus (CcoV, BAA02412.1), cat coronavirus (FCoV, CAA74228. 1), bovine coronavirus (BCoV, AAL40404. 1), etc (Table 1). Whole genome sequences among those host specific coronaviruses shared less than 30 percent similarities. In contrast, further sequence alignment analysis showed that the E protein sequences were nearly identical among the 42 MERS-CoV strains except one single point mutation from isoleucine to valine at amino acid position 9 (I9V) ( Fig. 1). Moreover, since isoleucine and valine residues both are hydrophobic, the I9V point mutation may not alter the antigenic behavior of the E protein. Together these results suggest that that E protein of MERS-CoV is highly conserved among different virus strains.
TABLE 1. Comparison with Other Coronavirus.
| Virus | Protein ID | Number of amino acids | Blast with MERS-CoV |
|---|---|---|---|
| SARS CoV | NP_828854 | 76 | 37.80% |
| HCoV | NP_073554.1 | 77 | 22.62% |
| CCoV | BAA02412.1 | 82 | 18.60% |
| FCoV | CAA74228.1 | 82 | 22.99% |
| BcoV | AAL40404.1 | 84 | 27.38% |
| RTCoV | AAF97741.1 | 88 | 25.00% |
| MHV | NP_068673 | 83 | 23.81% |
| IBV | AAO33465.1 | 103 | 15.60% |
3.2. The E Protein of MERS-CoV is Stable and Hydrophobic
ProtParam analysis of MERS-CoV E protein suggests that its theoretical pI value is 7.64 and the molecular weight is 9354.2 daltons. The computed instability index (II) is 33.00, suggesting that MERS-CoV E protein is classified as a stable protein (Protein with II values <40 is considered stable). Moreover, the grand average for hydropathicity (GRAVY) value is 0.795, indicating that E protein is hydrophobic (Protein with positive GRAVY value is hydrophobic while it is considered hydrophilic with negative GRAVY value).
3.3. Identification of Potential B-Cell Linear Epitopes
Various lengths of the polypeptides were set for epitope prediction by ABCpred program, as described in methods. Higher scores reflected a higher probability of being identified as a B-cell epitope. Eight linear B-cell epitopes in the MERS-CoV E protein showed scores above the threshold value of 0.8. Scores beyond 0.8 mean to be positive, as shown in Table 2. Meanwhile, one epitope peptide was identified by Bepipred 1.0 server, as shown in Table 3. We used 0.35 as the threshold. Scores beyond 0.35 account for potential epitote, and a higher score indicates a higer probability of the existing epitope. Although the resulting epitope peptides from ABCpred and Bepipred were not totally identical, the overlapping regions were considered as the B-cell line epitope areas. As shown in Table 3, epitopes 2, 5, and 7 were identified as the potential B-Cell line epitopes.
TABLE 2. Potential Epitopes (by ABCpred).
| Rank | Sequence | Position | Length | ABCpred score |
|---|---|---|---|---|
| 1 | VVCAITLLVCMAFLTA | 21–36 | 16 | 0.89 |
| 2 | TGRSVYVKFQDSKP | 61–74 | 14 | 0.88 |
| 3 | AFLTATRLCVQCMTGFNT | 32–49 | 18 | 0.84 |
| 4 | LFIVNFFIFTVVCAITLLVC | 11–28 | 20 | 0.82 |
| 5 | RSVYVKFQDSKPPLPP | 63–78 | 16 | 0.81 |
| 6 | LCVQCMTGFNTLLVQP | 39–54 | 16 | 0.81 |
| 7 | TGRSVYVKFQDSKPPLPP | 61–78 | 18 | 0.81 |
| 8 | AFLTATRLCVQCMT | 32–45 | 14 | 0.80 |
TABLE 3. Details of Bepipred Prediction.
| Rank | Sequence | Position location | Length | Mean residue score |
|---|---|---|---|---|
| 1 | QDSKPPLPPDEWV | 70–82 | 13 | 1.687 |
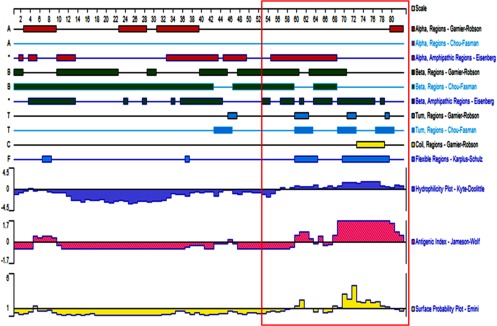
Structural analysis following the Kyte-Doolittle standard suggests that a large hydrophilic area was presented in the amino acids (aa) 58–82 region at the C-terminal of E protein. Further study of Jameson-Wolf antigenicity index showed that the highest indices were from aa 3–10, aa 58–64, and aa 68–82 regions. Moreover, analysis using Emini methods indicated that aa 67–82 epitope was possibly presented on the protein surface ( Fig. 2). Taken together, since the overlapped regions were considered as the potential epitopes, AA 58–82, with its amino acid sequence of LYNTGRSVYVKFQDSKPPLPPDEWV, was identified as a potential B cell linear epitope of the E protein.
Fig. 2.
Secondary structure and B cell line epitope presented by Protean program. Secondary structure was predicted using Garneier-Robson, Chou-Fasman, and Eisenberg methods. Red, green, and light blue regions represent alpha helix, beta lamella, and T-corner, respectively. Flexible region, hydrophilicity plot, antigen index, and surface possibility are highlighted with blue, pink and yellow, respectively. The area of the antigen epitope is marked by a red-frame box.
3.4. Secondary Structure of the E Protein of MERS-CoV
The secondary structures obtained using the Garneier-Robson, Chou-Fasman and Eisenberg methods were not consistent, as shown in Fig. 2. Various secondary structures (i.e., alpha helix, beta lamella, T-corner and coils) were analyzed using the Garneier-Robson method, the Chou-Fasman method and the Eisenberg method. Further study following the Karplus-Schulz method suggests that there are three flexible areas in the E protein, i.e., aa 6–12, aa 58–65 and aa 69–80 (Fig. 2). Those areas are predicted to be exposed on the surface of MERS-CoV, suggesting their roles as potential antigen epitopes, supporting our previous identification of aa 58–82 as a potential B cell linear epitope of the E protein.
3.5. Prediction of the 3D Structure of the E Protein of MERS-CoV
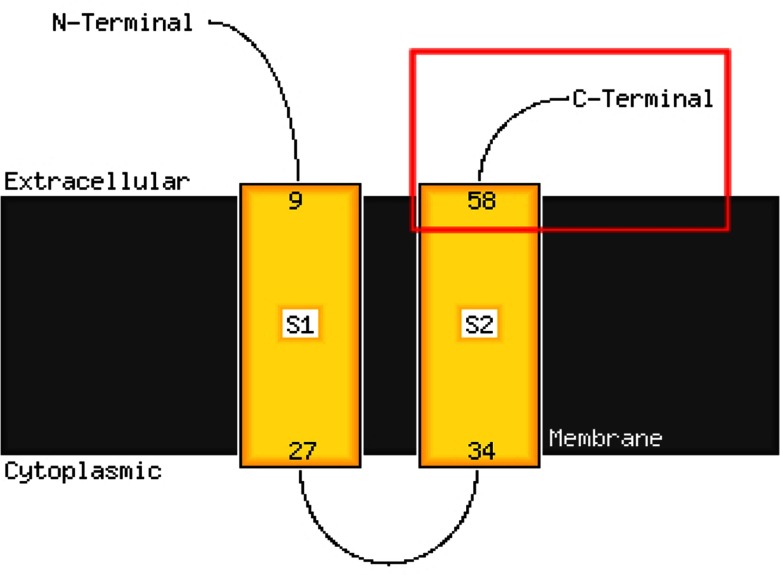
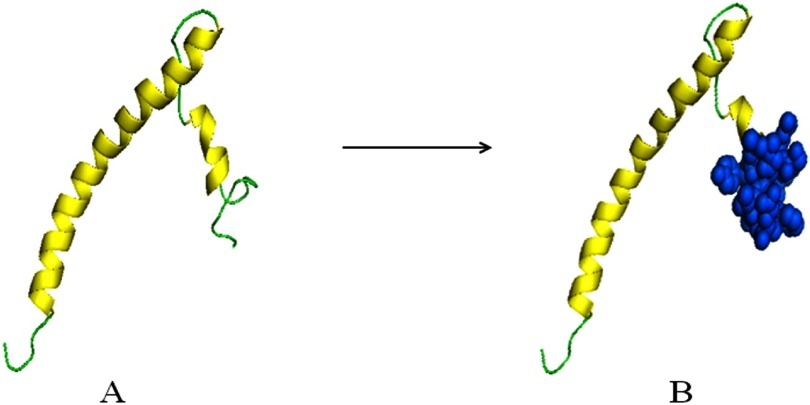
Since the 3D structure for the MERS-CoV E protein is unavailable in the PDB Molecule Database, PHYRE 2 server was used to predict and model multiple domains of E protein with high confidence based on the high scoring template. Predicted 3D structure of the E protein generated by PHYRE 2 server presented the Chain A of the E protein at the highest confidence level of 99.6 percent in the PDB Molecule Database. Further study using PHYRE 2 online server indicated that the N-terminal aa 1–9 and C-terminal aa 58–82 regions could possibly be located on the cell surface with two transmembrane domains S1 and S2 linked by a re-entrant helix, while the transmembrane helices were predicted to adopt the topology, as shown in Fig. 3. Final structure of the MERS-CoV E protein was generated by PyMOL, as shown in Fig. 4. The alpha helix was highlighted with yellow color while the loop was in green in model A, aa 58–82 regions was presented as marine blue spheres in model B.
Fig. 3.
Predicted transmembrane helix regions of the E protein by PHYRE 2. Cell membrane is presented as the gray shaded area. S1 and S2 are transmembrane domains of the E protein. Predicted B-cell line epitope was presented in the red-frame box.
Fig. 4.
Predicted protein modeling of the E protein. Helix is highlighted in yellow and loops is presented in green in model A. Amino acids 58–82 region is shown as marine blue spheres in model B.
4. Discussion
The full length natural antigens, particularly the hydrophobic membrane proteins, are hardly overexpressed and purified from prokaryotic cells with antigenicity. Previous studies suggest that protein antigenicity is generally determined by its specified epitopes instead of the full length sequence [18] . To identify the antigen epitopes, bioinformatics methods are used to predict their sequences. Predicted epitopes are further synthesized in vitro and validated with experiments. Dozens of B cell antigen epitopes have been discovered following various algorithms, offering us multiple reliable means of predicting the antigen's hydrophilic regions, accessibility, flexibility and antigenic solubility [19] . The ABCpred algorithm, which was used to predict the linear B-cell epitopes of MERS-CoV E protein in our study, has been previously shown to successfully predict epitopes with 65.93 percent accuracy ( http://www.imtech.res.in/raghava/abcpred/) [20]. Thus bioinformatics studies could provide reliable guidance in selecting specific immunogenic epitopes, which will be significant for vaccine design, epitope mapping and antibody studies. Further combination of various bioinformatics prediction methods could significantly increase the prediction accuracy.
Recent outbreak of MERS-CoV in South Korea was the largest one outside of Saudi Arabia in the world [21]. Because the mortality rate of the MERS-CoV is as high as 40 percent, it is considered as one of the most critical emerging pathogens threatening human health. Previous study showed that a recombinant SL-CoV containing a very small fragment of the SARS-CoV S gene was able to infect and cause disease in mice, highlighting its potential for pathogenicity in humans [22]. However, characterization of the immunogenic determinants for the MERS-CoV protein remains unresolved. Here in our study we found that the gene sequences of MERS-CoV were moderately conserved among different coronavirus strains although Cotten et al. studied MERS-CoV gene diversity and found numerous variations among different strains [23]. Nevertheless, at least two distinct lineages, including circulating and transmission patterns in the epidemic, are consistent in both human-to-human transmission and sporadic zoonotic events. The E protein, which is essential for virus packaging, is a moderately expressed small transmembrane protein and presented on the surface of virus envelope as well as the infected cells. Our further comparison studies showed that the E protein is highly conserved among different virus strains isolated from different species (e.g., Camelus dromedarius, Vespertilio superans and Homo sapiens) at different times and locations near the endemic areas [24]. Since sequence alignment revealed that the E protein from the 42 strains were nearly identical except a point mutation from isoleucine to valine. And Durai P [25] had stated that MERS-CoV E protein has striking similarities to SARS-CoV E protein, which has a resolved NMR structure. Therefore E protein was chosen as a potential antigenic target for the humoral immune responses, which might be significant for developing better diagnostic and research reagents in the future.
To further identify the potential antigen epitope regions of the E protein, ABCpred, Bepipred and the protean package in LaserGene software were used to predict the E protein secondary structure as well as B cell epitopes by overlapping the sequences generated from the three methods. Based on our results, aa 58–82 was identified as a potential antigen epitope region. Further online prediction and sequence comparison analysis revealed that this region was highly antigenicity and low variation region, supporting our prediction of AA 58–82 as an antigen epitope region. Since E protein structure is not available in the PDB molecular database, we used the PHYRE 2 server and PyMOL software to predict the 3D structure of E protein. The predicted structure was consistent with previous secondary structure prediction. AA 58–82 of the E protein was predicted to be a good candidate as the B cell epitope peptide area. Overall, using bioinformatics methods, we have successfully identified a potential B cell line epitope of MERS-CoV E protein, while experimental and clinical evidence are necessary to validate our studies. Future studies could also relate to other proteins in MERS-CoV and other coronavirus strains. Since currently there is no effective treatment or preventive vaccine available targeting MERS-CoV, the fusion B-cell epitopes of the E protein identified in this study might be the potential targets to design effective MERS-CoV vaccines and facilitate the development of rapid diagnostic methods in the future.
Acknowledgments
This study was partially supported by the Technologies R & D Program of Guangzhou (No. 201507020062) and National Natural Science Foundation of China (NO.31670168). Qian Xie, Xiaoyan He, Bao Zhang, and Wei Zhao contributed equally.
Biographies

Qian Xie received the BSc degree in preventive medicine from Southern Medical University, in 2013 and the masters degree, in 2016 . She studied pathogen biology. Now, she is a teaching assistant in the School of Public Health, Southern Medical University, Major Research Direction: Molecular virology. She has published five papers in journals and conferences during the graduate learning phase. And, she is also currently participating in two research projects.

Xiaoyan He received the BSc degree from the School of Distance Learning for Medical Education at Peking University, in 2010 and graduated in nursing. She published eight articles in specialized journals, three papers in conference proceedings, and two chapters of published books. Her current research interests include biotechnology, microbiology and hospital infection.

Fangji Yang received the BSc degree in clinical medicine from Southern Medical University, in 2008. He received the MSc degree majoring in infectious diseases from Sun Yat -sen University, in 2013. He is currently working toward the PhD degree in the Department Of Infectious Diseases of the Third Affiliated Hospital at Sun Yat-sen University. His current research interests include diagnosis, treatment and prevention of emerging infectious diseases.

Xuling Liu studied at Southern Medical University where she working toward the bachelor's degree in medicine. In 2016, she participated in three research projects, such as the Middle East respiratory syndrome-coronavirus, Dengue virus, and Zika virus. Her current research interests include bioinformatics analysis, the construction of animal models, and molecular biology.

Ying Li is currently working toward the bachelor's degree in preventive medicine at Southern Medical University. He is an assistant in this research.

Yujing Liu is studying at Southern Medical University, and her major is preventive medicine. In 2015, she initiated her research activities at the BSL-3 Laboratory of School of Public Health, Southern Medical University.

ZhengMeng Yang studied biopharmaceutics at the Southern Medical University. He will receive the bachelor degree from the school of biotechnology, in 2017. Since 2015, he has been working at the BSL-3 Laboratory in the School of Public Health, Southern Medical University. He is learning the experimental method and operation under the safety regulation. The main research interest is related to virology and biotechnology.

Jianhai Yu studied at Southern Medical University where he acquired his medical master. From 2011-2016, he participated in more than four research projects on the Middle East respiratory syndrome-coronavirus, Dengue virus, and Zika virus. His current research interests include virology, bioinformatics analysis, gene sequencing, molecular epidemiology, and animal models establishment.

Bao Zhang studied BSc degree biochemistry and molecular biology and received the master's degree, in 2000 and the PhD degree, in 2003 in biochemistry and molecular biology from Southern Medical University. He studies the mechanism of virus infection and viral evolution, such as MERS CoV, EV71, and H1N1.

Wei Zhao received the PhD degree from the Beijing Institute of Microbiology and Epidemiology, in 2001 and studied molecular virology at the Beijing Institute of Microbiology and Epidemiology. He is currently a professor in the School of Public Health, Southern Medical University, and also the director of the Lab of Biosafety Level 3. He has published more than 100 papers in journals and conferences, has been the PI of several funded projects, and supervised three PhD students. The main research interests are related to bioinformatics, mainly in the topics of evolutionary computation an omics data analysis and mining in virus.
Funding Statement
This study was partially supported by the Technologies R & D Program of Guangzhou (No. 201507020062) and National Natural Science Foundation of China (NO.31670168).
Contributor Information
Qian Xie, Email: 416900030@qq.com.
Xiaoyan He, Email: hexy@smu.edu.cn.
Fangji Yang, Email: 1227889117@qq.com.
Xuling Liu, Email: 443236702@qq.com.
Ying Li, Email: 408473708@qq.com.
Yujing Liu, Email: 898060471@qq.com.
ZhengMeng Yang, Email: 326166548@qq.com.
Jianhai Yu, Email: 1014056098@qq.com.
Bao Zhang, Email: Zhangb@smu.edu.cn.
Wei Zhao, Email: zhaowei@smu.edu.cn.
References
- [1]. Zaki A. M., et al. , “Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia,” New Engl. J. Med., vol. 367, no. 19, pp. 1814–1820, Nov, 8, 2012. [DOI] [PubMed] [Google Scholar]
- [2]. van Boheemen S., et al. , “Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans,” MBio., vol. 3, no. 6, 2012, Art. no. e00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. King A. M. Q., Virus Taxonomy, Classification and Nomenclature of Viruses. New York, NY, USA: Academic Press, 2000, pp. 835–849. [Google Scholar]
- [4]. Raamsman M. J., et al. , “Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E,” J. Virol., vol. 74, no. 5, pp. 2333–2342, Mar. 2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Peiris J. S., et al. , “Coronavirus as a possible cause of severe acute respiratory syndrome,” Lancet, vol. 361, no. 9366 , pp. 1319–1325, Apr. 19, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6]. Drosten C., et al. , “Identification of a novel coronavirus in patients with severe acute respiratory syndrome,” New Engl. J. Med., vol. 348, no. 20, pp. 1967–1976, May 15, 2003. [DOI] [PubMed] [Google Scholar]
- [7]. Shen X., et al. , “Small envelope protein E of SARS: Cloning, expression, purification, CD determination, and bioinformatics analysis,” Acta Pharmacol Sin, vol. 24, no. 6, pp. 505–511, Jun. 2003. [PubMed] [Google Scholar]
- [8]. El-Manzalawy Y., Dobbs D., and Honavar V., “Predicting linear B-cell epitopes using string kernels,” J. Mol. Recognit., vol. 21, no. 4, pp. 243–255, Jul-Aug 2008 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9]. Larsen J. E., Lund O., and Nielsen M., “Improved method for predicting linear B-cell epitopes,” Immunome Res., vol. 2 , 2006, Art. no. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]. Parker J. M., Guo D., and Hodges R. S., “New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites,” Biochemistry, vol. 25, no. 19, pp. 5425–5432 , Sep. 23, 1986. [DOI] [PubMed] [Google Scholar]
- [11]. Nishikawa K., “Assessment of secondary-structure prediction of proteins. Comparison of computerized Chou-Fasman method with others,” Biochim. Biophys. Acta. , vol. 748, no. 2, pp. 285–299, Oct. 28, 1983. [DOI] [PubMed] [Google Scholar]
- [12]. Kyte J. and Doolittle R. F., “A simple method for displaying the hydropathic character of a protein ,” J. Mol. Biol., vol. 157, no. 1, pp. 105–32, May 5, 1982. [DOI] [PubMed] [Google Scholar]
- [13]. Jameson B. A., and Wolf H., “The antigenic index: A novel algorithm for predicting antigenic determinants ,” Comput. Appl. Biosci., vol. 4, no. 1, pp. 181 –186, Mar. 1988. [DOI] [PubMed] [Google Scholar]
- [14]. Emini E. A., et al. , “Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide,” J. Virol., vol. 55 , no. 3, pp. 836–839, Sep. 1985. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. Kelley L. A., et al. , “The Phyre2 web portal for protein modeling, prediction and analysis,” vol. 10, no. 6, pp. 845 –858, Jun. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Liang M. P., et al. , “WebFEATURE: An interactive web tool for identifying and visualizing functional sites on macromolecular structures,” Nucleic Acids Res., vol. 31, no. 13, pp. 3324–3327 , Jul. 1, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]. W. D, “The PyMol Molecular Graphics System,” 2002. [Online]. Available: http://pymol.org/, Accessed on: 3 June 2015.
- [18]. Hopp T. P. and Woods K. R., “Prediction of protein antigenic determinants from amino acid sequences,” Proc. Natl. Academy Sci. United State America, vol. 78, no. 6, pp. 3824–3828, Jun. 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Van Regenmortel M. H. and Daney de Marcillac G., “An assessment of prediction methods for locating continuous epitopes in proteins,” Immunol Lett., vol. 17, no. 2 , pp. 95–107, Feb. 1988. [DOI] [PubMed] [Google Scholar]
- [20]. Saha S. and Raghava G. P. , “Prediction of continuous B-cell epitopes in an antigen using recurrent neural network ,” Proteins, vol. 65, no. 1, pp. 40 –48, Oct. 1, 2006. [DOI] [PubMed] [Google Scholar]
- [21]. Xie Q. , et al. , “Genomic sequencing and analysis of the first imported Middle East Respiratory Syndrome Coronavirus (MERS CoV) in China,” Sci. China. Life Sci. , vol. 58, no. 8, pp. 818–820, Aug. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22]. Zhou P., Han Z., Wang L. F., and Shi Z., “Identification of immunogenic determinants of the spike protein of SARS-like coronavirus,” Virol Sin., vol. 28 , no. 2, pp. 92–96, Apr. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Cotten M., et al. , “Transmission and evolution of the middle east respiratory syndrome coronavirus in Saudi Arabia: A descriptive genomic study,” Lancet , vol. 382, no. 9909, pp. 1993–2002, Dec. 14, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Surya W., Li Y., Verdia-Baguena C., Aguilella V. M. , and Torres J., “MERS coronavirus envelope protein has a single transmembrane domain that forms pentameric ion channels,” Virus Res., vol. 201, pp. 61–66, Apr. 2, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Durai P., Batool M., Shah M., and Choi S., “Middle East respiratory syndrome coronavirus: Transmission, virology and therapeutic targeting to aid in outbreak control,” Exp. Mol. Med., vol. 47, Aug. 28, 2015, Art. no. e181. [DOI] [PMC free article] [PubMed] [Google Scholar]