Abstract
A growing accumulation of plastic wastes has become a severe environmental and social issue. It is urgent to develop innovative approaches for the disposal of plastic wastes. In recent years, reports on biodegradation of synthetic plastics by microorganisms or enzymes have sprung up, and these offer a possibility to develop biological treatment technology for plastic wastes. In this review, we have comprehensively summarized the microorganisms and enzymes that are able to degrade a variety of generally used synthetic plastics, such as polyethylene (PE), polystyrene (PS), polypropylene (PP), polyvinyl chloride (PVC), polyurethane (PUR), and polyethylene terephthalate (PET). In addition, we have highlighted the microbial metabolic pathways for plastic depolymerization products and the current attempts toward utilization of such products as feedstocks for microbial production of chemicals with high value. Taken together, these findings will contribute to building a conception of bio-upcycling plastic wastes by connecting the biodegradation of plastic wastes to the biosynthesis of valuable chemicals in microorganisms. Last, but not least, we have discussed the challenges toward microbial degradation and valorization of plastic wastes.
Keywords: plastic wastes, biodegradation, valorization, depolymerase, protein engineering, synthetic biology
Introduction
Synthetic plastics, including polyethylene (PE), polystyrene (PS), polypropylene (PP), polyvinyl chloride (PVC), polyurethane (PUR), and polyethylene terephthalate (PET) (Table 1), have become fundamental to almost every aspect of our lives. According to the latest statistics of Plastics-Europe, the global yield of plastics reached 348 million tons in 2018 (Plastics Europe, 2018). China and the European Union account for 29.4 and 18.5%, ranking first and second in the world, of all the world’s plastic use, respectively (China Plastics Industry, 2017; Plastics Europe, 2018). Concomitant with the growing consumption of plastics, the generation of plastic wastes increases rapidly around the world. It is predicted that up to 26 billion tons of plastic wastes will be produced by 2050, and more than half will be thrown away into landfills and finally enter ecospheres, such as oceans and lakes, leading to serious environmental pollution (Jambeck et al., 2015; Lönnstedt and Eklöv, 2016; Geyer et al., 2017). As a result, plastic wastes have become a malevolent symbol of our wasteful society.
TABLE 1.
Types and properties of generally used synthetic plastics.
| Plastics | Abbreviation | Structure formula | Tm (°C)a | Tg (°C)b | XC (%)c | Recycling codes |
| High-density polyethylene | HDPE | 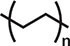 |
200–300 | −120 | 80–90 |  |
| Low-density polyethylene | LDPE | 160–260 | −120 | 45–65 |  |
|
| Polystyrene | PS | 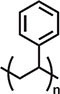 |
240 | 63–112 | – |  |
| Polypropylene | PP | 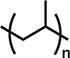 |
130 | −10–18 | 60–70 |  |
| Polyvinyl chloride | PVC | 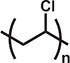 |
100–260 | 60–70 | – |  |
| Polyethylene terephthalate | PET | 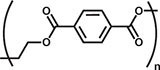 |
260 | 80 | 40–60 |  |
| Polyester polyurethane | Polyester PUR | 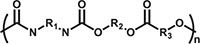 |
8–20 (soft) | −75 to −50 (soft) 185–205 (hard) | 40–50 |  |
| Polyether polyurethane | Polyether PUR | 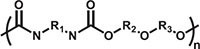 |
-95 (soft) 100 (hard) | −10 to 45 (soft) 190–240 (hard) |
aTm, melting temperature. bTg, glass transition temperature. cXC, crystallinity.
The current methods for disposing of plastic wastes mainly include landfilling, incineration, and mechanical and chemical recycling (Peng et al., 2018). In most countries, especially the developing countries, landfilling is the major method for plastic wastes disposal due to its operability and low cost. However, the accumulated plastic wastes have occupied a great amount of land. Incineration of plastic wastes can reduce the demand of landfills and recover heat energy, but we also need to reduce the environmental effects of secondary pollutants generated from the incinerating process, such as dioxins, carbon monoxide, nitrogen oxides, and so on. Although mechanical recycling has become the primary recycling method and is applied for reusing thermoplastic wastes, the properties of most recycled materials are significantly compromised after a number of processing cycles, and the resulting commercial values are thus limited. As an alternative, chemical recycling can recover the monomers and other chemicals from plastic wastes, but its success relies on the affordability of processes and the efficiency of catalysts (Rahimi and García, 2017). Nowadays, it is reported that only 9 and 12% of global plastic wastes is recycled and incinerated, while up to 79% is discarded into landfills or the natural environment, indicating that there is a great need for exploring innovative recycling methods to dispose of plastic wastes (Garcia and Robertson, 2017; Geyer et al., 2017).
In recent years, a number of studies have reported that several microorganisms and enzymes are capable of degrading synthetic plastics. Although numerous reviews and viewpoints on the topic of biodegradation of plastic have been published, they have mainly focused on the biodegradation of a single kind of plastic, such as PE (Restrepo-Flórez et al., 2014), PS (Ho et al., 2018), PP (Arutchelvi et al., 2008), PUR (Cregut et al., 2013; Peng et al., 2018; Magnin et al., 2019c), and PET (Wei and Zimmermann, 2017a; Kawai et al., 2019; Taniguchi et al., 2019). A comprehensive review into biodegradation of all main kinds of plastic is necessary (Wei and Zimmermann, 2017b). Moreover, a review focusing on not only the biodegradation but also the biological upcycling of plastic wastes is even more attractive (Wierckx et al., 2015; Salvador et al., 2019; Blank et al., 2020). In this review, we have summarized the microorganisms and enzymes that have been proven to be capable of degrading plastics, such as PE, PS, PP, PVC, PUR, and PET, as well as the microbial metabolic pathways of the plastic depolymerization products and the current attempts toward utilization of these products as feedstocks for microbial valorization. Based on the above understandings, we have attempted to develop a biologically upcycling conception for plastic wastes through building a metabolic link between biodegradation of plastic wastes and biosynthesis of valuable chemicals in microorganisms (Figure 1). Finally, we have discussed the existing knowledge gaps and challenges facing microbial degradation and valorization of plastic wastes.
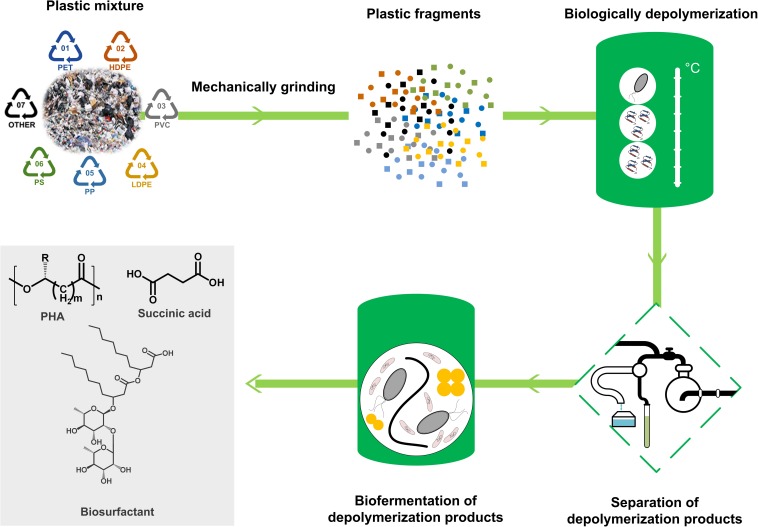
FIGURE 1.
The basic conception of bio-upcycling plastic wastes. A mixture of a variety of plastic wastes will be firstly mechanically grinded and biologically depolymerized by plastic-degrading microorganisms and enzymes. Then, the depolymerization products will be separated from the culture and utilized as feedstocks for microbial fermentation to produce chemicals with high value, such as polyhydroxyalkanoate (PHA), succinic acid, and biosurfactant.
Microbial Degradation of Synthetic Plastics
A number of microorganisms capable of degrading polyolefins (PE, PS, and PP), PVC, PUR, and PET have been isolated from the open environment, such as the soil of a plastic-dumping site, waste of mulch films, marine water, soil contaminated by crude oil, sewage sludge, landfills, and the guts of plastic-eating worms (Tables 2–7). The screening of plastic-degrading microorganisms is crucial for identifying the depolymerases and other key enzymes involved in plastic degradation.
TABLE 2.
Bacteria, fungi, and enzymes associated with polyethylene (PE) biodegradation.
| Strain/Enzyme | Isolated source | Tested PE | Incubation time, d | Weight loss, % | Molecular weight | Degradation products | References |
| Rhodococcus ruber C208 | Soil of disposal site | LDPE film | 30 | 4 | – | – | Orr et al., 2004 |
| Bacillus sphericus Alt; Bacillus cereus BF20 | Marine water | LDPE film | 180 | 2.5–10 | – | – | Sudhakar et al., 2008 |
| Arthrobacter sp. GMB5; Pseudomonas sp. GMB7 | Plastic waste dumpsites | HDPE film | 30 | 12–15 | – | – | Balasubramanian et al., 2010 |
| Pseudomonas sp. E4 | Soil | LMWPE | 80 | – | – | – | Yoon et al., 2012 |
| Pseudomonas sp. AKS2 | Waste dumping soil | LDPE film | 45 | 5 | – | – | Tribedi and Sil, 2013 |
| Bacillus subtilis H1584 | Marine water | LDPE film | 30 | 1.75 | – | – | Harshvardhan and Jha, 2013 |
| Enterobacter asburiae YT1; Bacillus sp. YP1 | Gut of waxworm | LDPE film | 60 | 6–11 | Decreased | Detected | Yang et al., 2014 |
| Serratia marcescens | Ground soil | LLDPE film | 70 | 36 | – | – | Azeko et al., 2015 |
| Achromobacter xylosoxidans | Soil | HDPE film | 150 | 9.38 | – | – | Kowalczyk et al., 2016 |
| Zalerion maritimum | Marine environment | PE pellets | 28 | – | – | – | Paço et al., 2017 |
| Phormidium lucidum; Oscillatoria subbrevis | Domestic sewage water | LDPE film | 42 | – | – | – | Sarmah and Rout, 2018 |
| Alcanivorax borkumensis | Mediterranean Sea | LDPE film | 7 | 3.5 | – | – | Delacuvellerie et al., 2019 |
| manganese peroxidase | Phanerochaete chrysosporium | PE film | 12 | – | Decreased | – | Iiyoshi et al., 1998 |
| soybean peroxidase | Soybean | HDPE film | 2 h | – | – | – | Zhao et al., 2004 |
| laccase | Rhodococcus ruber C208 | LDPE film | 30 | 2.5 | Decreased | – | Santo et al., 2013 |
| alkB gene | Pseudomonas sp. E4 | LMWPE sheet | 80 | 19.3 | – | – | Yoon et al., 2012 |
| alkB1, alkB2 gene | Pseudomonas aeruginosa E7 | LMWPE film | 50 | 19.6–27.6 | – | – | Jeon and Kim, 2016a |
TABLE 7.
Enzymes associated with polyethylene terephthalate (PET) biodegradation.
| Enzyme | Isolated source | Tested PET | Crystallinity, % | Reaction temperature, °C | Incubation time, d | Weight loss, % | References |
| TfH | Thermobifida fusca | PET bottle and pellets | 9 | 55 | 21 | 54.2 | Müller et al., 2005 |
| HiC; PmC; PsC | Humicola insolens; Pseudomonas mendocina; Fusarium solani | Low-crystallinity PET film | 7 | 70 | 6 | 97% | Ronkvist et al., 2009 |
| LC-cutinase | Compost metagenomic library | Low-crystallinity PET film | 8.4 | 50 | 7 | 50 | Sulaiman et al., 2012 |
| Cut190 | Saccharomonospora viridis | Low-crystallinity PET film | 8.4 | 63 | 3 | 27 | Kawai et al., 2014 |
| IsPETase | Ideonella sakaiensis | Low-crystallinity PET film | 1.9 | 30 | 0.75 | – | Yoshida et al., 2016 |
| IsPETase | Ideonella sakaiensis | Low-crystallinity PET film | – | 30 | 1 | 1 | Wei et al., 2019b |
| TfCut2 | Thermobifida fusca | Low-crystallinity PET chip | 7 | 70 | 5 | 97 | Wei et al., 2019a |
PE
As early as the 1970s, Albertsson carried out an experiment on microbial degradation of 14C-labeled PE (average weight molecular weight of 300,000 Da) by using three different soil microbiotas as inocula (Albertsson, 1978). In terms of the release of 14CO2, the microbial degradation rate of PE was calculated to be in the range of 0.36–0.39% after 2 years (Albertsson, 1978). When the 14C-labeled PE was extracted with cyclohexane to get rid of its low molecular weight components (average weight molecular weight of 1,000 Da), the microbial degradation rate dropped to 0.16% (Albertsson, 1980). Therefore, it was concluded that the release of 14CO2 was mainly derived from the microbial degradation of the low molecular weight PE fraction, which was similar to the microbial degradation of straight-chain n-alkanes (Albertsson, 1980). After that, Kawai et al. claimed that the upper limit of molecular weight for PE degradation by microorganisms was about 2,000 Da based on the results of a numerical simulation (Kawai et al., 1999, 2002, 2004; Watanabe et al., 2003, 2004).
Although the high molecular weight was considered as a key factor impeding the microbial degradation of PE, the physicochemical pretreatments, including UV irradiation (Albertsson et al., 1987; Albertsson and Karlsson, 1988, 1990), chemical oxidizing agents (Brown et al., 1974), and thermo-oxidation (Lee et al., 1991), could facilitate the microbial degradation of long-chain PE since these pretreatments led to the depolymerization of long-chain PE as well as the formation of low molecular weight products (Albertsson et al., 1995, 1998; Erlandsson et al., 1998; Hakkarainen and Albertsson, 2004). Consequently, it was assumed that the environmental degradation of long-chain PE could be achieved by the synergistic actions of photo- or thermo-oxidation and the biological activity of microorganisms (Hakkarainen and Albertsson, 2004).
Nevertheless, it was intriguing to figure out whether the long-chain PE (molecular weight > 2,000 Da) could be degraded by microorganisms from nature. A number of strains capable of degrading un-pretreated PE have been isolated from a variety of environments, including mulch films, marine water, soil contaminated by crude oil, sewage sludge, and landfills (Table 2; Orr et al., 2004; Sivan et al., 2006; Sudhakar et al., 2008; Balasubramanian et al., 2010; Yoon et al., 2012; Tribedi and Sil, 2013; Harshvardhan and Jha, 2013; Yang et al., 2014; Azeko et al., 2015; Kowalczyk et al., 2016; Paço et al., 2017; Sarmah and Rout, 2018; Delacuvellerie et al., 2019). Some of these strains showed the ability to utilize un-pretreated PE as a carbon source based on the characterizations of biofilm formation on PE films, weight loss of PE materials, surface deterioration, and changes in the mechanical and thermal properties of PE (Table 2). For example, it was reported that the weight loss of un-pretreated PE degraded by a strain Serratia marcescens reached 36% in an incubation period of 70 days (Azeko et al., 2015). Moreover, two cyanobacteria, Phormidium lucidum and Oscillatoria subbrevis, exhibited the capability of degrading 30% of the initial weight of tested PE over a 42-day period (Sarmah and Rout, 2018). However, these promising reports of PE degradation based on weight loss are less convincing since there is no additional evidence to support that the weight loss is caused by the degradation of the long-chain PE other than the low molecular weight components in PE.
Notably, a few studies reported that the waxworms, possessing an inherent ability to feed on and digest beeswax, could chew, and eat PE films (Yang et al., 2014; Bombelli et al., 2017; Chalup et al., 2018; Kundungal et al., 2019). The biodegradation of PE has been detected through contact with the homogenate of the waxworm Galleria mellonella (Bombelli et al., 2017) or after passage through the gut of the lesser waxworm Achroia grisella (Kundungal et al., 2019), according to the changes in chemical compositions characterized by the analyses of Fourier transform infrared spectroscopy (FTIR) and nuclear magnetic resonance (NMR). However, further investigations are necessitated in order to determine whether the depolymerization of PE has occurred in the waxworm gut.
As intestinal microbial symbionts have been recognized as indispensable for the digestion of insects (Engel and Moran, 2013), we have hypothesized that the microbial symbionts in the waxworm gut also play an important part in the degradation of PE (Yang et al., 2014). Two bacterial strains, Enterobacter asburiae YT1 and Bacillus sp. YP1, were isolated from the gut of waxworm Plodia interpunctella, and their PE-degrading capability was documented within a limited incubation period of 60 days based on the characterizations of biofilm formation, changes in the PE physical properties (tensile strength and surface topography), chemical structure (hydrophobicity and appearance of carbonyl groups), molecular weight (accompanied by the formation of daughter products), and weight loss (Table 2). These findings indicated that the bacteria from waxworms could be a promising source for the further screening PE-degrading microbes (Yang et al., 2014; Yang et al., 2015a).
Although a diverse range of PE-degrading microbes has been reported, only four microbial enzymes have been shown to be responsible for PE degradation (Table 2). Iiyoshi et al. (1998) found that manganese peroxidase (MnP), from lignin-degrading fungi Phanerochaete chrysosporium, could decrease the tensile strength and average molecular weight of PE film. Zhao et al. (2004) also found that the combination of soybean peroxidase (SBP) and hydrogen peroxide could oxidize the surface of PE film and diminish the surface hydrophobicity. Santo et al. (2013) showed that the extracellular laccase secreted by the PE-degrading bacterium, Rhodococcus ruber C208, could oxidize the PE films to generate carbonyl groups and decrease the molecular weight. While these past studies have identified the above peroxidase and laccase to be capable of catalyzing the degradation of PE, their catalytic mechanisms in the process of microbial degradation of PE remained unclear. In addition, three alkane hydroxylase genes, alkB, alkB1, and alkB2, were cloned in Escherichia coli and the resulting recombinant strains were found to be able to degrade low molecular-weight PE (Table 2; Yoon et al., 2012; Jeon and Kim, 2015, 2016a). These results indicated that the alkB, alkB1, or alkB2 played a key role in the degradation of low molecular-weight PE. Additionally, a recent study based on quantum mechanics calculations also suggested that the enzymatic cleavage of carbon–carbon bonds of polyolefins (PE and PS) by oxidases or oxygenases was possible (Xu et al., 2019). However, future efforts are required to characterize the biochemical functions of the oxidases or oxygenases, such as the enzymes encoded by the genes alkB, alkB1, or alkB2, within the biodegradation of PE.
PS
Guillet et al. (1974) first used two types of 14C-PS (α- and β-14C) as substrates to assess microbial degradation of PS in both soil and activated sewage sludge, and they showed that less than 0.01% could be degraded to 14CO2 in the course of 8 weeks. Afterward, 14C-labeled PS was also used as a substrate to determine the degradation of PS by soil microbiota, 17 lignin-degrading fungi, and five mixed floras (Sielicki et al., 1978; Kaplan et al., 1979). According to the release of 14CO2, the degradation rate was only 1.5∼3.0% during 16 weeks, up to 0.24% within 5 weeks, and 0.04∼0.57% within 11 weeks (Sielicki et al., 1978; Kaplan et al., 1979).
Besides the mixed flora, researchers have also tried to isolate PS-degrading microbes from different environment samples (Table 3). Eisaku et al. (2003) reported that three soil microorganisms, Xanthomonas sp., Sphingobacterium sp., and Bacillus sp. STR-YO, could degrade PS. Mor and Sivan (2008) found that an actinomycete, Rhodococcus ruber C208, was able to utilize PS as its sole carbon source to grow, and this led to a weight loss of 0.8% within 8 weeks. In addition, three fungi and three bacteria were isolated from the soil-buried expanded PS films, and they could adhere and grow on PS (Table 3; Atiq et al., 2010; Atiq, 2011). However, the reported biodegradation rates of PS by these strains was quite low, and there was no evidence of changes in either the physical or chemical properties of its long-chain PS molecules after microbial degradation.
TABLE 3.
Bacteria, fungi, and enzymes associated with polystyrene (PS) biodegradation.
| Strain/Enzyme | Isolated source | Tested PS | Incubation time, d | Weight loss, % | Molecular weight | Degradation products | References |
| Xanthomonas sp.; Sphingobacterium sp.; Bacillus sp. STR-YO | Field soil | PS film | 8 | 40–56 | – | – | Eisaku et al., 2003 |
| Rhodococcus ruber C208 | Soil of disposal site | PS film | 56 | 0.8 | – | – | Mor and Sivan, 2008 |
| Microbacterium sp. NA23; Paenibacillus urinalis NA26; Bacillus sp. NB6; Pseudomonas aeruginosa NB26 | Soil buried expanded PS film | PS film | 56 | – | – | Detected | Atiq et al., 2010 |
| Rhizopus oryzae NA1; Aspergillus terreus NA2; Phanerochaete chrysosporium NA3 | Soil buried expanded PS film | PS film | 56 | – | Increased | Detected | Atiq, 2011 |
| Exiguobacterium sp. YT2 | Mealworm’s gut | PS film | 60 | 7.5% | Decreased | Detected | Yang et al., 2015c |
| hydroquinone peroxidase | Azotobacter beijerinckii HM121 | PS film | 20 min | – | Decreased | Detected | Nakamiya et al., 1997 |
Extraordinarily, mealworms (larvae of Tenebrio molitor) were reported to be able to eat and rapidly degrade up to 50% of ingested Styrofoam (trade name of PS foam) during 24 h, and this was supported by the change in chemical composition, reduction in molecular weight, and the isotopic trace after passage through the intestinal tract (Yang et al., 2015b). With the same protocols, the PS-degrading capability was also documented in a broader range of mealworms from 12 different locations worldwide, indicating that PS degradation in mealworms is ubiquitous (Yang et al., 2018). This discovery also inspired researchers to explore more insect species, such as dark mealworms (Tenebrio obscurus) (Peng et al., 2019) and superworms (Zophobas atratus) (Yang et al., 2020), that also could eat and degrade PS.
We wondered whether the microbial symbionts associated with mealworms and superworms contributed to the degradation of PS. While the gut microbial symbionts were suppressed with antibiotics, the PS-degrading capacity of mealworms or superworms was impaired. This result indicated that gut microbial symbionts played an important role in the biodegradation of ingested Styrofoam (Yang et al., 2015c, Yang et al., 2020). Furthermore, one strain of Exiguobacterium sp. YT2, isolated from the gut of Tenebrio molitor, was proven to be capable of degrading 7.5% weight of PS in vitro within 60 days, while the decrease in molecular weight of the residual PS pieces and the release of water-soluble daughter products were also detected (Yang et al., 2015c; Table 3). At the time of writing, more bacteria have been isolated from the gut of plastic-eating mealworms or superworms, and their potential for PS degradation is still under assessment (Xia et al., 2019).
With respect to the PS-degrading enzymes, only hydroquinone peroxidase, secreted by a lignin-degrading bacterium Azotobacter beijerinckii HM121, was able to depolymerize PS into low molecular products in the presence of non-aqueous medium of dichloromethane (Nakamiya et al., 1997).
PP
In 1993, microbial degradation of PP was firstly assessed by cultures enriched from sandy soils containing PE wastes (Cacciari et al., 1993). After an incubation period of 175 days, the amount of degradation products, which were extracted with methylene chloride, accounted for 40% of the initial weight of tested PP. However, 90% of the extracted products were identified as aromatic esters, which were derived from the plasticizers, a chemical added especially into plastic to adjust the flexibility, workability, or stretchability. Meanwhile, only 10% of the extracted products were identified as hydrocarbons (C10H22 to C31H64) that may be derived from the degradation of PP itself. This result indicated that the plasticizers, other than the PP itself, were prone to be degraded by the sandy soil microorganisms (Cacciari et al., 1993).
From that time on, several microorganisms from different environmental samples have been tested for their potential to degrade PP (Table 4). For example, when PP films were incubated with soil microbiota from a plastic-dumping site, 0.4% weight loss and 33% increase in the crystallinity of residual PP were observed after 12 months, implying that the amorphous parts of PP could be degraded by soil microbiota (Arkatkar et al., 2009). Additionally, it was found that three bacteria and two fungal strains (Table 4), isolated from the soil of a plastic-dumping site, could utilize PP as their carbon source for growth and degrade 0.05–5% of PP after incubation for 12 months (Arkatkar et al., 2010; Jeyakumar et al., 2013). Mixed consortia of four bacterial isolates, from waste management landfills and sewage treatment plants, could also degrade the PP strips and pellets with a weight loss of 44.2–56.3% after 140 days (Skariyachan et al., 2018). Moreover, two marine bacteria of Bacillus sp. strain 27 and Rhodococcus sp. strain 36, isolated from mangrove environments, were also able to grow in aqueous synthetic media containing PP microplastics and caused a weight loss of 4.0–6.4% after 40 days (Auta et al., 2018). However, it is hard to determine whether the weight loss caused by the reported microbes above was attributed to the depolymerization of the long-chain PP or the degradation of the low molecular weight components, as the analyses of changes in molecular weight were absent.
TABLE 4.
Bacteria, fungi, and enzymes associated with polypropylene (PP) biodegradation.
| Strain/Enzyme | Isolated source | Tested PP | Incubation time, d | Weight loss, % | Molecular weight | Degradation products | References |
| Pseudomonas stutzeri; Bacillus subtilis; Bacillus flexus | Plastic-dumping site | PP film | 365 | – | – | Detected | Arkatkar et al., 2010 |
| Phanerochaete chrysosporium; Engyodontium album | Plastic-dumping site | PP film | 365 | 4–5 | – | Detected | Jeyakumar et al., 2013 |
| Stenotrophomonas panacihumi | Soil of waste storage yard | PP film | 90 | – | Increased | – | Jeon and Kim, 2016b |
| Aneurinibacillus aneurinilyticus; Brevibacillus agri; Brevibacillus sp.; Brevibacillus brevis | Landfills and sewage | PP film and pellets | 140 | 22.8–27.0 | – | Detected | Skariyachan et al., 2018 |
| Bacillus sp. strain 27; Rhodococcus sp. strain 36 | Mangrove environments | PP microplastic | 40 | 4–6.4 | – | – | Auta et al., 2018 |
A mesophilic strain, Stenotrophomonas panacihumi PA3-2, isolated from the soil of an open storage yard for municipal solid waste, was reported to be able to degrade two kinds of low molecular weight PP (Mn: 2,800, 3,600 Da) and one high molecular weight PP (Mn: 44,000 Da) with a biodegradability of 12.7–20.3% in terms of CO2 release and an increase in the molecular weight after 90 days (Jeon and Kim, 2016b). The results indicated that this strain could only degrade the low molecular weight fractions rather than the long-chain PP.
Until now, there are no enzymes reported to be capable of degrading PP, and little knowledge is available for the mechanism of microbial degradation of PP (Arutchelvi et al., 2008). However, similar to PE, it was found that the physicochemical pretreatments, including γ-irradiation (Alariqi et al., 2006), UV irradiation (Jeyakumar et al., 2013), thermo-oxidation (Jeyakumar et al., 2013), and blend with degradable additives, could facilitate the microbial degradation of PP (Jeyakumar et al., 2013; Jain et al., 2018).
PVC
Among all main kinds of synthetic plastics, PVC possesses the highest proportion of plasticizer (up to 50%). As plasticizers can be utilized by many fungi or bacteria as sources of nutrient carbons, plasticized PVC is usually susceptible to fungal or bacterial attack (Berk, 1950; Berk et al., 1957; Bessems, 1988; Kurane, 1988; Gumargalieva et al., 1999; Webb et al., 1999). For instance, a number of plasticized PVC bathroom items, such as bathtub lids, bath mats, and shower curtains, were found to be damaged by a variety of fungi (Table 5; Moriyama et al., 1993). Several fungal isolates (Table 5) from various environmental samples, such as atmosphere (Webb et al., 2000), plasticized PVC sheets buried in the grassland soil (Sabev et al., 2006; Ali et al., 2014), and plastic wastes disposal sites (Khatoon et al., 2019), also exhibited the ability to deteriorate the plasticized PVC. In addition, a number of bacterial strains (Table 5), isolated from garden soil, landfill leachate, waste disposal sites, and marine environments, have also been reported to be able to degrade the plasticized PVC (Nakamiya et al., 2005; Latorre et al., 2012; Anwar et al., 2016; Kumari et al., 2019; Giacomucci et al., 2019). However, these abovementioned plasticized PVC-degrading microorganisms just metabolized a component of the plasticizer [such as bis (2-ethylhexyl) phthalate, DEHP] rather than the backbone of PVC. Microorganisms capable of degrading both PVC and plasticizers have not been discovered so far. Thus, the key enzymes involved in the microbial degradation of PVC are still unknown.
TABLE 5.
Bacteria, fungi, and enzymes associated with polyvinyl chloride (PVC) biodegradation.
| Strain/Enzyme | Isolated source | Tested PVC | Incubation time, d | Weight loss,% | Molecular weight | Degradation products | References |
| Alternaria sp. TOF-46 | Japanese bathrooms | Plasticized PVC rim | 180 | – | – | – | Moriyama et al., 1993 |
| Poliporus versicolor; Pleurotus sajor caju | Lignocellulosic waste | PVC film | 30 | – | – | Detected | Klrbas et al., 1999 |
| Aureobasidium pullulans | Leaf/wood surfaces | Plasticized PVC | 7 | – | – | – | Webb et al., 1999 |
| Aspergillus niger | PVC wires | Plasticized PVC film | 365 | – | – | – | Gumargalieva et al., 1999 |
| Aureobasidium pullulans | Atmosphere | Plasticized PVC film | 42 | 3.7 | – | – | Webb et al., 2000 |
| Penicillium janthinellum | PVC buried in soil | Plasticized PVC sheet | 300 | – | – | – | Sabev et al., 2006 |
| Mycobacterium sp. NK0301 | Garden soil | Plasticized PVC film | 3 | – | – | Detected | Nakamiya et al., 2005 |
| Chryseomicrobium imtechense; Lysinibacillus fusiformis; Acinetobacter calcoaceticus; Stenotrophomonas pavanii | Landfill leachate | Plasticized PVC curtain | 34 | – | – | – | Latorre et al., 2012 |
| Phanerochaete chrysosporium; Lentinus tigrinus; Aspergillus niger; Aspergillus sydowii | PVC film buried in soil | PVC film | 300 | – | Decreased | Detected | Ali et al., 2014 |
| Acanthopleurobacter pedis; Bacillus cereus; Pseudomonas otitidis; Bacillus aerius; | Plastic disposal sites | PVC film | 90 | – | Decreased | Detected | Anwar et al., 2016 |
| Bacillus sp. AIIW2 | Marine | Un-plasticized PVC film | 90 | 0.26 | – | Detected | Kumari et al., 2019 |
| Phanerocheate chrysosporium | Plastic disposal site | PVC film | 28 | 31 | – | Detected | Khatoon et al., 2019 |
| Pseudomonas citronellolis | Soil | Plasticized PVC film | 45 | 13 | Decreased | – | Giacomucci et al., 2019 |
In future screening experiments, it is important to characterize the ability of strains to depolymerize the long-chain molecules of PVC by using virgin plastic in which low molecular weight components (monomers, oligomers, and plasticizers) were extracted by use of a suitable solvent or determining the decrease in the average molecular weight and the broadening of the molecular weight distribution of the residues after degradation.
PUR
PUR is the universal nomenclature for the plastic derived from the condensation of polyisocyanates and polyols with the linkages of intramolecular urethane bonds (Table 1). Depending on the chemical structures of the polyols used, PUR synthesized from polyester polyol is designated as polyester PUR, while that synthesized from polyether polyol is termed as polyether PUR.
In 1968, the initial research into microbial degradation of PUR was made by Darby and Kaplan. They found that seven fungi can grow on the surface of solid polyester PUR (Table 6; Darby and Kaplan, 1968). Since then, a number of fungi have been proven to be able to degrade polyester PUR (Table 6; Crabbe et al., 1994; Cosgrove et al., 2007; Russell et al., 2011; Mathur and Prasad, 2012; Álvarez-Barragán et al., 2016; Khan et al., 2017; Osman et al., 2017; Magnin et al., 2019a). In addition to fungi, many bacteria also have been demonstrated to be capable of degrading polyester PUR (Table 6; Kay et al., 1991, 1993; Nakajima-Kambe et al., 1995, 1997; Howard and Blake, 1998; Ii et al., 1998; Howard et al., 1999, 2001a,b; Rowe and Howard, 2002; Oceguera-Cervantes et al., 2007; Gautam et al., 2007; Nair and Kumar, 2007; Shah et al., 2008, 2013a,b, 2016; Howard and Burks, 2012; Peng et al., 2014; Nakkabi et al., 2015a, b; Pérez-Lara et al., 2016).
TABLE 6.
Bacteria, fungi, and enzymes associated with polyurethane (PUR) biodegradation.
| Strain/Enzyme | Isolated source | Tested PUR | Incubation time, d | Weight loss, % | Molecular weight | Degradation products | References |
| Chaetomium globosum | Soil | Polyester/polyether PUR film | 21 | – | – | – | Darby and Kaplan, 1968 |
| Curvularia senegalensis | Soil | Impranil DLN | 7 | – | – | – | Crabbe et al., 1994 |
| Geomyces pannorum | Acidic soil | Impranil DLN | 150 | – | – | – | Cosgrove et al., 2007 |
| Alternaria sp. PURDK2 | Environment | Polyether PUR film | 70 | 27.5 | – | Detected | Matsumiya et al., 2010 |
| Pestalotiopsis microspora | Plant stems | Impranil DLN, | 14 | – | – | – | Russell et al., 2011 |
| Aspergillus flavus | Plastic disposal sites | Polyester PUR film | 30 | 60.6 | – | – | Mathur and Prasad, 2012 |
| Cladosporium tenuissimum | Garden soil | Impranil DLN; polyether varnish | 14 | 65 | – | Detected | Álvarez-Barragán et al., 2016 |
| Aspergillus tubingensis | Waste disposal site | Polyester PUR beads | 20 | – | – | – | Khan et al., 2017 |
| Aspergillus sp. S45 | Waste-dumping site | Polyester PUR film | 28 | 15–20 | – | Detected | Osman et al., 2017 |
| Penicillium sp. | PUR wastes | Impranil DLN; polyester/polyether PUR film | 60 | 8.9 | Decreased | – | Magnin et al., 2019a |
| Corynebacterium sp., BI2; Pseudomonas aeruginosa | Soil | Polyester PUR foam | 84 | 1.2–17.7 | – | – | Kay et al., 1991 |
| Comamonas acidovorans | Soil | Polyester PUR film | 7 | – | – | Detected | Nakajima-Kambe et al., 1995 |
| Bacillus sp. | Soil | Impranil DLN | 4 | – | – | – | Ii et al., 1998 |
| Pseudomonas fluorescens | Soil | Impranil DLN | ND | – | – | – | Howard and Blake, 1998 |
| Pseudomonas chlororaphis | Soil | Impranil DLN | ND | – | – | – | Howard et al., 1999 |
| Bacillus subtilis | Soil | Impranil DLN | ND | – | – | – | Rowe and Howard, 2002 |
| Acinetobacter gerneri | Soil | Impranil DLN | ND | – | – | – | Howard and Burks, 2012 |
| Alicycliphilus sp. BQ1 | Decomposed soft foam | Polyester PUR film | 100 | – | – | Detected | Oceguera-Cervantes et al., 2007 |
| Bacillus pumilus | PUR-contaminated water | Impranil DLN | 3 | – | – | – | Nair and Kumar, 2007 |
| Pseudomonas chlororaphis | Soil | Ester PUR foam | 12 | – | – | – | Gautam et al., 2007 |
| Bacillus sp. AF8; Pseudomonas sp. AF9; Micrococcus sp. 10; Arthrobacter sp. AF11; Corynebacterium sp. AF12 | Soil | Polyester PUR film | 28 | – | – | – | Shah et al., 2008 |
| Bacillus subtilis; Pseudomonas aeruginosa | Soil | Polyester PUR pellets | 20 | – | – | Detected | Shah et al., 2016 |
| Pseudomonas putida | Soil | Impranil DLN | 8 | – | – | – | Peng et al., 2014 |
| Bacillus safensis | Cedar wood | Impranil DLN; | 7 | – | – | – | Nakkabi et al., 2015a, b |
| Aspergillus niger; Cladosporium herbarum | Natural humid conditions | Polyether PUR foam | 70 | – | – | – | Filip, 1979 |
| Staphylococcus epidermidis | An intravenous catheter | Polyether PUR film | 30 | – | – | – | Jansen et al., 1991 |
| Alternaria tenuissima | Infected leaves | Polyether PUR film | 60 | – | – | – | Oprea et al., 2018 |
| Pseudomonas denitrificans, Pseudomonas fluorescens, Bacillus subtilis, Yarrowia lipolytica | Soil | Polyether PUR film | 150 | 2.8–10.5 | – | – | Stepien et al., 2017 |
| esterase | Curvularia senegalensis | Impranil DLN | 21 | – | – | – | Crabbe et al., 1994 |
| pudA | Comamonas acidovorans | Polyester PUR film | 2 | – | – | – | Akutsu et al., 1998 |
| lipase | Bacillus subtilis | Impranil DLN | 1 | – | – | – | Rowe and Howard, 2002 |
| pulA | Pseudomonas fluorescens | Impranil DLN | ND | – | – | – | Ruiz and Howard, 1999 |
| pueA | Pseudomonas chlororaphis | Impranil DLN | 6 h | – | – | – | Stern and Howard, 2000 |
| pueB | Pseudomonas chlororaphis | Impranil DLN | 20 h | – | – | – | Howard et al., 2001b |
| LC cutinase; TfCut2; Tcur1278; Tcur0390 | Compost metagenomic library; Thermobifida fusca | Impranil DLN; polyester PUR cubes | 100 h | 0.3–3.2 | decreased | – | Schmidt et al., 2017 |
| Esterase E3576 | Protéus (France) | Polyester/polyether PUR film | 51 | 33 | – | Detected | Magnin et al., 2019b |
With regard to polyether PUR (Table 6), it was much less susceptible to microbial degradation in comparison to the polyester PUR (Darby and Kaplan, 1968). Notwithstanding, in 1979, Filip observed growth of Aspergillus niger and Cladosporium herbarum in shake cultures with polyether PUR resilient foam as the sole nutrient source (Filip, 1979). Afterward, Jansen et al. isolated a strain of Staphylococcus epidermidis KH11 from an infected catheter and demonstrated its capacity to utilize polyether PUR in the absence of any organic nutrients (Jansen et al., 1991). In 2010, a fungus, Alternaria sp. PURDK2, was reported to be able to degrade 27.5% of the weight of tested polyether PUR foam in the Luria-Bertani (LB) glucose agar after 70 days. Furthermore, this fungus was also capable of degrading two small molecule analogs of PUR, ethylphenylcarbamate (EPC) and diphenylmethane-4,4′-dibutylurea (D-MDI), into aniline and ethanol, indicating that the fungus could secret urethane-bond–degrading enzymes (Matsumiya et al., 2010). In 2016, eight fungal strains (Table 6) were showed be able to grow in mineral medium with a polyether PUR varnish as the sole carbon source and degrade 65% of solid polyether PUR foams in 50% potato dextrose broth (PDB) over 21 days (Álvarez-Barragán et al., 2016). Stepien et al. found that three bacteria and one yeast (Table 6) could degrade commercial polyether PUR films (Tecoflex®) and cause a weight loss of 2.8–10.5% within 5 months (Stepien et al., 2017). Oprea et al. assessed the biodegradability of pyridine-based polyether PUR elastomers by a fungus Alternaria tenuissima, and found that the fungus could decrease the mechanical properties and deteriorate the surface morphology after 60 days (Oprea et al., 2018).
The genes and enzymes contributing to microbial degradation of polyester PUR have been widely investigated (Table 6). In 1994, Crabbe et al. purified an esterase from a polyester PUR-degrading fungus, Curvularia senegalensis, and showed that this esterase can cleavage the ester bonds in the soft segments of polyester PUR (Crabbe et al., 1994). While Akutsu et al. purified a cell surface-bond polyester PUR-degrading esterase from the polyester PUR-degrading bacterium Comamonas acidovorans TB-35, Nomura et al. cloned a gene pudA encoding polyester PUR-degrading esterase in this strain (Akutsu et al., 1998; Nomura et al., 1998). Howard et al. purified a protease from Pseudomonas fluorescens (Vega et al., 1999), an esterase from Comamonas acidovorans (Allen et al., 1999), three esterases from Pseudomonas chlororaphis (Howard et al., 1999; Ruiz et al., 1999), and a lipase from Bacillus subtilis (Rowe and Howard, 2002). All the purified serine hydrolases above have the same hydrolytic capacity to emulsify polyester PUR. In addition, they also cloned a gene named pulA from Pseudomonas fluorescens (Ruiz and Howard, 1999) and two genes, pueA and pueB, from Pseudomonas chlororaphis (Stern and Howard, 2000; Howard et al., 2001b). These genes encoded three different esterases involved in the microbial degradation of emulsified polyester PUR by Pseudomonas fluorescens and Pseudomonas chlororaphis. In 2017, Schmidt et al. found that four polyester hydrolases, LC cutinase, TfCut2, Tcur1278, and Tcur0390, were able to degrade emulsified polyester PUR (Schmidt et al., 2017). Among these three cutinase, LC cutinase caused weight losses of up to 4.9 and 4.1% of two commercial polyester PUR elastomers of Elastollan B85A-10 and C85A-10, respectively, within a reaction time of 200 h at 70°C. Recently, an esterase (E3576), screened from 50 commercially available hydrolases, was shown to be able to hydrolyze a waterborne polyester PUR dispersion and degrade a solid polycaprolactone polyol-based polyester PUR with weight loss of 33% after 51 days (Magnin et al., 2019b). However, this esterase (E3576) cannot degrade poly(hexamethylene adipate) diol-based polyester PUR films, indicating that the chemical structures of the polyol segments significantly affect the biodegradability of polyester PUR (Kim and Kim, 1998; Magnin et al., 2019b).
Although the above reported lipases or esterases were able to rapidly degrade the emulsified polyester PUR (Impranil DLN) by cleaving the ester bonds in the polyester polyols segments, they exhibited a weak capability of degrading the solid polyester PUR substrates, such as PUR film, foam, and elastomer (Schmidt et al., 2017). The degradation products were not identified, and the biochemical mechanism was still unclear. Moreover, no specific depolymerases have been reported to be able to degrade the polyether PUR and cleave the urethane bones in both polyester and polyether PUR.
PET
The purpose of initial efforts to find out the hydrolases capable of hydrolyzing PET was to modify the surface wettability of PET fabrics (Hsieh and Cram, 1998; Yoon et al., 2002; Gübitz and Paulo, 2003; Alisch et al., 2004; Fischer-Colbrie et al., 2004; O’Neill and Cavaco-Paulo, 2004; Zhang et al., 2004; Silva et al., 2005; Vertommen et al., 2005). In the process of enzymatic surface modification, ester linkages on the surface of PET were hydrolyzed to produce polar hydroxyl and carboxylic groups, but the inner bulk of PET was not degraded. In a recent review focusing on enzymatic degradation of PET, Kawai et al. defined those hydrolases with moderate surface-hydrolyzing capability as PET surface-modifying enzymes (Kawai et al., 2019). By contrast, the hydrolases with significant capability of hydrolyzing the inner bulk of PET (causing at least 10% weight loss) were termed as PET hydrolases (Kawai et al., 2019). Hereinafter, only the reported PET hydrolases were reviewed (Table 7).
In 2005, Müller et al. (2005) reported that a cutinase-like hydrolase TfH, from an actinomycete Thermobifida fusca, can effectively degrade up to 50% of the initial weight of low-crystallinity PET (lcPET, 9%) at 55°C for 3 weeks. This is the first report on the enzymatic degradation of the inner bulk of PET films that opens the door for enzymatic PET recycling in the future (Müller, 2006). Thereafter, Ronkvist et al. compared the PET-hydrolyzing activities of three cutinases from different microorganisms, Humicola insolens (HiC, now named Thermomyces insolens), Pseudomonas mendocina (PmC), and Fusarium solani (FsC), using lcPET films (7%) and high-crystallinity biaxially oriented PET films (hcPET, 35%) as substrates (Ronkvist et al., 2009). Results showed that HiC caused a 97% weight loss of lcPET film (7%) at 70°C within 96 h, while PmC or FsC only led to a weight loss up to 5%. Thus, HiC can be designated as PET hydrolase, while PmC and FsC should be ascribed to PET surface-modifying enzymes. However, the three cutinases could hardly hydrolyze the hcPET films (35%). After that, Sulaiman et al. (2012) found that a LC-cutinase, encoded by one gene from the metagenomic library of leaf-branch compost, can efficiently hydrolyze low-crystallinity PET package film (lcPET-P, 8.4%) at 50°C and generate up to 50% weight loss over 7 days. In addition, Kawai et al. found that a cutinase Cut190, from Saccharomonospora viridis AHK190, can hydrolyze the lcPET (7%) and lcPET-P (8.4%) at 63°C, resulting in a weight loss of 13.5 and 27.0% for lcPET and lcPET-P, respectively, over 3 days (Kawai et al., 2014). It was recently shown that the recombinant Thermobifida fusca cutinase TfCut2 expressed by B. subtilis could degrade the lcPET films (7%) with a weight loss up to 97.0% and two low-crystallinity PET samples from postconsumer packages (AP-PET, 5%; CP-PET, 6%) with maximum weight losses of 50.5 and 56.6%, respectively, within 120 h at 70°C (Wei et al., 2019a).
Remarkably, Yoshida et al. found a bacterium, Ideonella sakaiensis 201-F6, capable of degrading lcPET films (1.9%) at an ambient temperature, and they identified a PET-hydrolyzing enzyme, termed as IsPETase, from this bacterium (Yoshida et al., 2016). The IsPETase was heat-labile (20∼45°C) and exhibited greater PET degradation activity than the above reported PET hydrolases at a mesophilic temperature of 30°C (Yoshida et al., 2016; Taniguchi et al., 2019). Nevertheless, the degradation rate of lcPET film (7%) by IsPETase at 30°C over a 24 h-incubation period was only 1% (weight loss), which was markedly lower than that caused by the above reported PET hydrolases at a thermophilic temperature (50∼70°C) (Wei et al., 2019a, b). Moreover, the hydrolytic activity of IsPETase against lcPET films (1.9%) was obviously higher than that for hcPET films (30∼40%) (Yang et al., 2016; Yoshida et al., 2016).
Overall, the above reported PET hydrolases are prone to degrade the lcPET (<10%) but not the hcPET (Vertommen et al., 2005; Ronkvist et al., 2009; Yang et al., 2016; Yoshida et al., 2016; Wei et al., 2019a). The effect of different crystallinity on the enzymatic degradation could be explained by the changes in the macromolecular aggregate structures of the polymer. Polymer molecules generally pack together in a non-uniform way with a mixture of ordered regions (crystalline-like) and disordered domains (amorphous). As the polymer chains in the amorphous domains are less densely packed than those in the crystalline domains, the lcPET, comprising a high proportion of amorphous domains, is more susceptible to enzymatic degradation. However, the high-crystallinity PET (30∼40%) represents the most abundant types of postconsumer plastic, and methods for lowering the crystallinity of PET to enhance the enzymatic degradation are highly sought.
Additionally, the enzymatic hydrolytic reactions of PET are inclined to take place under the temperature close to the glass transition temperature of PET (Tg, 65∼75°C). Under such a thermophilic temperature, the polymer chains in the amorphous PET domains can gain enough mobility to access the active sites of PET hydrolases (Ronkvist et al., 2009; Wei and Zimmermann, 2017a; Kawai et al., 2019). As a result, it suggests that efficient enzymatic degradation of PET requires thermostable PET depolymerases. Approaches of glycosylation (Shirke et al., 2018) and rational protein engineering, such as the optimization of surface salt bridge (Shirke et al., 2016), mutation of Ca2+ and Mg2+ binding sites (Then et al., 2015), introduction of disulfide bridge (Then et al., 2016), stabilization of a β6-β7 connecting loop, and extension of subsite IIc (Son et al., 2019), have been applied to improve the thermostability of these PET hydrolases. Notwithstanding, there is room for increasing the half-life of PET hydrolases above 65°C.
Microbial Valorization of Plastic Wastes
The initial step of the microbial degradation process is to secrete depolymerases to break down the long-chain polymers into low molecular weight oligomers or monomers, which can be further assimilated into microbial cells or metabolized into CO2. According to the principle of circular economy, these depolymerization products could be exploited for the biosynthesis of high-value chemicals through specific metabolic pathways, which could be considered as a way of valorizing plastic wastes (Wierckx et al., 2015).
From TPA, EG, and 6-Hydroxyhexanoate to Succinic Acids and Polyhydroxyalkanoate (PHA)
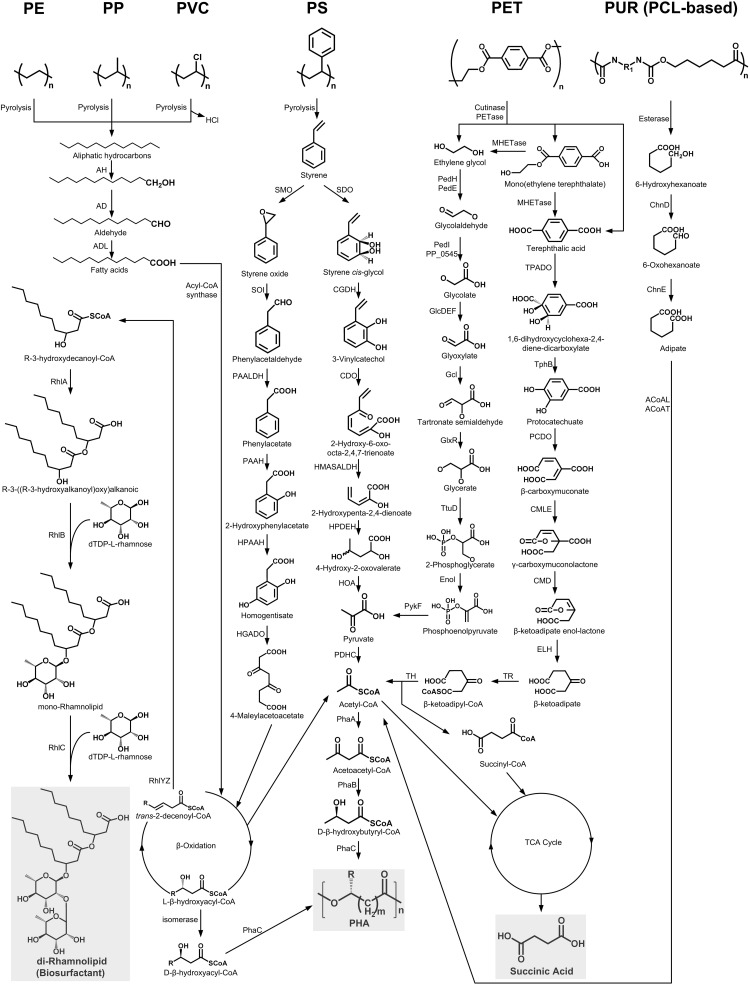
Enzymatic hydrolysis of PET could release constituent monomers ethylene glycol (EG), terephthalic acid (TPA), mono(ethylene terephthalate) (MHET), and bis(2-hydroxyethyl)TPA (BHET) by cleaving the ester bond (Ronkvist et al., 2009; Yoshida et al., 2016). Among these products, MHET could be further degraded into TPA and EG by the action of MHETase (Figure 2; Yoshida et al., 2016; Palm et al., 2019). In addition, the ester linkages in polycaprolactone polyol-based PUR (PCL-based PUR) could be hydrolyzed by an esterase (E3576) to generate 6-hydroxyhexanoate (Magnin et al., 2019b). These products could be further metabolized by specific microorganisms through different metabolic pathways (Figure 2; Brzostowicz et al., 2003; Yoshida et al., 2016).
FIGURE 2.
The metabolic pathways of depolymerization products of six kinds of plastics. Plastics: PE, polyethylene; PS, polystyrene; PP, polypropylene; PVC, polyvinyl chloride; PUR, polyurethane; PCL, polycaprolactone diol; PET polyethylene terephthalate. Enzymes: AH, alkane hydroxylase; AD, alcohol dehydrogenase; ALD, aldehyde dehydrogenase; RhlYZ, R-specific enoyl-CoA hydratase; RhlA, HAA synthetase; RhlB, rhamnosyltransferase 1; RhlC, rhamnosyltransferase 2; SMO, styrene monooxgenase; SOI styrene oxide isomerase; PAALDH, phenacetaldehyde dehydrogenase; PAAH, phenylacetate hydroxylase; HPAAH, 2-hydroxyphenylacetate hydroxylase; HGADO, homogentisate 1,2-dioxygenase; SDO, styrene dioxygenase; CGDH, cis-glycol dehydrogenase; CDO, catechol 2,3-dioxygenase; HMASALDH, 2-hydroxymuconic acid semialdehyde hydrolase; HPDEH, 2-hydroxypenta-2,4-dienoate hydratase; HOA, 4-hydroxy-2-oxovalerate aldolase; PDHC, pyruvate dehydrogenase complex; PhaA, β-ketothiolase; PhaB acetoacetyl-CoA reductase; PhaC, PHA synthase; PedH, quinoprotein alcohol dehydrogenase; PedE, quinoprotein alcohol dehydrogenase; PedI, aldehyde dehydrogenase family protein; PP_0545, aldehyde dehydrogenase family protein; GlcDEF, glycolate oxidase; Gcl glyoxylate carboligase; GlxR, tartronate semialdehyde reductase; TtuD, hydroxypyruvate reductase; PykF, pyruvate kinase; TPADO, TPA dioxygenase; TphB, 1,2-dihydroxy-3,5-cyclohexadiene-1,4-dicarboxylate dehydrogenase; PCDO, protocatechuate 3,4-dioxygenase; CMLE, β-carboxy-cis,cis-muconate lactonizing enzyme; CMD, β-carboxymuconolactone decarboxylase; ELH, enollactone hydrolase; TR, β-ketoadipate:succinyl-CoA transferase; TH, β-ketoadipyl-CoA thiolase; ChnD, 6-hydroxycaproate dehydrogenase; ChnE, 6-oxohexanoic dehydrogenase; ACoAL, adipate-CoA ligase; ACoAT, acetyl-CoA C-acyltransferase.
In bacterial species, a TPA transporter is implicated in the transport of TPA into the cell (Hosaka et al., 2013). Once inside the cell, the TPA can be transformed into 1,6-dihydroxycyclohexa-2,4-diene-dicarboxylate (DCD) by the activity of the TPA dioxygenase (TPADO). DCD is further oxidized by the 1,2-dihydroxy-3,5-cyclohexadiene-1,4-dicarboxylate dehydrogenase (TphB) to form protocatechuate (PCA) (Figure 2; Wang et al., 1995; Choi et al., 2005; Sasoh et al., 2006). The PCA could be degraded by the ortho-, meta-, and para-cleavage pathways under the catalysis of 3,4-dioxygenase (PCDO), 4,5-dioxygenase, and 2,3-dioxygenase, respectively (Harwood and Parales, 1996). Of these, the ortho-cleavage pathway has been thoroughly studied. The resulting metabolite, β-carboxymuconate (CM), will eventually be converted into acetyl-CoA and succinyl-CoA, which can enter the tricarboxylic acid (TCA) cycle to generate succinic acid (Figure 2).
In 2008, Kenny et al. first isolated three microorganisms, Pseudomonas putida GO16, Pseudomonas putida GO19, and Pseudomonas frederiksbergensis GO23, which could utilize TPA for not only growth but also accumulation of medium chain length PHA (mclPHA). Subsequently, they used the TPA fraction from PET pyrolysis as the feedstock for microbial production of mclPHA by these Pseudomonas species. The maximal production rate of PHA reached approximately 8.4 mg⋅l–1⋅h–1 (Kenny et al., 2008). When TPA and glycerol waste from biodiesel manufacture were co-supplied to Pseudomonas putida GO16 in a fed-batch bioreactor, the production rate of PHA reached approximately 108.8 mg⋅l–1⋅h–1 (Kenny et al., 2012).
EG could be metabolized by many kinds of microorganisms through two different pathways (Figure 2). In the pathway of acetogens, EG is degraded to ethanol and acetaldehyde, which is eventually transformed to acetate via acetyl-CoA (Trifunović et al., 2016). In contrast, through the pathway of Pseudomonas aeruginosa, EG is initially oxidized into glycolate by a series of dehydrogenases, and the generated glycolate will be further transformed into glyoxylate by the glycolate oxidase (GlcDEF). Glyoxylate could be converted into glycerate, which finally forms pyruvate (Figure 2; Child and Willetts, 1978; Kataoka et al., 2001).
P. putida KT2440 could accumulate mclPHA under nitrogen-limiting conditions but could not efficiently utilize EG as its sole carbon source. Through adaptive laboratory evolution, one mutant of P. putida KT2440 that could utilize EG as its sole carbon source was isolated. Comparative genomic analyses between the wild strain and the mutant revealed that a transcriptional regulator, GclR, played a central role in repressing the glyoxylate carboligase pathway (Li et al., 2019). With this knowledge, Franden et al. (2018) demonstrated that the overexpression of a combination of the glyoxylate carboligase (Gcl) operon with the glycolate oxidase (GlcDEF) operon endowed P. putida KT2440 with the ability to utilize EG as a sole carbon source for growth and accumulate in nitrogen-limiting M9 medium.
As for 6-hydroxyhexanoate, the hydrolysis product of PCL-based PUR, it is first converted to 6-oxohexanoic by the 6-hydroxyhexanoate dehydrogenase (ChnD). 6-oxohexanoic is eventually transformed into adipate by the action of the 6-oxohexanoic dehydrogenase (ChnE). After ligation with CoA, the adipate will be further converted to 3-oxoadipyl-CoA, which finally forms succinyl-CoA and acetyl-CoA that enter the TCA cycle with the production of succinic acid (Figure 2; Brzostowicz et al., 2003).
From Aromatic Hydrocarbons to Succinic Acids and PHA
Styrene, the aromatic monomer of PS, could be generated from the PS pyrolysis in the absence of air (Kaminsky and Kim, 1999) and is directly utilized as a carbon source by many microorganisms via two different catabolic pathways (Figure 2; O’Leary et al., 2002).
The first one is the direct aromatic ring cleavage pathway (Figure 2). In this pathway, the aromatic ring of styrene is firstly hydroxylated to styrene cis-glycol by styrene dioxygenase (SDO). Styrene cis-glycol is then further oxidized by a cis-glycol dehydrogenase (CGDH) to form 3-vinylcatechol. This product is degraded into pyruvate, which is further converted to acetyl-CoA by the pyruvate dehydrogenase complex (PDHC). Acetyl-CoA will finally enter the TCA cycle to generate succinic acid or be transformed into acetoacetyl-CoA, which could form β-D-Hydroxybutyryl-CoA or could be converted to PHA by an acetoacetyl-CoA reductase (PhaB) or a PHA synthase (PhaC), respectively (Anderson and Dawes, 1990). The other styrene metabolism pathway involves vinyl side-chain oxidation (Figure 2). Styrene is first converted into phenylacetic acid (PAA) by several enzymes, such as styrene monooxygenase (SMO), styrene oxide isomerase (SOI), and phenylacetaldehyde dehydrogenase (PAALDH). PAA is further hydroxylated and passed through the β-oxidation process to yield acetyl-CoA, which will then enter the TCA cycle or be converted into PHA (O’Leary et al., 2005; Oelschlägel et al., 2018).
In 2005, Ward et al. first found that Pseudomonas putida CA-3 could convert the metabolite of styrene, PAA, into polyhydroxyalkanoate (PHA) when a limiting concentration of nitrogen was added to the growth medium. Their finding built the metabolic link between styrene degradation and PHA accumulation in P. putida CA-3 and found a trail for the microbial valorization of PS waste into valuable chemicals (Ward et al., 2005; Nikodinovic-Runic et al., 2011).
Afterward, Ward et al. (2006) used the styrene oil, the pyrolysis products of PS waste at 520°C in a fluidized bed reactor, as the sole source of carbon and energy to support the growth and PHA accumulation of P. putida CA-3 in the shake flask experiments. In a run, the transformation rate from PS waste to PHA was 10%. In order to improve the conversion rate, Goff et al. performed a batch fermentation of P. putida CA-3 grown on styrene oil in a stirred tank reactor with an optimized nitrogen feeding strategy (Goff et al., 2007).
From Aliphatic Hydrocarbons to Fatty Acids, PHA, and Biosurfactants
Although PE, PP, and PVC, with similar carbon–carbon backbone chains, have been shown to be degraded by a number of microorganisms, the key depolymerases involved in the degradation process and the resulting depolymerization products remain unknown. However, pyrolysis in the absence of air could be an alternative method that can effectively depolymerize those plastic wastes into low molecular weight aliphatic hydrocarbons (Aguado et al., 2002).
It has been reported that the pyrolytic hydrocarbons of PE can be degraded via a terminal oxidation process similar to the microbial degradation pathway of n-alkane (Figure 2; Yoon et al., 2012; Jeon and Kim, 2015, 2016a). This process starts by the oxidation of a terminal methyl group by an alkane hydroxylase (AH) to generate a primary alcohol, which is further oxidized by an alcohol dehydrogenase (AD) to the corresponding aldehyde and finally converted into fatty acids by an aldehyde dehydrogenase (ADL) (Rojo, 2009). Fatty acids are then conjugated to CoA by an acyl-CoA synthase and further processed by β-oxidation to produce acetyl-CoA, L-β-hydroxyacyl-CoA, and trans-2-decenoyl-CoA (Figure 2). Acetyl-CoA can enter the TCA cycle to generate succinic acid or acetoacetyl-CoA, which can be finally converted to PHA under the appropriate condition (Anderson and Dawes, 1990). The L-β-hydroxyacyl-CoA is isomerized into D-β-hydroxyacyl-CoA, which can finally be converted into PHA through a PHA synthase (PhaC) (Sabirova et al., 2006). In addition, the trans-2-decenoyl-CoA is hydrated by the R-specific enoyl-CoA hydratase (RhlYZ) to form R-3-hydroxydecanoyl-CoA, which then acts as the direct lipid precursor used by the R-3-((R-3-hydroxyalkanoyl)oxy) alkanoic acids (HAA) synthase (RhlA) for the synthesis of HAAs. HAA, combined with dTDP-L-rhamnoses, can be converted into rhamnolipid biosurfactants by the rhamnosyltransferase 1 (RhlB) and the rhamnosyltransferase 2 (RhlC) (Abdel-Mawgoud et al., 2014).
Guzik et al. (2014) first used the pyrolytic hydrocarbons of PE as the starting material for microbial fermentation to produce PHA. Pseudomonas aeruginosa PAO-1, tested from 23 bacterial strains capable of degrading hydrocarbons or producing PHA, was reported to be able to accumulate PHA with almost 25% of cell dry weight when supplied with PE pyrolytic hydrocarbons and biosurfactants. Another bacterial strain, Ralstonia eutropha H16 (previously known as Cuprivadus necator or Wausternia eutropha), also exhibited PHA accumulation when supplied with non-oxygenated PE pyrolytic hydrocarbons as a carbon source in a nitrogen-rich tryptone soya broth (TSB) growth medium (Johnston et al., 2017). In contrast to PE pyrolysis in the absence of air, pyrolysis in the presence of air would not only cleave the long chains of PE but also introduce the carbonyl and hydroxyl groups into the backbone of pyrolytic hydrocarbons, which could improve the bioavailability of pyrolytic hydrocarbons as a carbon source for microbial fermentation to produce PHA by the strain Ralstonia eutropha H16 (Radecka et al., 2016).
In addition, PP could also be depolymerized into branched chain fatty alcohols and alkenes by pyrolysis. In 2019, Mihreteab et al. reported that strain Yarrowia lipolytica 78-003 was able to convert such depolymerization products to value-added fatty acids when mixed with biosurfactants and trace nutrients. During a period of 312 h, Y. lipolytica 78-003 assimilated more than 80% of the substrate and produced up to 492 mg L–1 lipids mainly composed of C16-C18 unsaturated fatty acids (Mihreteab et al., 2019). Johnston et al. (2019) found that R. eutropha H16 could utilize oxidized PP fragments as an additional carbon source to produce PHA in TSB medium.
As for PVC, although the pyrolysis at 300°C in the N2 flow has been showed to be able to depolymerize PVC into hydrocarbons along with the dechlorination in the form of HCl (Yuan et al., 2014), there are no reports about microbial strains that can utilize PVC pyrolysis products as carbon source so far. However, as these products are of similar chemical compositions to those of PE and PP, it is justifiable to believe that the strains of P. aeruginosa, R. eutropha H16, and Yarrowia lipolytica 78-003, which are able to assimilate the pyrolysis hydrocarbons of PE and PP, could also utilize PVC pyrolysis products to produce valuable chemicals.
Microbial growth on hydrocarbons is often associated with the production of biosurfactant, which can emulsify the hydrophobic hydrocarbons in aqueous media to increase the bioavailability of hydrocarbons to the cells. For instance, the strain Renibacterium salmoninarum 27BN was found to be able to produce rhamnolipids when grown on n-hexadecane (Christova et al., 2004). An oil-degrading bacterium Dietzia maris As-13-3, isolated from deep sea hydrothermal field, could also produce di-rhamnolipid as a biosurfactant, while tetradecane, n-hexadecane, and pristine were utilized as sole carbon sources (Wang et al., 2014). These results imply that pyrolysis hydrocarbons of PE, PP, and PVC could also be utilized as feedstocks to produce biosurfactants by these known hydrocarbon-degrading and biosurfactant-producing microorganisms.
Concluding Remarks and Future Prospects
As described above, a number of plastic-degrading microorganisms and enzymes have been sourced from the environment. However, an understanding of depolymerases contributing to the breakdown of plastics remains scarce. Therefore, future efforts should be devoted to identifying more depolymerases from the plastic-degrading microorganisms. In addition, enhancing the efficiency of enzymatic degradation is a big challenge. On the one hand, the macromolecular aggregate structures of plastics, such as the crystalline structures and cross-linking networks, impede the enzymatic degradation. The development of physical pretreatments, such as mechanical grinding and γ-irradiations, may help disorder these macromolecular aggregate structures and improve enzymatic degradation (Alariqi et al., 2006). On the other hand, approaches of rational protein engineering and direction evolution are necessary to improve the activity and stability of depolymerases, which will benefit the enhancement of enzymatic degradation efficiency.
While the long-chain polymer molecules could have been effectively depolymerized into small subunits (monomers or oligomers) by depolymerases, these small depolymerization products would be incorporated into cells as the feedstocks for metabolism (Table 8). Based on the advances in the understanding of the depolymerases and the microbial metabolic pathways of depolymerization products, it is fascinating to apply synthetic biology to build microbial cell factories that could depolymerize plastic wastes and utilize the small depolymerization products to produce chemicals with high value (Wierckx et al., 2015; Salvador et al., 2019; Blank et al., 2020). If this is manageable, it would not only contribute to the disposal of plastic wastes but also establish an improved cyclic utilization of plastics.
TABLE 8.
Strains for the valorization of depolymerization products of plastics.
| Plastics | Depolymerization methods | Depolymerization products | Strains | Metabolites | Yields | References |
| PET | Pyrolysis at 450°C | TPA | Pseudomonas putida GO16, Pseudomonas putida GO19, Pseudomonas frederiksbergensis GO23 | PHA | 8.4 mg⋅l–1⋅h–1 | Kenny et al., 2008 |
| PET | Pyrolysis | TPA | Pseudomonas putida GO16 | PHA | 108.8 mg⋅l–1⋅h–1 | Kenny et al., 2012 |
| PET | – | EG | Pseudomonas putida KT2440 | PHA | 0.06 g PHA per g EG | Franden et al., 2018 |
| PS | – | Styrene | Pseudomonas putida CA-3 | PHA | 0.11 g PHA per g carbon | Ward et al., 2005 |
| PS | – | Styrene | Pseudomonas putida CA-3 | PHA | 3.36 g⋅l–1 | Nikodinovic-Runic et al., 2011 |
| PS | Pyrolysis at 520°C | Styrene | Pseudomonas putida CA-3 | PHA | 62.5 mg PHA per g styrene | Ward et al., 2006 |
| PS | Pyrolysis | Styrene | Pseudomonas putida CA-3 | PHA | 0.28 g PHA per g styrene | Goff et al., 2007 |
| PE | Pyrolysis | Paraffins from C8 to C32 | Pseudomonas aeruginosa PAO-1 | PHA | 25% of the cell dry weight | Guzik et al., 2014 |
| PE | Pyrolysis | Hydrocarbons | Ralstonia eutropha H16 | PHA | 0.46 g⋅l–1 | Johnston et al., 2017 |
| PE | Pyrolysis in air | Oxidized hydrocarbons | Ralstonia eutropha H16 | PHA | 1.24 g⋅l–1 | Radecka et al., 2016 |
| PP | Pyrolysis at 540°C | Branched chain fatty alcohols and alkenes | Yarrowia lipolytica 78-003 | Fatty acids | 492 mg⋅l–1 over 312 h | Mihreteab et al., 2019 |
| PP | Thermal oxidation at 80–100°C in the oxygen–ozone mixture | Oxidized PP fragments | Ralstonia eutropha H16 | PHA | 1.36 g⋅l–1 | Johnston et al., 2019 |
| – | – | n-hexadecane | Renibacterium salmoninarum 27BN | Rhamnolipid | 0.92 g⋅l–1 | Christova et al., 2004 |
| – | – | n-hexadecane | Dietzia maris As-13-3 | Di-rhamnolipid | 120 mg⋅l–1 | Wang et al., 2014 |
Author Contributions
YY generated the idea and designed the project. YH contributed to the analysis and discussion. JR and YY prepared the figures and tables. YY and JR wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by grants from National Natural Science Foundation of China (Nos. 31961133015 and 51603004), the Young Elite Scientist Sponsorship Program of the China Association of Science and Technology (No. 2017QNRC001), and the Beijing Institute of Technology Research Fund Program for Young Scholars (No. 3160011181804).
References
- Abdel-Mawgoud A. M., Lépine F., Déziel E. (2014). A stereospecific pathway diverts β-oxidation intermediates to the biosynthesis of rhamnolipid biosurfactants. Chem. Biol. 21 156–164. 10.1016/j.chembiol.2013.11.010 [DOI] [PubMed] [Google Scholar]
- Aguado R., Olazar M., San José M. J., Gaisán B., Bilbao J. (2002). Wax formation in the pyrolysis of polyolefins in a conical spouted bed reactor. Energ. Fuel. 16 1429–1437. 10.1021/ef020043w [DOI] [Google Scholar]
- Akutsu Y., Nakajima-Kambe T., Nomura N., Nakahara T. (1998). Purification and properties of a polyester polyurethane-degrading enzyme from Comamonas acidovorans TB-35. Appl. Environ. Microbiol. 64 62–67. 10.1128/aem.64.1.62-67.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alariqi S. A., Kumar A. P., Rao B. S. M., Singh R. P. (2006). Biodegradation of γ-sterilised biomedical polyolefins under composting and fungal culture environments. Polym. Degrad. Stab. 91 1105–1116. 10.1016/j.polymdegradstab.2005.07.004 [DOI] [Google Scholar]
- Albertsson A. C. (1978). Biodegradation of synthetic polymers. II. A limited microbial conversion of 14C in polyethylene to 14CO2 by some soil fungi. J. Appl. Polym. Sci. 22 3419–3433. 10.1002/app.1978.070221207 [DOI] [Google Scholar]
- Albertsson A. C. (1980). Microbial and oxidative effects in degradation of polyethene. J. Appl. Polym. Sci. 25 1655–1671. 10.1002/app.1980.070250813 [DOI] [Google Scholar]
- Albertsson A. C., Andersson S. O., Karlsson S. (1987). The mechanism of biodegradation of polyethylene. Polym. Degrad. Stab. 18 73–87. 10.1016/0141-3910(87)90084-x [DOI] [Google Scholar]
- Albertsson A. C., Barenstedt C., Karlsson S., Lindberg T. (1995). Degradation product pattern and morphology changes as means to differentiate abiotically and biotically aged degradable polyethylene. Polymer 36 3075–3083. 10.1016/0032-3861(95)97868-g [DOI] [Google Scholar]
- Albertsson A. C., Erlandsson B., Hakkarainen M., Karlsson S. (1998). Molecular weight changes and polymeric matrix changes correlated with the formation of degradation products in Biodegraded Polyethylene. J. Environ. Polym. Degrad. 6 187–195. [Google Scholar]
- Albertsson A. C., Karlsson S. (1988). The three stages in degradation of polymers—polyethylene as a model substance. J. Appl. Polym. Sci. 35 1289–1302. 10.1002/app.1988.070350515 [DOI] [Google Scholar]
- Albertsson A. C., Karlsson S. (1990). The influence of biotic and abiotic environments on the degradation of polyethylene. Prog. Polym. Sci. 15 177–192. 10.1016/0079-6700(90)90027-x [DOI] [Google Scholar]
- Ali M. I., Ahmed S., Robson G., Javed I., Ali N., Atiq N., et al. (2014). Isolation and molecular characterization of polyvinyl chloride (PVC) plastic degrading fungal isolates. J. Basic Microbiol. 54 18–27. 10.1002/jobm.201200496 [DOI] [PubMed] [Google Scholar]
- Alisch M., Feuerhack A., Müller H., Mensak B., Andreaus J., Zimmermann W. (2004). Biocatalytic modification of polyethylene terephthalate fibres by esterases from actinomycete isolates. Biocatal. Biotrans. 22 347–351. 10.1080/10242420400025877 [DOI] [Google Scholar]
- Allen A. B., Hilliard N. P., Howard G. T. (1999). Purification and characterization of a soluble polyurethane degrading enzyme from Comamonas acidovorans. Int. Biodeter. Biodegr. 43 37–41. [Google Scholar]
- Álvarez-Barragán J., Domínguez-Malfavón L., Vargas-Suárez M., Gonzalez-Hernandez R., Aguilar-Osorio G., Loza-Tavera H. (2016). Biodegradative activities of selected environmental fungi on a polyester polyurethane varnish and polyether polyurethane foams. Appl. Environ. Microbiol. 82 5225–5235. 10.1128/AEM.01344-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A. J., Dawes E. A. (1990). Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Mol. Biol. Res. 54 450–472. 10.1128/mmbr.54.4.450-472.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar M. S., Kapri A., Chaudhry V., Mishra A., Ansari M. W., Souche Y., et al. (2016). Response of indigenously developed bacterial consortia in progressive degradation of polyvinyl chloride. Protoplasma 253 1023–1032. 10.1007/s00709-015-0855-9 [DOI] [PubMed] [Google Scholar]
- Arkatkar A., Arutchelvi J., Bhaduri S., Uppara P. V., Doble M. (2009). Degradation of unpretreated and thermally pretreated polypropylene by soil consortia. Int. Biodeter. Biodegr. 63 106–111. 10.1016/j.ibiod.2008.06.005 [DOI] [Google Scholar]
- Arkatkar A., Juwarkar A. A., Bhaduri S., Uppara P. V., Doble M. (2010). Growth of Pseudomonas and Bacillus biofilms on pretreated polypropylene surface. Int. Biodeter. Biodegr. 64 530–536. 10.1016/j.ibiod.2010.06.002 [DOI] [Google Scholar]
- Arutchelvi J., Sudhakar M., Arkatkar A., Doble M., Bhaduri S., Uppara P. V. (2008). Biodegradation of polyethylene and polypropylene. Indian J. Biotechnol. 7 9–22. [Google Scholar]
- Atiq N. (2011). Biodegradability of Synthetic Plastics Polystyrene and Styrofoam by Fungal Isolates. Islamabad: Quaid-i-Azam University Press. [Google Scholar]
- Atiq N., Garba A., Ali M. I., Andleeb S., Khan N. A., Robson G. D. (2010). Isolation and identification of polystyrene biodegrading bacteria from soil. Afr. J. Microbiol. Res. 4 1537–1541. [Google Scholar]
- Auta H. S., Emenike C. U., Jayanthi B., Fauziah S. H. (2018). Growth kinetics and biodeterioration of polypropylene microplastics by Bacillus sp. and Rhodococcus sp. isolated from mangrove sediment. Mar. Pollut. Bull. 127 15–21. 10.1016/j.marpolbul.2017.11.036 [DOI] [PubMed] [Google Scholar]
- Azeko S. T., Etuk-Udo G. A., Odusanya O. S., Malatesta K., Anuku N., Soboyejo W. O. (2015). Biodegradation of linear low density polyethylene by Serratia marcescens subsp. marcescens and its cell free extracts. Waste Biomass Valor. 6 1047–1057. 10.1007/s12649-015-9421-0 [DOI] [Google Scholar]
- Balasubramanian V., Natarajan K., Hemambika B., Ramesh N., Sumathi C. S., Kottaimuthu R., et al. (2010). High–density polyethylene (HDPE)-degrading potential bacteria from marine ecosystem of Gulf of Mannar, India. Lett. Appl. Microbiol. 51 205–211. 10.1111/j.1472-765X.2010.02883.x [DOI] [PubMed] [Google Scholar]
- Berk S. (1950). Effect of fungus growth on the tensile strength of polyvinyl chloride films plasticized with three plasticizers. ASTM Bull. 168 53–55. [Google Scholar]
- Berk S., Ebert H., Teitell L. (1957). Utilization of plasticizers and related organic compounds by fungi. Ind. Eng. Chem. 49 1115–1124. 10.3390/ijerph16132441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessems E. (1988). The biodeterioration of plasticized PVC and its prevention. J. Vinyl Technol. 10 3–6. 10.1002/vnl.730100103 [DOI] [Google Scholar]
- Blank L. M., Narancic T., Mampel J., Tiso T., O’Connor K. (2020). Biotechnological upcycling of plastic waste and other non-conventional feedstocks in a circular economy. Curr. Opin. Biotechnol. 62 212–219. 10.1016/j.copbio.2019.11.011 [DOI] [PubMed] [Google Scholar]
- Bombelli P., Howe C. J., Bertocchini F. (2017). Polyethylene bio-degradation by caterpillars of the wax moth Galleria mellonella. Curr. Biol. 27 R292–R293. 10.1016/j.cub.2017.02.060 [DOI] [PubMed] [Google Scholar]
- Brown B. S., Mills J., Hulse J. M. (1974). Chemical and biological degradation of waste plastics. Nature 250 161–163. 10.1038/250161a0 [DOI] [PubMed] [Google Scholar]
- Brzostowicz P. C., Walters D. M., Thomas S. M., Nagarajan V., Rouviere P. E. (2003). mRNA differential display in a microbial enrichment culture: simultaneous identification of three cyclohexanone monooxygenases from three species. Appl. Environ. Microbiol. 69 334–342. 10.1128/aem.69.1.334-342.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacciari I., Quatrini P., Zirletta G., Mincione E., Vinciguerra V., Lupattelli P., et al. (1993). Isotactic polypropylene biodegradation by a microbial community: physicochemical characterization of metabolites produced. Appl. Environ. Microbiol. 59 3695–3700. 10.1128/aem.59.11.3695-3700.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalup A., Ayup M. M., Monmany Garzia A. C., Malizia A., Martin E., De Cristóbal R., et al. (2018). First report of the lesser wax moth Achroia grisella F. (Lepidoptera: Pyralidae) consuming polyethylene (silo-bag) in northwestern Argentina. J. Apic. Res. 57 569–571. 10.1080/00218839.2018.1484614 [DOI] [Google Scholar]
- Child J., Willetts A. (1978). Microbial metabolism of aliphatic glycols bacterial metabolism of ethylene glycol. Biochim. Biophys. Acta 538 316–327. 10.1016/0304-4165(78)90359-8 [DOI] [PubMed] [Google Scholar]
- China Plastics Industry (2017). Data from: In 2017, the Total Output of China’s Plastic Products was 751.155 Million Tons, an Increase of 3.4% Year-on-Year (EB/OL). Available online at: http://www.iplast.cn/shownews.asp?id=6625 (accessed October 2, 2019). [Google Scholar]
- Choi K. Y., Kim D., Sul W. J., Chae J. C., Zylstra G. J., Kim Y. M., et al. (2005). Molecular and biochemical analysis of phthalate and terephthalate degradation by Rhodococcus sp. strain DK17. FEMS Microbiol. Lett. 252 207–213. 10.1016/j.femsle.2005.08.045 [DOI] [PubMed] [Google Scholar]
- Christova N., Tuleva B., Lalchev Z., Jordanova A., Jordanov B. (2004). Rhamnolipid biosurfactants produced by Renibacterium salmoninarum 27BN during growth on n-hexadecane. Z. Naturforsch. C 59 70–74. 10.1515/znc-2004-1-215 [DOI] [PubMed] [Google Scholar]
- Cosgrove L., McGeechan P. L., Robson G. D., Handley P. S. (2007). Fungal communities associated with degradation of polyester polyurethane in soil. Appl. Environ. Microbiol. 73 5817–5824. 10.1128/aem.01083-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe J. R., Campbell J. R., Thompson L., Walz S. L., Schultz W. W. (1994). Biodegradation of a colloidal ester-based polyurethane by soil fungi. Int. Biodeter. Biodegr. 33 103–113. 10.1016/0964-8305(94)90030-2 [DOI] [Google Scholar]
- Cregut M., Bedas M., Durand M. J., Thouand G. (2013). New insights into polyurethane biodegradation and realistic prospects for the development of a sustainable waste recycling process. Biotechnol. Adv. 31 1634–1647. 10.1016/j.biotechadv.2013.08.011 [DOI] [PubMed] [Google Scholar]
- Darby R. T., Kaplan A. M. (1968). Fungal susceptibility of polyurethanes. Appl. Microbiol. 16 900–905. 10.1128/aem.16.6.900-905.1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacuvellerie A., Cyriaque V., Gobert S., Benali S., Wattiez R. (2019). The plastisphere in marine ecosystem hosts potential specific microbial degraders including Alcanivorax borkumensis as a key player for the low-density polyethylene degradation. J. Hazard. Mater. 380:120899. 10.1016/j.jhazmat.2019.120899 [DOI] [PubMed] [Google Scholar]
- Eisaku O., Linn K., Takeshi E., Taneaki O., Yoshinobu I. (2003). Isolation and characterization of polystyrene degrading microorganisms for zero emission treatment of expanded polystyrene. Environ. Eng. Res. 40 373–379. [Google Scholar]
- Engel P., Moran N. A. (2013). The gut microbiota of insects–diversity in structure and function. FEMS Microbiol. Rev. 37 699–735. 10.1111/1574-6976.12025 [DOI] [PubMed] [Google Scholar]
- Erlandsson B., Karlsson S., Albertsson A. C. (1998). Correlation between molar mass changes and CO2 evolution from biodegraded 14C-labeled ethylene-vinyl alcohol copolymer and ethylene polymers. Acta Polym. 49 363–370. [DOI] [Google Scholar]
- Filip Z. (1979). Polyurethane as the sole nutrient source for Aspergillus niger, and Cladosporium herbarum. Appl. Microbiol. Biotechnol. 7 277–280. 10.1007/bf00498022 [DOI] [Google Scholar]
- Fischer-Colbrie G., Heumann S., Liebminger S., Almansa E., Cavaco-Paulo A., Guebitz G. M. (2004). New enzymes with potential for PET surface modification. Biocatal. Biotrans. 22 341–346. 10.1080/10242420400024565 [DOI] [Google Scholar]
- Franden M. A., Jayakody L. N., Li W. J., Wagner N. J., Cleveland N. S., Michener W. E., et al. (2018). Engineering Pseudomonas putida KT2440 for efficient ethylene glycol utilization. Metab. Eng. 48 197–207. 10.1016/j.ymben.2018.06.003 [DOI] [PubMed] [Google Scholar]
- Garcia J. M., Robertson M. L. (2017). The future of plastics recycling. Science 358 870–872. [DOI] [PubMed] [Google Scholar]
- Gautam R., Bassia S., Yanful E. K., Cullen E. (2007). Biodegradation of automotive waste polyester polyurethane foam using Pseudomonas chlororaphis ATCC55729. Int. Biodeterior. Biodegrad. 60 245–249. 10.1016/j.ibiod.2007.03.009 [DOI] [Google Scholar]
- Geyer R., Jambeck J., Law K. L. (2017). Production, use, and fate of all plastics ever made. Sci. Adv. 3:e1700782. 10.1126/sciadv.1700782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomucci L., Raddadi N., Soccio M., Lotti N., Fava F. (2019). Polyvinyl chloride biodegradation by Pseudomonas citronellolis and Bacillus flexus. New Biotechnol. 52 35–41. 10.1016/j.nbt.2019.04.005 [DOI] [PubMed] [Google Scholar]
- Goff M., Ward P. G., O’Connor K. E. (2007). Improvement of the conversion of polystyrene to polyhydroxyalkanoate through the manipulation of the microbial aspect of the process: a nitrogen feeding strategy for bacterial cells in a stirred tank reactor. J. Biotechnol. 132 283–286. 10.1016/j.jbiotec.2007.03.016 [DOI] [PubMed] [Google Scholar]
- Gübitz G. M., Paulo A. C. (2003). New substrates for reliable enzymes: enzymatic modification of polymers. Curr. Opin. Biotechnol. 14 577–582. 10.1016/j.copbio.2003.09.010 [DOI] [PubMed] [Google Scholar]
- Guillet J. E., Regulski T. W., McAneney T. B. (1974). Biodegradability of photodegraded polymers. II. Tracer studies of biooxidation of Ecolyte PS polystyrene. Environ. Sci. Technol. 8 923–925. 10.1016/j.chemosphere.2008.07.035 [DOI] [PubMed] [Google Scholar]
- Gumargalieva K. Z., Zaikov G. E., Semenov S. A., Zhdanova O. A. (1999). The influence of biodegradation on the loss of a plasticiser from poly (vinyl chloride). Polym. Degrad. Stab. 63 111–112. 10.1016/s0141-3910(98)00071-8 [DOI] [Google Scholar]
- Guzik M. W., Kenny S. T., Duane G. F., Casey E., Woods T., Babu R. P., et al. (2014). Conversion of post consumer polyethylene to the biodegradable polymer polyhydroxyalkanoate. Appl. Microbiol. Biotechnol. 98 4223–4232. 10.1007/s00253-013-5489-2 [DOI] [PubMed] [Google Scholar]
- Hakkarainen M., Albertsson A. C. (2004). “Environmental degradation of polyethylene,” in Long Term Properties of Polyolefins ed. Albertsson A. C. (Berlin: Springer; ), 177–200. 10.1007/b13523 [DOI] [Google Scholar]
- Harshvardhan K., Jha B. (2013). Biodegradation of low–density polyethylene by marine bacteria from pelagic waters, Arabian Sea, India. Mar. Pollut. Bull. 77 100–106. 10.1016/j.marpolbul.2013.10.025 [DOI] [PubMed] [Google Scholar]
- Harwood C. S., Parales R. E. (1996). The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50 553–590. 10.1146/annurev.micro.50.1.553 [DOI] [PubMed] [Google Scholar]
- Ho B. T., Roberts T. K., Lucas S. (2018). An overview on biodegradation of polystyrene and modified polystyrene: the microbial approach. Crit. Rev. Biotechnol. 38 308–320. 10.1080/07388551.2017.1355293 [DOI] [PubMed] [Google Scholar]
- Hosaka M., Kamimura N., Toribami S., Mori K., Kasai D., Fukuda M., et al. (2013). Novel tripartite aromatic acid transporter essential for terephthalate uptake in Comamonas sp. strain E6. Appl. Environ. Microbiol. 79 6148–6155. 10.1128/AEM.01600-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard G. T., Blake R. C. (1998). Growth of Pseudomonas fluorescens on a polyester–polyurethane and the purification and characterization of a polyurethanase–protease enzyme. Int. Biodeterior. Biodegrad. 42 213–220. 10.1016/s0964-8305(98)00051-1 [DOI] [Google Scholar]
- Howard G. T., Burks T. (2012). Growth of Acinetobacter gerneri P7 on polyurethane and the purification and characterization of a polyurethanase enzyme. Biodegradation 23 561–573. 10.1007/s10532-011-9533-6 [DOI] [PubMed] [Google Scholar]
- Howard G. T., Ruiz C., Hilliard N. P. (1999). Growth of Pseudomonas chlororaphis on a polyester–polyurethane and the purification and characterization of a polyurethanase–esterase enzyme. Int. Biodeterior. Biodegrad. 43 7–12. 10.1016/s0964-8305(98)00057-2 [DOI] [Google Scholar]
- Howard G. T., Vicknair J., Mackie R. I. (2001a). Sensitive plate assay for screening and detection of bacterial polyurethanase activity. Lett. Appl. Microbiol. 32 211–214. 10.1046/j.1472-765x.2001.00887.x [DOI] [PubMed] [Google Scholar]
- Howard G. T., Crother B., Vicknair J. (2001b). Cloning, nucleotide sequencing and characterization of a polyurethanase gene (pueB) from Pseudomonas chlororaphis. Int. Biodeterior. Biodegrad. 47 141–149. 10.1016/s0964-8305(01)00042-7 [DOI] [Google Scholar]
- Hsieh Y. L., Cram L. A. (1998). Enzymatic hydrolysis to improve wetting and absorbency of polyester fabrics. Text. Res. J. 68 311–319. 10.1177/004051759806800501 [DOI] [Google Scholar]
- Ii R. C. B., Norton W. N., Howard G. T. (1998). Adherence and growth of a Bacillus species on an insoluble polyester polyurethane. Int. Biodeterior. Biodegrad. 42 63–73. 10.1016/s0964-8305(98)00048-1 [DOI] [Google Scholar]
- Iiyoshi Y., Tsutumi Y., Nishida T. (1998). Polyethylene degradation by lignin-degrading fungi and manganese peroxidase. J. Wood Sci. 44 222–229. 10.1007/bf00521967 [DOI] [Google Scholar]
- Jain K., Bhunia H., Sudhakara Reddy M. (2018). Degradation of polypropylene–poly-L-lactide blend by bacteria isolated from compost. Bioremediat. J. 22 73–90. 10.1080/10889868.2018.1516620 [DOI] [Google Scholar]
- Jambeck J. R., Geyer R., Wilcox C., Siegler T. R., Perryman M., Andrady A., et al. (2015). Marine pollution. Plastic waste inputs from land into the ocean. Science 347 768–771. 10.1126/science.1260352 [DOI] [PubMed] [Google Scholar]
- Jansen B., Schumacher-Perdreau F., Peters G., Pulverer G. (1991). Evidence for degradation of synthetic polyurethanes by Staphylococcus epidermidis. Zentralbl. Bakteriol. 276 36–45. 10.1016/s0934-8840(11)80216-1 [DOI] [PubMed] [Google Scholar]
- Jeon H. J., Kim M. N. (2015). Functional analysis of alkane hydroxylase system derived from Pseudomonas aeruginosa E7 for low molecular weight polyethylene biodegradation. Int. Biodeter. Biodegr. 103 141–146. 10.1016/j.ibiod.2015.04.024 [DOI] [Google Scholar]
- Jeon H. J., Kim M. N. (2016a). Comparison of the functional characterization between alkane monooxygenases for low-molecular-weight polyethylene biodegradation. Int. Biodeter. Biodegr. 114 202–208. 10.1016/j.ibiod.2016.06.012 [DOI] [Google Scholar]
- Jeon H. J., Kim M. N. (2016b). Isolation of mesophilic bacterium for biodegradation of polypropylene. Int. Biodeter. Biodegr. 115 244–249. 10.1016/j.ibiod.2016.08.025 [DOI] [Google Scholar]
- Jeyakumar D., Chirsteen J., Doble M. (2013). Synergistic effects of pretreatment and blending on fungi mediated biodegradation of polypropylenes. Bioresour. Technol. 148 78–85. 10.1016/j.biortech.2013.08.074 [DOI] [PubMed] [Google Scholar]
- Johnston B., Jiang G., Hill D., Adamus G., Kwiecień I., Ziêba M., et al. (2017). The molecular level characterization of biodegradable polymers originated from polyethylene using non-oxygenated polyethylene wax as a carbon source for polyhydroxyalkanoate production. Bioengineering 4:73. 10.3390/bioengineering4030073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston B., Radecka I., Chiellini E., Barsi D., Ilieva V. I., Sikorska W., et al. (2019). Mass spectrometry reveals molecular structure of polyhydroxyalkanoates attained by bioconversion of oxidized polypropylene waste fragments. Polymers 11:1580. 10.3390/polym11101580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky W., Kim J. S. (1999). Pyrolysis of mixed plastics into aromatics. J. Anal. Appl. Pyrol. 51 127–134. 10.1016/s0165-2370(99)00012-1 [DOI] [Google Scholar]
- Kaplan D. L., Hartenstein R., Sutter J. (1979). Biodegradation of polystyrene, poly(methyl methacrylate), and phenol formaldehyde. Appl. Environ. Microbiol. 38 551–553. 10.1128/aem.38.3.551-553.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka M., Sasaki M., Hidalgo A.-R. G. D., Nakano M., Shimizu S. (2001). Glycolic acid production using ethylene glycol-oxidizing microorganisms. Biosci. Biotechnol. Biochem. 65 2265–2270. 10.1271/bbb.65.2265 [DOI] [PubMed] [Google Scholar]
- Kawai F., Kawabata T., Oda M. (2019). Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields. Appl. Microbiol. Biotechnol. 103 4253–4268. 10.1007/s00253-019-09717-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai F., Oda M., Tamashiro T., Waku T., Tanaka N., Yamamoto M., et al. (2014). A novel Ca2+-activated, thermostabilized polyesterase capable of hydrolyzing polyethylene terephthalate from Saccharomonospora viridis AHK190. Appl. Microbiol. Biotechnol. 98 10053–10064. 10.1007/s00253-014-5860-y [DOI] [PubMed] [Google Scholar]
- Kawai F., Shibata M., Yokoyama S., Maeda S., Tada K., Hayashi S. (1999). Biodegradability of scott-gelead photodegradable polyethylene and polyethylene wax by microorganisms. Macromol. Symp. 144 73–84. 10.1002/masy.19991440108 [DOI] [Google Scholar]
- Kawai F., Watanabe M., Shibata M., Yokoyama S., Sudate Y. (2002). Experimental analysis and numerical simulation for biodegradability of polyethylene. Polym. Degrad. Stab. 76 129–135. 10.1016/s0141-3910(02)00006-x [DOI] [Google Scholar]
- Kawai F., Watanabe M., Shibata M., Yokoyama S., Sudate Y., Hayashi S. (2004). Comparative study on biodegradability of polyethylene wax by bacteria and fungi. Polym. Degrad. Stab. 86 105–114. 10.1016/j.polymdegradstab.2004.03.015 [DOI] [Google Scholar]
- Kay M. J., Mccabe R. W., Morton L. H. G. (1993). Chemical and physical changes occurring in polyester polyurethane during biodegradation. Int. Biodeter. Biodegr. 31 209–225. 10.1016/0964-8305(93)90006-n [DOI] [Google Scholar]
- Kay M. J., Morton L. H. G., Prince E. L. (1991). Bacterial degradation of polyester polyurethane. Int. Biodeter. Biodegr. 27 205–222. 10.1016/0265-3036(91)90012-g [DOI] [Google Scholar]
- Kenny S. T., Runic J. N., Kaminsky W., Woods T., Babu R. P., Keely C. M., et al. (2008). Up-cycling of pet (polyethylene terephthalate) to the biodegradable plastic pha (polyhydroxyalkanoate). Environ. Sci. Technol. 42 7696–7701. 10.1021/es801010e [DOI] [PubMed] [Google Scholar]
- Kenny S. T., Runic J. N., Kaminsky W., Woods T., Babu R. P., O’Connor K. E. (2012). Development of a bioprocess to convert PET derived terephthalic acid and biodiesel derived glycerol to medium chain length polyhydroxyalkanoate. Appl. Microbiol. Biotechnol. 95 623–633. 10.1007/s00253-012-4058-4 [DOI] [PubMed] [Google Scholar]
- Khan S., Nadir S., Shah Z. U., Shah A. A., Karunarathna S. C., Xu J., et al. (2017). Biodegradation of polyester polyurethane by Aspergillus tubingensis. Environ. Pollut. 225 469–480. 10.1016/j.envpol.2017.03.012 [DOI] [PubMed] [Google Scholar]
- Khatoon N., Jamal A., Ali M. I. (2019). Lignin peroxidase isoenzyme: a novel approach to biodegrade the toxic synthetic polymer waste. Environ. Technol. 40 1366–1375. 10.1080/09593330.2017.1422550 [DOI] [PubMed] [Google Scholar]
- Kim Y. D., Kim S. C. (1998). Effect of chemical structure on the biodegradation of polyurethanes under composting conditions. Polym. Degrad. Stab. 62 343–352. 10.1016/s0141-3910(98)00017-2 12005513 [DOI] [Google Scholar]
- Klrbas N., Keskin N., Güner A. (1999). Biodegradation of polyvinylchloride (PVC) by white rot fungi. B. Environ. Contam. Tox. 63 335–342. 10.1007/s001289900985 [DOI] [PubMed] [Google Scholar]
- Kowalczyk A., Chyc M., Ryszka P., Latowski D. (2016). Achromobacter xylosoxidans as a new microorganism strain colonizing high-density polyethylene as a key step to its biodegradation. Environ. Sci. Pollut. Res. Int. 23 11349–11356. 10.1007/s11356-016-6563-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A., Chaudhary D. R., Jha B. (2019). Destabilization of polyethylene and polyvinylchloride structure by marine bacterial strain. Environ. Sci. Pollut. Res. Int. 26 1507–1516. 10.1007/s11356-018-3465-1 [DOI] [PubMed] [Google Scholar]
- Kundungal H., Gangarapu M., Sarangapani S., Patchaiyappan A., Devipriya S. P. (2019). Efficient biodegradation of polyethylene (HDPE) waste by the plastic-eating lesser waxworm (Achroia grisella). Environ. Sci. Pollut. Res. Int. 26 18509–18519. 10.1007/s11356-019-05038-9 [DOI] [PubMed] [Google Scholar]
- Kurane R. (1988). Biodegradation of phthalate ester in Japan. Biosci. Ind. 46 3173–3177. [Google Scholar]
- Latorre I., Hwang S., Montalvo-Rodriguez R. (2012). Isolation and molecular identification of landfill bacteria capable of growing on di-(2-ethylhexyl) phthalate and deteriorating PVC materials. J. Environ. Health Sci. Part A 47 2254–2262. 10.1080/10934529.2012.707549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B., Pometto A. L., Fratzke A., Bailey T. B. (1991). Biodegradation of degradable plastic polyethylene by Phanerochaete and Streptomyces species. Appl. Environ. Microbiol. 57 678–685. 10.1128/aem.57.3.678-685.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. J., Jayakody L. N., Franden M. A., Wehrmann M., Daun T., Hauer B., et al. (2019). Laboratory evolution reveals the metabolic and regulatory basis of ethylene glycol metabolism by Pseudomonas putida KT2440. Environ. Microbiol. 21 3669–3682. 10.1111/1462-2920.14703 [DOI] [PubMed] [Google Scholar]
- Lönnstedt O. M., Eklöv P. (2016). Environmentally relevant concentrations of microplastic particles influence larval fish ecology. Science 352 1213–1216. 10.1126/science.aad8828 [DOI] [PubMed] [Google Scholar]
- Magnin A., Hoornaert L., Pollet E., Laurichesse S., Phalip V., Avérous L. (2019a). Isolation and characterization of different promising fungi for biological waste management of polyurethanes. Microb. Biotechnol. 12 544–555. 10.1111/1751-7915.13346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnin A., Pollet E., Phalip V., Avérous L. (2019c). Evaluation of biological degradation of polyurethanes. Biotechnol. Adv. 39:107457. 10.1016/j.biotechadv.2019.107457 [DOI] [PubMed] [Google Scholar]
- Magnin A., Pollet E., Perrin R., Ullmann C., Persillon C., Phalip V., et al. (2019b). Enzymatic recycling of thermoplastic polyurethanes: synergistic effect of an esterase and an amidase and recovery of building blocks. Waste Manag. 85 141–150. 10.1016/j.wasman.2018.12.024 [DOI] [PubMed] [Google Scholar]
- Mathur G., Prasad R. (2012). Degradation of polyurethane by Aspergillus flavus (ITCC 6051) isolated from soil. Appl. Biochem. Biotechnol. 167 1595–1602. 10.1007/s12010-012-9572-4 [DOI] [PubMed] [Google Scholar]
- Matsumiya Y., Murata N., Tanabe E., Kubota K., Kubo M. (2010). Isolation and characterization of an ether-type polyurethane-degrading microorganism and analysis of degradation mechanism by Alternaria sp. J. Appl. Microbiol. 108 1946–1953. [DOI] [PubMed] [Google Scholar]
- Mihreteab M., Stubblefield B. A., Gilbert E. S. (2019). Microbial bioconversion of thermally depolymerized polypropylene by Yarrowia lipolytica for fatty acid production. Appl. Microbiol. Biotechnol. 103 7729–7740. 10.1007/s00253-019-09999-2 [DOI] [PubMed] [Google Scholar]
- Mor R., Sivan A. (2008). Biofilm formation and partial biodegradation of polystyrene by the actinomycete Rhodococcus ruber. Biodegradation 19 851–858. 10.1007/s10532-008-9188-0 [DOI] [PubMed] [Google Scholar]
- Moriyama Y., Kimura N., Inoue R., Kawaguchi A. (1993). Examination of fungal deterioration on plasticized polyvinyl chloride by cryo-scanning electron microscopy. Int. Biodeter. Biodegr. 31 231–239. 10.1016/0964-8305(93)90008-p [DOI] [Google Scholar]
- Müller R. J. (2006). Biological degradation of synthetic polyesters—Enzymes as potential catalysts for polyester recycling. Process Biochem. 41 2124–2128. 10.1016/j.procbio.2006.05.018 [DOI] [Google Scholar]
- Müller R. J., Schrader H., Profe J., Dresler K., Deckwer W. D. (2005). Enzymatic degradation of poly (ethylene terephthalate): rapid hydrolyse using a hydrolase from T. fusca. Macromol. Rapid Commun. 26 1400–1405. 10.1002/marc.200500410 [DOI] [Google Scholar]
- Nair S., Kumar P. (2007). Molecular characterization of a lipase-producing Bacillus pumilus strain (NMSN-1d) utilizing colloidal water-dispersible polyurethane. World J. Microbiol. Biotechnol. 23 1441–1449. 10.1007/s11274-007-9388-5 [DOI] [Google Scholar]
- Nakajima-Kambe T., Onuma F., Akutsu Y., Nakahara T. (1997). Determination of the polyester polyurethane breakdown products and distribution of the polyurethane degrading enzyme of Comamonas acidovorans, strain TB-35. J. Food Sci. Technol. 83 456–460. 10.1016/s0922-338x(97)83000-0 [DOI] [Google Scholar]
- Nakajima-Kambe T., Onuma F., Kimpara N., Nakahara T. (1995). Isolation and characterization of a bacterium which utilizes polyester polyurethane as a sole carbon and nitrogen source. FEMS Microbiol. Lett. 129 39–42. 10.1111/j.1574-6968.1995.tb07554.x [DOI] [PubMed] [Google Scholar]
- Nakamiya K., Hashimoto S., Ito H., Edmonds J. S., Yasuhara A., Morita M. (2005). Microbial treatment of bis (2-ethylhexyl) phthalate in polyvinyl chloride with isolated bacteria. J. Biosci. Bioeng. 99 115–119. 10.1263/jbb.99.115 [DOI] [PubMed] [Google Scholar]
- Nakamiya K., Ooi T., Kinoshita S. (1997). Non-heme hydroquinone peroxidase from Azotobacter beijerinckii HM121. J. Ferment. Bioeng. 84 14–21. 10.1016/s0922-338x(97)82780-8 [DOI] [Google Scholar]
- Nakkabi A., Sadiki M., Fahim M., Ittobane N., Saad I. K., Barkai H., et al. (2015a). Biodegradation of poly(ester urethane)s by Bacillus subtilis. Int. J. Environ. Res. 9 157–162. [Google Scholar]
- Nakkabi A., Sadiki M., Ibnssouda S., Fahim M. (2015b). Biological degradation of polyurethane by a newly isolated wood bacterium. Int. J. Recent Adv. Multidiscip. Res. 2 222–225. [Google Scholar]
- Nikodinovic-Runic J., Casey E., Duane G. F., Mitic D., Hume A. R., Kenny S. T., et al. (2011). Process analysis of the conversion of styrene to biomass and medium chain length polyhydroxyalkanoate in a two-phase bioreactor. Biotechnol. Bioeng. 108 2447–2455. 10.1002/bit.23187 [DOI] [PubMed] [Google Scholar]
- Nomura N., Shigeno-Akutsu Y., Nakajima-Kambe T., Nakahara T. (1998). Cloning and sequence analysis of a polyurethane esterase of Comamonas acidovorans TB-35. J. Ferment. Bioeng. 86 339–345. 10.1016/s0922-338x(99)89001-1 [DOI] [Google Scholar]
- Oceguera-Cervantes A., Carrillo-García A., López N., Bolaños-Nuñez S., Cruz-Gómez M. J., Wacher C., et al. (2007). Characterization of the polyurethanolytic activity of two Alicycliphilus sp. strains able to degrade polyurethane and n-methylpyrrolidone. Appl. Environ. Microbiol. 73 6214–6223. 10.1128/aem.01230-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelschlägel M., Zimmerling J., Tischler D. (2018). A review: The styrene metabolizing cascade of side-chain oxygenation as biotechnological basis to gain various valuable compounds. Front. Microbiol. 9:490. 10.3389/fmicb.2018.00490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary N. D., O’Connor K. E., Goff M., Dobson A. D. (2002). Biochemistry, genetics and physiology of microbial styrene degradation. FEMS Microbiol. Rev. 26 403–417. 10.1111/j.1574-6976.2002.tb00622.x [DOI] [PubMed] [Google Scholar]
- O’Leary N. D., O’Connor K. E., Ward P., Goff M., Dobson A. D. (2005). Genetic characterization of accumulation of polyhydroxyalkanoate from styrene in Pseudomonas putida CA-3. Appl. Environ. Microbiol. 71 4380–4387. 10.1128/aem.71.8.4380-4387.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill A., Cavaco-Paulo A. (2004). Monitoring biotransformations in polyesters. Biocatal. Biotrans. 22 353–356. 10.1080/10242420400025760 [DOI] [Google Scholar]
- Oprea S., Potolinca V. O., Gradinariu P., Oprea V. (2018). Biodegradation of pyridine-based polyether polyurethanes by the Alternaria tenuissima fungus. J. Appl. Polym. Sci. 135:46096 10.1002/app.46096 [DOI] [Google Scholar]
- Orr I. G., Hadar Y., Sivan A. (2004). Colonization, biofilm formation and biodegradation of polyethylene by a strain of Rhodococcus ruber. Appl. Microbiol. Biotechnol. 65 97–104. [DOI] [PubMed] [Google Scholar]
- Osman M., Satti S. M., Luqman A., Hasan F., Shah Z., Shah Z. Z. (2017). Degradation of polyester polyurethane by Aspergillus sp. strain S45 isolated from soil. J. Polym. Environ. 26 301–310. 10.1007/s10924-017-0954-0 [DOI] [Google Scholar]
- Paço A., Duarte K., da Costa J. P., Santos P. S., Pereira R., Pereira M. E., et al. (2017). Biodegradation of polyethylene microplastics by the marine fungus Zalerion maritimum. Sci. Total Environ. 586 10–15. 10.1016/j.scitotenv.2017.02.017 [DOI] [PubMed] [Google Scholar]
- Palm G. J., Reisky L., Böttcher D., Müller H., Michels E. A., Walczak M. C., et al. (2019). Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate. Nat. Commun. 10:1717. 10.1038/s41467-019-09326-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B. Y., Su Y., Chen Z., Chen J., Zhou X., Benbow M. E., et al. (2019). Biodegradation of Polystyrene by Dark (Tenebrio obscurus) and Yellow (Tenebrio molitor) Mealworms (Coleoptera: Tenebrionidae). Environ. Sci Technol. 53 5256–5265. 10.1021/acs.est.8b06963 [DOI] [PubMed] [Google Scholar]
- Peng R. T., Xia M. L., Ru J. K., Huo Y. X., Yang Y. (2018). Microbial degradation of polyurethane plastics. Chin. J. Biotechnol. 34 1398–1409. [DOI] [PubMed] [Google Scholar]
- Peng Y. H., Shih Y. H., Lai Y. C., Liu Y. Z., Liu Y. T., Lin N. C. (2014). Degradation of polyurethane by bacterium isolated from soil and assessment of polyurethanolytic activity of a Pseudomonas putida strain. Environ. Sci. Pollut. Res. Int. 21 9529–9537. 10.1007/s11356-014-2647-8 [DOI] [PubMed] [Google Scholar]
- Pérez-Lara L. F., Vargas-Suárez M., López-Castillo N. N., Cruz-Gómez M. J., Loza-Tavera H. (2016). Preliminary study on the biodegradation of adipate/phthalate polyester polyurethanes of commercial type by Alicycliphilus sp. BQ8. J. Appl. Polym. Sci. 133:42992. [Google Scholar]
- Plastics Europe (2018). Data From: Plastics - The Facts 2018: An Analysis of European Plastics Production, Demand and Waste Data (EB/OL). Available online at: http://www.plasticseurope.org (accessed October 2, 2019). [Google Scholar]
- Radecka I., Irorere V., Jiang G., Hill D., Williams C., Adamus G., et al. (2016). Oxidized polyethylene wax as a potential carbon source for PHA production. Materials. 9:367. 10.3390/ma9050367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi A., García J. M. (2017). Chemical recycling of waste plastics for new materials production. Nat. Rev. Chem. 1:0046. [Google Scholar]
- Restrepo-Flórez J. M., Bassi A., Thompson M. R. (2014). Microbial degradation and deterioration of polyethylene–A review. Int. Biodeter. Biodegr. 88 83–90. 10.1016/j.ibiod.2013.12.014 [DOI] [Google Scholar]
- Rojo F. (2009). Degradation of alkanes by bacteria. Environ. Microbiol. 11 2477–2490. 10.1111/j.1462-2920.2009.01948.x [DOI] [PubMed] [Google Scholar]
- Ronkvist Å. M., Xie W., Lu W., Gross R. A. (2009). Cutinase-catalyzed hydrolysis of poly(ethylene terephthalate). Macromolecules 42 5128–5138. 10.1021/ma9005318 [DOI] [Google Scholar]
- Rowe L., Howard G. T. (2002). Growth of Bacillus subtilis on polyurethane and the purification and characterization of a polyurethanase-lipase enzyme. Int. Biodeter. Biodegr. 50 33–40. 10.1016/s0964-8305(02)00047-1 [DOI] [Google Scholar]
- Ruiz C., Howard G. T. (1999). Nucleotide sequencing of a polyurethanase gene (pulA) from Pseudomonas fluorescens. Int. Biodeter. Biodegr. 44 127–131. 10.1016/s0964-8305(99)00074-8 [DOI] [Google Scholar]
- Ruiz C., Main T., Hilliard N. P., Howard G. T. (1999). Purification and characterization of two polyurethanase enzymes from Pseudomonas chlororaphis. Int. Biodeter. Biodegr. 43 43–47. 10.1016/s0964-8305(98)00067-5 [DOI] [Google Scholar]
- Russell J. R., Huang J., Anand P., Kucera K., Sandoval A. G., Dantzler K. W., et al. (2011). Biodegradation of polyester polyurethane by endophytic fungi. Appl. Environ. Microbiol. 77 6076–6084. 10.1128/AEM.00521-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabev H. A., Handley P. S., Robson G. D. (2006). Fungal colonization of soil-buried plasticized polyvinyl chloride (pPVC) and the impact of incorporated biocides. Microbiology 152 1731–1739. 10.1099/mic.0.28569-0 [DOI] [PubMed] [Google Scholar]
- Sabirova J. S., Ferrer M., Lünsdorf H., Wray V., Kalscheuer R., Steinbüchel A., et al. (2006). Mutation in a “tesB-like” hydroxyacyl-coenzyme A-specific thioesterase gene causes hyperproduction of extracellular polyhydroxyalkanoates by Alcanivorax borkumensis SK2. J. Bacteriol. 188 8452–8459. 10.1128/jb.01321-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador M., Abdulmutalib U., Gonzalez J., Kim J., Smith A. A., Faulon J. L., et al. (2019). Microbial genes for a circular and sustainable bio-PET economy. Genes 10:373. 10.3390/genes10050373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo M., Weitsman R., Sivan A. (2013). The role of the copper-binding enzyme–laccase–in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. Int. Biodeter. Biodegr. 84 204–210. 10.1016/j.ibiod.2012.03.001 [DOI] [Google Scholar]
- Sarmah P., Rout J. (2018). Efficient biodegradation of low-density polyethylene by cyanobacteria isolated from submerged polyethylene surface in domestic sewage water. Environ. Sci. Pollut. Res. Int. 25 33508–33520. 10.1007/s11356-018-3079-7 [DOI] [PubMed] [Google Scholar]
- Sasoh M., Masai E., Ishibashi S., Hara H., Kamimura N., Miyauchi K., et al. (2006). Characterization of the terephthalate degradation genes of Comamonas sp. strain E6. Appl. Environ. Microbiol. 72 1825–1832. 10.1128/aem.72.3.1825-1832.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J., Wei R., Oeser T., Dedavid E., Silva L. A., Breite D., et al. (2017). Degradation of polyester polyurethane by bacterial polyester hydrolases. Polymers 9:65. 10.3390/polym9020065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A. A., Hasan F., Akhter J. I., Hameed A., Ahmed S. (2008). Degradation of polyurethane by novel bacterial consortium isolated from soil. Ann. Microbiol. 58 381–386. 10.1007/bf03175532 [DOI] [Google Scholar]
- Shah Z., Gulzar M., Hasan F., Shah A. A. (2016). Degradation of polyester polyurethane by an indigenously developed consortium of Pseudomonas and Bacillus species isolated from soil. Polym. Degrad. Stab. 134 349–356. 10.1016/j.polymdegradstab.2016.11.003 [DOI] [Google Scholar]
- Shah Z., Krumholz L., Aktas D. F., Hasan F., Khattak M., Shah A. A. (2013a). Degradation of polyester polyurethane by a newly isolated soil bacterium, Bacillus subtilis strain MZA-75. Biodegradation 24 865–877. 10.1007/s10532-013-9634-5 [DOI] [PubMed] [Google Scholar]
- Shah Z., Hasan F., Krumholz L., Aktas D. F., Shah A. A. (2013b). Degradation of polyester polyurethane by newly isolated Pseudomonas aeruginosa, strain MZA-85 and analysis of degradation products by GC–MS. Int. Biodeter. Biodegr. 77 114–122. 10.1016/j.ibiod.2012.11.009 [DOI] [PubMed] [Google Scholar]
- Shirke A. N., Basore D., Butterfoss G. L., Bonneau R., Bystroff C., Gross R. A. (2016). Toward rational thermostabilization of Aspergillus oryzae cutinase: insights into catalytic and structural stability. Proteins 84 60–72. 10.1002/prot.24955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirke A. N., White C., Englaender J. A., Zwarycz A., Butterfoss G. L., Linhardt R. J., et al. (2018). Stabilizing leaf and branch compost cutinase (LCC) with glycosylation: mechanism and effect on PET hydrolysis. Biochemistry 57 1190–1200. 10.1021/acs.biochem.7b01189 [DOI] [PubMed] [Google Scholar]
- Sielicki M., Focht D. D., Martin J. P. (1978). Microbial degradation of (14C) polystyrene and 1,3-diphenylbutane. Can. J. Microbiol. 24 798–803. 10.1139/m78-134 [DOI] [PubMed] [Google Scholar]
- Silva C. M., Carneiro F., O’Neill A., Fonseca L. P., Cabral J. S., Guebitz G., et al. (2005). Cutinase—a new tool for biomodification of synthetic fibers. J. Polym. Sci. Pol. Chem. 43 2448–2450. 10.1002/pola.20684 [DOI] [Google Scholar]
- Sivan A., Szanto M., Pavlov V. (2006). Biofilm development of the polyethylene-degrading bacterium Rhodococcus ruber. Appl. Microbiol. Biotechnol. 72 346–352. 10.1007/s00253-005-0259-4 [DOI] [PubMed] [Google Scholar]
- Skariyachan S., Patil A. A., Shankar A., Manjunath M., Bachappanavar N., Kiran S. (2018). Enhanced polymer degradation of polyethylene and polypropylene by novel thermophilic consortia of Brevibacillus sps. and Aneurinibacillus sp. screened from waste management landfills and sewage treatment plants. Polym. Degrad. Stab. 149 52–68. 10.1016/j.polymdegradstab.2018.01.018 [DOI] [Google Scholar]
- Son H. F., Cho I. J., Joo S., Seo H., Sagong H. Y., Choi S. Y., et al. (2019). Rational protein engineering of thermo-stable PETase from Ideonella sakaiensis for highly efficient PET degradation. ACS Catal. 9 3519–3526. 10.1021/acscatal.9b00568 [DOI] [Google Scholar]
- Stepien A. E., Zebrowski J., Piszczyk Ł., Boyko V. V., Riabov S. V., Dmitrieva T., et al. (2017). Assessment of the impact of bacteria Pseudomonas denitrificans, Pseudomonas fluorescens, Bacillus subtilis and yeast Yarrowia lipolytica on commercial poly (ether urethanes). Polym. Test. 63 484–493. 10.1016/j.polymertesting.2017.08.038 [DOI] [Google Scholar]
- Stern R. V., Howard G. T. (2000). The polyester polyurethanase gene (pueA) from Pseudomonas chlororaphis encodes a lipase. FEMS Microbiol. Lett. 185 163–168. 10.1111/j.1574-6968.2000.tb09056.x [DOI] [PubMed] [Google Scholar]
- Sudhakar M., Doble M., Murthy P. S., Venkatesan R. (2008). Marine microbe-mediated biodegradation of low- and high-density polyethylenes. Int. Biodeter. Biodegr. 61 203–213. 10.1016/j.ibiod.2007.07.011 [DOI] [Google Scholar]
- Sulaiman S., Yamato S., Kanaya E., Kim J. J., Koga Y., Takano K., et al. (2012). Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl. Environ. Microbiol. 78 1556–1562. 10.1128/AEM.06725-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi I., Yoshida S., Hiraga K., Miyamoto K., Kimura Y., Oda K. (2019). Biodegradation of PET: current status and application aspects. ACS Catal. 9 4089–4105. 10.1021/acscatal.8b05171 [DOI] [Google Scholar]
- Then J., Wei R., Oeser T., Barth M., Belisário-Ferrari M. R., Schmidt J., et al. (2015). Ca2+ and Mg2+ binding site engineering increases the degradation of polyethylene terephthalate films by polyester hydrolases from Thermobifida fusca. Biotechnol. J. 10 592–598. 10.1002/biot.201400620 [DOI] [PubMed] [Google Scholar]
- Then J., Wei R., Oeser T., Gerdts A., Schmidt J., Barth M., et al. (2016). A disulfide bridge in the calcium binding site of a polyester hydrolase increases its thermal stability and activity against polyethylene terephthalate. FEBS Open Bio 6 425–432. 10.1002/2211-5463.12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribedi P., Sil A. K. (2013). Low-density polyethylene degradation by Pseudomonas sp, AKS2 biofilm. Environ. Sci. Pollut. Res. Int. 20 4146–4153. 10.1007/s11356-012-1378-y [DOI] [PubMed] [Google Scholar]
- Trifunović D., Schuchmann K., Müller V. (2016). Ethylene glycol metabolism in the acetogen Acetobacterium woodii. J. Bacteriol. 198 1058–1065. 10.1128/JB.00942-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega R. E., Main T., Howard G. T. (1999). Cloning and expression in Escherichia coli of a polyurethane-degrading enzyme from Pseudomonas fluorescens. Int. Biodeter. Biodegr. 43 49–55. 10.1016/s0964-8305(98)00068-7 [DOI] [Google Scholar]
- Vertommen M. A., Nierstrasz V. A., Van Der Veer M., Warmoeskerken M. M. (2005). Enzymatic surface modification of poly(ethylene terephthalate). J. Biotechnol. 120 376–386. 10.1016/j.jbiotec.2005.06.015 [DOI] [PubMed] [Google Scholar]
- Wang W., Cai B., Shao Z. (2014). Oil degradation and biosurfactant production by the deep sea bacterium Dietzia maris As-13-3. Front. Microbiol. 5:711. 10.3389/fmicb.2014.00711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Z., Zhou Y., Zylstra G. J. (1995). Molecular analysis of isophthalate and terephthalate degradation by Comamonas testosteroni YZW-D. Environ. Health Perspect. 103 9–12. 10.1289/ehp.95103s49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P. G., de Roo G., O’Connor K. E. (2005). Accumulation of polyhydroxyalkanoate from styrene and phenylacetic acid by Pseudomonas putida CA-3. Appl. Environ. Microbiol. 71 2046–2052. 10.1128/aem.71.4.2046-2052.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P. G., Goff M., Donner M., Kaminsky W., O’Connor K. E. (2006). A twostep chemo-biotechnological conversion of polystyrene to a biodegradable thermoplastic. Environ. Sci. Technol. 40 2433–2437. 10.1021/es0517668 [DOI] [PubMed] [Google Scholar]
- Watanabe M., Kawai F., Shibata M., Yokoyama S., Sudate Y. (2003). Computational method for analysis of polyethylene biodegradation. J. Comput. Appl. Math. 161 133–144. 10.1016/s0377-0427(03)00551-x [DOI] [Google Scholar]
- Watanabe M., Kawai F., Shibata M., Yokoyama S., Sudate Y., Hayashi S. (2004). Analytical and computational techniques for exogenous depolymerization of xenobiotic polymers. Math. Biosci. 192 19–37. 10.1016/j.mbs.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Webb J. S., Nixon M., Eastwood I. M., Greenhalgh M., Robson G. D., Handley P. S. (2000). Fungal colonization and biodeterioration of plasticized polyvinyl chloride. Appl. Environ. Microbiol. 66 3194–3200. 10.1128/aem.66.8.3194-3200.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb J. S., Van der Mei H. C., Nixon M., Eastwood I. M., Greenhalgh M., Read S. J., et al. (1999). Plasticizers increase adhesion of the deteriogenic fungus Aureobasidium pullulans to polyvinyl chloride. Appl. Environ. Microbiol. 65 3575–3581. 10.1128/aem.65.8.3575-3581.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R., Breite D., Song C., Gräsing D., Ploss T., Hille P., et al. (2019a). Biocatalytic degradation efficiency of postconsumer polyethylene terephthalate packaging determined by their polymer microstructures. Adv. Sci. 6:1900491. 10.1002/advs.201900491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R., Song C., Gräsing D., Schneider T., Bielytskyi P., Böttcher D., et al. (2019b). Conformational fitting of a flexible oligomeric substrate does not explain the enzymatic PET degradation. Nat. Commun. 10:5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R., Zimmermann W. (2017a). Biocatalysis as a green route for recycling the recalcitrant plastic polyethylene terephthalate. Microb. Biotechnol. 10 1302–1307. 10.1111/1751-7915.12714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R., Zimmermann W. (2017b). Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: how far are we? Microb. Biotechnol. 10 1308–1322. 10.1111/1751-7915.12710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierckx N., Prieto M. A., Pomposiello P., de Lorenzo V., O’Connor K., Blank L. M. (2015). Plastic waste as a novel substrate for industrial biotechnology. Microb. Biotechnol. 8 900–903. 10.1111/1751-7915.12312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M., Wang J., Huo Y. X., Yang Y. (2019). Mixta tenebrionis sp. nov., isolated from the gut of the plastic-eating mealworm Tenebrio molitor L. Int. J. Syst. Evol. Microbiol. 70 790–796. 10.1099/ijsem.0.003826 [DOI] [PubMed] [Google Scholar]
- Xu J., Cui Z., Nie K., Cao H., Jiang M., Xu H., et al. (2019). A quantum mechanism study of the cc bond cleavage to predict the bio-catalytic polyethylene degradation. Front. Microbiol. 10:489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Yang Y., Wu W. M., Zhao J., Jiang L. (2014). Evidence of polyethylene biodegradation by bacterial strains from the guts of plastic-eating waxworms. Environ. Sci. Technol. 48 13776–13784. 10.1021/es504038a [DOI] [PubMed] [Google Scholar]
- Yang S. S., Wu W. M., Brandon A. M., Fan H. Q., Receveur J. P., Li Y., et al. (2018). Ubiquity of polystyrene digestion and biodegradation within yellow mealworms, larvae of Tenebrio molitor Linnaeus (Coleoptera: Tenebrionidae). Chemosphere 212 262–271. 10.1016/j.chemosphere.2018.08.078 [DOI] [PubMed] [Google Scholar]
- Yang Y., Chen J., Wu W. M., Zhao J., Yang J. (2015a). Complete genome sequence of Bacillus sp. YP1, a polyethylene-degrading bacterium from waxworm’s gut. J. Biotechnol. 200 77–78. 10.1016/j.jbiotec.2015.02.034 [DOI] [PubMed] [Google Scholar]
- Yang Y., Yang J., Wu W. M., Zhao J., Song Y., Gao L., et al. (2015b). Biodegradation and mineralization of polystyrene by plastic-eating mealworms. 1. chemical and physical characterization and isotopic tests. Environ. Sci. Technol. 49 12080–12086. 10.1021/acs.est.5b02661 [DOI] [PubMed] [Google Scholar]
- Yang Y., Yang J., Wu W. M., Zhao J., Song Y., Gao L., et al. (2015c). Biodegradation and mineralization of polystyrene by plastic-eating mealworms. 2. role of gut microorganisms. Environ. Sci. Technol. 49 12087–12093. 10.1021/acs.est.5b02663 [DOI] [PubMed] [Google Scholar]
- Yang Y., Wang J., Xia M. (2020). Biodegradation and mineralization of polystyrene by plastic-eating superworms Zophobas atratus. Sci. Total Environ. 708:135233. 10.1016/j.scitotenv.2019.135233 [DOI] [PubMed] [Google Scholar]
- Yang Y., Yang J., Jiang L. (2016). Comment on “A bacterium that degrades and assimilates poly (ethylene terephthalate)”. Science 353:759. 10.1126/science.aaf8305 [DOI] [PubMed] [Google Scholar]
- Yoon M. G., Jeon H. J., Kim M. N. (2012). Biodegradation of polyethylene by a soil bacterium and AlkB cloned recombinant cell. J. Bioremediat. Biodegrad. 3:145. [Google Scholar]
- Yoon M.-Y., Kellis J., Poulose A. J. (2002). Enzymatic modification of polyester. AATCC Rev. 2 33–36. [Google Scholar]
- Yoshida S., Hiraga K., Takehana T., Taniguchi I., Yamaji H., Maeda Y., et al. (2016). A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351 1196–1199. 10.1126/science.aad6359 [DOI] [PubMed] [Google Scholar]
- Yuan G., Chen D., Yin L., Wang Z., Zhao L., Wang J. Y. (2014). High efficiency chlorine removal from polyvinyl chloride (PVC) pyrolysis with a gas-liquid fluidized bed reactor. Waste Manag. 34 1045–1050. 10.1016/j.wasman.2013.08.021 [DOI] [PubMed] [Google Scholar]
- Zhang J., Wang X., Gong J., Gu Z. (2004). A study on the biodegradability of polyethylene terephthalate fiber and diethylene glycol terephthalate. J. Appl. Polym. Sci. 93 1089–1096. 10.1002/app.20556 [DOI] [Google Scholar]
- Zhao J., Guo Z., Ma X., Liang G., Wang J. (2004). Novel surface modification of high-density polyethylene films by using enzymatic catalysis. J. Appl. Polym. Sci. 91 3673–3678. 10.1002/app.13619 [DOI] [Google Scholar]