Abstract
The first step in differentiation of pluripotent stem cell toward endoderm-derived cell/organ is differentiation to definitive endoderm (DE) which is the central issue in developmental biology. Based on several evidences, we hypothesized that activin-A optimization as well as replacement of fetal bovine serum (FBS) with knockout serum replacement (KSR) is important for differentiation of induced pluripotent stem cell (iPSC) line into DE. Therefore, a stepwise differentiation protocol was applied on R1-hiPSC1 cell line. At first, activin-A concentration (30, 50, 70 and 100 ng/ml) was optimized. Then, substitution of FBS with KSR was evaluated across four treatment groups. The amount of differentiation of iPSC toward DE was determined by quantitative gene expression analyses of pluripotency (NANOG and OCT4), definitive endoderm (SOX17 and FOXA2) and endoderm-derived organs (PDX1, NEUROG3, and PAX6). Based on gene expression analyses, the more decrease in concentrations of activin-A can increase the differentiation of iPSC into DE, therefore, 30 ng/ml activin-A was chosen as the best concentration for the differentiation of R1-hiPSC1 line toward endoderm-derived organ. Moreover, complete replacement of FBS with gradually increased KSR improved the differentiation of iPSC toward DE. For this reason, the addition of 0% KSR at day 1, 0.2% at day 2 and 2% for the next 3 days was the best optimal protocol of the differentiation of iPSC toward DE. Overall, our results demonstrate that optimization of activin-A is important for differentiation of iPSC line. Furthermore, the replacement of FBS with KSR can improve the efficiency of iPSC differentiation toward DE.
Keywords: hiPSC, Differentiation, Definitive endoderm, Activin-A, KSR
Introduction
Various methods have been established for the differentiation of pluripotent stem cells into several endoderm-derived organs in vitro (Han et al. 2017; Memon et al. 2018). These stepwise differentiation methods are based on tight and temporal activation and inhibition of distinct signaling pathways including TGFβ, WNT/β-catenin, SHH, FGF, and NOTCH in order to direct pluripotent stem cells to commit toward specific cells/organs (Hoveizi et al. 2015; Huggins et al. 2017; Bogacheva et al. 2018). Moreover, maturity and functionality of differentiated cells/organs highly rely upon the efficiency of each step of the differentiation protocol (Bogacheva et al. 2018). This efficiency considerably relies on distinctive cytokines, growth factors and small molecules as well as their concentrations in each step.
The more interesting concept about all of these protocols is the necessity to differentiate into DE as the first interphase step for further differentiation into specific cell/organs (Han et al. 2017). This step of differentiation defined by induction and expression of various transcription factors such as SOX17 and FOXA2 (Bogacheva et al. 2018). The DE differentiation involves induction of signaling pathways such as nodal and wnt by using activin-A, nodal, Wnt-3a, CHIR99021, and serum (Pauklin and Vallier 2015). It was well known that activin-A, a member of TGFβ superfamily and inducer of nodal signaling, participate in broad range of biological processes including: stem cell renewal, proliferation, differentiation and apoptosis as well as cell fate decision, organogenesis and homeostasis (Pauklin and Vallier 2015; Bogacheva et al. 2018). It has been reported that activin-A exerts its activity through concentration and gradient to ordain positional information during development, which finally directs cell fate (Brennan et al. 2001). Furthermore, time and duration of nodal signaling are important for cell fate decision and generation of different cell types (Hagos and Dougan 2007).
It is well shown that activin-A conducts its apoptotic activity through activation of caspase in a dose- and time-dependent manner, and the Smad pathway plays an important role in activin-A-induced programed cell death (Chen et al. 2002). Furthermore, activin-A has inductive and direct effects on blocking differentiation and maintaining pluripotency. In addition, Smad2/3, as downstream effector of activin/nodal signaling, bind to NANOG and OCT4 and then interact with a variety of promoters necessary for maintaining of pluripotency (Vallier et al. 2009). Moreover, activin-A can induce mesoderm and endoderm differentiation in a dose-dependent manner during normal embryonic development (D’Amour et al. 2006; Bogacheva et al. 2018). It has been found that Activin-A signals through Smad-2/3 and in collaboration with wnt signaling control mesoderm and endoderm differentiation. WNT/β-catenin collaborates with Smad2/3 and targets the SOX17 gene and in combination with SOX17 activates FOXA2 gene expression to promote endoderm development (Pauklin and Vallier 2015). Furthermore, it was shown that the concentration of activin-A below 30 ng/ml generates mesoderm while concentrations between 30–100 ng/ml induce endoderm (Sulzbacher et al. 2009). Although most of the researchers preferred to use the highest concentration of activin-A in their differentiation protocols (D’Amour et al. 2006; Memon et al. 2018; Bogacheva et al. 2018), we assumed that activin-A concentration needs appropriate optimization according to its numerous functions. Additionally, there was evidence that growth factors and hormones present in FBS can inhibit the induction effect of activin-A (Sulzbacher et al. 2009). Therefore, the use of other serum replacement materials was suggested. To this aim, we examined the effect of different concentrations of activin-A together with the effect of FBS and KSR on DE differentiation of our iPSC line.
Material and methods
In vitro differentiation
The R1-hiPSC1 cell line (RSCB0042) was taken from the Royan Institute (Tehran, Iran) and cultured on MEF (mouse embryonic fibroblasts) layer, which was inactivated by mitomycin C (M7949, Sigma, Germany) and fed with iPSC medium (Shaer et al. 2016). Contamination with Mycoplasma spp. was checked using PCR with universal primers (Molla Kazemiha et al. 2009). The R1-hiPSC1 cell line was differentiated into DE by using a two-step protocol (Fig. 1).
Step 1. Embryoid body formation The embryoid body (EB) was generated from iPSC using static suspension method (Rungarunlert et al. 2009). Briefly, the iPSC was mechanically detached from flasks by using a cell scraper (SPL, South Korea) and then resuspended into an ultra-low attachment 6-well plate (SPL, South Korea) and incubated for 2–3 days in order to form spherical EB. Next, EBs were collected using their self-sedimentation characteristic in 15-ml conical tubes (SPL, South Korea) and transferred onto 2% gelatin-coated (G1890, Sigma, Germany) 35-mm plate and incubated for 1–2 days. During these days, cells were fed with DMEM/F12 medium containing 20% FBS (10270106, Gibco, USA), 100 Unit/ml penicillin, and 100 µg/ml streptomycin (15140122, Gibco, USA).
Step 2. Definitive-endoderm Established EBs were subjected to differentiate by treating with activin-A (30, 50, 70 and 100 ng/ml; H4666, Sigma, Germany) and 3 µM CHIR99021 (SML1046, Sigma, Germany) in RPMI1640 medium (21875034, Gibco, USA) for 1 day, which encourage mesendoderm induction. Then, cells were treated with activin-A (30, 50, 70 and 100 ng/ml) in RPMI 1640 medium with 0.2% FBS for 1 day. Next, cells were subjected to treatment with activin-A (30, 50, 70 and 100 ng/ml) in RPMI 1640 medium with 2% FBS for a further 3 days.
Fig. 1.
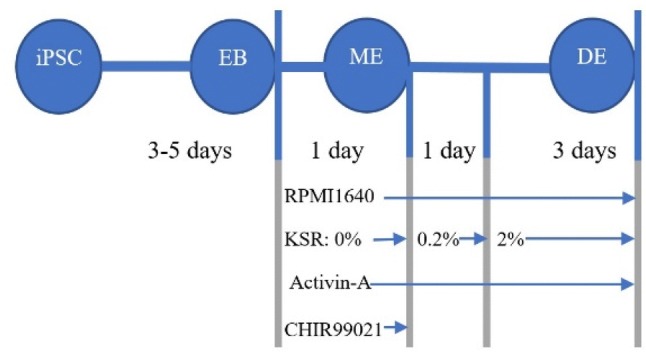
Schematic representation of R1-hiPSC1 differentiation toward DE. Factors involved in this process and duration of each step were represented under each step. As indicated, all the steps were performed between 8 and 10 days. iPSCs induced pluripotent stem cells, EB embryoid body, ME mesendoderm, DE definitive endoderm
It should be noted that at first, cells were subjected to treatment with different concentrations of activin-A. For evaluation of distinctive DE efficiency (produced from various concentrations of activin-A), DE cells further differentiated into posterior foregut (Pezzolla et al. 2015) and gene expression of PDX1, NEUROG3 and PAX6 was evaluated. Then, the best concentration of activin-A was subjected to optimization of FBS or KSR (10828010, Gibco, USA) by definition of 4 groups including: group 1—treatment with 0.2% FBS at days 1 and 2 and 2% FBS for the next 3 days; group 2—0% FBS at day 1, 0.2% FBS at day 2 and 2% FBS for the next 3 days; group 3—0.2% KSR at days 1 and 2 and 2% KSR for the next 3 days; group 4—0% KSR at day 1, 0.2% KSR at day 2 and 2% KSR for the next 3 days.
Gene expression analysis
Total RNA was extracted from each well by using Trizol reagent (15596026, Invitrogen, USA) according to the manufacturer's protocol. Five-hundred ng RNA was adhered to complementary DNA synthesis by using PrimeScript First Strand cDNA Synthesis Kit (6110A, Takara, Japan). 1 μl of cDNA was applied into quantitative real-time PCR (qPCR) to analyze gene expression changes after the differentiation process. The qPCR was carried out in the StepOnePlus instrument (Applied Biosystem, USA) and performed using SYBR® Premix EX Taq™ II kit (RR820A, Takara, Japan) and primers presented in Table 1. All reactions were performed in triplicate and GAPDH was used as the reference gene. All quantifications were expressed as fold change in comparison with undifferentiated hiPSC. All reactions were analyzed by LinRegPCR (version 2017.1) software and only highly efficient reactions (95% < E > 105% and 0.99 < r2) were applied for further analysis. Quality control analysis of qPCR, fold change calculation and statistical analysis were performed by GenEX v.6.1 software. Relative gene expression analysis was performed using 2−ΔΔCT analysis.
Table 1.
Oligonucleotide sequences used for gene expression analysis
| Target | Seq. (5′ → 3′) | Tm | Product size |
|---|---|---|---|
| OCT4 |
F: GAGAACCGAGTGAGAGGCAACC R: CATAGTCGCTGCTTGATCGCTTG |
64 | 167 |
| NANOG |
F: GTCCCGGTCAAGAAACAGAAG R: GTCTTCACCTGTTTGTAGCTG |
58 | 152 |
| SOX17* |
F: CGCTTTCATGGTGTGGGCTAAGGACG R: TAGTTGGGGTGGTCCTGCATGTGCTG |
60 | 186 |
| FOXA2* |
F: AGCGAGTTAAAGTATGCTGG R: GTAGCTGCTCCAGTCGGA |
60 | 67 |
| PDX1 |
F: GAAGTCTACCAAAGCTCACG R: CGTGAGATGTACTTGTTGAATAGG |
60 | 153 |
| NEUROG3 |
F: CGGTAGAAAGGATGACGC R: CAGGTCACTTCGTCTTCC |
58 | 103 |
| PAX6 |
F: TAGTAAACCGAGAGTAGCGAC R: GTTTATTGATGACACGCTTGG |
60 | 154 |
| GAPDH** |
F: GGACTCATGACCACAGTCCA R: CCAGTAGAGGCAGGGATGAT |
60 | 119 |
Statistical analysis
All experiments were performed in triplicate, and data presented as a mean ± SEM. One-way ANOVA was used for comparison between the control group and treated groups. Tukey–Kramer’s test was used as a post hoc test. p value was set as statistically significant at the 0.05 level. Statistical analyses were performed by GenEX v.6.1 software. Descriptive statistics were performed by EXCEL v.2017 and graph design were done by GraphPad Prism v7.01.
Results
Optimization of activin-A
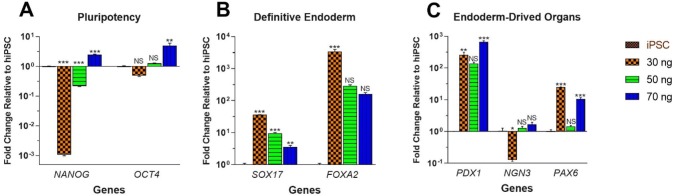
The iPSC was successfully produced EB after 2 days and attached well to the gelatin-coated dish and expand during 1–2 days. Then, EBs were subjected to optimization of activin-A. Treatment of EBs with 100 ng/ml treatment of activin-A represented a high level of mortality among the EBs after 3 days and did not generate adequate cell for analysis. However, lower concentrations of activin-A produced sufficient cell for gene expression analysis. Pluripotency genes (NANOG and OCT4) analyses revealed that 30 ng/ml of activin-A remarkably down-regulated NANOG (0.001 ± 3.43 × 10–6; p < 0.001) and OCT4 (0.48 ± 0.02; p = 0.88) genes during differentiation. In contrast, 70 ng/ml of activin-A significantly increased gene expression of both NANOG (2.48 ± 0.10; p < 0.001) and OCT4 (5.02 ± 0.99; p = 0.002), while 50 ng/ml activin-A significantly down-regulated NANOG (0.21 ± 0.005; p < 0.001) and slightly increased OCT4 gene expression (1.25 ± 0.04; p = 0.98) (Fig. 2a). Therefore, the more decrease in activin-A concentrations, the more the down-regulation of pluripotency genes.
Fig. 2.
The effect of activin-A concentrations on differentiation of iPSC toward DE. The iPSC was first differentiated into EB followed by treatment with 30, 50 and 70 ng/ml of activin-A, then pluripotency genes and DE-specific gens were evaluated. DE cells further differentiated into posterior foregut and endoderm-derived organ-specific genes were analyzed. Fold changes (log 10) of each treatment were compared with iPSCs. All error bars indicate standard error of mean (SEM) of three biological replicates. a Gene expression analyses of pluripotency genes (NANOG and OCT4) was performed at DE stage and 8th day of differentiation; b gene expression analyses of DE-specific genes (SOX17 and FOXA2) was performed at DE stage and 8th day of differentiation; c gene expression analyses of PDX1, NEUROG3 (NGN3), and PAX6 which play a role in endoderm-derived organs differentiation was performed at posterior foregut stage and 14th day of differentiation. *p < 0.05; **p < 0.01; ***p < 0.001; NS not significant
DE-specific genes (SOX17 and FOXA2) analyses showed that a decrease in activin-A concentration results in more increases in both SOX17 and FOXA2 gene expressions (Fig. 2b). Interestingly, 30 ng/ml activin-A meaningfully increased the expression of SOX17 and FOXA2 genes by 36.26 ± 0.43 (p < 0.001) and 3406.15 ± 632.31 (p < 0.001) fold, respectively. Although 50 and 70 ng/ml activin-A considerably improved gene expression of SOX17 (9.55 ± 0.57; p < 0.001 and 3.62 ± 0.37; p = 0.008, respectively), but they did not significantly increase FOXA2 gene expression (292.04 ± 19.83; p > 0.05 and 161.76 ± 16.12; p > 0.05, respectively). Therefore, 30 ng/ml activin-A brings better differentiation of iPSC toward DE.
Additionally, gene expressions of PDX1, NEUROG3, and PAX6, in posterior foregut stage of differentiation, which participate in the differentiation of endoderm-derived organs were evaluated. It was shown that 30 ng/ml activin-A considerably improved expression of PDX1 to the level (258.66 ± 56.90; p = 0.005) that substantially silenced NEUROG3 (0.12 ± 0.01; p = 0.04) and properly regulated PAX6 by 25.11 ± 0.30 (p < 0.001) fold increase; while 50 ng/ml activin-A could not increase the expression of PDX1 (138.15 ± 14.86; p > 0.05) to the value that silence NEUROG3 (1.28 ± 0.17; p > 0.05) and regulate PAX6 (1.45 ± 0.08; p > 0.05). Although 70 ng/ml activin-A significantly enhanced PDX1 (675.59 ± 45.88; p < 0.001) and PAX6 (10.98 ± 1.14; p < 0.001) gene expression, it did not supress NEUROG3 (1.68 ± 0.28; p > 0.05) gene expression (Fig. 2c).
According to gene expression analyses on pluripotency, DE and endoderm, 30 ng/ml activin-A was selected as the best concentration for differentiation of R1-hiPSC toward endoderm-derived organs.
Optimization of serum
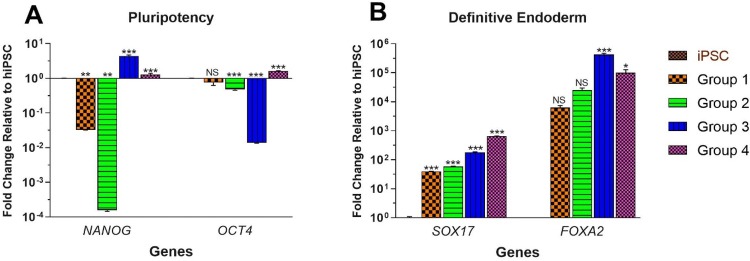
Following activin-A optimization in 30 ng/ml as the best concentration, optimization of FBS or KSR was performed. Expression of NANOG was substantially decreased by 0.03 ± 0.00 (p = 0.007) and 0.00 ± 0.00 (p = 0.005) among FBS-treated groups 1 and 2 and meaningfully increased by 4.41 ± 0.32 (p < 0.001) and 1.29 ± 0.11 (p < 0.001) among KSR-treated groups 3 and 4, correspondingly. Although expression of OCT4 was not considerably decreased in group 1 (0.75 ± 0.12; p > 0.05), its expression was importantly decreased in FBS-treated group 2 (0.48 ± 0.02; p < 0.001) and significantly decreased by 0.01 ± 0.00 (p < 0.001) and remarkably increased by 1.62 ± 0.04 (p < 0.001) among KSR-treated groups 3 and 4, respectively (Fig. 3a).
Fig. 3.
The effect of serum optimization on gene expression of pluripotency and DE-specific genes in four treatment groups. Group 1: cells treatment with 0.2% FBS at days 1 and 2 and 2% FBS for the next 3 days, group 2: 0% FBS at day 1, 0.2% FBS at day 2 and 2% FBS for the next 3 days, group 3: 0.2% KSR at days 1 and 2 and 2% KSR for the next 3 days, group 4: 0% KSR at day 1, 0.2% KSR at day 2 and 2% KSR for the next 3 days. Gene expression analyses were performed at DE stage and 8th day of differentiation. Fold changes (log 10) of each treatment were compared with iPSCs. All error bars indicate standard error of mean (SEM) of three biological replicates. a Gene expression analyses of pluripotency genes (NANOG and OCT4); b gene expression analyses of DE-specific genes (SOX17 and FOXA2). *p < 0.05; **p < 0.01; ***p < 0.001; NS not-significant
Furthermore, the expression of DE-specific gene, SOX17, was significantly increased by 39.76 ± 0.16 (p < 0.001), 59.31 ± 0.83 (p < 0.001), 182.80 ± 4.39 (p < 0.001), and 658.73 ± 7.91 (p < 0.001) among group 1 to group 4, respectively. Conversely, expression of FOXA2 was not greatly increased in FBS-treated group 1 (6314.22 ± 1099.10; p > 0.05) and group 2 (25,365.75 ± 4708.84; p > 0.05), while its expression was significantly increased among KSR-treated group 3 (429,469.66 ± 39,404.81; p < 0.001) and group 4 (101,278.77 ± 27,196.45; p = 0.047) (Fig. 3b). According to these expression profiles, group 4 was the best group regarding serum optimization.
Discussion
Development of pluripotent stem cell into specific lineage or organ embraces the transition from several developmental stages. These developmental stages are under precise regulation and crosstalk between signaling pathways (Deol et al. 2017). The first step in the development of pluripotent stem cells is differentiation to the three germ layers which is highly dependent on the concentrations of activin-A. We found that exposure to high concentration (100 ng/ml) of activin-A led to elevated mortality of our cell line, while decreasing its concentrations to 30–70 ng/ml rescued them from early death. It is well shown that a high concentration of activin-A (90 ng/ml) inhibits cell growth followed by an increase in apoptosis in HepG2 hepatoma cell. Moreover, it is indicated that inhibitory effect of activin-A is time- and dose-dependent as 1 ng/ml activin-A can decrease 40% of total cells population, while 100 ng/ml of activin-A decreases 80% of total cell population of LNCaP cell line (Chen et al. 2002).
To determine the best activin-A concentration, the gene expression profiles of three groups, including: pluripotency and DE which applied on DE stage, and 3 endoderm genes, which applied on posterior foregut stage were used. Our results indicated that the expressions of NANOG and OCT4 were highly increased in the presence of 70 ng/ml activin-A. On the contrary, the effect of 30 ng/ml activin-A decreased the expression of NANOG and OCT4 to 0.0 and 0.5, respectively. Previous reports showed that the expressions of NANOG and OCT4 were slightly increased (⁓ 1.5-fold) during first day of differentiation toward DE. During second day of differentiation, expressions of NANOG and OCT4 were reduced to basal level (⁓ onefold) comparable to undifferentiated cells. Afterward, expressions of NANOG and OCT4 were more decreased to 0.5 during the third day of differentiation toward DE. These changes lead to an increased expression of DE markers SOX17 and FOXA2 on the second day, followed by a gradual increase in their expressions during endoderm development (Teo et al. 2011).
Further analyses of DE markers show that gene expression of SOX17 and FOXA2 were increased by decreasing of activin-A concentration. Additionally, high level of pluripotency gene expressions clearly shows that they are required for germ layer specification. OCT4 in association with SOX2 and other factors blocks primitive streak formation (Teo et al. 2011). Further, NANOG induces EOMES expression and EOMES in turn interacts with Smad2/3 (effectors of nodal signaling) and promotes mesendoderm specification, which further developed into DE (Teo et al. 2011; Pauklin and Vallier 2015). Therefore, not only downregulation of NANOG and OCT4 does not favor DE induction, but also their expression maintenance is essential for DE development.
The third group of genes evaluated for the behavior of different EBs in posterior foregut stage were PDX1, NEUROG3 and PAX6. As previously reported, PDX1 gene expression is increased during the early stage of embryonic development toward pancreas, liver, etc. (Zhu et al. 2017) and its initial increase result in down-regulation of NEUROG3 (Pezzolla et al. 2015; Zhu et al. 2017). Interestingly, we observed that decreasing of activin-A concentration led to a high increase in PDX1 which by its turn results in full silencing of NEUROG3 at 30 ng/ml activin-A. Furthermore, it is reported that PAX6 has two functions as an ectoderm marker during the patterning of three germ layers (Sulzbacher et al. 2009; Huggins et al. 2017) as well as a transcription factor which in association with PDX1 and NKX2-2 promote pancreatic islet differentiation (Shaer et al. 2016; Huggins et al. 2017). Therefore, it is important to regulate the expression of PAX6 during the first step of iPSC differentiation. Although increased expression of PAX6 drives differentiation toward ectoderm, endoderm markers (SOX17 and FOXA2) did not increase to a high level when treated with 50 and 70 ng/ml activin-A. However, tight regulation of PAX6 expression (not too high and not too low) directs differentiation of iPSC toward pancreatic islets when treated with 30 ng/ml activin-A.
Serum optimization and replacement of FBS with KSR improved expressions of both NANOG and OCT4 to 1.29 ± 0.11 and 1.62 ± 0.04-fold change in group 4, respectively, which bore more resemblance to those reported in other studies (Shaer et al. 2016; Huggins et al. 2017). At the same time, the expressions of NANOG and OCT4 did not rise in the FBS-treated groups 1 and 2. Moreover, the high level of NANOG gene expression and full silencing of OCT4 in group 3 were not consistent with the minimum gene expression amounts needed for the induction of DE markers as described above. Furthermore, when FBS was replaced with KSR, the gene expressions of FOXA2 and SOX17 were significantly increased in the KSR-treated groups 3 and 4. Although expression of FOXA2 in group 3 was higher than those in group 4, overall coordination of gene expression between pluripotency and DE-specific genes leads to choose group 4 as the best treatment group for differentiation of iPSC to DE.
The reason behind the different effects of FBS and KSR is that the presence of insulin/IGF in FBS can activate PI3K through IGF signaling pathway, in which PI3K antagonizes activin-A-dependent differentiation toward DE (McLean et al. 2007; Sulzbacher et al. 2009). As a result, the addition of PI3K inhibitors (e.g., LY 294002 and AKT1-II) to FBS-containing medium or replacement of FBS with KSR (without IGF but contain insulin) or BSA can increase the efficiency of differentiation toward DE (Sulzbacher et al. 2009).
The second important outcome of this research is elimination of high cell confluency which is crucial for initiation of DE differentiation. Most of the researches done on differentiation of pluripotent stem cells into DE started with 70–90% global confluency on their dish followed by treatment by 100 ng/ml activin-A (Rezania et al. 2014; Oh et al. 2015; Memon et al. 2018), while our protocol required 50–60% local confluency (around each EB) for initiation of differentiation followed by 30 ng/ml activin-A. These findings result in economizing our research regarding both providing cell and purchasing activin-A. It is important to note that the studied gene expression analysis should have been confirmed at the protein level, which is the limitation of our study. Thus, further studies are needed to elucidate this point.
In conclusion, we showed that activin-A optimization was important for iPSC differentiation and 30 ng/ml activin-A was the best optimal concentration for differentiation of R1-hiPSC1 line toward DE. Furthermore, replacement of FBS with KSR in the first stage of differentiation protocols can increase the accuracy of regulation of genes involved in differentiation of iPSC toward endoderm-derived organs.
Acknowledgements
The authors are grateful to the Organ Transplant Research Center, Shiraz University of medical sciences and University of Sistan and Baluchestan, for their executive and financial support of this project.
Author contribution
Conceptualization was done by NA. Laboratory experiments was done by SG-D, SL and AS. Statistical analyses were done by SG-D and MS. Funding acquisition was done by NA and MHS. The first draft of the article was written by SG-D and NA. Methodology and investigation were done by HS-L, RY and IHA-A. All authors reviewed, edited and approved the manuscript.
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Contributor Information
Negar Azarpira, Email: negarazarpira@yahoo.com.
Hamid Reza Soleimanpour-Lichaei, https://www.researchgate.net/profile/Hamid_Reza_Soleimanpour-Lichaei.
Shahrokh Lorzadeh, https://www.linkedin.com/in/shahrokh-lorzadeh-086a706b/.
Alice Sabet, https://www.linkedin.com/in/alice-sabet-00240158/.
References
- Bogacheva MS, Khan S, Kanninen LK, Yliperttula M, Leung AW, Lou Y-R. Differences in definitive endoderm induction approaches using growth factors and small molecules. J Cell Physiol. 2018;233(4):3578–3589. doi: 10.1002/jcp.26214. [DOI] [PubMed] [Google Scholar]
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RSP, Robertson EJ. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411(6840):965–969. doi: 10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- Chen YG, Lui HM, Lin SL, Lee JM, Ying SY. Regulation of cell proliferation, apoptosis, and carcinogenesis by activin. Exp Biol Med. 2002;227(2):75–87. doi: 10.1177/153537020222700201. [DOI] [PubMed] [Google Scholar]
- D’Amour KA, Bang AG, Fau-Eliazer S, Kelly OG, Agulnick AD, Smart NG, Moorman MA, Kroon E, Carpenter MK, Baetge EE. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- Deol GSJ, Cuthbert TN, Gatie MI, Spice DM, Hilton LR, Kelly GM. Wnt and hedgehog signaling regulate the differentiation of F9 cells into extraembryonic endoderm. Front Cell Dev Biol. 2017;5:93–93. doi: 10.3389/fcell.2017.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagos EG, Dougan ST. Time-dependent patterning of the mesoderm and endoderm by Nodal signals in zebrafish. BMC Dev Biol. 2007;7:1–18. doi: 10.1186/1471-213X-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y-J, Kang Y-H, Shivakumar SB, Bharti D, Son Y-B, Choi Y-H, Park W-U, Byun J-H, Rho G-J, Park B-W. Stem cells from cryopreserved human dental pulp tissues sequentially differentiate into definitive endoderm and hepatocyte-like cells in vitro. Int J Med Sci. 2017;14(13):1418–1429. doi: 10.7150/ijms.22152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoveizi E, Massumi M, Ebrahimi-barough S, Tavakol S, Ai J. Differential effect of Activin A and WNT3a on definitive endoderm differentiation on electrospun nanofibrous PCL scaffold. Cell Biol Int. 2015;39(5):591–599. doi: 10.1002/cbin.10430. [DOI] [PubMed] [Google Scholar]
- Huggins IJ, Bos T, Gaylord O, Jessen C, Lonquich B, Puranen A, Richter J, Rossdam C, Brafman D, Gaasterland T, Willert K. The WNT target SP5 negatively regulates WNT transcriptional programs in human pluripotent stem cells. Nat Commun. 2017;8(1):1034–1034. doi: 10.1038/s41467-017-01203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi M, Azarpira N, Shamdani S, Hojjat-Assari S, Naserian S, Karimi MH. Differentiation of umbilical cord derived mesenchymal stem cells to hepatocyte cells by transfection of miR-106a, miR-574-3p, and miR-451. Gene. 2018;667:1–9. doi: 10.1016/j.gene.2018.05.028. [DOI] [PubMed] [Google Scholar]
- McLean AB, D'Amour KA, Jones KL, Krishnamoorthy M, Kulik MJ, Reynolds DM, Sheppard AM, Liu H, Xu Y, Baetge EE, Dalton S. Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells. 2007;25(1):29–38. doi: 10.1634/stemcells.2006-0219. [DOI] [PubMed] [Google Scholar]
- Memon B, Karam M, Al-Khawaga S, Abdelalim EM. Enhanced differentiation of human pluripotent stem cells into pancreatic progenitors co-expressing PDX1 and NKX6.1. Stem Cell Res Ther. 2018;9(1):15–15. doi: 10.1186/s13287-017-0759-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla Kazemiha V, Shokrgozar MA, Arabestani MR, Shojaei Moghadam M, Azari S, Maleki S, Amanzadeh A, Jeddi Tehrani M, Shokri F. PCR-based detection and eradication of mycoplasmal infections from various mammalian cell lines: a local experience. Cytotechnology. 2009;61(3):117–124. doi: 10.1007/s10616-010-9252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh BJ, Oh SH, Choi JM, Jin SM, Shim WY, Lee MS, Lee MK, Kim KW, Kim JH. Co-culture with mature islet cells augments the differentiation of insulin-producing cells from pluripotent stem cells. Stem Cell Rev Rep. 2015;11(1):62–74. doi: 10.1007/s12015-014-9554-8. [DOI] [PubMed] [Google Scholar]
- Pauklin S, Vallier L. Activin/Nodal signalling in stem cells. Development. 2015;142(4):607–619. doi: 10.1242/DEV.091769. [DOI] [PubMed] [Google Scholar]
- Pezzolla D, López-Beas J, Lachaud CC, Domínguez-Rodríguez A, Smani T, Hmadcha A, Soria B. Resveratrol ameliorates the maturation process of β-cell-like cells obtained from an optimized differentiation protocol of human embryonic stem cells. PLoS ONE. 2015;10(3):e0119904–e0119904. doi: 10.1371/journal.pone.0119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezania A, Bruin JE, Arora P, Rubin A, Batushansky I, Asadi A, O'Dwyer S, Quiskamp N, Mojibian M, Albrecht T, Yang YH, Johnson JA-O, Kieffer TJ. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol. 2014;32(11):1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- Rungarunlert S, Techakumphu M, Pirity MK, Dinnyes A. Embryoid body formation from embryonic and induced pluripotent stem cells: benefits of bioreactors. World J Stem Cells. 2009;1(1):11–21. doi: 10.4252/wjsc.v1.i1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaer A, Azarpira N, Karimi MH, Soleimani M, Dehghan S. Differentiation of human-induced pluripotent stem cells into insulin-producing clusters by microRNA-7. Exp Clin Transplant. 2016;14(5):555–563. doi: 10.6002/ect.2014.0144. [DOI] [PubMed] [Google Scholar]
- Sulzbacher S, Schroeder IS, Truong TT, Wobus AM. Activin A-induced differentiation of embryonic stem cells into endoderm and pancreatic progenitors-the influence of differentiation factors and culture conditions. Stem Cell Rev. 2009;5(2):159–173. doi: 10.1007/s12015-009-9061-5. [DOI] [PubMed] [Google Scholar]
- Teo AKK, Arnold SJ, Trotter MWB, Brown S, Ang LT, Chng Z, Robertson EJ, Dunn NR, Vallier L. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011;25(3):238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L, Mendjan S, Brown S, Chng Z, Teo A, Smithers LE, Trotter MWB, Cho CHH, Martinez A, Rugg-Gunn P, Brons G, Pedersen RA. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development. 2009;136(8):1339–1349. doi: 10.1242/dev.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zare A, Rashki A, Ghahari S, Ghayoori B. The analysis of correlation between IL-12 gene expression and hepatitis B virus in the affected patients. Virusdisease. 2015;26(3):196–199. doi: 10.1007/s13337-015-0261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Liu Q, Zhou Z, Ikeda Y. PDX1, Neurogenin-3, and MAFA: critical transcription regulators for beta cell development and regeneration. Stem Cell Res Ther. 2017;8(1):240–240. doi: 10.1186/s13287-017-0694-z. [DOI] [PMC free article] [PubMed] [Google Scholar]