Abstract
Neuroblastoma is an aggressive pediatric malignancy of the neural crest with suboptimal cure rates and a striking predilection for widespread metastases, underscoring the need to identify novel therapeutic vulnerabilities. We recently identified the RNA binding protein LIN28B as a driver in high-risk neuroblastoma and demonstrated it promotes oncogenic cell proliferation by coordinating a RAN-Aurora kinase A network. Here, we demonstrate that LIN28B influences another key hallmark of cancer, metastatic dissemination. Using a murine xenograft model of neuroblastoma dissemination, we show that LIN28B promotes metastasis. We demonstrate that this is in part due to the effects of LIN28B on self-renewal and migration, providing an understanding of how LIN28B shapes the metastatic phenotype. Our studies reveal that the let-7 family, which LIN28B inhibits, decreases self-renewal and migration. Next, we identify PDZ Binding Kinase (PBK) as a novel LIN28B target. PBK is a serine/threonine kinase that promotes the proliferation and self-renewal of neural stem cells and serves as an oncogenic driver in multiple aggressive malignancies. We demonstrate that PBK is both a novel direct target of let-7i and that MYCN regulates PBK expression, thus elucidating two oncogenic drivers that converge on PBK. Functionally, PBK promotes self-renewal and migration, phenocopying LIN28B. Taken together, our findings define a role for LIN28B in neuroblastoma metastasis and define the targetable kinase PBK as a potential novel vulnerability in metastatic neuroblastoma.
Abbreviations: PBK, PDZ Binding Kinase; GALNT14, N-acetyl-galactosaminyltransferase; AURKA, Aurora kinase A; TARGET, Therapeutically Applicable Research to Generate Effective Treatments project
Keywords: Neuroblastoma, LIN28B, Let-7, PDZ binding kinase, Metastasis
Introduction
Neuroblastoma is an aggressive pediatric tumor of the developing peripheral sympathetic nervous system that remains a substantial challenge in pediatric oncology [1]. At diagnosis, patients with high-risk disease present with striking metastatic burden, underscoring the clinical aggression of this disease [1]. However, our understanding of the molecular mechanisms that drive neuroblastoma metastasis remain incompletely understood, as, consequently, do therapies for metastatic disease.
We [2] and others [3], [4] have demonstrated that LIN28B, an RNA binding protein, is highly expressed in neuroblastoma subsets and that this expression is associated with higher stage neuroblastoma and inferior patient outcome. LIN28B, with its paralog LIN28A, plays pivotal roles in regulating multiple processes that shape normal development, including the cell cycle and apoptosis, self-renewal, glycolysis, and oxidative phosphorylation, among others [5]. These same processes are often subverted in a variety of tumors; accordingly, LIN28B is deregulated in multiple tumor histotypes, including cancers of the colon [6] and ovary [7], Wilms tumor [8], and hepatocellular carcinomas [9]. Targeted LIN28B overexpression in respective murine tissues leads to the development of neuroblastoma [3], colon cancer [10], and liver cancer [9], credentialing LIN28B as a bona fide oncogene. Mechanistically, LIN28B and LIN28A inhibit the let-7 family of microRNAs and also bind directly to a variety of RNA species, including mRNAs, snoRNAs, and long non-coding RNAs [5]. In humans, there are multiple let-7 family members that have been shown to repress a number of targets implicated in cell proliferation and self-renewal, including expression of RAS, MYC, and HMGA2 [5].
We and others previously demonstrated that LIN28B promotes neuroblastoma proliferation, in part through regulating the expression of RAN GTPase and Aurora kinase A [11], [12], [3]. While alteration of the cell cycle is a seminal characteristic of cancer, multiple hallmarks comprise the malignant phenotype, including metastatic dissemination [13]. Given the positive association of high LIN28B expression with advanced stage disease and poorer outcome [2], along with the fact that LIN28B promotes metastasis in the context of esophageal cancer [14] and colon cancer [6], we investigated whether LIN28B and let-7 act similarly in the context of neuroblastoma metastasis.
Materials and methods
Cell culture
Neuroblastoma cell lines (SKNDZ, Kelly, IMR5, NGP, NB-1643, all MYCN-amplified) were obtained from the Children’s Hospital of Philadelphia (CHOP) neuroblastoma cell line bank and 293 T cells from System Biosciences (LV900A-1); genomic annotations and culture conditions are as previously described [11], [15]. The Emory Genomics Core authenticated cell lines for use. Cell lines were discarded after 20–30 passages. Mycoplasma testing was performed every 3–6 months using the Mycoplasma test kit (PromoCell, PK-CA91-1024) and on an ad hoc basis.
Plasmid and lentiviral preparation
All shRNA constructs were purchased from Sigma (pLK0.1 lentiviral backbone) and catalog numbers are listed in Supplementary Table S1. Dr. David Barrett’s laboratory (Children’s Hospital of Philadelphia) provided the lentiviral GFP/luciferase plasmid used in the neuroblastoma dissemination model [16]. Mature let-7i (sequence obtained from http://www.mirbase.org/) was custom cloned into a lentiviral vector (pLenti-H1-GFP) by ViGene Biosciences. The plasmids for the PBK 3′UTR (pLightswitch_3UTR vector) and the PBK promoter (approximately 900 base pairs of promoter; pLightswitch_Prom vector) were purchased from Switchgear Genomics and catalog numbers are listed in Supplementary Table S1. Using site-directed mutagenesis, the Emory Integrated Genomics Core (EIGC) generated the PBK 3′UTR let-7 binding site mutant, with primers described in Supplementary Table S1. The EIGC generated pcDNA3.1-MYCN using the primers in Supplementary Table S1.
Lentiviral production and transduction were performed as previously described by us and others [11], [17], [18], [19]. To prepare lentiviruses, we utilized FuGENE6 to transfect various shRNA/expression constructs, along with pMD2.G (encoding envelope plasmid VSV-G) and psPAX2 (packaging plasmid), into HEK293T cells, as previously described by us and other investigators. We collected viral supernatant 48 and 72 hours post transfection and filtered with 0.45 µm nitrocellulose membranes. For transduction, cells were seeded on day 0, such that they were approximately 70% confluent on day 1. On day 1, 3 mL of virus was added to cells, along with 8 µg/mL polybrene (Sigma), optimizing transduction. On day 1, 6 hours post addition of virus, 3 mL media was added. On day 2, media was changed and, on day 4, puromycin (Sigma) was added for at least 72 hours prior to plating for assays. Of note, single cell cloning was not performed; thus cell cultures represent an amalgamation of multiple clones.
In vivo tumor dissemination model
Female NOD-scidIL2rγnull (NSG) mice (6–7 weeks old; The Jackson Laboratory) were housed at the Emory University HSRB Animal Facility in sterile cages in 12-h/12-h light–dark cycles. All experimental procedures were Emory IACUC approved. We infected SKNDZ cells with GFP/luciferase virus and flow sorted cells, generating the SKNDZ-GFP/luciferase model. We subsequently infected SKNDZ-GFP/luciferase cells with control or shLIN28B lentiviruses, creating stable SKNDZ-GFP-Con, SKNDZ-GFP-shLIN28B-1, and SKNDZ-GFP-shLIN28B-3 models. After acclimatizing, mice were randomized to Con, shLIN28B-1, and shLIN28B-3 groups (n = 10 for all). Respective groups received 1 million Con, shLIN28B-1, or shLIN28B-3 cells via tail vein injection. Starting 14 days post injection, bioluminescence imaging was performed twice a week. For imaging, 150 mg/kg luciferin was intraperitoneally injected into mice 10 minutes prior to imaging with IVIS Spectrum Imaging Systems (Perkinelmer). Imaging settings remained the same throughout the study and luminescence intensity was measured using Living Image Software (Perkinelmer).
Real-Time PCR analysis and Western blotting
RNA was isolated from cells and Real-Time PCR analysis performed as previously noted [11]. We utilized TaqMan and TaqMan microRNA assays (Life Technologies), as shown in Supplementary Table S1. Western blotting was carried out as previously detailed, with antibodies in Supplementary Table S1 [11].
Tumorsphere and cell proliferation assays
As previously described [20], single-cell suspensions of cells were detached, filtered through a 100 µm cell strainer, and plated in Tumorsphere medium (DMEM/F12 (Gibco) supplemented with 20 ng/ml human recombinant epidermal growth factor (EGF, Corning), 40 ng/ml human recombinant basic fibroblast growth factor (bFGF, Corning), 1×B27 (Gibco), 1×N2 (Gibco), 0.1 mM beta-mercaptoethanol (Sigma), 2 µg/ml heparin (Stem Cell Technologies), and 1% antibiotic-antimycotic (Gemini). We plated 30,000–40,000 cells per well in 6-well ultra-low attachment plates, and importantly, cells did not aggregate under these conditions [20]. Medium was replenished 3–4 days after plating. Seven to nine days after plating, tumorspheres were dissociated and counted.
CellTiter-Glo Luminescent Cell Viability assays (Promega) were performed to determine the effects of LIN28B or PBK knockdown or let-7i overexpression on cell proliferation. On day 0, 2000 cells were seeded with 5–6 replicates into RPMI-containing medium in 96-well plates. On day 4, luminescence substrate was added to each well and processed per the manufacturer’s protocol.
Wound migration assays
Approximately 100,000 cells per well were seeded in 96-well ImageLock plates, yielding 90–100% confluency 24 hours after plating. To inhibit proliferation, cells were treated with 2 µg/ml mitomycin C (Sigma) for 1 hour and wounds generated using the Incucyte wound maker [19]. After rinsing with phosphate-buffered saline 3 times, cells were further cultured in the IncuCyte® Scratch Wound Cell Migration and Invasion System (Essen BioScience) and images were taken every hour during the 24 hour incubation. The relative percentage of wound area occupied by migrated cells was calculated relative to the original wound area using IncuCyte ZOOM analysis software (Essen BioScience).
3′UTR and promoter luciferase reporter assays
Assays were carried out with the Lightswitch Luciferase Assay System (Switchgear Genomics) as previously detailed. We utilized control and let-7i microRNA mimetics (Dharmacon; listed in Supplementary Table S1) and the effects of control and let-7i microRNA mimetics on the PBK 3′UTR were normalized to effects on actin (which does not contain let-7 binding sites) [11]. PBK promoter luciferase assays were carried out as previously described, with PBK promoter luciferase values normalized to CellTiter-Glo luminescence assay values [18].
Neuroblastoma patient datasets
To investigate novel gene-gene correlations and to perform Kaplan-Meier analyses, we utilized the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl). Individual databases used to investigate correlations and survival analyses are found within the R2: Genomics Analysis and Visualization Platform, with appropriate citations in the text and notation of individual databases in the figure legend. To quantify gene expression at different stages of neuroblastoma, the paired-end reads were aligned with STAR aligner v2.5.2b and expression was quantified in terms of Fragments Per Kilobase of transcript per Million mapped reads (FPKM) with RSEM v1.2.28 using hg38 as the reference genome and GENCODE [3] v23 gene annotation. Boxplots were generated using the R package ggpubr and represent PBK gene level expression on the y-axis and International Neuroblastoma Staging System (INSS Stage) on the x-axis. ANOVA P-value denotes significance of PBK expression differences stratified by INSS stage.
Chromatin immunoprecipitation sequencing (ChIP-Seq)
Kelly MYCN ChIP-Seq was performed and analyzed as previously described [18]. For NB-1643 and NGP MYCN and all H3K27Ac ChIP-Seq, cell lines were grown in a 150 mm dish to 60–80% confluency, fixed, and pelleted according to the Active Motif protocol (htts://www.activemotif.com/documents/1848.pdf). Immunoprecipitations were performed using 30 μg of cell line chromatin and 6 μg of N-Myc antibody (Active Motif # 61185) or 4 μg of H3K27Ac antibody (Active Motif #39133). Libraries were prepared by Active Motif and sequenced on a NextSeq 500 to a depth of ∼50 M reads (Jefferson University Genomics Laboratory) and data were analyzed as described previously [18].
Statistical analyses
Statistical analyses were performed using GraphPad Prism software version 7. One-way ANOVA with Tukey’s post-hoc test was performed to compare the differences among groups. Unpaired student t-test (two tailed) was used when two groups were compared. Data are presented as mean ± standard error. Survival analyses in the animal study were performed using the methods of Kaplan and Meier.
Results
LIN28B promotes neuroblastoma metastatic dissemination
To investigate the impact of LIN28B on neuroblastoma metastasis, we employed a tail vein model previously used to help identify oncogenic pathways that contribute to neuroblastoma metastasis [21], [22]. Using SKNDZ, a neuroblastoma cell line we have previously shown has high LIN28B expression [2], [11], we generated control and LIN28B-depleted models that express luciferase/GFP, allowing in vivo imaging.
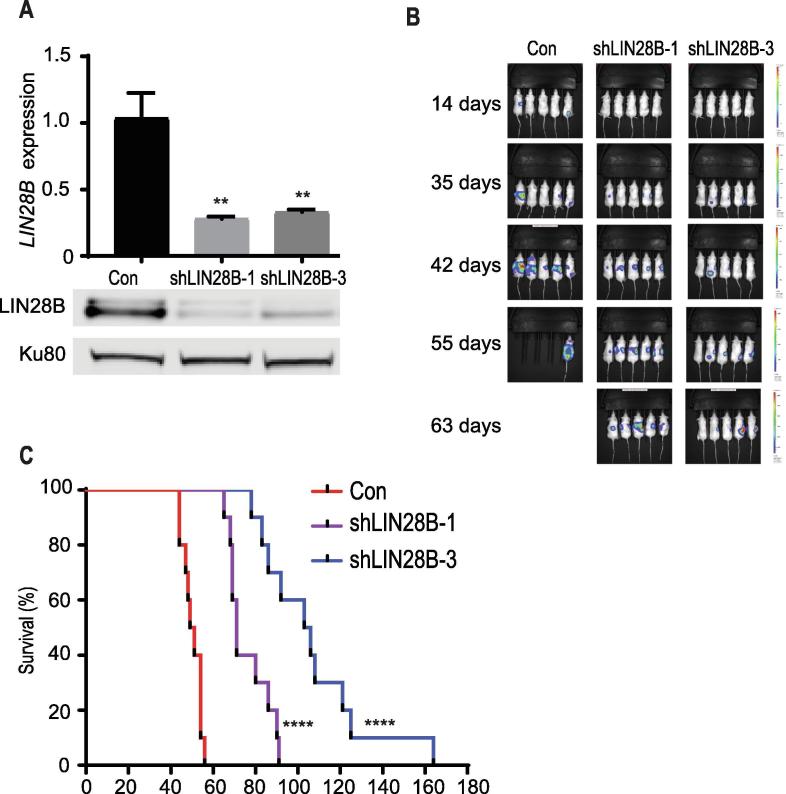
As shown in Fig. 1A, in comparison to control, LIN28B-depleted cell lines expressed lower levels of LIN28B mRNA and protein. We injected control and LIN28B-depleted cell lines into the tail vein of NSG mice, employing two independent shRNAs directed against LIN28B [21], [3]. Mice were followed over time via IVIS imaging and those within the control cohort first displayed evidence of metastatic outgrowth at day 14, with significant tumor burden noted in the majority of control mice by day 42. At comparable time points, we observed significantly lower tumor burden in mice bearing LIN28B-depleted tumors, compared to the control cohort (Fig. 1B and Supplementary Fig. S1A). By day 63, all mice in the control cohort had died, with lower tumor burden in cohorts bearing LIN28B-depleted tumors (Fig. 1B). Control mice died by day 56, compared to mice bearing tumors in which LIN28B was depleted (days 91 and 164 for shLIN28B-1 and shLIN28B-3, respectively; Fig. 1C). Following death, subsets of mice underwent a limited necropsy. Almost all mice developed widespread liver and mesenteric metastases, sites often observed within neuroblastoma patients [1]. In addition, we observed some mice with occasional skull/brain and lung metastases, sites sometimes seen in neuroblastoma patients, and associated with worse prognoses (data not shown) [23]. To assess whether LIN28B-depleted cell line models maintained lower levels of LIN28B expression, we isolated representative control and LIN28B-depleted tumors that had grown in the liver and performed RT-PCR and Western blotting. In comparison to control tumors, LIN28B-depleted tumors maintained knockdown of LIN28B RNA/protein (Supplementary Fig. S1B and S1C). Collectively, these studies demonstrate that LIN28B promotes neuroblastoma metastasis in the in vivo setting and that shRNA-mediated depletion of LIN28B diminishes tumor dissemination, significantly prolonging survival.
Fig. 1.
LIN28B promotes neuroblastoma metastatic dissemination. (A) RT-PCR and Western blot demonstrating LIN28B expression in the GFP-luciferase expressing SKNDZ control and LIN28B-depleted cell line models. Two independent shRNAs were used to deplete LIN28B (shLIN28B-1 and shLIN28B-3). Western blotting demonstrates knockdown of LIN28B, with Ku80 as a loading control. (B) IVIS imaging of mice bearing control tumors or tumors in which LIN28B was depleted, at various time points. (C) Kaplan-Meier survival curves for mice bearing control tumors or tumors in which LIN28B was depleted. **p < 0.01, ****p < 0.0001. See also Supplementary Fig. S1.
LIN28B positively influences neuroblastoma self-renewal
We and others showed that LIN28B expression is high in MYCN-amplified neuroblastoma [2], [3] and therefore employed three independent, MYCN-amplified neuroblastoma cell lines with high LIN28B expression (SKNDZ, Kelly, and IMR5) [2], [11]. We and others had demonstrated that LIN28B promotes cell proliferation, a finding we again confirmed (Supplementary Fig. S2) [3], [2], [11]. We sought to mechanistically dissect how LIN28B impacts the metastatic cascade and first examined the impact of LIN28B on anoikis resistance, but did not observe effects of LIN28B on this property (data not shown). In addition to anoikis resistance, many metastatic cells exhibit enhanced self-renewal, the continuous expansion of cells that possess long-term growth potential [24], [25]. Given the role of LIN28B and let-7 in regulating self-renewal in normal development, we hypothesized that LIN28B might promote self-renewal, thus driving neuroblastoma metastasis.
We first asked whether LIN28B regulates genes that support self-renewal, including NANOG, OCT4, and SOX2, as well as NESTIN, which has been shown to be associated with neural crest stem-like cells and neuroblastoma aggression [26]. Neuroblastoma cells were cultured under non-adherent, serum-free conditions and formed tumorspheres. We effectively depleted LIN28B (Fig. 2A) and observed decreased levels of NANOG, OCT4, SOX2, and NESTIN (Fig. 2B–E) mRNA; similar changes were noted at the level of protein (Fig. 2F). The influence of LIN28B on these markers of self-renewal strongly suggests a role for LIN28B in mediating self-renewal. To directly investigate the influence of LIN28B on self-renewal, we employed tumorsphere assays, providing a functional readout of self-renewal. We successfully depleted LIN28B in three neuroblastoma cell lines and noted decreased self-renewal in all neuroblastoma cell lines (Fig. 2G-I). Collectively, these results suggest that LIN28B influences self-renewal as one means of promoting metastasis.
Fig. 2.
LIN28B positively influences neuroblastoma self-renewal. (A–E) Quantitation of LIN28B (A), NANOG (B), OCT4 (C), SOX2 (D), and NESTIN mRNA levels (E), respectively, in SKNDZ cells grown under tumorsphere conditions. The mRNA level in the control group was normalized to 1 and the fold change of mRNA level in each experimental group compared to control group is presented. (F) Western blots of corresponding protein levels. (G–I) Tumorsphere quantitation in control and LIN28B-depleted neuroblastoma cell lines SKNDZ (G), Kelly (H), and IMR5 (I). Cell lines were infected with control lentiviruses or lentiviruses expressing two independent shRNAs directed against LIN28B, and grown under tumorsphere conditions. Western blotting of LIN28B and Ku80 in corresponding cell lines. Results are representative of at least two independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, ****, p < 0.0001. See also Supplementary Fig. S2.
LIN28B promotes the migration of neuroblastoma cells
Another key property that metastatic cells acquire is enhanced migratory ability. In two cell line models, we performed wound migration assays at an early time point (24 hours) and observed that LIN28B promotes neuroblastoma migration (Supplementary Fig. S3A-B). These effects were not due to its influence on proliferation as we detected no differences in cell number between control and LIN28B-depleted cell lines when assessed at 24 hours (Supplementary Fig. S3C). To further ensure that the influence of LIN28B on proliferation did not account for effects on cell migration, we treated cells with mitomycin C, thus inhibiting proliferation [19], and again observed that LIN28B depletion impedes neuroblastoma migration (Fig. 3A–C).
Fig. 3.
LIN28B promotes the migration of neuroblastoma cells. (A–C) Depicted are representative images from wound closure assays of control and LIN28B-depleted neuroblastoma cell lines SKNDZ (A), Kelly (B), and IMR5 (C). Cell lines were infected with control lentiviruses or lentiviruses expressing two independent shRNAs directed against LIN28B. In addition, cells were treated with mitomycin C. Images were acquired at 0 and 24 hours. The relative percentage of wound area occupied by migrated cells 24 hours after scratch was calculated relative to the original wound area using IncuCyte ZOOM analysis software. The percentage of wound closure in control group was normalized to 1, and the fold change of wound closure in experimental groups was calculated. Of note, the same cell line models utilized in Fig. 2 were used in these assays, with relative LIN28B levels as shown in Fig. 2G–I. Results are representative of at least two independent experiments. *p < 0.05, **p < 0.01, ****, p < 0.0001. See also Supplementary Fig. S3.
Let-7i inhibits both neuroblastoma self-renewal and migration
One of the most well characterized functions of LIN28B is its inhibition of the maturation and processing of let-7 microRNAs [5]. Other investigators previously found that let-7 diminishes cell proliferation [3], a finding we replicated (Supplementary Fig. S4). We asked whether let-7 also inhibits self-renewal and migration, similar to depletion of LIN28B. For let-7 studies, we used let-7i, as we and other investigators previously showed that LIN28B negatively regulates let-7i in neuroblastoma [3], [11]. We generated neuroblastoma cell lines expressing mature let-7i (bypassing the inhibitory effect of LIN28B on let-7 processing) and confirmed let-7i overexpression (Fig. 4A). Similar to depletion of LIN28B, let-7i overexpression leads to significantly decreased neuroblastoma self-renewal (Fig. 4B–C) and migration (Fig. 4D–E).
Fig. 4.
Let-7 inhibits both neuroblastoma self-renewal and migration. RT-PCR demonstrates expression of let-7i in SKNDZ and IMR5 cell lines. Cell lines were infected with control lentiviruses or lentiviruses expressing let-7i. The mRNA level in control group was normalized to 1, and the fold change of mRNA levels in each experimental group compared to control group was calculated. (B-C) Tumorsphere quantitation of control and let-7i expressing neuroblastoma cell lines SKNDZ (B) and IMR5 (C). (D-E) Given the influence of let-7i on cell proliferation, cells were treated with mitomycin C. The relative percentage of wound area occupied by migrated cells 24 hours after scratch was calculated relative to the original wound area using IncuCyte ZOOM analysis software. The percentage of wound closure in control group was normalized to 1, and the fold change of wound closure in experimental groups was calculated. Results are representative of at least two independent experiments. **p < 0.01, ****, p < 0.0001. See also Supplementary Fig. S4.
PDZ binding kinase is a novel LIN28B-influenced kinase
We next sought to identify novel LIN28B-influenced genes that might be therapeutically targetable. Given our discovery of Aurora kinase A (AURKA) as a novel LIN28B target [11], we hypothesized that LIN28B might coordinate the expression of additional oncogenic kinases to promote neuroblastoma aggression [1]. Therefore, we first took a candidate approach, perturbing LIN28B levels and surveying effects on candidate kinases that shape the malignant phenotype. We did not observe effects of LIN28B on p44/42 MAPK or phospho-p44/42 MAPK (Thr202/Tyr204), a key node in MAPK signaling (data not shown). Additionally, LIN28B did not affect phospho-p38 (Thr180/Tyr182) expression (data not shown). Finally, LIN28B did not influence the expression of the p110α and p110β catalytic subunits of PI3K (data not shown).
Given these negative findings, we interrogated the Therapeutically Applicable Research to Generate Effective Treatments project (TARGET; https://ocg.cancer.gov/programs/target) dataset [27], previously utilized in our discovery of RAN and AURKA as novel LIN28B-influenced genes [11]. We evaluated the top 10 kinases significantly and positively correlated with high LIN28B expression (listed in Supplementary Table S2). We chose to focus our studies on PDZ Binding kinase (PBK), ranked 4/10 of the top correlated kinases, for the following reasons: 1) Its possible role in neuroblastoma, or indeed, in pediatric tumors, was undefined; 2) PBK, also known as T-LAK cell-originated protein kinase (TOPK), is a serine/threonine kinase that promotes the proliferation and self-renewal of neural stem cells [28]; 3) PBK is overexpressed in diverse adult histotypes and implicated in multiple hallmarks of cancer, including cell cycle regulation, apoptosis, and metastasis [29]; 4) PBK inhibitors have demonstrated preclinical efficacy in aggressive adult tumors, including metastatic colon cancer [30] and ovarian cancer [31], [32].
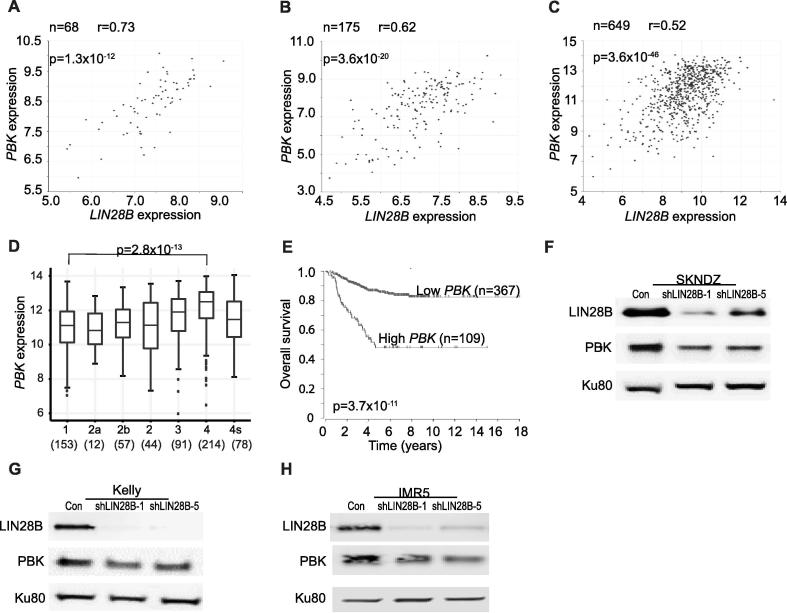
We found LIN28B and PBK expression to be positively correlated in neuroblastoma, in both the MYCN-amplified (Fig. 5A) and non-MYCN-amplified setting (Fig. 5B) [27]. To strengthen this observation, we investigated the LIN28B-PBK correlation in additional neuroblastoma datasets and noted similar correlations (Fig. 5C and Supplementary Fig. S5A) [33], [34]. In further support of a possible oncogenic role for PBK in neuroblastoma, we demonstrated that its expression was associated with higher stage neuroblastoma (Fig. 5D, Supplementary Fig. S5B) [33], [34] and lower overall survival (Fig. 5E, Supplementary Fig. S5C) [33], [34]. While these data demonstrate a correlation between LIN28B and PBK expression, they do not demonstrate whether LIN28B influences the expression of PBK. We depleted LIN28B and noted decreased levels of PBK (Fig. 5F-H), demonstrating that LIN28B promotes PBK expression.
Fig. 5.
PBK is a novel LIN28B-influenced kinase. (A-B) Correlation between LIN28B and PBK expression in 68 MYCN-amplified (A) and 175 MYCN non-amplified (B) primary neuroblastomas. Log2 of LIN28B expression is depicted on the x axis, with log2 of PBK expression on the y axis. Neuroblastoma datasets obtained from the TARGET consortium. (C) Correlation between LIN28B and PBK expression in 649 primary neuroblastomas. Log2 of LIN28B expression is depicted on the x axis, with log2 of PBK expression on the y axis. Data obtained from Kocak dataset. (D) Expression of PBK in primary neuroblastoma tumors, as shown for International Neuroblastoma Staging System stages 1 through 4, with 4S. PBK expression was quantified in terms of Fragments Per Kilobase of transcript per Million mapped reads (FPKM). The number of tumors is depicted in parentheses. Data obtained from Kocak dataset. (E) Kaplan-Meier analysis curves, with individuals grouped by low and high PBK expression. Data obtained from Kocak dataset. (F-H) LIN28B and PBK protein levels in SKNDZ (F), Kelly (G), and IMR5 (H) cell lines in which LIN28B was depleted using two independent shRNAs. Ku80 serves as a loading control. P values and r values listed. See also Supplementary Fig. S5 and Table S2.
Two neuroblastoma oncogenes, LIN28B and MYCN, regulate the expression of PBK
We next dissected the mechanisms by which LIN28B influences PBK expression. As let-7 is a key downstream effector of LIN28B, and as we showed that let-7i inhibits self-renewal and migration, we first investigated whether let-7i influences PBK expression. We engineered three neuroblastoma cell lines to overexpress let-7i, verified significant let-7i overexpression (Fig. 4A and Supplementary Fig. S6A), and demonstrated that let-7i expression downregulates PBK (Fig. 6A–C). Various microRNA target prediction programs [35], [36] predict that PBK has one let-7 binding site in its 3′UTR and thus we speculated that PBK might be a novel, direct let-7 target. We performed 3′UTR reporter assays and showed that treatment with let-7i inhibits PBK 3′UTR-driven luciferase activity (Fig. 6D). We then mutated the let-7 binding site in the 3′UTR of PBK and demonstrated that this relieved the inhibitory effect of let-7i. Collectively, these results argue that PBK is a direct let-7i target.
Fig. 6.
Both LIN28B/let-7i and MYCN directly regulate the expression of PBK. (A–C) PBK protein levels in SKNDZ (A), Kelly (B), and IMR5 (C) cell lines in which let-7i is overexpressed. Ku80 serves as a loading control. (D) 3′UTR assays showing the effect of let-7i on PBK 3′UTR activity. Control microRNA and mature let-7i were transfected into 293 T cells and the effect on wild-type PBK 3′UTR and mutated PBK (PBK mut) 3′UTR activity was quantitated. The luminescence intensity in control microRNA was normalized to 1, and the fold change of luminescence intensity in the let-7i group in the presence of PBK 3′UTR or muted PBK (PBK mut) is depicted. (E-G) Bar graphs depict mRNA levels of PBK expression in SKNDZ (E), Kelly (F), and IMR5 (G) cell lines in which MYCN was depleted using two independent shRNAs. The mRNA level in control group was normalized to 1, and the fold change of mRNA levels in each experimental group compared to control group was calculated and presented in the bar graphs. Immunoblots depict expression of MYCN and PBK protein, with Ku80 serving as a loading control. (H) Plot depicting binding of MYCN (black) and H3K27Ac binding (gray) to PBK. ChIP-Seq performed in the MYCN-amplified cell line models Kelly, NB-1643, and NGP. (I) PBK promoter assays showing the effect of MYCN on PBK promoter activity. Control and MYCN were transfected into 293 T cells and the effect on PBK promoter activity was quantitated. *p < 0.05, **p < 0.01, ***p < 0.001, ****, p < 0.0001. See also Supplementary Fig. S6.
We and others have previously shown that LIN28B expression is high in MYCN-amplified neuroblastoma [3], [11]. LIN28B has been shown to promote MYCN expression [3], and, reciprocally, MYCN binds the LIN28B promoter, positively regulating its expression [37]. Interestingly, PBK is overexpressed in lymphomas and it was recently reported that MYC positively regulates PBK expression by binding to its promoter [38]. We observed that MYCN and PBK expression are positively correlated in neuroblastoma tumors (Supplementary Fig. S6B–C) [27], [33] and since MYCN and MYC share many of the same transcriptional targets, we hypothesized that MYCN might directly regulate PBK expression [39], [40]. To determine whether MYCN directly influences PBK, we depleted MYCN and confirmed effective knockdown of MYCN protein (Fig. 6E–G). In three neuroblastoma cell line models, depletion of MYCN led to decreased PBK mRNA and protein levels (Fig. 6E–G).
Analysis of ChIP-Seq data demonstrated that MYCN binds the PBK promoter and is accompanied by the active enhancer histone mark, H3K27Ac (Fig. 6H) [18], suggesting direct transcriptional upregulation of PBK by MYCN. We performed PBK promoter reporter assays and showed that MYCN promotes PBK-driven luciferase activity (Fig. 6I). Taken together, these findings establish that two neuroblastoma oncogenes, LIN28B and MYCN, regulate the expression of PBK, and illustrate a novel convergence of LIN28B/let-7, MYCN, and PBK signaling.
PBK promotes neuroblastoma proliferation, self-renewal and migration, phenocopying the effects of LIN28B
If one of the major ways by which LIN28B shapes neuroblastoma aggression and metastasis is by positively regulating PBK, then PBK depletion would be expected to phenocopy LIN28B depletion. To assess this hypothesis, we successfully depleted PBK in neuroblastoma cell line models (Fig. 7A–C) and observed that, similar to LIN28B depletion, depletion of PBK led to decreased proliferation (Supplementary Fig. 7A–B), self-renewal (Fig. 7A–C), and cell migration (Fig. 7D–F).
Fig. 7.
PBK promotes neuroblastoma self-renewal and migration. (A–C) Tumorsphere quantitation in control and PBK-depleted neuroblastoma cell lines SKNDZ (A), Kelly (B), and IMR5 (C). Cell lines were infected with control lentiviruses or lentiviruses expressing three independent shRNAs directed against PBK. Western blotting of PBK and Ku80 in cell lines in corresponding cell lines. (D-F) Depicted are representative images from wound migration assays of control and PBK-depleted neuroblastoma cell lines SKNDZ (D), Kelly (E), and IMR5 (F). Cell lines were infected with control lentiviruses or lentiviruses expressing shRNAs directed against PBK. In addition, cells were treated with mitomycin C. Images were acquired at 0 and 24 hours. The relative percentage of wound area occupied by migrated cells 24 hours after scratch was calculated relative to the original wound area using IncuCyte ZOOM analysis software. The percentage of wound closure in control group was normalized to 1, and the fold change of wound closure in experimental groups was calculated. **p < 0.01, ***p < 0.001, ****, p < 0.0001. See also Supplementary Fig. S7.
Discussion
These findings expand upon studies showing that targeted expression of LIN28B within the developing murine neural crest leads to the development of neuroblastoma [3]. We show that LIN28B enhances the ability of disseminated human neuroblastoma cells to initiate and sustain metastatic colonization and outgrowth. This is due in part to the influence of LIN28B on the self-renewal of neuroblastoma cells, potentially allowing tumor cells to repopulate indefinitely, as well as on neuroblastoma migration, perhaps allowing cells to exit from the primary tumor and disseminate elsewhere. We demonstrate that let-7i overexpression acts similarly to LIN28B depletion and define PBK as a novel LIN28-influenced kinase. Functionally, our studies reveal that PBK promotes neuroblastoma proliferation, self-renewal, and migration, phenocopying LIN28B. Interestingly, the neuroblastoma oncogene LMO1 promotes neuroblastoma proliferation [2], migration, and metastasis [19]. Moreover, cyclin D1, well known for its role in cell cycle regulation, promotes self-renewal [41], invasion, and metastasis [42]. Similarly, our results demonstrate that LIN28B and PBK shape multiple hallmarks of cancer.
PBK has been shown to be expressed at high levels in multiple aggressive tumors seen primarily in adults, including head and neck cancers, esophageal cancer, liver cancer, colon cancer, and prostate cancer [29]. PBK is a tractable therapeutic target, against which clinically relevant inhibitors, primarily targeting the kinase activity of PBK, exist. In preclinical models of colon [30] and ovarian cancer [31], PBK inhibition led to decreased metastatic dissemination. Future studies will focus on elucidating the role of PBK in neuroblastoma and additional pediatric malignancies.
Mechanistically, our data indicate that LIN28B and MYCN signaling intertwine and influence PBK expression by two different mechanisms. First, our studies reveal PBK to be a novel and direct let-7 target. Second, our data demonstrate that MYCN binds the PBK promoter and promotes MYCN expression. This is reminiscent of Aurora kinase A, which we previously identified as a LIN28B/let-7 target and other investigators have shown interacts with MYCN to stabilize MYCN protein [43]. While drugging LIN28B/let-7 is still in its infancy, investigators have developed small molecule inhibitors that disrupt the repression of let-7 by LIN28B, allowing restoration of let-7 levels [44]. Aurora kinase A inhibitors, such as alisertib, have undergone Phase 2 testing in neuroblastoma and were found to be well tolerated and to demonstrate activity, primarily in the non-MYCN-amplified context [45]. Additionally, BET inhibition provides a means of targeting MYC/MYCN and has demonstrated efficacy in some preclinical neuroblastoma models [46], [47]. Moreover, others have shown that MYCN influences and collaborates with epigenetic machinery, including the PRC2 complex, providing additional therapeutic opportunities [48], [49]. Finally, PBK inhibition has demonstrated in vivo efficacy against aggressive adult histotypes [29].
Due to the inherent heterogeneity within primary tumor/metastatic sites, as well as the heterogeneity shaped by multimodal neuroblastoma therapies, developing a robust compendium of therapeutic agents for combinatorial regimens will likely be necessary to optimize neuroblastoma therapy. It will be of substantial interest to determine whether regimens targeting LIN28B/let-7, PBK, MYCN, and/or AURKA might improve the treatment of metastatic neuroblastoma.
Author’s contributions
Conception and design: D. Chen, J. Cox, R.W. Schnepp.
Development of methodology: D. Chen, J. Cox, J. Annam, R.W. Schnepp.
Acquisition of data: D. Chen, J. Cox, J. Annam, M. Weingart, G. Essien, P. Khurana, S.M. Cuya, A. Pilgrim, D. Li, C. Shields, O. Laur, R.W. Schnepp.
Analysis and interpretation of data (biostatistics, statistical analysis, interpretation of clinical data and genomic datasets): D. Chen, K.S. Rathi, J. L. Rokita, K. Bosse, A. Pilgrim, R.W. Schnepp.
Writing, review and/or revision of the manuscript: D. Chen, R.W. Schnepp.
Administrative, technical, or material support: J.M. Maris, R.W. Schnepp.
Study supervision: R.W. Schnepp.
Acknowledgments
Acknowledgments
This work was supported in part by NIH Grant K08-7K08CA194162-02 (R.W.S.), NIH Grant R35 CA220500 (J.M.M.), NIH Grant R01 CA124709 (J.M.M.), the Alex’s Lemonade Stand Foundation (J.L.R., K.R.B., R.W.S.), the Damon Runyon Cancer Research Foundation (PST-07-16; K.R.B.), Hyundai Hope on Wheels (R.W.S.), Andrew McDonough B + Foundation (R.W.S.), Team Connor Foundation (R.W.S.), Rally Foundation for Childhood Cancer Research (R.W.S.), the Winship Cancer Institute American Cancer Society Institutional Research Grant (R.W.S.), the Aflac Cancer and Blood Disorders Center Trust (R.W.S.) , and the William Woods, M.D., Aflac Clinical Investigator Chair (R.W.S.).
In addition, we thank Dr. David Barrett, the Children’s Hospital of Philadelphia, for the lentiviral GFP/Luciferase plasmid. Of note, the Emory Integrated Genomics Core is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Additional support was provided by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR000454. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2020.04.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Related to Fig. 1. LIN28B promotes neuroblastoma metastatic dissemination. (A) Mean bioluminescence intensities of mice from control and shLIN28B-depleted cohorts. (B) RT-PCR demonstrating LIN28B expression in the tumor tissue of control mice or mice with tumors in which LIN28B was depleted. The LIN28B mRNA level in control group was normalized to 1, and the fold changes of LIN28B mRNA level in each experimental group compared to control group presented. Of note, we isolated 3 control tumors, 2 tumors from the shLIN28B-1 cohort, and 2 tumors from the shLIN28B-3 cohort. (C) Quantitation of LIN28B protein level in the tumor tissues of control mice or mice with tumors in which LIN28B was depleted. The LIN28B protein level in control group was normalized to 1, and the fold changes of LIN28B protein level in each experimental group compared to control group are presented. Western blots below. * p < 0.05.
Related to Fig. 2. LIN28B positively influences neuroblastoma self-renewal. The SKNDZ cell line was infected with control lentiviruses or lentiviruses directed against LIN28B and cell proliferation quantified using CellTiter-Glo. Western blotting of LIN28B and Ku80 as shown in Fig. 2G. ****, p < 0.0001.
Related to Fig. 3. LIN28B promotes the migration of neuroblastoma cells. (A-B) Quantitation of wound closure over 24 hours for SKNDZ (A) and Kelly (B) cells infected with control lentiviruses or lentiviruses expressing shRNAs directed against LIN28B. Of note, cells were not treated with mitomycin. Error bars represent SEM. (C) The SKNDZ cell line was infected with control lentiviruses or lentiviruses directed against LIN28B and cells counted 24 hours post seeding. The number of control cells was normalized to 1, and fold change of cell number in experimental groups compared to control is presented. Western blotting of LIN28B and Ku80 as shown in Fig. 2G.
Related to Fig. 4. Let-7 inhibits both neuroblastoma self-renewal and migration. The SKNDZ cell line was infected with control or let-7i expressing lentiviruses and cell proliferation quantified using CellTiter-Glo. Expression of let-7i as shown in Fig. 4A. ****, p < 0.0001.
Related to Fig. 5. PBK is a novel LIN28B-influenced kinase. (A) Correlation between LIN28B and PBK expression in 498 primary neuroblastomas. Log2 of LIN28B expression is depicted on the x axis, with log2 of PBK expression on the y axis. (B) Expression of PBK in primary neuroblastoma tumors, as shown for International Neuroblastoma Staging System stages 1 through 4, with 4S. The number of tumors is depicted in parentheses. PBK expression was quantified in terms of Fragments Per Kilobase of transcript per Million mapped reads (FPKM). C) Kaplan-Meier analysis curves, with individuals grouped by low and high PBK expression. Data obtained from the Su dataset. P values and r values listed.
Related to Fig. 6. Both LIN28B/let-7 and MYCN directly regulate the expression of PBK. (A) RT-PCR demonstrates expression of let-7i in Kelly cell lines. Cell lines were infected with control lentiviruses or lentiviruses expressing let-7i. The let7i RNA level in control group was normalized to 1, and the fold changes of let7i RNA level in each experimental group compared to control group were calculated and presented in the bar graphs. (B) Correlation between MYCN and PBK expression in 249 primary neuroblastomas. Log2 of MYCN expression is depicted on the x axis, with log2 of PBK expression on the y axis. Data obtained from the TARGET dataset. (C) Correlation between MYCN and PBK expression in 649 primary neuroblastomas. Log2 of MYCN expression is depicted on the x axis, with log2 of PBK expression on the y axis. Data obtained from the Kocak dataset. P values and r values listed.
Related to Fig. 7. PBK promotes neuroblastoma self-renewal and migration. (A-B) Cell proliferation quantitated by CellTiter-Glo in control and PBK-depleted neuroblastoma cell lines SKNDZ (A), and IMR5 (B). Expression of PBK as shown in Fig. 7A and C. ****, p < 0.0001.
This table lists plasmids, shRNA constructs, antibodies, and TaqMan assays.
This table depicts the kinases correlated with LIN28B expression in the TARGET database. P values and r values are listed.
References
- 1.Matthay K.K., Maris J.M., Schleiermacher G., Nakagawara A., Mackall C.L., Diller L. Neuroblastoma. Nat Rev Dis Primers. 2016;2:16078. doi: 10.1038/nrdp.2016.78. [DOI] [PubMed] [Google Scholar]
- 2.Diskin S.J., Capasso M., Schnepp R.W., Cole K.A., Attiyeh E.F., Hou C. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat Genet. 2012;44:1126–1130. doi: 10.1038/ng.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molenaar J.J., Domingo-Fernandez R., Ebus M.E., Lindner S., Koster J., Drabek K. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet. 2012;44:1199–1206. doi: 10.1038/ng.2436. [DOI] [PubMed] [Google Scholar]
- 4.Powers J.T., Tsanov K.M., Pearson D.S., Roels F., Spina C.S., Ebright R. Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma. Nature. 2016;535:246–251. doi: 10.1038/nature18632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shyh-Chang N., Daley G.Q. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013;12:395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King C.E., Cuatrecasas M., Castells A., Sepulveda A.R., Lee J.S., Rustgi A.K. LIN28B promotes colon cancer progression and metastasis. Cancer Res. 2011;71:4260–4268. doi: 10.1158/0008-5472.CAN-10-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin X., Shen J., Dan P., He X., Xu C., Chen X. RNA-binding protein LIN28B inhibits apoptosis through regulation of the AKT2/FOXO3A/BIM axis in ovarian cancer cells. Signal Transduct Target Ther. 2018;3:23. doi: 10.1038/s41392-018-0026-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urbach A., Yermalovich A., Zhang J., Spina C.S., Zhu H., Perez-Atayde A.R. Lin28 sustains early renal progenitors and induces Wilms tumor. Genes Dev. 2014;28:971–982. doi: 10.1101/gad.237149.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen L.H., Robinton D.A., Seligson M.T., Wu L., Li L., Rakheja D. Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell. 2014;26:248–261. doi: 10.1016/j.ccr.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madison B.B., Liu Q., Zhong X., Hahn C.M., Lin N., Emmett M.J. LIN28B promotes growth and tumorigenesis of the intestinal epithelium via Let-7. Genes Dev. 2013;27:2233–2245. doi: 10.1101/gad.224659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnepp R.W., Khurana P., Attiyeh E.F., Raman P., Chodosh S.E., Oldridge D.A. A LIN28B-RAN-AURKA signaling network promotes neuroblastoma tumorigenesis. Cancer Cell. 2015;28:599–609. doi: 10.1016/j.ccell.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnepp R.W., Diskin S.J. LIN28B: an orchestrator of oncogenic signaling in neuroblastoma. Cell Cycle. 2016;15:772–774. doi: 10.1080/15384101.2015.1137712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Hamano R., Miyata H., Yamasaki M., Sugimura K., Tanaka K., Kurokawa Y. High expression of Lin28 is associated with tumour aggressiveness and poor prognosis of patients in oesophagus cancer. Br J Cancer. 2012;106:1415–1423. doi: 10.1038/bjc.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harenza J.L., Diamond M.A., Adams R.N., Song M.M., Davidson H.L., Hart L.S. Transcriptomic profiling of 39 commonly-used neuroblastoma cell lines. Sci Data. 2017;4 doi: 10.1038/sdata.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett D.M., Seif A.E., Carpenito C., Teachey D.T., Fish J.D., June C.H. Noninvasive bioluminescent imaging of primary patient acute lymphoblastic leukemia: a strategy for preclinical modeling. Blood. 2011;118:e112–e117. doi: 10.1182/blood-2011-04-346528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu F., Zhang X., Weisberg E., Chen S., Hur W., Wu H. Discovery of a selective irreversible BMX inhibitor for prostate cancer. ACS Chem Biol. 2013;8:1423–1428. doi: 10.1021/cb4000629. [DOI] [PubMed] [Google Scholar]
- 18.Bosse K.R., Raman P., Zhu Z., Lane M., Martinez D., Heitzeneder S. Identification of GPC2 as an oncoprotein and candidate immunotherapeutic target in high-risk neuroblastoma. Cancer Cell. 2017;32(295–309) doi: 10.1016/j.ccell.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu S., Zhang X., Weichert-Leahey N., Dong Z., Zhang C., Lopez G. LMO1 Synergizes with MYCN to promote neuroblastoma initiation and metastasis. Cancer Cell. 2017;32(310–23) doi: 10.1016/j.ccell.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahller Y.Y., Williams J.P., Baird W.H., Mitton B., Grossheim J., Saeki Y. Neuroblastoma cell lines contain pluripotent tumor initiating cells that are susceptible to a targeted oncolytic virus. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sartelet H., Durrieu L., Fontaine F., Nyalendo C., Haddad E. Description of a new xenograft model of metastatic neuroblastoma using NOD/SCID/Il2rg null (NSG) mice. Vivo. 2012;26:19–29. [PubMed] [Google Scholar]
- 22.Seong B.K., Fathers K.E., Hallett R., Yung C.K., Stein L.D., Mouaaz S. A metastatic mouse model identifies genes that regulate neuroblastoma metastasis. Cancer Res. 2017;77:696–706. doi: 10.1158/0008-5472.CAN-16-1502. [DOI] [PubMed] [Google Scholar]
- 23.Morgenstern D.A., London W.B., Stephens D., Volchenboum S.L., Simon T., Nakagawara A. Prognostic significance of pattern and burden of metastatic disease in patients with stage 4 neuroblastoma: a study from the International Neuroblastoma Risk Group database. Eur J Cancer. 2016;65:1–10. doi: 10.1016/j.ejca.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Celia-Terrassa T., Kang Y. Distinctive properties of metastasis-initiating cells. Genes Dev. 2016;30:892–908. doi: 10.1101/gad.277681.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song K.H., Park M.S., Nandu T.S., Gadad S., Kim S.C., Kim M.Y. GALNT14 promotes lung-specific breast cancer metastasis by modulating self-renewal and interaction with the lung microenvironment. Nat Commun. 2016;7:13796. doi: 10.1038/ncomms13796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas S.K., Messam C.A., Spengler B.A., Biedler J.L., Ross R.A. Nestin is a potential mediator of malignancy in human neuroblastoma cells. J Biol Chem. 2004;279:27994–27999. doi: 10.1074/jbc.M312663200. [DOI] [PubMed] [Google Scholar]
- 27.Pugh T.J., Morozova O., Attiyeh E.F., Asgharzadeh S., Wei J.S., Auclair D. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dougherty J.D., Garcia A.D., Nakano I., Livingstone M., Norris B., Polakiewicz R. PBK/TOPK, a proliferating neural progenitor-specific mitogen-activated protein kinase kinase. J Neurosci. 2005;25:10773–10785. doi: 10.1523/JNEUROSCI.3207-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herbert K.J., Ashton T.M., Prevo R., Pirovano G., Higgins G.S. T-LAK cell-originated protein kinase (TOPK): an emerging target for cancer-specific therapeutics. Cell Death Dis. 2018;9:1089. doi: 10.1038/s41419-018-1131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D.J., Li Y., Reddy K., Lee M.H., Kim M.O., Cho Y.Y. Novel TOPK inhibitor HI-TOPK-032 effectively suppresses colon cancer growth. Cancer Res. 2012;72:3060–3068. doi: 10.1158/0008-5472.CAN-11-3851. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda Y., Park J.H., Miyamoto T., Takamatsu N., Kato T., Iwasa A. T-LAK cell-originated protein kinase (TOPK) as a prognostic factor and a potential therapeutic target in ovarian cancer. Clin Cancer Res. 2016;22:6110–6117. doi: 10.1158/1078-0432.CCR-16-0207. [DOI] [PubMed] [Google Scholar]
- 32.Zykova T., Zhu F., Wang L., Li H., Lim D.Y., Yao K. Targeting PRPK function blocks colon cancer metastasis. Mol Cancer Ther. 2018;17:1101–1113. doi: 10.1158/1535-7163.MCT-17-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kocak H., Ackermann S., Hero B., Kahlert Y., Oberthuer A., Juraeva D. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis. 2013;4 doi: 10.1038/cddis.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su Z., Fang H., Hong H., Shi L., Zhang W., Zhang W. An investigation of biomarkers derived from legacy microarray data for their utility in the RNA-seq era. Genome Biol. 2014;15:523. doi: 10.1186/s13059-014-0523-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 36.Betel D., Wilson M., Gabow A., Marks D.S., Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beckers A., Van Peer G., Carter D.R., Gartlgruber M., Herrmann C., Agarwal S. MYCN-driven regulatory mechanisms controlling LIN28B in neuroblastoma. Cancer Lett. 2015;366:123–132. doi: 10.1016/j.canlet.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu F., Gartenhaus R.B., Zhao X.F., Fang H.B., Minkove S., Poss D.E. c-Myc and E2F1 drive PBK/TOPK expression in high-grade malignant lymphomas. Leuk Res. 2013;37:447–454. doi: 10.1016/j.leukres.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 39.Fredlund E., Ringner M., Maris J.M., Pahlman S. High Myc pathway activity and low stage of neuronal differentiation associate with poor outcome in neuroblastoma. Proc Natl Acad Sci USA. 2008;105:14094–14099. doi: 10.1073/pnas.0804455105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valentijn L.J., Koster J., Haneveld F., Aissa R.A., van Sluis P., Broekmans M.E. Functional MYCN signature predicts outcome of neuroblastoma irrespective of MYCN amplification. Proc Natl Acad Sci USA. 2012;109:19190–19195. doi: 10.1073/pnas.1208215109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeselsohn R., Brown N.E., Arendt L., Klebba I., Hu M.G., Kuperwasser C. Cyclin D1 kinase activity is required for the self-renewal of mammary stem and progenitor cells that are targets of MMTV-ErbB2 tumorigenesis. Cancer Cell. 2010;17:65–76. doi: 10.1016/j.ccr.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuste N.P., Fernandez-Hernandez R., Cemeli T., Mirantes C., Pedraza N., Rafel M. Cytoplasmic cyclin D1 regulates cell invasion and metastasis through the phosphorylation of paxillin. Nat Commun. 2016;7:11581. doi: 10.1038/ncomms11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otto T., Horn S., Brockmann M., Eilers U., Schuttrumpf L., Popov N. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15:67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 44.Roos M., Pradere U., Ngondo R.P., Behera A., Allegrini S., Civenni G. A Small-Molecule Inhibitor of Lin28. ACS Chem Biol. 2016;11:2773–2781. doi: 10.1021/acschembio.6b00232. [DOI] [PubMed] [Google Scholar]
- 45.DuBois S.G., Mosse Y.P., Fox E., Kudgus R.A., Reid J.M., McGovern R. Phase II trial of alisertib in combination with irinotecan and temozolomide for patients with relapsed or refractory neuroblastoma. Clin Cancer Res. 2018;24:6142–6149. doi: 10.1158/1078-0432.CCR-18-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puissant A., Frumm S.M., Alexe G., Bassil C.F., Qi J., Chanthery Y.H. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henssen A., Althoff K., Odersky A., Beckers A., Koche R., Speleman F. Targeting MYCN-driven transcription By BET-bromodomain inhibition. Clin Cancer Res. 2016;22:2470–2481. doi: 10.1158/1078-0432.CCR-15-1449. [DOI] [PubMed] [Google Scholar]
- 48.He S., Liu Z., Oh D.Y., Thiele C.J. MYCN and the epigenome. Front Oncol. 2013;3:1. doi: 10.3389/fonc.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen L., Alexe G., Dharia N.V., Ross L., Iniguez A.B., Conway A.S. CRISPR-Cas9 screen reveals a MYCN-amplified neuroblastoma dependency on EZH2. J Clin Invest. 2018;128:446–462. doi: 10.1172/JCI90793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Fig. 1. LIN28B promotes neuroblastoma metastatic dissemination. (A) Mean bioluminescence intensities of mice from control and shLIN28B-depleted cohorts. (B) RT-PCR demonstrating LIN28B expression in the tumor tissue of control mice or mice with tumors in which LIN28B was depleted. The LIN28B mRNA level in control group was normalized to 1, and the fold changes of LIN28B mRNA level in each experimental group compared to control group presented. Of note, we isolated 3 control tumors, 2 tumors from the shLIN28B-1 cohort, and 2 tumors from the shLIN28B-3 cohort. (C) Quantitation of LIN28B protein level in the tumor tissues of control mice or mice with tumors in which LIN28B was depleted. The LIN28B protein level in control group was normalized to 1, and the fold changes of LIN28B protein level in each experimental group compared to control group are presented. Western blots below. * p < 0.05.
Related to Fig. 2. LIN28B positively influences neuroblastoma self-renewal. The SKNDZ cell line was infected with control lentiviruses or lentiviruses directed against LIN28B and cell proliferation quantified using CellTiter-Glo. Western blotting of LIN28B and Ku80 as shown in Fig. 2G. ****, p < 0.0001.
Related to Fig. 3. LIN28B promotes the migration of neuroblastoma cells. (A-B) Quantitation of wound closure over 24 hours for SKNDZ (A) and Kelly (B) cells infected with control lentiviruses or lentiviruses expressing shRNAs directed against LIN28B. Of note, cells were not treated with mitomycin. Error bars represent SEM. (C) The SKNDZ cell line was infected with control lentiviruses or lentiviruses directed against LIN28B and cells counted 24 hours post seeding. The number of control cells was normalized to 1, and fold change of cell number in experimental groups compared to control is presented. Western blotting of LIN28B and Ku80 as shown in Fig. 2G.
Related to Fig. 4. Let-7 inhibits both neuroblastoma self-renewal and migration. The SKNDZ cell line was infected with control or let-7i expressing lentiviruses and cell proliferation quantified using CellTiter-Glo. Expression of let-7i as shown in Fig. 4A. ****, p < 0.0001.
Related to Fig. 5. PBK is a novel LIN28B-influenced kinase. (A) Correlation between LIN28B and PBK expression in 498 primary neuroblastomas. Log2 of LIN28B expression is depicted on the x axis, with log2 of PBK expression on the y axis. (B) Expression of PBK in primary neuroblastoma tumors, as shown for International Neuroblastoma Staging System stages 1 through 4, with 4S. The number of tumors is depicted in parentheses. PBK expression was quantified in terms of Fragments Per Kilobase of transcript per Million mapped reads (FPKM). C) Kaplan-Meier analysis curves, with individuals grouped by low and high PBK expression. Data obtained from the Su dataset. P values and r values listed.
Related to Fig. 6. Both LIN28B/let-7 and MYCN directly regulate the expression of PBK. (A) RT-PCR demonstrates expression of let-7i in Kelly cell lines. Cell lines were infected with control lentiviruses or lentiviruses expressing let-7i. The let7i RNA level in control group was normalized to 1, and the fold changes of let7i RNA level in each experimental group compared to control group were calculated and presented in the bar graphs. (B) Correlation between MYCN and PBK expression in 249 primary neuroblastomas. Log2 of MYCN expression is depicted on the x axis, with log2 of PBK expression on the y axis. Data obtained from the TARGET dataset. (C) Correlation between MYCN and PBK expression in 649 primary neuroblastomas. Log2 of MYCN expression is depicted on the x axis, with log2 of PBK expression on the y axis. Data obtained from the Kocak dataset. P values and r values listed.
Related to Fig. 7. PBK promotes neuroblastoma self-renewal and migration. (A-B) Cell proliferation quantitated by CellTiter-Glo in control and PBK-depleted neuroblastoma cell lines SKNDZ (A), and IMR5 (B). Expression of PBK as shown in Fig. 7A and C. ****, p < 0.0001.
This table lists plasmids, shRNA constructs, antibodies, and TaqMan assays.
This table depicts the kinases correlated with LIN28B expression in the TARGET database. P values and r values are listed.