Abstract
Heterotrimeric G proteins mediate a variety of signaling processes by coupling G protein–coupled receptors to intracellular effector molecules. In Drosophila, the Gαq gene encodes several Gαq splice variants, with the Gαq1 isoform protein playing a major role in fly phototransduction. However, Gαq1 null mutant flies still exhibit a residual light response, indicating that other Gαq splice variants or additional Gq α subunits are involved in phototransduction. Here, we isolated a mutant fly with no detectable light responses, decreased rhodopsin (Rh) levels, and rapid retinal degeneration. Using electrophysiological and genetic studies, biochemical assays, immunoblotting, real-time RT-PCR, and EM analysis, we found that mutations in the Gαq gene disrupt light responses and demonstrate that the Gαq3 isoform protein is responsible for the residual light response in Gαq1 null mutants. Moreover, we report that Gαq3 mediates rhodopsin synthesis. Depletion of all Gαq splice variants led to rapid light-dependent retinal degeneration, due to the formation stable Rh1-arrestin 2 (Arr2) complexes. Our findings clarify essential roles of several different Gαq splice variants in phototransduction and retinal integrity in Drosophila and reveal that Gαq3 functions in rhodopsin synthesis.
Keywords: phototransduction, G protein, rhodopsin, retinal degeneration, cell signaling, calcium homeostasis, G protein-coupled receptor signaling, light response, splice variant
Introduction
Heterotrimeric G proteins and G protein–coupled receptors play pivotal roles in mediating a variety of extracellular signals to intracellular signaling pathways, such as hormones, neurotransmitters, peptides, and sensory stimuli (1, 2). In the Drosophila visual system, light stimulation activates the major rhodopsin (Rh1) to form metarhodopsin, which in turn activates heterotrimeric G proteins and norpA gene-encoded phospholipase C (PLCβ)4 (3). Activated PLC catalyzes phosphatidylinositol 4,5-bisphosphate to generate diacylglycerol (DAG) and inositol 1,4,5-triphosphate (IP3) (4). IP3 induces the release of Ca2+ from intracellular Ca2+ stores, whereas both DAG and IP3 may trigger extracellular Ca2+ influx by opening transient receptor potential (Trp) and transient receptor potential-like (TrpL) channels on the cell membrane (5–8). The Gαq gene encodes several Gαq splice variants, among which the Gαq-RD variant generates the Gαq1 isoform protein, and other splice variants generate the Gαq3 isoform protein (9). Although both strong alleles of norpA and trpl;trp double mutants show completely abolished photoresponses (4, 10, 11), the Gαq1 isoform null mutant allele (Gαq961) still displays a residual light response (12). These data indicate that other Gαq splice variants, or the Gq α subunits encoded by additional genes, contribute to the residual light responses in Gαq1 null mutants.
Intracellular Ca2+ homeostasis controlled by Gq signaling is also essential for photoreceptor cell survival (13). Mutations in phototransduction cascade components, such as those in trp and norpA, prevent normal light-induced Ca2+ influx, resulting in stable Rh1/Arr2 complex formation and severe rapid light-dependent retinal degeneration (14, 15). Disruption of stable Rh1/Arr2 complexes by genetic removal of Arr2 or suppression of Rh1 endocytosis can suppress the retinal degeneration either in norpA or trp mutant flies (15, 16). Rh1/Arr2 complex formation is thought to attribute to impaired Ca2+ influx-activated CaM kinase II, which usually phosphorylates Arr2 to release Arr2 from Rh1 (17, 18). However, neither Gαq1 nor Gαq961 mutants undergo rapid retinal degeneration (12, 19), exhibiting only slight retinal degeneration after keeping them in 12-h light/12-h dark cycles for 21 days (12). The disagreeing retinal degeneration phenotype between Gαq and norpA mutant is therefore unclear.
Here, we isolate a mutant fly with no detectable light responses and reveal that mutations in the Gαq gene cause the defective light responses. We demonstrate that Gαq3 is responsible for the residual light response in Gαq1 null mutants and show that depletion of all Gαq splice variants results in rapid light-dependent retinal degeneration due to formation of stable Rh1/Arr2 complexes. In addition, we reveal that Gαq3 plays essential roles in Rh1 synthesis. Our study clarify the essential role of different Gαq splice variants in fly phototransduction, retinal degeneration, and rhodopsin synthesis.
Results
Isolation of a fly mutant with no detectable responses to light stimulation
To characterize the components of the phototransduction machinery, we obtained a collection of transgenic transposon p[GawB] homozygous viable strains and performed an electroretinogram (ERG)-based screen for additional genes in fly phototransduction. We isolated a mutant fly, which showed no detectable ERG responses to saturated light stimulations (Fig. 1A). Using the inverse PCR technique, we identified that the p[GawB] element was inserted into the 18C3 chromosomal region located on the X chromosome. To eliminate extra mutations in the genetic background, we backcrossed the mutant with the WT w1118 strain (based on the ERG phenotype) for eight generations and refer to the out-crossed mutant as nlr (no detectable light response) mutant. Unexpectedly, nlr mutants did not contain any p[GawB] element insertions, indicating the abolished ERG response in the nlr mutants is due to mutations in the genetic background, and not p[GawB] element insertion.
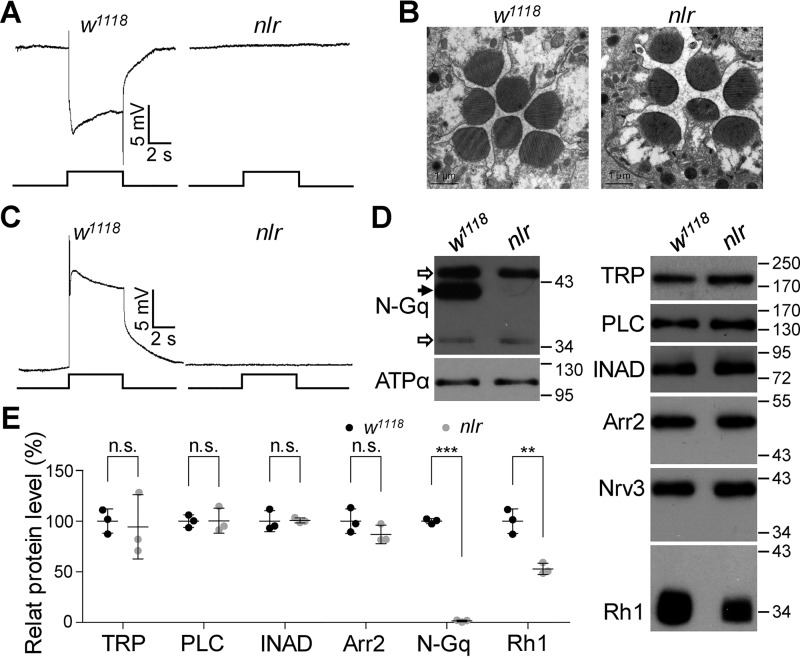
Figure 1.
nlr mutants exhibit no response to light stimulation. A, representative ERG traces of WT and nlr mutant flies. For all ERG traces, event markers represent 5-s light pulses, and scale bars are 5 mV. B, EM images show a normal rhabdomeral structure in 1-day-old nlr mutant flies. C, intracellular recordings of light responses in photoreceptors. The scale bar and light pulse is 5 mV and 5 s, respectively. D, Western blots of Gαq protein levels recognized by anti-N-Gαq antibodies and other visual molecules in nlr mutants. Each lane was loaded with half of the isolated retina. The specific Gαq band is indicated with a black arrow and nonspecific band with open arrows. ATPα and Nrv3 were used as loading controls. E, quantification of protein levels of visual molecules in w1118 and nlr mutants.
The significantly reduced ERG response in nlr mutants could be due to a defective rhabdomere structure or reflect deficits in the phototransduction cascade. To distinguish between these possibilities, we first performed an electron microscopy (EM) study to examine the rhabdomere structure of newly enclosed adult flies. However, EM images did not reveal any morphological defects in nlr mutant rhabdomeres (Fig. 1B). Intracellular recording found that light stimulation was unable to evoke any detectable responses in nlr mutant photoreceptors (Fig. 1C). These data indicated that the defective light response in nlr mutants was due to abnormalities in the phototransduction cascade. We performed Western blot analysis to examine the protein levels of components and regulators in the phototransduction cascade, including major rhodopsin (Rh1), Gαq, PLC, TRP, INAD, and Arr2. Interestingly, protein levels for all Gαq isoforms recognized by anti-Gαq-N antibodies were significantly reduced (100 ± 2.4 versus 1.4 ± 0.9%, p < 0.0001, t test; Fig. 1D–E). Meanwhile, a partial reduction of Rh1 (100 ± 12.2 versus 38.3 ± 5.5%, p = 0.0014, t test) was found in nlr mutants, whereas the other visual molecules examined were comparable with WT flies (Fig. 1D–E). These results suggest that the defective light responses in nlr mutants might be due to the absence of Gαq protein.
Mutations in Gαq gene are responsible for defective light responses in nlr mutants
To identify the mutations in nlr flies that are responsible for the defective light-response phenotype, we first mapped the mutations to the 49B5-49B12 chromosomal region based on the ERG phenotype covered by the deficient chromosome Df(2R)Exel7121 (missing 49B5 to 49B12, Fig. 2, A and B). This result further supports the conclusion that the abnormal ERG phenotype in nlr mutants is not due to p[GawB] element insertion. Next, we further narrowed the mutation to the 49B8-49B10 region based on the ERG phenotype covered by the deficient chromosome Df(2R)Gαq1.3, which removes Gαq, CG30054, CG17760, muskelin, and part of the CG33792 genes (Fig. 2, A and B). Next, we generated clones of Df(2R)Gαq1.3-covered gene nulls in the retina through ey-FLP-induced FRT recombination in the deficiency line, Df(2R)Gαq1.3. We found that this fly lacked Gαq protein and showed an abolished light response (Fig. 2, C and D), indicating that the defective light response observed in nlr mutants is contributed to by the mutations located in the deficient chromosome Df(2R)Gαq1.3-covered region.
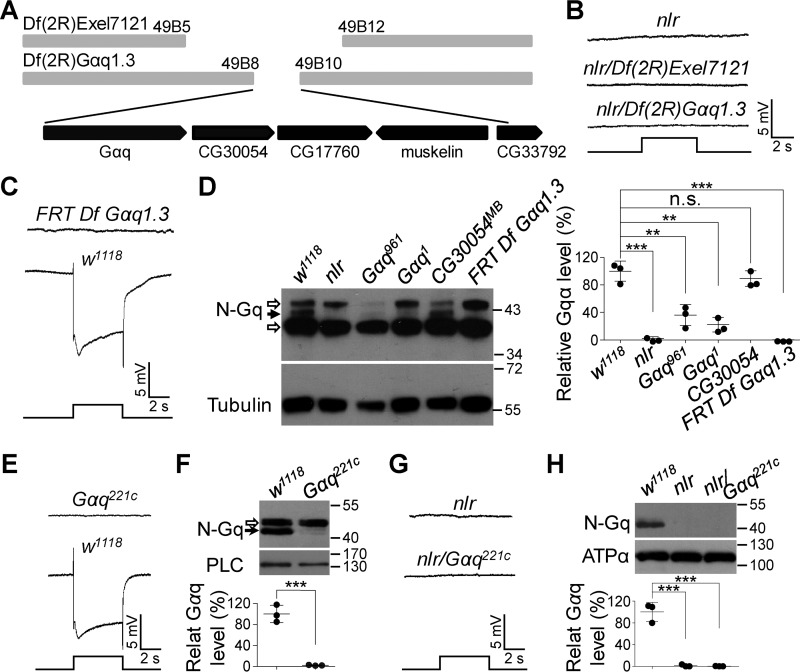
Figure 2.
Gαq gene mutations cause the abolished light responses in nlr mutants. A, annotated deletion region of the deficient chromosome Df(2R)Exel7121 and Df(2R)Gαq1.3. Annotated genes covered by Df(2R)Gαq1.3 are presented in the bottom panel. B, nlr over either Df(2R)Exel7121 (nlr/Df(2R)Exel7121) or Df(2R)Gαq1.3 (nlr/Df(2R)Gαq1.3) show no photoresponses. Event markers represent 5-s light pulses, and scale bars are 5 mV. C, ERG response of the clones of Df(2R)Gαq1.3-covered gene nulls in the retina. D, Western blots of Gαq protein levels in the various mutants. Each lane was loaded with half of the isolated retina. Gαq961 and Gαq1 mutants were used as negative controls, whereas CG30054MB mutants were used as positive controls. The specific Gαq band and nonspecific bands are marked with a black arrow and open arrows, respectively. Quantification of Gαq levels for each genotype is presented in the right panel. E, ERG response of Gαq221c null mutant clones in the retina. Event markers represent 5-s light pulses, and scale bars are 5 mV. F, Western blots of Gαq protein levels in w1118 and Gαq221c null mutant photoreceptor cells. The specific Gαq band and nonspecific bands are marked with a black arrow and an open arrow, respectively. The eye-specific protein PLC was used as a loading control. Quantification of Gαq levels for each genotype is presented in the lower panel. G, nlr/Gαq221c flies show no photoresponses. Event markers represent 5-s light pulses, and scale bars are 5 mV. H, Western blots of Gαq protein levels in the various mutants. Each lane was loaded with half of the isolated retina. Quantification of Gαq levels for each genotype is presented in the lower panel.
Among the genes covered by the deficient chromosome Df(2R)Gαq1.3, the Gαq gene plays an essential role in Drosophila phototransduction (12, 20). To test whether Gαq gene mutations contribute to the significantly reduced light responses in nlr mutants, we obtained a Gαq221c mutant allele that removes a 359-bp fragment around the translation start site of all Gαq splice variants (21). Because Gαq221c homozygous mutations are lethal, we generated Gαq221c null mutant clones in the retina through ey-FLP-induced FRT recombination. This fly was also absent of Gαq protein (100 ± 16.4 versus 2.1 ± 0.7%, p = 0.0005, t test) and displayed an abolished light response (Fig. 2, E and F). Next, we recombined nlr with Gαq221c mutant flies and found that nlr/Gαq221c flies showed no detectable responses to light stimulus and an absence of Gαq protein (Fig. 2, G and H). These data demonstrate that mutations in the Gαq gene are responsible for the defective light responses in nlr mutants.
Identification of new Gαq1 isoform mutation in nlr mutant
The Gαq gene encodes several Gαq splice variants (Fig. 3A), and the splice variant Gαq1 (also named as Gαq-PD, AAM68631) has been shown to play a major role in Drosophila phototransduction (12, 20). Thus, we wondered whether nlr mutants contain any mutations in the Gαq gene. Indeed, subsequent DNA sequencing revealed a mutation (5501T/A) in exon 7 of the Gαq gene in nlr mutant flies (Fig. 3A), which is within the Gαq1 isoform but not included in other Gαq splice variants. This mutation corresponds to a missense mutation (303V/D) in the GTPase domain of Gαq1 (amino acids 247–359).
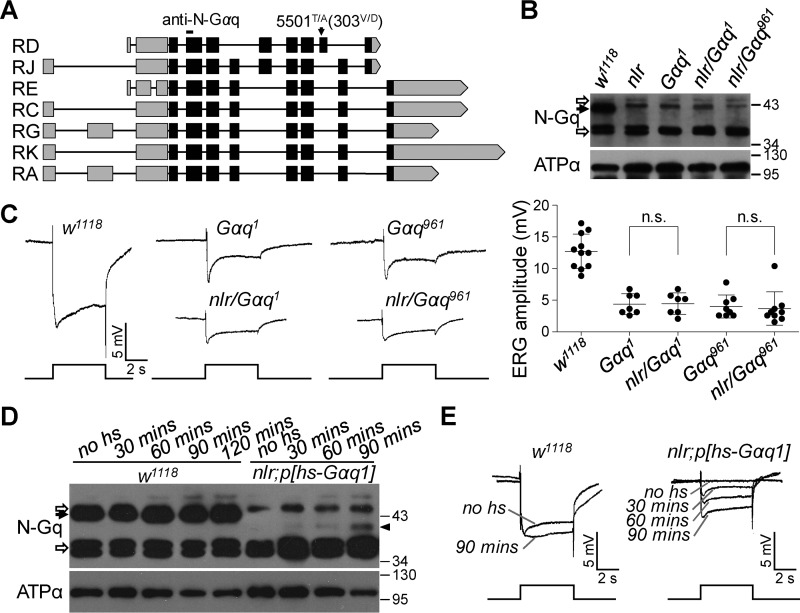
Figure 3.
GαqV303D mutation of the Gαq1 isoform largely contributes to the abolished light response in nlr mutants. A, annotated transcripts of the Gαq gene. Position of the point mutation in the nlr allele is marked with a black arrow. The anti-N-Gαq antibody-recognized site is labeled at the top. B, Western blots of Gαq protein levels in each genotype. Each lane was loaded with half of the isolated retina. The specific Gαq band is indicated with a black arrow and nonspecific bands were indicated with open arrows. C, light response in nlr/Gαq1 and nlr/Gαq961 heterozygous flies. For all ERG traces, event markers represent 5-s light pulses, and scale bars are 5 mV. Quantification of ERG amplitudes for each genotype is presented in the right panel. D, Western blots show Gαq protein levels with different treatments. nlr;P[HS::Gαq] adults reared at 21 °C were exposed to different heat shock times at 37 °C, and all flies were collected and analyzed 24 h after heat shock. The specific Gαq band and nonspecific bands are marked with a black arrow and open arrows, respectively. E, ERG traces show light responses with different treatments. Gαq protein was induced as described in D. For all ERG traces, event markers represent 5-s light pulses, scale bars are 5 mV, and the treatment conditions for each trace are marked.
To examine whether this mutation (5501T/A) disrupts Gαq1 function, we combined the nlr mutant allele with a Gαq hypomorphic allele (Gαq1), which changes a Gly to Ala in a splice acceptor site causing the use of a cryptic splice site 9 nucleotides downstream and an in-frame deletion of 3 codons encompassing amino acid residues 154–156 (20). Meanwhile, we combined the nlr mutant allele with a Gαq1 isoform null mutant allele (Gαq961), which contains a mutation (961C/T) in exon 4 and causes a nonsense mutation (Arg-117 to stop codon) of Gαq1 exclusively (12). Western blotting showed that Gαq protein levels were significantly reduced in both nlr/Gαq1 and nlr/Gαq961 flies (Fig. 3B). ERG recording further revealed that both nlr/Gαq1 and nlr/Gαq961 flies exhibited significantly reduced light responses to saturated light stimulation, similar to that of either Gαq1 (Fig. 3C; 4.3 ± 1.6 versus 4.4 ± 1.7 mV, p = 0.94, t test) or Gαq961 (Fig. 3C; 4.0 ± 1.8 versus 3.7 ± 2.6 mV, p = 0.77, t test) flies. These results indicated that the 5501T/A Gαq gene mutation largely contributed to the abolished light response in the nlr mutant. Furthermore, the above data excluded the possibility that abolished light responses in nlr mutants were due to the dominant suppression of the GαqV303D mutant protein.
To further validate that the GαqV303D mutation in Gαq1 largely contributed to the abolished light response in nlr mutants, we obtained p[HS::Gαq1] transgenic flies and performed rescue experiments. Convincingly, Gαq1 expression largely restored the abolished ERG responses in nlr mutants (Fig. 3, D and E). Taken together, these data demonstrate that the 5501T/A Gαq gene mutation largely contributed to the abolished light response in nlr mutants and excludes the possibility that the abolished light responses in nlr mutants were due to the dominant suppression of GαqV303D mutant protein.
Gαq3 is responsible for residual light responses in Gαq1 isoform null mutants
Given that mutations in the Gαq gene are responsible for the abolished light responses in nlr mutants, and the Gαq gene encodes several Gαq splice variants, we suspected that other Gαq splice variants might contribute to the residual light responses in Gαq1 null mutant flies. To test this hypothesis, we generated Gαq1/Gαq221c and Gαq961/Gαq221c flies, and found that although these flies responded to light stimulus, ERG amplitudes were smaller than that of either Gαq1 (Fig. 4A; 4.3 ± 1.6 versus 0.8 ± 0.4 mV, p < 0.0001, t test) or Gαq961 (Fig. 4A; 4.0 ± 1.8 versus 0.7 ± 0.3 mV, p = 0.0008, t test) flies. In Drosophila, splice variants Gαq-RE, Gαq-RC, Gαq-RG, Gαq-RK, and Gαq-RA encode the same Gαq3 isoform (Fig. 3A). These data indicate that the residual light response observed in Gαq1 null mutants was contributed to by Gαq3.
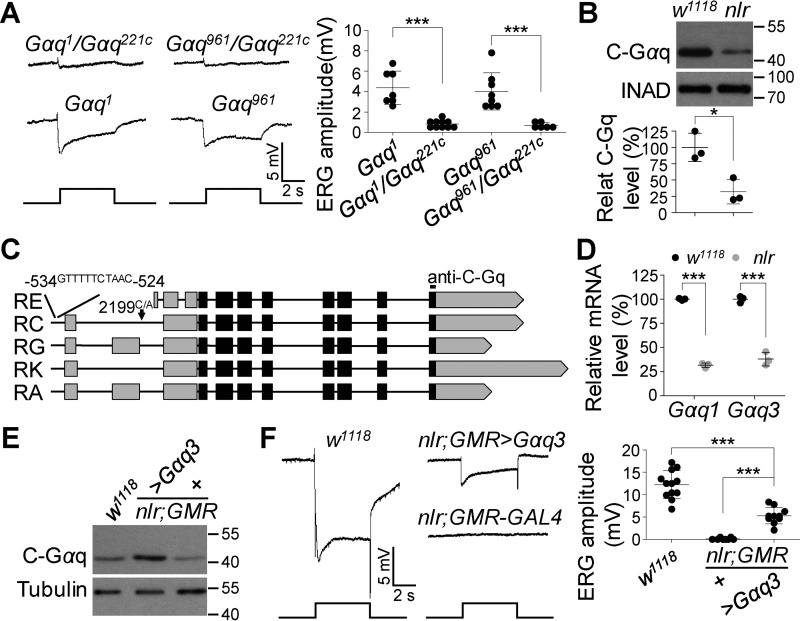
Figure 4.
Gαq3 isoform contributes to the residual photoresponse in Gαq1 isoform null mutants. A, representative ERG traces of Gαq1/Gαq221c and Gαq961/Gαq221c flies. For all ERG traces, event markers represent 5-s light pulses, and scale bars are 5 mV. Quantification of ERG amplitudes for each genotype is presented in the right panel. B, Western blots of Gαq3 protein levels in w1118 and nlr mutant photoreceptor cells. The eye-specific protein INAD was used as a loading control. Quantification of Gαq3 levels for each genotype is presented in the lower panel. C, additional mutations in the Gαq gene. The point mutation in the nlr allele is marked with a black arrow, and the sequence inserted upstream of the transcription start site is listed at the top. The anti-C-Gαq antibody-recognized site is labeled at the top. D, mRNA levels of Gαq1 and Gαq3 in w1118 and nlr mutant retina. Total RNA was extracted from isolated adult retina. Rp49 was used as an internal control. Data are presented as mean ± S.D. from three independent experiments. E, Western blots show Gαq3 protein levels in rescued flies. F, representative ERG traces of w1118, nlr;GMR-GAL4, and nlr;GMR-GAL4/UAS::Gαq3 flies. For all ERG traces, event markers represent 5-s light pulses, and scale bars are 5 mV. Quantification of ERG amplitudes for each genotype is presented in the right panel.
To examine Gαq3 protein levels in nlr mutants, we conducted Western blot analysis using anti-Gαq-C antibodies that can recognize Gαq3 specifically. Protein levels of Gαq3 were significantly reduced in nlr mutants (Fig. 4B; 100 ± 21.8 versus 31.9 ± 18.7%, p = 0.015, t test), suggesting additional mutations in nlr mutants affect the expression of Gαq3 isoforms. Next, we conducted DNA sequencing in the whole Gαq gene region. We failed to identify additional mutations in Gαq3 isoform-coding regions of nlr mutant flies. However, in the promotor region of the Gαq gene in nlr mutants, we identified an 11-bp sequence insertion (GTTTTTCTAAC) at the −534 to −524 position that was absent in w1118 flies, as well as a mutation (2199C/A) in an 11-bp sequence (2193–2203, CTAATTCGATT) conserved in the promoter region of several photoreceptor cell-specific genes (22, 23) (Fig. 4C). Moreover, qRT-PCR analysis validated that the mRNAs of Gαq3 isoforms, as well as Gαq1, were significant reduced in nlr mutants (Fig. 4D; Gαq1: 100 ± 1.0 versus 33.7 ± 2.4%, p < 0.0001, t test; Gαq3: 100 ± 3.3 versus 35.8 ± 7.0%, p < 0.0001, t test), demonstrating that these mutations in nlr mutants affect Gαq gene transcription.
To further confirm that Gαq3 mediates phototransduction, we generated p[UAS::Gαq3] transgenic flies and performed rescue experiments. Consistently, Gαq3 expression in nlr mutants generated clear ERG responses (0.13 ± 0.2 versus 5.3 ± 1.8 mV, p < 0.0001, t test), and the amplitude of ERG responses was comparable with that in either Gαq1 or Gαq961 mutant flies (Fig. 4, E and F). Taken together, these data demonstrate that Gαq3 also mediates phototransduction, and further indicates that the residual light response observed in Gαq1 null mutants is contributed to by Gαq3.
Depletion of all Gαq splice variants results in severe retinal degeneration
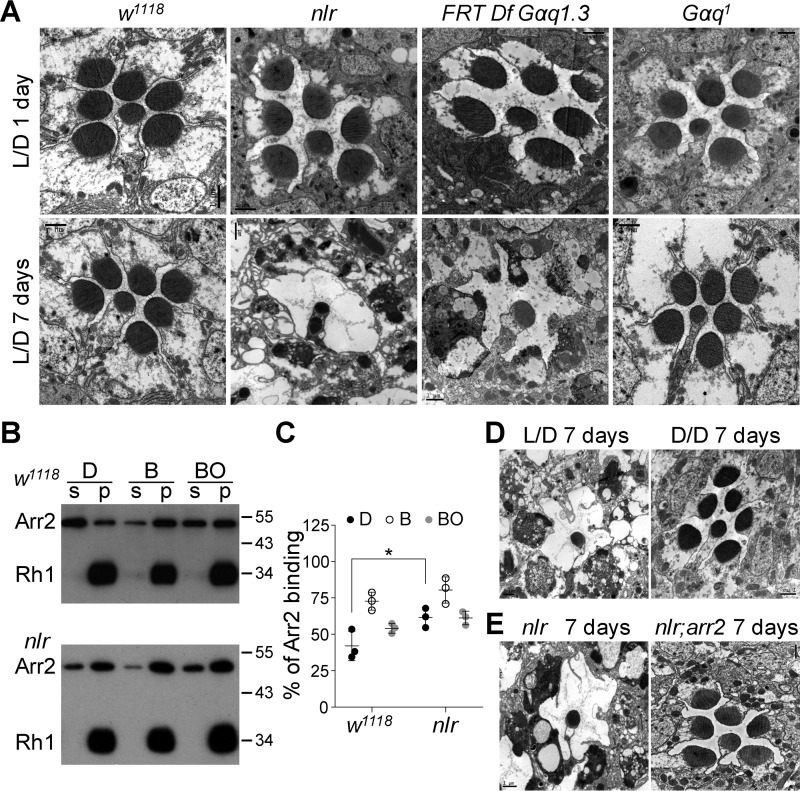
In Drosophila, mutations in genes that prevent normal Ca2+ influx after light stimulation, such as those in trp and norpA, usually cause stable Rh1/Arr2 complex formation and light-dependent retinal degeneration (13–15). In contrast, neither 6-day-old Gαq1 nor Gαq961 mutants show obvious retinal degeneration, and Gαq961 mutants show only slight accumulation of stable Rh1/Arr2 complexes after light exposure (12, 19). The residual light responses in either Gαq1 or Gαq961 mutants might be able to trigger sufficient Ca2+ influx to activate CaM kinase II, which subsequently phosphorylates Arr2 to release Arr2 from Rh1 (17, 18). If true, stable Rh1/Arr2 complexes should accumulate with severe retinal degeneration after the depletion of all Gαq splice variants. Indeed, EM images revealed obvious retinal degeneration in 7-day-old nlr and Df(2R)Gαq1.3-covered gene null mutants raised under regular 12-h light/12-h dark cycles, but not in Gαq1 mutants raised in the same conditions (Fig. 5A).
Figure 5.
Depletion of all Gαq splicing variants leads to rapid retinal degeneration. A, EM analysis reveals nlr mutants and the clones of Df(2R)Gαq1.3-covered gene nulls underwent rapid retinal degeneration. Flies were raised in 12-h light/12-h dark conditions for the indicated time. Each picture shows a single ommatidium. Scale bar, 1 μm. B, Western blotting of fractions of w1118 and nlr mutant heads. Arr2-Rh1–binding assays were performed as described under “Experimental procedures.” C, quantification of the percentage of Arr2 bound to rhodopsin-containing membranes in the dark (D), after treatment with blue light (B), or after treatment with blue light followed by orange light (BO). D, EM images show normal rhabdomeral structure in 7-day-old dark-reared nlr mutant flies. Flies were raised in constant darkness. Scale bar, 1 μm. E, EM images show normal rhabdomeral structure in 7-day-old nlr;arr2 double mutants. Flies were raised in 12-h light/12-h dark conditions for 7 days. Scale bar, 1 μm.
Next, we examined whether retinal degeneration in nlr mutants was due to the accumulation of stable Rh1/Arr2 complexes. With 480-nm blue light exposure, Rh1 is photoconverted to metarhodopsin and induces binding with Arr2. Metarhodopsin can be photoconverted to inactivated rhodopsin by an additional 580-nm orange light exposure, which leads to the release of Arr2 (15). In WT flies, blue light exposure caused about 74% binding between Arr2 and Rh1, and ∼49% release of Arr2 from Rh1 following exposure to orange light (Fig. 5, B and C). In contrast, blue light exposure triggered approximately 88% binding between Arr2 and Rh1, and less than 32% release of Arr2 from Rh1 following exposure to orange light in nlr mutants (Fig. 5, B and C). These data imply that depleting all Gαq splice variants results in stable Rh1/Arr2 complexes accumulation.
To provide further support that stable Rh1/Arr2 complex formation triggers retinal degeneration in nlr mutants, we conducted an EM study in 7-day-old dark-reared nlr mutants. Interestingly, 7-day-old dark-reared nlr mutants displayed intact rhabdomere structures (Fig. 5D). Furthermore, genetic removal of arr2 in the nlr mutant background suppressed retinal degeneration in nlr mutant flies (Fig. 5E). These data demonstrate that depleting all Gαq splice variants stabilizes Rh1/Arr2 complex formation, triggering severe light-dependent retinal degeneration.
Gαq3 mediates Rh1 synthesis
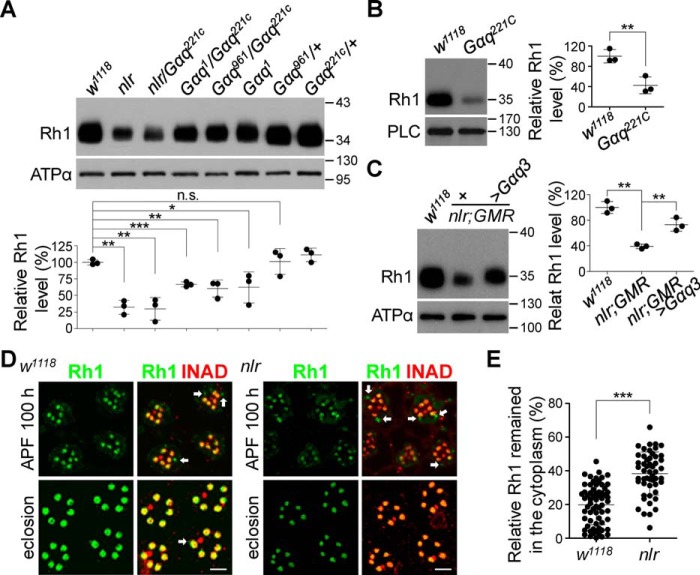
Except for undetectable responses to light stimuli, 1-day-old nlr mutants also exhibited reduced Rh1 levels (Fig. 1, D and E; Fig. 6A). Convincingly, Gαq221c null mutant clones in the retina also showed a significant reduction in Rh1 levels (Fig. 6B; 100 ± 13.1 versus 42.4 ± 16.6%, p = 0.0092, t test). Furthermore, Western blot analysis showed that nlr/Gαq221c recombined flies displayed a significant reduction in Rh1 levels, whereas Gαq1, Gαq1/Gαq221c, and Gαq961/Gαq221c flies displayed a partial Rh1 reduction (Fig. 6A). These results indicate that other Gαq splice variants, but not Gαq1, largely contribute to Rh1 synthesis. Next, we carried out rescue experiments and found that Gαq3 expression in nlr mutants largely restored Rh1 levels (Fig. 6C; 73.1 ± 10.3 versus 39.1 ± 3.6%, p = 0.0057, t test). These data demonstrate that Gαq3 plays essential roles in Rh1 synthesis.
Figure 6.
Gαq3 isoform regulates rhodopsin synthesis. A, Western blots of Rh1 levels in each genotype. Rh1 quantification for each genotype is presented in the lower panel. B, Western blots of Rh1 levels in w1118 and Gαq221c mutant flies. Rh1 quantification for each genotype is presented in the right panel. C, Western blots of Rh1 levels in w1118, nlr;GMR-GAL4, and nlr;GMR-GAL4/UAS::Gαq3 flies. Rh1 quantification for each genotype is presented in the right panel. D, Rh1 distribution in developing w1118 (left) and nlr mutant retina (right). Fly heads were collected at 100 h after pupal formation and eclosion, and sections were prepared and stained with anti-Rh1 (green) and anti-INAD (red) antibodies. The Rh1 signal remaining in the cytoplasm is indicated with arrows. Scale bar, 5 μm. E, quantification of Rh1 remaining in the cytoplasm at 100 h after pupal formation for each genotype.
To explore the role of Gαq3 in regulating Rh1 synthesis, we monitored the distribution of Rh1 in developing photoreceptors. During pupal development, nascent Rh1 in WT pupae was gradually loaded into rhabdomeres, and most Rh1 was successfully loaded into the rhabdomeres by fly eclosion (Figs. 6D). In contrast, a large fraction of Rh1 remained in the cytoplasm in nlr mutant pupae (Fig. 6, D and E). These results indicate that proper loading of Rh1 into rhabdomeres requires Gαq3.
Discussion
Gαq3 isoforms mediate residual light responses in Gαq1 null mutants
In Drosophila photoreceptors, G proteins are essential to activate the phototransduction cascade (20, 24). The Gαq gene encodes several Gαq splice variants, and Gαq1 has been shown to function as the predominant G protein in fly phototransduction (12, 20). In this study, we identified a mutation (5501T/A) in the Gαq gene, which specifically mutates Val to Asp at residue 303 in Gαq1 but not Gαq3 isoforms. Although Val is replaced with Ile at residue 303 in vertebrate Gαq proteins, the hydrophobicity at this position is evolutionally conserved. Structural analyses have shown that the Val-303 region localizes to the interface between Gα proteins and its downstream effector PLC (25–27). The change of a hydrophobic residue to a polar one may affect the interaction between these two proteins. A recent study has shown that the GαqV303D mutant protein is unable to activate PLC in vivo (28).
Although the 5501T/A Gαq gene mutation largely contributes to abolished light responses, this mutation is not fully responsible for the abolished light responses in nlr mutants because both nlr/Gαq1 and nlr/Gαq961 flies still exhibited a residual light response similar to Gαq1 and Gαq961 mutants (12). These data also excluded the possibility that the GαqV303D mutant protein dominantly suppresses the function of Gαq protein. Gαq1 expression in nlr mutants largely recovers the light response, and further excluded the possibility that abolished light responses in nlr mutants are due to the dominant suppression of GαqV303D mutant protein.
The Gαq gene encodes several Gαq splice variants, and Gαq221c mutants disrupt the expression of all Gαq splice variants (21). Our ERG recording revealed that Gαq221c null mutant clones showed no light responses. Previous whole-cell voltage-clamp recordings showed that the photoresponse of Gαq1 homozygous cells is larger than that of Gαq1 heterozygous cells (20). These results indicate that other Gαq splice variants might contribute to the residual light response in Gαq1 null mutants. In this study, we demonstrate that Gαq3 contributes to the residual light response in Gαq1 null mutants.
The Gαq gene encodes several Gαq splice variants. Originally, two cDNAs resulting from different Gαq gene splicing were isolated (23). These two cDNAs encode Gαq1 and Gαq2 isoform proteins, respectively. Functional studies demonstrated that Gαq1 mediates the light response, whereas Gαq2 has no effect on phototransduction (20). Subsequently, two additional Gαq splice variants were isolated (9). Now, seven total Gαq splice variants have been annotated in flybase, and these splice variants encode three different isoform proteins, including Gαq1, Gαq3, and Gαq4. In this study, we demonstrated that Gαq3 also mediate phototransduction. Overexpression of Gαq3 in nlr mutants induced detectable light responses but failed to fully restore the light response. Interestingly, the rescue flies exhibited comparable ERG trace amplitude and dynamics with that of Gαq1 and Gαq961 flies. These results indicate that different Gαq isoform proteins play different roles in phototransduction.
Gαq mediates retinal degeneration
Mutations in most genes encoding components of the phototransduction cascade result in rapid retinal degeneration, except for Gαq hypomorphic allele Gαq1 and Gαq1 isoform null mutant allele Gαq961 (12, 13, 19, 29). Previous studies have shown that both Gαq1 and Gαq961 mutants underwent slow light-dependent retinal degeneration due to slow accumulation of stable Rh1/Arr2 complexes (12, 29). In these Gαq mutants, the small residual photoresponse may reduce Ca2+ influx, which partially activates CaM kinase II and leads to the slow release of Arr2 from Rh1. In this study, we showed that nlr mutants underwent rapid light-dependent retinal degeneration similar to that observed in norpA mutants (15, 30). We showed that disruption of stable Rh1/Arr2 complex formation prevented retinal degeneration in the mutants. Under normal conditions, the interaction between Arr2 and Rh1 is transient, because light-triggered Ca2+ influx may activate CaM kinase II, which subsequently phosphorylates Arr2 to release Arr2 from Rh1 (17, 18). In nlr mutants, photoresponses were completely abolished so that the normal rise in Ca2+ after light stimulation was blocked, causing stable Rh1/Arr2 complex formation and retinal degeneration. These observations and explanations are consistent with mutations such as trp and norpA.
Gαq3 isoforms regulate Rh1 synthesis
In this study, we showed the first evidence that Gαq3 regulates Rh1 synthesis. Rh1 is transported to the plasma membrane by vesicular transport mechanisms regulated by a large number of trafficking proteins (31–35). Previous studies have shown that Gαq homologue CG30054 regulates inositol 1,4,5,-trisphosphate receptor (IP3R) to mediate calcium mobilization from intracellular stores and promote calcium-regulated secretory vesicle exocytosis (36). Given that Gαq3 shows high sequence identity to CG30054, they may regulate Rh1 synthesis through promoting calcium-regulated secretory vesicle exocytosis.
Experimental procedures
Fly genetics
p[GawB] strain collections obtained from Yi Rao's lab were originally generated by Ulrike Heberlein's lab (37) and nlr mutant flies were out-crossed for eight generations with the w1118 strain to eliminate the genetic background. The Gαq1 mutant and p[HS::dGαq] transgenic fly were obtained from Charles S. Zuker's laboratory. All other flies used in this work were from Bloomington stock center. Genetic mosaics of the deficiency line P{neoFRT}42D,Df(2R)Gαq1.3 and Gαq221c were induced by the FLP-FRT,p[GMR-hid] technique with an ey-FLP driver to generate mitotic clones of a single genotype in the eye (38). The genotype of WT flies is w1118 and the mutant alleles used for each gene in this work are Gαq961, Gαq1, and arr25. To avoid light-dependent retinal degeneration, all flies were reared at 22 °C in dark and examined at 1–2 days old.
Generation of transgenic flies
To generate p[UAS::Gαq3] transgenic flies, Gαq3 cDNA was subcloned into the pUAST-attB vector and injected into y1,w67c23;P{CaryattP2} flies. The transgene was subsequently crossed into the nlr mutant background and proteins were specifically expressed in the eye using binary expression systems (39).
Antibodies
The Nrv3 antibody generated by GenScript (Nanjing, China) was raised in rabbits against GST-Nrv3 (96–176 amino acids) recombinant protein. The antibody was purified through an affinity column coupled with His-Nrv3 fusion protein. The sources of other antibodies were Developmental Studies Hybridoma Bank (Rh1, RRID: AB528451; ATPα, RRID: AB2166869), Millipore (C-Gαq, RRID: AB310221), Abcam (N-Gαq, ab190082), Sigma (tubulin, RRID: AB477593), Hong-sheng Li (TRP), and Craig Montell (PLC).
Inverse PCR
Inverse PCR was performed as previously described (40). Briefly, genomic DNA was first digested with HpaII, and the resulting fragments were circulated using DNA ligase. The circular products were used as templates in PCR amplifications with primers flanking the 5′ end or 3′ end of the known p-element sequence. The PCR products were sequenced and aligned to the fly reference genome to locate p-element insertion sites.
Electrophysiological recordings
ERG recordings were performed as previously described (41). Briefly, recording and reference electrodes filled with Ringer's solution were placed on the surfaces of the fly eye and shoulder. Fly eyes were stimulated with 5-s light pulses (4000 Lux). For each genotype and condition, more than eight flies were examined. To quantitate the light response, the amplitude of ERG response was measured and the standard deviation was calculated.
Intracellular recordings were performed as previously described (42). Briefly, a low-resistance glass microelectrode filled with 2 m KCl was placed into a small hole on the compound eye. A reference electrode was filled with Ringer's solution and placed at the retina layer. The microelectrode placed on the eye was gradually inserted into the opening until light-induced membrane depolarization was observed. The signals were amplified and recorded using a Warner IE210 Intracellular Electrometer.
Western blots
After immersing fly heads in precooled phosphate-buffered saline (PBS), retinas were separated from brains with fine tweezers and homogenized in SDS-sample buffer. Proteins were fractionated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Pall) in Tris glycine buffer. Blots were probed with anti-Rh1 (1:3,000), anti-C-Gαq (1:1,000), anti-N-Gαq (1:1,000), anti-Arr2 (1:1,000), anti-INAD (1:1,000), anti-PLC (1:1,000), anti-TRP (1:1,000), anti-ATPα (1:2,000), anti-Nrv3 (1:2,500), and anti-tubulin (1:10,000) antibodies. Blots were subsequently probed with either anti-rabbit or anti-mouse IgG conjugated with peroxidase (GE Healthcare), and signals were detected using ECL reagents (Amersham Biosciences).
Real-time RT-PCR
Total RNA was extracted from isolated retina using TRIzol reagent (Invitrogen). Real-time RT-PCR was conducted using One-step SYBR Primer Script RT-PCR kit (TAKARA) with primer pairs for Gαq1 (5′-GATCACGCTGCGGCCAAACA-3′/5′-GTTTTCGGTATCTGTGGC-3′), Gαq3 (5′-CAGATTTAGAATGGTAGACG-3′/5′-CCTCCATTCGATTCTCATTATC-3′), and rp49 (5′-AACGTTTACAAATGTGTATTCCGACC-3′/5′-ATGACCATCCGCCCAGCATACAGG-3′). Rp49 was used as an internal control, and relative mRNA levels were calculated by setting WT Gαq1 and Gαq3 mRNA levels as 100%.
EM
Electron microscopy (EM) was performed as described previously (43). Fly heads were immerged in 2.5% glutaraldehyde, 0.1 m sodium cacodylate (pH 7.2) at 4 °C for 12 h. After rinsing with 0.1 m sodium cacodylate three times, fixed fly heads were stained with 1% osmium tetroxide for 1 h at room temperature. A series of standard ethanol dehydrations were conducted, and fly heads were immersed in two 10-min washes of propylene oxide. Fly heads were then embedded with standard procedures. For EM, 100-nm thin sections were cut and collected on copper support grids. After staining with uranyl acetate, sections were stained with lead citrate. Micrographs were taken at 80 KV on Hitachi-7650.
Arr2-binding assays
Arr2-binding assays were carried out as described previously (16). For each group and condition, 10 adult flies were collected and adapted in complete darkness for 2 h. After exposure with 60 s of pure blue light (480 ± 10 nm), fly heads were isolated by liquid nitrogen and homogenized in the dark. After centrifugation at 14,600 × g for 5 min, the pellet and supernatant were separated for Western blot analysis. Arr2-release assays were performed in the same manner, except that flies were exposed to pure blue light for 60 s, followed by pure orange light exposure for 2 min (580 ± 10 nm). All operations were conducted under very dim red light.
Experimental design and statistical analysis
Genetic studies and electrophysiological recordings on fly eyes were conducted to explore the function of Gαq splice variants and Gαq homology in phototransduction and Rh1 endocytosis. Biochemical studies and genetic studies on fly eyes, etc. were conducted to explore the detailed mechanism.
Western blotting was analyzed by ImageJ software (National Institute of Health, USA), and data from three independent experiments were averaged. Statistical analysis was performed using Prism 6.0 (GraphPad software). All data are presented as mean ± S.D. For quantification of ERG amplitudes, more than eight flies were recorded for each genotype and ERG amplitudes were averaged to obtain a mean. All data are presented as mean ± S.D. Two-tailed Student's t tests were used to compare between genotypes. Statistical significance was set as p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), and no significance (n.s.).
Data availability
All data are contained within the manuscript.
Author contributions
Q. G. and J. W. data curation; Q. G. formal analysis; Q. G. and J. W. investigation; Y. T., S. C., Z. C. Z., and J. H. funding acquisition; Y. T., S. C., and Z. C. Z. methodology; Y. T., S. C., and Z. C. Z. writing-review and editing; J. H. conceptualization; J. H. supervision; J. H. writing-original draft; J. H. project administration.
Acknowledgments
We thank Ulrike Heberlein and Yi Rao for the p[GawB] strain collection; Charles S. Zuker for Gαq1 mutant flies and p[HS::Gαq1] transgenic flies; Craig Montell for anti-PLC antibody; Hong-sheng Li for anti-TRP antibody; Bloomington Stock Center for the flies; Wen Hu for the ERG screen; and members of the Han laboratory for critical comments on the manuscript.
This work was supported by National Natural Science Foundation of China Grant 31771171 (to J. H.), and Jiangsu Natural Science Foundation for outstanding young scientists Grants BK20170080 (to Z. C. Z.), BK20180061 (to Y. T.), and BK20170703 (to S. C.). The authors declare that they have no conflicts of interest with the contents of this article.
- PLC
- phospholipase C
- IP3
- inositol 1,4,5-triphosphate
- Trp
- transient receptor potential
- TrpL
- transient receptor potential-like
- DAG
- diacylglycerol
- ERG
- electroretinogram.
References
- 1. Pierce K. L., Premont R. T., and Lefkowitz R. J. (2002) Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 3, 639–650 10.1038/nrm908 [DOI] [PubMed] [Google Scholar]
- 2. Neer E. J. (1995) Heterotrimeric G proteins: organizers of transmembrane signals. Cell 80, 249–257 10.1016/0092-8674(95)90407-7 [DOI] [PubMed] [Google Scholar]
- 3. Tian Y., Hu W., Tong H., and Han J. (2012) Phototransduction in Drosophila. Sci. China. Life Sci. 55, 27–34 10.1007/s11432-011-4512-4, 10.1007/s11427-012-4272-4 [DOI] [PubMed] [Google Scholar]
- 4. Bloomquist B. T., Shortridge R. D., Schneuwly S., Perdew M., Montell C., Steller H., Rubin G., and Pak W. L. (1988) Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell 54, 723–733 10.1016/S0092-8674(88)80017-5 [DOI] [PubMed] [Google Scholar]
- 5. Montell C., and Rubin G. M. (1989) Molecular characterization of the Drosophila trp locus: a putative integral membrane protein required for phototransduction. Neuron 2, 1313–1323 10.1016/0896-6273(89)90069-X [DOI] [PubMed] [Google Scholar]
- 6. Hardie R. C., and Minke B. (1992) The trp gene is essential for a light-activated Ca2+ channel in Drosophila photoreceptors. Neuron 8, 643–651 10.1016/0896-6273(92)90086-S [DOI] [PubMed] [Google Scholar]
- 7. Phillips A. M., Bull A., and Kelly L. E. (1992) Identification of a Drosophila gene encoding a calmodulin-binding protein with homology to the trp phototransduction gene. Neuron 8, 631–642 10.1016/0896-6273(92)90085-R [DOI] [PubMed] [Google Scholar]
- 8. Ranganathan R., Harris G. L., Stevens C. F., and Zuker C. S. (1991) A Drosophila mutant defective in extracellular calcium-dependent photoreceptor deactivation and rapid desensitization. Nature 354, 230–232 10.1038/354230a0 [DOI] [PubMed] [Google Scholar]
- 9. Ratnaparkhi A., Banerjee S., and Hasan G. (2002) Altered levels of Gq activity modulate axonal pathfinding in Drosophila. J. Neurosci. 22, 4499–4508 10.1523/JNEUROSCI.22-11-04499.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Niemeyer B. A., Suzuki E., Scott K., Jalink K., and Zuker C. S. (1996) The Drosophila light-activated conductance is composed of the two channels TRP and TRPL. Cell 85, 651–659 10.1016/S0092-8674(00)81232-5 [DOI] [PubMed] [Google Scholar]
- 11. Reuss H., Mojet M. H., Chyb S., and Hardie R. C. (1997) In vivo analysis of the Drosophila light-sensitive channels, TRP and TRPL. Neuron 19, 1249–1259 10.1016/S0896-6273(00)80416-X [DOI] [PubMed] [Google Scholar]
- 12. Hu W., Wan D., Yu X., Cao J., Guo P., Li H. S., and Han J. (2012) Protein Gq modulates termination of phototransduction and prevents retinal degeneration. J. Biol. Chem. 287, 13911–13918 10.1074/jbc.M112.339895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang T., and Montell C. (2007) Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 454, 821–847 10.1007/s00424-007-0251-1 [DOI] [PubMed] [Google Scholar]
- 14. Wang T., Jiao Y., and Montell C. (2005) Dissecting independent channel and scaffolding roles of the Drosophila transient receptor potential channel. J. Cell Biol. 171, 685–694 10.1083/jcb.200508030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alloway P. G., Howard L., and Dolph P. J. (2000) The formation of stable rhodopsin-arrestin complexes induces apoptosis and photoreceptor cell degeneration. Neuron 28, 129–138 10.1016/S0896-6273(00)00091-X [DOI] [PubMed] [Google Scholar]
- 16. Mu Y., Tian Y., Zhang Z. C., and Han J. (2019) Metallophosphoesterase regulates light-induced rhodopsin endocytosis by promoting an association between arrestin and the adaptor protein AP2. J. Biol. Chem. 294, 12892–12900 10.1074/jbc.RA119.009602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alloway P. G., and Dolph P. J. (1999) A role for the light-dependent phosphorylation of visual arrestin. Proc. Natl. Acad. Sci. U.S.A. 96, 6072–6077 10.1073/pnas.96.11.6072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matsumoto H., Kurien B. T., Takagi Y., Kahn E. S., Kinumi T., Komori N., Yamada T., Hayashi F., Isono K., and Pak W. L. (1994) Phosrestin I undergoes the earliest light-induced phosphorylation by a calcium/calmodulin-dependent protein kinase in Drosophila photoreceptors. Neuron 12, 997–1010 10.1016/0896-6273(94)90309-3 [DOI] [PubMed] [Google Scholar]
- 19. Kiselev A., Socolich M., Vinós J., Hardy R. W., Zuker C. S., and Ranganathan R. (2000) A molecular pathway for light-dependent photoreceptor apoptosis in Drosophila. Neuron 28, 139–152 10.1016/S0896-6273(00)00092-1 [DOI] [PubMed] [Google Scholar]
- 20. Scott K., Becker A., Sun Y., Hardy R., and Zuker C. (1995) Gqα protein function in vivo: genetic dissection of its role in photoreceptor cell physiology. Neuron 15, 919–927 10.1016/0896-6273(95)90182-5 [DOI] [PubMed] [Google Scholar]
- 21. Banerjee S., Joshi R., Venkiteswaran G., Agrawal N., Srikanth S., Alam F., and Hasan G. (2006) Compensation of inositol 1,4,5-trisphosphate receptor function by altering sarco-endoplasmic reticulum calcium ATPase activity in the Drosophila flight circuit. J. Neurosci. 26, 8278–8288 10.1523/JNEUROSCI.1231-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mismer D., and Rubin G. M. (1989) Definition of cis-acting elements regulating expression of the Drosophila melanogaster ninaE opsin gene by oligonucleotide-directed mutagenesis. Genetics 121, 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee Y. J., Dobbs M. B., Verardi M. L., and Hyde D. R. (1990) dgq: a Drosophila gene encoding a visual system-specific Gα molecule. Neuron 5, 889–898 10.1016/0896-6273(90)90349-K [DOI] [PubMed] [Google Scholar]
- 24. Lee Y. J., Shah S., Suzuki E., Zars T., O'Day P. M., and Hyde D. R. (1994) The Drosophila dgq gene encodes a G alpha protein that mediates phototransduction. Neuron 13, 1143–1157 10.1016/0896-6273(94)90052-3 [DOI] [PubMed] [Google Scholar]
- 25. Noel J. P., Hamm H. E., and Sigler P. B. (1993) The 2.2 Å crystal structure of transducin-α complexed with GTPγS. Nature 366, 654–663 10.1038/366654a0 [DOI] [PubMed] [Google Scholar]
- 26. Lambright D. G., Noel J. P., Hamm H. E., and Sigler P. B. (1994) Structural determinants for activation of the α-subunit of a heterotrimeric G protein. Nature 369, 621–628 10.1038/369621a0 [DOI] [PubMed] [Google Scholar]
- 27. Lambright D. G., Sondek J., Bohm A., Skiba N. P., Hamm H. E., and Sigler P. B. (1996) The 2.0 Å crystal structure of a heterotrimeric G protein. Nature 379, 311–319 10.1038/379311a0 [DOI] [PubMed] [Google Scholar]
- 28. Cao J., Bollepalli M. K., Hu Y., Zhang J., Li Q., Li H., Chang H., Xiao F., Hardie R. C., Rong Y. S., and Hu W. (2018) A single residue mutation in the Gαq subunit of the G protein complex causes blindness in Drosophila. G3 8, 363–371 10.1534/g3.117.300340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Iakhine R., Chorna-Ornan I., Zars T., Elia N., Cheng Y., Selinger Z., Minke B., and Hyde D. R. (2004) Novel dominant rhodopsin mutation triggers two mechanisms of retinal degeneration and photoreceptor desensitization. J. Neurosci. 24, 2516–2526 10.1523/JNEUROSCI.5426-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meyertholen E. P., Stein P. J., Williams M. A., and Ostroy S. E. (1987) Studies of the Drosophila norpA phototransduction mutant: II. photoreceptor degeneration and rhodopsin maintenance. J. Comp. Physiol. A 161, 793–798 10.1007/BF00610221 [DOI] [PubMed] [Google Scholar]
- 31. Schopf K., and Huber A. (2017) Membrane protein trafficking in Drosophila photoreceptor cells. Eur. J. Cell Biol. 96, 391–401 10.1016/j.ejcb.2016.11.002 [DOI] [PubMed] [Google Scholar]
- 32. Shetty K. M., Kurada P., and O'Tousa J. E. (1998) Rab6 regulation of rhodopsin transport in Drosophila. J. Biol. Chem. 273, 20425–20430 10.1074/jbc.273.32.20425 [DOI] [PubMed] [Google Scholar]
- 33. Satoh A. K., O'Tousa J. E., Ozaki K., and Ready D. F. (2005) Rab11 mediates post-Golgi trafficking of rhodopsin to the photosensitive apical membrane of Drosophila photoreceptors. Development 132, 1487–1497 10.1242/dev.01704 [DOI] [PubMed] [Google Scholar]
- 34. Xiong B., and Bellen H. J. (2013) Rhodopsin homeostasis and retinal degeneration: lessons from the fly. Trends Neurosci. 36, 652–660 10.1016/j.tins.2013.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xiong B., Bayat V., Jaiswal M., Zhang K., Sandoval H., Charng W. L., Li T., David G., Duraine L., Lin Y. Q., Neely G. G., Yamamoto S., and Bellen H. J. (2012) Crag is a GEF for Rab11 required for rhodopsin trafficking and maintenance of adult photoreceptor cells. PLoS Biol. 10, e1001438 10.1371/journal.pbio.1001438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamanaka N., Marqués G., and O'Connor M. B. (2015) Vesicle-mediated steroid hormone secretion in Drosophila melanogaster. Cell 163, 907–919 10.1016/j.cell.2015.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Corl A. B., Berger K. H., Ophir-Shohat G., Gesch J., Simms J. A., Bartlett S. E., and Heberlein U. (2009) Happyhour, a Ste20 family kinase, implicates EGFR signaling in ethanol-induced behaviors. Cell 137, 949–960 10.1016/j.cell.2009.03.020 [DOI] [PubMed] [Google Scholar]
- 38. Stowers R. S., and Schwarz T. L. (1999) A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 152, 1631–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guo C., Pan Y., and Gong Z. (2019) Recent advances in the genetic dissection of neural circuits in Drosophila. Neurosci. Bull. 35, 1058–1072 10.1007/s12264-019-00390-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dunne C. R., Cillo A. R., Glick D. R., John K., Johnson C., Kanwal J., Malik B. T., Mammano K., Petrovic S., Pfister W., Rascoe A. S., Schrom D., Shapiro S., Simkins J. W., Strauss D., et al. (2014) Structured inquiry-based learning: Drosophila GAL4 enhancer trap characterization in an undergraduate laboratory course. PLoS Biol. 12, e1002030 10.1371/journal.pbio.1002030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Han J., Gong P., Reddig K., Mitra M., Guo P., and Li H.-S. (2006) The fly CAMTA transcription factor potentiates deactivation of rhodopsin, a G protein-coupled light receptor. Cell 127, 847–858 10.1016/j.cell.2006.09.030 [DOI] [PubMed] [Google Scholar]
- 42. Hu W., Wang T., Wang X., and Han J. (2015) Ih channels control feedback regulation from amacrine cells to photoreceptors. PLoS Biol. 13, e1002115 10.1371/journal.pbio.1002115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tian Y., Li T., Sun M., Wan D., Li Q., Li P., Zhang Z. C., Han J., and Xie W. (2013) Neurexin regulates visual function via mediating retinoid transport to promote rhodopsin maturation. Neuron 77, 311–322 10.1016/j.neuron.2012.11.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the manuscript.