Abstract
Actinobacillus pleuropneumoniae (App) is the etiological agent of acute porcine pneumonia and responsible for severe economic losses worldwide. The capsule polymer of App serotype 1 (App1) consists of [4)-GlcNAc-β(1,6)-Gal-α-1-(PO4-] repeating units that are O-acetylated at O-6 of the GlcNAc. It is a major virulence factor and was used in previous studies in the successful generation of an experimental glycoconjugate vaccine. However, the application of glycoconjugate vaccines in the animal health sector is limited, presumably because of the high costs associated with harvesting the polymer from pathogen culture. Consequently, here we exploited the capsule polymerase Cps1B of App1 as an in vitro synthesis tool and an alternative for capsule polymer provision. Cps1B consists of two catalytic domains, as well as a domain rich in tetratricopeptide repeats (TPRs). We compared the elongation mechanism of Cps1B with that of a ΔTPR truncation (Cps1B-ΔTPR). Interestingly, the product profiles displayed by Cps1B suggested processive elongation of the nascent polymer, whereas Cps1B-ΔTPR appeared to work in a more distributive manner. The dispersity of the synthesized products could be reduced by generating single-action transferases and immobilizing them on individual columns, separating the two catalytic activities. Furthermore, we identified the O-acetyltransferase Cps1D of App1 and used it to modify the polymers produced by Cps1B. Two-dimensional NMR analyses of the products revealed O-acetylation levels identical to those of polymer harvested from App1 culture supernatants. In conclusion, we have established a protocol for the pathogen-free in vitro synthesis of tailored, nature-identical App1 capsule polymers.
Keywords: vaccine development, processivity, nuclear magnetic resonance (NMR), polysaccharide, virulence factor, oligosaccharide, Actinobacillus pleuropneumoniae, animal vaccines, bacterial capsules, enzyme engineering, tetratricopeptide repeats, glycoconjugate vaccine, processivity
Introduction
Actinobacillus pleuropneumoniae (App)2 is an encapsulated, Gram-negative, pig-specific bacterial pathogen that colonizes the upper respiratory tract and causes severe economic losses globally (1). App can be grouped into 18 different serotypes based on its surface capsule polymer composition (2), and serotypes can be identified by PCR-based typing methods (3). All serotypes are pathogenic, but predominance of strains and virulence of serotypes can vary geographically and temporally (1). The capsule polymer represents one of the main virulence factors of App and enables it to evade the hosts' immune response by masking underlying structures and by preventing the efficient formation of the membrane attack complex and thus complement mediated killing (4, 5).
So far, general treatment of App infection includes the use of a broad variety of antibiotics, which can result in the increase of bacterial strains harboring antibiotic resistances (1). Commercial vaccines against App are currently based on toxins, attenuated strains, and subunit vaccines or combinations of the above (1, 6). They were shown to significantly reduce clinical symptoms in vaccinated pigs, but they fail to achieve total protection and, most importantly, fail to protect against transmission, highlighting the importance for the development of new vaccines (1, 6). Effective vaccines against other encapsulated pathogens, such as Neisseria meningitidis and Haemophilus influenzae, are so called glycoconjugate vaccines, consisting of processed capsule polymers coupled to a carrier protein (7). Initial studies reporting the generation of a glycoconjugate vaccine against App serotype 1 (App1) using polymer harvested from bacterial cultures reported reduced mortality rates, diminished lung lesions, and greater weight gain in vaccinated pigs (8, 9). However, the vaccine was never brought to market, presumably due to the high costs associated with glycoconjugate production.
Recently, it was shown that labor-intensive, biohazardous polymer purification from pathogen cultures can be avoided by utilizing recombinant capsule polymerases for the in vitro synthesis of polymers used in glycoconjugate vaccines against N. meningitidis (10–13). Of special biotechnological value in this context are polymerases deploying a distributive mode of chain elongation, because they allow the generation of product pools of uniform chain length (Gaussian-shaped distribution) as well as control over the average degree of polymerization (DP) of the products. In contrast to processive enzymes, which catalyze multiple transfers without releasing the growing polymer (and thus produce a heterogeneous product profile), strictly distributive polymerases transfer only a single sugar moiety at a time and dissociate afterward, allowing the stoichiometry between donor and acceptor substrate to dictate chain length (14–16).
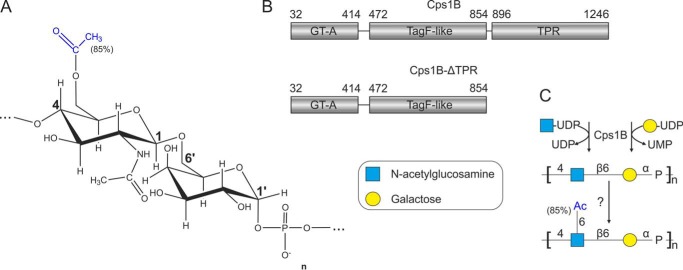
We recently described the recombinant production of Cps1B, the capsule polymerase of App serotype 1 (17). Cps1B belongs to the TagF-like polymerase family, a group of enzymes assembling teichoic acid–like capsule polymers in Gram-negative bacteria expressing a group 2 capsule biosynthesis system. Cps1B assembles a polymer consisting of [4)-GlcNAc-β(1,6)-Gal-α-1-[PO4−] repeating units (Fig. 1, A and C). Like many group 2 capsule polymerases (11, 18–20), it can initiate chain elongation even in the absence of acceptor substrates (hereafter referred to as de novo activity), only requiring the nucleotide-activated donor substrates UDP-Gal and UDP-GlcNAc (17). The mode of elongation (distributive versus processive) utilized by the enzyme has not been analyzed yet. In vivo, the polymer generated by Cps1B is partly (85%) O-acetylated at O-6 of the GlcNAc by an as yet unidentified O-acetyltransferase (21).
Figure 1.
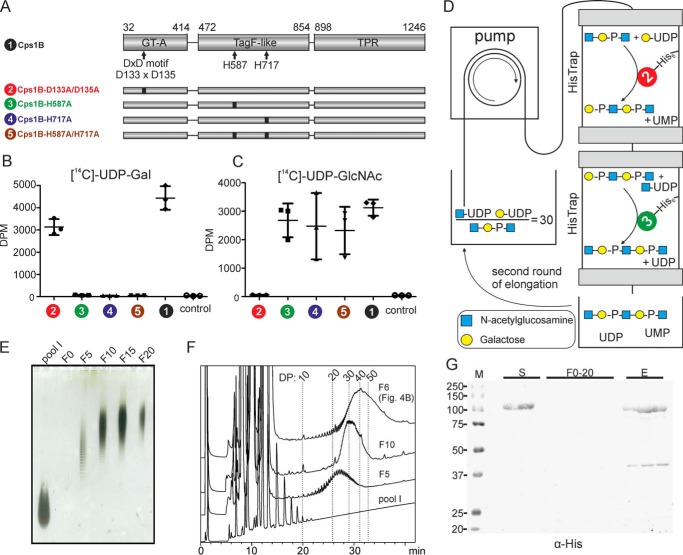
Capsule polymer and capsule polymerase of App1. A, structure of the O-acetylated capsule polymer of App1. B, domain architecture of soluble Cps1B constructs as reported previously (17). GT-A, glycosyltransferase A-folded domain; TagF-like, domain modeled onto teichoic acid synthase TagF; TPR, domain rich in TPRs. C, enzymatic synthesis of the O-acetylated capsule polymer of App1. The linear polymer backbone is assembled by the capsule polymerase Cps1B. The O-acetyl group at O-6 of GlcNAc is transferred by a yet unknown enzyme.
Cps1B was shown to consist of two catalytically active domains, a glycosyltransferase domain adopting a GT-A fold and a hexose-phosphate transferase domain similar to the teichoic acid synthase TagF (17, 22) (Fig. 1B). In addition, a domain rich in tetratricopeptide repeats (TPRs) was identified (17, 23). Interestingly, a truncation of the enzyme lacking the first 31 amino acid residues (ΔN31) was shown to be considerably better-expressed than the full-length version (17) and is from here on referred to as Cps1B (see Fig. 1B). A variant lacking the TPR domain (Fig. 1B, Cps1B-ΔTPR) was previously introduced into Cps1B, leading to Cps1B-ΔTPR. The subsequent introduction of single-domain mutations of the GT-A folded domain (D133A and D135A of the DXD motif) and the TagF-like domain (H587A and H717A) yielded the constructs Cps1B-D133A/D135A-ΔTPR, Cps1B-H587A-ΔTPR, and Cps1B-H717A-ΔTPR, of which only one domain remained active (17).
To optimize Cps1B as a tool for vaccine synthesis, in this study, we investigated the elongation mode of Cps1B and Cps1B-ΔTPR. In addition, we immobilized single-domain mutants on HisTrap columns, separating the two catalytic activities of Cps1B and creating an experimental setup that yielded product populations of low dispersity, which meet the standards set by state-of-the-art glycoconjugate vaccines. The identification of the O-acetyltransferase Cps1D and its utilization for O-acetylating the products assembled by Cps1B allowed the synthesis of nature-identical App1 capsule poly- and oligomers.
Results
Generation and characterization of oligosaccharide acceptors
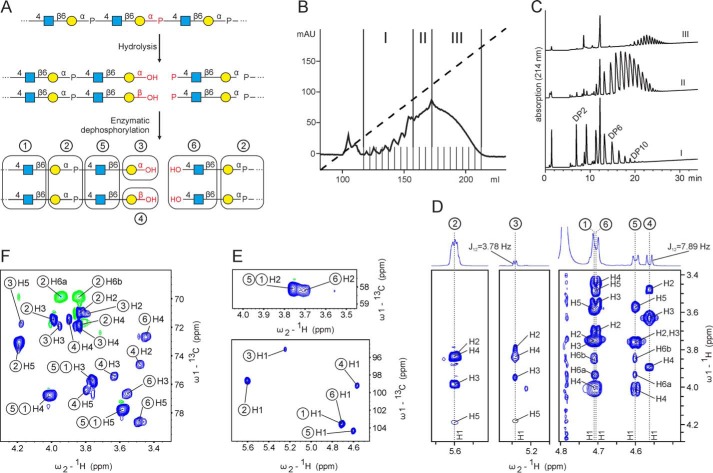
In previous studies, the elongation mechanism of capsule polymerases was analyzed by varying the donor/acceptor (d/a) ratio in in vitro reactions (14). In such an experiment, distributive enzymes would generate a Gaussian-shaped product pool with low dispersity, and the average degree of polymerization (avDP) of the products would correspond to the d/a ratio. In contrast, processive enzymes would generate heterogeneous product pools containing predominantly polymers with an avDP larger than the d/a ratio together with nonused acceptors. Ideal acceptors to prime the reaction of group 2 capsule polymerases are short oligosaccharides obtained through hydrolysis of long polymers (11, 12, 14). To generate such acceptors, we first exploited the de novo activity of Cps1B (17) to synthesize long polymers as suitable starting material for hydrolysis. As previously described for other phosphate-containing group 2 polymers (11, 14, 19), the purified material was then subjected to mild acidic hydrolysis (Fig. 2A). The resulting oligosaccharides were separated by anion-exchange chromatography and divided into three pools (Fig. 2B). The termini of the fragments were freed from phosphomonoesters by enzymatic dephosphorylation to make them accessible to elongation by Cps1B (Fig. 2A) (note that group 2 capsule polymerases elongate the nascent chain at the nonreducing end (5)). Each pool was analyzed by HPLC-AEC (Fig. 2C). The pool containing the smallest oligosaccharides (pool I, Fig. 2, B and C) was further analyzed by 1D and 2D NMR spectroscopy, which showed the occurrence of six resonances in the anomeric region (Fig. 2, D and E (bottom)). The spin systems belonging to these anomeric signals are labeled with circled numbers in Fig. 2 (D–F), and the residues assigned to each individual spin system are indicated in Fig. 2A. As expected, the two dominant spin systems, ➀ and ➁, were identical to previously published spectra of non-O-acetylated App1 polymer (17, 21) and thus were assigned to internal GlcNAc and Gal moieties, respectively (Fig. 2A). Because hydrolytic cleavage of a phosphodiester linkage is favored between the anomeric carbon and the phosphate (24, 25) (Fig. 2A), we hypothesized that the reducing-end sugar of the oligosaccharides is galactose. Indeed, spin systems ➂ and ➃ showed C1 chemical shifts (derived from an 13C-1H HSQC experiment; see Fig. 2E (bottom) and Table 1) and H1-H2 coupling constants (obtained through 1D 1H NMR; see Fig. 2D) that were similar to those in previously published spectra of oligosaccharides carrying a reducing-end galactose in α or β configuration (26), respectively. The chemical shifts of C2 of both spin systems (71.0 ppm for spin system ➂, 74.6 ppm for spin system ➃; see Fig. 2F and Table 1) clearly identified them as galactoses rather than GlcNAc moieties, for which C2 chemical shifts of ∼58 ppm would be expected (compare C2 of spin systems ➂ and ➃ (Fig. 2F) with C2 of GlcNAc spin systems ➀, ➄, and ➅ (Fig. 2E (top) and Table 1). Resonances for positions 1–5 of ➂ and ➃ were assigned based on 1H-1H COSY (not shown), 1H-1H TOCSY, and 1H-13C HSQC experiments (Fig. 2 (D–F) and Table 1) and are in perfect agreement with chemical shift data of reducing-end galactose residues in α and β configuration substituted at O6 (β1,6 linkage) that were deposited in the Glycosciences.de database (27, 28) under LinucsID accession number 1471. According to LinucsID 1471, 13C chemical shifts of C6 of ➂ and ➃ should be expected at 71.9 and 71.4 ppm, respectively (offset due to referencing to 2,2-dimethyl-2-silapentanesulfonic acid (DSS) included). Unfortunately, the C6-H6 and C6-H6′ correlations of ➂ and ➃ could not be identified in the multiplicity-edited 1H-13C HSQC spectrum due to overlap of the negative CH2 resonances with positive CH signals. However, in an 1H-13C HMBC experiment, which is able to correlate protons and carbons via three bonds and thus ideal to establish glycosidic linkages, the H1 of the GlcNAc spin system ➄ showed a strong correlation to this region (71.4 ppm), indicating that ➄ represents a GlcNAc moiety linked to C6 of ➂ and ➃, or, in other words, the second last reducing-end sugar (spin system ➄, Fig. 2A). This finding is corroborated by the chemical shift prediction program CASPER (29), which predicts 13C chemical shifts for C6 (Gal) of 6P-GlcNAcβ1,6GalαOH and 6P-GlcNAcβ1,6GalβOH of 71.9 and 71.7 ppm, respectively (referenced to DSS). In agreement with these observations, all other GlcNAc spin systems detected in pool I showed correlations to C6 of internal Gal (spin system ➁, 69.8 ppm). As expected, despite its distinct anomeric signal (Fig. 2, D and E (bottom)), all other resonances belonging to ➄ were identical to the ones of internal GlcNAc (spin system ➀) (Fig. 2 (D–F) and Table 1).
Figure 2.
Acceptor oligosaccharide synthesis and characterization. A, schematic representation of the hydrolytic cleavage of the phosphodiester linkage present in the polymer synthesized by Cps1B. Because the linkage between the anomeric carbon of the galactose and the phosphate is more prone to hydrolysis, we hypothesized that the phosphate remains attached to GlcNAc at the nonreducing end of the resulting fragments. Treatment with alkaline phosphatase removes the phosphate and makes the nonreducing end accessible for enzymatic elongation. Residues causing distinct spin systems detectable by NMR analyses are highlighted by boxes and organized throughout the entire figure using circled numbers. B, polymer synthesized by Cps1B was hydrolyzed, purified using anion-exchange chromatography, fractionated into three pools (I–III), and dephosphorylated. C, analysis of the pooled fractions using HPLC-based anion-exchange chromatography with UV detection (214 nm). The assignment of the DP to the eluted oligosaccharides is based on the assumption that the smallest DP retained by the AEC column requires at least one negatively charged phosphate, which is the case for a terminally dephosphorylated DP2 (GlcNAc-Gal-P-GlcNAc-Gal). The assignment is in good agreement with the avDP of 4.5 determined for pool I by 1H NMR. D, 1H-1H TOCSY showing all anomeric resonances (H1) detected in oligosaccharide pool I on the ω2 axis and correlations to the other protons of the indicated spin system on the ω1 axis. E and F, corresponding 1H-13C HSQC spectra showing the chemical shifts of protons on the ω2 axis and the chemical shift of carbons to which they are connected on the ω1 axis.
Table 1.
1H and 13C chemical shifts observed in pool I, referenced to DSS
| Spin system/atom | ➀ (internal GlcNAc) | ➁ (internal Gal) | ➂ (reducing-end α-Gal) | ➃ (reducing-end β-Gal) | ➄ (GlcNAc) | ➅ (nonreducing-end GlcNAc) |
|---|---|---|---|---|---|---|
| H1 | 4.707 | 5.602 | 5.245 | 4.565 | 4.602 | 4.706 |
| H2 | 3.761 | 3.839 | 3.791 | 3.481 | 3.749 | 3.702 |
| H3 | 3.751 | 3.989 | 3.952 | 3.628 | 3.763 | 3.558 |
| H4 | 4.008 | 3.839 | 3.839a | 3.897 | 4.011 | 3.447 |
| H5 | 3.584 | 4.191 | 4.177 | 3.798 | 3.572 | 3.487 |
| H6a | 3.934 | 3.945 | NDb | NDb | 3.933 | 3.933 |
| H6b | 3.847 | 3.840 | NDb | NDb | 3.850 | 3.760 |
| C1 | 103.5 | 98.7 | 95.1 | 99.2 | 104.3 | 103.5c |
| C2 | 58.1 | 71.0 | 71.1 | 74.6 | 58.1c | 58.2 |
| C3 | 75.8 | 71.5 | 71.8 | 75.4 | 75.7c | 76.7 |
| C4 | 76.8 | 71.8 | 71.8a | 71.4 | 76.8c | 72.7 |
| C5 | 77.8 | 73.1 | 71.8 | 76.4 | 77.8c | 78.6 |
| C6 | 63.2 | 69.9 | (71.4)d | (71.4)d | 63.2c | 63.4 |
| C9 (NAc) | 177.5 | 177.5c | 177.5c | |||
| H10 (NAc) | 2.068 | 2.055c | 2.068c | |||
| C10 (NAc) | 25.0 | 25.0c | 25.0c | |||
| P | −4.92 |
a Overlap with spin system ➁.
b ND, not determined due to overlap of negative CH2 signals with positive CH signals in the multiplicity-edited 1H-13C HSQC.
c Overlap with spin system ➀.
d Derived from an H1➄-C6➂/➃ correlation in the 1H-13C HMBC spectrum.
The remaining spin system ➅ displaying the characteristic C2 chemical shift of a GlcNAc (Fig. 2E, top) could be assigned to a GlcNAc positioned at the nonreducing end of the oligosaccharides. This finding agrees with the expected site of hydrolysis (Fig. 2A). The successful removal of the phosphate from position 4 of this residue (Fig. 2A) was confirmed by the absence of a monophosphate signal in the 31P and 31P-1H HMBC experiments (data not shown) and further corroborated by the upfield shifted resonances of H4 and C4 as established from 1H-1H COSY (not shown) and 1H-1H TOCSY and 13C-1H HSQC experiments (Fig. 2 (D and F), compare C4 and H4 chemical shifts of spin system ➅ with the H4 and C4 chemical shifts of spin systems ➀ and ➄). Again, all resonances of spin system ➅ agree with published spectra of nonreducing-end GlcNAc moieties in the Glycosciences.de database (LinucsID 384) (27, 28). Finally, it is important to note that the integrals of the isolated anomeric signals of the internal spin system ➁ and the reducing-end spin system ➄ can be conveniently used to calculate the average degree of polymerization (avDP) of pool I to be avDP = 4.5, using (H1Int/H1Ter) + 1, where H1Int is the signal intensity of the anomeric signal of spin system ➁ and H1Ter is the signal intensity of the anomeric signal of spin system ➄.
Analysis of the elongation mechanism of Cps1B and Cps1B-ΔTPR
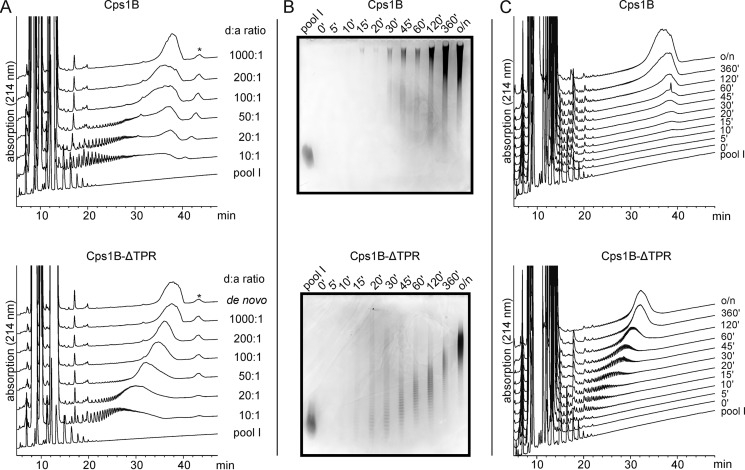
Next, we used the oligosaccharides of pool I to analyze the product profiles generated by Cps1B at varying d/a ratios. Because previous studies have shown that the truncation of the TPR domain influences the oligomerization state of Cps1B (17), and because oligomerization can be a prerequisite for processivity (16), we included Cps1B-ΔTPR in the study. In overnight reactions, Cps1B and Cps1B-ΔTPR were incubated with donor substrates (UDP-Gal and UDP-GlcNAc) and varying oligosaccharide acceptor (pool I) concentrations (d/a of 10:1–1,000:1). The products were visualized by HPLC-based AEC and Alcian blue/silver-stained PAGE. Even at high acceptor concentrations (see Fig. 3A (top), d/a of 10:1), Cps1B generates long chains, and the product pool is of high dispersity, which can be considered an indication for processive elongation. In contrast, and although the product pool generated by Cps1B-ΔTPR is not strictly Gaussian-shaped and homogeneous, the dispersity is considerably lower and corresponds much better to the d/a ratio (see Fig. 3A, bottom).
Figure 3.
Product profiles generated by Cps1B and Cps1B-ΔTPR. A, polymer synthesized by Cps1B and Cps1B-ΔTPR at varying d/a ratios after overnight incubation was analyzed via HPLC-based anion-exchange chromatography. Pool I was used as marker for low-molecular-weight material, and a de novo reaction containing only donor substrates was utilized as marker for very long chains (polymer synthesized under de novo conditions can be considered to be of maximum chain length). Material eluting >40 min (*)—and thus even later than de novo synthesized polymer—could not be visualized on an Alcian blue/silver-stained gel (not shown) and thus appears not to be of carbohydrate origin. B and C, polymer synthesis was monitored in a time course experiment. Pool I was used as acceptor (d/a ratio of 100:1), and samples were taken at the indicated time points. The products were visualized via Alcian blue/silver-stained PAGE (25%) (B) and HPLC-based anion-exchange chromatography (C). The high-intensity signals recorded during the first 15 min of each HPLC run are caused by the reaction products UMP and UDP.
Under in vitro conditions, it has been reported that the products of distributive enzymes grow in size over time, whereas processive enzymes predominantly release long polymers, leading to an accumulation of long polymers even at early time points of the reaction (14, 15). Consequently, we analyzed the time course of acceptor elongation by Cps1B and Cps1B-ΔTPR. Catalytic amounts of Cps1B and Cps1B-ΔTPR were incubated at a d/a ratio of 100:1 to allow the synthesis of long chains. The product profile was monitored over time, and samples were taken as indicated and analyzed using HPLC-AEC and Alcian blue/silver-stained PAGE (Fig. 3, B and C). Even after short incubation times, Cps1B predominantly generated long polymers, corroborating a processive mode of elongation. In contrast, the polymer synthesized by Cps1B-ΔTPR continuously grew over time, indicating distributive elongation. In summary, the above-presented results demonstrate that Cps1B-ΔTPR is favored over Cps1B for biotechnological applications because it offers the possibility to better control the length of the produced products either by adjusting the d/a ratio or by controlling the time of the reaction.
On-column synthesis of App1 oligosaccharides
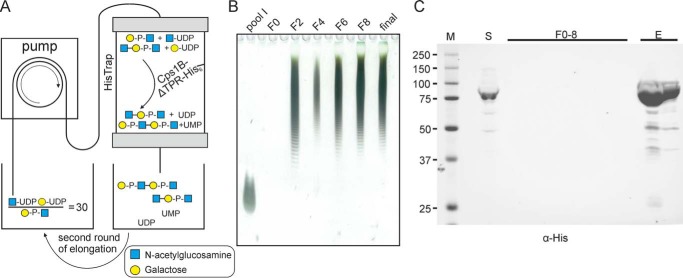
In a recent study, our group established a protocol for the tailored, enzymatic solid-phase synthesis of capsule polymers from N. meningitidis serogroups A and X, which allowed the upscaling of the reaction and the potential reuse of the enzyme-loaded matrices (14). To establish a similar experimental setup for the synthesis of App1 oligosaccharides, Cps1B-ΔTPR was expressed in Escherichia coli; the bacteria were lysed, and Cps1B-ΔTPR was immobilized on a column via its C-terminal His tag. After thorough washing to remove nonspecifically bound proteins, the reaction mix containing the substrates (UDP-GlcNAc and UDP-Gal) as well as the acceptor molecules in a ratio of 30:1 was circulated over the column (Fig. 4A). The dispersity of the products was visualized by Alcian blue/silver-stained PAGE showing pool I as a size control (Fig. 4B). Already after the second round of circulation (Fig. 4B, F2), the acceptor was elongated to an oligosaccharide pool that barely changed during further rounds of circulation (Fig. 4B). Importantly, the lack of detectable protein in the sampled fraction analyzed by Western blot analysis demonstrated that Cps1B-ΔTPR was stably coupled to the column throughout the course of the reaction and could be eluted afterward using imidazole (Fig. 4C, F0–8).
Figure 4.
Solid-phase synthesis of App1 oligosaccharides. A, schematic overview of the experimental setup. The acceptors are depicted in a simplified form. Cps1B-ΔTPR-His was immobilized on a HisTrap column, and the reaction mix containing the substrates (UDP-Gal and UDP-GlcNAc) and the acceptor oligosaccharides (pool I) was circulated for several consecutive rounds. B, Alcian blue/silver-stained PAGE (25%) of collected fractions (F = round of circulation). Pool I was used as marker material (because small fragments can diffuse from the gel, the lane was heavily overloaded to allow sufficient signal intensity). C, Western blot analysis of the loaded supernatant (S), collected fractions (F0–8), and eluted protein (E), stained against the C-terminal His tag of Cps1B-ΔTPR. No protein contamination could be detected in the reaction mixture throughout the experiment, indicating stable binding of Cps1B-ΔTPR to the column. After the experiment, Cps1B-ΔTPR could be eluted from the column using imidazole (E).
The two-domain architecture of Cps1B can be exploited to reduce product dispersity
Whereas the solid-phase coupling of Cps1B-ΔTPR was successful (Fig. 4), the synthesized product pool was of considerable dispersity. To further reduce the heterogeneity of the products, an experimental setup allowing the separation of the catalytic activities of Cps1B was designed (Fig. 5D), comprising one column loaded with a mutant having only galactose-1-phosphate transferase activity and a second column loaded with a mutant exhibiting solely GlcNAc transferase activity. Because allocation of the catalytic activity of the GT-A– and TagF–like folded domains of Cps1B has so far only been based on bioinformatics predictions (17), we first confirmed the substrate specificity of each domain. Therefore, we introduced the previously described single-domain mutants of Cps1B-ΔTPR (17) into Cps1B (we chose Cps1B instead of Cps1B-ΔTPR to exclude any influence of the missing TPR domain on substrate recognition) (Fig. 5A) and incubated them with radioactively labeled donor substrates. Enzymatically synthesized App1 polymer was used as acceptor substrate, because it consists of a mixture of chains terminating with either Gal or GlcNAc at the nonreducing end, allowing both transferase domains to add their respective substrates. Fig. 5 (B and C) demonstrates that Cps1B-D133A/D135A (construct 2, inactive GT-A fold; Fig. 5A) only shows transferase activity in the presence of UDP-Gal, whereas Cps1B-H587A, Cps1B-H717A, and Cps1B-H587A/H717A (constructs 3–5, inactive TagF-like domain; Fig. 5A) are exclusively active in the presence of UDP-GlcNAc. WT Cps1B (construct 1) was used as positive control and was shown to utilize both substrates.
Figure 5.
Size-controlled App1 oligosaccharide synthesis using spatial separation of the catalytic activities of Cps1B. A, schematic overview of the domain structure and single-domain mutants of Cps1B. The first and last amino acid of each modeled domain and catalytically relevant amino acids are indicated. B and C, the activity of WT Cps1B (1) and single-domain mutants (2–5) was analyzed in a radioactive incorporation assay using [14C]UDP-Gal (B) and [14C]UDP-GlcNAc (C) as donor substrates and purified App1 polymer as acceptor. A reaction without enzyme was used as control. D, schematic overview of the solid-phase synthesis setup allowing domain separation of Cps1B. Cps1B-D133A/D135A and Cps1B-H587A were each immobilized on a HisTrap column, and a reaction mix with a d/a ratio of 30 was circulated through the setup. E, separation and detection of synthesized oligosaccharides by Alcian blue/silver-stained PAGE (25%). The acceptor oligosaccharides (pool I) were used as size control (because small fragments can diffuse from the gel, the lane was heavily overloaded to allow sufficient signal intensity). F20 contains the final oligosaccharide pool, which comprises the volume required to wash the setup; it is thus less concentrated. F, HPLC-based anion-exchange chromatography coupled to UV detection (214 nm) was used to compare representative fractions collected in this experiment (F5 and F10) and in the experiment shown in Fig. 4 (F6). Pool I functioned as size marker and could be used as a starting point to assign a chain length (DP) to each observed peak in the chromatogram. Separation could be achieved for oligomers < DP50. G, Western blot analysis of the loaded supernatant (S), collected fractions (F0–20), and eluted protein (E) developed against the C-terminal His tag. No protein contamination of the polymer fractions could be detected, and Cps1B mutants could be eluted after the experiment by the addition of imidazole.
Next, a mutant with galactose-1-phosphate transferase activity (Cps1B-D133A/D135A, construct 2) and a mutant with GlcNAc transferase activity (Cps1B-H587A, construct 3) were expressed, each mutant was coupled to a separate HisTrap column, and both columns were thoroughly washed to remove nonspecifically bound E. coli protein. The columns were connected in series, and the reaction mixture containing the substrates and the acceptor pool I in a d/a ratio of 30:1 was circulated through the setup (Fig. 5D). Note that the galactose-1-phosphate transferase domain was encountered first by the reaction mixture, allowing the transfer of Gal-1P onto the nonreducing end of GlcNAc of the acceptors (pool I) to happen first. After every fifth round of circulation, samples were taken and analyzed on an Alcian blue/silver-stained PAGE compared with pool I, which was used as size control. After five rounds of circulation, the products were of considerable size and grew further with additional rounds of elongation (Fig. 5E). As expected, on an Alcian blue/silver-stained PAGE, the product population appeared less disperse if compared with the material synthesized by Cps1B-ΔTPR (compare Figs. 4B and 5E). This finding was corroborated by a direct comparison (by HPLC-AEC) of the products generated using both setups (Fig. 5F): F10 obtained using the setup shown in Fig. 5 comprises oligosaccharides with DP20–DP50, whereas F6 produced using the setup shown in Fig. 4 is considerably more disperse (Fig. 5F). It is of note that the average DP of F10 agrees with the preset d/a ratio of 30 (Fig. 5F). Again, no protein contamination could be detected in the collected fractions (Fig. 5G).
Cps1D is the O-acetyltransferase of App1
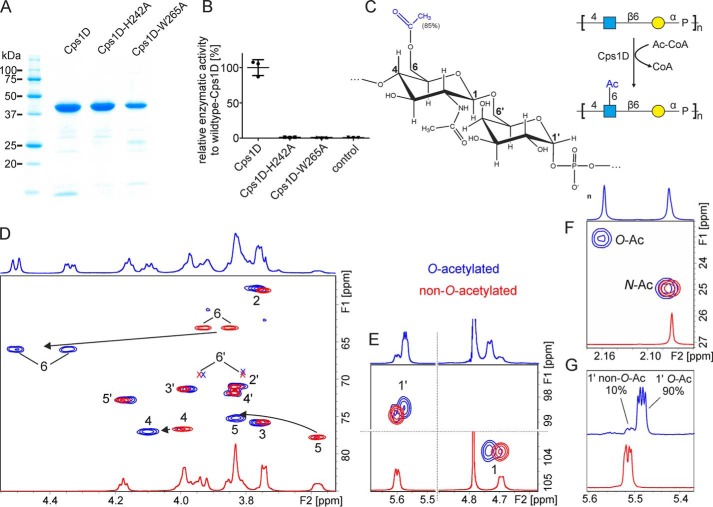
Natural capsule polymer harvested from App1 culture is partly modified by O-acetylation (85%) in position C6 of GlcNAc (Fig. 1A) (21). Previous studies predicted the gene product cps1D from the capsule biosynthesis gene cluster of App1 to encode for an O-acetyltransferase (30). To investigate the enzymatic function of Cps1D, we performed homology modeling using the structure prediction software PHYRE2 (31). Cps1D was modeled with 100% confidence onto the crystal structure of the polysialic acid specific O-acetyltransferase NeuO from E. coli K1, which belongs to the left-handed β-helix family of O-acetyltransferases (32). Importantly, amino acids His-147 und Trp-171, critical for NeuO activity, aligned with His-242 and Trp-265 of Cps1D. Consequently, WT Cps1D as well as Cps1D-H242A and Cps1D-W265A mutant constructs were designed, cloned, expressed, and successfully purified from E. coli expression cultures via their C-terminal His tag (Fig. 6A). Cps1D activity was tested according to a previously published spectrophotometric assay in which free SH-groups of CoA, released from acetyl-CoA after each O-acetyl transfer (Fig. 6, B and C), react with 5,5′-dithiobis(2-nitrobenzoic acid) to 5-thionitrobenzoic acid, which can be detected through absorption (33). As expected, WT Cps1D demonstrated O-acetyltransferase activity in the spectrophotometric assay, whereas no activity could be observed for the single-amino acid mutants Cps1D-H242A and Cps1D-W265A (Fig. 6B).
Figure 6.
Functional characterization of the O-acetyltransferase Cps1D. A, Coomassie-stained SDS-PAGE of purified Csp1D, Cps1D-H242A, and Cps1D-W265A. B, activity of WT Cps1D and single-amino acid mutants (Cps1D-H242A and Cps1D-W265A) was evaluated in a spectrophotometric assay. WT activity was set to 100%. A reaction without enzyme was used as negative control to monitor spontaneous hydrolysis of acetyl-CoA (control). C, structure of O-acetylated App1 polymer and schematic overview of the reaction catalyzed by Cps1D. Acetyl-CoA is used as donor substrate for the transfer of an acetyl group onto OH-6 of GlcNAc. D–F, 1H-13C HSQC NMR analysis of enzymatically O-acetylated (blue) and non-O-acetylated (red) App1 capsule polymer. Cps1D-mediated O-acetylation of GlcNAc O6 leads to a shift of the correlations for GlcNAc H4-C4, H5-C5, and 6H-C6 (D) and GlcNAc H1-C1, and Gal H1-C1 (E) and to the occurrence of a new correlation in the acetyl region (F). Correlations only visible at lower contour levels are indicated by X. G, in addition to a long polymer, Cps1D can be used to O-acetylate the chains of lower-molecular-weight (avDP30) obtained by solid-phase synthesis (see Fig. 5) by up to 90%.
To perform NMR analyses, the Cps1D reaction was upscaled. 2 mg of non-O-acetylated polymer backbone (synthesized by Cps1B and purified) was incubated with acetyl-CoA in molar ratios of 1:0.5, 1:1, and 1:2, respectively. The resulting products were purified by anion-exchange chromatography and analyzed by 1D and 2D NMR. Spectra were compared with non-O-acetylated polymer (Fig. 6 and Table 2). Whereas the majority of correlations observed in a 1H-13C HSQC experiment were widely unaffected by the enzymatic addition of the O-acetyl group, the C6-H6, C5-H5, and C4-H4 cross-peaks of GlcNAc were markedly shifted downfield (Fig. 6D, see arrows and compare red (non-O-acetylated) and blue (O-acetylated) spectra). Interestingly, the chemical shifts of both anomeric protons of the repeating unit are affected as well, allowing a rough estimation and a convenient visualization of the degree of O-acetylation (Fig. 6E). As expected, an additional correlation can be observed in the region that is characteristic for acetyl groups (Fig. 6F). It is important to note that H6a/b O-Ac are isolated in the 1H NMR, allowing the quantification of the level of O-acetylation from the integrals of H6a or H6b and H1 (e.g. H6aOAc/(H1O-Ac + H1non-O-Ac). In addition, the well-separated signals of the N-acetyl group and the O-acetyl group are even better suited for quantification (Fig. 6F). The level of O-acetylation in reactions with a donor/acceptor ratio of 0.5:1, 1:1, and 2:1 was 47, 83, and 95%, respectively, which is in very good agreement with the level observed for the natural polymer (85% O-acetylation (21)). To investigate whether Cps1D is also able to O-acetylate shorter oligosaccharide fragments, the material produced by solid-phase synthesis (Fig. 5E) was incubated with Cps1D and acetyl-CoA at a donor/acceptor ratio of 1:1. It is important to note that, due to the well-isolated N-acetyl and O-acetyl groups, the material did not have to be purified, and a quantification of the O-acetylation level was possible despite the presence of remaining constituents of the reaction mixture, demonstrating that 1H NMR represents a suitable tool to follow the progress of the reaction. Again, the level of O-acetylation achieved (90%) was in good agreement with published results and is best visualized in the anomeric region of a 1D 1H NMR (Fig. 6G). Altogether, our data show that the pathogen-free in vitro synthesis of nature-identical App1 capsule polymers with defined length is possible using recombinant enzymes.
Table 2.
1H and 13C chemical shifts observed in O-acetylated App1, referenced to DSS
| Spin system/atom | GlcNAc | Gal |
|---|---|---|
| H1 | 4.735 | 5.577 |
| H2 | 3.765 | 3.827 |
| H3 | 3.763 | 3.976 |
| H4 | 4.101 | 3.835 |
| H5 | 3.831 | 4.164 |
| H6a | 4.506 | 3.927 |
| H6b | 3.847 | 3.796 |
| C1 | 103.6 | 98.4 |
| C2 | 57.9 | 71.0 |
| C3 | 75.8 | 71.3 |
| C4 | 77.0 | 71.8 |
| C5 | 75.2 | 72.8 |
| C6 | 66.0 | 69.7 |
| C7 (Ac) | 176.8 | |
| H8 (Ac) | 2.16 | |
| C8 (Ac) | 23.1 | |
| C9 (NAc) | 177.5 | |
| H10 (NAc) | 2.075 | |
| C10 (NAc) | 25.0 | |
| P | −5.46 |
Discussion
App is responsible for huge economic losses; the development of an effective vaccine that can both protect individuals and prevent transmission within the herd is of utmost importance (1). Based on a comprehensive biochemical characterization, this report presents a protocol exploiting the capsule polymerase Cps1B and the O-acetyltransferase Cps1D from A. pleuropneumoniae serotype 1 for the in vitro synthesis of nature-identical oligosaccharides.
Our findings indicate that the TPR domain of Cps1B contributes to a processive elongation mechanism. Previous studies demonstrated that the truncation of the TPR domain changed the oligomerization state of Cps1B from dimeric to monomeric (17). Oligomerization was described as the structural basis for the processivity of various classes of enzymes (16). In contrast, extended polymer-binding sites were shown to mediate processivity in carbohydrate polymerases (14–16, 34). Although this study was focused on the biotechnological use of Cps1B, we performed initial limited proteolysis and surface plasmon resonance experiments to clarify whether the TPR domain is involved in the binding of non-O-acetylated App1 polymer (data not shown). However, the results were inconclusive, presumably due to the large size of the TPR domain and nonspecific binding to charged surfaces in the SPR, respectively. It is also worth noting that the product pools generated by Cps1B-ΔTPR are not strictly Gaussian-shaped, indicating the existence of other processivity-promoting features within the enzyme.
The majority of TagF-like polymerases contain a TPR domain either at the C terminus (e.g. Cps1B) or at the N terminus (e.g. Cps7D (App7)) (17). Regardless of whether this domain mediates processivity through oligomerization or polymer binding, it can be speculated that its deletion could be an ideal strategy to change the elongation mechanism of this entire family of enzymes. Unfortunately, the first attempts to truncate the TPR domain from Cps7D abolished expression (17), indicating that it has a more critical influence on stability and folding when located at the N terminus.
Given its considerable size, the TPR domain likely adopts additional functions in the in vivo context, such as mediating interactions with the ABC transporter of the capsule biosynthesis complex or with sugar-activating enzymes (5). A comprehensive biochemical characterization of this domain is necessary to understand its complex functions in the TagF-like protein family.
In agreement with previous bioinformatics studies (17), we present for the first time experimental evidence that the GT-A folded domain harbors the GlcNAc transferase activity, whereas the TagF-like folded domain transfers Gal, most likely as Gal 1-phosphate. Interestingly, TagF-like enzymes usually transfer polyol- rather than hexose-phosphates (35). Consequently, we searched for similarities between the TagF-like folded domain of Cps1B and other important sugar-phosphate transferases. Examples are (i) the polyisoprenyl-phosphate N-acetyl-aminosugar/hexose-1-phosphate transferases (e.g. WecA, TagO, MraY, and Wbpa (36, 37)) generating the undecaprenyl-PP-sugar primers for the synthesis of diverse oligo- and polysaccharides and (ii) stealth proteins (38), which were shown to synthesize the phosphate containing capsular polysaccharides of N. meningitidis serogroups A and X (11, 18, 39) and the mannose-6-phosphate lysosomal recognition marker (40). Multiple-sequence alignments performed in this study (data not shown) using Clustal Omega (41) showed little sequence identity (9–18%) between the TagF-like domain of Cps1B and the above mentioned phosphotransferases, and no conserved motifs could be found. However, the characteristic motifs of the TagF-like protein family are clearly present, corroborating that Cps1B is indeed part of the TagF-like protein family (17).
We decided to exploit single-domain mutants of Cps1B to reduce the heterogeneity of the product population. This approach was previously utilized by DeAngelis et al. (42) for the hyaluronan synthase from Pasteurella multocida, an enzyme that generates a polymer with a [4)-GlcUA-β(1,3)-GlcNAc-β(1] repeating unit. DeAngelis et al. (42) circulated the reaction mix through an enzyme reactor containing the mutant with active GlcNAc transferase domain until all of the acceptor was elongated by one residue. Subsequently, the mixture was transferred to the second enzyme reactor containing the mutant with GlcUA transferase activity and circulated until the addition of the second residue was complete. Although highly precise, the time required for this procedure increases with chain length (DP20 was produced within 2 days). Rather than producing monodisperse products, our aim was (i) to demonstrate that domain separation combined with solid-phase coupling was possible using Cps1B mutants and (ii) to generate a product pool with a dispersity and chain length that agrees with the material used in glycoconjugate vaccines (10, 12, 14). Consequently, we chose to simplify the experimental setup. Although this led to a considerable increase in dispersity (when compared with DeAngelis et al. (42)), the obtained product pool comprised only 30 product species (ranging between DP20 and DP50), which is less disperse than the material used for vaccines against, for example, N. meningitidis, which contain oligosaccharides ranging from DP10 to DP60 (10, 12, 14, 43). With DP30 being the most prominent oligosaccharide species (see Fig. 5F), the obtained avDP corresponded to the d/a ratio of 30 utilized in the experiment, indicating that an average of 25 repeating units was added to the starting material (pool I, avDP 4.5) and corroborating that the experimental setup successfully prevented processive elongation and offered sufficient size control.
We identified Cps1D as the O-acetyltransferase of App1 and successfully provided proof of principle for its use in biotechnological applications. Future studies will show whether Cps1D can be used in a solid-phase setup together with the Cps1B mutants to further streamline the synthesis protocol.
Glycoconjugate vaccines consisting of capsule polymer conjugated to a carrier protein have been demonstrated to prevent transmission of important human pathogens like H. influenzae, N. meningitidis, and Streptococcus pneumoniae (7). Although experimental glycoconjugate vaccines against App and other porcine pathogens were tested with promising outcomes, the experience with these substances for App prevention is limited (4, 8, 9, 44). The upsides of glycoconjugate vaccines are their extreme efficacy and safety and the fact that they induce immunoglobulin isotype switching (7). The downsides of conventionally produced glycoconjugate vaccines are their specificity for only one serotype and their comparatively expensive production (45). In this context, it is advantageous that the presented protocols exclusively require low-cost laboratory equipment, which makes the process affordable and easy to implement. The use of E. coli safety strains for protein expression and pure chemicals as building blocks for oligomer synthesis avoids biohazard and keeps purification efforts to a minimum. An additional advantage of generating oligosaccharides enzymatically rather than harvesting polymer from pathogen cultures is that conjugatable linkers can be introduced in the priming acceptor (10, 13). The resulting oligosaccharides can be readily coupled to the carrier protein, and the risk of undesired chemical modification of the final antigen, which can occur during linker introduction and activation, is reduced.
As an alternative to in vitro enzymatic synthesis, assembling entire glycoconjugates in engineered E. coli strains has emerged as a promising and potentially cost-effective alternative for vaccine provision (46–48). However, the bacterial enzymatic machinery has to be altered and adapted to each serotype, and it remains to be seen whether these systems will be available for a broad variety of pathogens. For both technologies, a profound understanding of the enzymatic mechanism of Cps1B and Cps1D is of utmost importance, and our study paves the way for the biotechnological use of the enzymes.
A recent publication (49) defined the challenges for future glycoconjugate vaccine development and pointed out that one of the main reasons for failure of clinical trials in the past has been the inadequate understanding of the mechanisms by which the human immune system interacts with these complex antigens. The authors emphasized the importance of developing more suitable animal models and platforms that would allow the precise control of parameters like oligosaccharide length, loading of carrier protein with antigen, and modification of the epitope (among others) (49). The technology presented herein offers a toolkit for altering parameters like chain length and the degree of O-acetylation of the App1 capsular antigen. It could thus be used to study the influence of such parameters on the immune response in pigs as a natural App host organism, omitting the use of a surrogate animal model. This might not only improve our understanding of the as yet poorly understood cell-mediated immune responses against an App infection (1, 50), but can also contribute to a better understanding of the mechanism of action of glycoconjugate vaccines in general.
Experimental procedures
General cloning
The generation of plasmids pcps1B32–1246-His6 (tac) and pcps1B32–858-His6 encoding ΔN31-cps1B-His6 and ΔN31-cps1B-ΔTPR-His6 was described previously (17). Published single amino acid mutations were introduced into ΔN31-cps1B-His6 according to (51) using the primer pairs shown in Table 3. cps1D (GenBankTM accession numbers CP029003.1 and AWG96004.1) was amplified by PCR from heat-inactivated bacterial lysates using the primers given in Table 3 and cloned via restriction-free cloning (52) into the plasmid pΔN37-cslB-His6 (19), replacing the cslB sequence. All constructs used in this study are listed in Table 4.
Table 3.
Primers used in this study
| Primer designation or category | Primer sequence |
|---|---|
| Primer pairs used for generating the underlined mutations in cps1B | |
| D133A/D135A | |
| CL162 | 5′-TTACCTTTATTGCGCCAGCGGATTTTCTTAG-3′ |
| CL163 | 5′-CCCATTCTGTTTGTACGTATTTTAGTCC-3′ |
| H587A | |
| IB06 | 5′-ATTTTTACAGGCGGGTATAACTAAAGATGATTTATCTCAATGGTTCAATAC-3′ |
| IB07 | 5′-GTTATACCCGCCTGTAAAAATATAAATTTTTTACTAAAATCATAATTGTCGCC-3′ |
| H717A | |
| IB08 | 5′-AATATTAGGCGCTGGTGCAAAAATAACTTTATATCCTAAATTTTTAATT-3′ |
| IB09 | 5′-TTGCACCAGCGCCTAATATTGAACCATATTTAAATGAGTTTAACATCCCCC-3′ |
| Primers for the amplification of cps1D | |
| IB30 | 5′-GGTGGTGGTGGTGGTGCTCGAGCTCTGTTTCCATAGTATTTTTACAG-3′ |
| IB31 | 5′-CGAGCACTTCACCAACAAGGACCATAGCATATGAATATTAAAGAAATATATAAAAAGATAAAGAATG-3′ |
| Primer pairs used for generating the underlined mutations in cps1D | |
| H242A | |
| IB32 | 5′-ATGATGCTGCTCCAATTTACGATGTAAACACAGGC-3′ |
| IB33 | 5′-AATTGGAGCAGCATCATCAGTACGAATTTGATTATTTGTTG-3′ |
| W265A | |
| IB34 | 5′-ACCATGTTGCGATTGGATATGGAGCAACAATTTTATCTGG-3′ |
| IB35 | 5′-ATCCAATCGCAACATGGTCGCCTATTTGAATATCTTTAG-3′ |
Table 4.
Constructs used in this study
| Construct | Molecular mass | Source |
|---|---|---|
| kDa | ||
| Cps1B constructs | ||
| ΔN31-Cps1B-His6 | 144.2 | Ref. 17 |
| ΔN31-Cps1B-D133A/D135A-His6 | 144.2 | This study |
| ΔN31-Cps1B-H587A-His6 | 144.2 | This study |
| ΔN31-Cps1B-H717A-His6 | 144.2 | This study |
| ΔN31-Cps1B-H587A/H717A-His6 | 144.2 | This study |
| ΔN31-Cps1B-ΔC388-His6 | 99.3 | Ref. 17 |
| Cps1D constructs | ||
| Cps1D-His6 | 40.9 | This study |
| Cps1D-H242A-His6 | 40.9 | This study |
| Cps1D-W265A-His6 | 40.9 | This study |
Protein expression and purification
Expression and purification of recombinant Cps1B and Cps1D was performed as described (17) with minor modifications: buffers for the affinity chromatography and size-exclusion chromatography of Cps1D contained 300 and 150 mm sodium chloride, respectively.
Enzymatic reactions and analyses via HPLC and PAGE
Reactions shown in Fig. 3A were carried out with 1 μm enzyme in a total volume of 75 μl of assay buffer (20 mm Tris, pH 8, 10 mm MgCl2, 1 mm DTT) containing an 8 mm concentration of each substrate (UDP-Gal and UDP-GlcNAc) and acceptor concentrations resulting in the indicated d/a ratios. Reaction mixtures were incubated at 37 °C overnight. HPLC-AEC analyses were performed on a Prominence UFLC-XR system (Shimadzu) equipped with a CarboPac PA-100 (Thermo Scientific) column as described (17) with minor adjustments: H2O and 1 m NaCl were used as mobile phases M1 and M2, and polymer was separated using a linear gradient of 0–60% M2 over 44 min. Additionally, samples were analyzed by 15 and 25% PAGE according to Ref. 53.
For the experiment shown in Fig. 3 (B and C), 20 and 5 nm concentrations of Cps1B and Cps1B-ΔTPR, respectively, were incubated in a total volume of 900 μl with a d/a ratio of 100:1 at 37 °C. Samples were taken at the indicated time points, snap-frozen, and heat-inactivated at 60 °C for 5 min prior to HPLC-AEC analysis.
On-column assay
Proteins were expressed according to published protocols (17). 250 and 125 ml of E. coli M15 p[Rep4] expression culture of Cps1B single-domain mutants and Cps1B-ΔTPR, respectively, were pelleted and lysed, and protein was coupled to a 1-ml HisTrap column (GE Healthcare). After thoroughly washing with binding (50 mm Tris, pH 8.0, 500 mm NaCl, 20 mm imidazole, 1 mm DTT) and equilibration buffer (20 mm Tris, pH 8.0, 10 mm MgCl2, 1 mm DTT), 5 ml of reaction mixture (equilibration buffer containing 8 mm UDP-GlcNAc and 8 mm UDP-Gal (Carbosynth or Roche Applied Science) and acceptor in a d/a ratio of 30) were circulated with a flow rate of 0.3 ml/min. Fractions were taken as indicated in Figs. 4 and 5. After the experiment was finished, protein was eluted through the addition of elution buffer (500 mm NaCl, 50 mm Tris, pH 8, 500 mm imidazole), and samples were analyzed by SDS-PAGE, Alcian blue/silver-stained PAGE, and HPLC-AEC as described below.
De novo synthesis and purification of App1 polymer
For App1 polymer synthesis, 5 μm Cps1B was incubated in reaction buffer (20 mm Tris, pH 8, 10 mm MgCl2, 1 mm DTT) containing an 8 mm concentration of each donor substrate at 37 °C overnight. Purification was achieved through anion-exchange chromatography (Mono Q 10/100 GL, GE Healthcare) using a sodium chloride gradient (0–150 mm NaCl over 3 column volumes, 150–1,000 mm of NaCl over 28.5 column volumes). Polymer-containing fractions were pooled, dialyzed against water (10,000 molecular weight cut-off, Zellutrans; Roth), and freeze-dried.
Preparation of App1 oligosaccharides
App1 oligosaccharides were generated through mild acidic hydrolysis of long polymer (2.5 mg/ml) in 50 mm acetic acid, pH 4.75, at 80 °C for 6 h. Oligosaccharides were purified by anion-exchange chromatography (Mono Q 10/100 GL, GE Healthcare) using a linear sodium chloride gradient from 0 to 700 mm NaCl over 28.5 column volumes. Fractions were divided into three pools, dialyzed (1,000 molecular weight cut-off, Zellutrans; Roth), and freeze-dried, and oligosaccharides were dephosphorylated with alkaline phosphatase (calf intestinal, New England Biolabs) according to the manufacturer's guidelines. Removal of alkaline phosphatase was achieved with Amicon centrifugal devices (30,000 molecular weight cut-off).
Activity testing of Cps1B by use of a radioactive assay system
Enzymatic activity of the Cps1B domains was analyzed using an adapted version of the radioactive incorporation assay previously described for the capsule polymerase of N. meningitidis serogroups X, A, and L (11, 18, 19). Briefly, reactions were carried out with 0.01 nmol of purified enzyme in a total volume of 20 μl of assay buffer (20 mm Tris, pH 8, 10 mm MgCl2, 5 μg of purified App1 polymer, and 1 mm DTT). The reactions were started by the addition of either 0.267 nmol of UDP-[14C]GlcNAc or 1.45 nmol of UDP-[14C]Gal (both 2.96 kBq, American Radiolabeled Chemicals). Samples were incubated at 37 °C, and 5 μl aliquots were spotted onto Whatman 3 MM Chr paper after 0 and 15 min. Free radioactivity was eluted by descending paper chromatography using a mixture of 30% 1 m ammonium acetate, pH 7.5, and 70% ethanol (v/v). Chromatographically immobile 14C-labeled material was quantified by scintillation counting.
In vitro assay for the determination of Cps1D activity
In a total volume of 100 μl of reaction buffer (50 mm Tris, pH 7.5, 50 mm NaCl, 20% glycerol (w/v)) containing 2 mm dithiobis(2-nitrobenzoic acid) (Calbiochem), 1 mm acetyl-CoA (lithium salt, Sigma-Aldrich) and 0.3 mg/ml purified App1 polysaccharide, the Cps1D reaction was initiated through the addition of 32 pmol of purified enzyme. Absorption was measured continuously at 30 °C using a PowerWave 340 microplate spectrophotometer (BioTek) at 405 nm and half-area 96-well plates (Greiner). The mean was calculated from three repeated measurements using different aliquots of the same enzyme preparation. SDS-PAGE and immunoblotting were performed as described (19).
NMR spectroscopy
All data were recorded on a 600-MHz Avance III HD spectrometer (Bruker Biospin, Rheinstetten, Germany) equipped with a 1H/13C/15N/31P QXI probe. Lyophilized samples were dissolved in D2O (100% D, Armar, Leipzig, Germany) and measured in 5-mm TA tubes (Armar) at 298 K. Spectra were referenced to DSS using a sealed external sample of 2 mm sucrose, 0.5 mm DSS in H2O/D2O (Bruker standard sample for water suppression). The 13C and 31P axis were indirectly referenced using the recommended scaling factors Ξ of 0.251449530 and 0.404808636, respectively, according to IUPAB (54).
1H NMR spectra were typically measured with a spectral width of 20 ppm, 65,536 data points, and 64 transients; 31P NMR spectra were typically measured with a spectral width of 50 ppm, 16,000 data points, and 64 transients. 2D 1H-13C HSQC spectra were collected using the Bruker pulse sequence hsqcedetgpsisp2.2 with 2,048 data points and 16-ppm spectral width for 1H and 230 data points and 100.4-ppm spectral width using 32 scans, a recycle delay of 1.5 s resulting in a measurement time of 3.5 h. 2D 1H-1H TOCSY spectra were measured with 2,048 data points and 8.3-ppm spectral width for 1H and 512 data points and 8.3-ppm spectral width using 4 scans, a mixing time of 80 ms, a recycle delay of 2 s resulting in a measurement time of 1.5 h. 2D 1H-1H COSY spectra were recorded using cosygpppqf with 2,048 data points and 8.3-ppm spectral width for 1H and 128 data points and 8.3-ppm spectral width using 32 scans, a recycle delay of 2 s resulting in a measurement time of 2 h 40 min. 2D 1H-31P HMBC spectra were measured using the pulse sequence hmbclpndqf with 4,096 data points and 10-ppm spectral width for 1H and 64 data points and 30.5-ppm spectral width using 32 scans, a recycle delay of 1.5 s resulting in a measurement time of 1 h 12 min. 2D 1H-1H NOESY spectra were recorded with 2,048 data points and 8.3-ppm spectral width for 1H and 700 data points and 8.3 ppm spectral width using 32 scans, a mixing time of 120 ms, a recycle delay of 1 s resulting in a measurement time of 8 h 20 min. 2D 1H-13C HMBC spectra were collected using the pulse sequence hmbclpndqf with 4,096 data points and 20-ppm spectral width for 1H and 512 data points and 222-ppm spectral width using 64 scans, optimized for a JCH long-range coupling of 8 Hz, a recycle delay of 2 s resulting in a measurement time of 20.5 h. Data were processed with Topspin 3.6 (Bruker Biospin), and chemical shift assignment was achieved using the software Sparky 3.115 (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco).
Data availability
All data are contained within the paper. NMR chemical shifts have additionally been deposited in the glycosciences.de database (LinucsID 3258 and 29812).
Author contributions
I. B. and T. F. conceptualization; I. B., C. L., M. S., and T. F. formal analysis; I. B., C. L., J. I. F., R. G.-S., M. S., and T. F. validation; I. B., C. L., M. S., and T. F. investigation; I. B., M. S., and T. F. visualization; I. B., C. L., M. S., and T. F. methodology; I. B., M. S., and T. F. writing-original draft; I. B., C. L., J. I. F., R. G.-S., M. S., and T. F. writing-review and editing; C. L. and T. F. supervision; R. G.-S. resources; R. G.-S. and T. F. funding acquisition; T. F. project administration.
Acknowledgments
We thank Monika Berger and Andrea Bethe (both from Hannover Medical School) for excellent technical assistance, Jochen Meens (University of Veterinary Medicine Hannover) for providing App lysates, Sonja Kunstmann (Technical University of Denmark) for performing initial surface plasmon resonance experiments, and Thomas Lütteke (GIP GmbH) for help depositing the NMR data into the Glycosciences.de database.
This study was supported by the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) Project 412824531 and LOM (impact-oriented funding) funds to the Institute of Clinical Biochemistry from Medizinische Hochschule Hannover (Hannover Medical School). C. L., T. F., and R. G.-S. have submitted patent applications in the field of vaccine development.
- App
- Actinobacillus pleuropneumoniae
- avDP
- average degree of polymerization
- AEC
- anion-exchange chromatography
- Cps1B
- capsule polymerase of App serotype 1
- COSY
- correlation spectroscopy
- Cps1D
- O-acetyltransferase of App serotype 1
- HMBC
- heteronuclear multiple bond correlation
- HSQC
- heteronuclear single quantum coherence
- TagF
- wall teichoic acid synthase from Staphylococcus epidermis
- TOCSY
- total correlated spectroscopy
- TPR
- tetratricopeptide repeat
- DP
- degree of polymerization
- d/a
- donor/acceptor
- 1D and 2D
- one- and two-dimensional, respectively
- DSS
- 2,2-dimethyl-2-silapentanesulfonic acid
- F
- fraction.
References
- 1. Sassu E. L., Bossé J. T., Tobias T. J., Gottschalk M., Langford P. R., and Hennig-Pauka I. (2018) Update on Actinobacillus pleuropneumoniae—knowledge, gaps and challenges. Transbound. Emerg. Dis. 65, 72–90 10.1111/tbed.12739 [DOI] [PubMed] [Google Scholar]
- 2. Perry M. B., Altman E., Brisson J. R., Beynon L. M., and Richards J. C. (1990) Structural characteristics of the antigenic capsular polysaccharides and lipopolysaccharides involved in the serological classification of Actinobacillus (Haemophilus) pleuropneumoniae strains. Serodiagn. Immunother. Infect. Dis. 4, 299–308 [Google Scholar]
- 3. Bossé J. T., Li Y., Fernandez Crespo R., Lacouture S., Gottschalk M., Sárközi R., Fodor L., Casas Amoribieta M., Angen Ø Nedbalcova K, Holden M. T. G., Maskell D. J., Tucker A. W., Wren B. W., Rycroft A. N., Langford P. R., and BRaDP1T consortium. (2018) Comparative sequence analysis of the capsular polysaccharide loci of Actinobacillus pleuropneumoniae serovars 1–18, and development of two multiplex PCRs for comprehensive capsule typing. Vet. Microbiol. 220, 83–89 10.1016/j.vetmic.2018.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dubreuil J. D., Jacques M., Mittal K. R., and Gottschalk M. (2000) Actinobacillus pleuropneumoniae surface polysaccharides: their role in diagnosis and immunogenicity. Anim. Heal. Res. Rev. 1, 73–93 10.1017/s1466252300000074 [DOI] [PubMed] [Google Scholar]
- 5. Willis L. M., and Whitfield C. (2013) Structure, biosynthesis, and function of bacterial capsular polysaccharides synthesized by ABC transporter-dependent pathways. Carbohydr. Res. 378, 35–44 10.1016/j.carres.2013.05.007 [DOI] [PubMed] [Google Scholar]
- 6. Loera-Muro A., and Angulo C. (2018) New trends in innovative vaccine development against Actinobacillus pleuropneumoniae. Vet. Microbiol. 217, 66–75 10.1016/j.vetmic.2018.02.028 [DOI] [PubMed] [Google Scholar]
- 7. Vella M., and Pace D. (2015) Glycoconjugate vaccines: an update. Expert Opin. Biol. Ther. 15, 529–546 10.1517/14712598.2015.993375 [DOI] [PubMed] [Google Scholar]
- 8. Byrd W., Harmon B. G., and Kadis S. (1992) Protective efficacy of conjugate vaccines against experimental challenge with porcine Actinobacillus pleuropneumoniae. Vet. Immunol. Immunopathol. 34, 307–324 10.1016/0165-2427(92)90172-m [DOI] [PubMed] [Google Scholar]
- 9. Byrd W., and Kadis S. (1992) Preparation, characterization, and immunogenicity of conjugate vaccines directed against Actinobacillus pleuropneumoniae virulence determinants. Infect. Immun. 60, 3042–3051 10.1128/iai.60.8.3042-3051.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oldrini D., Fiebig T., Romano M. R., Proietti D., Berger M., Tontini M., De Ricco R., Santini L., Morelli L., Lay L., Gerardy-Schahn R., Berti F., and Adamo R. (2018) Combined chemical synthesis and tailored enzymatic elongation provide fully synthetic and conjugation-ready Neisseria meningitidis serogroup X vaccine antigens. ACS Chem. Biol. 13, 984–994 10.1021/acschembio.7b01057 [DOI] [PubMed] [Google Scholar]
- 11. Fiebig T., Freiberger F., Pinto V., Romano M. R., Black A., Litschko C., Bethe A., Yashunsky D., Adamo R., Nikolaev A., Berti F., and Gerardy-Schahn R. (2014) Molecular cloning and functional characterization of components of the capsule biosynthesis complex of Neisseria meningitidis serogroup A: toward in vitro vaccine production. J. Biol. Chem. 289, 19395–19407 10.1074/jbc.M114.575142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fiebig T., Romano M. R., Oldrini D., Adamo R., Tontini M., Brogioni B., Santini L., Berger M., Costantino P., Berti F., and Gerardy-Schahn R. (2016) An efficient cell free enzyme-based total synthesis of a meningococcal vaccine candidate. NPJ Vaccines 1, 16017 10.1038/npjvaccines.2016.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCarthy P. C., Saksena R., Peterson D. C., Lee C.-H., An Y., Cipollo J. F., and Vann W. F. (2013) Chemoenzymatic synthesis of immunogenic meningococcal group C polysialic acid-tetanus Hc fragment glycoconjugates. Glycoconj. J. 30, 857–870 10.1007/s10719-013-9490-x [DOI] [PubMed] [Google Scholar]
- 14. Fiebig T., Litschko C., Freiberger F., Bethe A., Berger M., and Gerardy-Schahn R. (2018) Efficient solid-phase synthesis of meningococcal capsular oligosaccharides enables simple and fast chemoenzymatic vaccine production. J. Biol. Chem. 293, 953–962 10.1074/jbc.RA117.000488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Keys T. G., Fuchs H. L. S., Ehrit J., Alves J., Freiberger F., and Gerardy-Schahn R. (2014) Engineering the product profile of a polysialyltransferase. Nat. Chem. Biol. 10, 437–442 10.1038/nchembio.1501 [DOI] [PubMed] [Google Scholar]
- 16. Yakovlieva L., and Walvoort M. T. C. (2020) Processivity in bacterial glycosyltransferases. ACS Chem. Biol. 15, 3–16 10.1021/acschembio.9b00619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Litschko C., Oldrini D., Budde I., Berger M., Meens J., Gerardy-Schahn R., Berti F., Schubert M., and Fiebig T. (2018) A new family of capsule polymerases generates teichoic acid-like capsule polymers in Gram-negative pathogens. MBio 9, e00641–18 10.1128/mBio.00641-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fiebig T., Berti F., Freiberger F., Pinto V., Claus H., Romano M. R., Proietti D., Brogioni B., Stummeyer K., Berger M., Vogel U., Costantino P., and Gerardy-Schahn R. (2014) Functional expression of the capsule polymerase of Neisseria meningitidis serogroup X: a new perspective for vaccine development. Glycobiology 24, 150–158 10.1093/glycob/cwt102 [DOI] [PubMed] [Google Scholar]
- 19. Litschko C., Romano M. R., Pinto V., Claus H., Vogel U., Berti F., Gerardy-Schahn R., and Fiebig T. (2015) The capsule polymerase CslB of Neisseria meningitidis serogroup L catalyzes the synthesis of a complex trimeric repeating unit comprising glycosidic and phosphodiester linkages. J. Biol. Chem. 290, 24355–24366 10.1074/jbc.M115.678094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jing W., and DeAngelis P. L. (2004) Synchronized chemoenzymatic synthesis of monodisperse hyaluronan polymers. J. Biol. Chem. 279, 42345–42349 10.1074/jbc.M402744200 [DOI] [PubMed] [Google Scholar]
- 21. Altman E., Brisson J. R., and Perry M. B. (1986) Structural studies of the capsular polysaccharide from Haemophilus pleuropneumoniae serotype 1. Biochem. Cell Biol. 64, 707–716 10.1139/o86-097 [DOI] [PubMed] [Google Scholar]
- 22. Lovering A. L., Lin L. Y.-C., Sewell E. W., Spreter T., Brown E. D., and Strynadka N. C. J. (2010) Structure of the bacterial teichoic acid polymerase TagF provides insights into membrane association and catalysis. Nat. Struct. Mol. Biol. 17, 582–589 10.1038/nsmb.1819 [DOI] [PubMed] [Google Scholar]
- 23. D'Andrea L. D., and Regan L. (2003) TPR proteins: the versatile helix. Trends Biochem. Sci. 28, 655–662 10.1016/j.tibs.2003.10.007 [DOI] [PubMed] [Google Scholar]
- 24. Ricci S., Bardotti A., D'Ascenzi S., and Ravenscroft N. (2001) Development of a new method for the quantitative analysis of the extracellular polysaccharide of Neisseria meningitidis serogroup A by use of high-performance anion-exchange chromatography with pulsed-amperometric detection. Vaccine 19, 1989–1997 10.1016/S0264-410X(00)00427-8 [DOI] [PubMed] [Google Scholar]
- 25. Berti F., Romano M. R., Micoli F., Pinto V., Cappelletti E., Gavini M., Proietti D., Pluschke G., MacLennan C. A., and Costantino P. (2012) Relative stability of meningococcal serogroup A and X polysaccharides. Vaccine 30, 6409–6415 10.1016/j.vaccine.2012.08.021 [DOI] [PubMed] [Google Scholar]
- 26. Odonmazig P., Ebringerová A., Machová E., and Alföldi J. (1994) Structural and molecular properties of the arabinogalactan isolated from Mongolian larchwood (Larix dahurica L.). Carbohydr. Res. 252, 317–324 10.1016/0008-6215(94)90028-0 [DOI] [PubMed] [Google Scholar]
- 27. Loss A., and Lütteke T. (2015) Using NMR data on GLYCOSCIENCES.de. Methods Mol. Biol. 1273, 87–95 10.1007/978-1-4939-2343-4_6 [DOI] [PubMed] [Google Scholar]
- 28. Lütteke T., Bohne-Lang A., Loss A., Goetz T., Frank M., and von der Lieth C.-W. (2006) GLYCOSCIENCES.de: an Internet portal to support glycomics and glycobiology research. Glycobiology 16, 71R–81R 10.1093/glycob/cwj049 [DOI] [PubMed] [Google Scholar]
- 29. Lundborg M., and Widmalm G. (2011) Structural analysis of glycans by NMR chemical shift prediction. Anal. Chem. 83, 1514–1517 10.1021/ac1032534 [DOI] [PubMed] [Google Scholar]
- 30. Xu Z., Chen X., Li L., Li T., Wang S., Chen H., and Zhou R. (2010) Comparative genomic characterization of Actinobacillus pleuropneumoniae. J. Bacteriol. 192, 5625–5636 10.1128/JB.00535-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. E. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schulz E. C., Bergfeld A. K., Ficner R., and Mühlenhoff M. (2011) Crystal structure analysis of the polysialic acid specific O-acetyltransferase NeuO. PLoS ONE 6, e17403 10.1371/journal.pone.0017403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bergfeld A. K., Claus H., Lorenzen N. K., Spielmann F., Vogel U., and Mühlenhoff M. (2009) The polysialic acid-specific O-acetyltransferase OatC from Neisseria meningitidis serogroup C evolved apart from other bacterial sialate O-acetyltransferases. J. Biol. Chem. 284, 6–16 10.1074/jbc.M807518200 [DOI] [PubMed] [Google Scholar]
- 34. Forsee W. T., Cartee R. T., and Yother J. (2006) Role of the carbohydrate binding site of the Streptococcus pneumoniae capsular polysaccharide type 3 synthase in the transition from oligosaccharide to polysaccharide synthesis. J. Biol. Chem. 281, 6283–6289 10.1074/jbc.M511124200 [DOI] [PubMed] [Google Scholar]
- 35. Fitzgerald S. N., and Foster T. J. (2000) Molecular analysis of the tagF gene, encoding CDP-glycerol: poly(glycerophosphate) glycerophosphotransferase of Staphylococcus epidermidis ATCC 14990. J. Bacteriol. 182, 1046–1052 10.1128/JB.182.4.1046-1052.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liston S. D., Mann E., and Whitfield C. (2017) Glycolipid substrates for ABC transporters required for the assembly of bacterial cell-envelope and cell-surface glycoconjugates. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1394–1403 10.1016/j.bbalip.2016.10.008 [DOI] [PubMed] [Google Scholar]
- 37. Tytgat H. L. P., and Lebeer S. (2014) The sweet tooth of bacteria: common themes in bacterial glycoconjugates. Microbiol. Mol. Biol. Rev. 78, 372–417 10.1128/MMBR.00007-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sperisen P., Schmid C. D., Bucher P., and Zilian O. (2005) Stealth proteins: in silico identification of a novel protein family rendering bacterial pathogens invisible to host immune defense. PLoS Comput. Biol. 1, e63 10.1371/journal.pcbi.0010063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Muindi K. M., McCarthy P. C., Wang T., Vionnet J., Battistel M., Jankowska E., and Vann W. F. (2014) Characterization of the meningococcal serogroup X capsule N-acetylglucosamine-1-phosphotransferase. Glycobiology 24, 139–149 10.1093/glycob/cwt091 [DOI] [PubMed] [Google Scholar]
- 40. Tiede S., Storch S., Lübke T., Henrissat B., Bargal R., Raas-Rothschild A., and Braulke T. (2005) Mucolipidosis II is caused by mutations in GNPTA encoding the α/β GlcNAc-1-phosphotransferase. Nat. Med. 11, 1109–1112 10.1038/nm1305 [DOI] [PubMed] [Google Scholar]
- 41. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. DeAngelis P. L., Oatman L. C., and Gay D. F. (2003) Rapid chemoenzymatic synthesis of monodisperse hyaluronan oligosaccharides with immobilized enzyme reactors. J. Biol. Chem. 278, 35199–35203 10.1074/jbc.M306431200 [DOI] [PubMed] [Google Scholar]
- 43. Bröker M., Dull P. M., Rappuoli R., and Costantino P. (2009) Chemistry of a new investigational quadrivalent meningococcal conjugate vaccine that is immunogenic at all ages. Vaccine 27, 5574–5580 10.1016/j.vaccine.2009.07.036 [DOI] [PubMed] [Google Scholar]
- 44. Goyette-Desjardins G., Calzas C., Shiao T. C., Neubauer A., Kempker J., Roy R., Gottschalk M., and Segura M. (2016) Protection against Streptococcus suis serotype 2 infection using a capsular polysaccharide glycoconjugate vaccine. Infect. Immun. 84, 2059–2075 10.1128/IAI.00139-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Plotkin S., Robinson J. M., Cunningham G., Iqbal R., and Larsen S. (2017) The complexity and cost of vaccine manufacturing: an overview. Vaccine 35, 4064–4071 10.1016/j.vaccine.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tytgat H. L. P., Lin C. W., Levasseur M. D., Tomek M. B., Rutschmann C., Mock J., Liebscher N., Terasaka N., Azuma Y., Wetter M., Bachmann M. F., Hilvert D., Aebi M., and Keys T. G. (2019) Cytoplasmic glycoengineering enables biosynthesis of nanoscale glycoprotein assemblies. Nat. Commun. 10, 5403 10.1038/s41467-019-13283-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harding C. M., and Feldman M. F. (2019) Glycoengineering bioconjugate vaccines, therapeutics, and diagnostics in E. coli. Glycobiology 29, 519–529 10.1093/glycob/cwz031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Micoli F., Del Bino L., Alfini R., Carboni F., Romano M. R., and Adamo R. (2019) Glycoconjugate vaccines: current approaches towards faster vaccine design. Expert Rev. Vaccines 18, 881–895 10.1080/14760584.2019.1657012 [DOI] [PubMed] [Google Scholar]
- 49. Avci F., Berti F., Dull P., Hennessey J., Pavliak V., Prasad A. K., Vann W., Wacker M., and Marcq O. (2019) Glycoconjugates: what it would take to master these well-known yet little-understood immunogens for vaccine development. mSphere 4, e00520–19 10.1128/mSphere.00520-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ramjeet M., Deslandes V., Gouré J., and Jacques M. (2008) Actinobacillus pleuropneumoniae vaccines: from bacterins to new insights into vaccination strategies. Anim. Health Res. Rev. 9, 25–45 10.1017/S1466252307001338 [DOI] [PubMed] [Google Scholar]
- 51. Liu H., and Naismith J. H. (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91 10.1186/1472-6750-8-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bond S. R., and Naus C. C. (2012) RF-Cloning.org: an online tool for the design of restriction-free cloning projects. Nucleic Acids Res. 40, W209–W213 10.1093/nar/gks396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Min H., and Cowman M. K. (1986) Combined alcian blue and silver staining of glycosaminoglycans in polyacrylamide gels: application to electrophoretic analysis of molecular weight distribution. Anal. Biochem. 155, 275–285 10.1016/0003-2697(86)90437-9 [DOI] [PubMed] [Google Scholar]
- 54. Markley J. L., Bax A., Arata Y., Hilbers C. W., Kaptein R., Sykes B. D., Wright P. E., and Wüthrich K. (1998) Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB Inter-Union Task Group on the Standardization of Data Bases of Protein and Nucleic Acid Structures Determined by NMR Spectroscopy. J. Biomol. NMR 12, 1–23 10.1023/A:1008290618449 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the paper. NMR chemical shifts have additionally been deposited in the glycosciences.de database (LinucsID 3258 and 29812).