Abstract
γ-aminobutyric acid (GABA) was the first molecule that was edited with MEGA-PRESS. GABA edited spectroscopy is challenged by limited selectivity of editing pulses. Coediting of resonances from macromolecules (MM) is the greatest single limitation of GABA edited spectroscopy. In this contribution, relative signal contributions from GABA, MM and homocarnosine to the total MEGA-PRESS edited signal at ~3 ppm, i.e., GABA+, are simulated at 3 tesla using several acquisition schemes. The base scheme is modeled after those currently supplied by vendors: it uses typical pulse shapes and lengths, it minimizes the first echo time (TE), and the delay between the editing pulses is kept at TE/2. Edited spectra are simulated for imperfect acquisition parameters such as incorrect frequency, larger chemical shift displacement, incorrect transmit B1-field calibration for localization and editing pulses, and longer TE. An alternative timing scheme and longer editing pulses are also considered. Additional simulations are performed for symmetric editing around the MM frequency to suppress the MM signal. The relative influences of these acquisition parameters on the constituents of GABA+ are examined from the perspective of modern experimental designs for investigating brain GABA concentration differences in healthy and diseased humans. Other factors that influence signal contributions, such as T1 and T2 relaxation times are also considered.
Keywords: editing, GABA, lysine, macromolecules, homocarnosine, magnetic resonance spectroscopy
Graphical abstract
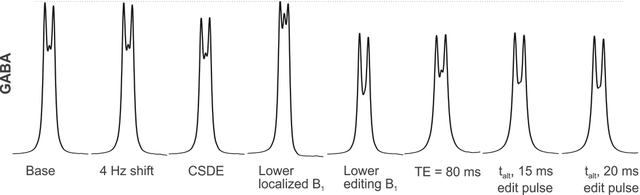
The influences of acquisition parameters on edited GABA signal at 3.02 ppm were investigated using simulation of MEGA-PRESS sequence at 3T. Signal contributions from macromolecule and homocarnosine to the edited signal were also considered.
1. INTRODUCTION
γ-aminobutyric acid (GABA) editing sequences are hampered by coediting of macromolecules (MM) and homocarnosine (Hcar) due to the limited selectivity of the editing pulses. In the original work on GABA editing, MM contribution was characterized alongside that of GABA1,2, and the measured signal was designated as total signal as opposed to pure GABA signal. Rothman et al. estimated the MM contribution to GABA* to be 60% at 2.1 T1. In the next several applications2,3 at 4 T, MM signal was found to be approximately 50% of the total signal, which was abbreviated GABA+. Fifty percent became the accepted approximation of MM signal contribution4 to the combined edited GABA, MM and Hcar resonances, which continues to be abbreviated as GABA+. Several early approaches reduced3 or eliminated5,6 the MM contribution to GABA edited spectra by increasing the frequency selectivity of the editing pulse (i.e., by optimizing pulse shape3, using a long pulse1, or scanning at higher field) or by introducing a symmetric pulsing scheme5 to suppress MM contribution. Use of lysine (Lys) was introduced to mimic MM for development purposes5, since Lys residues are probably the predominant contributors to the J-coupled MM signals at 3.0 ppm and 1.7 ppm in the brain7. An inversion recovery approach was introduced to measure MM contamination in vivo6. Hcar was known from an early stage to coedit with GABA and to have a concentration of ~0.5 mM in the occipital lobe8, compared to a GABA concentration of ~1 mM1–6.
The earliest GABA edited experiments were attentive to diligent setting of acquisition parameters. Pulse frequencies were set carefully, including the chemical shift of the editing pulse1,5 and the center frequency of the localization pulses. Frequency drift was watched over, since a misset frequency would cause a notable change in signal6,9. Short, high bandwidth localization pulses were used to minimize chemical shift displacement error (CSDE), but required high transmit B11, which was generally achieved via surface radiofrequency (RF) coils1–3,6 . Short localization pulses also left sufficient time in the optimal 1/2J echo time (TE) (68 ms) for long editing pulses with high frequency selectivity. It was acknowledged that the estimated GABA concentration was dependent on the T1 and T2 relaxation times of GABA and the reference signal1–3,6.
In spite of widespread use of MEGA-PRESS for GABA editing, the effect of imperfect acquisition parameters on GABA+ signal and MM co-editing is often overlooked. Acquisition parameters are often set using approaches that have been adapted from imaging protocols, and those approaches are not optimal for spectroscopy. Vendor-specific differences in acquisition parameters (i.e., differences in pulse sequence timing, pulse shapes and lengths, and MM suppression schemes) are the current focus of optimizing test-retest repeatability and minimizing site-to-site variance10–12. However, the following aspects of optimization are frequently not reported. 1) The procedure used to calculate and set the frequency of the editing pulse is frequently not reported. For example, it could be calculated from the water frequency or from the frequency of a strong singlet in a non-edited spectrum. 2) At what time the frequency of the editing pulse is set in the protocol, and whether the frequency is checked and/or updated during acquisition of the edited spectrum is frequently not reported. This becomes important if there is drift during the acquisition. 3) The length and bandwidth of localization pulses are frequently not reported. Maximum system B1 limits how short localization pulses can be and still achieve excitation, which limits the ability to minimize CSDE. Lengthening the pulses makes the bandwidth smaller and the CSDE greater. The procedure used to calibrate RF power is often not described, perhaps because RF power is not calibrated. 4) T1 and T2 relaxation times of GABA10–20 are often not considered. 5) MM coediting is usually mentioned whereas that of Hcar is not.
The goal of this paper is to provide comparative data on the influences of acquisition parameters on edited GABA and MM. The extent to which Hcar contributes to the GABA+ resonance is also considered. Reported measures of GABA+ in healthy and diseased humans are cast In light of influences of acquisition parameters and coedited compounds.
2. EXPERIMENTAL DETAILS
2.1. Simulation methods
Metabolite spectra were simulated in MATLAB (The MathWorks Inc, Natick, MA) based on density matrix formalism21. All simulations were performed at 3 T using a spectral width of 6 kHz and 4096 complex points during signal acquisition.
The PRESS sequence was simulated by taking into account the RF shapes, durations and interpulse timings. 2D localization22 consisting of 40×40 spatial points23 was achieved by sweeping the frequency of each pair of refocusing pulses along each spatial dimension from -FWHM to FWHM, where FWHM is the bandwidth in Hz of the refocusing pulses as previously described23. The carrier frequency was set to 3.0 ppm. Resulting spectra are presented as if all compounds have the same concentration.
The edited signals for GABA, Lys and Hcar were integrated within a ±25 Hz region around 3.01 ppm after applying an exponential decay function to match their linewidths to in vivo condition of 8 Hz, performing three times zero-filling and baseline correction.
2.2. Selection of acquisition schemes
Figure 1 illustrates the MEGA-PRESS pulse sequence used to simulate edited GABA, Lys, and Hcar resonances under the experimental conditions of interest at 3 T. The base sequence was designed as a compromise among those currently available from the major vendors18. Non-proprietary and accessible pulse shapes and lengths were used, including a 3.6 ms Hamming sinc for excitation (B1 = 14.3 μT, bandwidth = 2.47 kHz measured at FWHM), 5.8 ms Mao pulses for refocusing (B1 = 22 μT, bandwidth = 0.99 kHz at FWHM), and 15 ms sinc-gauss pulses (bandwidth = 81 Hz at FWHM) set at 1.90 ppm for edit ON and at 7.46 ppm for edit OFF conditions. No RF pulse used more than 24 μT for B1 maximum, in accordance with capabilities of currently available hardware on clinical systems. The spacing between the middle of the editing pulses (τ3 + τ4) was kept at TE/2 (TE = 68 ms) and the first spin echo (τ1) of PRESS was kept as short as most of the vendors are able to achieve12, i.e., 14.7 ms. τ2 and τ5 were set to maintain the desired TE with τ2 = τ5. Several acquisition conditions were investigated using the following acquisition parameters that were different from those mentioned above:
Figure 1:
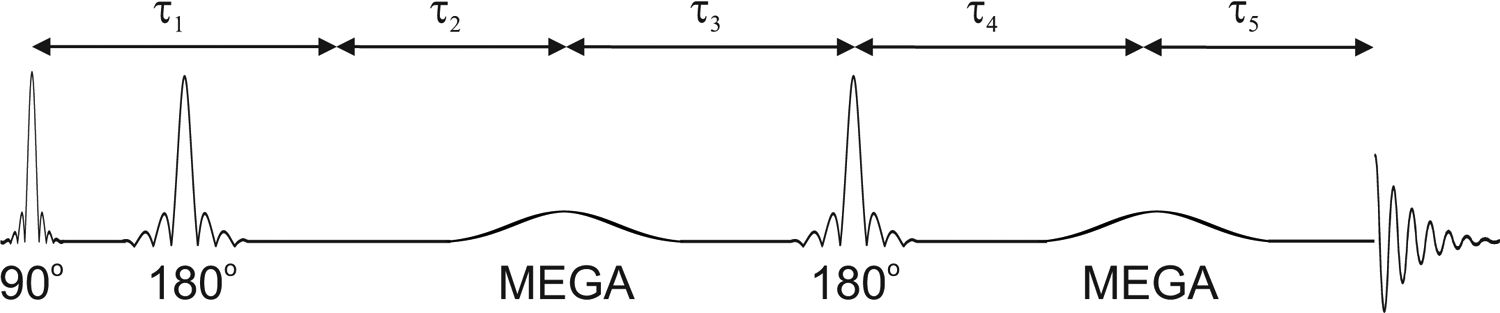
Diagram of the MEGA-PRESS sequence timing used in simulations. Pulse shapes for: PRESS excitation, Hamming sinc; PRESS refocusing, Mao; and MEGA editing, sinc-gauss. The spacing between the middle of the editing pulses (τ3 + τ4) was kept at TE/2. Symmetry was kept with τ3 = τ4 and τ2 = τ5. Details for the pulse lengths and powers, timing and settings of chemical shift used to simulate several acquisition scenarios are detailed in the text.
Frequency misset and drift were simulated for GABA and Lys by shifting the editing pulse carrier frequency to ±2, ±4, ±10, and ±20 Hz from 1.9 ppm. This was based on our experience that respiration can cause frequency to vary by about 4 Hz, but it is not exceptional for us to see frequency shifts as high as 10 Hz, and cooling after sequences with high gradient demand can cause up to 20 Hz drift10 over a typical MEGA-PRESS edited acquisition.
Larger chemical shift displacement was simulated for GABA by increasing the length of the refocusing pulses from 5.8 ms to 9.45 ms (and decreasing B1 from 22 μT to 13.5 μT) while keeping the delay between the editing pulses at TE/2 i.e., 34 ms.
The effects of miscalibrated B1 for PRESS localization (excitation and refocusing pulses) and editing pulses were simulated by using 25% lower B1 than the optimal B1.
The echo time was increased to TE = 80 ms while keeping the spacing between the middle of the editing pulses (τ3 + τ4) at TE/2 = 40 ms and the first spin echo (τ1) of PRESS at 14.7 ms.
An alternative timing (talt) in which the two editing pulses are separated by less than TE/2 as used by some vendors was simulated. In our simulation, we used τ2 = τ3 = τ4 = τ5 = 13.325 ms to simulate GABA at TE = 68 ms (the delay between editing pulses was 26.65 ms). GABA was simulated at this talt using editing pulse lengths of 15 ms and 20 ms.
The MM-suppressed editing sequence, i.e., placing the “off” pulse symmetric about the problematic coupling partner (as opposed to symmetric about water) was simulated for GABA and Lys. Since Lys was used to imitate MM, the symmetric pulse was placed at 1.52 ppm in this case, i.e., symmetric about the 1.71 ppm of Lys 5CH2 relative to the 1.90 ppm that was used for GABA editing.
2.3. Selection of coupling systems
The GABA system is yet to be fully worked out24, though our pulse-acquire simulations suggest that the missing information is inconsequential at in vivo line widths. We used the system specified in Govind et al.25 because it used phantom (i.e., pure) spectra that were stated to have been measured at physiologic pH and temperature, and it included geminal couplings.
The MM coupling system relevant to editing GABA 2CH2 at 3.01 ppm via coupling to 3CH2 at 1.89 ppm and specified tentatively in the literature was used7,26, i.e., Lys 6CH2 at 3.02 ppm via coupling to Lys 5CH2 at 1.71 ppm. Robin de Graaf provided the Lys coupling system in Table 1, which he derived from human plasma samples scanned at 25°C and pH of 7.227. Since the 4CH2 - 5CH2 - 6CH2 moiety is most pertinent to MEGA editing of the 3.01 ppm resonance of GABA, we only simulated these protons. Note that it would take undue computational time to simulate the whole molecule with localization since Lys has nine protons. We used pulse-acquire simulation at short TE and non-localized PRESS at long TE (i.e., 68 ms) to affirm that differences in the Lys resonance at 3.02 ppm between our abbreviated 6-proton simulation and the full 9-proton simulation were inconsequential at in vivo line widths, which is in agreement with the literature28.
Table 1.
Lys coupling system used for simulations. Derived for ref27 from human plasma samples at 25°C and pH 7.2.
| Spin | Chemical shift (ppm) | J-couplings(Hz) | Connectivity |
|---|---|---|---|
| 2CH | 3.746 | 6.09 | 2–3 |
| 6.09 | 2–3’ | ||
| 3CH2 | 1.881 | −15.27 | 3–3’ |
| 5.67 | 3–4 | ||
| 10.51 | 3–4’ | ||
| 3’CH2 | 1.898 | 10.13 | 3’−4 |
| 6.1 | 3’−4’ | ||
| 4CH2 | 1.431 | −12 | 4–4’ |
| 6 | 4–5 | ||
| 8 | 4–5’ | ||
| 4’CH2 | 1.4929 | 10 | 4’−5 |
| 6 | 4’−5’ | ||
| 5CH2 | 1.712 | −12 | 5–5’ |
| 6 | 5–6 | ||
| 8 | 5–6’ | ||
| 5’CH2 | 1.713 | 10 | 5’−6 |
| 6.5 | 5’−6’ | ||
| 6CH2 | 3.018 | −10 | 6–6’ |
| 6’CH2 | 3.0198 |
Parameters for the Hcar system were taken from de Graaf29. The spectrum used to derive this system was measured at 25°C and phosphate-buffered (50 mM, pH 7.0).
3. RESULTS
Figure 2 and Table 2 show that the edited GABA signal is impacted 8% or less over the frequency misset range from −4 to +10 Hz. However, the impact on GABA signal at a ±20 Hz frequency misset is on the order of 50%. Editing for Lys at 0 Hz from 1.90 ppm is suboptimal as expected, given that the coupling partner for Lys is at 1.71 ppm. The edited Lys signal is impacted as much as 38% over the GABA-defined frequency misset range of ±10 Hz.
Figure 2.
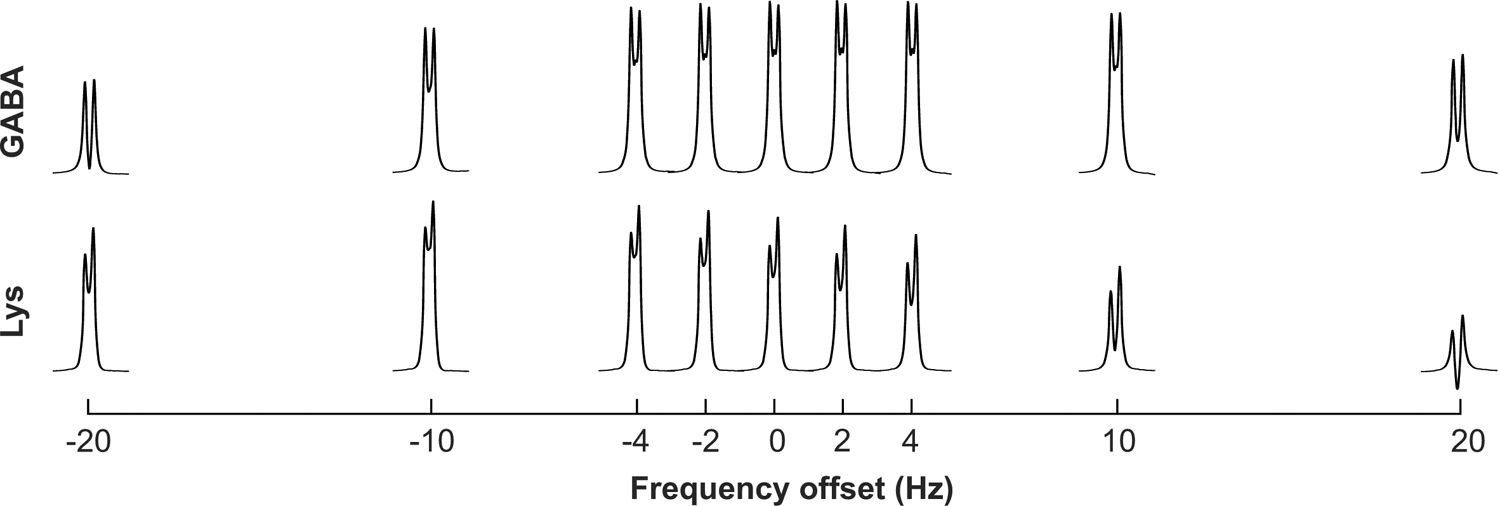
Impact of frequency misset on MEGA-PRESS edited GABA and Lys resonances near 3.01 ppm. GABA (top) and Lys (bottom) were simulated with the base sequence with editing pulses placed at 1.9 ppm (i.e., 0 Hz frequency offset) and shifted ±2, ±4, ±10, and ±20 Hz from 1.9 ppm.
Table 2.
Integrals of edited GABA and Lys 3.01 ppm resonances under ideal (0 Hz shift) and the misset frequencies illustrated in Figure 2. GABA and Lys values are normalized to the area under the GABA resonance measured with the base sequence. Lys values are additionally presented normalized to placement of the editing pulse at 1.9 ppm (i.e., 0 Hz frequency offset).
| Frequency shift from 1.9 ppm (Hz) | GABA | Lys | Normalized Lys |
|---|---|---|---|
| −20 | 0.46 | 0.70 | 0.91 |
| −10 | 0.82 | 0.88 | 1.13 |
| −4 | 0.96 | 0.85 | 1.09 |
| −2 | 0.98 | 0.82 | 1.05 |
| 0 | 1.00 | 0.77 | 1.00 |
| 2 | 1.00 | 0.72 | 0.94 |
| 4 | 1.00 | 0.67 | 0.86 |
| 10 | 0.92 | 0.48 | 0.62 |
| 20 | 0.62 | 0.23 | 0.29 |
Figure 3 and Table 3 demonstrate that chemical shift displacement errors and utilization of miscalibrated B1 for PRESS localization or editing pulses impact the GABA signal in the range 7% to 26% whereas typically encountered drift (4 Hz) has less than 4% impact on GABA signal (but as much as 14% on Lys signal). Going to the longer TE (80 ms) leads to a loss of 8% of signal (without taking T2 losses into account). Different sequence timing (alternative timing) has a 12% impact, i.e., a smaller impact than miscalibration of editing pulse power and comparable to miscalibration of PRESS pulse power. Using the longer, more selective editing pulses leads to a loss of only 3% of GABA signal beyond the loss due to the alternative timing.
Figure 3.
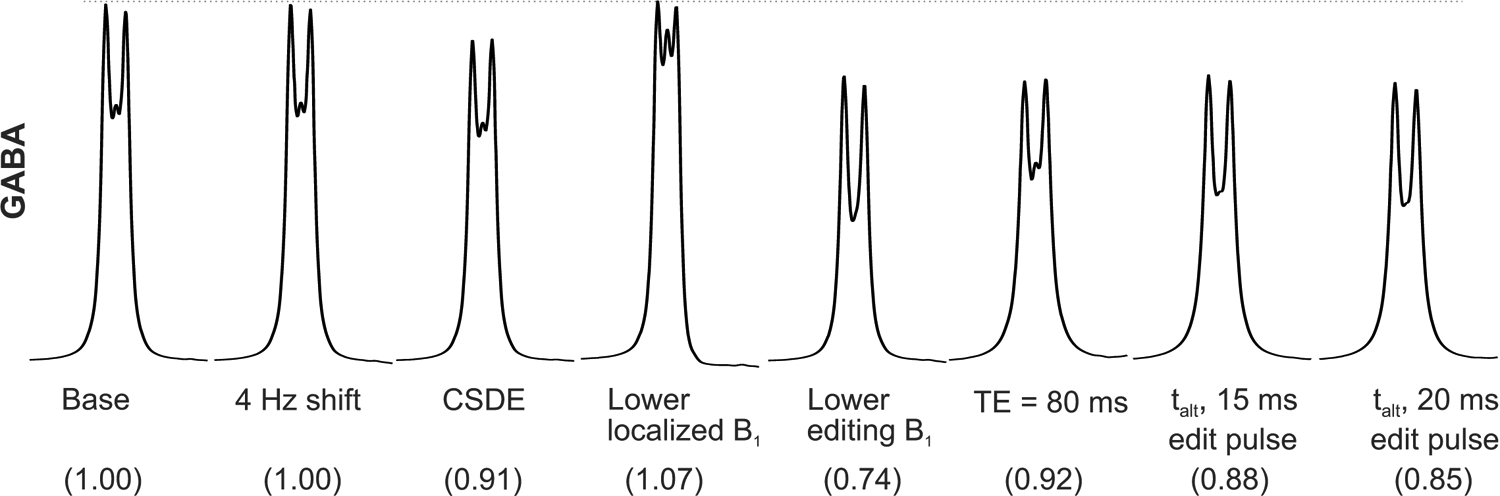
Relative impacts on MEGA-PRESS edited 2CH2 GABA of frequency misset (+4 Hz offset), large CSDE, suboptimal powers of localization (25% lower localized B1) and editing (25% lower editing B1), longer echo time (TE = 80 ms), and alternative timings for TE = 68 ms with 15 and 20 ms editing pulses. The dotted line is a point of reference to compare to the base sequence. The normalized integral of GABA resonance at 3.03ppm relative to the base sequence is given in parenthesis.
Table 3.
Integrals of edited GABA resonances under acquisition circumstances of Figure 3. All values were normalized to the area under the GABA resonance measured with the base sequence. talt = alternative timings.
| Simulation | Normalized Integral |
|---|---|
| Base | 1.00 |
| 4 Hz shift | 1.00 |
| CSDE | 0.91 |
| Miscalibrated (lower) PRESS 90° and 180° B1 | 1.07 |
| Miscalibrated (lower) editing pulse B1 | 0.74 |
| TE = 80 ms | 0.92 |
| talt | 0.88 |
| talt, 20 ms edit pulse | 0.85 |
Figure 4 illustrates how under-powering the localization pulses leads to a counterintuitive 7% increase (table 3) in edited GABA signal. The sub-optimally powered pulse has a different FWHM and excites more signal outside the nominal volume-of-interest (VOI). Sub-optimal power has influences on both the edit-on and edit-off experiments, leading to an increase in the area under the edited GABA resonance outside the nominal VOI.
Figure 4.
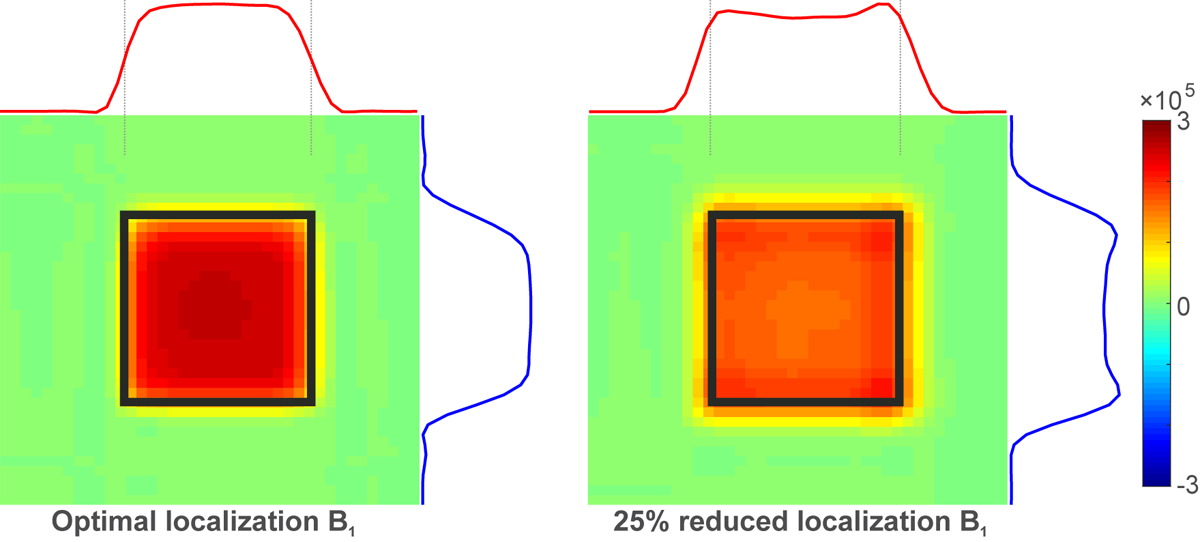
Spatial 2-dimensional representation of simulation of MEGA-PRESS edited 2CH2 GABA with the base sequence at optimal and suboptimal PRESS localization pulse powers (75% of the power needed for full inversion). Warm to cool colors represent the area under the edited GABA resonance. Black squares are the same for both panels and illustrate the intended nominal localization boundaries.
Figure 5 shows the well-known reduction in editing efficiency that occurs when using symmetric pulsing to eliminate MM contamination. Importantly, Figure 5 shows that even under optimal frequency setting, a noteworthy amount of Lys signal remains when the MM-suppressed scheme is used with MEGA-PRESS and 15 ms editing pulses at 3 T, as previously shown28.
Figure 5.

GABA and Lys 3.01 ppm resonances when MEGA-PRESS editing is simulated with the symmetric pulsing scheme (editing pulses at 1.9 and 1.52 ppm, blue) for elimination of MM contribution relative to the base scheme (editing pulses at 1.9 and 7.46 ppm, black). For symmetric pulsing, instead of setting the “off” pulse of the editing symmetric about water, they were set symmetric about the problematic coupling partner. Since Lys was used to imitate MM, the symmetric pulse was placed at 1.52 ppm, i.e., symmetric about the 1.71 ppm chemical shift of Lys 5CH2 relative to the 1.9 ppm used for GABA editing.
Figure 6 illustrates the MEGA-PRESS simulation of Hcar coedited with GABA (edit ON pulse at 1.9 ppm). The edited GABA resonance is shown for comparison. The simulations were done with equal concentrations of Hcar and GABA. The area under the Hcar resonance is 91% that of GABA.
Figure 6.
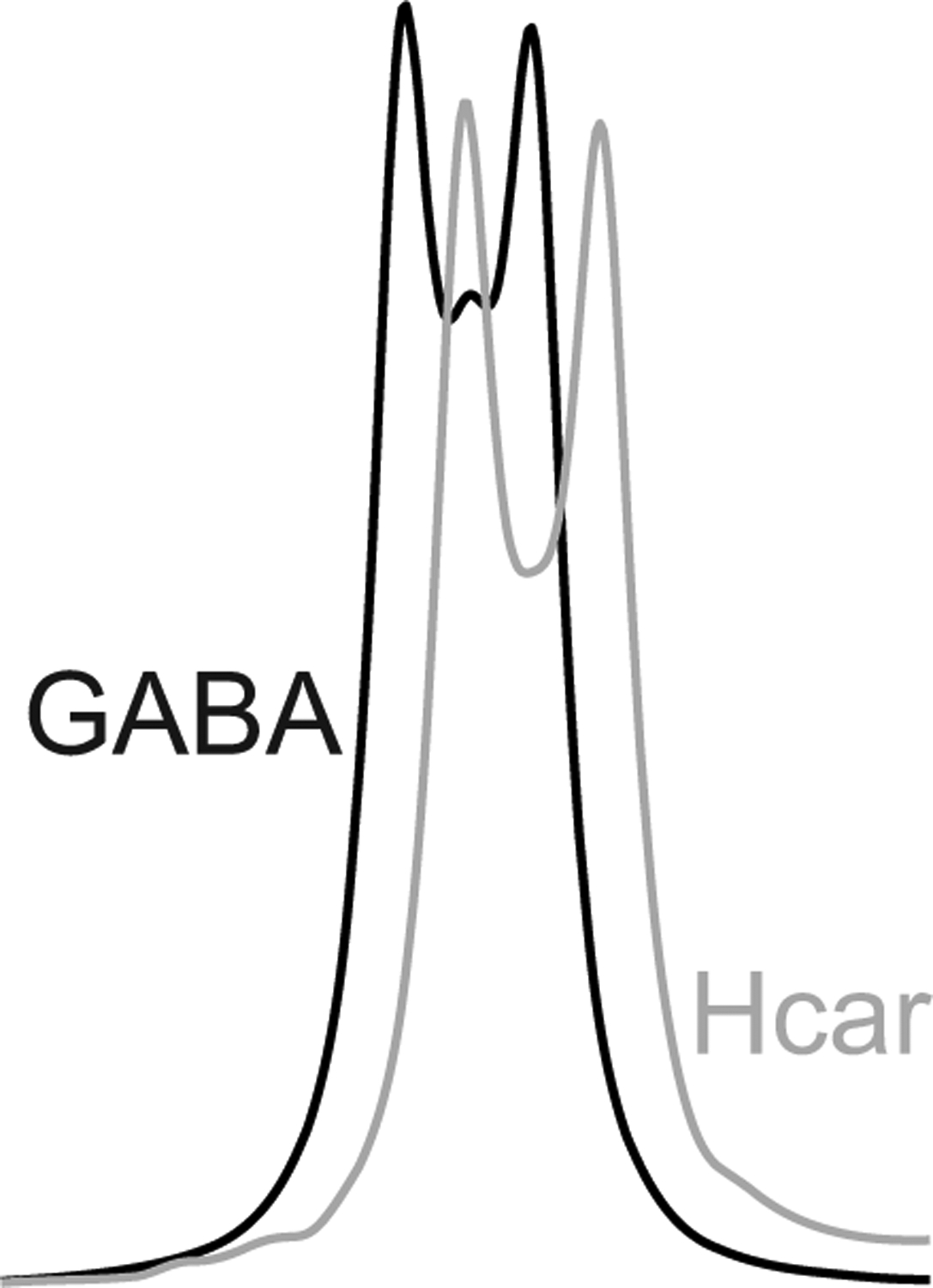
Coediting of Hcar resonance with GABA simulated with the base sequence and setting the “on” editing pulses at 1.9 ppm. The black line shows the edited GABA resonance and the gray line shows the coedited Hcar resonance. GABA and Hcar were simulated at equal concentration.
4. DISCUSSION
Whereas most of the influences on GABA+ signal examined herein have already been reported, the goal of this study was to provide a systematic characterization of the influence of acquisition parameters. The reported outcomes agree with the literature10,12,28,30. Whereas many previous reports estimated MM contribution by using a frequency shifted GABA spectrum, we used Lys as a surrogate.
Recent publications report GABA or GABA+ signal differences in the range of 3% to 30% between healthy and diseased humans31–35. In comparison, frequency missets within 10 Hz, which are likely to occur in participants who move during the scan or if time elapses between setting the frequency and running MEGA-PRESS acquisition, have little influence on GABA signal but up to 38% influence on MM signal (Table 2). Assuming 1:1 GABA:MM signal, a 10 Hz frequency misset could cause a 21% change in GABA+ signal.
Another source of uncertainty in GABA quantification is possible variability in MM concentration with participant characteristics such as disease and age. Human brain MM concentration has been reported to vary in disease as much as 100%36,37. A recent study on human brain MM reported an ~10% higher MM level in older than young adults38. Assuming 1:1 GABA:MM signal, 10% and 100% higher MM would cause 5% and 50% higher GABA+ signal, respectively.
Our findings (Figure 5) agree with the literature that using the MM-suppressed symmetric pulsing scheme with MEGA-PRESS at 3 T reduces the GABA signal and does not eliminate all of the MM signal (mimicked through Lys signal). Imperfect suppression of MM is likely due to the asymmetry or strong coupling in Lys as previously shown28. The effectiveness of the MM-suppressing symmetric pulsing scheme improves with longer editing pulses with better frequency selectivity5, but this is difficult to achieve with MEGA-PRESS. The effectiveness of the symmetric pulsing scheme also improves at higher fields (e.g., at 7 T there is no loss in GABA-editing efficiency and MM are almost completely suppressed even with 15 ms editing pulses28).
The current findings agree with recent literature10 in that vendor-specific differences in MEGA-PRESS implementation cause editing efficiency for GABA to vary by up to 10%. However, miscalibrated transmit B1 can cause differences of similar size or greater (Table 3). Also, if the maximum transmit B1 is insufficient for a given individual, and localization pulses are lengthened to accommodate lower maximum B1 (as in the Siemens implementation), signal can change by ~10% due to increased CSDE. Our simulation of the two-dimensional editing efficiency profile (Figure 4) is in agreement with literature30.
Changes in relaxation times could also affect GABA signal. Scant literature on GABA suggests that T1 is in the vicinity of 1.3 s39 and T2 of 88 ms40 at 3 T. Commonly used repetition time (TR) and TE are 2000 and 68 ms, respectively11. If T1 were to become 20% longer or T2 20% shorter41, then GABA signal would change by ~20%.
The normal concentration of Hcar has been reported to range from 0.3 to 0.6 mM42 in postmortem brain and at 0.47 mM in in vivo brain using Hcar resonances that are partially resolved at 7.05 and 8.02 ppm in short TE spectra at 2.1 T when macromolecule resonances are removed by subtraction8. The 3.0 ppm resonances of GABA and Hcar cannot be separated using editing based on the coupling partners at ~1.90 ppm, even at high field, due the close proximity of chemical shifts. Normal, medication and disease-associated human brain Hcar levels have been reported to vary in the range of 2 to 4 times8,42–44. Based on reported in vivo contributions and relative editing yields (Figure 6), Hcar signal in GABA+ is not negligible.
In addition to acquisition parameters examined in this study, robust normalization and quantification must be considered to achieve practical utility. Spectral width and number of discrete points, which were held constant in this work, can influence aliasing and peak definition and thus quantification. Frequency and phase corrections of individual shots and surveillance for absence of subtraction artifact are other important aspects of quantification that were not considered in the present simulated data. Other acquisition parameters that are important for in vivo application but were not simulated in this project are B0 shims, eddy current correction, and consistency of VOI placement.
To acquire the most physiologically relevant data possible, some acquisition parameters have obvious optimal settings. Because each human participant loads the RF coil to a different extent, B1 calibration of all pulses for each experiment is warranted. It is also helpful to keep PRESS localization pulses short to minimize CSDE45. Frequency can be set immediately before each acquisition and locked at best, or tracked and reset when drift approaches 10 Hz if locking is not possible. Choice of other acquisition parameters depends on circumstances. It is crucial that users understand that MM coediting is present and that an informed decision on whether to use a MM suppressing approach (and if so, which one) needs to be made based on the study hypotheses (i.e., whether MM are likely to confound) and limitations (i.e., SNR and scan time). It is also crucial for users to understand that frequency-associated errors in relative GABA and MM signal acquisition cannot be corrected with the frequency and phase correction step of post-processing. In addition to those presented herein, other MM-reduction approaches based on increasing the length of the editing pulses have been proposed4,46,47. Scanning at longer TE, generally to accommodate longer, more selective editing pulses, makes the measure more susceptible to T2 variance, in addition to the loss of GABA signal to T2 outright. While scanning at the short TR of 2 s increases SNR per unit time, it also makes the measure more susceptible to condition-specific variance in T1 than if TR were longer.
One way to remove MM contribution with substantially less loss of MEGA-PRESS edited GABA signal (TE=68 ms) is to scan at ultra-high field6. However, difficulty achieving sufficient B1 to limit CSDE currently confines this approach to the periphery of the brain. Another possibility is to forgo editing and rely on meticulous fitting of ultra-short TE ultra-high field spectra, which have the additional advantage of reduced sensitivity to T2. Such meticulous fitting involves robust characterization of the MM contribution.
5. CONCLUSIONS
Acquisition parameters that are not usually mentioned in recent literature have impact on GABA and GABA+ signal that is comparable to those that are current areas of focus for development toward clinical trials. Given that individual parameter missets cause GABA+ signal errors of magnitude comparable to physiologically interesting differences, combinations of missets could obscure interpretation.
Several matters could be addressed to advance the field of MEGA-PRESS editing of GABA+. There is need for full specification of the coupling systems of GABA, Lys and Hcar at physiologic temperature and pH. There is also need to measure human brain GABA T1 and T2 in healthy and diseased humans. Knowledge on MM T1 and T2, as well as the extent to which MM and Hcar contributions vary is needed. Better understanding of why symmetric pulsing does not fully remove coedited Lys signal could lead to optimized MM-suppression schemes at 3 T. It might be possible to characterize GABA in non-edited spectra at 3 T, although baseline contributions and many realistic experimental conditions48 would make this challenging. New pulse sequences could be developed that allow longer editing pulses. Spectral variability and the effects of frequency drift could be minimized with real-time motion and shim correction and frequency navigators49.
ACKNOWLEDGMENT
We thank Robin de Graaf for providing the Lys coupling system.
Sponsors: This work was supported by the National Institutes of Health grant number P41 EB015894 and P30 NS076408.
List of abbreviations
- CSDE
chemical shift displacement error
- FWHM
full-width-half-maximum
- GABA
γ-aminobutyric acid
- GABA+
total edited signal at 3 ppm
- Hcar
homocarnosine
- Lys
lysine
- MM
macromolecules
- RF
radiofrequency
- talt
alternative timing
- TE
echo time
- TR
repetition time
- VOI
volume-of-interest
REFERENCES
- 1.Rothman DL, Petroff OA, Behar KL, Mattson RH. Localized 1H NMR measurements of gamma-aminobutyric acid in human brain in vivo. Proc Natl Acad Sci U S A. 1993;90(12):5662–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11(6):266–272. [DOI] [PubMed] [Google Scholar]
- 3.Hetherington HP, Newcomer BR, Pan JW. Measurements of human cerebral GABA at 4.1 T using numerically optimized editing pulses. Magn Reson Med. 1998;39(1):6–10. [DOI] [PubMed] [Google Scholar]
- 4.Near J, Simpson R, Cowen P, Jezzard P. Efficient gamma-aminobutyric acid editing at 3T without macromolecule contamination: MEGA-SPECIAL. NMR Biomed. 2011;24(10):1277–1285. [DOI] [PubMed] [Google Scholar]
- 5.Henry PG, Dautry C, Hantraye P, Bloch G. Brain GABA editing without macromolecule contamination. Magn Reson Med. 2001;45(3):517–520. [DOI] [PubMed] [Google Scholar]
- 6.Terpstra M, Ugurbil K, Gruetter R. Direct in vivo measurement of human cerebral GABA concentration using MEGA-editing at 7 Tesla. Magn Reson Med. 2002;47(5):1009–1012. [DOI] [PubMed] [Google Scholar]
- 7.Behar KL, Ogino T. Characterization of macromolecule resonances in the 1H NMR spectrum of rat brain. Magn Reson Med. 1993;30(1):38–44. [DOI] [PubMed] [Google Scholar]
- 8.Petroff OAC, Hyder F, Mattson RH, Rothman DL. Topiramate increases brain GABA, homocarnosine, and pyrrolidinone in patients with epilepsy. Neurology. 1999;52(3):473. [DOI] [PubMed] [Google Scholar]
- 9.Henry PG, van de Moortele PF, Giacomini E, Nauerth A, Bloch G. Field-frequency locked in vivo proton MRS on a whole-body spectrometer. Magn Reson Med. 1999;42(4):636–642. [DOI] [PubMed] [Google Scholar]
- 10.Harris AD, Glaubitz B, Near J, et al. Impact of frequency drift on gamma-aminobutyric acid-edited MR spectroscopy. Magn Reson Med. 2014;72(4):941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikkelsen M, Barker PB, Bhattacharyya PK, et al. Big GABA: Edited MR spectroscopy at 24 research sites. Neuroimage. 2017;159:32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullins PG, McGonigle DJ, O’Gorman RL, et al. Current practice in the use of MEGA-PRESS spectroscopy for the detection of GABA. Neuroimage. 2014;86:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bogner W, Hangel G, Esmaeili M, Andronesi OC. 1D-spectral editing and 2D multispectral in vivo(1)H-MRS and (1)H-MRSI - Methods and applications. Anal Biochem. 2017;529:48–64. [DOI] [PubMed] [Google Scholar]
- 14.Da Silva T, Hafizi S, Rusjan PM, et al. GABA levels and TSPO expression in people at clinical high risk for psychosis and healthy volunteers: a PET-MRS study. J Psychiatry Neurosci. 2019;44(2):111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao F, Edden RA, Li M, et al. Edited magnetic resonance spectroscopy detects an age-related decline in brain GABA levels. Neuroimage. 2013;78:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mikkelsen M, Harris AD, Edden RAE, Puts NAJ. Macromolecule-suppressed GABA measurements correlate more strongly with behavior than macromolecule-contaminated GABA+measurements. Brain Res. 2018;1701:204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mikkelsen M, Rimbault DL, Barker PB, et al. Big GABA II: Water-referenced edited MR spectroscopy at 25 research sites. Neuroimage. 2019;191:537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saleh MG, Rimbault D, Mikkelsen M, et al. Multi-vendor standardized sequence for edited magnetic resonance spectroscopy. Neuroimage. 2019;189:425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shungu DC, Mao X, Gonzales R, et al. Brain gamma-aminobutyric acid (GABA) detection in vivo with the J-editing (1) H MRS technique: a comprehensive methodological evaluation of sensitivity enhancement, macromolecule contamination and test-retest reliability. NMR Biomed. 2016;29(7):932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tapper S, Tisell A, Helms G, Lundberg P. Retrospective artifact elimination in MEGA-PRESS using a correlation approach. Magn Reson Med. 2019;81(4):2223–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henry PG, Marjańska M, Walls JD, Valette J, Gruetter R, Ugurbil K. Proton-observed carbon-edited NMR spectroscopy in strongly coupled second-order spin systems. Magn Reson Med. 2006;55(2):250–257. [DOI] [PubMed] [Google Scholar]
- 22.Branzoli F, Di Stefano AL, Capelle L, et al. Highly specific determination of IDH status using edited in vivo magnetic resonance spectroscopy. Neuro Oncol. 2018;20(7):907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maudsley AA, Govindaraju V, Young K, et al. Numerical simulation of PRESS localized MR spectroscopy. J Magn Reson. 2005;173(1):54–63. [DOI] [PubMed] [Google Scholar]
- 24.Kreis R, Bolliger CS. The need for updates of spin system parameters, illustrated for the case of gamma-aminobutyric acid. NMR Biomed. 2012;25(12):1401–1403. [DOI] [PubMed] [Google Scholar]
- 25.Govind V, Young K, Maudsley AA. Corrigendum: proton NMR chemical shifts and coupling constants for brain metabolites. Govindaraju V, Young K, Maudsley AA, NMR Biomed. 2000; 13: 129–153. NMR in biomedicine. 2015;28(7):923–924. [DOI] [PubMed] [Google Scholar]
- 26.Behar KL, Rothman DL, Spencer DD, Petroff OA. Analysis of macromolecule resonances in 1H NMR spectra of human brain. Magn Reson Med. 1994;32(3):294–302. [DOI] [PubMed] [Google Scholar]
- 27.de Graaf RA, Prinsen H, Giannini C, Caprio S, Herzog RI. Quantification of (1)H NMR Spectra from Human Plasma. Metabolomics. 2015;11(6):1702–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Graaf RA, Rothman DL. Spectral Editing. In: Bottomley PA, Griffiths JR, eds. Handbook of Magnetic Resonance Spectroscopy In Vivo. West Sussex: John Wiley & Sons; 2016. [Google Scholar]
- 29.De Graaf RA. In Vivo NMR Spectroscopy. 3 ed. Sussex West: John Wiley & Sons Ltd; 2019. [Google Scholar]
- 30.Kaiser LG, Young K, Off DJM, Mueller SG, Matson GB. A detailed analysis of localized J-difference GABA editing: theoretical and experimental study at 4T. Nmr in Biomedicine. 2008;21(1):22–32. [DOI] [PubMed] [Google Scholar]
- 31.Elmaki EEA, Gong T, Nkonika DM, Wang G. Examining alterations in GABA concentrations in the basal ganglia of patients with Parkinson’s disease using MEGA-PRESS MRS. Jpn J Radiol. 2018;36(3):194–199. [DOI] [PubMed] [Google Scholar]
- 32.Grewal M, Dabas A, Saharan S, Barker PB, Edden RA, Mandal PK. GABA quantitation using MEGA-PRESS: Regional and hemispheric differences. J Magn Reson Imaging. 2016;44(6):1619–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu B, Yang H, Gao F, et al. Investigation of brain GABA+ in primary hypothyroidism using edited proton MR spectroscopy. Clin Endocrinol (Oxf). 2017;86(2):256–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nantes JC, Proulx S, Zhong J, et al. GABA and glutamate levels correlate with MTR and clinical disability: Insights from multiple sclerosis. Neuroimage. 2017;157:705–715. [DOI] [PubMed] [Google Scholar]
- 35.Rowland LM, Krause BW, Wijtenburg SA, et al. Medial frontal GABA is lower in older schizophrenia: a MEGA-PRESS with macromolecule suppression study. Mol Psychiatry. 2016;21(2):198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang JH, Graham GD, Behar KL, Alger JR, Prichard JW, Rothman DL. Short echo time proton magnetic resonance spectroscopic imaging of macromolecule and metabolite signal intensities in the human brain. Magn Reson Med. 1996;35(5):633–639. [DOI] [PubMed] [Google Scholar]
- 37.Mader I, Seeger U, Weissert R, et al. Proton MR spectroscopy with metabolite-nulling reveals elevated macromolecules in acute multiple sclerosis. Brain : a journal of neurology. 2001;124(Pt 5):953–961. [DOI] [PubMed] [Google Scholar]
- 38.Marjanska M, McCarten JR, Hemmy LS, Terpstra M. Altered macromolecular pattern in aging brain. Paper presented at: International Society of Magnetic Resonance in Medicine2015; Toronto. [Google Scholar]
- 39.Puts NA, Barker PB, Edden RA. Measuring the longitudinal relaxation time of GABA in vivo at 3 Tesla. J Magn Reson Imaging. 2013;37(4):999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edden RAE, Intrapiromkul J, Zhu H, Cheng Y, Barker PB. Measuring T2 in vivo with J-difference editing: Application to GABA at 3 tesla. J Magn Reson Imaging. 2012;35(1):229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marjanska M, Emir UE, Deelchand DK, Terpstra M. Faster metabolite 1H transverse relaxation in the elder human brain. PloS one. 2013;8(10):e77572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13(3):129–153. [DOI] [PubMed] [Google Scholar]
- 43.Kish SJ, Perry TL, Hansen S. Regional distribution of homocarnosine, homocarnosine-carnosine synthetase and homocarnosinase in human brain. J Neurochem. 1979;32(6):1629–1636. [DOI] [PubMed] [Google Scholar]
- 44.Petroff OA, Mattson RH, Behar KL, Hyder F, Rothman DL. Vigabatrin increases human brain homocarnosine and improves seizure control. Ann Neurol. 1998;44(6):948–952. [DOI] [PubMed] [Google Scholar]
- 45.Edden RAE, Barker PB. Spatial effects in the detection of γ-aminobutyric acid: Improved sensitivity at high fields using inner volume saturation. Magn Reson Med. 2007;58(6):1276–1282. [DOI] [PubMed] [Google Scholar]
- 46.Edden RA, Puts NA, Barker PB. Macromolecule-suppressed GABA-edited magnetic resonance spectroscopy at 3T. Magn Reson Med. 2012;68(3):657–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu M, Hurd R, Noeske R, et al. GABA editing with macromolecule suppression using an improved MEGA-SPECIAL sequence. Magn Reson Med. 2018;79(1):41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Near J, Andersson J, Maron E, et al. Unedited in vivo detection and quantification of gamma-aminobutyric acid in the occipital cortex using short-TE MRS at 3 T. NMR Biomed. 2013;26(11):1353–1362. [DOI] [PubMed] [Google Scholar]
- 49.Deelchand DK, Joers JM, Auerbach EJ, Henry P- G. Prospective motion and B0 shim correction for MR spectroscopy in human brain at 7T. Magn Reson Med. 2019;0(0). [DOI] [PMC free article] [PubMed] [Google Scholar]


