Abstract
Objective
To explore the transcriptomic differences between patients with hypertrophic cardiomyopathy (HCM) and controls.
Patients and Methods
RNA was extracted from cardiac tissue flash frozen at therapeutic surgical septal myectomy for 106 patients with HCM and 39 healthy donor hearts. Expression profiling of 37,846 genes was performed using the Illumina Human HT-12v3 Expression BeadChip. All patients with HCM were genotyped for pathogenic variants causing HCM. Technical validation was performed using quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot. This study was started on January 1, 1999, and final analysis was completed on April 20, 2020.
Results
Overall, 22% of the transcriptome (8443 of 37,846 genes) was expressed differentially between HCM and control tissues. Analysis by genotype revealed that gene expression changes were similar among genotypic subgroups of HCM, with only 4% (1502 of 37,846) to 6% (2336 of 37,846) of the transcriptome exhibiting differential expression between genotypic subgroups. The qRT-PCR confirmed differential expression in 92% (11 of 12 genes) of tested transcripts. Notably, in the context of coronavirus disease 2019 (COVID-19), the transcript for angiotensin I converting enzyme 2 (ACE2), a negative regulator of the angiotensin system, was the single most up-regulated gene in HCM (fold-change, 3.53; q-value =1.30×10−23), which was confirmed by qRT-PCR in triplicate (fold change, 3.78; P=5.22×10−4), and Western blot confirmed greater than 5-fold overexpression of ACE2 protein (fold change, 5.34; P=1.66×10−6).
Conclusion
More than 20% of the transcriptome is expressed differentially between HCM and control tissues. Importantly, ACE2 was the most up-regulated gene in HCM, indicating perhaps the heart’s compensatory effort to mount an antihypertrophic, antifibrotic response. However, given that the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) uses ACE2 for viral entry, this 5-fold increase in ACE2 protein may confer increased risk for COVID-19 manifestations and outcomes in patients with increased ACE2 transcript expression and protein levels in the heart.
Abbreviations and Acronyms: ΔCt, transcript of interest minus GAPDH control; ACE2, angiotensin I converting enzyme 2; ACEi, angiotensin-converting enzyme inhibitor; ARB, angiotensin receptor blocker; AT1R, angiotensin type 1 receptor; BP, blood pressure; cDNA, complementary DNA; CHF, congestive heart failure; COVID-19, coronavirus disease 2019; ECG, electrocardiogram; GTP, guanosine triphosphate; HCM, hypertrophic cardiomyopathy; hrsACE2, human recombinant soluble angiotensin I converting enzyme 2; HTN, hypertension; ICU, intensive care unit; IQR, interquartile range; LV, left ventricular; MIG, maximum instantaneous gradient; mRNA, messenger RNA; MYBPC3, myosin binding protein C; MYH7, beta myosin heavy chain; NA, not available; NS, not significant; NYHA, New York Heart Association; qRT-PCR, quantitative real-time polymerase chain reaction; RAAS, renin-angiotensin-aldosterone system; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SCD, sudden cardiac death; UTR, untranslated region
Hypertrophic cardiomyopathy (HCM) affects approximately 1 in 500 individuals1 and is among the leading causes of identifiable sudden cardiac death (SCD) in the young.2 HCM is often a genetic disease, typically with autosomal dominant inheritance, that is defined clinically as cardiac hypertrophy without physiologic explanation. Hundreds of pathogenic variants in many HCM-susceptibility genes have been identified, most of which encode components of the sarcomere.3, 4, 5, 6, 7, 8, 9, 10, 11 However, genetic tests are negative in approximately 50% of all unrelated patients with HCM that are diagnosed by clinical studies.4 Additionally, the transcriptional changes that cause and result from HCM, with and without pathogenic variants, remain largely unknown because prior studies analyzed data from small numbers of patients.5 , 6 To better identify common transcriptional changes that represent fundamental and heretofore unrecognized pathogenic responses of human HCM, we performed transcriptome analysis of human HCM tissues.
Patients and Methods
We designed a case-control study to identify the messenger RNAs (mRNAs) differentially expressed in HCM-affected myocardium (n=106) vs control myocardium (n=39). All patients signed informed consent, and protocols were approved by Mayo Clinic Institutional Review Board or the Human Research Ethics Committee of the University of Sydney. This study was started on January 1, 1999, and final analysis was completed on April 20, 2020.
All patients undergoing therapeutic surgical septal myectomy for symptomatic relief of obstructive HCM between January 1, 1999, and December 31, 2010, were eligible for inclusion in this study. The diagnosis of HCM was made by an experienced cardiologist from Mayo Clinic HCM Clinic based on physical examination, electrocardiogram (ECG), and echocardiogram/cardiac magnetic resonance imaging findings. Diagnosis was corroborated by histologic examination of the patient’s surgical septal myectomy specimen. A representative portion of myectomy specimen was flash frozen at the time of excision and subsequently stored at −80°C.
Data for patient age, sex, age at diagnosis, New York Heart Association (NYHA) classification, blood pressure, heart rate, family history of HCM, and family history of SCD were extracted from each patient’s electronic medical record. Echocardiographic parameters were extracted from each patient’s preoperative echocardiography study. Degree of endocardial and interstitial fibrosis was assessed semi-quantitatively at the time of resection by a cardiovascular pathologist (J.J.M.).
A cohort of control tissue (n=39) was procured from the University of Sydney consisting of donor hearts for which there was not a suitable transplant recipient. A normal phenotype had been confirmed by means of cardiac examination, ECG, and echocardiogram obtained within 24 hours before explantation.
DNA Extraction
DNA was extracted from the HCM myectomy and control donor heart tissues using the Qiagen PureGene DNA Purification Kit (Qiagen, Inc) according to the manufacturer’s protocol. Briefly, cells were lysed with detergent, RNA was removed using an RNase enzyme, proteins were removed by salt precipitation, and DNA was recovered with alcohol precipitation.
Genotyping
Damaging variants in 10 genes implicated in sarcomeric HCM (ACTC1, MYBPC3, MYH6, MYH7, MYL2, MYL3, TNNC1, TNNI3, TNNT2, and TPM1; for expansion of gene symbols, use search tool at www.genenames.org) and 3 genes known to mimic HCM (GLA, LAMP2, and PRKAG2) were studied using filter-based hybridization capture, as described previously.7 In brief, genomic DNA extracted from the cardiac myectomy tissue was used to construct barcoded (3 base pairs) genomic DNA libraries. Ten or 20 barcoded genomic DNA libraries were then combined into each sample pool. DNA concatemers consisting of target gene segments were amplified and bound to 25-mm nitrocellulose membrane filters (Millipore). Sample genomic library pools were enriched by hybridization to filter-bound DNA concatemers and subjected to either single- or paired-end sequencing using either Genome Analyzer II (Illumina) or HiSeq (Illumina). Sequences were aligned with Novoalign (Novocraft Technologies) and analyzed using The Genome Analysis Toolkit (Broad Institute).8
Tissue RNA Extraction and Quality Assessment
Total RNA was extracted from each tissue sample using the Qiagen miRNeasy Kit (Qiagen, Inc) according to the manufacturer’s protocol. The technique uses a phenol/chloroform extraction protocol and RNA purification columns. RNA quality was assessed by the Mayo Clinic Advanced Genomic Technology Center Microarray Shared Resource using the 2100 Bioanalyzer (Agilent) to obtain electropherograms from which an RNA integrity number9 was calculated. Only RNA samples with high enough quality (RNA integrity number ≥6.0) were used for transcriptome analyses.
Microarray Hybridization
Complementary DNA (cDNA) corresponding to case and control mRNA was produced by reverse transcription. The cDNA was converted to biotin-labeled complementary RNA and hybridized to the Human HT-12 v3 Expression BeadChip (Illumina) to quantify the expression level of 48,804 mRNA transcripts representing 37,846 genes by streptavidin-Cy3 staining and laser excitation fluoroscopy. The HCM and donor samples were randomly assigned to ensure that the groups were balanced over batches (BeadChips) to avoid potential confounding from batches. Laboratory quality control measures included simultaneously hybridizing housekeeping controls, hybridization controls, negative controls, and technical replicates.
Quantitative Real-Time Polymerase Chain Reaction and Western Blot Validation
Quantitative real-time polymerase chain reaction (qRT-PCR) was performed on randomly selected samples from each comparison group (18 HCM cases and 13 controls) to validate some of the microarray results. The cDNA was generated from each RNA sample using the iScript cDNA Synthesis Kit (Bio-Rad) according to the manufacturer’s protocol. TaqMan Gene Expression Assays (Applied Biosystems) for the genes of interest and the endogenous control GAPDH were used per the manufacturer’s protocol and were run on the ABI Prism 7900HT Real Time System (Applied Biosystems). Each reaction was run in triplicate. Protein expression of ACE2 was confirmed by Western blot analysis with loading normalized for actin.
Immunohistochemistry
Tissue sections were deparaffinized in xylene, dipped in decreasing concentrations of ethyl alcohol, and rehydrated in distilled water. Antigen retrieval for ACE2 was performed by placing slides in preheated citrate as the retrieval solution in a steamer at 98°C for 40 minutes. The staining procedure was carried out in the Dako Autostainer Plus as follows. Tissue sections were treated with Peroxidase Blocking Reagent (Dako) for 15 minutes, washed with 1× Wash Buffer (Dako), and treated with Protein Block SNIPER (Biocare Medical LLC) for 10 minutes. The primary antibody for ACE2 (R&D Systems) was diluted 1:400 in Background Reducing Diluent (Dako) and incubated for 60 minutes at room temperature. After rinsing with wash buffer, sections were incubated in secondary antibody and tertiary reagent from the Goat HRP Kit (Biocare Medical LLC) for 15 minutes. Betazoid diaminobenzidine chromogenic substrate system (Biocare Medical LLC) was used for colorimetric visualization. Counterstaining with hematoxylin followed by dehydration in increasing concentrations of ethyl alcohol and xylene were performed before permanent coverslipping.
All slides were graded by a cardiac pathologist (J.J.M.) and graded for intensity (0 = no staining, 1+ = dot-like sarcoplasmic staining, and 2+=diffuse sarcoplasmic staining) and distribution (scoring 0 = 0% of cells, 1 = <25%, 2 = 26%-75%, and 3 = ≥75% of cells) of ACE2 staining.
Analytical and Statistical Methods
Specimens were allocated randomly to arrays using randomized block methods to avoid confounding of biological and experimental effects. Illumina BeadStudio, version 3.1.3, with gene expression module 3.4 was used to process raw data without background correction and normalization. The gene level expression data were exported and analyzed in R (The R Foundation; http://www.r-project.org/). Briefly, the un-normalized raw data were first log2 transformed and evaluated for potential outlier samples and bead chip effects by graphic and dimension reduction approaches (density plot, MA plot, and principal components analysis).
Outlier samples were excluded for further analyses and the remaining good samples were normalized together using fastlo,10 a model-based intensity-dependent normalization method that produces results essentially the same as those from cyclic loess10 but in a fraction of the time. Gene-level expression was compared between HCM and normal tissue or between genotype subgroups overall followed by pairwise contrasts through analysis of variance linear models together with false discovery rates (FDRs).11 Genes with a false discovery rate q-value less than 0.05 were considered statistically significant for HCM and normal tissue comparison.
For the pairwise comparisons between different genotype subgroups of HCM, P<.05 was considered statistically significant. A less stringent cutoff was used here in an effort to elucidate differences between 2 similar disease conditions, acknowledging that the chance of a false-positive result is higher. A cutoff for biological significance was set as an absolute fold change greater than 1.5 between genotype subsets of HCM.
The qRT-PCR data were analyzed after calculating 2−ΔCt for the average ΔCt value (transcript of interest minus GAPDH control) for the triplicate replicates of each sample. A 1-tailed t test was used. A fold change was calculated by the 2−ΔΔCt method,12 taking 2−ΔCt for the overall average of all cases divided by 2−ΔCt for the overall average of all controls.
Results
Cohorts
All participants with HCM (n=121) provided written informed consent to participate in this Mayo Clinic Institutional Review Board–approved study. Among 121 tissue samples, 106 samples passed RNA and microarray quality controls. The HCM samples were obtained from mostly white individuals (54 men and 52 women) aged 9 to 78 years. Clinical manifestations of HCM and control participants are summarized in Table 1 . For the 106 HCM cases, median age at diagnosis was 43 years (interquartile range [IQR], 27 to 55 years), median age at myectomy was 51 years (IQR, 32 to 60 years), 77% (n=82) had NYHA class III to IV symptoms, 27% (n=29) had a family history of HCM, and 13% (n=14) had a family history of SCD. Median left ventricular mass index was 171 g/m2 (IQR, 139 to 225 g/m2), median left ventricular wall thickness was 22 mm (IQR, 18 to 26 mm), and median left ventricular outflow tract maximum instantaneous gradient was 68 mm Hg (IQR, 29 to 100 mm Hg). Histopathologic examination of myectomy samples showed moderate or severe interstitial fibrosis in 27% (n=29) and moderate or severe endocardial fibrosis in 63% (n=67) of patients.
Table 1.
Demographics of HCM and Control Cohorts
| Cases | Controls | |
|---|---|---|
| No. | 106 | 39 |
| Sex (male/female), no. | 54/52 | 19/20 |
| Age, diagnosis (y), median (IQR) | 43 (27-55) | NA |
| Age, myectomy (y), median (IQR) | 51 (32-60) | N/A |
| Age, death (y), median (IQR) | N/A | 38 (23-48) |
| NYHA class III-IV, no. (%) | 82 (77) | NA |
| Family history of HCM, no. (%) | 29 (27) | NA |
| Family history of SCD, no. (%) | 14 (13) | NA |
| Systolic BP (mm Hg), median (IQR) | 120 (109-130) | NA |
| Diastolic BP (mm Hg), median (IQR) | 70 (62-80) | NA |
| Heart rate (beats/min), median (IQR) | 68 (60-76) | NA |
| Ejection fraction (%), median (IQR) | 74 (68-77) | NA |
| LV mass index (g/m2), median (IQR) | 171 (139-225) | NA |
| LV wall thickness (mm), median (IQR) | 22 (18-26) | NA |
| LV outflow tract MIG (mm Hg), median (IQR) | 68 (29-100) | NA |
| Interstitial fibrosis score of moderate or severe, no. (%) | 29 (27) | NA |
| Endocardial fibrosis score of moderate or severe, no. (%) | 67 (63) | NA |
BP = blood pressure; HCM = hypertrophic cardiomyopathy; IQR = interquartile range; LV = left ventricular; MIG = maximum instantaneous gradient; N/A = not applicable; NA = not available; NYHA = New York Heart Association; SCD = sudden cardiac death.
From the donor hearts, 39 of 44 RNA samples passed RNA and microarray quality control and were used for microarray analyses. These 39 donors included 19 men and 20 women, with an average age of 37±15 years, approximately 10 years younger than HCM cases (Table 1).
Pathogenic variants were identified in 55% (n=58 of 106) of the HCM cohort. In 45% (48/106) of participants, the 13-gene test panel did not identify the cause for HCM. As anticipated, mutations in MYBPC3 (encoding cardiac myosin binding protein C) and MYH7 (encoding beta myosin heavy chain) genes were the most commonly identified (23/106 [22%] and 17/106 [16%,] respectively). Clinical parameters of patients with HCM, based on the 2 largest genotype groups (MYBPC3-positive and MYH7-positive) are shown in Table 2 . These 2 groups were similar but differed from genotype-negative patients with HCM by younger age at diagnosis and myectomy, and higher prevalence of familial HCM, similarly to prior larger studies.4 , 13, 14, 15 There was no difference in NYHA classification, systolic blood pressure, heart rate, ejection fraction, left ventricular mass index, left ventricular wall thickness, left ventricular outflow tract maximum instantaneous gradient, or interstitial or endocardial fibrosis among the 3 primary genotypic subgroups (MHY7-positive, MYBPC3-positive, and genotype-negative HCM).
Table 2.
Baseline Characteristics by Major Genotype Subgroups
| MYBPC3-HCM | MYH7-HCM | Genotype Negative | P | |
|---|---|---|---|---|
| No. | 23 | 17 | 48 | |
| Sex (male/female) | 13/10 | 7/10 | 27/21 | NS |
| Age, diagnosis (y), median (IQR) | 37 (23-45) | 37 (13-45) | 52 (33-64) | .003 |
| Age, myectomy (y), median (IQR) | 38 (30-53) | 43 (16-52) | 57 (44-67) | .003 |
| NYHA class III-IV, no. (%) | 19 (83) | 12 (71) | 35 (73) | NS |
| Family history of HCM, no. (%) | 11 (48) | 7 (41) | 8 (17) | .01 |
| Family history of SCD, no. (%) | 6 (26) | 1 (6) | 5 (10) | NS |
| Systolic BP (mm Hg), median (IQR) | 120 (110-122) | 118 (102-127) | 125 (113-137) | NS |
| Diastolic BP (mm Hg), median (IQR) | 70 (62-75) | 66 (57-70) | 74 (64-82) | .01 |
| Heart rate (beats/min), median (IQR) | 68 (60-73) | 70 (60-78) | 68 (59-76) | NS |
| Ejection fraction (%), median (IQR) | 70 (65-76) | 75 (75-80) | 73 (70-76) | NS |
| LV mass index (g/m2), median (IQR) | 192 (144-238) | 183 (130-258) | 171 (134-217) | NS |
| LV wall thickness (mm), median (IQR) | 24 (18-27) | 25 (18-29) | 21 (16-24) | NS |
| LV outflow tract MIG (mm Hg), median (IQR) | 40 (16-106) | 81 (70-121) | 64 (30-94) | NS |
| Interstitial fibrosis score moderate or severe, no. (%) | 5 (22) | 4 (24) | 13 (27) | NS |
| Endocardial fibrosis score moderate or severe, no. (%) | 15 (65) | 13 (76) | 30 (63) | NS |
BP = blood pressure; HCM = hypertrophic cardiomyopathy; IQR = interquartile range; LV = left ventricular; MIG = maximum instantaneous gradient; MYBPC3 = myosin binding protein C; MYH7 = beta myosin heavy chain; NS = not significant; NYHA = New York Heart Association; SCD = sudden cardiac death.
Microarray Results
Cases vs Controls
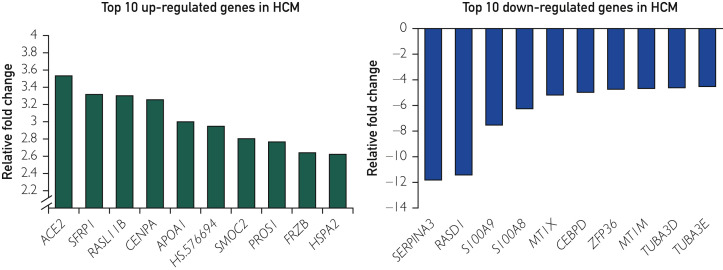
Overall, 37,846 genes targeted by 48,804 probes were analyzed for all samples (Gene Expression Omnibus accession number GSE36961), and 8443 genes (22% of the transcriptome) were expressed differentially between HCM and controls based on a false discovery rate q-value less than 0.05. These differentially expressed genes represent 1075 molecular functions and 4272 biological processes as defined by the Gene Ontology Consortium. The top 10 up-regulated (2.5- to 3.5-fold increased expression compared with controls) and top 10 down-regulated genes (4.5- to 11.8-fold decreased expression compared with controls) along with fold changes and Gene Ontology classification are listed in Figure 1 and Tables 3 and 4 .
Figure 1.
Top up-regulated and down-regulated genes in hypertrophic cardiomyopathy (HCM). Bar diagram shows the top 10 up-regulated (left) and down-regulated (right) genes in tissues of patients with obstructive HCM compared with controls.
Table 3.
Top 10 Differentially Expressed Messenger RNA Transcripts Up-Regulated in HCM Compared With Controls
| Gene | Official Full Name | Fold Change | q-Value | Gene Ontology Biological Process Term(s) | Gene Ontology Molecular Function Term(s) |
|---|---|---|---|---|---|
| ACE2 | Angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 | +3.53 | 1.30×10−23 | Angiotensin catabolic process in blood; regulation of inflammatory response; regulation of vasoconstriction | Peptidase activity; glycoprotein binding; zinc ion binding |
| SFRP1 | Secreted frizzled-related protein 1 | +3.32 | 5.93×10−21 | Regulation of cell growth; canonical Wnt receptor signaling pathway | Cysteine-type endopeptidase activity; Wnt-protein binding |
| RASL11B | RAS-like, family 11, member B | +3.30 | 3.44×10−22 | Small GTPase-mediated signal transduction | GTPase activity |
| CENPA | Centromere protein A | +3.25 | 3.37×10−16 | Nucleosome assembly | Chromatin binding; DNA binding; protein binding |
| APOA1 | Apolipoprotein A-I | +3.00 | 1.17×10−12 | Lipid metabolic process | Cholesterol transport activity |
| HS.576694 | Not available | +2.95 | 2.06×10−16 | Not available | Not available |
| SMOC2 | SPARC-related modular calcium binding 2 | +2.80 | 7.56×10−27 | Extracellular matrix organization | Calcium ion binding |
| PROS1 | Protein S (alpha) | +2.77 | 3.06×10−29 | Blood coagulation | Calcium ion binding |
| FRZB | Frizzled-related protein | +2.64 | 2.75×10−24 | Negative regulation of cell growth; negative regulation of Wnt receptor signaling pathway | Wnt-activated receptor activity; Wnt-protein binding |
| HSPA2 | Heat shock 70 kDa protein 2 | +2.62 | 1.07×10−16 | Positive regulation of cyclin-dependent protein kinase activity involved in G2/M | Adenosine triphosphate binding |
HCM = hypertrophic cardiomyopathy; GTPase = hydrolase enzyme that binds to the nucleotide guanosine triphosphate; RAS = rat sarcoma.
Table 4.
Top 10 Differentially Expressed mRNA Transcripts, Down-Regulated in HCM Compared With Controls
| Gene | Official Full Name | Fold Change q-Value | q-Value | Gene Ontology Biological Process Term(s) | Gene Ontology Molecular Function Term(s) |
|---|---|---|---|---|---|
| SERPINA3 | Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 | −11.81 | 1.73×10−37 | Inflammatory response; regulation of proteolysis; regulation of lipid metabolic process | Peptidase inhibitor activity; DNA binding; protein binding |
| RASD1 | RAS, dexamethasone-induced 1 | −11.43 | 2.26×10−40 | Small GTPase-mediated signal transduction | GTP binding; GTPase activity |
| S100A9 | S100 calcium-binding protein A9 | −7.54 | 7.18×10−36 | Cell-cell signaling; leukocyte chemotaxis; actin cytoskeleton reorganization | Calcium ion binding; protein binding; signal transducer activity |
| S100A8 | S100 calcium-binding protein A8 | −6.27 | 1.23×10−30 | Inflammatory response; response to zinc ion; response to ethanol | Calcium ion binding; protein binding |
| MT1X | Metallothionein 1X | −5.18 | 1.02×10−30 | Response to metal ion | Zinc ion binding |
| CEBPD | CCAAT/enhancer binding protein (C/EBP), delta | −4.95 | 1.13×10−40 | Transcription from RNA polymerase II promoter | Protein dimerization activity; sequence-specific DNA binding |
| ZFP36 | Zinc finger protein 36, C3H type, homolog (mouse) | −4.72 | 2.07×10−40 | 3'-UTR-mediated mRNA stabilization | DNA binding; mRNA binding; protein binding; zinc ion binding |
| MT1M | Metallothionein 1M | −4.67 | 2.49×10−35 | Negative regulation of growth | Zinc ion binding |
| TUBA3D | Tubulin, alpha 3d | −4.61 | 1.75×10−39 | Microtubule-based movement | GTP binding; GTPase activity; structural molecule activity |
| TUBA3E | Tubulin, alpha 3e | −4.51 | 0.00 | Microtubule-based movement | GTP binding; GTPase activity; structural molecule activity |
HCM = hypertrophic cardiomyopathy; GTP = guanosine triphosphate; GTPase = hydrolase enzyme that binds to the nucleotide guanosine triphosphate; mRNA = messenger RNA; RAS = rat sarcoma; UTR = untranslated region.
Quantitative RT-PCR
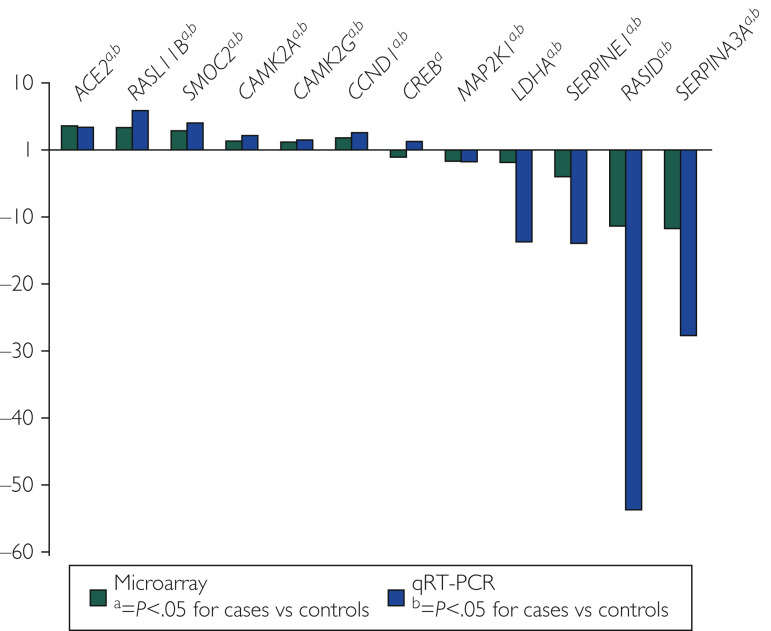
Using qRT-PCR, mRNA results were validated for 12 select genes with differential expression (fold change, >1.5) in HCM and control samples. Genes were selected based on potential functional roles in cardiac hypertrophy and included ACE2, CAMK2A, CAMK2G, CCND1, CREB1, LDHA, MAP2K1, RASD1, RASL11B, SERPINE1, SERPINA3, and SMOC2. With the exception of CREB1, all other genes (n=11 of 12; 92%) showed similar magnitude and directionality of differential expression by both techniques (Figure 2 ).
Figure 2.
Technical validation of the microarray technique. Microarray fold change and quantitative real-time polymerase chain reaction (qRT-PCR) fold change are plotted side by side for the 12 genes tested. The qRT-PCR data validated the microarray data for 11 of 12 genes (92%).
Overexpression of ACE2 in HCM
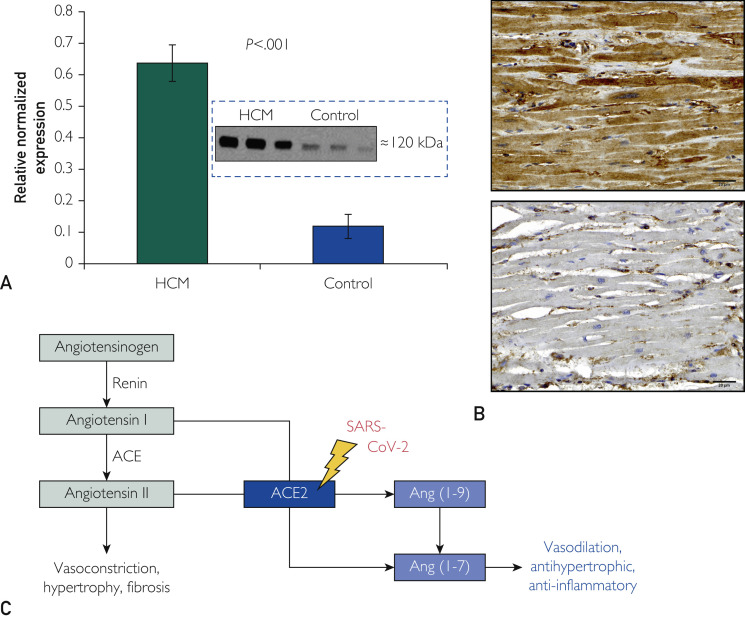
Notably, the ACE2-encoded angiotensin I/angiotensin II converting enzyme subtype 2, an important counter-regulator of the renin-angiotensin-aldosterone system (RAAS) involved in hypertrophy, fibrosis, and vasoconstriction, was the most up-regulated gene in HCM tissues (3.53-fold increase vs controls, confirmed by qRT-PCR; Figures 1 and 2). Western blot analyses indicated 5.34-fold increased ACE2 protein expression compared with control (P<.001; Figure 3 A). Additionally, immunohistochemistry cardiomyocyte staining of cardiac myectomy tissue from 14 patients with HCM showed significantly increased ACE2-antibody staining intensity (P=.002) and distribution (P<.001) of ACE2 protein compared with 8 control samples (Figure 3B).
Figure 3.
Marked accentuation of angiotensin-converting enzyme type 2 (ACE2) in hypertrophic cardiomyopathy (HCM). Bar diagram shows: (A) 5.3-fold increase in ACE2 protein in HCM patients compared with controls by Western blot analysis and (B) significant staining of ACE2 antibody in the myectomy specimen from a patient with obstructive HCM. Flow chart shows (C) the role of ACE2 in converting angiotensin (ang) I to ang (1-9) and angiotensin II to ang (1-7) to counter the effects of angiotensin II. SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2.
Interestingly, ACE2 is located on the X-chromosome and therefore sex differences could be expected. Thus, we performed a sex-corrected analysis of our expression data for ACE2 and observed that there was still 3.60-fold higher expression of ACE2 in HCM cases vs controls (P=2.66E-27). Further, after adding age as an adjustment variable to account for a younger control cohort, ACE2 overexpression in cases still persisted (3.66-fold; P=1.01E-26). Overall, although there was no significant difference in ACE2 expression between males and females in the control cohort (fold change, −1.01; P=.95), ACE2 transcript levels were 1.33-fold higher in female cases (P<.01).
Finally, because there are several RAAS-pathway antagonists, such as ACE inhibitors (ACEis) and angiotensin receptor blockers (ARBs), that are commonly used in the treatment program for various cardiovascular diseases, we examined whether the presence of such a drug before surgical myectomy affected ACE2 expression. Overall, there were 13 patients treated with either an ACEi or ARB before their surgical myectomy. Although there was slightly lower ACE2 expression among treated patients (−1.32-fold change), this did not reach statistical significance (P=.082; q-value =1).
Genotype Subgroup Analyses
Subgroup analyses were performed comparing the 3 largest genotypic subsets; MYBPC3-positive, MYH7-positive, and genotype-negative HCM. Pairwise comparisons of MYH7-positive and MYBPC3-positive, MYH7-positive and genotype-negative HCM, and MYBC3-positive and genotype-negative HCM were performed. There were no gene expression changes that met a false discovery rate q-value < 0.1, suggesting a high probability of false-positive findings.
However, given that the disease states under comparison are, as previously documented, phenotypically indistinguishable (at clinical, gross anatomic, and microscopic levels), we hypothesized that gene expression changes due to genotype subgroup might be subtle. Therefore, we accepted a higher false-positive rate to reveal potentially important differences in gene expression. After adjusting to meet P<.05, there were approximately 1000 to 2000 gene expression changes in each subgroup comparison, totaling approximately 4% (1502 of 37,846) to 6% (2336 of 37,846) of genes tested. Most differentially expressed genes had low absolute-fold changes and high false discovery rate q-values. The overall 3-way comparison of gene expression among these 3 genotypic subgroups of HCM revealed that 94% (35,510 of 37,846) of gene expression changes were shared. Because additional differences between genotypic subgroups were subtle, we have summarized them in the Supplemental Data (available online at http://www.mayoclinicproceedings.org).
Discussion
The Human HCM mRNA Transcriptome
Our analysis of 106 HCM myectomy tissues and 39 control tissues identified 8443 differentially expressed genes, 22% of all genes analyzed. These genes participate in 1075 molecular functions and 4272 processes as defined by the Gene Ontology Consortium. Remarkably, the most differentially expressed genes were not previously identified in hypertrophic pathways. Whether this reflects the relatively small sizes of study groups, variability in age and treatments, background genotypes, or other factors is unknown. Previous genotype-phenotype studies have not identified a gene-specific profile.16, 17, 18 Nevertheless, we suggest that the newly identified differentially expressed genes warrant further investigation.
ACE2 and HCM
Given the current and devastating coronavirus disease 2019 (COVID-19) pandemic (>2.5 million confirmed cases) that has claimed more than 172,000 lives worldwide in less than 4 months (April 21, 2020), it was noteworthy that the most up-regulated gene in HCM samples was ACE2 (3.53-fold; q-value=1.30×10−23). ACE2 protein was also increased more than 5-fold by Western blot analyses (P<.001). ACE2 encodes angiotensin-converting enzyme subtype 2, which has important compensatory roles in modulating excessive activation of the RAAS as occurs in hypertension (HTN), congestive heart failure (CHF), and atherosclerosis. In its soluble form, ACE2 acts as a carboxypeptidase cleaving the prohypertrophic polypeptides angiotensin I and angiotensin II to angiotensin-(1-9) and angiotensin-(1-7), respectively, thereby producing counter-regulating, vasodilating, and potentially antihypertrophic/antifibrotic polypeptides (Figure 3C).19
Accordingly, we speculate that up-regulation of both ACE2 transcript and ACE2 protein levels might be a compensatory counter-regulatory signaling response (pathoresponsive) in patients with obstructive HCM. This was echoed in a recent article by Liu et al20 in which the investigators studied the transcriptome of HCM mouse models and found that profibrotic pathways initiated by an increase of endothelin-1 were the main drivers of HCM pathogenesis in mice through microRNA-29 and transforming growth factor β signaling. However, using our preliminary microarray data that were derived from the patients in our study and made publicly available (GSE36961), this differential expression of transforming growth factor β–signaling genes was not observed. Instead, the increased transcript levels of ACE2 were noted, prompting the speculation that ACE2 overexpression might be a compensatory response.20
Last, ACE2, located in the X-chromosome, was upregulated significantly in female patients with HCM (1.33-fold compared with males; P<.01) and although only 1 gene, findings like these could start to shed light and form the basis to understanding some of the underlying (epigenetic) contributions to the significantly different outcomes that are observed for women with HCM.21, 22, 23
ACE2 and Its Possible Role in COVID-19–Related Morbidity and Mortality
Beyond the potential relevance of ACE2 expression in HCM hearts and its disease pathogenesis, in its membrane-bound state, ACE2 plays an important role as a functional receptor required for viral entry and subsequent viral replication for the severe acute respiratory syndrome coronavirus (SARS-CoV) family of viruses24 , 25 and thereby may contribute to the increased morbidity and mortality from SARS-CoV-2 in adult patients with a variety of heart diseases.26 The currently endemic SARS-CoV-2 is a member of the SARS family of coronaviruses that bind to membrane-bound ACE2 through its viral spike protein.27 Recently, Zhou et al27 assessed virus infectivity in HeLa cells transfected with ACE2 and demonstrated preferential binding to ACE2 over other coronavirus receptors, such as aminopeptidase N or dipeptidyl peptidase 4. After binding to ACE2, cleavage of the spike protein (possibly by the transmembrane protease-2 serine enzyme) primes viral internalization by endocytosis. This viral internalization may result in loss of membrane ACE2 and a subsequent increase in the angiotensin II to angiotensin-(1-7) ratio, which in turn allows excessive angiotensin II and unopposed angiotensin type 1 receptor–mediated lung injury in patients with COVID-19 infection and the development of severe acute respiratory distress syndrome.24 , 25
ACE2 is expressed in many other tissues, including the intestinal and vascular epithelium, kidneys, and heart. However, expression in cardiomyocytes is low. In a study of cardiac cell samples from donor hearts, both ACE2 and transmembrane protease-2 serine showed the highest expression in the heart tissue’s pericyte subpopulation rather than in cardiomyocytes.28 Nevertheless, widespread ACE2 expression may contribute to the multiorgan dysfunction seen in patients with COVID-19 infection.29 , 30
The marked 5-fold increase in ACE2 protein in HCM may provide a mechanism to explain higher rates of severe outcomes in COVID-19–infected patients who also have cardiovascular comorbid conditions, as well as the direct cardiac damage caused by SARS-CoV-2 infections. Although the incidence of COVID-19 infection seems highest in the elderly or immunocompromised, a large number of affected patients and those requiring hospitalization have significant comorbid conditions, including highly prevalent cardiovascular diseases such as HTN or CHF. A large meta-analysis of more than 46,000 patients in China showed that the most common comorbid conditions were HTN (17%±7%; 95% CI, 14% to 22%), diabetes mellitus (8%±6%; 95% CI, 6% to 11%), and cardiovascular disease (5%±4%; 95% CI, 4% to 7%).31 These percentages were much higher in patients requiring hospitalization or even intensive care unit (ICU) admission. In 2 separate inpatient studies, preexisting HTN was present in 30% (and up to 60% for ICU or nonsurviving patients), as were concomitant cardiovascular disease (15%, increased to 13% to 25% in ICU or nonsurviving patients).32 , 33
There is mounting evidence for cardiotropism with SARS-CoV-2 infection and direct cardiac toxicity.32 In early studies, 7.2% of all COVID-19–infected patients and 22% of patients admitted to ICUs showed evidence of myocardial injury with elevated high-sensitivity troponin I levels or new ECG abnormalities with clinical manifestations of myocardial ischemia or myocarditis. Based on the significantly increased expression of membrane-bound ACE2 in HCM hearts, we speculate that obstructive HCM, and perhaps other ACE2-generating heart diseases, sensitizes the myocardium to increased SARS-CoV-2 viral entry and subsequent viral replication, while the subsequent decrease of surface ACE2 (after viral internalization) leads to an increase of damaging angiotensin II and angiotensin type 1 receptor activity and loss of the protective effects of angiotensin-(1-7) (Figure 3). Just recently, Liu et al34 proposed a similar hypothesis, describing the resulting unopposed increase in angiotensin II and subsequent downstream increase in detrimental inflammation, reactive oxygen species, vasoconstriction, and thrombosis as the basis for cardiac damage stemming from SARS-CoV-2 infection. Similarly, Chen et al35 describe this as a potential mechanism, illustrated by ACE2 overexpression in tissue from heart failure patients.
The effect of ACEis or ARBs is under active investigation. Although initial reports suggested potentially worse outcomes in patients with COVID-19 infection who were using ACEis,36 , 37 a subsequent review article concluded that there was insufficient evidence for this claim, prompting all major cardiac societies to advise that patients with heart disease treated with these medications should continue to use them as prescribed.38 However, ARBs could attenuate the impact of SARS-CoV-2 by blocking the damaging effects resulting from a viral-mediated decrease in ACE2 and subsequent increase in damaging angiotensin II level. Experimental studies of the related virus, SARS-CoV, showed that down-regulation of ACE2 exacerbated lung injury, and treatment with the ARB losartan mitigated these effects.24 Furthermore, a large study among 1128 patients with HTN and diagnosed with COVID-19 infection showed that the unadjusted mortality rate was significantly lower in those for whom HTN was treated with ACEi/ARBs (3.7% vs 9.8%; P=.001). This risk remained consistently lower when performed as a propensity score–matched analysis with adjustment of imbalanced variables such as age, sex, comorbid conditions, and in-hospital medications (adjusted hazard ratio, 0.37; 95% CI, 0.15-0.89; P=.03).39
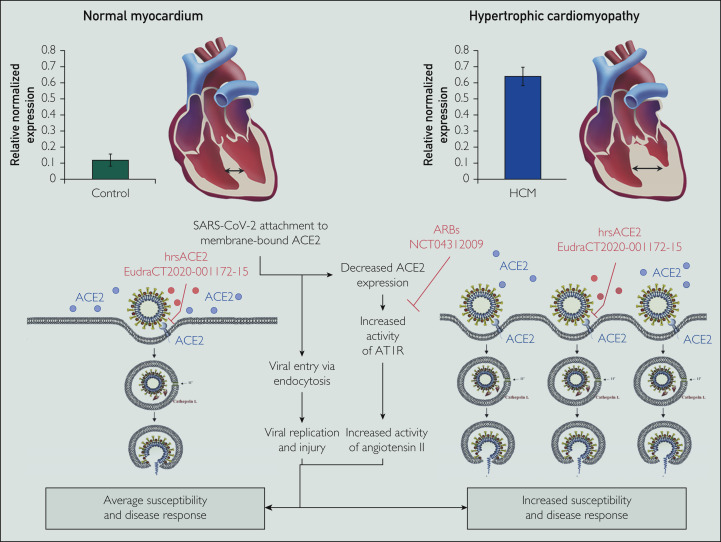
In light of these findings, a clinical trial has been launched testing the effect of losartan in study-eligible patients with COVID-19 infection (NCT04312009; Figure 4 ). In addition, clinical-grade human recombinant soluble ACE2 (hrsACE2) can block early stages of SARS-CoV-2 infection significantly by preventing the virus from entering the cell,40 highlighting the crucial and dual role of ACE2 in health and disease (Figure 4). A clinical trial to test hrsACE2 in patients was commenced recently in Europe (EudraCT2020-001172-15).
Figure 4.
Angiotensin-converting enzyme type 2 (ACE2) overexpression and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Central illustration shows possible mechanism behind ACE2 overexpression and SARS-CoV-2 infection picturing normal ACE2 expression (left) and ACE2 protein overexpression in obstructive hypertrophic cardiomyopathy (HCM; right). The SARS-CoV-2 virus hijacks membrane-bound ACE2 for cellular entry. Aside from allowing cellular invasion and viral replication, internalization of the SARS-CoV-2–ACE2 complex causes a decrease in surface ACE2. Loss of surface ACE2: (1) increases the angiotensin II to ang (1-7) ratio and (2) increases angiotensin type 1 receptor (AT1R) activity with a resultant increase in damaging angiotensin II activity. Shown are potential therapeutic targets (and clinical trials) using either angiotensin receptor blockers (ARBs; losartan specifically) or human recombinant soluble ACE2 (hrsACE2). For patients with ACE2-accentuating heart diseases such as obstructive HCM, the speculated increase in viral infectivity of the heart muscle remains to be proven. (Portion of figure adapted from: Simmons G, Zmora P, Gierer S, Heurich A, Pöhlman S. Proteolytic activation of the SARS-coronavirus spike protein: Cutting enzymes at the cutting edge of antiviral research. Antivir Res. 2013;100(3):605-614 with permission from Elsevier, license number 4814880904484).
Study Limitations
We studied only patients with HCM undergoing therapeutic surgical septal myectomy, representing 1 phenotypic subset of HCM. However, this study design has therefore an inherent bias because this procedure provides the only means to ethically obtain heart tissue. It therefore remains to be determined whether ACE2 elevation was a marker of this state of the disease alone (obstructive HCM) and whether it persisted following the relief of left ventricular outflow tract obstruction. Whether ACE2 elevation is present in the hearts of patients with nonobstructive HCM, other cardiomyopathies, HTN, or other forms of acquired heart disease is unknown.
To overcome the logistical and ethical issue of obtaining healthy heart tissue, we used a common source for healthy heart tissue41, 42, 43, 44, 45 as controls, both due to the scarcity of this reagent and the need for these control tissues to be procured and flash-frozen to preserve RNA (immediate flash-freezing close to the time of death). Although confounding of specimen source and case/control status must be considered as a possible explanation for the results herein, because these are patients from a different ethnic background (although likely both Euro-Caucasian), this collaboration provided us with the best source of flash frozen cardiac tissue, guaranteeing preserved high-quality RNA necessary for these types of analyses.
Conclusions
Using a high-throughput gene expression profiling technology, we discovered that greater than 20% of the transcriptome is expressed differentially between HCM and control heart tissue, whereas 5% of transcriptomic changes differ within the 3 most common HCM genotypes. Importantly, the single most up-regulated gene in the cardiac transcriptome for patients with obstructive HCM was ACE2. Further, the proven 5-fold increase in ACE2 protein levels in the heart may shed light on the increased morbidity and mortality in COVID-19–infected patients with underlying cardiovascular diseases. However, it remains to be demonstrated whether nonobstructive HCM, other cardiomyopathies, other acquired cardiovascular diseases, or HTN precipitate overexpression of ACE2 protein in the heart as shown here for obstructive HCM. Whether an ARB such as losartan or delivery of an ACE2 decoy with hrsACE2 might be effective therapeutic strategies for COVID-19–infected patients with ACE2-accentuating diseases such as obstructive HCM warrants further investigation.
Acknowledgments
Drs Bos and Hebl contributed equally to this manuscript and should be considered co-equal first authors.
Footnotes
Dr Hebl is now with Intermountain Heart Institute, Intermountain Healthcare, Salt Lake City, UT. Dr Herman is now with Department of Pathology and Laboratory Medicine, University of Pennsylvania, Philadelphia. Dr Teekakirikul is now with Department of Medicine and Therapeutics, Prince of Wales Hospital, Shatin, New Territories, Hong Kong.
Grant support: This work is supported by the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program, the Paul and Ruby Tsai and Family Hypertrophic Cardiomyopathy Research Fund, the National Institutes of Health (HL084553 and HL133165 to C.E.S. and J.G.S.), and the Howard Hughes Medical Institute (C.E.S.).
Potential Competing Interests: Dr Ackerman is a consultant for Abbott, Audentes Therapeutics, Boston Scientific, Daiichi Sankyo, Invitae, LQT Therapeutics, Medtronic, MyoKardia Inc, and UpToDate. Drs Noseworthy, Friedman, and Ackerman and Mayo Clinic have financial interest in AliveCor. Drs C.E. Seidman and J.G. Seidman are founders and own shares in Myokardia Inc, a startup company that is developing therapeutics that target the sarcomere. None of these entities were involved in this study in any manner. The remaining authors report no competing interests.
Supplemental material can be found online at http://www.mayoclinicproceedings.org. Supplemental material attached to journal articles has not been edited, and the authors take responsibility for the accuracy of all data.
Supplemental Online Material
References
- 1.Maron B.J., Gardin J.M., Flack J.M., Gidding S.S., Kurosaki T.T., Bild D.E. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92(4):785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 2.Maron B.J., Roberts W.C., McAllister H.A., Rosing D.R., Epstein S.E. Sudden death in young athletes. Circulation. 1980;62(2):218–229. doi: 10.1161/01.cir.62.2.218. [DOI] [PubMed] [Google Scholar]
- 3.Poetter K., Jiang H., Hassanzadeh S. Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle. Nat Genet. 1996;13(1):63–69. doi: 10.1038/ng0596-63. [DOI] [PubMed] [Google Scholar]
- 4.Morner S., Richard P., Kazzam E. Identification of the genotypes causing hypertrophic cardiomyopathy in northern Sweden. J Mol Cell Cardiol. 2003;35(7):841–849. doi: 10.1016/s0022-2828(03)00146-9. [DOI] [PubMed] [Google Scholar]
- 5.Lim D.S., Roberts R., Marian A.J. Expression profiling of cardiac genes in human hypertrophic cardiomyopathy: insight into the pathogenesis of phenotypes. J Am Coll Cardiol. 2001;38(4):1175–1180. doi: 10.1016/s0735-1097(01)01509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang J.J., Allen P.D., Tseng G.C. Microarray gene expression profiles in dilated and hypertrophic cardiomyopathic end-stage heart failure. Physiol Genomics. 2002;10(1):31–44. doi: 10.1152/physiolgenomics.00122.2001. [DOI] [PubMed] [Google Scholar]
- 7.Herman D.S., Hovingh G.K., Iartchouk O. Filter-based hybridization capture of subgenomes enables resequencing and copy-number detection. Nat Methods. 2009;6(7):507–510. doi: 10.1038/nmeth.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKenna A., Hanna M., Banks E. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schroeder A., Mueller O., Stocker S. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballman K.V., Grill D.E., Oberg A.L., Therneau T.M. Faster cyclic loess: normalizing RNA arrays via linear models. Bioinformatics. 2004;20(16):2778–2786. doi: 10.1093/bioinformatics/bth327. [DOI] [PubMed] [Google Scholar]
- 11.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57(1):289–300. [Google Scholar]
- 12.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Erdmann J., Daehmlow S., Wischke S. Mutation spectrum in a large cohort of unrelated consecutive patients with hypertrophic cardiomyopathy. Clin Genet. 2003;64(4):339–349. doi: 10.1034/j.1399-0004.2003.00151.x. [DOI] [PubMed] [Google Scholar]
- 14.Richard P., Charron P., Carrier L., EUROGENE Heart Failure Project Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107(17):2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 15.Van Driest S.L., Ommen S.R., Tajik A.J., Gersh B.J., Ackerman M.J. Yield of genetic testing in hypertrophic cardiomyopathy. Mayo Clin Proc. 2005;80(6):739–744. doi: 10.1016/S0025-6196(11)61527-9. [DOI] [PubMed] [Google Scholar]
- 16.Bos J.M., Will M.L., Gersh B.J., Kruisselbrink T.M., Ommen S.R., Ackerman M.J. Characterization of a phenotype-based genetic test prediction score for unrelated patients with hypertrophic cardiomyopathy. Mayo Clin Proc. 2014;89(6):727–737. doi: 10.1016/j.mayocp.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Driest S.L., Ommen S.R., Tajik A.J., Gersh B.J., Ackerman M.J. Sarcomeric genotyping in hypertrophic cardiomyopathy. Mayo Clin Proc. 2005;80(4):463–469. doi: 10.1016/S0025-6196(11)63196-0. [DOI] [PubMed] [Google Scholar]
- 18.Van Driest S.L., Vasile V.C., Ommen S.R. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44(9):1903–1910. doi: 10.1016/j.jacc.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 19.Vickers C., Hales P., Kaushik V. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J Biol Chem. 2002;277(17):14838–14843. doi: 10.1074/jbc.M200581200. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y., Afzal J., Vakrou S. Differences in microRNA-29 and pro-fibrotic gene expression in mouse and human hypertrophic cardiomyopathy. Front Cardiovasc Med. 2019;6:170. doi: 10.3389/fcvm.2019.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geske J.B., Ong K.C., Siontis K.C. Women with hypertrophic cardiomyopathy have worse survival. Eur Heart J. 2017;38(46):3434–3440. doi: 10.1093/eurheartj/ehx527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nijenkamp L.L.A.M., Bollen I.A.E., van Velzen H.G. Sex differences at the time of myectomy in hypertrophic cardiomyopathy. Circ Heart Fail. 2018;11(6):e004133. doi: 10.1161/CIRCHEARTFAILURE.117.004133. [DOI] [PubMed] [Google Scholar]
- 23.Rowin E.J., Maron M.S., Wells S., Patel P.P., Koethe B.C., Maron B.J. Impact of sex on clinical course and survival in the contemporary treatment era for hypertrophic cardiomyopathy. J Am Heart Assoc. 2019;8(21):e012041. doi: 10.1161/JAHA.119.012041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuba K., Imai Y., Rao S. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11(8):875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W., Moore M.J., Vasilieva N. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi S., Qin M., Shen B. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China [published online ahead of print March 25, 2020] https://doi.org/10.1001/jamacardio.2020.0950 JAMA Cardiol. [DOI] [PMC free article] [PubMed]
- 27.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Litviňuková M., Talavera- López C., Maatz H. Cells and gene expression programs in the adult human heart [published online ahead of print April 5, 2020] https://doi.org/10.1101/2020.04.03.024075 Bioarchives.
- 29.Tikellis C., Thomas M.C. Angiotensin-converting enzyme 2 (ACE2) is a key modulator of the renin angiotensin system in health and disease. Int J Pept. 2012;2012:256294. doi: 10.1155/2012/256294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J., Zheng Y., Gou X. Prevalence of comorbidities in the novel Wuhan coronavirus (COVID-19) infection: a systematic review and meta-analysis. Int J Infect Dis. 2020;94:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D., Hu B., Hu C. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China [published online ahead of print February 7, 2020] https://doi.org/10.1001/jama.2020.1585 JAMA. [DOI] [PMC free article] [PubMed]
- 33.Zou Y., Song L., Wang Z. Prevalence of idiopathic hypertrophic cardiomyopathy in China: a population-based echocardiographic analysis of 8080 adults. Am J Med. 2004;116(1):14–18. doi: 10.1016/j.amjmed.2003.05.009. [DOI] [PubMed] [Google Scholar]
- 34.Liu P.P., Blet A., Smyth D., Li H. The science underlying COVID-19: implications for the cardiovascular system [published online ahead of print April 15, 2020] https://doi.org/10.1161/circulaionaha.120.047549 Circulation. [DOI] [PubMed]
- 35.Chen Liang, Li Xiangjie, Chen Mingquan, Feng Yi, Xiong Chenglong. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc Res. 2020;116(1):1097–1100. doi: 10.1093/cvr/cvaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burrell L.M., Risvanis J., Kubota E. Myocardial infarction increases ACE2 expression in rat and humans. Eur Heart J. 2005;26(4):369–375. doi: 10.1093/eurheartj/ehi114. discussion 322-324. [DOI] [PubMed] [Google Scholar]
- 37.Ferrario C.M., Jessup J., Chappell M.C. Effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockers on cardiac angiotensin-converting enzyme 2. Circulation. 2005;111(20):2605–2610. doi: 10.1161/CIRCULATIONAHA.104.510461. [DOI] [PubMed] [Google Scholar]
- 38.South A.M., Tomlinson L., Edmonston D., Hiremath S., Sparks M.A. Controversies of renin-angiotensin system inhibition during the COVID-19 pandemic [published online ahead of print April 3, 2020] https://doi.org/10.1038/s41581-020-0279-4 Nat Rev Nephrol. [DOI] [PMC free article] [PubMed]
- 39.Zhang P., Zhu L., Cai J. Association of inpatient use of angiotensin converting enzyme inhibitors and angiotensin II receptor blockers with mortality among patients with hypertension hospitalized with COVID-19 [published online ahead of print April 18, 2020] https://doi.org/10.1161/circresaha.120.317134 Circ Res. [DOI] [PMC free article] [PubMed]
- 40.Monteil V., Kwon H., Prado P. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble ACE2 [published online ahead of print April 17, 2020] https://doi.org/10.1016/j.cell/2020.04.004 Cell. [DOI] [PMC free article] [PubMed]
- 41.Messer A.E., Gallon C.E., McKenna W.J., Dos Remedios C.G., Marston S.B. The use of phosphate-affinity SDS-PAGE to measure the cardiac troponin I phosphorylation site distribution in human heart muscle. Proteomics Clin Appl. 2009;3(12):1371–1382. doi: 10.1002/prca.200900071. [DOI] [PubMed] [Google Scholar]
- 42.Hoskins A.C., Jacques A., Bardswell S.C. Normal passive viscoelasticity but abnormal myofibrillar force generation in human hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2010;49(5):737–745. doi: 10.1016/j.yjmcc.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fermin D.R., Barac A., Lee S. Sex and age dimorphism of myocardial gene expression in nonischemic human heart failure. Circ Cardiovasc Genet. 2008;1(2):117–125. doi: 10.1161/CIRCGENETICS.108.802652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Dijk S.J., Paalberends E.R., Najafi A. Contractile dysfunction irrespective of the mutant protein in human hypertrophic cardiomyopathy with normal systolic function. Circ Heart Fail. 2012;5(1):36–46. doi: 10.1161/CIRCHEARTFAILURE.111.963702. [DOI] [PubMed] [Google Scholar]
- 45.Krüger M., Kötter S., Grützner A. Protein kinase G modulates human myocardial passive stiffness by phosphorylation of the titin springs. Circ Res. 2009;104(1):87–94. doi: 10.1161/CIRCRESAHA.108.184408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.