Abstract
Statistical experimental designs were used to optimize conditions for recombinant Burkholderia pyrrocinia feruloyl esterase (BpFae) production in bacteria under lactose induction. After optimization by single factor design, Plackett–Burman design, steepest ascent design and the response surface method, the optimal conditions for BpFae production were: 6 g/L lactose, pH 5.5, pre-induced period 5 h, 23 °C, shaker rotational speed of 240 rpm, medium volume of 50 mL/250 mL, inoculum size 0.2% (v/v), and a post-induced period of 32 h in a Luria–Bertani culture. The produced BpFae activity was 7.43 U/mL, which is 2.92 times higher than that obtained under optimal conditions using IPTG as the inducer. BpFae activity was 4.82 U/mL in a 5 L fermenter under the abovementioned optimal conditions. BpFae produced a small amount of ethyl acetate but had no effect on the synthesis of other important esters in Baijiu. The results underpin further investigations into BpFae characterization and potential applications.
Keywords: Burkholderia pyrrocinia, Esterification, Feruloyl esterase, Lactose, Optimization
Introduction
The efficient use of renewable lignocellulosic biomass by biological methods has become an important research topic because of our increasing awareness of sustainable development (Xu et al. 2019b). The architecture of lignocellulosic biomass is complex and heterogeneous. Thus, a key step for effective lignocellulosic biomass use is the conversion of this material into low molecular weight carbon sources (Phuengmaung et al. 2019; Xu et al. 2019b). The complex lignocellulosic cross-linking network structure is composed of cellulose, hemicelluloses, pectin, lignins and hydroxycinnamates (such as ferulic acid (FA) and p-coumaric acid), and degradation of this lignocellulosic biomass requires the synergistic action of a series of cellulases, hemicellulases and accessory enzymes (Meng et al. 2019; Phuengmaung et al. 2019; Wei et al. 2019; Xu et al. 2019b). Previous work has focused primarily on cellulases and hemicellulases, whereas the accessory enzymes remain poorly characterized (Meng et al. 2019). Among the various accessory enzymes, research on feruloyl esterases (FAEs) that catalyze the hydrolysis of an ester linkage between polysaccharides and hydroxycinnamates has increased dramatically because of their important role in de-esterification of covalent interactions between cellulose, hemicelluloses, pectin, lignins and hydroxycinnamates (Mackenzie et al. 1987; Phuengmaung et al. 2019). After de-esterification, opportunities for other lignocellulolytic enzymes to digest lignocellulosic biomass improve with the release of oligosaccharides or FA with strong antioxidant activity (Meng et al. 2019; Phuengmaung et al. 2019). For example, blends of FAEs, cellulase, xylanase and glucosidase were efficiently combined to hydrolyze steam-pretreated sugarcane bagasse (Fortes Gottschalk et al. 2010). In addition, FAEs can be used for other potential applications, such as synthesizing hydroxycinnamate fatty esters, use in pulp and paper industries, as an addition in feed, and production of bioactive phenolic components in pharmaceutical industries (Koseki et al. 2009; Topakas et al. 2007). Thus, FAEs have received significant attention recently because of their potential use in various applications.
FAEs are distributed in plants, fungi and bacteria, with most FAEs of microbial origin (Wu et al. 2019a). Although previous studies reported FAEs produced by filamentous fungi or bacteria in submerged fermentation or solid-state fermentation, titers of very low activity were obtained from these studies and were usually less than 1 U/mL of culture medium (Gopalan et al. 2015). This low activity limits the application of FAEs. Screening of new strains with high FAE production, mutation of native strains, optimization of FAE production conditions and gene heterologous cloning and expression are commonly used to enhance the production of FAEs, which is the primary focus in solving the bottlenecks for FAE applications. The results of Asther et al. showed that solid-state fermentation was better than a submerged culture when methyl caffeate and methyl p-coumarate were used as substrates (Asther et al. 2002). Compared with production using a parent strain, the activity (18 nkat/mL) of FAEs were 16-times higher in an FAE-transformed Aspergillus niger strain, i.e., the faeB gene was inserted into an expression vector under the control of the gpd promoter and expression was induced by sugar beet pulp (Levasseur et al. 2004). Over the past few years, studies have revealed that particular fungi and bacteria can produce FAEs, and some FAE-encoding genes have also been cloned (Hassan and Hugouvieux-Cotte-Pattat 2011; Rashamuse et al. 2007). Unfortunately, since the first identification in the 1980s the number of FAE-producing strains has remained low, with only ~ 100 FAEs being reported. Thus, increasing the screening rate for identification of new FAE-producing strains is important. Moreover, identified FAE genes can be cloned for recombinant FAE production. Identification of new FAE-producing strains that produce higher levels of FAEs or better characterization should resolve the current application bottlenecks.
Baijiu is one of six distilled spirits in the world and is produced by a unique brewing method (Wang et al. 2019). The production process depends on natural fermentation environments and the enrichment of various microorganisms, which produces a variety of enzymes and metabolites that affect the quality of Baijiu (Wang et al. 2019). The Baijiu producing environment is a natural microbial bank, which contains bacteria, fungi, yeast, actinomycetes and archaebacteria. In recent years, many new microbial strains have been screened and identified from such an environment (Xu et al. 2019a). Among these newly identified microorganisms, there are many important strains producing enzyme resources. In our previous studies, a novel lipase-producing strain, Burkholderia pyrrocinia (B. pyrrocinia), was identified from the Baijiu producing environment, and is likely to be important for the synthesis of esters in Baijiu (Li et al. 2018). By analyzing the whole genome of B. pyrrocinia, we found that it contains two novel FAEs, among which BpFae has been successfully expressed in Escherichia coli (E. coli). In previous studies, we used isopropyl-β-d-thiogalactopyranoside (IPTG) to induce an E. coli recombinant strain to produce BpFae. The results showed that this recombinant strain has good FAE production ability with FAE activity reaching a value of 2.54 U/mL after optimization. Lactose may be a suitable inducer and better than IPTG, because lactose is cost efficient, safe and can simultaneously function as an inducer and carbon/energy source (Gombert and Kilikian 1998; Kilikian et al. 2000). In this report, we optimized the expression of a recombinant strain to produce BpFAE using lactose as an inducer. We also explored the recombinant BpFae capacity to synthesize some important esters that affect the quality of Baijiu.
Materials and methods
Materials, reagents and media preparation
Burkholderia pyrrocinia B1213 was isolated and preserved at the China General Microbiological Culture Collection Center (CGMCC, No: 12806) (Li et al. 2018). Ferulic acid methyl ester, FA, ethyl acetate, ethyl lactate, ethyl butyrate, ethyl valerate, ethyl hexanoate, ethyl caprylate, isoamyl acetate, ethyl caprate, ethyl heptanoate and ethyl pelargonate were purchased from Sigma-Aldrich (St. Louis, MO, USA). Ampicillin, kanamycin and bovine serum albumin (BSA) were purchased from Takara (Tokyo, Japan). All other chemicals were analytical grade and commercially available unless otherwise stated. E. coli cells were grown in Luria–Bertani (LB, 5 g/L yeast extract, 10 g/L peptone and 10 g/L NaCl) medium for gene cloning and protein overexpression. Other medium, including Terrific broth medium (TB), LB medium with magnesium (LBBM), LB medium with magnesium and extra NaCl (LBBNM), LB medium with magnesium and glycerol (LBBMG), LB medium with magnesium, glycerol and sorbitol (LBBSMG), and medium for expression (MX) were prepared as reported previously (Golotin et al. 2016).
Cultivation and FAE production by B. pyrrocinia B1213
The basal medium of the flask culture for FAE production contained 100 g/L wheat bran, 1.3 g/L (NH4)2SO4, 0.37 g/L KH2PO4, 0.25 g/L MgSO4·7H2O, 0.07 g/L CaCl2·2H2O, 0.02 g/L FeCl3, 1.0 g/L yeast extract. The initial pH of the medium was adjusted to 6.0 and no further adjustment was made (Kumar et al. 2011). Experiments were carried out in 250 mL Erlenmeyer flasks containing 50 mL medium by liquid-state fermentation (LSF). Each flask was inoculated with activated B. pyrrocinia B1213 at an inoculum size of 4% (v/v). Cultures were incubated at 37 °C with a shaker speed of 200 rpm for 5 days. Each flask was withdrawn periodically every 24 h, the debris removed by centrifugation at 13,751 × g for 10 min at 4 °C, and the supernatant used as the crude enzyme for assaying pH, protein concentration and FAE activity.
BpFae selection and production
We reported previously the detailed procedure for producing BpFae (Fu et al. 2019). Briefly, the BpFae gene was subcloned into BamH I and Sal I sites of the pGEX-4T-1 vector and the gene was in-frame with the downstream hexa-His-tag. The recombinant pGEX-4T-1-BpFae plasmid was transformed into E. coli BL21 (DE3) cells using the standard calcium chloride method. A positive clone was confirmed by gene sequencing (BGI Tech, Beijing, China) and cultivated in LB medium for 6 h. The bacterial suspension was then stored in 20% (v/v) glycerol at − 80 °C for long-term use. The positive transformants were cultured at 37 °C in LB medium including 40 µg/mL ampicillin by shaking at 180 rpm for 5 h. Then, 4 g/L of lactose was added to the culture to induce BpFae expression at 20 °C for 15 h with shaking at 200 rpm and an incubation ratio of 0.2%.
Optimization of the production conditions by single factor design
Various cultivation conditions, including medium type, lactose concentration, incubation ratio, initial pH, post-induction temperature, shaker rotational speed, pre-induction period (before adding lactose into the culture), medium volume and post-induction period (after adding lactose into the culture) were investigated for BpFae production using single factor design (Table 1).
Table 1.
Factors and levels of single factor design
| Factor | Level |
|---|---|
| Medium types | LBBMG, LB, TB, SOB, MX, LBBM, TB-GN, LBBSMG and LBBNM |
| Lactose concentration (g/L) | 0, 2, 4, 6, 8, 10 and 12 |
| Incubation size (%, v/v) | 0.1, 0.2, 0.4, 0.8, 1.6 and 3.2 |
| Initial pH | 4.0, 5.0, 5.5, 6.0, 7.0, 8.0 and 9.0 |
| Post-induction temperature (°C) | 16, 20, 24, 28 and 32 |
| Shaker rotational speed (rpm) | 80, 120, 160, 200 and 240 |
| Pre-induction period (h) | 2, 4, 5, 6, 8 and 10 |
| Medium volume (mL/250 mL) | 12.5, 25, 50, 75, 100 and 125 |
| Post-induction period (h) | 4, 8, 12, 16, 20, 24 and 28 |
Optimization of the production conditions by Plackett–Burman (PB) design
The PB design for seven variables, including initial pH (X1), post-induction temperature (X2), shaker rotational speed (X3), pre-induction period (X4), post-induction period (X5), medium volume (X6) and lactose concentration (X7) at two levels were used for screening, according to the single factor results (Table 2). Each variable is represented at two levels, high and low, which are denoted by (+ 1) and (− 1), respectively (Table 2). The design was run in a single block and the order of the experiments was fully randomized. The PB design is shown in Table 3 and designed by Minitab software 17.1 (Minitab, Inc. State College, PA, USA). A regression model was obtained based on the experimental data. The statistical significance was determined by F value analysis, and the proportion of variance explained by the model obtained was given by the multiple coefficient of determination, R2.
Table 2.
Levels of the variables and statistical analysis in PB design for BpFae activity
| Code | Variables | Low level (− 1) | High level (+ 1) | Effect (EXi) | F values | P values | Significance |
|---|---|---|---|---|---|---|---|
| X1 | pH | 4.5 | 6.5 | − 0.00742 | 4.16 | 0.076 | |
| X2 | Post-induction temperature (°C) | 24 | 32 | − 0.03727 | 104.88 | 0.000 | ** |
| X3 | Shaker rotational speed (rpm) | 120 | 200 | 0.02631 | 52.27 | 0.000 | ** |
| X4 | Pre-induction period (h) | 3 | 7 | 0.00406 | 1.25 | 0.297 | |
| X5 | Post-induction period (h) | 20 | 28 | 0.03163 | 75.54 | 0.000 | ** |
| X6 | Medium volume (mL/250 mL) | 50 | 100 | − 0.01265 | 12.08 | 0.008 | ** |
| X7 | Lactose concentration (g/L) | 2 | 6 | − 0.01155 | 10.08 | 0.013 | * |
*Significant at 5% level (P < 0.05), **significant at 1% level (P < 0.01)
Table 3.
PB design matrix for evaluating factors influencing BpFae activity
| Test number | X1 | X2 (°C) | X3 (rpm) | X4 (h) | X5 (h) | X6 (mL/250 mL) | X7 (g/L) | BpFae activity (U/mL) |
|---|---|---|---|---|---|---|---|---|
| 1 | + 1 | + 1 | − 1 | + 1 | − 1 | − 1 | − 1 | 1.14 |
| 2 | + 1 | − 1 | + 1 | − 1 | − 1 | − 1 | + 1 | 3.16 |
| 3 | − 1 | + 1 | − 1 | − 1 | − 1 | + 1 | + 1 | 0.69 |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.10 |
| 5 | − 1 | + 1 | + 1 | + 1 | − 1 | + 1 | + 1 | 1.73 |
| 6 | + 1 | + 1 | − 1 | + 1 | +1 | − 1 | + 1 | 1.96 |
| 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.15 |
| 8 | + 1 | − 1 | − 1 | − 1 | + 1 | + 1 | + 1 | 2.92 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.85 |
| 10 | − 1 | − 1 | + 1 | + 1 | + 1 | − 1 | + 1 | 5.89 |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.76 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.86 |
| 13 | + 1 | + 1 | + 1 | − 1 | + 1 | + 1 | − 1 | 2.49 |
| 14 | − 1 | − 1 | − 1 | + 1 | + 1 | + 1 | − 1 | 4.24 |
| 15 | − 1 | − 1 | − 1 | − 1 | − 1 | − 1 | − 1 | 2.86 |
| 16 | − 1 | + 1 | + 1 | − 1 | + 1 | − 1 | − 1 | 4.37 |
| 17 | + 1 | − 1 | + 1 | + 1 | − 1 | + 1 | − 1 | 3.28 |
Optimization of the production conditions by steepest ascent path design
The regression model obtained from the PB design screened important variables and these were used to construct the steepest ascent path. The direction of the steepest ascent path is determined from the regression results. Experiments were conducted along the steepest ascent path and using practical experience until the BpFae activity showed no further increase (Table 4). A particular point where the BpFae activity was highest by steepest ascent path design would be near the optimal point and could be used as the center point of RSM.
Table 4.
Experimental designs and the results of steepest ascent for BpFae activity
| Test count | X1 (°C) | X2 (rpm) | X3 (h) | X4 (mL/250 mL) | X5 (g/L) | BpFae activity (U/mL) |
|---|---|---|---|---|---|---|
| 1 | 32 | 120 | 20 | 80 | 10 | 1.98 |
| 2 | 28 | 160 | 24 | 65 | 8 | 4.78 |
| 3 | 24 | 200 | 28 | 50 | 6 | 6.72 |
| 4 | 20 | 240 | 32 | 35 | 4 | 2.71 |
| 5 | 16 | 280 | 36 | 20 | 2 | 2.71 |
Optimization of the production conditions by response surface methodology (RSM)
RSM was used to optimize the screened variables for enhanced BpFae activity using the Box–Behnken experimental design (BBD, Design-Expert Software 11.0, StatEase Inc., Minneapolis, MN, USA). The variables and center point were identified and determined as the statistically significant influence on BpFae activity by PB design and steepest ascent path design. The design listed in Table 5 was composed of three factors [post-induction temperature (A), post-induction period (B) and shaker rotational speed (C)] and three levels, including three replicates at the center point. Each experiment was performed in triplicate and the average BpFae activity was taken as the response.
Table 5.
BBD design and the responses of the dependent variables
| Test number | Post-induction temperature (°C) | Post-induction period (h) | Shaker rotational speed (rpm) | BpFae activity (U/mL) | |||
|---|---|---|---|---|---|---|---|
| A | Code A | B | Code B | C | Code C | Y | |
| 1 | 24 | 0 | 32 | + 1 | 240 | + 1 | 7.38 |
| 2 | 20 | − 1 | 32 | + 1 | 200 | 0 | 6.53 |
| 3 | 24 | 0 | 32 | + 1 | 160 | − 1 | 7.16 |
| 4 | 20 | 0 | 28 | 0 | 240 | + 1 | 6.04 |
| 5 | 28 | + 1 | 24 | − 1 | 200 | 0 | 5.35 |
| 6 | 24 | 0 | 28 | 0 | 200 | 0 | 6.65 |
| 7 | 20 | − 1 | 24 | − 1 | 200 | 0 | 5.71 |
| 8 | 28 | + 1 | 28 | 0 | 240 | + 1 | 5.65 |
| 9 | 20 | − 1 | 28 | 0 | 160 | − 1 | 5.74 |
| 10 | 28 | + 1 | 28 | 0 | 160 | − 1 | 5.05 |
| 11 | 24 | 0 | 24 | − 1 | 160 | − 1 | 6.15 |
| 12 | 24 | 0 | 28 | 0 | 200 | 0 | 6.74 |
| 13 | 24 | 0 | 28 | 0 | 200 | 0 | 6.81 |
| 14 | 24 | 0 | 24 | − 1 | 240 | + 1 | 7.04 |
| 15 | 28 | + 1 | 32 | + 1 | 200 | 0 | 5.39 |
Culture amplification using a bioreactor
The optimal conditions obtained through the above optimization processes were used to scale production in a 5 L INFORS Minifors Benchtop bioreactor (INFORS, Switzerland). LB medium was used and the lactose solution used to induce BpFae production was autoclaved separately. The initial pH of the fermentation medium was adjusted to 5.5. For seed preparation, recombinant BL21 (DE3) E. coli cells harboring the pGEX-4T-1-BpFae construct were inoculated in 50 mL LB medium with 40 μg/mL ampicillin as the selection marker, and the flask was incubated 12 h at 37 °C, 180 rpm. The cultivated seed was added to 3.0 L working volume at an inoculation ratio of 0.2% (v/v). The cultivation temperature was 37 °C, the rotation speed of the bioreactor was 180 rpm and the airflow rate was 0.5 the vessel volume per minute (airflow rate/operating volume, vvm) at the pre-induction stage. The optical density at 600 nm (OD600), pH and dissolved oxygen (DO) were measured every hour. When the OD600 reached 2.5, the cells were induced using a 6 g/L autoclaved lactose solution, and the cultivation temperature and rotation speed were adjusted to 23 °C and 240 rpm, respectively, at the start of the induction phase. Samples of cultures were taken in situ every 4 h to monitor OD600, pH, DO, BpFae activity and protein concentration.
Expression analysis of BpFae
The induced cells were harvested by centrifugation at 13,751 × g for 10 min at 4 °C, resuspended in 50 mmol/L sodium phosphate buffer (pH 7.0) and disrupted by ultrasonication in an ice water bath for 10 min with a 2 s-on/4 s-off program. The suspension was harvested by centrifugation at 13,751 × g for 10 min at 4 °C. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), BpFae activity and protein analyses were performed to evaluate the expression of BpFae in E. coli.
FAE activity assay and protein concentration determination
FAE activity was assayed according to the method of Fu et al. (2019). Briefly, a suitably diluted enzyme solution (50 µL) was added to 450 µL methyl ferulic acid (MFA) substrate solution (1 mmol/L, dissolved in DMSO) dissolved in 50 mmol/L sodium phosphate buffer (pH 7.0). The mixture was incubated at 37 °C for 10 min and 500 µL acetonitrile was added to the mixture to terminate the reaction. The products were filtered through a 0.22-µm filter and analyzed by high performance liquid chromatography (HPLC) with a UV detector (Agilent, Santa Clara, CA, USA) and a ZORBAX Eclipse Plus C-18 column (Agilent). The column was maintained at 35 °C and eluted with a mixture of acetonitrile, water and acetic acid (70:29.7:0.3, v/v/v) at a flow rate of 0.6 mL/min. The column eluent was monitored at 320 nm. One unit (U) of enzyme activity was defined as the enzyme amount required for the release of 1 µmol ferulic acid in 1 min under the standard conditions provided above. The protein concentration was estimated using a BCA (bicinchoninic acid) protein assay kit (Beijing Solarbio Science and Technology Co., Ltd., Beijing, China) according to the manufacturer’s instructions. BSA was used as the standard protein.
SDS-PAGE
SDS-PAGE was performed under denaturing conditions as described by Laemmli using a 12.5% separating gel and a 4.5% stacking gel, and proteins were visualized by staining with 0.25% Coomassie brilliant blue R250 (Laemmli 1970). Briefly, 30 μL of a protein sample was added to 7.5 μL sample buffer, boiled in a water bath for 5 min, cooled to room temperature and loaded onto the gel.
Ability of BpFae to synthesize important esters for Baijiu
The synthesis of esters was carried out in a closed 20-mL conical flask with a stopper. Reaction mixtures containing 0.9 mol/L of one kind of alcohol (ethyl alcohol and isoamyl alcohol) and 0.3 mol/L of one kind of acid, including acetic acid, lactic acid, butyrate, hexanoic, octoic acid, capric acid, valeric acid, heptanoic acid and nonanoic acid, with 1 U/mL of BpFae in 10 mL of a n-hexane system were incubated at 40 °C for 6 h. The reaction was initiated by adding the enzyme to the substrate mixture with stirring at 180 rpm. For the control sample, BpFae was replaced by the supernatant of BL21 (DE3) E. coli cells harboring pGEX-4T-1. Each reaction was performed in triplicate and the average ester concentrations were taken.
An Agilent 7890B GC with a FID (Agilent) and a HP-INNOWAX column (30 m × 0.32 mm × 0.50 μm, Agilent) were used to assay the esters. The following analytical conditions were used: the injection temperature was 280 °C; splitless mode was adopted; the carrier gas was nitrogen; and the detector temperature was 280 °C. The column temperature was held at 40 °C for 5 min, increased to 170 °C at a rate of 8 °C/min, held at 170 °C for 10 min, and finally increased to 240 °C at a rate of 8 °C/min and maintained there for 5 min. The response factor of each ester was acquired by analyzing the standard mix, and the esters content was calculated as grams of esters produced per mL of sample.
Statistical analysis
Statistical differences of the assessed strategies were analyzed using a one-way ANOVA (P < 0.05) with the Tukey test. Assays have been conducted at least in triplicate and the reported values correspond to the mean value and its standard deviation. Minitab 17.1 (Minitab Inc.), SPSS 24.0 (IBM Corp., New York, USA), OriginPro 9.1 (OriginLab, Northampton, MA, USA), Design-Expert 11 (Stat-Ease, Inc. USA) and Excel 2016 (Microsoft, USA) were used to analyze the data.
Results and discussion
Production of FAE by B. pyrrocinia B1213
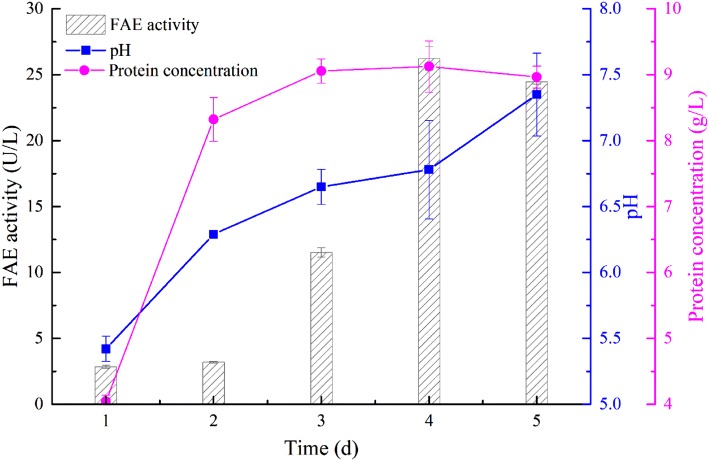
Burkholderia pyrrocinia B1213 was grown in a liquid culture for 5 days using wheat bran as the carbon source. FAE activities were monitored as a function of the incubation time. Enzyme activities increased during the early stages of growth, reached the highest level after 4 days of culture (26.24 U/L) and remained static until day 5 (Fig. 1). The change in protein concentration for the fermentation broth was consistent with that of FAE activity. Although there are many strains that can produce FAEs, the FAE activity by native strains is generally lower than 1 U/mL (Topakas et al. 2007), and FAE activity from B. pyrrocinia B1213 is also low. This may be because the cinnamic acid derivatives content in nature is far lower than xylan and glucan. From the perspective of microbial evolution, secretion levels of FAEs by native strains are far lower than those of xylanases and glucanases. However, cinnamic acid derivatives such as ferulic or coumaric acids are covalently bound to xylan, glucan and other high molecular materials via ester linkages, which increases the complexity of lignocellulose structures and makes their degradation challenging (Oliveira et al. 2019). FAEs can release FA and other hydroxycinnamic acids from lignocellulose, which then undergoes degradation by lignocellulolytic enzymes such as xylanase and glucanase. Therefore, FAEs play an important role in the degradation of lignocellulose.
Fig. 1.
Time course of FAE production by B. pyrrocinia B1213 with wheat bran as the carbon source
The main raw materials for Baijiu brewing are sorghum, wheat, corn and other raw grain materials. Effective degradation of the cell wall of these raw materials facilitates bioconversion of starch into ethanol and flavor components in Baijiu. Clearly, the effective degradation of the cell wall of these raw materials requires FAEs. Additionally, the presence of FAEs in a Baijiu brewing system will also contribute to the FA content in Baijiu, which may help to reduce potential health issues from alcohol in Baijiu because of its excellent antioxidant activity (Wu et al. 2019b). Burkholderia pyrrocinia B1213 was selected from the Baijiu brewing environment and produces glucanase, FAEs and other important lignocellulose degradation enzymes. Thus, it plays an important role in the degradation of raw materials and production of FA in the Baijiu brewing process. In addition, B. pyrrocinia B1213 has good plasticizer degradation activity, which may also be related to the production of FAEs (Li et al. 2019). Currently, most FAEs have been isolated from fungal rather than bacterial sources and production of an FAE produced by B. pyrrocinia has not been reported. Therefore, we cloned and expressed the FAE from B. pyrrocinia B1213 in an effort to improve the activity of an FAE using molecular biology tools. This work provides the foundation for subsequent analysis of the enzymatic properties of this FAE and its potential applications (Fu et al. 2019).
Optimization of the production conditions by single factor design
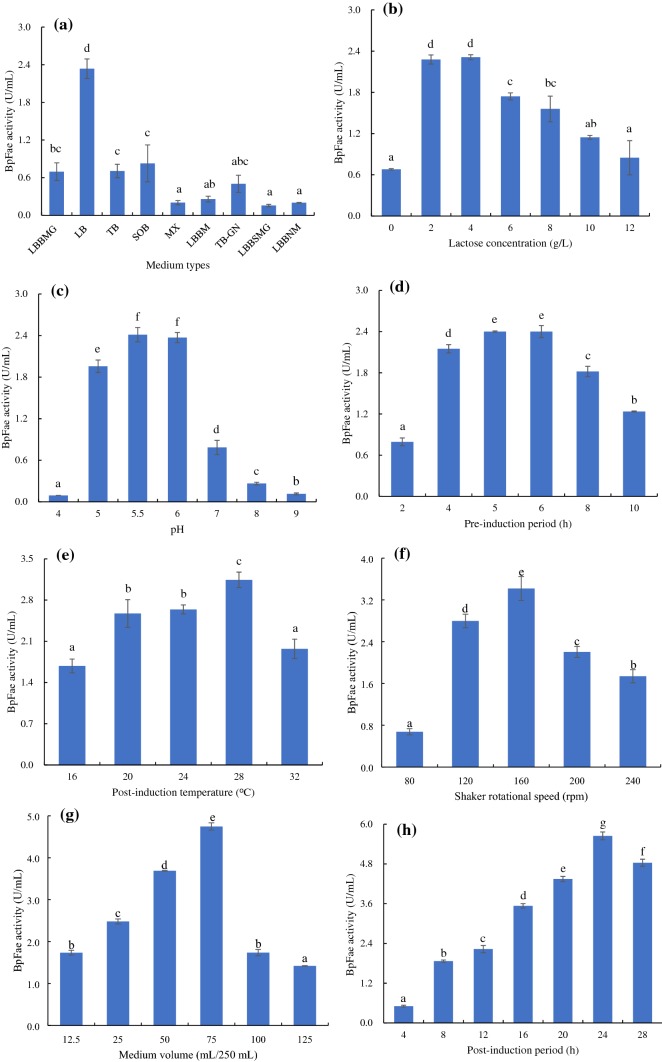
Nine standard culture media, i.e., LB, TB, MX, SOB, LBBM, LBBMG, LBBNM, LBBSMG and TB-GN, were used to overexpress BpFae in E. coli. As shown in Fig. 2a, LB medium was the best expression medium for BpFae overexpression induced by the addition of lactose, which differed to a previous study, where IPTG was used to induce overexpression (the best medium was SOB medium) (Fu et al. 2019). Although IPTG is a lactose analogue and has similar efficacy in inducing E. coli to express heterologous proteins, the results showed that different levels of FAE overexpression were observed when using IPTG and lactose. This may be because lactose is both an inducer and a carbon source for fermentation, and thus participates in the metabolic pathways of E. coli (Gombert and Kilikian 1998), whereas IPTG is not a carbon source.
Fig. 2.
Effect of medium types (LBBMG, LB, TB, SOB, MX, LBBM, TB-GN, LBBSMG and LBBNM) (a), lactose concentration (0, 2, 4, 6, 8, 10 and 12 g/L) (b), pH (4.0, 5.0, 5.5, 6.0, 7.0, 8.0 and 9.0) (c), pre-induction period (2, 4, 5, 6, 8 and 10 h) (d), post-induction temperature (16, 20, 24, 28 and 32 °C) (e), shaker rotational speed (80, 120, 160, 200 and 240 rpm) (f), medium volume (12.5, 25, 50, 75, 100 and 125 mL/250 mL) (g) and post-induction period (4, 8, 12, 16, 20, 24 and 28 h) (h) on BpFae activity
Lactose is a low-cost, non-toxic inducer used with pGEX-4T-1 expression systems (Su et al. 2019). Normally, protein overexpression using the pGEX-4T-1 vector can be fully induced using high concentrations of lactose (Su et al. 2019). However, when lactose is used as an inducer, protein expression and cell growth should remain balanced to obtain the highest protein production levels (Kilikian et al. 2000). In this study, seven different lactose concentrations (0, 2, 4, 6, 8, 10 and 12 g/L) were tested. As shown in Fig. 2b, when the lactose concentration was low (2–4 g/L) the activity of the recombinant BpFae was high, whereas at higher lactose concentrations, a reduction in BpFae activity was observed. The highest BpFae activity, 2.31 U/mL, was obtained at a lactose concentration of 4 g/L. This activity is approximately 2.0-fold higher than the result using the optimal IPTG concentration (i.e., 1.15 U/mL) (Fu et al. 2019). The results (data not shown) showed that the biomass and the content of BpFae in the supernatant after cell disruption (the content of soluble protein) were higher when using lactose rather than IPTG as the inducer, and there was minimal amount of insoluble BpFae in the cell debris (no inclusion bodies) when using lactose to induce overexpression (there were a few inclusion bodies when IPTG was used as the inducer) (Su et al. 2019). Soluble expression of the target protein can be achieved by increasing the cell biomass and reducing the expression rate of the target protein, which may explain why lactose induction is better than IPTG induction in this study (Monteiro et al. 2000). In addition, higher lactose concentrations (> 10 g/L) decreased the final BpFae activity, especially 12 g/L. This phenomenon may have been caused by high osmotic pressure on cell growth and metabolic burden (Su et al. 2019).
The optimal pH for BpFae production was examined over the pH range of 4–9. The activity of BpFae first increased and then decreased with increasing pH. The activity of BpFae was highest over the pH range of 5.5–6.0 (Fig. 2c), which is slightly higher when compared with that observed with IPTG induction (the optimal pH was 5.0) (Fu et al. 2019). pH is a key factor to examine, because pH affects the expression of membrane and periplasmic proteins, metabolic enzymes, the activity of bacterial proteases and secretory production of proteins (Choi and Lee 2004; Stancik et al. 2002). In general, the optimal pH for protein expression is 4.4–9.2, which is consistent with the optimal growth pH of E. coli (Stancik et al. 2002). Different heterologous proteins have different optimal pH values when overexpressed, which may be due to the effect of heterologous proteins on E. coli cells and cell metabolism.
The pre-induction period has an important effect on the expression of heterologous proteins. Although, short pre-induction periods enable E. coli to express heterologous proteins earlier, it often leads to a low cell biomass and average protein yields because of the earlier or longer toxicity of particular heterologous protein to the host E. coli cells. While long pre-induction periods that enable E. coli cells to grow to higher densities prior to induction can still result in low expression levels of the heterologous protein, because nutrients have been depleted in the medium and other metabolites that inhibit overexpression have accumulated. Therefore, it is important to obtain an ideal pre-induction period. The addition of inducing agents is usually carried out when the cells are in the mid-to-late logarithmic growth stage (Li et al. 2017; Shoja Alsadati et al. 2008). In this study, the optimal pre-induction period was 4–6 h (OD600 was ~ 2.5), at which point cells were in a mid-log growth phase (Fig. 2d). This result differed slightly to results obtained when using IPTG as the inducer (Fu et al. 2019). This may be because lactose is a carbon source nutrient, which promotes further growth of E. coli cells (Kilikian et al. 2000).
The post-induction temperature is also a critical factor that influences the secretion and solubility of overexpressed protein, the yield of protein production and the activity of cellular enzymes (Gadgil et al. 2005). Reducing the temperature can facilitate expression of the target protein in the soluble form. In this study, the optimum post-induction temperature for expression of BpFae was found to be 28 °C (Fig. 2e). In contrast, in previous reports the optimal post-induction temperature for overexpression of heterologous proteins in E. coli cells was ~ 30 °C (Derakhshani et al. 2019). The post-induction temperature affects the induction rate and cell growth rate, so different heterologous proteins and expression systems will have different optimal induction temperatures. The best temperature for post-induction is often when the induction rate and cell growth rate are balanced.
A particular problem for bioprocesses is ensuring the correct level of oxygen input. This problem has significant effects on the metabolism of microorganisms and is exacerbated by rapid increases in cell density (Kumar et al. 1991). A shift to no dissolved oxygen for a short period has been shown to cause strong segregation instability to the plasmid system in E. coli. These periods can also potentially alter bacterial metabolism, which affects both growth and recombinant protein production, and thus lowers cell productivity (Hopkins et al. 1987; Phue and Shiloach 2005). There are many reports describing recombinant protein expression with oxygen limitations (Qoronfleh 1999; Zare et al. 2019). Different levels of dissolved oxygen must be evaluated for each protein overexpression system. We investigated the shaker rotational speed and medium volume to study the effect of dissolved oxygen on the production of BpFae. The results showed that the activity of BpFae first increased and then decreased with increased shaker rotational speed and medium volume (Fig. 2f, g). As other reports have shown, there is competition between replication and expression of chromosomal DNA and plasmid DNA for limited cellular resources (Ryan et al. 1989). Thus, BpFae activity is highest at an intermediate dissolved oxygen level. Previous studies have indicated that operating at high dissolved oxygen levels is beneficial for cell mass formation; however, such operation is not necessarily optimal for overexpression of the recombinant protein (Ryan et al. 1989). In addition, it is also possible that the rate of lactose induction is slower than that of IPTG, so the amount of dissolved oxygen required when using lactose to induce protein overexpression is likely to be lower when compared with the oxygen level requirements under IPTG induction (Fu et al. 2019).
The inoculum size affects the microbial biomass when the inducer is added, and biomass is often linked to the amount of heterologous protein produced by the bacteria (Shoja Alsadati et al. 2008). Thus, the effect of the inoculum size on BpFae activity was studied. The results showed that there was no significant difference in BpFae activity under different inoculum sizes [0.1% (v/v) to 3.2% (v/v)], but the highest value was observed when the inoculum size was 0.2% (v/v) with the BpFae activity reaching 2.13 U/mL (data not shown). The results showed that the cells were in the mid-to-late log growth phase when lactose was added under the investigated inoculum size, and differences in cell biomass were not significant when lactose was added at different points in this growth phase. This was also observed for IPTG induction.
The post-induction period affects protein expression and/or solubility (Sadeghian-Rizi et al. 2019). Although, long incubation periods often result in higher protein yields, the optimum incubation time for overexpression is target protein specific (Sadeghian-Rizi et al. 2019). BpFae activity increased continuously as the post-induction period was increased (Fig. 2h). The BpFae activity reached a maximum at a post-induction period of 24 h. At longer periods, the activity decreased, which may be due to degradation of the target protein by proteases released after cell death (Baneyx and Georgiou 1990; Maurizi 1992).
Optimization of the production conditions by PB design
BpFae activity was evaluated using PB design and the results are presented in Table 3. Changes in BpFae activity ranged from 0.69 to 5.89 U/mL. Based on the coefficient of regression, five out of seven variables, i.e., X2, X5, X3, X6 and X7, significantly influenced BpFae activity at the 5% level of significance. X3 and X5 had positive coefficients of 0.02631 and 0.03163, respectively, whereas the other three variables showed negative coefficients (Table 2). Thus, X3 and X5 should be enhanced, and the increase of X2, X6 and X7 should be avoided in subsequent experiments, because this is more conducive for improving BpFae activity. The above five parameters should be studied further, whereas X1 and X4 were found to have an insignificant effect on the BpFae activity and should be omitted in subsequent experiments (Table 2). In the following experiments, X1 and X4 were set to 5.5 and 5 h, respectively, according to the single factor results described above.
Optimization of the production conditions by steepest ascent path design
Steepest ascent path design was undertaken to determine the optimal regions of these five significant factors (Zhou et al. 2011). The proper direction of changing variables was based on the above regression analysis of the PB design. To obtain the maximum BpFae activity, the design increased the values of variables X2 and X3 while decreasing the values of variables X1, X4 and X5 (Table 4). The results showed that run three with the highest response of 6.72 U/mL gave the maximum yield. Thus, the third set of tests was used as the central point of the response surface.
Optimization of the production conditions by RSM design
Based on the results of PB design and steepest ascent path design, a three-factor (post-induction temperature, post-induction period and shaker rotational speed) and three-level BBD were used to optimize the best fermentation conditions for BpFae production. Table 5 shows the maximum and minimum levels of variables chosen for trials in the design and the experimental responses for the 15 experimental runs. The results in Table 5 show that there is considerable variation in BpFae activity, and this variability is dependent on the different culture conditions. Maximum BpFae activity was achieved in test number 1 (7.38 U/mL), whereas the minimum BpFae activity was observed in run number 10 (5.05 U/mL). The center point in the design was repeated three times for estimating the error.
By applying multiple regression analysis on the experimental data, the following second order polynomial coded equation was found to explain BpFae production regardless of the significance of the terms:
| 1 |
where Y is the predicted response (BpFae activity).
The statistical significance of Eq. (1) was checked by F test analysis, and the analysis of variance (ANOVA) for the response surface quadratic model is shown in Table 6. The model fits well to the experimental data. A lower value of the coefficient of variation (CV = 1.91%) indicates better reliability of the experiments performed. The determination coefficient (R2) implies that the sample variation of 99.06% for BpFae production is attributed to the independent variables, and only about 0.94% of the total variation cannot be explained by the model. The correlation coefficient (R = 0.9953) for Eq. (1) is close to one, indicating a strong correlation between the experimental results and the theoretical values. Linear and quadratic terms were both significant at the 1% level and the cross product was significant at the 5% level.
Table 6.
Regression coefficients and their significances for BpFae activity from the results of the BDD
| Source | Sum of squares | df | Mean square | F value | P value | Significant |
|---|---|---|---|---|---|---|
| Model | 7.40 | 9 | 0.8219 | 58.34 | 0.0002 | ** |
| A-Incubation temperature | 0.8321 | 1 | 0.8321 | 59.06 | 0.0006 | ** |
| B-Shaker rotational speed | 0.6105 | 1 | 0.6105 | 43.33 | 0.0012 | ** |
| C-Incubation time | 0.5050 | 1 | 0.5050 | 35.85 | 0.0019 | ** |
| AB | 0.1521 | 1 | 0.1521 | 10.80 | 0.0218 | * |
| AC | 0.0225 | 1 | 0.0225 | 1.60 | 0.2620 | |
| BC | 0.1122 | 1 | 0.1122 | 7.97 | 0.0370 | * |
| A2 | 4.89 | 1 | 4.89 | 346.86 | < 0.0001 | ** |
| B2 | 0.0970 | 1 | 0.0970 | 6.89 | 0.0469 | * |
| C2 | 0.0051 | 1 | 0.0051 | 0.3604 | 0.5745 | |
| Residual | 0.0704 | 5 | 0.0141 | |||
| Lack of fit | 0.0576 | 3 | 0.0192 | 2.98 | 0.2611 | Not significant |
| Pure error | 0.0129 | 2 | 0.0064 | |||
| Cor total | 7.47 | 14 | ||||
| R2= 0.9906 | = 0.9736 | = 0.8728 | CV = 1.91% |
*Significant at 5% level (P < 0.05), **significant at 1% level (P < 0.01)
The F value and the corresponding P value are given in Table 6. The corresponding P values suggest that among the independent variables, A (post-induction temperature), B (post-induction period) and C (shaker rotational speed) had a significant effect on BpFae activity. The positive coefficients for B and C indicated a linear effect that causes an increase in BpFae activity, whereas the negative coefficient for A indicates a linear effect that causes a decrease in BpFae activity, which was consistent with the PB design results. The quadric term of A and B also had a significant effect. Moreover, two interactions between (A, B) and (B, C) were also found to contribute to the response at a significant level. Model terms are significant when P values are less than 0.05. In this case, A, B, C, AB, BC, A2 and B2 were the significant model terms.
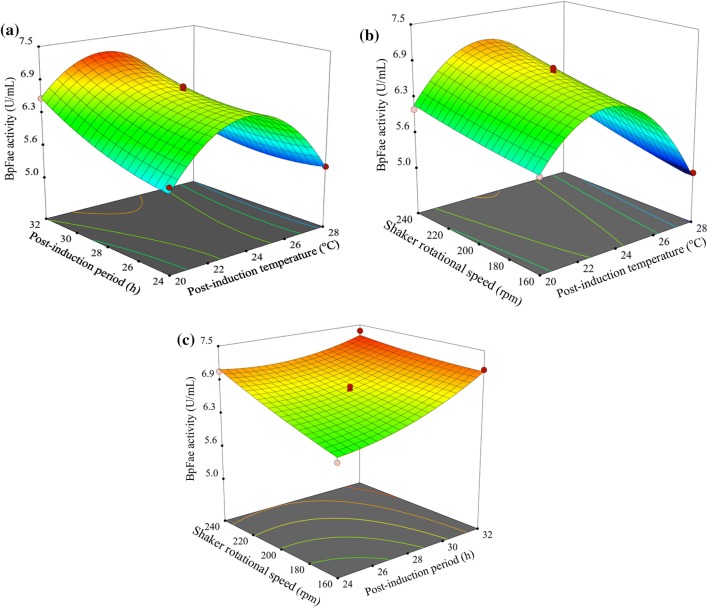
The 3D response surface plots for BpFae production described by the above model were drawn to illustrate the effects of the independent variables and to depict the interactions between two variables by keeping the third variables at their zero levels (Fig. 3). The results in Fig. 3a indicate that BpFae production yields increased gradually as the post-induction period increased. This phenomenon was more pronounced when the post-induction temperature was set at a mid-value, resulting in a change in BpFae production from 6.22 to 7.22 U/mL. This suggests that increasing the post-induction period with an appropriate post-induction temperature is beneficial to BpFae production. Increasing the post-induction temperature caused a significant increase in BpFae activity, but beyond a certain temperature the activity decreased. The analysis of Fig. 3a reveals that the optimal ranges of post-induction temperature and post-induction period for BpFae production were 22.5–25.1 °C and 30–32 h, respectively. The effects of A (post-induction temperature) and C (shaker rotational speed) on BpFae production are shown in Fig. 3b. Clearly, post-induction temperature was more important than shaker rotational speed on BpFae production with enzyme activity ranging between 5.05 and 7.03 U/mL as the post-induction temperature was increased, whereas BpFae production was not affected significantly by changes in the shaker rotational speed. The balance among cell number, activity, heterologous protein expression rate and correct protein folding is key to substantial protein production, and the post-induction temperature achieves high-efficiency heterologous protein expression by effectively adjusting the balance between them. The effects of B (post-induction period) and C (shaker rotational speed) on BpFae production are presented in Fig. 3c. BpFae production increased rapidly as the post-induction period and shaker rotational speed increased. Increasing the shaker rotational speed improves dissolved oxygen levels, thus promoting an increase in biomass. At the same time, extending the post-induction time affords induced E. coli cells to produce larger quantities of the target enzyme.
Fig. 3.
3D surface interaction of variable on the BpFae activity response using the BBD (post-induction temperature, post-induction period and shaker rotational speed). a Interaction of post-induction temperature and post-induction period; b interaction of post-induction temperature and shaker rotational speed; c interaction of post-induction period and shaker rotational speed
The optimal values of the variables giving the highest yield of BpFae production were derived by the Design-expert 11 software with the following critical values: A (post-induction temperature) = 23.2 °C, B (post-induction period) = 32 h and C (shaker rotational speed) = 240 rpm. The maximum predicted value of Y was 7.33 U/mL. According to results of the statistically designed experiments, the optimized process parameters were: 6 g/L lactose, pH 5.5, pre-induction period 5 h, 23 °C, shaker rotational speed 240 rpm, medium volume of 50 mL/250 mL, inoculum size 0.2% (v/v) and a post-induction period of 32 h in an LB culture.
A repeat fermentation of BpFae under optimal conditions was carried out for verification of the optimized parameters. The maximal FAE level obtained was 7.43 U/mL, which closely matches the predicted value. Optimization resulted in a 1.92-fold increase in BpFae production when compared with that of 2.54 U/mL obtain by IPTG induction.
Batch fermentation result
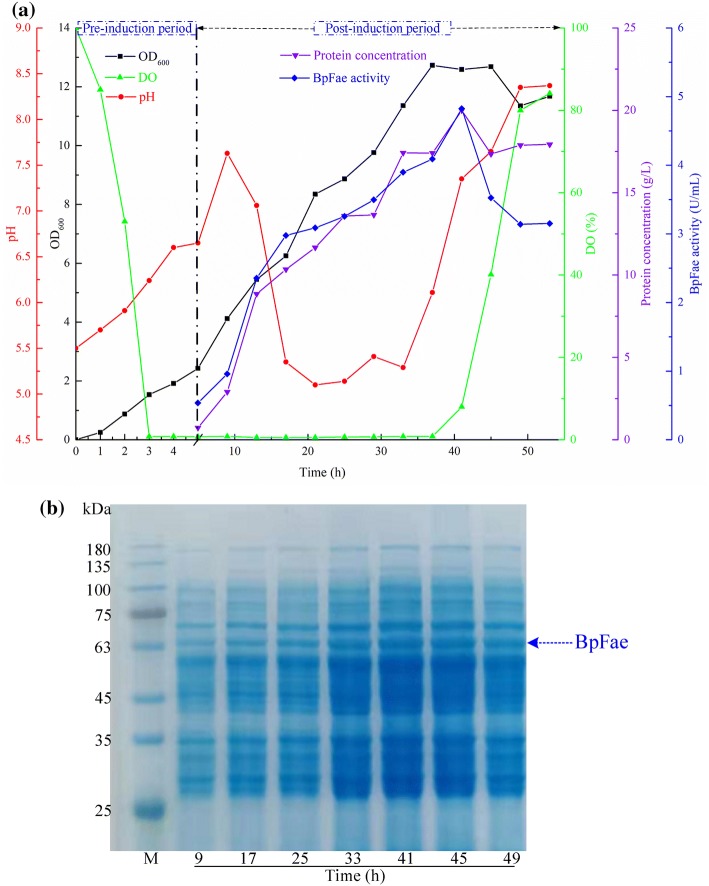
The feasibility of the regression models was examined in a 5-L fermenter. Figure 4 shows the batch profile of BpFae production and the protein concentration profile in the fermenter under optimized conditions. As shown in Fig. 4a, minor BpFae activity was detected during the first 9 h [pre-induction period (5 h) and during the early stage of induction (4 h)], and a significant increase in BpFae activity was observed after 13 h cultivation. The maximum BpFae activity reached 4.82 U/mL after 41 h cultivation, and a slight decrease in BpFae activity was observed after that time, which is consistent with the change in protein concentration and target protein (~ 65 kDa) content observed by SDS-PAGE analysis (Fig. 4b). This result has been repeatedly verified in a 5 L fermenter. The results showed that BpFae activity in the fermenter was lower than that in a flash shaker. This can be attributed to differences in optimal fermentation parameters between a flask shaker and the large-scale fermentation process, such as dissolved oxygen, cell viability, metabolites and biomass (Li et al. 2007).
Fig. 4.
Time course profile of recombinant BpFae secretory production in E. coli in 5 L fermenter (a) and SDS–PAGE analysis (b). Symbols: protein content of BpFae (filled down pointing triangle), BpFae activity (filled diamond), OD600 (filled square), DO (filled up pointing triangle), pH (filled circle). Lane M, low molecular weight standard protein markers. The time course indicated samples after 9, 17, 25, 33, 41, 45 and 49 h, respectively
Synthesis of esters using BpFae
The synthesis of several important ester compounds, which have an important influence on Baijiu quality, by BpFae in n-hexane was studied. The results are shown in Table 7. The results showed that BpFae has negligible effect on the esterification of other important esters except for the synthesis of a small amount of ethyl acetate. Primary structure analysis of FAEs reveals that these enzymes have a Ser–His–Asp catalytic triad at their active site, with an α/β hydrolase fold, which is very similar to that of lipases (Hatzakis and Smonou 2005). Although some reports showed that FAEs have some esterification activity, few studies have been reported the potential of FAEs for the transesterification of alcohols (Bonzom et al. 2018; Hatzakis and Smonou 2005; Kikugawa et al. 2012; Zeng et al. 2014). Our results differ to those of Hatzakis et al., which may be due to the different substrate specificity of FAEs and the different alcohols selected (Hatzakis and Smonou 2005). Our study focused on the role of BpFae in the catalytic synthesis of important flavor esters in Baijiu. This is the first report describing the synthesis of important esters in Baijiu catalyzed by FAEs. Although the effect of the synthesized esters in Baijiu by BpFae was minimal, it may improve the utilization of starch content in sorghum mainly by cooperating with other enzymes in Baijiu brewing systems. In a follow-up study, we will carry out cooperative treatment of sorghum with xylanase to improve the utilization of sorghum by Saccharomyces cerevisiae.
Table 7.
Ester synthesis capacity of BpFae
| Esters | Yield of ester (g/L) | Esters | Yield of ester (g/L) |
|---|---|---|---|
| Ethyl acetate | 0.25 | Ethyl heptanoate | – |
| Ethyl lactate | – | Ethyl caprylate | – |
| Ethyl butyrate | – | Ethyl pelargonate | – |
| Ethyl valerate | – | Ethyl caprate | – |
| Ethyl hexanoate | – | Isoamyl acetate | – |
– no detection
Conclusion
In this report, an optimization strategy that included single factor design, PB design, steepest ascent path design and RSM design, was used to improve recombinant BpFae overexpression in E. coli using lactose as the inducer. After optimization, the highest BpFae activity obtained was 7.43 U/mL, which was an improvement of 1.92-fold when compared with results obtained using IPTG as the inducer. The culture was then scaled in a 5 L fermenter according to the optimal conditions identified. Although BpFae activity was lower than that in shaking flasks induced with lactose, it was 1.9 times higher than that observed for E. coli cultures in shaking flasks induced by IPTG. This study improved the expression of BpFae and provides sufficient enzyme resources for a follow-up study examining enzyme properties and applications.
Acknowledgements
We thank Andrew Dingley and Emma Barratt for insightful discussions and providing language help. This research was supported by the National Natural Science Foundation of China (nos. 31701592, 31830069, and 31671798), General Project of Scientific Research Program of Beijing Municipal Education Commission (Grant number KM201910011006/PXM2019_014213_000007) and Quality Construction of Talents Training/First-class Speciality Construction/Food Science and Engineering (PXM2019_014213_000010).
Compliance with ethical standards
Conflict of interest
The authors declare no competing financial interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Guangsen Fan and Yuting Zhu contributed equally to this work.
References
- Asther M, Haon M, Roussos S, Record E, Delattre M, Lesage-Meessen L, Labat M, Asther M. Feruloyl esterase from Aspergillus niger a comparison of the production in solid state and submerged fermentation. Process Biochem. 2002;38:685–691. doi: 10.1016/S0032-9592(02)00196-6. [DOI] [Google Scholar]
- Baneyx F, Georgiou G. In vivo degradation of secreted fusion proteins by the Escherichia coli outer membrane protease OmpT. J Bacteriol. 1990;172:491–494. doi: 10.1128/jb.172.1.491-494.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonzom C, Schild L, Gustafsson H, Olsson L. Feruloyl esterase immobilization in mesoporous silica particles and characterization in hydrolysis and transesterification. BMC Biochem. 2018 doi: 10.1186/s12858-018-0091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Lee SY. Secretory and extracellular production of recombinant proteins using Escherichia coli. Appl Microbiol Biotechnol. 2004;64:625–635. doi: 10.1007/s00253-004-1559-9. [DOI] [PubMed] [Google Scholar]
- Derakhshani A, Keshavarz KF, Banadkoki SB, Shirazi FH, Barati M, Fereidouni M, Safarpour H. Optimization of induction parameters, structure quality assessment by ATR-FTIR and in silico characterization of expressed recombinant polcalcin in three different strains of Escherichia coli. Int J Biol Macromol. 2019;138:97–105. doi: 10.1016/j.ijbiomac.2019.07.078. [DOI] [PubMed] [Google Scholar]
- Fortes Gottschalk LM, Oliveira RA, Da Silva Bon EP. Cellulases, xylanases, beta-glucosidase and ferulic acid esterase produced by Trichoderma and Aspergillus act synergistically in the hydrolysis of sugarcane bagasse. Biochem Eng J. 2010;51:72–78. doi: 10.1016/j.bej.2010.05.003. [DOI] [Google Scholar]
- Fu ZL, Fan GS, Zhu YT, Sun BG, Teng C, Li H, Liu Q, Yang R, Li XT (2019) Soluble expression of a novel feruloyl esterase from Burkholderia pyrrocinia B1213 in Escherichia coli and optimization of feruloyl esterase production. Biotechnol Biotechnol Equip (Accepted)
- Gadgil M, Kapur V, Hu WS. Transcriptional response of Escherichia coli to temperature shift. Biotechnol Progr. 2005;21:689–699. doi: 10.1021/bp049630l. [DOI] [PubMed] [Google Scholar]
- Golotin VA, Balabanova LA, Noskova YA, Slepchenko LV, Bakunina IY, Vorobieva NS, Terenteva NA, Rasskazov VA. Optimization of cold-adapted alpha-galactosidase expression in Escherichia coli. Protein Expr Purif. 2016;123:14–18. doi: 10.1016/j.pep.2016.03.006. [DOI] [PubMed] [Google Scholar]
- Gombert AK, Kilikian BV. Recombinant gene expression in Escherichia coli cultivation using lactose as inducer. J Biotechnol. 1998;60:47–54. doi: 10.1016/S0168-1656(97)00185-5. [DOI] [PubMed] [Google Scholar]
- Gopalan N, Rodriguez-Duran LV, Saucedo-Castaneda G, Nampoothiri KM. Review on technological and scientific aspects of feruloyl esterases: a versatile enzyme for biorefining of biomass. Bioresour Technol. 2015;193:534–544. doi: 10.1016/j.biortech.2015.06.117. [DOI] [PubMed] [Google Scholar]
- Hassan S, Hugouvieux-Cotte-Pattat N. Identification of two feruloyl esterases in Dickeya dadantii 3937 and induction of the major feruloyl esterase and of pectate lyases by ferulic acid. J Bacteriol. 2011;193:963–970. doi: 10.1128/JB.01239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzakis NS, Smonou I. Asymmetric transesterification of secondary alcohols catalyzed by feruloyl esterase from Humicola insolens. Bioorg Chem. 2005;33:325–337. doi: 10.1016/j.bioorg.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Hopkins DJ, Betenbaugh MJ, Dhurjati P. Effects of dissolved oxygen shock on the stability of recombinant Escherichia coli containing plasmid pKN401. Biotechnol Bioeng. 1987;29:85–91. doi: 10.1002/bit.260290113. [DOI] [PubMed] [Google Scholar]
- Kikugawa M, Tsuchiyama M, Kai K, Sakamoto T. Synthesis of highly water-soluble feruloyl diglycerols by esterification of an Aspergillus niger feruloyl esterase. Appl Microbiol Biotechnol. 2012;95:615–622. doi: 10.1007/s00253-012-4056-6. [DOI] [PubMed] [Google Scholar]
- Kilikian BV, Suarez ID, Liria CW, Gombert AK. Process strategies to improve heterologous protein production in Escherichia coli under lactose or IPTG induction. Process Biochem. 2000;35:1019–1025. doi: 10.1016/S0032-9592(00)00137-0. [DOI] [Google Scholar]
- Koseki T, Fushinobu S, Shirakawa H, Komai M. Occurrence, properties, and applications of feruloyl esterases. Appl Microbiol Biotechnol. 2009;84:803–810. doi: 10.1007/s00253-009-2148-8. [DOI] [PubMed] [Google Scholar]
- Kumar PK, Maschke HE, Friehs K, Schugerl K. Strategies for improving plasmid stability in genetically modified bacteria in bioreactors. Trends Biotechnol. 1991;9:279–284. doi: 10.1016/0167-7799(91)90090-5. [DOI] [PubMed] [Google Scholar]
- Kumar CG, Kamle A, Mongolla P, Joseph J. Parametric optimization of feruloyl esterase production from Aspergillus terreus strain GA2 isolated from tropical agro-ecosystems cultivating sweet sorghum. J Microbiol Biotechnol. 2011;21:947–953. doi: 10.4014/jmb.1104.04014. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levasseur A, Benoit I, Asther M, Asther M, Record E. Homologous expression of the feruloyl esterase B gene from Aspergillus niger and characterization of the recombinant enzyme. Protein Expr Purif. 2004;37:126–133. doi: 10.1016/j.pep.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Li Y, Cui FJ, Liu ZQ, Xu YY, Zhao H. Improvement of xylanase production by Penicillium oxalicum ZH-30 using response surface methodology. Enzyme Microb Technol. 2007;40:1381–1388. doi: 10.1016/j.enzmictec.2006.10.015. [DOI] [Google Scholar]
- Li J, Qi JS, Wang T, Mao Q, Han C, Li YJ. Effects of IPTG addition time on fermentation of α-ketobutyric acid in Escherichia coli. Bull Ferment Sci Technol. 2017;46:77–82. doi: 10.16774/j.cnki.issn.1674-2214.2017.02.003. [DOI] [Google Scholar]
- Li JL, Shen WJ, Fan GS, Li XT. Screening, purification and characterization of lipase from Burkholderia pyrrocinia B1213. 3 Biotech. 2018 doi: 10.1007/s13205-018-1414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JL, Zhang JF, Yadav MP, Li XT. Biodegradability and biodegradation pathway of di-(2-ethylhexyl) phthalate by Burkholderia pyrrocinia B1213. Chemosphere. 2019;225:443–450. doi: 10.1016/j.chemosphere.2019.02.194. [DOI] [PubMed] [Google Scholar]
- Mackenzie CR, Bilous D, Schneider H, Johnson KG. Induction of cellulolytic and xylanolytic enzyme systems in Streptomyces spp. Appl Environ Microbiol. 1987;53:2835–2839. doi: 10.1002/bit.260300813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurizi MR. Proteases and protein degradation in Escherichia coli. Experientia. 1992;48:178–201. doi: 10.1007/BF01923511. [DOI] [PubMed] [Google Scholar]
- Meng Z, Yang QZ, Wang JZ, Hou YH. Cloning, characterization, and functional expression of a thermostable type B feruloyl esterase from thermophilic Thielavia terrestris. Appl Biochem Biotechnol. 2019;189:1304–1317. doi: 10.1007/s12010-019-03065-3. [DOI] [PubMed] [Google Scholar]
- Monteiro RA, Souza EM, Yates MG, Pedrosa FO, Chubatsu LS. Use of lactose to induce expression of soluble NifA protein domains of Herbaspirillum seropedicae in Escherichia coli. Can J Microbiol. 2000;46:1087–1090. doi: 10.1139/cjm-46-11-1087. [DOI] [PubMed] [Google Scholar]
- Oliveira DM, Mota TR, Oliva B, Segato F, Marchiosi R, Ferrarese-Filho O, Faulds CB, Dos Santos WD. Feruloyl esterases: biocatalysts to overcome biomass recalcitrance and for the production of bioactive compounds. Bioresour Technol. 2019;278:408–423. doi: 10.1016/j.biortech.2019.01.064. [DOI] [PubMed] [Google Scholar]
- Phue JN, Shiloach J. Impact of dissolved oxygen concentration on acetate accumulation and physiology of E. coli BL21, evaluating transcription levels of key genes at different dissolved oxygen conditions. Metab Eng. 2005;7:353–363. doi: 10.1016/j.ymben.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Phuengmaung P, Sunagawa Y, Makino Y, Kusumoto T, Handa S, Sukhumsirichart W, Sakamoto T. Identification and characterization of ferulic acid esterase from Penicillium chrysogenum 31B: de-esterification of ferulic acid decorated with l-arabinofuranoses and d-galactopyranoses in sugar beet pectin. Enzyme Microb Technol. 2019;131:109380. doi: 10.1016/j.enzmictec.2019.109380. [DOI] [PubMed] [Google Scholar]
- Qoronfleh MW. Dissolved oxygen concentration affects the accumulation of HIV-1 recombinant proteins in Escherichia coli. Appl Biochem Biotechnol. 1999;80:107–120. doi: 10.1385/ABAB:80:2:107. [DOI] [PubMed] [Google Scholar]
- Rashamuse KJ, Burton SG, Cowan DA. A novel recombinant ethyl ferulate esterase from Burkholderia multivorans. J Appl Microbiol. 2007;103:1610–1620. doi: 10.1111/j.1365-2672.2007.03394.x. [DOI] [PubMed] [Google Scholar]
- Ryan W, Parulekar SJ, Stark BC. Expression of beta-lactamase by recombinant Escherichia coli strains containing plasmids of different sizes–effects of pH, phosphate, and dissolved oxygen. Biotechnol Bioeng. 1989;34:309–319. doi: 10.1002/bit.260340306. [DOI] [PubMed] [Google Scholar]
- Sadeghian-Rizi T, Behdani M, Naghavi-al-hosseini F, Dakhilpour SS, Khanahmad H, Jahanian-Najafabadi A. Optimization of anti-CXCL10 nanobody expression using response surface methodology and evaluation of its anti-metastatic effect on breast cancer cells. Int J Pept Res Ther. 2019 doi: 10.1007/s10989-019-09941-0. [DOI] [Google Scholar]
- Shoja Alsadati SA, Varedi Kolaei SM, Babaeipour VA, Farnoud AM. Recent advances in high cell density cultivation for production of recombinant protein. Iran J Biotechnol. 2008;6:63–84. [Google Scholar]
- Stancik LM, Stancik DM, Schmidt B, Barnhart DM, Yoncheva YN, Slonczewski JL. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J Bacteriol. 2002;184:4246–4258. doi: 10.1128/JB.184.15.4246-4258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LQ, Wu SX, Feng JY, Wu J. High-efficiency expression of Sulfolobus acidocaldarius maltooligosyl trehalose trehalohydrolase in Escherichia coli through host strain and induction strategy optimization. Bioprocess Biosyst Eng. 2019;42:345–354. doi: 10.1007/s00449-018-2039-4. [DOI] [PubMed] [Google Scholar]
- Topakas E, Vafiadi C, Christakopoulos P. Microbial production, characterization and applications of feruloyl esterases. Process Biochem. 2007;42:497–509. doi: 10.1016/j.procbio.2007.01.007. [DOI] [Google Scholar]
- Wang MY, Yang JG, Zhao QS, Zhang KZ, Su C. Research progress on flavor compounds and microorganisms of maotai flavor baijiu. J Food Sci. 2019;84:6–18. doi: 10.1111/1750-3841.14409. [DOI] [PubMed] [Google Scholar]
- Wei YQ, Wu D, Wei D, Zhao Y, Wu JQ, Xie XY, Zhang RJ, Wei ZM. Improved lignocellulose-degrading performance during straw composting from diverse sources with actinomycetes inoculation by regulating the key enzyme activities. Bioresour Technol. 2019;271:66–74. doi: 10.1016/j.biortech.2018.09.081. [DOI] [PubMed] [Google Scholar]
- Wu SL, Nan F, Jiang JW, Qiu JR, Zhang YQ, Qiao BB, Li S, Xin ZH. Molecular cloning, expression and characterization of a novel feruloyl esterase from a soil metagenomic library with phthalate-degrading activity. Biotechnol Lett. 2019;41:995–1006. doi: 10.1007/s10529-019-02693-3. [DOI] [PubMed] [Google Scholar]
- Wu ZY, He F, Qin D, Li HH, Sun JY, Sun XT, Sun BG. Determination of phenolic compounds in alcoholic fermentation materials and spent grains by ultrasound-assisted alkali alcohol extraction coupled with HPLC. Anal Methods. 2019;11:5366–5375. doi: 10.1039/c9ay01739a. [DOI] [Google Scholar]
- Xu PX, Chai LJ, Qiu T, Zhang XJ, Lu ZM, Xiao C, Wang S, Shen CH, Shi JS, Xu ZH. Clostridium fermenticellae sp. Nov., isolated from the mud in a fermentation cellar for the production of the Chinese liquor, baijiu. Int J Syst Evol Microbiol. 2019;69:859–865. doi: 10.1099/ijsem.0.003254. [DOI] [PubMed] [Google Scholar]
- Xu ZS, Wang T, Zhang SS. Extracellular secretion of feruloyl esterase derived from Lactobacillus crispatus in Escherichia coli and its application for ferulic acid production. Bioresour Technol. 2019 doi: 10.1016/j.biortech.2019.121526. [DOI] [PubMed] [Google Scholar]
- Zare H, Mir Mohammad Sadeghi H, Akbari V. Optimization of fermentation conditions for reteplase expression by Escherichia coli using response surface methodology. Avicenna J Med Biotechnol. 2019;11:162–168. [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yin X, Wu MC, Yu T, Feng F, Zhu TD, Pang QF. Expression of a novel feruloyl esterase from Aspergillus oryzae in Pichia pastoris with esterification activity. J Mol Catal B Enzym. 2014;110:140–146. doi: 10.1016/j.molcatb.2014.10.002. [DOI] [Google Scholar]
- Zhou JY, Yu XJ, Ding C, Wang ZP, Zhou QQ, Pao H, Cai WM. Optimization of phenol degradation by Candida tropicalis Z-04 using Plackett–Burman design and response surface methodology. J Environ Sci China. 2011;23:22–30. doi: 10.1016/S1001-0742(10)60369-5. [DOI] [PubMed] [Google Scholar]