Abstract
The Mycobacterium tuberculosis virulence factor EsxA and its chaperone EsxB are secreted as a heterodimer (EsxA:B) and are crucial for mycobacterial escape from phagosomes and cytosolic translocation. Current findings support the idea that for EsxA to interact with host membranes, EsxA must dissociate from EsxB at low pH. However, the molecular mechanism by which the EsxA:B heterodimer separates is not clear. In the present study, using liposome-leakage and cytotoxicity assays, LC-MS/MS–based proteomics, and CCF-4 FRET analysis, we obtained evidence that the Nα-acetylation of the Thr-2 residue on EsxA, a post-translational modification that is present in mycobacteria but absent in Escherichia coli, is required for the EsxA:B separation. Substitutions at Thr-2 that precluded Nα-acetylation inhibited the heterodimer separation and hence prevented EsxA from interacting with the host membrane, resulting in attenuated mycobacterial cytosolic translocation and virulence. Molecular dynamics simulations revealed that at low pH, the Nα-acetylated Thr-2 makes direct and frequent “bind-and-release” contacts with EsxB, which generates a force that pulls EsxB away from EsxA. In summary, our findings provide evidence that the Nα-acetylation at Thr-2 of EsxA facilitates dissociation of the EsxA:B heterodimer required for EsxA membrane permeabilization and mycobacterial cytosolic translocation and virulence.
Keywords: Mycobacterium tuberculosis, bacterial pathogenesis, virulence factor, post-translational modification (PTM), membrane, protein-protein interaction, chaperone, ESAT-6, EsxA, Nα-acetylation
Introduction
Mycobacterium tuberculosis (Mtb)3 is the causative agent for tuberculosis, one of the leading infectious diseases in the world with 10 million people falling ill in 2017, and ∼1.6 million deaths (1, 2). It is believed that after the Mtb-containing aerosolized droplets are inhaled into the lung, Mtb is encountered by alveolar macrophages and internalized into the phagosome, where Mtb manages to survive through arresting phagosome maturation, including inhibition of vATPase-mediated acidification (3–6). Recent compelling evidence supports that Mtb penetrates the phagosome and translocates into the cytosol (termed cytosolic translocation), where Mtb replicates and undergoes cell-to-cell spreading (7). The ability of Mtb to arrest phagosome maturation and to translocate from the phagosome to the cytosol has been attributed, at least in part, to the type VII secretion system, named ESX-1 and the secreted virulence factors EsxA (ESAT-6) and EsxB (CFP-10). The Mtb mutants with either gene deletions or defects in secretion of EsxA and/or EsxB were not able to translocate into the cytosol and showed significant reduction in host-cell lysis and cell-to-cell spreading (7–19).
In our previous studies, we have found that Mtb EsxA exhibits a unique membrane-permeabilizing activity that is not present in the homologous EsxA ortholog from nonpathogenic Mycobacterium smegmatis (Ms) (20). EsxA undergoes pH-dependent conformational changes, inserts into the membrane, and forms a membrane-spanning complex (21). The essential role of EsxA membrane-permeabilizing activity in phagosome rupture and cytosolic translocation is further confirmed by a recent study in which single-residue mutations at Gln-5 of EsxA up- or down-regulated the membrane-permeabilizing activity and consequently up- or down-regulated mycobacterial cytosolic translocation and virulence in cultured cells and in zebra fish (22).
The genes encoding EsxA and EsxB are located in the same operon within the ESX-1 locus. EsxA and EsxB are co-expressed and co-secreted as a heterodimer (23). Our earlier study has demonstrated that EsxA, but not EsxB, has the membrane-permeabilizing activity, and EsxB is believed to function as a chaperone (20). Current studies support a model that the heterodimer is dissociated at low pH to allow EsxA to penetrate the membranes (24). However, the data regarding to the heterodimer dissociation are conflicting. The native heterodimer extracted from Mtb culture filtrate was found to be dissociated at low pH (24). Surprisingly, however, the studies using the recombinant proteins prepared from Escherichia coli suggest that the heterodimer was not dissociated by acidification. This is evidenced by one of our earlier studies that the heterodimer prepared from E. coli (hereafter termed “Ec-heterodimer”) was inactive in membrane disruption. In the absence of lipid membranes, EsxA formed aggregates in the acidic solution because of increased solvent-exposed hydrophobicity. In contrast, the heterodimer showed little aggregation at pH 4.0, suggesting that EsxB remains bound to EsxA at low pH and prevents EsxA from forming aggregates, which otherwise would be observed if EsxA was released from EsxB (20). Our data are consistent with an earlier CD analysis showing that the Ec-heterodimer is not dissociated at low pH (25).
We hypothesized that the mycobacteria-produced proteins contain unique features (e.g. post-translational modifications) that are required for heterodimer dissociation at low pH. In line with this hypothesis, the native EsxA protein isolated from the culture filtrate of Mtb was displayed as multiple spots in 2-D SDS-PAGE, and some of the spots contained a Nα-acetylation at residue Thr-2 (26). Moreover, the heterodimer produced from a Ms strain was found to have a Nα-acetylation on the Thr-2 residue of EsxA (27). Interestingly, EsxB preferred to bind the nonacetylated EsxA, but not the acetylated form in a 2-D overlay assay (26). Deletion of the Nα-acetyltransferase in Mycobacterium marinum (Mm) disrupted the homeostasis of EsxA Nα-acetylation and attenuated the virulence (28). Together, these studies suggest that the Nα-acetylation of EsxA plays an important role in mycobacterial virulence through facilitating heterodimer dissociation at low pH. In the present study, we have obtained the evidence showing that the Nα-acetylation at Thr-2 of EsxA is required for EsxA membrane permeabilization, mycobacterial cytosolic translocation and virulence through facilitating heterodimer dissociation.
Results
The Ms-produced Mtb heterodimer, but not Ec-produced heterodimer, disrupted liposomal membrane at low pH
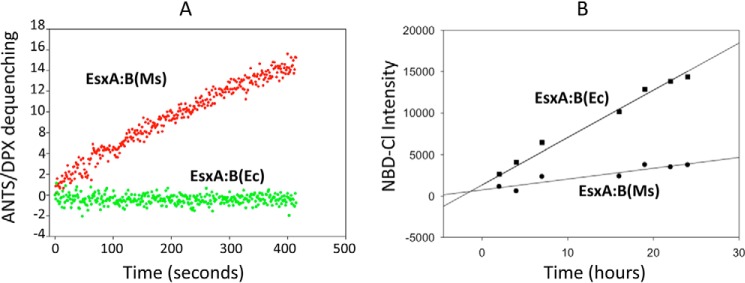
We hypothesized that Ms-heterodimer, but not Ec-heterodimer, dissociates at low pH and permeabilizes the liposomal membrane. The membrane-permeabilizing activity of Ms-heterodimer and Ec-heterodimer was tested with the ANTS/DPX fluorescence dequenching assay. As expected, Ms-heterodimer permeabilized the membrane at low pH, whereas Ec-heterodimer was not active (Fig. 1A). 4-Chloro-7-nitrobenzofurazan (NBD-Cl) only reacts with free N-terminal α-amino group in nonacetylated proteins and emits fluorescence, but it does not react with Nα-acetylated proteins because of a lack of free N-terminal amino group. Thus, we used NBD-Cl to test the states of Nα-acetylation for Ms-heterodimer and Ec-heterodimer. As expected, the Ms-heterodimer exhibited a significantly lower NBD-Cl fluorescence, compared with Ec-heterodimer, indicating that Ms-heterodimer, but not Ec-heterodimer, is Nα-acetylated (Fig. 1B).
Figure 1.
The EsxA:B heterodimer produced in Ms, but not in E. coli, permeabilized the liposomes at low pH, implicating the role of Nα-acetylation in heterodimer dissociation. A, the heterodimers EsxA:B purified from M. smegmatis (Ms) or E. coli (Ec) were tested in three independent experiments with ANTS/DPX dequenching assay at pH 4.0. B, equal amounts of the heterodimer EsxA:B(Ms) or EsxA:B(Ec) were incubated with NBD-Cl at room temperature. NBD-Cl is a fluorescent dye that only reacts with a free N-terminal site without Nα-acetylation was recorded at 535 nm (excitation at 480 nm) and plotted as a function of time.
The mutations at Thr-2 abolished the membrane-permeabilizing activity of the Ms heterodimers through blocking separation of EsxA and EsxB
The Gln and Ala residues have been used to functionally mimic acetylation of amino group of an internal Lys residue, whereas an Arg serves as a nonacetylated control (29). Thus, we generated T2A, T2Q, and T2R mutations and tested the effects of these mutations on the heterodimer membrane–permeabilizing activity.
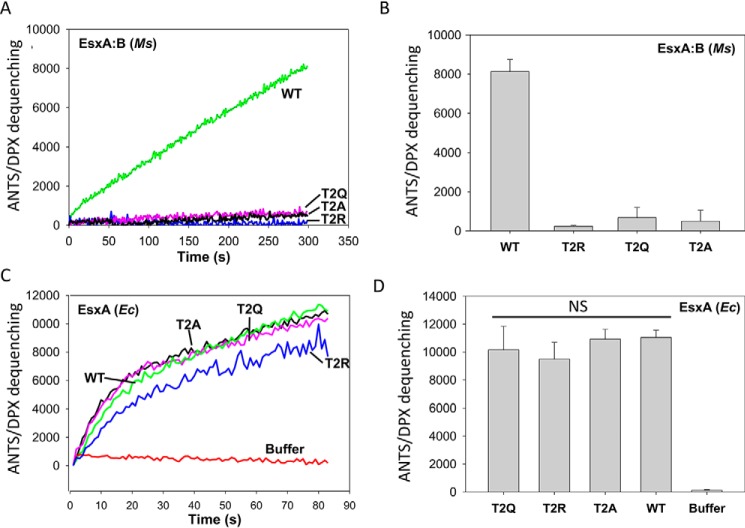
Unexpectedly, all of the mutations abolished the Ms-heterodimer's membrane-permeabilizing activity (Fig. 2, A and B). The result suggests that either the mutations blocked the heterodimer separation or abolished the EsxA membrane-permeabilizing activity. To test whether the mutations abolished the membrane-permeabilizing activity, we purified the EsxA proteins containing the same mutations from E. coli and applied them to ANTS/DPX dequenching assay. The result showed that the mutations did not affect EsxA membrane-permeabilizing activity (Fig. 2, C and D), suggesting that the mutations at Thr-2 blocked the heterodimer separation at low pH.
Figure 2.
The mutations at Thr-2 of EsxA diminished the membrane-permeabilizing activity of EsxA:B heterodimer. A, the EsxA:B heterodimer proteins (WT and the mutants carrying mutations T2A, T2Q, and T2R) were purified from Ms. The membrane-permeabilizing activity of the purified heterodimer proteins was tested by ANTS/DPX fluorescence dequenching assay. The representative ANTS/DPX fluorescent dequenching curves were shown. B, the average end-point fluorescence intensities from at least three independent experiments were calculated. The results represent the averages of three replicates, and error bars represent S.D. C, the EsxA proteins (WT and T2A, T2Q, and T2R mutants) were purified from Ec. The membrane-permeabilizing activity of the Ec-EsxA proteins purified from Ec was tested by ANTS/PDX assay. Representative curves are shown. D, the average end-point fluorescence intensities from at least three independent experiments were calculated, with error bars denoting S.D. NS, not significant.
The mutations that block EsxA and EsxB separation do not have Nα-acetylation
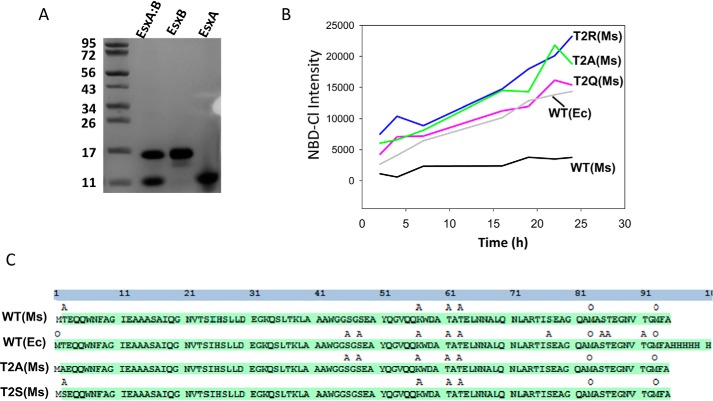
To validate the acetylation state of EsxA WT and the mutants, we developed a protocol to isolate EsxA and EsxB from the Ms-heterodimer proteins (Fig. 3A). Then the presence of Nα-acetylation on the EsxA proteins was measured by NBD-Cl (Fig. 3B). Consistent with the results in Fig. 1B, Ec-EsxA(WT) had a significantly higher fluorescence signal than Ms-EsxA(WT). Similar to Ec-EsxA(WT), the Ms-EsxA mutants (T2A, T2Q, and T2R) emitted significantly higher fluorescence signals than Ms-EsxA(WT), suggesting that the Ms-EsxA mutants were not Nα-acetylated. Next, we applied Ms-EsxA(WT), Ms-EsxA(T2A), and Ec-EsxA(WT) to LC-MS/MS to further confirm the Nα-acetylation states. In addition, Ms-EsxA(T2S), a mutant equivalent to WT, was included in the LC-MS/MS analysis. The results showed that both Ms-EsxA(WT) and Ms-EsxA(T2S) had the first Met residue removed and the second residue (either Thr-2 or Ser2) acetylated (Fig. 3C). Although Ms-EsxA(T2A) had the first Met residue removed, the second Ala residue was not acetylated. The Ec-EsxA(WT) still has the first Met residue. Interestingly, the LC-MS/MS detected multiple acetylation and oxygenation modifications in the internal sequences of both Ms-EsxA and Ec-EsxA, and the roles of these modifications are currently unknown.
Figure 3.
Detection of Nα-acetylation of EsxA by NBD-Cl and MS. A, the EsxA:B heterodimer purified from Ms was separated by 6 m guanidine, after which EsxA and EsxB were purified separately via nickel affinity as described under “Experimental procedures.” B, the indicated EsxA proteins, purified from Ms, were incubated with NBD-Cl. At the indicated times, the fluorescence intensity of NBD-Cl was measured. C, the indicated EsxA proteins, purified from Ms or Ec as indicated, were analyzed via LC-MS/MS to identify the post-translational modifications. The residues with acetylation are labeled as A, and the residues with oxidation are labeled as O.
EsxB preferred to bind nonacetylated EsxA to inhibit the membrane-permeabilizing activity
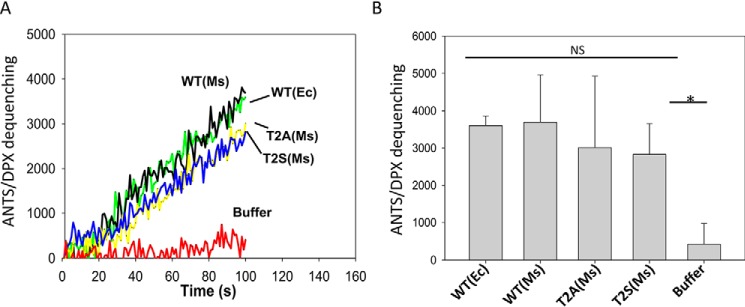
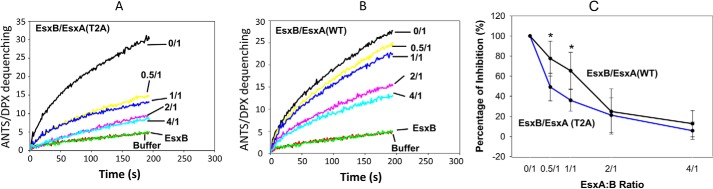
An earlier study has shown that EsxB preferred to bind nonacetylated EsxA rather than acetylated EsxA in a 2-D overlay assay (26). Thus, we hypothesize that EsxB will prefer to inhibit the membrane-permeabilizing activity of the nonacetylated Ms-EsxA(T2A) than that of the acetylated Ms-EsxA(WT). First, we tested and confirmed that the proteins Ms-EsxA(WT) and Ms-EsxA(T2A) that were isolated from the heterodimers had similar membrane-permeabilizing activity to Ec-EsxA(WT), which once again confirms that the states of Nα-acetylation do not affect membrane-permeabilizing activity (Fig. 4, A and B). Then Ms-EsxA(WT) and Ms-EsxA(T2B) were incubated with EsxB at a series of EsxB/EsxA molar ratios. As expected, EsxB preferred to inhibit Ms-EsxA(T2A) rather than Ms-EsxA(WT) in membrane permeabilization, especially at the lower EsxB/EsxA ratios (Fig. 5, A–C).
Figure 4.
The Nα-acetylation of EsxA did not affect the membrane-permeabilizing activity of EsxA. A, the indicated Ms-EsxA proteins isolated from the Ms-heterodimer were tested for membrane-permeabilizing activity using ANTS/DPX assay. The Ec-EsxA (WT) protein, purified from Ec, was used as a control. The representative curves from at least three independent experiments are shown. B, the average end-point fluorescence intensity from at least three independent experiments was calculated and is shown (p < 0.05).
Figure 5.
EsxB preferentially inhibited nonacetylated EsxA(T2A) over its Nα-acetylated counterpart. The Ms-produced EsxA(WT) and EsxA(T2A) proteins were incubated with various concentrations of EsxB as the indicated molar ratios. The mixtures were tested in triplicate by ANTS/DPX assay for membrane-permeabilizing activity. The representative curves are shown in A and B, respectively. The relative inhibition from at least three independent experiments for a total n = 9 was summarized in C. For EsxA:B 0.5:1.0 ratio, p = 0.0015 and for EsxA:B 1:1 ratio p = 0.0012. Error bars represent S.D.
The mutations without Nα-acetylation attenuated mycobacterial virulence and inhibited cytosolic translocation
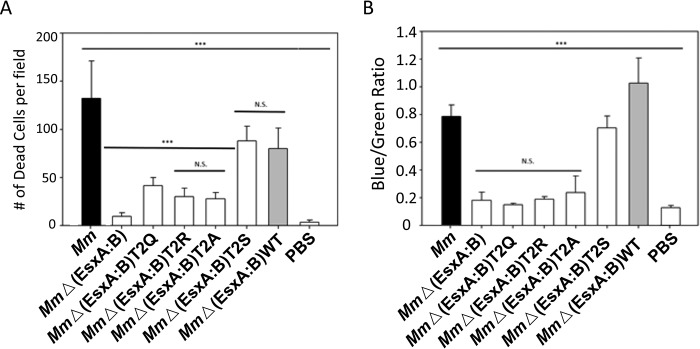
Here, we investigated the effects of the T2X mutations in mycobacterial pathogenesis. The genes carrying T2X mutations were expressed in the MmΔEsxA:B strain, in which the endogenous esxB-esxA operon was deleted. We found that the T2X mutations did not affect the expression and secretion of EsxA and EsxB in the Mm strains (Fig. S1). As expected, the Mm strains carrying the nonacetylated mutations T2A, T2Q, and T2R had a significantly lower cytotoxicity than the strain carrying the acetylated mutation T2S (Fig. 6A). Mm WT and MmΔEsxA:B were used as the positive control and negative control, respectively. Furthermore, using the previously established CCF4-FRET assay, we found that the nonacetylated mutations T2A, T2Q, and T2R abolished mycobacterial cytosolic translocation, whereas T2S maintained a similar activity as the WT (Fig. 6B). The data are consistent to a previous report indicating that deletion of a Nα-acetyltransferase in Mm disrupted the homeostasis of EsxA acetylation and attenuated the virulence (28).
Figure 6.
The non-Nα-acetylate EsxA diminished mycobacterial virulence and cytosolic translocation in macrophages. A, RAW263.4 cells were infected with the indicated Mm strains at MOI of 10. The cytotoxicity was measured by using the live/dead assay. Dead cells were counted in random fields (***, n = 22, p < 0.0001). B, mycobacterial cytosolic translocation was monitored by using CCF4-AM as a FRET reporter. The blue/green ratio was measured by comparing emissions at 450 and 530 nm with excitation at 409 nm. The data were calculated from at least three independent experiments (***, n = 3, p < 0.0001).
Molecular dynamic simulation detects frequent “bind-and-release” contacts between the acetylated Thr-2(Ac) and EsxB
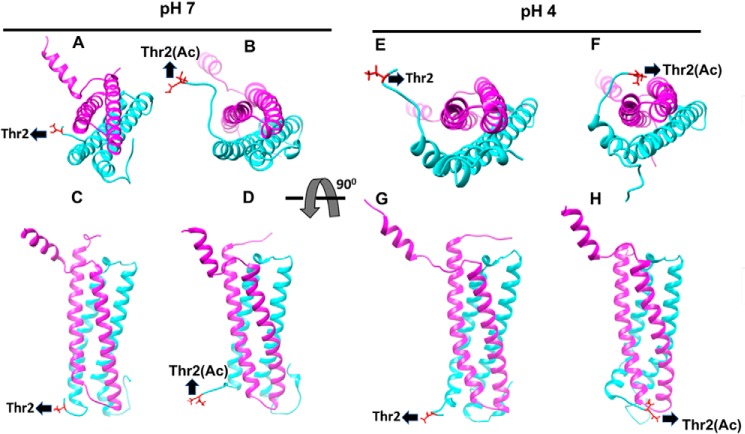
The reported solution structure of EsxA:B heterodimer does not have the Nα-acetylation on Thr-2, and the Thr-2 residue is distal from the contact interface between EsxA and EsxB. It is not clear how the acetylation at Thr-2 affects the heterodimer separation at low pH. Thus, we performed (MD) simulation on the heterodimers with and without Nα-acetylation of Thr-2 at pH 7 and 4, respectively (Movie S1 and Fig. 7). At pH 7 the nonacetylated Thr-2 comes in a close vicinity of EsxB but is unable to make a direct contact to EsxB (Fig. 7, A and C). Compared with nonacetylated Thr-2, the acetylated Thr-2(Ac) moves further away from EsxB at pH 7 (Fig. 7, B and D). Interestingly, at pH 4 the nonacetylated N-terminal loop of EsxA has no direct contact with EsxB (Fig. 7, E and G), but the acetylated N-terminal loop is able to make direct contacts with EsxB in a frequent bind-and-release mode (Movie S1 and Fig. 7, F and H). The electrostatic force was calculated between two sets of residues as demonstrated in Fig. S2 using DelPhiForce program. A contact is defined as two residues that contain at least a pair of atoms within 4 Å distance. The magnitude of the force is 43.959 pN, and its direction is shown by the orange arrow (Fig. S2). Therefore, when the N terminus of EsxA is moving away from EsxB, it pulls EsxB away in the direction of the arrow. Hence, the MD simulation data support that the acetylated Thr-2 residue plays a significant role in the dissociation process of the complex.
Figure 7.
Molecular dynamic simulation detects the acetylated Thr-2(Ac) interacts with EsxB in a bind-and-release mode. The structures of Mtb EsxA:B heterodimers with/out Nα-acetylation were analyzed by molecular dynamic simulation. The figures were generated from snapshots of 20-ns MD simulations at pH 7 and pH 4. EsxA, EsxB, and the Thr-2 residue are shown in cyan, pink, and red, respectively. The structures of EsxA:B heterodimer with nonacetylated Thr-2 at pH 7 and pH 4 are shown in A (pH 7, top view), C (pH 7, side view), E (pH 4, top view), and G (pH 4, side view), respectively. The structures of EsxA:B heterodimer with acetylated Thr-2(Ac) are shown in B (pH 7, top view), D (pH 7, side view), F (pH 4, top view), and H (pH 4, side view), respectively.
Discussion
EsxA and EsxB are co-expressed and co-secreted as a heterodimer in mycobacteria. The roles of ESX-1, EsxA, and EsxB in mycobacterial cytosolic translocation and virulence have been confirmed in a series of studies (7, 17–19). Earlier biochemical studies have demonstrated that EsxA has a pH-dependent membrane-permeabilizing activity, whereas EsxB appears to function as a chaperone for EsxA (20). Current studies support a model that the EsxA:B heterodimer is dissociated at low pH, which allows EsxA to permeabilize the membranes (24). However, the mechanism of the heterodimer separation is not known. For the first time, the present study tested the heterodimers containing EsxA with or without Nα-acetylation and obtained the evidence that the Nα-acetylation at Thr-2 of EsxA facilitates the heterodimer separation at low pH, which allows EsxA to permeabilize liposomal membrane in vitro, as well as mediate mycobacterial phagosome escape and cytosolic translocation in mycobacteria-infected macrophages.
As discussed above, the essentiality of EsxA and EsxB in mycobacterial pathogenesis has been well-documented in a series of reports. Genetic manipulations that either deleted the gene of esxA or esxB or abolished the secretion of EsxA and EsxB have attenuated mycobacterial virulence and inhibited the phagosome rupture, cytosolic translocation, and cell-to-cell spreading (7, 10, 17–19). Moreover, the biochemical characterizations have demonstrated that EsxA possesses a unique membrane-permeabilizing activity that is not present in its ortholog in nonpathogenic M. smegmatis (20). Thus, it is reasonable to believe that during infection, the secreted EsxA exerts its membrane-permeabilizing activity to penetrate the phagosome membranes and facilitate mycobacterial cytosolic translocation. We have reported that the mutations at the Gln-5 residue of EsxA (e.g. Q5V and Q5K) have resulted in up- or down-regulation of EsxA membrane-permeabilizing activity in vitro. Moreover, these mutations up- or down-regulated the mycobacterial virulence and cytosolic translocation accordingly, demonstrating the specific and accurate correlation between EsxA membrane-permeabilizing activity and mycobacterial virulence, as well as the ability to penetrate phagosome membrane (22). Once again, the present study provides new evidence that the Nα-acetylation at Thr-2 of EsxA is required for mycobacterial virulence and cytosolic translocation through facilitating the heterodimer separation.
Because Thr-2 has no any contact with EsxB as shown in the reported solution structure of EsxA:B heterodimer, how Nα-acetylation on Thr-2 affects heterodimer separation had become a puzzle. Here, the MD simulation result provides an interesting model, in which the acetylated Thr-2(Ac) has frequent bind-and-release contacts with EsxB at low pH, generating a dragging force to pull EsxB away from EsxA.
Nα-Acetylation is a common modification in eukaryotes and plays important roles in protein–protein interaction, protein activity and stability, and cell growth and cell cycle, etc. (30, 31). Although Nα-acetylation in eukaryotes has been well-studied, little is known about it in bacteria, including mycobacteria. Over 100 proteins in Mtb have been found to be Nα-acetylated, including EsxA (32), and protein acetylation has been correlated to pathogenesis (33, 34). Currently, three Nα-acetyltransferases in E. coli, RimI, RimL, and RimJ, have been identified to acetylate ribosomal proteins at Ser-18, Leu-12, and Ser-5, respectively (34–36). A biochemical study has found that RimI in Mtb has a relaxed substrate specificity, but little is known about the physiological substrates of the enzyme (37), which warrants further investigation.
A recent study has shown that the recombinant EsxA does not lyse cell membranes, and the lytic activity previously attributed to EsxA is due to residual ASB-14 detergent in the preparation (38). In fact, we had the similar observations that addition of the recombinant EsxA protein to the surface of lung epithelial cell lines WI-26 and A549 did not lyse the cells. Moreover, Conrad et al. (40) showed that blocking phagosomal acidification by bafilomycin did not decrease the ESX-1–mediated phagosomal permeabilization, suggesting that acidification is not required for membrane permeabilization. Recently, Lienard et al. (39) employed a collection of Mm ESX-1 transposon mutants, including the mutants that disrupt EsxA secretion, to infect macrophages and showed that the factors independent of EsxAB play a role in cytosolic translocation. It is not clear how the discrepancy arises and what is the broken link between the EsxA pH-dependent membrane-permeabilizing activity in liposomal membrane and the ability of mycobacteria to rupture phagosome membrane during infection. Other factors from mycobacteria and host cells, even including properties of target membranes, may be involved in this process, which warrants further investigation.
Materials and methods
Generation of T2X mutations on the Mtb esxA gene for expression in E. coli, Ms, and Mm
For expression in E. coli
Using the previously reported plasmid pET22b–esxA–His6 as the template (20–22, 41), the mutations T2A, T2Q, T2R, and T2S were introduced into the esxA gene by PCR using the primers listed in Table S1. All of the mutations were confirmed by DNA sequencing. The resulting plasmids were transformed into BL21 (DE3) cells for expression. The cells were grown at 37 °C while shaking at 250 rpm until A600 reached 0.6–0.8. Protein expression was induced by adding 1 mm isopropyl-β-d-1-thiogalactopyranoside for 3–8 h at 37 °C. The cells were harvested, and the proteins were purified as previously described (20–22, 41). Typical yield averaged between 30 and 60 mg/liter of culture.
For expression in Ms
The pMyNT plasmid containing the Mtb esxB-esxA operon (27) was used as the template. The mutations T2A, T2Q, T2R, and T2S were introduced by overlapping PCR using the primers listed in Table S1. All of the mutations were confirmed by DNA sequencing. The pMyNT plasmids carrying various T2X mutations were electroporated into Ms mc2155 strain (voltage, 2,500 V; capacitance, 25 microfarads; resistance, 1,000 Ω). The Ms cultures were grown at 37 °C overnight or until A600 reached 2.0. Protein expression was induced by adding 0.2% (w/v) acetamide for 12–16 h. The cells were harvested. The soluble heterodimer proteins were purified with immobilized metal ion affinity chromatography by passing through a Ni2+ column, followed by a size-exclusion chromatography as previously described (27, 42–45). Typical yield averaged between 20 and 40 mg/liter of culture.
For expression in Mm
The T2X mutations (T2A, T2R, T2Q, and T2S) were generated by site-directed mutagenesis (Agilent QuikChange kit) using the pMH406 plasmid containing esxB-esxA operon as a template. The mutations were confirmed by DNA sequencing. The mutated plasmids were electroporated into MmΔEsxA:B as previously described (44).
Liposome leakage assay
The liposome leakage assay was performed as previously described (20, 21, 41, 46, 47). Briefly, 20 mg of 1,2-dioleoyl-sn-glycero-3-phosphocholine was dried with nitrogen air and left in a vacuum overnight. The samples were rehydrated with 1 ml of the buffer (5 mm HEPES, 50 mm 8-aminonapthalene-1,3,6 trisulfonic acid (ANTS), and 50 mm p-xylene-bis-pyridinium bromide (DPX)). The suspension was subjected to six freeze-thaw cycles and extruded via a 0.2-μm membrane filter 20 times. The liposomes were then desalted to remove excess ANTS and DPX using a HiTrap desalting column. The desalted liposomes were mixed with 150 mm NaCl, 100 mm NaAc at pH 4.0. Three separate reaction mixtures were generated and individually excited at 350 nm, and emissions were recorded at 520 nm in an ISS K2 phase modulation fluorometer. 100 μg of the tested protein was injected into the solution after ∼30 s of the assay, and fluorescence was observed.
Isolation of Mtb EsxA proteins from the Ms-produced Mtb EsxA:B heterodimer
The Mtb EsxA:B heterodimer purified from Ms was incubated in a solution containing 6 m guanidine at 4 °C overnight. The proteins were then passed through a HisTrap column (GE Healthcare). The His-tagged EsxB protein was bound to the column, and the untagged EsxA was collected in flow through. The His-tagged EsxB protein was eluted by an imidazole gradient. Both EsxA and EsxB were subjected to an extensive dialysis using a 3000 molecular weight cutoff membrane. The samples were then concentrated and passed through gel filtration for a complete buffer exchange.
Detection of Nα-acetylation by NBD-Cl
NBD-Cl only reacts with free N-terminal α-amino group in nonacetylated proteins and emits fluorescence, and it does not react with Nα-acetylated proteins because of the lack of free N-terminal amino group (48). Proteins (6 μm) were incubated with 0.5 mm NBD-Cl at room temperature. At different hours of postincubation, the samples were subjected to fluorescence measurement with excitation at 460 nm and emission at 535 nm.
Sample preparation for mass spectrometry
The concentration of four protein samples, EsxA protein purified from E. coli and the EsxA, EsxA(T2A), and EsxA proteins purified from Ms were determined via bicinchoninic acid assay (BCA) according to manufacturer instructions (Pierce BCA protein assay kit, catalog no. 23227). Then 100 μg of each protein sample was used for protein digestion via FASP protein digestion (Expedeon, catalog no. 44250), using either trypsin (catalog no. Sigma, catalog no. T6567) or pepsin (Sigma, catalog no. P7012). In summary, 100 μg of each sample was resuspended in 200 μl of 12.48 m urea Tris-HCl solution (urea solution). Then 10 mg of dl-DTT (Sigma, catalog no. D0632-5G) was added to each sample and placed in a nutating mixer for 45 min. The samples were then transferred to a 30-kDa filter and centrifuged for 15 min at 14,000 rpm. 200 μl of urea solution was added to each spin filter and centrifuged at 14,000 × g for 15 min and repeated once more. Then 100 μl of iodoacetamide 1× solution (iodoacetamide prepared at a 1:10 ratio in urea solution) was added to each spin filter and incubated without mixing for 20 min in the dark. Afterward, spin filters were centrifuged at 14,000 rpm for 10 min. The spin filters were washed twice by adding 100 μl of urea solution and centrifuging at 14,000 rpm for 15 min. Urea was removed from the samples by adding 100 μl of 50 mm ammonium bicarbonate solution to spin filters and centrifuged at 14,000 rpm for 15 min. This step was repeated twice for a total of three times. Spin filters were transferred to new spin filter tubes, and 100 μl of digestion solution at 0.02 μg/μl (either with trypsin or pepsin) was added to each sample and incubated at 37 °C for 18 h. 200 μl of 0.1% formic acid water solution was added to each sample and spin filters containing resultant peptides and centrifuged at 14,000 × g for 10 min. Filtrate containing digested proteins were frozen at −80 °C for 2 h and lyophilized for 12 h. The samples were resuspended with 100 μl of 0.1% formic acid water solution at a final concentration of 1 μg/μl.
LC-MS/MS and bioinformatic method
Resultant complex peptide mixtures were analyzed via technical duplicates by LC-MS/MS for 2 h to 1 day with the QE Orbitrap (Thermo Fisher Scientific) along with the Dionex UltiMate 3000 RSLCnano UHPLC system (Thermo Fisher Scientific) using the two digestive protocols (trypsin or pepsin). The samples were loaded in line onto a C18 PicoChip column (75-μm inner diameter × 15-μm tip packed with 10.5 cm of Reprosil PUR C18 3 μm 120 Å; 25 μm × 50 cm fused-silica tail, New Objective) that was previously equilibrated with solvent A (95% water, 5% acetonitrile, 0.1% formic acid) and solvent B (5% water, 95% acetonitrile, 0.1% formic acid). The column was conditioned for 10 min with a flowrate of 0.5 μl/min with 95% solvent A, 5% solvent B. The sample was injected and loaded onto column and conditioned for 10 min with solvent A. Elution of peptides was completed by running a linear gradient to 40% solvent B for 95 min, followed by 10 min of 95% solvent B. The sample was re-equilibrated for 10 min with 5% solvent B. Full-scan spectra were collected via Xcalibur (Thermo Fisher Scientific). Two blank injections were run in between samples using a 30-min interval with seesaw washes using 5–80% ACN gradients. QE Orbitrap settings are as follows: full MS resolution of 70,000, AGC target of 1e6, scan range from 400 to 1600 m/z; MS/MS were run with a resolution of 17,500, AGC target of 2e6, scan range from 200 to 2000, and 3 m/z isolation window. The spectra were searched using Proteome Discoverer 2.1.1.21 (Thermo Fisher Scientific) and filtered via Sequest HT with an estimated false-discovery rate of 0.01 against sequences from Ms M1552, E. coli BL21, human, bovine, human keratin, and porcine trypsin. A 20-ppm precursor and a 0.02 fragment mass tolerance were used. Cysteine carbamidomethylation, methionine, oxidation, and acetylation were set fixed and variable modifications, respectively. The output files were then manually analyzed and collectively used to generate representative data.
Western blotting
Mm recombinant strains with respective EsxA insertions were cultured in 7H9 medium and grown to mid-log phase. They were washed with PBS and transferred to Sauton's medium while normalizing all cultures to A600 = 0.8. The bacteria were cultured for 2 days until harvest. The bacterial cells were collected by centrifugation. The proteins in the culture supernatant were precipitated by TCA. The bacterial cells were resuspended in 1 ml of PBS containing a mixture of protease inhibitors (Thermo Fisher Scientific) and sonicated at 30% amplitude for five cycles of 30-s pulse and 60-s rest. The culture filtrates and total bacterial lysates were applied to SDS-PAGE and transferred transferring onto polyvinylidene difluoride membrane. Western blots were performed to detect EsxA using anti-EsxA antibody (sc-57730, Santa Cruz). As controls, Ag85 (secreted in culture filtrate) and GroEL (only in cell lysate) were also detected by anti-Ag85 (NR-13800, BEI) and anti-GroEl antibodies (NR-13813, BEI), respectively.
Live/dead cytotoxicity assay
RAW264.7 cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum with penicillin and streptomycin (100 units/ml) at 37 °C and 5% CO2. Raw 264.7 macrophages were plated in a 24-well plate with a density of 5 × 105/well for infection on the following day. The Mm strains were prepared with a single cell preparation protocol as previously described (22, 49). RAW264.7 cells were infected with various Mm strains at a multiplicity of infection (MOI) of 10 for 1 h. The macrophages were washed three times with PBS to remove free mycobacteria and incubated for another 3 h. The macrophages were stained using calcein-AM and ethidium homodimer (Life Sciences) for 30 min, enabling visualization under a fluorescence microscope for green cells (live) and red cells (dead). The numbers of dead cells were quantified from dozens of random fields from each sample.
CCF-4 FRET assay
Mycobacterial cytosolic translocation was measured by CCF-4 FRET assay as previously described (17, 22, 50). Briefly, RAW264.7 cells were plated in triplicate in a 6-well plate at a density of 2.5 × 106 cells/well. The macrophages were then incubated with CCF4-AM according to the manufacturer's protocol (Liveblazer B/G loading kit, Life Sciences). The cells were infected with recombinant Mm strains at a MOI of 10 for 2 h. Following infection, the macrophages were washed three times using PBS. Dulbecco's modified Eagle's medium with 10% fetal bovine serum was added to the cells and incubated for ∼2 days. The samples were then excited at 409 nm, and the emissions were measured at 450 and 535 nm. The blue/green ratio was calculated as I450/I535.
Molecular dynamic simulation
The structure of EsxA:B heterodimer was downloaded from Protein Data Bank with Protein Data Bank code 1WA8 (51). DelPhiPka web server (52) was used to obtain the protonation states of ionizable residues at pH 4 and pH 7 and assign the respective states with visual molecular dynamics (VMD) (53). Nα-Acetylation of Thr-2 was performed on VMD after removal of Met-1. The four structures, i.e. nonacetylated and acetylated at pH 4 and pH 7 of EsxA:B heterodimer, were then solvated in water box with TIP3 (54) water model and ionized with 150 mm NaCl in VMD. The final systems were then simulated with the MD simulation program NAMD (55). Each simulation was performed for 20 ns employing force field CHARMM27 (56). The temperature was set as 300 K, and the pressure was 1 atm. The snapshots from the simulations were taken to study the behaviors of the N-terminal loop of EsxA with and without Nα-acetylation of Thr-2 at pH 4 and pH 7.
Data availability statement
All data are contained within the manuscript.
Author contributions
J. A., C. B. K., S. V. R., Q. Z., C. D. A., and J. S. data curation; J. A., C. B. K., S. V. R., and Q. Z. formal analysis; J. A., C. B. K., and L. L. methodology; J. A., C. B. K., L. L., I. E., B. I. G., and J. S. writing-original draft; L. L. and J. S. conceptualization; L. L., I. E., B. I. G., H. O., and J. S. writing-review and editing; S. V. R., H. O., and J. S. resources; J. S. supervision; J. S. funding acquisition; J. S. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Matthias Wilmanns for providing the pMyNT plasmid. The following reagents were obtained through BEI Resources, NIAID, National Institutes of Health: polyclonal anti-M. tuberculosis antigen 85 complex (FbpA/FbpB/FbpC; genes Rv3804c, Rv1886c, and Rv0129c; antiserum, rabbit), NR-13800; and monoclonal anti-M. tuberculosis GroEL2 (gene Rv0440), clone CS-44 (produced in vitro), NR-13813.
This work was supported by NIGMS, National Institutes of Health Grant SC1GM095475 (to J. S.) and National Institutes of Health Grant R25GM069621 (to J. A.) via the RISE Program for graduate students. This work National Center for Research Resources Grant 5G12RR008124 and National Institute on Minority Health and Health Disparities Grant G12MD007592. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Movie S1, Table S1, and Figs. S1 and S2.
- Mtb
- M. tuberculosis
- Ms
- M. smegmatis
- Mm
- M. marinum
- Ec
- E. coli
- Ec-heterodimer
- heterodimer prepared from E. coli
- Ms-heterodimer
- heterodimer prepared from M. smegmatis
- MD
- molecular dynamic
- ANTS
- 8-aminonapthalene-1,3,6 trisulfonic acid
- DPX
- p-xylene-bis-pyridinium bromide
- NBD-Cl
- 4-chloro-7-nitrobenzofurazan
- MOI
- multiplicity of infection
- VMD
- visual molecular dynamics.
References
- 1. Zumla A., Raviglione M., Hafner R., and von Reyn C. F. (2013) Current concepts. N. Engl. J. Med. 368, 745–755 10.1056/NEJMra1200894 [DOI] [PubMed] [Google Scholar]
- 2. Global Tuberculosis Report 2018 (2018) World Health Organization, Geneva, Switzerland [Google Scholar]
- 3. Orme I. (2004) Adaptive immunity to mycobacteria. Curr. Opin. Microbiol. 7, 58–61 10.1016/j.mib.2003.11.002 [DOI] [PubMed] [Google Scholar]
- 4. Kang P. B., Azad A. K., Torrelles J. B., Kaufman T. M., Beharka A., Tibesar E., DesJardin L. E., and Schlesinger L. S. (2005) The human macrophage mannose receptor directs Mycobacterium tuberculosis lipoarabinomannan-mediated phagosome biogenesis. J. Exp. Med. 202, 987–999 10.1084/jem.20051239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pizarro-Cerdá J., and Cossart P. (2006) Bacterial adhesion and entry into host cells. Cell 124, 715–727 10.1016/j.cell.2006.02.012 [DOI] [PubMed] [Google Scholar]
- 6. Tan T., Lee W. L., Alexander D. C., Grinstein S., and Liu J. (2006) The ESAT-6/CFP-10 secretion system of Mycobacterium marinum modulates phagosome maturation. Cell Microbiol. 8, 1417–1429 10.1111/j.1462-5822.2006.00721.x [DOI] [PubMed] [Google Scholar]
- 7. van der Wel N., Hava D., Houben D., Fluitsma D., van Zon M., Pierson J., Brenner M., and Peters P. J. (2007) M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 129, 1287–1298 10.1016/j.cell.2007.05.059 [DOI] [PubMed] [Google Scholar]
- 8. Behr M. A., Wilson M. A., Gill W. P., Salamon H., Schoolnik G. K., Rane S., and Small P. M. (1999) Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284, 1520–1523 10.1126/science.284.5419.1520 [DOI] [PubMed] [Google Scholar]
- 9. Pym A. S., Brodin P., Brosch R., Huerre M., and Cole S. T. (2002) Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol. Microbiol. 46, 709–717 10.1046/j.1365-2958.2002.03237.x [DOI] [PubMed] [Google Scholar]
- 10. Hsu T., Hingley-Wilson S. M., Chen B., Chen M., Dai A. Z., Morin P. M., Marks C. B., Padiyar J., Goulding C., Gingery M., Eisenberg D., Russell R. G., Derrick S. C., Collins F. M., Morris S. L., et al. (2003) The primary mechanism of attenuation of bacillus Calmette–Guérin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc. Natl. Acad. Sci. U.S.A. 100, 12420–12425 10.1073/pnas.1635213100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lewis K. N., Liao R., Guinn K. M., Hickey M. J., Smith S., Behr M. A., and Sherman D. R. (2003) Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette–Guérin attenuation. J. Infect. Dis. 187, 117–123 10.1086/345862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guinn K. M., Hickey M. J., Mathur S. K., Zakel K. L., Grotzke J. E., Lewinsohn D. M., Smith S., and Sherman D. R. (2004) Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol. Microbiol. 51, 359–370 10.1046/j.1365-2958.2003.03844.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gao L.-Y., Guo S., McLaughlin B., Morisaki H., Engel J. N., and Brown E. J. (2004) A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol. Microbiol. 53, 1677–1693 10.1111/j.1365-2958.2004.04261.x [DOI] [PubMed] [Google Scholar]
- 14. Sherman D. R., Guinn K. M., Hickey M. J., Mathur S. K., Zakel K. L., and Smith S. (2004) Mycobacterium tuberculosis H37Rv: delta RD1 is more virulent than M. bovis bacille Calmette–Guérin in long-term murine infection. J. Infect. Dis. 190, 123–126 10.1086/421472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. MacGurn J. A., and Cox J. S. (2007) A genetic screen for Mycobacterium tuberculosis mutants defective for phagosome maturation arrest identifies components of the ESX-1 secretion system. Infect. Immun. 75, 2668–2678 10.1128/IAI.01872-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koo I. C., Wang C., Raghavan S., Morisaki J. H., Cox J. S., and Brown E. J. (2008) ESX-1–dependent cytolysis in lysosome secretion and inflammasome activation during mycobacterial infection. Cell Microbiol. 10, 1866–1878 10.1111/j.1462-5822.2008.01177.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Simeone R., Bobard A., Lippmann J., Bitter W., Majlessi L., Brosch R., and Enninga J. (2012) Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 8, e1002507 10.1371/journal.ppat.1002507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Houben D., Demangel C., van Ingen J., Perez J., Baldeón L., Abdallah A. M., Caleechurn L., Bottai D., van Zon M., de Punder K., van der Laan T., Kant A., Bossers-de Vries R., Willemsen P., Bitter W., et al. (2012) ESX-1 mediated translocation to the cytosol controls virulence of Mycobacteria. Cell Microbiol. 14, 1287–1298 10.1111/j.1462-5822.2012.01799.x [DOI] [PubMed] [Google Scholar]
- 19. Simeone R., Sayes F., Song O., Gröschel M. I., Brodin P., Brosch R., and Majlessi L. (2015) Cytosolic access of Mycobacterium tuberculosis: critical impact of phagosomal acidification control and demonstration of occurrence in vivo. PLoS Pathog. 11, e1004650 10.1371/journal.ppat.1004650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. De Leon J., Jiang G., Ma Y., Rubin E., Fortune S., and Sun J. (2012) Mycobacterium tuberculosis ESAT-6 exhibits a unique membrane-interacting activity that is not found in its ortholog from non-pathogenic Mycobacterium smegmatis. J. Biol. Chem. 287, 44184–44191 10.1074/jbc.M112.420869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma Y., Keil V., and Sun J. (2015) Characterization of Mycobacterium tuberculosis EsxA membrane insertion: roles of N- and C-terminal flexible arms and central helix-turn-helix motif. J. Biol. Chem. 290, 7314–7322 10.1074/jbc.M114.622076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Q., Wang D., Jiang G., Liu W., Deng Q., Li X., Qian W., Ouellet H., and Sun J. (2016) EsxA membrane-permeabilizing activity plays a key role in mycobacterial cytosolic translocation and virulence: effects of single-residue mutations at glutamine 5. Sci. Rep. 6, 32618 10.1038/srep32618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fortune S. M., Jaeger A., Sarracino D. A., Chase M. R., Sassetti C. M., Sherman D. R., Bloom B. R., and Rubin E. J. (2005) Mutually dependent secretion of proteins required for mycobacterial virulence. Proc. Natl. Acad. Sci. U.S.A. 102, 10676–10681 10.1073/pnas.0504922102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Jonge M. I., Pehau-Arnaudet G., Fretz M. M., Romain F., Bottai D., Brodin P., Honoré N., Marchal G., Jiskoot W., England P., Cole S. T., and Brosch R. (2007) ESAT-6 from Mycobacterium tuberculosis dissociates from its putative chaperone CFP-10 under acidic conditions and exhibits membrane-lysing activity. J. Bacteriol. 189, 6028–6034 10.1128/JB.00469-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lightbody K. L., Ilghari D., Waters L. C., Carey G., Bailey M. A., Williamson R. A., Renshaw P. S., and Carr M. D. (2008) Molecular features governing the stability and specificity of functional complex formation by Mycobacterium tuberculosis CFP-10/ESAT-6 family proteins. J. Biol. Chem. 283, 17681–17690 10.1074/jbc.M800123200 [DOI] [PubMed] [Google Scholar]
- 26. Okkels L. M., Müller E.-C., Schmid M., Rosenkrands I., Kaufmann S. H., Andersen P., and Jungblut P. R. (2004) CFP10 discriminates between nonacetylated and acetylated ESAT-6 of Mycobacterium tuberculosis by differential interaction. Proteomics 4, 2954–2960 10.1002/pmic.200400906 [DOI] [PubMed] [Google Scholar]
- 27. Poulsen C., Holton S., Geerlof A., Wilmanns M., and Song Y.-H. (2010) Stoichiometric protein complex formation and over-expression using the prokaryotic native operon structure. FEBS Lett. 584, 669–674 10.1016/j.febslet.2009.12.057 [DOI] [PubMed] [Google Scholar]
- 28. Medie F. M., Champion M. M., Williams E. A., and DiGiuseppe Champion P. A. (2014) Homeostasis of N-α terminal acetylation of EsxA correlates with virulence in Mycobacterium marinum. Infect Immun. 82, 4572–4586 10.1128/IAI.02153-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mukherjee S., Hao Y.-H., and Orth K. (2007) A newly discovered post-translational modification–the acetylation of serine and threonine residues. Trends Biochem. Sci. 32, 210–216 10.1016/j.tibs.2007.03.007 [DOI] [PubMed] [Google Scholar]
- 30. Ree R., Varland S., and Arnesen T. (2018) Spotlight on protein N-terminal acetylation. Exp. Mol. Med. 50, 1–13 10.1038/s12276-018-0116-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Drazic A., Myklebust L. M., Ree R., and Arnesen T. (2016) The world of protein acetylation. Biochim. Biophys. Acta 1864, 1372–1401 10.1016/j.bbapap.2016.06.007 [DOI] [PubMed] [Google Scholar]
- 32. Liu F., Yang M., Wang X., Yang S., Gu J., Zhou J., Zhang X. E., Deng J., and Ge F. (2014) Acetylome analysis reveals diverse functions of lysine acetylation in Mycobacterium tuberculosis. Mol. Cell. Proteomics 13, 3352–3366 10.1074/mcp.M114.041962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bi J., Wang Y., Yu H., Qian X., Wang H., Liu J., and Zhang X. (2017) Modulation of central carbon metabolism by acetylation of isocitrate lyase in Mycobacterium tuberculosis. Sci. Rep. 7, 44826 10.1038/srep44826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee W., VanderVen B. C., Walker S., and Russell D. G. (2017) Novel protein acetyltransferase, Rv2170, modulates carbon and energy metabolism in Mycobacterium tuberculosis. Sci. Rep. 7, 72 10.1038/s41598-017-00067-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yoshikawa A., Isono S., Sheback A., and Isono K. (1987) Cloning and nucleotide sequencing of the genes rimI and rimJ which encode enzymes acetylating ribosomal proteins S18 and S5 of Escherichia coli K12. Mol. Gen. Genet. 209, 481–488 10.1007/BF00331153 [DOI] [PubMed] [Google Scholar]
- 36. Tanaka S., Matsushita Y., Yoshikawa A., and Isono K. (1989) Cloning and molecular characterization of the generimL wich encodes an enzyme acetylating ribosomal protein L12 of Escherichia coli K12. Mol. Gen. Genet. 217, 289–293 10.1007/BF02464895 [DOI] [PubMed] [Google Scholar]
- 37. Cumberlidge A. G., and Isono K. (1979) Ribosomal protein modification in Escherichia coli: I. A mutant lacking the N-terminal acetylation of protein S5 exhibits thermosensitivity. J. Mol. Biol. 131, 169–189 10.1016/0022-2836(79)90072-X [DOI] [PubMed] [Google Scholar]
- 38. Pathak D., Bhat A. H., Sapehia V., Rai J., and Rao A. (2016) Biochemical evidence for relaxed substrate specificity of Nα-acetyltransferase (Rv3420c/rimI) of Mycobacterium tuberculosis. Sci. Rep. 6, 28892 10.1038/srep28892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lienard J., Nobs E., Lovins V., Movert E., Valfridsson C., and Carlsson F. (2020) The Mycobacterium marinum ESX-1 system mediates phagosomal permeabilization and type I interferon production via separable mechanisms. Proc. Natl. Acad. Sci. U.S.A. 117, 1160–1166 10.1073/pnas.1911646117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Conrad W. H., Osman M. M., Shanahan J. K., Chu F., Takaki K. K., Cameron J., Hopkinson-Woolley D., Brosch R., and Ramakrishnan L. (2017) Mycobacterial ESX-1 secretion system mediates host cell lysis through bacterium contact-dependent gross membrane disruptions. Proc. Natl. Acad. Sci. U.S.A. 114, 1371–1376 10.1073/pnas.1620133114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peng X., Jiang G., Liu W., Zhang Q., Qian W., and Sun J. (2016) Characterization of differential pore-forming activities of ESAT-6 proteins from Mycobacterium tuberculosis and Mycobacterium smegmatis. FEBS Lett. 590, 509–519 10.1002/1873-3468.12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Noens E. E., Williams C., Anandhakrishnan M., Poulsen C., Ehebauer M. T., and Wilmanns M. (2011) Improved mycobacterial protein production using a Mycobacterium smegmatis groEL1ΔC expression strain. BMC Biotechnol. 11, 27 10.1186/1472-6750-11-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Poulsen C., Panjikar S., Holton S. J., Wilmanns M., and Song Y.-H. (2014) WXG100 protein superfamily consists of three subfamilies and exhibits an α-helical C-terminal conserved residue pattern. PLoS One 9, e89313 10.1371/journal.pone.0089313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu H., Bao Y., Wang L., Li X., and Sun J. (2019) Mycobacterium marinum down-regulates miR-148a in macrophages in an EsxA-dependent manner. Int. Immunopharmacol. 73, 41–48 10.1016/j.intimp.2019.04.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ray S., Vazquez Reyes S., Xiao C., and Sun J. (2019) Effects of membrane lipid composition on Mycobacterium tuberculosis EsxA membrane insertion: a dual play of fluidity and charge. Tuberculosis (Edinb.) 118, 101854 10.1016/j.tube.2019.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sun J., Vernier G., Wigelsworth D. J., and Collier R. J. (2007) Insertion of anthrax protective antigen into liposomal membranes: effects of a receptor. J. Biol. Chem. 282, 1059–1065 10.1074/jbc.M609869200 [DOI] [PubMed] [Google Scholar]
- 47. Sun J., Lang A. E., Aktories K., and Collier R. J. (2008) Phenylalanine-427 of anthrax protective antigen functions in both pore formation and protein translocation. Proc. Natl. Acad. Sci. U.S.A. 105, 4346–4351 10.1073/pnas.0800701105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bernal-Perez L. F., Prokai L., and Ryu Y. (2012) Selective N-terminal fluorescent labeling of proteins using 4-chloro-7-nitrobenzofurazan: a method to distinguish protein N-terminal acetylation. Anal. Biochem. 428, 13–15 10.1016/j.ab.2012.05.026 [DOI] [PubMed] [Google Scholar]
- 49. Takaki K., Davis J. M., Winglee K., and Ramakrishnan L. (2013) Evaluation of the pathogenesis and treatment of Mycobacterium marinum infection in zebrafish. Nat. Protoc. 8, 1114–1124 10.1038/nprot.2013.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Acosta Y., Zhang Q., Rahaman A., Ouellet H., Xiao C., Sun J., and Li C. (2014) Imaging cytosolic translocation of Mycobacteria with two-photon fluorescence resonance energy transfer microscopy. Biomed. Opt. Express. 5, 3990–4001 10.1364/BOE.5.003990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Renshaw P. S., Lightbody K. L., Veverka V., Muskett F. W., Kelly G., Frenkiel T. A., Gordon S. V., Hewinson R. G., Burke B., Norman J., Williamson R. A., and Carr M. D. (2005) Structure and function of the complex formed by the tuberculosis virulence factors CFP-10 and ESAT-6. EMBO J. 24, 2491–2498 10.1038/sj.emboj.7600732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang L., Zhang M., and Alexov E. (2016) DelPhiPKa web server: predicting pKa of proteins, RNAs and DNAs. Bioinformatics 32, 614–615 10.1093/bioinformatics/btv607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Humphrey W., Dalke A., and Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- 54. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., and Klein M. L. (1998) Comparison of simple potential functions for simulating liquid water. J. Chem. Physics 79, 926–935 10.1063/1.445869 [DOI] [Google Scholar]
- 55. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., and Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I., and Mackerell A. D. (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript.